Abstract
Peroxiredoxin 6 represents a widely distributed group of peroxiredoxins that contain a single conserved cysteine in the protein monomer (1-cys Prdx). The cys when oxidized to the sulfenic form is reduced with glutathione (GSH) catalyzed by the π isoform of GSH-S-transferase. Three enzymatic activities of the protein have been described:1) peroxidase with H2O2, short chain hydroperoxides, and phospholipid hydroperoxides as substrates; 2) phospholipase A2 (PLA2); and 3) lysophosphatidylcholine acyl transferase (LPCAT). These activities have important physiological roles in antioxidant defense, turnover of cellular phospholipids, and the generation of superoxide anion via initiation of the signaling cascade for activation of NADPH oxidase (type 2). The ability of Prdx6 to reduce peroxidized cell membrane phospholipids (peroxidase activity) and also to replace the oxidized sn-2 fatty acyl group through hydrolysis/reacylation (PLA2 and LPCAT activities) provides a complete system for the repair of peroxidized cell membranes.
Keywords: Phospholipid hydroperoxide glutathione, peroxidase, Phospholipase A2, Lysophospholipid acyl transferase, NADPH oxidase, Phospholipid remodeling, Anti-oxidant defense
1. Introduction
Peroxiredoxin 6 (Prdx6) was the 6th and final mammalian protein to be recognized as a member of the peroxiredoxin (Prdx) family of non-seleno peroxidases. Because of prior use of the abbreviation Prx to identify the paired-related homeobox gene family [1], I have used Prdx, instead of Prx, as a unique identifier for the peroxiredoxin family of proteins. The protein was originally isolated from the ciliary body of the bovine eye and characterized as unique by its N-terminal amino acid sequence [2]. The protein was subsequently isolated from rat and bovine lung and rat olfactory mucosa [3–6]. Around the same time, search of the GenBank and EMBL databases identified the human Prdx6 gene [7,8] that had been generated by random cloning [9] and subsequently the rat and mouse genes were described [6,10,11]. Several names had been attached to the identical protein prior to agreement to designate the family as peroxiredoxins [12]; these former names include non-selenium GSH peroxidase [2,13,14], acidic (or lysosomal) Ca2+-independent PLA2 (aiPLA2) [4,6,8], antioxidant protein 2 (AOP2) [10,15], Clara cell protein 26 (CC26) [16], and p29 [17]. The corresponding gene has been called ORF6 [9], LTW4 [18], and keratinocyte growth factor (KGF)–regulated gene 1 [14].
Prdx6 is a 25 kDa protein formed by 224 amino acids (Fig. 1). Proteins that are homologous to mammalian Prdx6 are widely distributed throughout all kingdoms and have been described in archaea, bacteria, yeast, plants, insects, mollusks, amphibians, and birds [7,19–22]. Prdx6 is sometimes called 1-cys Prdx, reflecting the catalytic mechanism that is based on a single conserved cysteine residue in the Prdx monomer as contrasted with 2-cys Prdx enzymes that function with 2 conserved cys moieties per monomer. However, this nomenclature has anomalies, primarily related to more primitive organisms, since a 1-cys catalytic mechanism occurs in some non-Prdx6 members of the Prdx family while a 2-cys mechanism can be seen in members that are structurally similar to Prdx6 [23–25]. Based on protein structural features, a more recent classification of peroxiredoxins from all phyla indentifies 6 subfamilies: Prx1 (Prdx1-4 in the other nomenclature); Prx5 (Prdx5); Prx6 (Prdx6); and 3 subfamilies present only in bacteria and other lower forms (AhpE, PrxQ, and Tpx) [25]. This review of Prdx6 will focus on mammalian enzymes using the older 1-cys and 2-cys terminology. Several reviews of Prdx6 have been published previously [15,19,26–28].
Fig. 1. Amino acid sequence for mouse Prdx6.
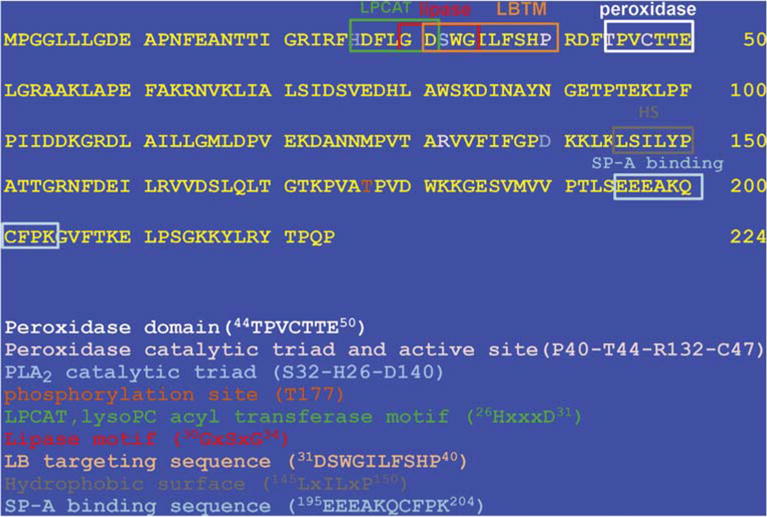
The full length sequence is shown. The consensus sequences are indicated for peroxidase and LPCAT activities, the PLA2 and peroxidase catalytic triads, the lipase motif, the motif (LBTM) for targeting of Prdx6 to the lamellar bodies, the hydrophobic surface (HS) for dimerization, the SP-A binding sequence, and the Prdx6 phosphorylation site.
2. Enzymatic activites of prdx6
2.1. Peroxidase activity
H2O2, short chain hydroperoxides such as t-butyl and cumene hydroperoxides, and peroxynitrite (ONOO−) are substrates for all of the Prdx enzymes including Prdx6 [22,29,30]. The catalytic cycle for reduction of hydroperoxides by peroxiredoxins involves 3 main chemical steps: peroxidation, resolution, and recycling [31]. The first step (peroxidation) reduces the hydroperoxide substrate resulting in oxidation of the reactive cys moiety to a sulfenic acid; the resolution step results in reduction of the sulfenic cys; and the recycling step leads to regeneration of the reduced cys sulfhydryl, i.e., the active enzyme. While the initial peroxidation step appears to be similar for all peroxiredoxins, the subsequent resolution and recycling steps vary between the 2-cys and 1-cys peroxiredoxins.
2.1.1. 2-cys vs 1-cys enzymes
As described above, the Prdx family members have been classified as either 2-cys (Prdx1-5) or 1- cys peroxiredoxins (Prdx6) based on the number of conserved cys per monomer. Prdx1-4 function as dimers in solution and use the N-terminal cys (peroxidatic cys) of one monomer to reduce hydroperoxides; the resultant oxidized (sulfenic,–SOH) cys interacts with the C-terminal cys of the other monomer (the resolving cys) to generate a protein di-sulfide. Prdx5 is classified as an atypical 2-cys Prdx with both the peroxidatic and the resolving cys on the same monomer forming an internal disulfide during the reaction cycle. In either case (typical or atypical), the resultant disulfide is reduced by thioredoxin [22].
2.1.2. Prdx6 peroxidatic mechanism
Similar to the mechanism for the 2-cys Prdx enzymes, reduction of hydroperoxides by 1-cys Prdx occurs through oxidation of its single conserved cys (located at amino acid 47 in Prdx6) [32]. The measured rates for reduction of H2O2 by Prdx6 are ~6 μmol/min/mg protein with Km 180 μM and rate constant (k1) ~3 × 107 M−1s−1 [2,30,33,34]. The rate constant for reduction of peroxynitrite is about 2 orders of magnitude lower [34]. The reaction of thiol with hydroperoxide is rapid so that the resolution phase rather than the reduction reaction is rate limiting for the peroxidatic function of the enzyme. The pKa of the reactive thiol in Prdx6 has been measured as 5.2 [31,34], similar to values for the 2-cys enzymes, so that the major fraction of Prdx6 in the cytoplasm (pH ~7) would be in the thiolate form (–S−) that is more reactive than the thiol (–SH).
2.1.3. Resolution
Resolution of the oxidized 2-cys enzymes is accomplished through formation of a disulfide by a relatively rapid reaction of the sulfenic with the resolving cys of the Prdx dimer. Unlike the 2-cys Prdx enzymes, Prdx6 does not have a second (resolving) cys, so the resolution of the sulfenic state requires interaction of the protein with an extraneous reductant. The physiological reductant that has been described for the mammalian enzyme is glutathione (GSH). GSH forms a mixed disulfide with oxidized Prdx6 (glutathionylation of Prdx6), analogous to the disulfide formed by interaction of the catalytic and resolving cys residues in the 2-cys enzymes [35–37]. (Protein glutathionylation also is seen in some 2-cys Prdx enzymes, but its role there is not related to their peroxidase activity [38]). Although Prdx6 functionally is a GSH peroxidase, it is classified as a peroxiredoxin based on the role of cys instead of selenocysteine as the redox-active group and its structural homology with other members of the peroxiredoxin family.
While the role of GSH in the reduction of oxidized 1-cys Prdx6 is now accepted, early studies were inconsistent and there were several negative reports [5,32,39]. It was subsequently found that the formation of the disulfide through reaction of the sulfenic 1-cys Prdx with GSH could be catalyzed by the π isoform of the enzyme GSH S-transferase (πGST) [36,37]. Thus, sulfenic Prdx6 can heterodimerize with GSH-loaded πGST followed by glutathionylation of the oxidized Prdx6 monomer [37,40]. This interaction is non-covalent. Prolonged interaction of Prdx6 with πGST can result in formation of a covalent disulfide bond between monomers [37] but, based on kinetic considerations, that is unlikely to be part of the normal reaction cycle. Confirmation of a role for πGST in the reduction of oxidized Prdx6 in intact cells was evaluated with the malignant MCF7 cell line that does not express πGST (or Prdx6). (Note that other studies have demonstrated the presence of some Prdx6 in MCF7 cells [41]; this expression may be strain-dependent). Intracellular delivery of πGST plus Prdx6 resulted in peroxidase activity while delivery of Prdx6 alone was ineffective [36]. The reversible heterodimerization of oxidized Prdx6 with πGST has been demonstrated in vivo by proximity ligation assay with fluorescence imaging of intact endothelial cells [42].
Although it is now clear that πGST can catalyse the reaction of GSH with oxidized Prdx6, several early studies demonstrated Prdx6 peroxidase activity in the presence of GSH alone. Those studies showing reduction by GSH that used native Prdx6 after isolation from various organs [2,4,5] may have inadvertently included πGST as a co-isolate with Prdx6, as shown with extraction of bovine lung [36]. However, our initial studies using recombinant Prdx6 from a single preparation showed effective resolution of the oxidized state by GSH in the absence of added πGST with activities in the range of 2–5.5 μmol/min/mg protein and a rate constant (k2) >106 M−1s−1 [13,30], not very different from subsequent assays with GSH plus πGST. The catalytic cys is contained within a narrow pocket that presumably does not permit access of GSH (in the absence of πGST) as needed for Prdx6 reduction [43]. The ability of GSH alone to reduce the oxidized cys in this one [6] preparation of Prdx6 [13,30] may have reflected a tertiary structural conformation that allowed access of GSH to the catalytic “pocket”.
2.1.4. Recycling
The final step (recycling) in the reaction pathway is to regenerate the active protein. For this, glutathionylated Prdx6 reacts with GSH to return to the fully reduced state (Figs. 2 and 3) [36,37]. This step appears to utilize free GSH as the reductant [36], although a possible role for πGST (or glutaredoxin) in mediating the interaction of the glutathionylated intermediate with GSH has not been excluded. Both oxidized GSH (GSSG) formed by reduction of 1-cys Prdx and oxidized thioredoxin formed by reduction of 2-cys Prdx are themselves reduced by NADPH-dependent enzymes (GSH and thioredoxin reductases, respectively). Thus, the reaction cycles for both the 1- and 2-cys Prdx enzymes comprise oxidation of the cys thiol to the sulfenic form followed by disulfide formation and then reduction to the “active” sulfhydryl with reducing equivalents ultimately derived from NADPH.
Fig. 2. Mechanism for Prdx6 peroxidase activity and its resolution.
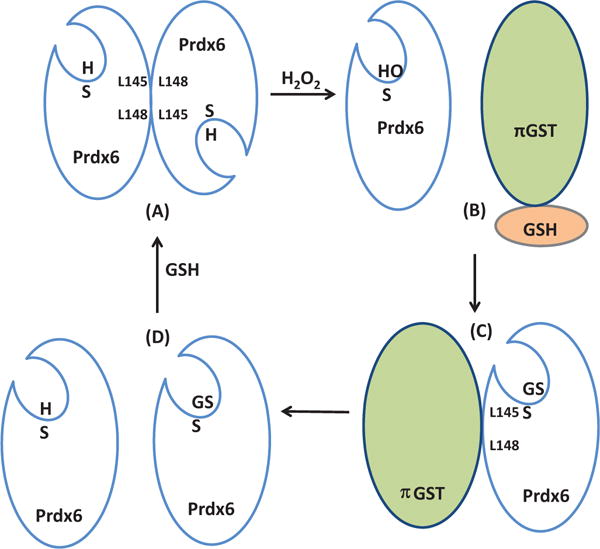
Clockwise from upper left. (A) In solution, Prdx6 exists as a homodimer through hydrophobic interactions surrounding the L145-L148 residues; the reduced peroxidatic sulfhydryl (SH) is at the bottom of a pocket. (B) Reduction of H2O2 (peroxidase activity) results in oxidation of the Prdx6 sulfhydryl to the sulfenic form (SOH) leading to dissociation of the homodimer. Interaction of the oxidized monomer of Prdx6 with GSH-loaded πGST results in: (C) heterodimerization via the L145-L148 hydrophobic interface of Prdx6 and glutathionylation of Prdx6 (SSG), followed by: (D) dissociation of the glutathionylated Prdx6 and πGST heterodimer. Interaction of the glutathionylated monomer with free GSH restores its normally reduced state allowing reformation of the homodimer.
Fig. 3. Reaction scheme for Prdx6 peroxidase activity with proposed formation of a sulfenylamide intermediate.
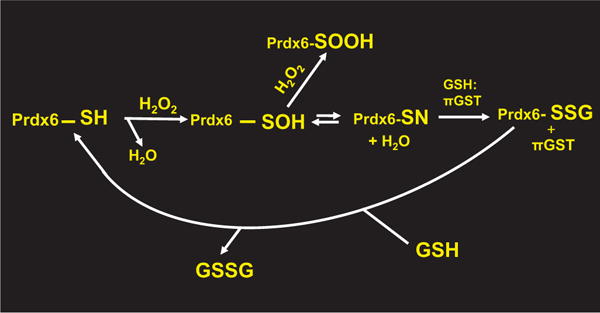
Interaction of Prdx6 with H2O2 generates the sulfenic state (Prdx6-SOH) of the reactive cysteine. The sulfenic cys is susceptible to further (irreversible) oxidation to the sulfinic form (Prdx6-SOOH). The formation of a reversible sulfenylamide (Prdx6-SN) could stabilize the sulfenic state and protect the protein from irreversible oxidation (see text). Interaction of the Prdx6 sulfenylamide with πGST:GSH results in Prdx6 glutathionylation (Prdx6-SSG) and subsequent resolution by interaction with GSH as described in Fig. 2. GSSG formed during the cycle is reduced by NADPH mediated by GSH reductase to regenerate 2 GSH (not shown).
2.1.5. Structure-function relationships
The motif surrounding the peroxidatic C47 in human, mouse, rat, and bovine Prdx6, 43TPVCTTE50 [28,30], and the corresponding peroxidase motif in 2-cys Prdxs, TFVCPTE [22], are largely homologous. Based on the tertiary structure of oxidized Prdx6, it was proposed that the adjacent H39 and R132 stabilize the C47 thiolate in its active form [43] but further study has shown a more complex relationship between the active site and adjacent amino acids. Peroxidatic activity for the 2-cysPrdx enzymes requires a triad (Pro,Thr,Arg) surrounding the catalytic cys [31]. The catalytic 47C in Prdx6 is surrounded by an identical triad (40P, 44T, 132R); the Pro, Thr, and peroxidatic cys (Cp) are found in a contiguous sequence (PXXXTXXCp) in Prdx6 as in the 2-cys enzymes [31]. The triad appears to play an important role in the activation of the peroxide substrate. Specific functions for the individual amino acids have been postulated: 1) the Pro shields the thiolate from unwanted reactions and positions the Thr and Arg for hydrogen bonding; 2) the Arg positions and activates the thiolate; and 3) the Thr and Arg position and activate one oxygen atom (the electrophilic center) of the peroxide substrate [31]. The relationship of the peroxidase motif to other important motifs in the Prdx6 amino acid sequence is shown in Fig. 1.
2.1.6. Prdx6 as a dimer
Although the catalytic cycle for Prdx6 does not involve an interaction of two monomers as with the typical 2-cys enzymes, the native (reduced) protein nonetheless is dimeric in crystal structure as well as in solution with the monomers arranged in an antiparallel state [35,44,45]. Dimerization is through hydrogen bonding involving the hydrophobic interface between the seven β-strands of each monomer, resulting in an extended 10-stranded β-sheet; the hydrophobic residues Leu145, Ile147, Leu148, and Pro150 help to stabilize the interface [35,43,44]. This configuration has been designated as a B-type dimer [25]. The crystal structure of Prdx6 shows the “thioredoxin fold” characteristic of peroxiredoxins and an extended C-terminus relative to most of the other Prdxs [43,46]. The formation of higher oligomers (e.g., decamers) of Prdx6 has been suggested [23], but the required experimental conditions and their possible physiological significance are unclear. The dimer affinity of Prdx6 (Kd < 20 nM) decreases upon its oxidation, providing oxidized monomer for heterodimerization with GSH-loaded πGST [35–37]. The heterodimer of Prdx6 and πGST forms only in the presence of GSH and removal of the thiol results in dissociation of the complex [37]. Interestingly, while the oxidized protein is largely monomeric in solution [35], it crystallized as a dimer rather than a monomer [43], possibly reflecting the use of conditions, such as high concentrations of protein, that favor the dimeric form.
2.1.7. πGST:GSH and other reductants
Reduction of oxidized Prdx6 as studied with recombinant enzyme is through glutathionylation mediated by πGST. Like Prdx6,πGST also exists in solution as a mixture of homodimers and monomers in equilibrium allowing heterodimerization of its monomer with an oxidized Prdx6 monomer [40]. Hetero-dimerization of oxidized Prdx6 with πGST utilizes the same hydrophobic interface as homodimerization of reduced Prdx6; this interface is centered on 145L-P150 of Prdx6 (Fig. 1), so that hetero-dimerization essentially represents a “swap” of a Prdx6 monomer for a πGST monomer [35,37]. The rate of heterodimerization has not been measured but presumably is rapid based on the overall k of >106 M−1s−1 cat for the coupled assay (see below). Polymorphisms associated with the πGST gene have been shown to influence the affinity of the protein for interaction with Prdx6 and presumably the rate constant for binding; πGST1-1A and C subtypes (Kd ~50 nM) have greater affinity for binding to Prdx6 as compared to πGST1-1B and D (Kd ~100 nM) [41].
GSTs are a large family of proteins and 13 different cytosolic as well as additional microsomal and mitochondrial isoforms have been identified in vertebrates [47]. Additional classes of GSTs have been described in non-vertebrates and plants, although documentation is still incomplete [48]. πGST is a widely distributed isoform of the GST family that is expressed in most mammalian organs and at especially high levels in lungs, heart, and brain [49]. The distribution of GSTs within organs can vary with cell type; for example, in the liver, hepatocytes express predominantly αGST while biliary tract cells express predominantly the π isoform [49]. Thus, hepatocytes that express Prdx6 at a relatively high level [50] have little to no expression of πGST. The mechanism for resolution of oxidized Prdx6 in the absence of πGST in these cells has not been adequately evaluated. Although preliminary studies from our laboratory have shown that the μ isoform of GST may catalyze the glutathionylation of Prdx6 (unpublished results), the possible role of this and other GST isoforms in resolution of oxidized 1-cys Prdx in cells that do not express πGST will require further study.
Other potential physiological reductants for Prdx6 have been proposed. Ascorbic acid was shown to reduce yeast mitochondrial 1-cys Prdx [51]; however, subsequent studies with the same protein have shown glutathionylation of the yeast enzyme (in the absence of πGST) rather than ascorbate-mediated reduction as the primary mechanism for resolution of the oxidized state [52,53]. Evaluation of this latter mechanism could provide insights into alternate means for glutathionylation of Prdx6 in mammalian systems that do not express πGST. Oxidized mammalian Prdx6 also can be reduced by ascorbate in vitro [51]; however, the rate constant for this reaction appears to be at least an order of magnitude less than that for GSH:πGST, suggesting that ascorbate is not the primary physiological reductant in mammals. Nonetheless, ascorbate-mediated reduction could be a pathway for the reduction of oxidized Prdx6 in the absence of GST or under special circumstances, e.g., depletion of GSH, and may be the primary reductant for 1-cys Prdx enzymes in plants [54]. Finally, cyclophilin A has been shown to bind to and to reduce Prdx6 in vitro [55] but this reaction is not specific and its possible physiological role as a reductant for oxidized Prdx6 has not been demonstrated.
2.1.8. Phospholipid hydroperoxide GSH peroxidase activity
Uniquely among the Prdxs, Prdx6 also is able to reduce phospholipid hydroperoxides (PLOOH); the activity responsible for PLOOH reduction has been called phospholipid hydroperoxide GSH peroxidase (PHGPx). Phosphatidylcholine hydroperoxide (PCOOH) has been the primary substrate used to measure PHGPx activity of Prdx6. The rate of reduction of PCOOH by recombinant Prdx6 is ~6 μmol/min/mg protein with Km 120–150 μM; the rate constant for reduction (k1) is ~107 M−1 s−1 with k2 (kcat) 2–5×106 M−1s−1 [13,36,37]. The reaction rate was similar for triton-dispersed PCOOH and oxidized phospholipids in liposomes (mol fraction: 0.5 dipalmitoyl phosphatidylcholine; 0.25 palmitoyl, linoleoyl phosphatidylcholine; 0.15 cholesterol; 0.10 phosphatidylglycerol) [33]. These reaction rates and kinetic constants for PCOOH are similar to those for H2O2 (see above).
The only protein besides Prdx6 for which significant PHGPx activity has been demonstrated is GSH peroxidase 4 (GPx4) [56,57] and none of the 2-cys Prdx enzymes have been shown to express this activity. (In this review, I have used PHGPx to describe the activity potentially shared by other enzymes and GPx4 as the name of a specific protein.) The reported activity for GPx4 in the reduction of peroxidized phospholipids is 20–30 μmol/min/mg protein with a rate constant (k2,kcat) ~1 × 107 M−1s−1 [57–59]; these values are only slightly greater than the corresponding values for Prdx6 indicated above. Although GPx4 may play an important role in the reduction of oxidized membrane lipids in some organs, its expression varies considerably among tissues [60–63]. For example, it is expressed at a relatively low level in lungs, an organ susceptible to oxidant stress based on its organismal location and physiological function. “Knock-out” of Prdx6 (Prdx6 null) diminished the PHGPx activity of lungs by 97% (Fig. 4), suggesting that the reduction of oxidized phospholipids in this organ largely reflects Prdx6 activity [64]. Homogenates from several organs obtained from heterozygotic mice that express only one allele for wild type GPx4 and the other allele for inactive GPx4 showed relatively little difference from control mice in their phospholipid hydroperoxide GSH peroxidase activity [63]. This result is compatible with, although not definitive for, a major role for Prdx6 with minimal input from GPx4 in the reduction of peroxidized phospholipids in these organs (heart, liver, kidney, spleen, lung). The structural basis for the activity of Prdx6 toward oxidized phospholipids is discussed below.
Fig. 4. Effect of Prdx6 “knock-out” on hydroperoxide reduction by mouse lung homogenate.
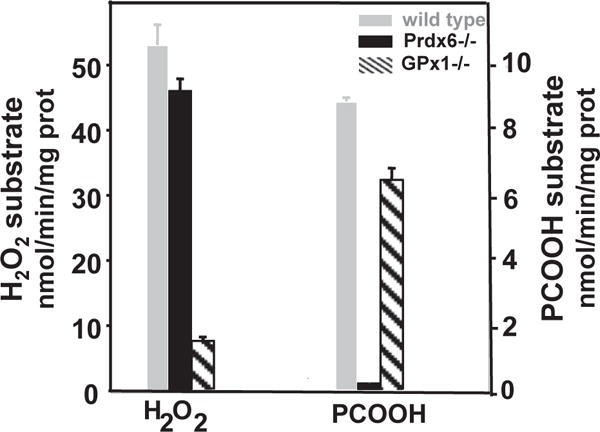
Knock-out of Prdx6 (Prdx6−/−) has only a minor effect on the GSH-mediated reduction of H2O2 (GPx activity, left y-axis) by the lung homogenate but results in a marked decrease in the GSH-mediated reduction of phosphatidylcholine hydroperoxide (PHGPx activity, right y-axis). Knock-out of GPx1 (GPx1−/−) resulted in the inverse with a large decrease in GPx activity but relatively little effect on PHGPx activity. PCOOH, phosphatidylcholine hydroperoxide. Reprinted from Ref. [64] with permission.
2.1.9. Sulfenylamide formation
In contrast to the 2-cys Prdxs that can be reduced from the sulfinic (–SOOH) state by sulfiredoxin, hyperoxidation of Prdx6 is irreversible [65]. Thus, preventing further oxidation of the sulfenic is essential for Prdx6 function. The πGST-catalyzed reaction of the sulfenic group with GSH to form a disulfide can stabilize the protein (pending its reduction) and prevent its hyperoxidation in vitro [66]. However, it is not clear that the πGST required for hetero-dimerization would be readily available intracellularly to prevent hyperoxidation of the protein under physiological conditions. Our mass spectroscopic evaluation of the recombinant protein following storage (that presumably was accompanied by auto-oxidation of the protein) indicated a molecular mass that was 2 Da less than the predicted mass of the reduced protein [35]. Based on this finding, we have proposed that a sulfenylamide forms through dehydration of oxidized Prdx6 (reduced protein +16 for “O” −18 for H2O =−2) [35]; an alternative possibility for the measured mass is a thioether, but that is extremely rare in nature [67]. As noted in the review by Forman in this volume, sulfenylamide formation of a protein in vivo requires the reactivity of its sulfhydryl group toward peroxides that is in the range demonstrated by Prdx6. Thus, sulfenylamide formation could represent a mechanism for stabilization of the oxidized thiol (Fig. 3) and this reaction could account for the surprising stability reported for oxidized (sulfenic) bovine Prdx6 [38,39]. The possible partner with C47 for sulfenylamide formation remains to be determined, but includes the adjacent V46 or T48 or, in the tertiary structure, H39 or R132. Although sulfenylamide formation was demonstrated for this protein in vitro, its formation and possible role for stabilization of Prdx6 has not yet been demonstrated under physiological conditions.
2.2. Phospholipase A2 activity
2.2.1. Mechanism for activity
Prdx6 also expresses a second major activity related to phospholipid metabolism as it is able to hydrolyze glycerophospholipids at the sn-2 position; thus, Prdx6 exhibits phospholipase A2 (PLA2) activity [4,6,8,30]. Properties of the enzyme have been studied with Prdx6 protein isolated from rat and bovine lungs [4,8] and recombinant rat and human protein [68–70]. Under optimal conditions, PLA2 activity of recombinant Prdx6 is ~100 nmol/min/mg protein with Km ~350 μM using radiolabeled dipalmitoyl phosphatidylcholine (PC) in mixed unilamellar liposomes (mol fraction: 0.5 dipalmitoyl PC, 0.25 egg PC, 0.15 cholesterol, 0.10 phosphatidylglycerol) as substrate; a rate constant for this reaction has not been reported. For comparison, the activity of secreted (type II) human platelet PLA2 is ~300 nmol/min/mg protein while Ca2+-independent PLA2 from a macrophage cell line is ~5 nmol/min/mg protein [71,72]. However, these comparisons have little meaning because of the varied substrate specificity and the complex conditions necessary for demonstration of in vitro activity. Prdx6 PLA2 has a decided preference for phosphatidylcholine as a component of liposomes as substrate with ~40% less activity toward phosphatidylethanolamine and even lower activity towards phospholipids with other head groups; there is no preference for the fatty acyl group in the sn-2 position although an alkyl substituent results in a decrease in activity by ~50% [4,6]. The active sites for PLA2 activity and peroxidase activity are distinct, making this a true “moonlighting” protein [30].
The PLA2 activity of recombinant Prdx6 as well as Prdx6 isolated from lungs is greatest under acidic conditions (~pH 4) and, unlike many other PLA2 enzymes, does not require the presence of Ca2+ [73–75]; thus, this activity initially was called either acidic calcium-independent PLA2 (aiPLA2) [4,6,73,75] or lysosomal PLA2 [8,30,74,76]. (Not to be confused with a 45 kDa protein that was identified subsequently and also called lysosomal PLA2 [77]). While the PLA2 activity of Prdx6 with the usual phospholipid substrates is much lower at cytosolic pH, activity at pH 7 towards phosphatidylcholines with an oxidized fatty acid in the sn-2 position, i.e., a phosphatidylcholine hydroperoxide, is similar to the level seen with this substrate under acidic conditions [78]. Unlike the case for peroxidase activity, PLA2 activity is present in the Prdx6 monomer and dimerization is not required [35].
aiPLA2 activity shows a distinctive response to chemical PLA2 inhibitors; it is insensitive to most of the inhibitors of other PLA2 enzymes but is inhibited competitively by 1-hexadecyl-3-trifluoroethylglycero-sn-2-phosphomethanol (MJ33), a tetrahedral mimic of the transition state [4,6,8,73,76,79]. (It should be noted that many purported PLA2 inhibitors function by making the substrate inaccessible to the enzyme, and in that sense are not classical enzyme inhibitors [80]). Inhibition by MJ33 is > 80% at a ratio of 1:100 inhibitor to substrate concentrations (mol:mol).
The motif in Prdx6 that is responsible for phospholipid hydrolysis is 32S-140D-26H [78], a catalytic triad that is commonly found in lipases and proteases [81,82]. This triad is present in all mammalian Prdx6 proteins for which the amino acid sequence has been determined and those examined have all shown PLA2 activity [28]. The 32S amino acid residue as well as the other members of the catalytic triad also are conserved in many non-mammalian 1-cys Prdx enzymes including those from avian, fish, amphibian, and Drosophila species [68], although whether that translates into PLA2 activity has not yet been demonstrated. A homologous triad is not present in the 2-cys Prdxs and none of these latter proteins has been reported with PLA2 activity.
2.2.2. Phosphorylation of Prdx6
The post-translational modification with the greatest effect on PLA2 activity is its phosphorylation at the T177 position. Activity of the phosphorylated enzyme is ~1.3 μmol/min/mg protein, a >10-fold increase compared to the non-phosphorylated protein; further, activity towards reduced phospholipid substrate is similar under acidic and neutral pH conditions [69]. Prdx6 is phosphorylated by MAP kinases (p38, ERK) as shown in vitro with recombinant protein and in cells by the use of MAP kinase inhibitors [69,83]. In silico analysis of Prdx6 based on the crystal structure shows accessibility of MAPK docking sites on the protein surface [69].
The site for phosphorylation, T177, in the crystal structure of Prdx6 is buried in the homodimer interface and would not be solvent accessible for phosphorylation in vitro [43]. However, analysis of the reduced protein in solution by zero length chemical crosslinking and homology modeling demonstrated considerable divergence from the crystal structure of the oxidized (sulfenic) protein, particularly in the C-terminal region [45]. Although these latter measurements indicate solvent accessibility to T177, that site is nonetheless far removed from the active site for PLA2 activity requiring a conformational change to influence substrate binding and/or enzymatic activity as described below.
2.3. Lysophospholipid acyl transferase activity
Prdx6 also demonstrates a third distinct activity, namely the ability to acylate lysophosphatidylcholine with a free fatty acid (CoA derivative); this activity is called lysoPC-acyl CoA transferase (LPCAT) [84]. Prdx6 exhibits one of the motifs, 26HxxxxD31, that characterize LPCAT activity [85,86] (Fig. 1). LPCAT activity is abolished by mutation of D31 but, surprisingly, was unaffected by mutation of H26 [84]. The activity expressed by Prdx6 has a marked preference for choline (lysoPC) as compared to other lysophospholipid head groups and also a preference for palmitoyl CoA as the sn-2 substituent fatty acid. Kinetic studies demonstrated that hydrolysis by Prdx6 (PLA2 activity) and reacylation (LPCAT activity) represent a continuous process without release of the lysoPC intermediate that is generated by the PLA2 reaction [84]. Thus, in the presence of palmitoyl CoA, the combined PLA2 and LPCAT activities essentially result in the generation of PC with palmitate in the sn-2 position. Prdx6 is one of 5 mammalian enzymes that have been described with LPCAT activity [85–87] but it is the only one to date that shows PLA2 and LPCAT activities in the same protein and also the only one with a predominantly cytosolic localization (see below).
2.4. Phospholipid binding
The three reactions described above that involve phospholipids, namely, the reduction of PLOOH (PHGPx activity), the hydrolysis of PC (PLA2 activity), and the acylation of lysoPC (LPCAT activity), all require the binding of Prdx6 to a phospholipid substrate. Binding of Prdx6 to liposomes as a model for cell membranes (Kd~ 230 nM) has been demonstrated using circular dichroism, microfiltration analysis, fluorescence spectrofluorometry, time-resolved fluorescence spectroscopy, isothermal titration calorimetry, and other techniques [33,78]. Binding is greatest with liposomes containing an anionic phospholipid and decreases by ~50% with non-charged (neutral) liposomes [78]. This means that the negative charge (e.g., associated with the presence of phosphatidylglycerol) facilitates binding of Prdx6 to the liposome although the subsequent hydrolytic activity is greatest towards phosphatidylcholine. Prdx6 binds to liposomal phospholipids at pH 4 but binds less well at pH 7, consistent with assays of its PLA2 activity. Also consistent with its PLA2 activity, phosphorylated Prdx6 binds avidly at both acidic and neutral pH while Prdx6 binds equally at both pHs to liposomes containing an oxidized phospholipid [33,69]. Thus, substrate binding appears to be a key element for understanding the lipid-related enzymatic activities of Prdx6.
Evaluation of Prdx6 by the biophysical techniques mentioned above provide evidence that the mechanism for the effect of phosphorylation on PLA2 activity is through increased binding associated with conformational change in the protein structure [88]. This latter study demonstrated that phosphorylation induces Prdx6 to assume a molten globule state that shows maximal enzymatic (PLA2) activity. This state is the configuration associated with activity for many proteins, allowing greater structural flexibility and a greater range of conformations for binding to substrates as compared with the fully folded native state [89–91]. The greater binding of Prdx6 to phospholipids at acidic pH and the increased binding to oxidized substrate also may be a consequence of altered protein conformation subsequent to thiol protonation (–SH) or oxidation (–SOH), although those possibilities have not yet been evaluated.
The amino acid residues 30G-G34 in Prdx6 (GDSWG) constitute what has been called a lipase domain (GxSxG) (Fig. 1); this motif is commonly observed in proteins that show lipolytic activity [92,93]. The amino acids of the lipase domain with extension to H26 are the key site for binding of Prdx6 to phospholipid substrates [33,78]. Based on analysis of the crystal structure [43], we have proposed that upon binding of phosphatidylcholine to the Prdx6 surface, the sn-2 fatty acyl group is positioned into a surface invagination (catalytic pocket) where it is susceptible to: a) reduction by the peroxidatic site centered on C47 that is located at the base of the pocket, or b) hydrolysis by the lipolytic site centered on S32 at the entrance to the pocket (Fig. 5). These structure-function relationships of Prdx6 are ultimately responsible for its unique enzymatic activities.
Fig. 5. Model for phospholipid-associated activities of Prdx6.
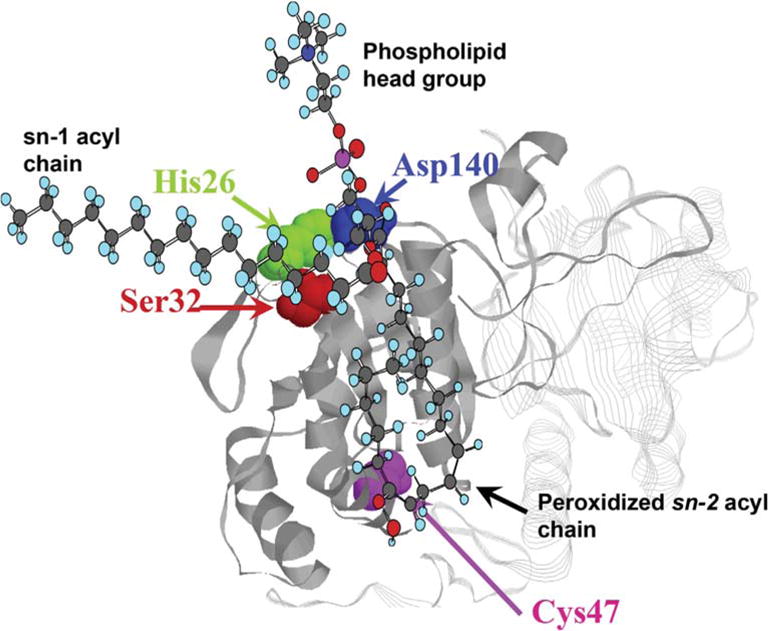
The model is based on the crystal structure of oxidized human Prdx6 [43]. Prdx6 binds to oxidized phosphatidylcholine at the lipid binding region associated with the lipase motif in the protein. We propose that the peroxidized sn-2 acyl chain inserts into the pocket containing the peroxidatic cys and is positioned at the surface-expressed PLA2 catalytic triad (32S-26H-140D). The sn-1 acyl chain and the phospholipid head group remain outside of the pocket. Thus, Prdx6 can reduce the oxidized acyl moiety (PHGPx activity) or hydrolyze the acyl bond (PLA2 activity) of the peroxidized phospholipid. This complex could be at the cytoplasmic face of a cell membrane at the left (not shown). The hydrophobic sn-1 fatty acyl chain and the hydrophilic head group would be within the membrane while the peroxidized sn-2 fatty acyl chain would be at the membrane surface facilitating interaction with Prdx6. Modified and reprinted with permission from Ref. [78].
3. Expression of Prdx6 protein and activities
3.1. In vitro expression
The multiple states of Prdx6 and its susceptibility to auto-oxidation have led to difficulty in isolation and storage of enzymatically active protein. We have developed conditions for generating active Prdx6 in relatively high yield and in the reduced state (<5% oxidized) using codon optimized E. coli for expression of the protein and ion-exchange chromatography for purification [45].
3.2. Cellular and subcellular localization
Prdx6 is widely distributed throughout all organs with especially high concentrations in the lung, brain, liver, kidney, and testis [6,27,94]. At present, no normal mammalian cells are known that lack Prdx6. The subcellular localization of Prdx6 is predominantly cytosolic. However, as demonstrated through subcellular fractionation, immunofluorescence, and immunogold electron microscopy, Prdx6 in lung alveolar type 2 epithelial cells is present in lysosomes and lamellar bodies (LB) as well as cystosol [52,70,95–97]. LB are lysosomal-related organelles (LROs) that, like lysosomes, maintain an acidic internal pH [98] and are the intracellular site for storage and metabolism of the lung surfactant [99–101]. Prdx6 plays an important role in the intra-LB metabolism of phospholipids (see below). The possible presence of Prdx6 in other LROs (e.g., melanosomes) has not been reported. Prdx6 was not detected in the lysosomal fraction isolated from human A431 epidermoid carcinoma cells [32].
The transport of Prdx6 from its site of synthesis in the endoplasmic reticulum to its localization in the luminal compartment of the LB of the type 2 alveolar epithelial cell proceeds via the vesicular pathway as shown by its inhibition with brefeldin A [96]. The amino acid sequence in Prdx6 between positions 31–40 (DSWGILFSHP) is required for LB targeting of the protein [96]. Within this sequence, the amino acids S32 and G34 appear to be critical while mutation of S38 has no effect on targeting; the importance of the other individual amino acids in the sequence has not been evaluated. Binding of Prdx6 to lipid is not required for targeting of the protein [96]. The cytosolic chaperone protein 14-3-3ε, after its activation by MAP kinase activity, binds to Prdx6 and is required for organellar targeting [95] although the precise mechanism for determining the distribution between cytosolic vs. organellar localization for the protein is not known. Mutation of S32 to threonine had no effect on lipid binding or enzymatic activities of Prdx6 but did abolish its binding to 14-3-3ε and its targeting to LB [68].
3.3. Regulation of expression
Prdx6 in rat lungs is expressed at relatively low levels in the fetus but is markedly increased immediately after birth and reaches adult levels at ~1–2 weeks of age [102]. Prdx6 expression postnatally is induced by oxidative stress as shown by treatment of lungs or cell lines with H2O2, hyperoxia, or paraquat [103–105]. Transcriptional activity for Prdx6 synthesis also is stimulated by KGF and by a glucocorticoid analogue (dexamethasone) independently of oxidants [14,102,105,106]. Both oxidant stress and KGF induce transcription though activation of the transcription factor Nrf2 that translocates to the nucleus and binds to an anti-oxidant (electrophilic) response element (ARE or ERE) located between 357 and 349 nucleotides upstream of the Prdx6 translational start site [104,106]. Dexamethasone activates Prdx6 transcription by binding to a glucocorticoid response element (GRF) located ~750 nucleotides upstream of the Prdx6 translational start site [106]. The effects of KGF and dexamethasone on Prdx6 protein expression are synergistic, consistent with their regulatory effect via separate but interactive elements in the Prdx6 DNA promoter [106]. Prdx6 expression also is regulated by the transcription factors CCAAT/enhancer protein β (C/EBPβ) and cAMP response element binding protein (CREB) [107].
3.4. Regulation of activity
Several potential physiological modulators of the PLA2 activity of Prdx6 have been shown, but no endogenous regulators of its peroxidase activity are known. As described above, phosphorylation of Prdx6 at position T177 mediated by MAP kinase leads to an increase in PLA2 activity of >10-fold [69]. Phosphorylation under physiological conditions has been shown with intact endothelial cells that were treated with angiotensin II [83]. With phosphorylation, cytosolic Prdx6 can bind to cell membranes, attaining substrate for its PLA2 activity [69].
Two specific cellular proteins have been shown to inhibit PLA2 activity. The first is surfactant protein A (SP-A), a protein that is present in the type 2 alveolar epithelial cells and the extracellular airspaces of lungs [97]. SP-A (monomeric mol wt ~26 kDa) binds to Prdx6 through hydrophobic interaction and H+-binding; the stoichiometry of binding is one Prdx6 monomer per SP-A trimer [70]. Inhibition of PLA2 activity is non-competitive with Ki~10 μg/ml (~130 nM for the SP-A trimer). The sites for interaction are 195E-K204 in Prdx6 and 83D-L99 in SP-A [108]; the role of specific amino acid residues in these sequences has not been investigated. Prdx6 PLA2 activity was significantly increased in lung homogenates from SP-A null mice [109] while LB that had been isolated from rat lungs showed a significant increase in PLA2 activity when treated with a chemical inhibitor of SP-A binding to Prdx6 [97]. These findings are compatible with a physiological role for SP-A in the regulation of Prdx6 PLA2 activity. Since Prdx6 and SP-A co-exist only in lung epithelial cells, lung LB, and the lung airway extracellular spaces, the regulatory role of SP-A on Prdx6 function likely would be limited to those compartments.
A second endogenous regulator of Prdx6 PLA2 activity is the cytosolic protein p67phox that is associated with the NADPH oxidase type 2 (NOX2) complex [17]. p67phox binds strongly to phosphorylated Prdx6 (Ki 65 nM) and inhibits its PLA2 activity while binding to non-phosphorylated Prdx6 is considerably weaker [110]. The NOX2 activation pathway requires phosphorylation of p67phox but this abolishes its binding to Prdx6 [110]. Prdx6 and p67phox co-exist in the cytoplasm of polymorphonuclear leukocytes, macrophages, endothelium, and possibly other cell types and their interaction in those cells may play a regulatory role related to NOX2 activation as discussed below.
4. Physiological roles of Prdx6
4.1. Prdx6 null mice
Prdx6 null mice have been generated by two different molecular strategies [111,112]. In both cases, mice appear to develop and reproduce normally with no overt phenotype. However, closer evaluation shows a characteristic phenotype including: 1) increased susceptibility to oxidant-mediated injury with altered repair of peroxidized cell membranes, 2) disturbances of sperm mobility with consequent effects on fertility associated with aging, 3) abnormalities related to lung lipid metabolism with age-related phospholipidosis and altered lung surfactant composition, and 4) greatly decreased activation of NADPH oxidase (type 2) with decreased generation and altered cell signaling. These abnormalities are now discussed with relevance to the physiological roles of Prdx6.
4.2. Antioxidant defense
4.2.1. Response to oxidative stress
There is much evidence that Prdx6 can protect against cell injury associated with oxidative stress. Studies have evaluated the effects of both over- and under-expression of Prdx6 in isolated cells, isolated organs, and whole animals. Cells in culture over-expressing Prdx6 have been obtained by their isolation from transgenic Prdx6 over-expressing mice, by generation of stable cell lines, and by use of TAT peptide as a protein delivery agent. Cells over-expressing Prdx6 have shown an increase in survival compared to control when oxidatively challenged with hydroperoxides, paraquat, UVB radiation, or OH generated by Cu2+/ascorbate [113–116]. Prx6 over-expression through adenoviral-mediated intratracheal delivery of a Prdx6 expression vector or in transgenic mice protected lungs from hyperoxic injury [117,118].
Prdx6 null mice showed increased lung or liver injury with exposure to hyperoxia, lipopolysaccharide (LPS), or paraquat [64,112,119,120]. Lung alveolar type 2 epithelial cells, lung pulmonary microvascular endothelial cells, peritoneal macrophages, and sperm that were isolated from Prdx6 null mice were more sensitive (increased injury) with exposure to paraquat or various hydroperoxides [64,121–123]. Prdx6 null mice as compared to wild type demonstrated increased age-related oxidative damage to their sperm chromatin [124] as discussed further below. Treatment of a lung epithelial cell line with a Prdx6 antisense oligonucleotide depressed Prdx6 expression and resulted in oxidative stress during routine cell culture with increased cytotoxicity on exposure to an •OH-generating system [125].
4.3. Mechanism for antioxidant protection
Based on the studies described above, there is a distinct correlation between the level of Prdx6 expression and susceptibility to a variety of agents that produce their effects through oxidant stress. However, the precise mechanism(s) for protection against oxidant injury by Prdx6 is not clear. One possibility would be the direct scavenging of H2O2 (or other low molecular weight hydroperoxides) by Prdx6. For this function, Prdx6 would compete with various cellular enzymes such as the GPx enzymes, other Prdxs, and catalase. A second possibility for the effect of Prdx6 is related to its ability to reduce PLOOH.
To gain an understanding of the relative roles for scavenging of H2O2 vs reduction of PLOOH in antioxidant protection by Prdx6, we compared the response of Prdx6 null and glutathione peroxidase 1 (GPx1) null mice to oxidant stress [64]. GPx1 is considered to be a major scavenger of cellularly-generated H2O2 and simple hydro-peroxides. Mice were exposed to hyperoxia, isolated lungs were exposed to t-BOOH or paraquat, and primarily isolated pulmonary microvascular endothelial cells also were exposed to t-BOOH. Lungs from the two types of mice showed similar levels for expression of mRNA or protein for antioxidant enzymes, except for the targeted protein. For all parameters investigated in each experimental model, there was relatively little injury in the GPx1 null mice while injury in the Prdx6 null was significantly greater [64]. The results of these comparative studies [64,121] suggest (with caveats, of course) that in these models of oxidant injury, the ability to reduce H2O2 has relatively minor importance while the ability of Prdx6 to either reduce or hydrolyze phospholipid hydroperoxides is crucial to its protective role.
4.3.1. Repair of peroxidized cell membranes
Lipid peroxidation, primarily affecting cell membranes, is recognized as one of the major mechanisms for cell injury associated with oxidant stress [126]. Lipid peroxidation occurs through proton abstraction, commonly from a phospholipid with an unsaturated fatty acid in the sn-2 position, resulting in a lipid radical that can initiate a chain reaction. Quenching of the chain reaction (for example, by α-tocopherol) can prevent further oxidation of lipids but does not alter the lipid hydroperoxides that are already formed; these can break down gradually to products including malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE) that have signaling or toxic effects and are commonly assayed to detect lipid peroxidation [126]. Peroxidation of cell membranes generally leads to cell death, primarily by apoptosis or by the recently described pathway of ferroptosis, although extensive injury can result in cellular necrosis as well [126,127]. Thus, the repair of peroxidized cell membranes is crucial for the survival of cells with oxidatively damaged cell membranes.
Two major reaction pathways have been proposed for the reduction of oxidized cell membrane phospholipids. The first is the direct reduction of the phospholipid hydroperoxides through PHGPx activity [128,129]. Prdx6 and GPx4 are the only two enzymes that have been demonstrated to reduce phospholipid hydroperoxides at a significant rate; both proteins are present in cytoplasm and use GSH as the physiological reductant. GPx4 also can reduce cholesterol hydroperoxides that may be present in oxidized cell membranes [59]; the possible reduction of that substrate by Prdx6 has not yet been evaluated. Loss of GPx4 (GPx4 null mice) results in fetal lethality and in cell death by ferroptosis, although whether those effects reflect primarily the loss of PHGPx activity is unclear [127,130]. Since the relative expression of Prdx6 and GPx4 may vary in different organs, their relative roles in cell membrane repair also may vary with cell type.
The other mechanism for “repair” of peroxidized phospholipids in cell membranes is the deacylation/reacylation pathway, also called a remodeling or the Lands pathway [129,131]. In this pathway, the peroxidized phospholipid is hydrolyzed to remove the oxidized sn-2 fatty acyl chain, generating a lysolipid that then is reacylated with a non-oxidized fatty acid; reacylation restores the normally reduced state of membrane lipids. The PLA2 activity of Prdx6 could catalyze the first step in this pathway while the LPCAT activity of Prdx6 could catalyze the second step of the deacylation/reacylation pathway. Thus, Prdx6 expresses all 3 activities that are known to be involved in the repair of peroxidized cell membranes (Fig. 6). In essence, the PHGPx plus the PLA2/LPCAT activities of Prdx6 can repair peroxidized cell membranes by either direct reduction, deacylation/reacylation, or both pathways acting in concert. Prdx6 thus represents a complete protein for the enzymatic repair of cell membrane lipid peroxidation.
Fig. 6. Role of Prdx6 in the repair of phospholipid hydroperoxides.
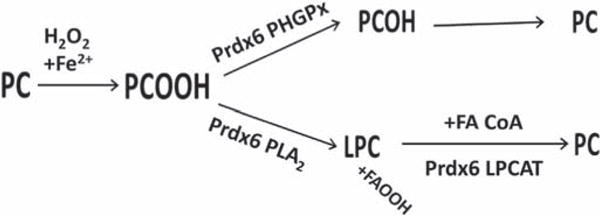
Phosphatidylcholine (PC) containing an sn-2 unsaturated fatty acid can be oxidized by H2O2 (+Fe2+) to PC hydroperoxide (PCOOH). To restore the normal state, the hydroperoxide can be reduced by the phospholipid hydroperoxide GSH peroxidase (PHGPx) activity of Prdx6 to the PC alcohol that is then reduced to the sulfhydryl. An alternative pathway for removal of PCOOH is hydrolysis at the sn-2 position mediated by Prdx6 PLA2 activity to generate lysoPC plus fatty acyl hydroperoxide; lysoPC can be reacylated with an acyl CoA substrate by the LPCAT activity of Prdx6 to regenerate the reduced phospholipid. Enzymes responsible for reduction of the PC alcohol (PCOH) and fatty acid hydroperoxide (FAOOH) that result from these reactions are not shown. Modified from Ref. [132] and reprinted with permission.
We have studied the role of Prdx6 in the repair of lipid peroxidation in lungs from mice exposed to hyperoxia and in isolated mouse lungs and isolated pulmonary microvascular endothelial cells exposed to tert-butyl hydroperoxide (t-BOOH). The content of lipid peroxidation products in wild type lungs and cells from these models of injury showed a gradual decrease over several hours following removal of the oxidant stress; however, there was essentially no repair in the absence of Prdx6 during the time frame studied [132] (Fig. 7). Restoration of either the PLA2 activity or the peroxidase activity of Prdx6 through transfection of the null cells with relevant mutants demonstrated that either activity was partially effective in reducing the degree of membrane phospholipid peroxidation and improved cell survival; transfection with Prdx6 constructs leading to expression of both activities restored the wild type phenotype [121,132]. Mutants that specifically lack LPCAT expression have not yet been evaluated. The results obtained using transfection with mutant proteins provide evidence that Prdx6 does indeed have an important role in the repair of peroxidized cell membranes, at least in lungs, and that both its PHGPx and PLA2 activities play important roles. Of note, GPx4 appeared to play no role in the repair of the peroxidized lung cell membranes in these models.
Fig. 7. Recovery of isolated perfused mouse lungs from lipid peroxidation induced by exposure to t-BOOH.
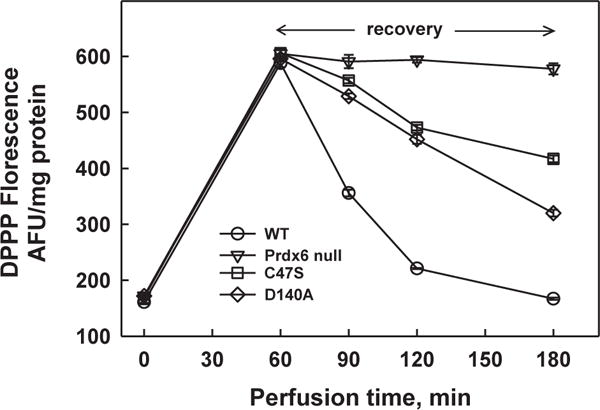
Lungs were exposed to 25 mM (wild type) or 15 mM (mutants) t-BOOH for 1 h followed by 2 h of perfusion with oxidant –free medium. Different concentrations of t-BOOH were used for wild type and mutant lungs to produce equal degrees of oxidant stress during the exposure period. Lung homogenate was analyzed for fluorescence of diphenyl-1-pyrenylphosphine (DPPP), an indicator for phospholipid hydroperoxides [122,163]. The increase of DPPP fluorescence indicates lipid peroxidation. Lungs were from wild type (red), Prdx6 null (blue), C47S Prdx6 knock-in (black), and D140A Prdx6 knock-in (green) mice. The C47S mice do not express the peroxidase activity of Prdx6; the D140A mice do not express the PLA2 activity of Prdx6. Each point is mean for n = 3, the small SE is not shown. Modified and reprinted with permission from Ref. [132].
Prdx6 is present in the cytoplasm of cells but is not a cell membrane protein. So, how does it act to reduce oxidized membrane phospholipids? As described above, Prdx6 in cytosol (pH 7) does not normally bind to phospholipids; however, it does bind with high affinity at cytosolic pH to oxidized phospholipids, thus becoming a membrane associated protein [33]. In the membrane-bound state, there is access to phospholipid hydroperoxide substrate for PHGPx activity of Prdx6 and also activation of its PLA2 activity. This oxidation-mediated translocation to the membranes allows Prdx6 to be inactive as a cytosolic protein (PLA2 activity under resting conditions generally would be undesirable) but to activate following oxidative stress as necessary for cell membrane repair (Fig. 8). Oxidative stress-induced translocation to the plasma membrane has been shown with A549 cells in culture [33]; oxidative stress to liver resulted in translocation of Prdx6 to mitochondria, possibly reflecting its binding intracellularly to oxidized mitochondrial membranes [133].
Fig. 8. Mechanism for repair of oxidized cell membranes.
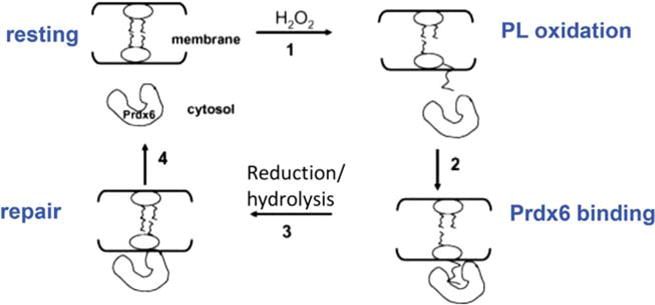
Prdx6 is cytosolic under normal resting conditions. Oxidation of a membrane phospholipid fatty acyl residue results in its increased hydrophilicity and its ‘flotation’ toward the membrane surface; interaction of Prdx6 with the oxidized lipid results in cell membrane association of the protein. Reduction and/or hydrolysis (plus acyl transferase) of the oxidized fatty acyl moiety ‘repairs’ the cell membrane phospholipid. Prdx6 then dissociates from the membrane to return to the resting state. Prdx6 exists as a dimer in the cytoplasm but only the peroxidatic monomer is shown. Modified from Ref. [33] and reprinted with permission.
4.3.2. Reproductive efficacy
A role for Prdx6 in reproduction was indicated by finding that the total number of litters, the litter size, and the total number of pups were significantly lower per Prdx6 null male as compared to WT controls [124]. Examination of sperm from 2-, 8-, and 20 month old male Prdx6 null mice showed age-dependent impairment of motility and maturation, increased DNA oxidation and fragmentation, and decreased DNA compaction and protamination [124]. These results suggest a protective role of Prdx6 in the age-associated decline in mouse sperm quality and fertility. This protective effect is presumably related to the anti-oxidant and the cell membrane repair roles of Prdx6 [123]. Interestingly, infertile men with low sperm motility and evidence of DNA damage have lower levels of Prdx6 in sperm as compared to control men [134].
4.4. Phospholipid synthesis and catabolism
Prdx6 is important in the metabolism of lung surfactant, a secreted product of lung epithelium that is essential for maintaining normal lung function. Lung surfactant comprises specific proteins (~10% of mass), phospholipids (~75%), the major fraction of which is dipalmitoyl phosphatidylcholine (DPPC), and neutral lipids (~15%). Surfactant components are synthesized in the endoplasmic reticulum, transported to the LB, and secreted by exocytosis into the alveolar airspaces [99]. After a short extracellular residence (1/2 time ~10 h), phospholipids (and proteins) are endocytosed by a receptor-mediated pathway and delivered to LB for either degradation or remodeling and resecretion [99,135,136]. The physiological role of this recycling pathway is not clear, although both quality as well as quantity control have been considered as possible “reasons”.
Prdx6 participates in both the degradation and remodeling pathways for phospholipid metabolism in the LB [73–76,79,136] (Fig. 9). The processes of degradation and remodeling both start with PLA2 activity to generate lysoPC. As the LB pH is ~5 [98], Prdx6 (non-phosphorylated) can bind to and hydrolyze native phospholipids. The degradative pathways result in liberation of free fatty acids and choline phosphate that can be reutilized for de novo phosphatidylcholine synthesis by the Kennedy pathway [131] or otherwise metabolized. The remodeling pathway is essentially the same as described for repair of peroxidized membrane phospholipids, although the phospholipid substrate in this case generally is not oxidized.
Fig. 9. Pathways for synthesis of dipalmitoyl phosphatidylcholine.
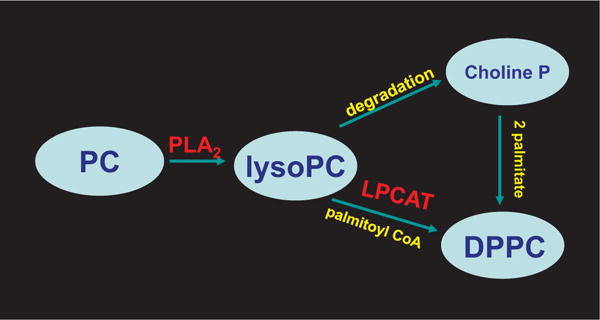
These pathways are important for the generation of the major surface active component of the lung surfactant. Lysophosphatidylcholine (lysoPC) is produced by hydrolysis of the sn-2 acyl bond of phosphatidylcholine (PC) through the PLA2 activity of Prdx6. LysoPC can be further degraded by various enzymes; the choline phosphate moiety is conserved and used along with two palmitates for DPPC synthesis by the de novo pathway. sn-1Palmitoyl lysoPC also can be re-acylated with a palmitoyl CoA by the LPCAT activity of Prdx6 (remodeling pathway) to generate DPPC.
Lung surfactant requires dipalmitoylphosphatidyl choline (DPPC) for its surface-active properties and this product can be provided by either de novo synthesis or by remodeling of phosphatidylcholines that do not contain palmitate in the sn-2 position [99,135]. The generation of DPPC is crucial to the proper function of the lung surfactant. The interaction of Prdx6 with SP-A, an endogenous regulator of the PLA2 activity of Prdx6 (see above), presumably plays an important role in the regulation of PC degradation and remodeling in lung LB, although the precise details of this regulation have not been investigated.
Lungs from Prdx6 null mice show a markedly decreased turnover rate of lung surfactant phospholipids with progressive accumulation (normalized to body wt) of phospholipids in the whole lung and alveolar air spaces as the mice age [74]. By contrast, the normalized phospholipid content of wild type lungs is stable following the neonatal period. Isolated lungs from Prdx6 null mice or with chemical (MJ33) inhibition of Prdx6 PLA2 activity in wild type lungs also show decreased lung surfactant phospholipid turnover [74,79]. The inverse, i.e., increased degradation of phospholipids, decreased phospholipid content and increased activity of the reacylation pathway of phospholipid metabolism is seen with lungs from transgenic mice overexpressing Prdx6 [137]. A pattern of altered lung surfactant phospholipid metabolism similar to Prdx6 null is seen in mice that express normal total lung Prdx6 but lack Prdx6 in the lamellar bodies due to a failure of its targeting [68,138]. This latter observation highlights the important role of the lamellar body and its Prdx6 content in lung surfactant metabolism.
4.5. Cell signaling
4.5.1. Activation of NOX2
NOX2, a member of the family of seven NOX/DuOX enzymes that generate or H2O2 as their primary product, has major roles in bacterial killing and cell signaling. NOX2 is normally inactive in the cell and is activated as required. PLA2 activity has long been suspected to have a role in the activation of NOX2, initially considered to be related to the generation of arachidonic acid as a substrate for the generation of eicosanoids [139]. However, an understanding of which one or more of the numerous PLA2 proteins that is involved in the activation process was elusive. Consistent with its known high specificity for the release of arachidonic acid, cytosolic PLA2 (cPLA2) was investigated as the enzyme responsible for NOX2 activation [140]. However, a careful study of the cPLA2 null mouse demonstrated that this enzyme has minimal effect on activation of NOX2 in response to agonists [141]. Further, the role of arachidonate in NOX2 activation proved to be primarily an in vitro phenomenon, although arachidonic acid (as well as other unsaturated fatty acids) may bind to components of the NOX2 complex in vivo to stabilize their expression [142]. Subsequent attention focused on lysophospholipid as the product of PLA2 activity that is required for NOX2 activation.
A possible role for Prdx6 in the activation of NOX2 was first suggested through co-immunoprecipitation of Prdx6 and p67phox from the cytosol of non-stimulated polymorphonuclear leukocytes (PMN) and increased generation by addition of Prdx6 to a reconstituted system containing PMN cell membranes [17]. The role of specific Prdx6 enzymatic activities in the activation process was not determined. Subsequent studies using chemical and molecular inhibition showed that the PLA2 activity of Prdx6 is absolutely required for NOX2 activation in a variety of cell types (endothelium, macrophages, PMN) as well as in several cell lines [83,143,144]. Activation of NOX2 is greatest with MAP kinase-mediated phosphorylation of Prdx6 in response to an appropriate agonist [83] (Fig. 10). Experimental agonists for activation of NOX2 include angiotensin II or phorbol ester as well as some disease models such as LPS administration or the response to ischemia; a requirement for Prdx6 PLA2 activity as a mediator in the activation process has been demonstrated for these specific agonists and interventions [83,144,145]. As noted above, phosphorylation of Prdx6 results in its binding to cell membranes at cytosolic pH and a marked increase in PLA2 activity [69]. PLA2 activity results in the generation of lysoPC plus a free fatty acid as its two products. LysoPC is then hydrolyzed to lysophosphatidic acid (LPA) that signals through its receptor leading to activation of rac [146], one of the co-factors (in addition to phosphorylated p47phox and p67phox) that are necessary for NOX2 activation [147] (Fig. 10). The proteins downstream from Prdx6 that are involved in the rac activation cascade are autotaxin (lysophospholipase D) and LPA receptor 1 (LPAR1), although a possible role for other proteins has not been excluded [146].
Fig. 10. Pathway for generation of lysophosphatidic acid and the activation of NADPH oxidase type 2 (NOX2).
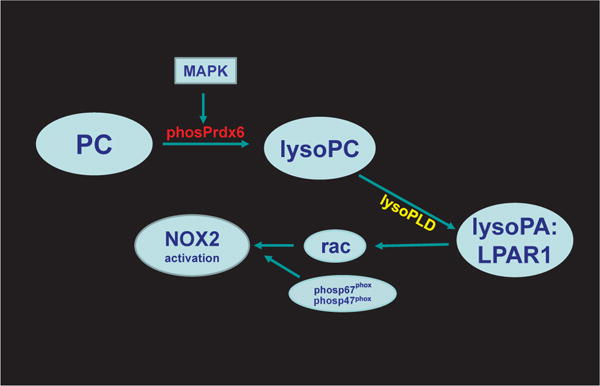
With an appropriate signal, Prdx6 is phosphorylated via mitogen activated protein kinase (MAPK) activity and binds to phospholipid in the cell membrane. LysoPC that is generated from PC by the PLA2 activity of Prdx6 can be metabolized via lysophospholipase D (lysoPLD) to lysophosphatidic acid (lysoPA). Interaction of lysoPA with its receptor (LPAR type 1) leads to release of rac (rac1 or 2 depending on cell type), one of the cytosolic protein co-factors required for NOX2 activation [146]. Other important co-factors for activation include phosphorylated p47phox and phosphorylated p67phox.
Based on its binding to phosphorylated Prdx6 with inhibition of its PLA2 activity (see above), we have postulated that p67phox (but not the phosphorylated protein) can terminate the NOX2 activation process [110]; however, the possible physiological role for this latter interaction needs further study. Prdx6 also has been suggested as required for activation of NOX1, possibly related to its PLA2 activity, but those studies are relatively preliminary and the mechanism for the effect has not been determined [148].
4.5.2. Prdx6 in endothelial cell mechanotransduction
Endothelium is constantly exposed to varying mechanical strain associated with shear stress; an increase or decrease in shear outside of the normal range results in a signaling response termed mechanotransduction. We have investigated this response to reduced shear (that is, simulated ischemia) using an isolated lung preparation and isolated endothelial cells that have been flow adapted in vitro [149,150]. The loss of shear is sensed by a cell membrane complex (mechanosome) that is localized to caveolae; the mechanosome consists of platelet endothelial cell adhesion molecule (PECAM), vascular endothelial growth factor receptor 2 (VEGFR2), vascular endothelial (VE)–cadherin, and possibly other elements [149,151]. The initial response to loss of shear is the inactivation of the inwardly-rectifying KATP channels that results in cell membrane depolarization [149,152]; these channels are normally maintained in the open configuration by shear stress. Cell membrane depolarization is the initiating signal for activation of kinases leading to phosphorylation of Prdx6 with initiation of the cascade that leads to activation of NOX2 (see above) and generation of [153,154]. (or, more likely, H2O2 formed by dismutation of ) is the mediator that signals for the liberation of vascular endothelial growth factor (VEGF) to promote neovascularization, important for repair of the vascular lesion [155]. Cell membrane depolarization also results in activation of voltage gated Ca2+ channels allowing Ca2+ influx into the cell, thereby activating eNOS to generate NO, a vasodilator [156]. These responses (vasodilation and neovascularization) represent homeostatic responses to the absence of shear (i.e., tissue ischemia) and conceptually can restore the impeded blood flow. This paradigm demonstrates a role for Prdx6-linked generation of H2O2 in an important cellular signaling function (Fig. 11). generated by NOX2 also plays an important role in cell motility and a variety of other cell biological processes including hypertrophy, proliferation and migration, and activation of various transcription factors [157], but the role of Prdx6 in the regulation of these activities, while probable, has not yet been studied.
Fig. 11. Scheme for endothelial mechanotransduction.
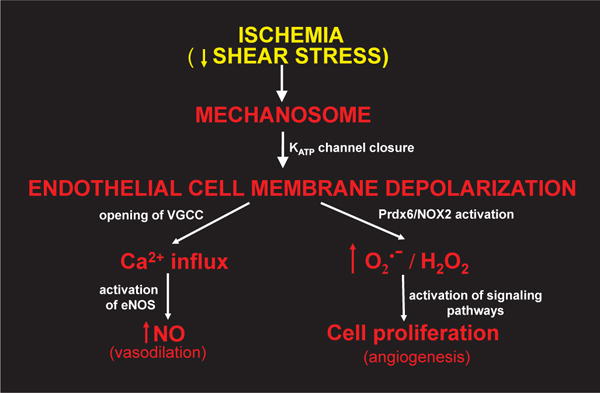
Decreased shear stress associated with stop of blood flow (ischemia) is sensed by the mechanosome leading to closure of inwardly rectifying KATP channels with resultant cell membrane depolarization, phosphorylation of Prdx6, and activation of NOX2 as shown in Fig. 10. The subsequent generation of results in release of angiogenic factors resulting in cellular proliferation and neovascularization. Cell membrane depolarization also leads to opening of voltage-gated Ca channels (VGCC) followed by Ca2+ influx into the cell and generation of the vasodilating agent NO via endothelial NO synthase (eNOS).
4.5.3. Prdx6 as a therapeutic target
In its normal physiological role, activation of NOX2 and the subsequent generation of is important for mediating various important homeostatic functions [158–160]. On the other hand, generation of as a major component of inflammation can be responsible for promoting cell injury in a variety of diseases [161]. One such condition is the syndrome of Acute Lung Injury (ALI) that is associated with sepsis and many other etiologies and has a mortality rate of ~40%. Based on its role in production of H2O2, inhibition of Prdx6 PLA2 activity has been proposed as a mechanism to prevent NOX2 activation in conditions characterized by inflammation. Treatment with an inhibitor of Prdx6 PLA2 activity (MJ33) protected against experimental ALI in mice subjected to intratracheal administration of lipopolysaccharide (LPS) or to a model for lung ischemia-reperfusion [144,145]. These relatively preliminary studies suggest a potential therapeutic role for the inhibition of Prdx6 PLA2 activity to prevent injury from generation associated with NOX2 activation. However, the antioxidant vs pro-oxidant roles of Prdx6 need to be considered when designing therapeutic strategies. An example of these opposing roles is the finding that inhibition of Prdx6 PLA2 activity protects against lung lipid peroxidation with hyperoxia [162] while the protein is required for the repair of peroxidized cell membranes [132]. In essence, the protein (Prdx6) is available to “clean up” its own mess generated by over-exuberant oxidant production by NOX2. Obviously, if lipid peroxidation is prevented, repair will not be necessary, but additional studies with Prdx6 PLA2 inhibitors will be required to sort out these issues.
Acknowledgments
I thank Dr. Mahendra Jain for helping to initiate the studies related to PLA2 activity and his important insights into the catalytic mechanisms for this enzyme. I also thank Dr. Henry Forman for his very helpful suggestions and Drs. David Speicher, Sheldon Fein-stein, Avinash Chander, Yefim Manevich, Shampa Chatterjee, Elena Sorokina, Chandra Dodia, Sandra Harper, Jose Vazquez-Medina and the many other collaborators who have helped to investigate the complexities of peroxidoxin 6 structure and function. Finally, I thank Dawn Williams for assistance in preparing the manuscript.
Abbreviations
- aiPLA2
acidic Ca2+-independent PLA2
- ALI
acute lung injury
- ARE
antioxidant response element
- t-BOOH
tert-butyl hydroperoxide
- DPPC
dipalmitoyl PC
- GSH
glutathione
- GSSG
oxidized GSH
- GPx
GSH peroxidase
- GST
GSH S-transferase
- KGF
keratinocyte growth factor
- LB
lamellar bodies
- LPCAT
lysophosphatidylcholine acyl transferase
- LRO
lysosomal related organelle
- NOX
NADPH oxidase
- PC
phosphatidylcholine
- PCOOH
PC hydroperoxide
- PHGPx
phospholipid hydroperoxide GSH peroxidase
- PLOOH
phospholipid hydroperoxide
- PMN
polymorphonuclear leukocytes
- Prdx
peroxiredoxin
- SP-A
surfactant protein A
References
- 1.Joly JS, Bourrat F, Nguyen V, Chourrout D. Ol-Prx 3, a member of an additional class of homeobox genes, is unimodally expressed in several domains of the developing and adult central nervous system of the medaka (Oryzias latipes) Proc Natl Acad Sci U S A. 1997;94:12987–12992. doi: 10.1073/pnas.94.24.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shichi H, Demar JC. Non-selenium glutathione peroxidase without gluta-thione S-transferase activity from bovine ciliary body. Experim Eye Res. 1990;50:513–520. doi: 10.1016/0014-4835(90)90040-2. [DOI] [PubMed] [Google Scholar]
- 3.Peshenko IV, Novoselov VI, Evdokimov VA, Yu Nikolaev V, Shuvaeva TM, Lipkin VM, Fesenko EE. Novel 28-kDa secretory protein from rat olfactory epithelium. FEBS Lett. 1996;381:12–14. doi: 10.1016/0014-5793(96)00071-3. [DOI] [PubMed] [Google Scholar]
- 4.Akiba S, Dodia C, Chen X, Fisher AB. Characterization of acidic Ca(2+)- independent phospholipase A2 of bovine lung. Comp Biochem Physiol B Biochem Mol Biol. 1998;120:393–404. doi: 10.1016/s0305-0491(98)10046-9. [DOI] [PubMed] [Google Scholar]
- 5.Peshenko IV, Novoselov VI, Evdokimov VA, Nikolaev YV, Kamzalov SS, Shuvaeva TM, Lipkin VM, Fesenko EE. Identification of a 28 kDa secretory protein from rat olfactory epithelium as a thiol-specific antioxidant. Free Radic Biol Med. 1998;25:654–659. doi: 10.1016/s0891-5849(98)00111-7. [DOI] [PubMed] [Google Scholar]
- 6.Kim TS, Dodia C, Chen X, Hennigan BB, Jain M, Feinstein SI, Fisher AB. Cloning and expression of rat lung acidic Ca(2+)-independent PLA2 and its organ distribution. Am J Physiol Lung Cell Mol Physiol. 1998;274:L750–L761. doi: 10.1152/ajplung.1998.274.5.L750. [DOI] [PubMed] [Google Scholar]
- 7.Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci U S A. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim TS, Sundaresh CS, Feinstein SI, Dodia C, Skach WR, Jain MK, Nagase T, Seki N, Ishikawa K, Nomura N, Fisher AB. Identification of a human cDNA clone for lysosomal type Ca2+-independent phospholipase A2 and properties of the expressed protein. J Biol Chem. 1997;272:2542–2550. doi: 10.1074/jbc.272.4.2542. [DOI] [PubMed] [Google Scholar]
- 9.Nagase T, Miyajima N, Tanaka A, Sazuka T, Seki N, Sato S, Tabata S, Ishikawa K, Kawarabayasi Y, Kotani H, Nomura N. Prediction of the coding sequences of unidentified human genes. III. The coding sequences of 40 new genes (KIAA0081-KIAA0120) deduced by analysis of cDNA clones from human cell line KG-1 (supplement) DNA Res. 1995;2:51–59. doi: 10.1093/dnares/2.1.51. [DOI] [PubMed] [Google Scholar]
- 10.Phelan SA, Johnson KA, Beier DR, Paigen B. Characterization of the murine gene encoding Aop2 (antioxidant protein 2) and identification of two highly related genes. Genomics. 1998;54:132–139. doi: 10.1006/geno.1998.5568. [DOI] [PubMed] [Google Scholar]
- 11.Lee TH, Yu SL, Kim SU, Kim YM, Choi I, Kang SW, Rhee SG, Yu DY. Characterization of the murine gene encoding 1-Cys peroxiredoxin and identification of highly homologous genes. Gene. 1999;234:337–344. doi: 10.1016/s0378-1119(99)00190-0. [DOI] [PubMed] [Google Scholar]
- 12.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Fisher AB, Dodia C, Manevich Y, Chen JW, Feinstein SI. Phospholipid hydroperoxides are substrates for non-selenium glutathione peroxidase. J Biol Chem. 1999;274:21326–21334. doi: 10.1074/jbc.274.30.21326. [DOI] [PubMed] [Google Scholar]
- 14.Frank S, Munz B, Werner S. The human homologue of a bovine non-selenium glutathione peroxidase is a novel keratinocyte growth factor-regulated gene. Oncogene. 1997;14:915–921. doi: 10.1038/sj.onc.1200905. [DOI] [PubMed] [Google Scholar]
- 15.Phelan SA. AOP2 (antioxidant protein 2): structure and function of a unique thiol-specific antioxidant. Antioxid Redox Signal. 1999;1:571–584. doi: 10.1089/ars.1999.1.4-571. [DOI] [PubMed] [Google Scholar]
- 16.Power JH, Nicholas TE. Immunohistochemical localization and characterization of a rat Clara cell 26-kDa protein (CC26) with similarities to glutathione peroxidase and phospholipase A2. Exp Lung Res. 1999;25:379–392. doi: 10.1080/019021499270141. [DOI] [PubMed] [Google Scholar]
- 17.Leavey PJ, Gonzalez-Aller C, Thurman G, Kleinberg M, Rinckel L, Ambruso DW, Freeman S, Kuypers FA, Ambruso DR. A 29-kDa protein associated with p67phox expresses both peroxiredoxin and phospholipase A2 activity and enhances superoxide anion production by a cell-free system of NADPH oxidase activity. J Biol Chem. 2002;277:45181–45187. doi: 10.1074/jbc.M202869200. [DOI] [PubMed] [Google Scholar]
- 18.Iakoubova OA, Pacella LA, Her H, Beier DR. LTW4 protein on mouse chromosome 1 is a member of a family of antioxidant proteins. Genomics. 1997;42:474–478. doi: 10.1006/geno.1997.4762. [DOI] [PubMed] [Google Scholar]
- 19.Nevalainen TJ. 1-Cysteine peroxiredoxin: a dual-function ezyme with peroxidase and acidic Ca++-independent phospholipase A2 activities. Biochimie. 2010;92:638, 634. doi: 10.1016/j.biochi.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 20.Nelson KJ, Knutson ST, Soito L, Klomsiri C, Poole LB, Fetrow JS. Analysis of the peroxiredoxin family: using active-site structure and sequence information for global classification and residue analysis. Protieins. 2011;79:947–964. doi: 10.1002/prot.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolomeo AM, Carraro A, Bakiu R, Toppo S, Place SP, Ferro D, Santovito G. Peroxiredoxin 6 from the Antarctic emerald rockcod: molecular characterization of its response to warming. J Comp Physiol B. 2015;186:59–71. doi: 10.1007/s00360-015-0935-3. [DOI] [PubMed] [Google Scholar]
- 22.Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- 23.Barranco-Medina S, Lazaro JJ, Dietz KJ. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009:1809–1816. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 24.Mizohata E, Sakai H, Fusatomi E, Terada T, Murayama K, Shirouzu M, Yokoyama S. Crystal structure of an archaeal peroxiredoxin from the aerobic hyperthermophilic crenarchaeon Aeropyrum pernix K1. J Mol Biol. 2005;354:317–329. doi: 10.1016/j.jmb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends biochem Sci. 2015;40:435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manevich Y, Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Schremmer B, Manevich Y, Feinstein SI, Fisher AB. Peroxiredoxins in the lung with emphasis on peroxiredoxin VI. Subcell Biochem. 2007;44:317–344. doi: 10.1007/978-1-4020-6051-9_15. [DOI] [PubMed] [Google Scholar]
- 28.Fisher AB. Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antiox Redox Signal. 2011;15:831–844. doi: 10.1089/ars.2010.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peshenko IV, Shichi H. Oxidation of active center cysteine of bovine 1-Cys peroxiredoxin to the cysteine sulfenic acid form by peroxide and peroxynitrite. Free Radic Biol Med. 2001;31:292–303. doi: 10.1016/s0891-5849(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 30.Chen JW, Dodia C, Feinstein SI, Jain MK, Fisher AB. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem. 2000;275:28421–28427. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- 31.Hall A, Nelson K, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid Redox Signal. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang SW, Baines IC, Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- 33.Manevich Y, Shuvaeva T, Dodia C, Kazi A, Feinstein SI, Fisher AB. Binding of peroxiredoxin 6 to substrate determines differential phospholipid hydroperoxide peroxidase and phospholipase A(2) activities. Arch Biochem Biophys. 2009;485:139–149. doi: 10.1016/j.abb.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toledo JC, Jr, Audi R, Ogusucu R, Monteiro G, Netto LE, Augusto O. Horseradish peroxidase compound I as a tool to investigate reactive protein-cysteine residues: from quantification to kinetics. Free Radic Biol Med. 2011;50:1032–1038. doi: 10.1016/j.freeradbiomed.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 35.Zhou S, Sorokina E, Harper S, Ralat L, Dodia C, Speicher D, Feinstein SI, Fisher A. Peroxiredoxin 6 homodimerization and heterodimerization with glutathione S-transferase pi are required for its peroxidase but not phospholipase A2 activity. Free Radic Biol Med. 2016;94:145–156. doi: 10.1016/j.freeradbiomed.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-Cys peroxiredoxin requires glutathionylation mediated by hetero-dimerization with pi GST. Proc Natl Acad Sci U S A. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ralat LA, Manevich Y, Fisher AB, Colman RF. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochemistry. 2006;45:360–372. doi: 10.1021/bi0520737. [DOI] [PubMed] [Google Scholar]
- 38.Chae HZ, Oubrahim H, Park JW, Rhee SG, Chock PB. Protein glutathionylation in the regulation of peroxiredoxins: a family of thiol-specific peroxidases that function as antioxidants, molecular chaperones, and signal modulators. Antioxid Redox Signal. 2012;16:506–523. doi: 10.1089/ars.2011.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peshenko IV, Singh AK, Shichi H. Bovine eye 1-Cys peroxiredoxin: expression in E. coli and antioxidant properties. J Ocul Pharmacol Ther. 2001;17:93–99. doi: 10.1089/108076801750125775. [DOI] [PubMed] [Google Scholar]
- 40.Ralat LA, Misquitta SA, Manevich Y, Fisher AB, Colman RF. Characterization of the complex of glutathione S-transferase pi and 1-cysteine peroxiredoxin. Arch Biochem Biophys. 2008;474:109–118. doi: 10.1016/j.abb.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manevich Y, Hutchens S, Tew KD, Townsend DM. Allelic variants of glutathione S-transferase P1-1 differentially mediate the peroxidase function of peroxiredoxin VI and alter membrane lipid peroxidation. Free Radic Biol Med. 2013;54:62–70. doi: 10.1016/j.freeradbiomed.2012.10.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou S, Lien YC, Shuvaeva T, DeBolt K, Feinstein SI, Fisher AB. Functional interaction of glutathione S-transferase pi and peroxiredoxin 6 in intact cells. Intern J Biochem Cell Biol. 2013;45:401–407. doi: 10.1016/j.biocel.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat Struct Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 44.Kim KH, Lee W, Kim EE. Crystal structures of human peroxiredoxin 6 in different oxidation states. Biochem Biophys Res Commun. 2016;477:717–722. doi: 10.1016/j.bbrc.2016.06.125. [DOI] [PubMed] [Google Scholar]
- 45.Rivera-Santiago RF, Harper SL, Zhou S, Sriswasdi S, Feinstein SI, Fisher AB, Speicher DW. Solution structure of the reduced form of human peroxiredoxin-6 elucidated using zero-length chemical cross-linking and homology modelling. Biochem J. 2015;468:87–98. doi: 10.1042/BJ20141463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poole LB, Nelson KJ. Distribution and features of the six classes of peroxiredoxins. Mol Cells. 2016;39:53–59. doi: 10.14348/molcells.2016.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Board PG, Menon D. Glutathione transferases, regulators of cellular metabolism and physiology. Biochim Biophys Acta. 2013;1830:3267–3288. doi: 10.1016/j.bbagen.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 48.da Fonseca RR, Johnson WE, O’Brien SJ, Vasconcelos V, Antunes A. Molecular evolution and the role of oxidative stress in the expansion and functional diversification of cytosolic glutathione transferases. BMC Evol Biol. 2010;10:281. doi: 10.1186/1471-2148-10-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terrier P, Townsend AJ, Coindre JM, Triche TJ, Cowan KH. An immunohistochemical study of pi class glutathione S-transferase expression in normal human tissue. Am J Pathol. 1990;137:845–853. [PMC free article] [PubMed] [Google Scholar]
- 50.Gallagher BM, Phelan SA. Investigating transcriptional regulation of Prdx6 in mouse liver cells. Free Radic Biol Med. 2007;42:1270–1277. doi: 10.1016/j.freeradbiomed.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 51.Monteiro G, Horta BB, Pimenta DC, Augusto O, Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci U S A. 2007;104:4886–4891. doi: 10.1073/pnas.0700481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greetham D, Grant CM. Antioxidant activity of the yeast mitochondrial one-Cys peroxiredoxin is dependent on thioredoxin reductase and glutathione in vivo. Mol Cell Biol. 2009;29:3229–3240. doi: 10.1128/MCB.01918-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedrajas JR, Padilla CA, McDonagh B, Barcena JA. Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in Saccharomyces cerevisiae. Antioxid Redox Signal. 2010;13:249–258. doi: 10.1089/ars.2009.2950. [DOI] [PubMed] [Google Scholar]
- 54.Dietz KJ. Peroxiredoxins in plants and cyanobacteria, Antioxid. Redox Signal. 2011;15:1129–1159. doi: 10.1089/ars.2010.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SP, Hwang YS, Kim YJ, Kwon KS, Kim HJ, Kim K, Chae HZ. Cyclophilin A binds to peroxiredoxins and activates its peroxidase activity. J Biol Chem. 2001;276:29826–29832. doi: 10.1074/jbc.M101822200. [DOI] [PubMed] [Google Scholar]
- 56.Fukuhara R, Kageyama T. Structure, gene expression, and evolution of primate glutathione peroxidases. Comp Biochem Physiol B Biochem Mol Biol. 2005;141:428–436. doi: 10.1016/j.cbpc.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 57.Maiorino M, Gregolin C, Ursini F. Phospholipid hydroperoxide glutathione peroxidase. Meth Enzymol. 1990;186:448–457. doi: 10.1016/0076-6879(90)86139-m. [DOI] [PubMed] [Google Scholar]
- 58.Han X, Fan Z, Yu Y, Liu S, Hao Y, Huo R, Wei J. Expression and characterization of recombinant human phospholipid hydroperoxide glutathione peroxidase. IUBMB Life. 2013;65:951–956. doi: 10.1002/iub.1220. [DOI] [PubMed] [Google Scholar]
- 59.Kernstock RM, Girotti AW. New strategies for the isolation and activity determination of naturally occurring type-4 glutathione peroxidase. Prot Expr Purif. 2008;62:216–222. doi: 10.1016/j.pep.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weitzel F, Ursini F, Wendel A. Phospholipid hydroperoxide glutathione peroxidase in various mouse organs during selenium deficiency and repletion. Biochim Biophys Acta. 1990;1036:88–94. doi: 10.1016/0304-4165(90)90018-r. [DOI] [PubMed] [Google Scholar]
- 61.Brigelius-Flohe R. Tissue-specific functions of individual glutathione per-oxidases. Free Radic Biol Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- 62.Baek IJ, Seo DS, Yon JM, Lee SR, Jin Y, Nahm SS, Jeong JH, Choo YK, Kang JK, Lee BJ, Yun YW, Nam SY. Tissue expression and cellular localization of phospholipid hydroperoxide glutathione peroxidase (PHGPx) mRNA in male mice. J Mol Histol. 2007;38:237–244. doi: 10.1007/s10735-007-9092-7. [DOI] [PubMed] [Google Scholar]
- 63.Brutsch SH, Wang CC, Li L, Stender H, Neziroglu N, Richter C, Kuhn H, Borchert A. Expression of inactive glutathione peroxidase 4 leads to embryonic lethality, and inactivation of the Alox15 gene does not rescue such knock-in mice. Antioxid Redox Signal. 2015;22:281–293. doi: 10.1089/ars.2014.5967. [DOI] [PubMed] [Google Scholar]
- 64.Liu G, Feinstein SI, Wang Y, Dodia C, Fisher D, Yu K, Ho YS, Fisher AB. Comparison of glutathione peroxidase 1 and peroxiredoxin 6 in protection against oxidative stress in the mouse lung. Free Radic Biol Med. 2010;49:1172–1181. doi: 10.1016/j.freeradbiomed.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woo HA, Jeong W, Chang TS, Park KJ, Park SJ, Yang JS, Rhee SG. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J Biol Chem. 2005;280:3125–3128. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 66.Claiborne A, Mallett TC, Yeh JI, Luba J, Parsonage D. Structural, redox, and mechanistic parameters for cysteine-sulfenic acid function in catalysis and regulation. Adv Prot Chem. 2001;58:215–276. doi: 10.1016/s0065-3233(01)58006-7. [DOI] [PubMed] [Google Scholar]
- 67.Lindgren J, Karlstrom A Eriksson. Intramolecular thioether crosslinking of therapeutic proteins to increase proteolytic stability. Chembiochem. 2014;15:2132–2138. doi: 10.1002/cbic.201400002. [DOI] [PubMed] [Google Scholar]
- 68.Sorokina EM, Dodia C, Zhou S, Tao JQ, Gao L, Raabe T, Feinstein SI, Fisher AB. Mutation of serine 32 to threonine in peroxiredoxin 6 preserves its structure and enzymatic function but abolishes its trafficking to lamellar bodies. J Biol Chem. 2016;291:9268–9280. doi: 10.1074/jbc.M115.698894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Y, Feinstein SI, Manevich Y, Chowdhury I, Pak JH, Kazi A, Dodia C, Speicher DW, Fisher AB. Mitogen-activated protein kinase-mediated phosphorylation of peroxiredoxin 6 regulates its phospholipase A(2) activity. Biochem J. 2009;419:669–679. doi: 10.1042/BJ20082061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu YZ, Manevich Y, Baldwin JL, Dodia C, Yu K, Feinstein SI, Fisher AB. Interaction of surfactant protein A with peroxiredoxin 6 regulates phospholipase A2 activity. J Biol Chem. 2006;281:7515–7525. doi: 10.1074/jbc.M504525200. [DOI] [PubMed] [Google Scholar]
- 71.Kramer RM, Johansen B, Hession C, Pepinsky RB. Structure and properties of a secretable phospholipase A2 from human platelets. Adv Exp Med Biol. 1990;275:35–53. doi: 10.1007/978-1-4684-5805-3_3. [DOI] [PubMed] [Google Scholar]
- 72.Ackermann EJ, Kempner ES, Dennis EA. Ca(2+)-independent cytosolic phospholipase A2 from macrophage-like P388D1+cells. Isolation and characterization. J Biol Chem. 1994;269:9227–9233. [PubMed] [Google Scholar]
- 73.Fisher AB, Dodia C. Role of phospholipase A2 enzymes in degradation of dipalmitoylphosphatidylcholine by granular pneumocytes. J Lipid Res. 1996;37:1057–1064. [PubMed] [Google Scholar]
- 74.Fisher AB, Dodia C, Feinstein SI, Ho YS. Altered lung phospholipid metabolism in mice with targeted deletion of lysosomal-type phospholipase A2. J Lipid Res. 2005;46:1248–1256. doi: 10.1194/jlr.M400499-JLR200. [DOI] [PubMed] [Google Scholar]
- 75.Fisher AB, Dodia C. Role of acidic Ca2+-independent phospholipase A2 in synthesis of lung dipalmitoyl phosphatidylcholine. Am J Physiol Lung Cell Mol Physiol. 1997;272:L238–L243. doi: 10.1152/ajplung.1997.272.2.L238. [DOI] [PubMed] [Google Scholar]
- 76.Fisher AB, Dodia C. Lysosomal-type PLA2 and turnover of alveolar DPPC. Am J Physiol Lung Cell Mol Physiol. 2001;280:L748–L754. doi: 10.1152/ajplung.2001.280.4.L748. [DOI] [PubMed] [Google Scholar]
- 77.Hiraoka M, Abe A, Shayman JA. Cloning and characterization of a lysosomal phospholipase A2, 1-O-acylceramide synthase. J Biol Chem. 2002;277:10090–10099. doi: 10.1074/jbc.M111977200. [DOI] [PubMed] [Google Scholar]
- 78.Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB. Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res. 2007;48:2306–2318. doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- 79.Fisher AB, Dodia C, Chander A, Jain M. A competitive inhibitor of phospholipase A2 decreases surfactant phosphatidylcholine degradation by the rat lung. Biochem J. 1992;288(Pt 2):407–411. doi: 10.1042/bj2880407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jain MK, Yu BZ, Rogers J, Ranadive GN, Berg OG. Interfacial catalysis by phospholipase A2: dissociation constants for calcium, substrate, products, and competitive inhibitors. Biochemistry. 1991;30:7306–7317. doi: 10.1021/bi00243a036. [DOI] [PubMed] [Google Scholar]
- 81.Fisher AB, Jain M. Encyclopedia of Life Sciences. John Wiley & Sons; Chichester: 2009. Phospholipases: degradation of phospholipids in membranes and emulsions. [Google Scholar]
- 82.Stryer L, Berg JM, Tymoczko J. Biochemistry. 5th. W H Freeman; San Francisco, USA: 2002. Chapt. 9.1. [Google Scholar]
- 83.Chatterjee S, Feinstein SI, Dodia C, Sorokina E, Lien YC, Nguyen S, Debolt K, Speicher D, Fisher AB. Peroxiredoxin 6 phosphorylation and subsequent phospholipase A2 activity are required for agonist-mediated activation of NADPH oxidase in mouse pulmonary microvascular endothelium and alveolar macrophages. J Biol Chem. 2011;286:11696–11706. doi: 10.1074/jbc.M110.206623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fisher AB, Dodia C, Sorokina EM, Li H, Zhou S, Raabe T, Feinstein SI. A novel lysoPhosphatidylcholine acyl transferase activity is expressed by peroxiredoxin 6. J Lipid Res. 2016;57:587–596. doi: 10.1194/jlr.M064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shindou H, Hishikawa D, Harayama T, Yuki K, Shimizu T. Recent progress on acyl CoA: lysophospholipid acyltransferase research, Suppl. J Lipid Res. 2009;50:S46–S51. doi: 10.1194/jlr.R800035-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen X, Hyatt BA, Mucenski ML, Mason RJ, Shannon JM. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc Natl Acad Sci U S A. 2006;103:11724–11729. doi: 10.1073/pnas.0604946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soupene E, Fyrst H, Kuypers FA. Mammalian acyl-CoA: lysophosphatidylcholine acyltransferase enzymes. Proc Natl Acad Sci U S A. 2008;105:88–93. doi: 10.1073/pnas.0709737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rahaman H, Zhou S, Dodia C, Feinstein SI, Huang S, Speicher D, Fisher AB. Increased phospholipase A2 activity with phosphorylation of peroxiredoxin 6 requires a conformational change in the protein. Biochemistry. 2012;51:5521–5530. doi: 10.1021/bi300380h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bemporad F, Gsponer J, Hopearuoho HI, Plakoutsi G, Stati G, Stefani M, Taddei N, Vendruscolo M, Chiti F. Biological function in a non-native partially folded state of a protein. EMBO J. 2008;27:1525–1535. doi: 10.1038/emboj.2008.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pervushin K, Vamvaca K, Vogeli B, Hilvert D. Structure and dynamics of a molten globular enzyme. Nat Struct Mol Biol. 2007;14:1202–1206. doi: 10.1038/nsmb1325. [DOI] [PubMed] [Google Scholar]
- 91.Uversky VN, Kutyshenko VP, Protasova N, Rogov VV, Vassilenko KS, Gudkov AT. Circularly permuted dihydrofolate reductase possesses all the properties of the molten globule state, but can resume functional tertiary structure by interaction with its ligands. Prot Sci. 1996;5:1844–1851. [Google Scholar]
- 92.Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343(Pt 1):177–183. [PMC free article] [PubMed] [Google Scholar]
- 93.Derewenda ZS, Sharp AM. News from the interface: the molecular structures of triacylglyceride lipases. Trends Biochem Sci. 1993;18:20–25. doi: 10.1016/0968-0004(93)90082-x. [DOI] [PubMed] [Google Scholar]
- 94.Fujii T, Fujii J, Taniguchi N. Augmented expression of peroxiredoxin VI in rat lung and kidney after birth implies an antioxidative role. Eur J Biochem. 2001;268:218–225. doi: 10.1046/j.1432-1033.2001.01843.x. [DOI] [PubMed] [Google Scholar]
- 95.Sorokina EM, Feinstein SI, Zhou S, Fisher AB. Intracellular targeting of peroxiredoxin 6 to lysosomal organelles requires MAPK activity and binding to 14-3-3epsilon. Am J Physiol Lung Cell Mol Physiol. 2011;300:C1430–C1441. doi: 10.1152/ajpcell.00285.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sorokina EM, Feinstein SI, Milovanova TN, Fisher AB. Identification of the amino acid sequence that targets peroxiredoxin 6 to lysosome-like structures of lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2009;297:L871–L880. doi: 10.1152/ajplung.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fisher AB, Dodia C, Chander A. Inhibition of lung calcium-independent phospholipase A2 by surfactant protein A. Am J Physiol Lung Cell Mol Physiol. 1994;267:L335–L341. doi: 10.1152/ajplung.1994.267.3.L335. [DOI] [PubMed] [Google Scholar]
- 98.Chander A, Johnson RG, Reicherter J, Fisher AB. Lung lamellar bodies maintain an acidic internal pH. J Biol Chem. 1986;261:6126–6131. [PubMed] [Google Scholar]
- 99.Fisher AB, Chander A. Intracellular processing of surfactant lipids in the lung. Ann Rev Physiol. 1985;47:789–802. doi: 10.1146/annurev.ph.47.030185.004041. [DOI] [PubMed] [Google Scholar]
- 100.Weaver TE, Na CL, Stahlman M. Biogenesis of lamellar bodies, lysosome-related organelles involved in storage and secretion of pulmonary surfactant. Semin Cell Dev Biol. 2002;13:263–270. doi: 10.1016/s1084952102000551. [DOI] [PubMed] [Google Scholar]
- 101.Fisher AB. Surfactant catabolism and recycling. In: Rooney SA, editor. Lung Surfactant: Clearance and Cellular Processing. Landes Bioscience; Austin, TX: 1998. pp. 165–189. Chapter 8. [Google Scholar]
- 102.Kim HS, Pak JH, Gonzales LW, Feinstein SI, Fisher AB. Regulation of 1-cys peroxiredoxin expression in lung epithelial cells. Am J Respir Cell Mol Biol. 2002;27:227–233. doi: 10.1165/ajrcmb.27.2.20010009oc. [DOI] [PubMed] [Google Scholar]
- 103.Kim HS, Manevich Y, Feinstein SI, Pak JH, Ho YS, Fisher AB. Induction of 1-cys peroxiredoxin expression by oxidative stress in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;285:L363–L369. doi: 10.1152/ajplung.00078.2003. [DOI] [PubMed] [Google Scholar]
- 104.Chowdhury I, Mo Y, Gao L, Kazi A, Fisher AB, Feinstein SI. Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radic Biol Med. 2009;46:146–153. doi: 10.1016/reeradbiomed.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sparling NE, Phelan SA. Identification of multiple transcripts for antioxidant protein 2 (Aop2): differential regulation by oxidative stress and growth factors. Redox Rep. 2003;8:87–94. doi: 10.1179/135100003125001404. [DOI] [PubMed] [Google Scholar]
- 106.Chowdhury I, Fisher AB, Christofidou-Solomidou M, Gao L, Tao JQ, Sorokina EM, Lien YC, Bates SR, Feinstein SI. Keratinocyte growth factor and glucocorticoid induction of human peroxiredoxin 6 gene expression occur by independent mechanisms that are synergistic. Antioxid Redox Signal. 2014;20:391–402. doi: 10.1089/ars.2012.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu X, Ji P, Zhang L, Bu G, Gu H, Wang X, Xiong Y, Zuo B. The expression of porcine Prdx6 gene is up-regulated by C/EBPbeta and CREB. PLoS One. 2015;10:e0144851. doi: 10.1371/journal.pone.0144851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krishnaiah SY, Dodia C, Sorokina EM, Li H, Feinstein SI, Fisher AB. Binding sites for interaction of peroxiredoxin 6 with surfactant protein A. Biochim Biophys Acta. 2016;1864:419–425. doi: 10.1016/j.bbapap.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jain D, Dodia C, Bates SR, Hawgood S, Poulain FR, Fisher AB. SP-A is necessary for increased clearance of alveolar DPPC with hyperventilation or secretagogues. Am J Physiol Lung Cell Mol Physiol. 2003;284:L759–L765. doi: 10.1152/ajplung.00200.2002. [DOI] [PubMed] [Google Scholar]
- 110.Krishnaiah SY, Dodia C, Feinstein SI, Fisher AB. p67phox terminates the phospholipase A2-derived signal for activation of NADPH oxidase (NOX2) FASEB J. 2013;27:2066–2073. doi: 10.1096/fj.12-222133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mo Y, Feinstein SI, Manevich Y, Zhang Q, Lu L, Ho YS, Fisher AB. 1-Cys peroxiredoxin knock-out mice express mRNA but not protein for a highly related intronless gene. FEBS Lett. 2003;555:192–198. doi: 10.1016/s0014-5793(03)01199-2. [DOI] [PubMed] [Google Scholar]
- 112.Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, Brown A, Lerner CP, Paigen B. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem. 2003;278:25179–25190. doi: 10.1074/jbc.M302706200. [DOI] [PubMed] [Google Scholar]
- 113.Dierick JF, Wenders F, Chainiaux F, Remacle J, Fisher AB, Toussaint O. Retrovirally mediated overexpression of peroxiredoxin VI increases the survival of WI-38 human diploid fibroblasts exposed to cytotoxic doses of tertbutylhydroperoxide and UVB. Biogerontology. 2003;4:125–131. doi: 10.1023/a:1024154024602. [DOI] [PubMed] [Google Scholar]
- 114.Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proc Natl Acad Sci U S A. 2002;99:11599–11604. doi: 10.1073/pnas.182384499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Singh SP, Chhunchha B, Fatma N, Kubo E, Singh SP, Singh DP. Delivery of a protein transduction domain-mediated Prdx6 protein ameliorates oxidative stress-induced injury in human and mouse neuronal cells. Am J Physiol Cell Physiol. 2016;310:C1–C16. doi: 10.1152/ajpcell.00229.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Walsh B, Pearl A, Suchy S, Tartaglio J, Visco K, Phelan SA. Overexpression of Prdx6 and resistance to peroxide-induced death in Hepa1-6 cells: Prdx suppression increases apoptosis. Redox Rep. 2009;14:275–284. doi: 10.1179/135100009X12525712409652. [DOI] [PubMed] [Google Scholar]
- 117.Wang Y, Manevich Y, Feinstein SI, Fisher AB. Adenovirus-mediated transfer of the 1-cys peroxiredoxin gene to mouse lung protects against hyperoxic injury. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1188–L1193. doi: 10.1152/ajplung.00288.2003. [DOI] [PubMed] [Google Scholar]
- 118.Wang Y, Phelan SA, Manevich Y, Feinstein SI, Fisher AB. Transgenic mice overexpressing peroxiredoxin 6 show increased resistance to lung injury in hyperoxia. Am J Respir Cell Mol Biol. 2006;34:481–486. doi: 10.1165/rcmb.2005-0333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB. Lung injury and mortality with hyperoxia are increased in peroxiredoxin 6 gene-targeted mice. Free Radic Biol Med. 2004;37:1736–1743. doi: 10.1016/j.freeradbiomed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 120.Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB. Peroxiredoxin 6 gene-targeted mice show increased lung injury with paraquat-induced oxidative stress. Antioxid Redox Signal. 2006;8:229–237. doi: 10.1089/ars.2006.8.229. [DOI] [PubMed] [Google Scholar]
- 121.Lien YC, Feinstein SI, Dodia C, Fisher AB. The roles of peroxidase and phospholipase A2 activities of peroxiredoxin 6 in protecting pulmonary microvascular endothelial cells against peroxidative stress. Antioxid Redox Signal. 2012;16:440–451. doi: 10.1089/ars.2011.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Y, Feinstein SI, Fisher AB. Peroxiredoxin 6 as an antioxidant enzyme: protection of lung alveolar epithelial type II cells from H2O2-induced oxidative stress. J Cell Biochem. 2008;104:1274–1285. doi: 10.1002/jcb.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ozkosem B, Feinstein SI, Fisher AB, O’Flaherty C. Absence of peroxiredoxin 6 amplifies the effect of oxidant stress on mobility and SCSA/CMA3 defined chromatin quality and impairs fertilizing ability of mouse spermatozoa. Biol Reprod. 2016;94:68. doi: 10.1095/biolreprod.115.137646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ozkosem B, Feinstein SI, Fisher AB, O’Flaherty C. Advancing age increases sperm chromatin damage and impairs fertility in peroxiredoxin 6 null mice. Redox Biol. 2015;5:15–23. doi: 10.1016/j.redox.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pak JH, Manevich Y, Kim HS, Feinstein SI, Fisher AB. An antisense oligonucleotide to 1-cys peroxiredoxin causes lipid peroxidation and apoptosis in lung epithelial cells. J Biol Chem. 2002;277:49927–49934. doi: 10.1074/jbc.M204222200. [DOI] [PubMed] [Google Scholar]
- 126.Ayala A, Munoz MF, Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73:2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ursini F, Maiorino M, Gregolin C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim Biophys Acta. 1985;839:62–70. doi: 10.1016/0304-4165(85)90182-5. [DOI] [PubMed] [Google Scholar]
- 129.Sevanian A, Muakkassah-Kelly SF, Montestruque S. The influence of phospholipase A2 and glutathione peroxidase on the elimination of membrane lipid peroxides. Arch Biochem Biophys. 1983;223:441–452. doi: 10.1016/0003-9861(83)90608-2. [DOI] [PubMed] [Google Scholar]
- 130.Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moessinger C, Klizaite K, Steinhagen A, Philippou-Massier J, Shevchenko A, Hoch M, Ejsing CS, Thiele C. Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 2014;15:43. doi: 10.1186/s12860-014-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li H, Benipal B, Zhou S, Dodia C, Chatterjee S, Tao JQ, Sorokina EM, Raabe T, Feinstein SI, Fisher AB. Critical role of peroxiredoxin 6 in the repair of peroxidized cell membranes following oxidative stress. Free Radic Biol Med. 2015;87:356–365. doi: 10.1016/j.freeradbiomed.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eismann T, Huber N, Shin T, Kuboki S, Galloway E, Wyder M, Edwards MJ, Greis KD, Shertzer HG, Fisher AB, Lentsch AB. Peroxir-edoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266–G274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gong S, San Gabriel MC, Zini A, Chan P, O’Flaherty C. Low amounts and high thiol oxidation of peroxiredoxins in spermatozoa from infertile men. J Androl. 2012;33:1342–1351. doi: 10.2164/jandrol.111.016162. [DOI] [PubMed] [Google Scholar]
- 135.Goss V, Hunt AN, Postle AD. Regulation of lung surfactant phospholipid synthesis and metabolism. Biochim Biophys Acta. 2013;1831:448–458. doi: 10.1016/j.bbalip.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 136.Chander A, Reicherter J, Fisher AB. Degradation of dipalmitoyl phosphatidylcholine by isolated rat granular pneumocytes and reutilization for surfactant synthesis. J Clin Invest. 1987;79:1133–1138. doi: 10.1172/JCI112929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fisher AB, Dodia C, Yu K, Manevich Y, Feinstein SI. Lung phospholipid metabolism in transgenic mice overexpressing peroxiredoxin 6. Biochim Biophys Acta. 2006;1761:785–792. doi: 10.1016/j.bbalip.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 138.Kook S, Wang P, Young LR, Schwake M, Saftig P, Weng X, Meng Y, Neculai D, Marks MS, Gonzales L, Beers MF, Guttentag S. Impaired lysosomal integral membrane protein 2-dependent peroxiredoxin 6 delivery to lamellar bodies accounts for altered alveolar phospholipid content in adaptor protein-3-deficient pearl mice. J Biol Chem. 2016;291:8414–8427. doi: 10.1074/jbc.M116.720201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bromberg Y, Pick E. Activation of NADPH-dependent superoxide production in a cell-free system by sodium dodecyl sulfate. J Biol Chem. 1985;260:13539–13545. [PubMed] [Google Scholar]
- 140.Lowenthal A, Levy R. Essential requirement of cytosolic phospholipase A(2) for activation of the H(+) channel in phagocyte-like cells. J Biol Chem. 1999;274:21603–21608. doi: 10.1074/jbc.274.31.21603. [DOI] [PubMed] [Google Scholar]
- 141.Rubin BB, Downey GP, Koh A, Degousee N, Ghomashchi F, Nallan L, Stefanski E, Harkin DW, Sun C, Smart BP, Lindsay TF, Cherepanov V, Vachon E, Kelvin D, Sadilek M, Brown GE, Yaffe MB, Plumb J, Grinstein S, Glogauer M, Gelb MH. Cytosolic phospholipase A2-alpha is necessary for platelet-activating factor biosynthesis, efficient neutrophil-mediated bacterial killing, and the innate immune response to pulmonary infection: cPLA2-alpha does not regulate neutrophil NADPH oxidase activity. J Biol Chem. 2005;280:7519–7529. doi: 10.1074/jbc.M407438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matonao R, Miyano K, Kiyohara T, Sumimoto H. Arachidonic acid induces direct interaction of the p67(phox)-Rac complex with the phagocyte oxidase Nox2, leading to superoxide production. J Biol Chem. 2014;289:24874–24884. doi: 10.1074/jbc.M114.581785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ellison MA, Thurman GW, Ambruso DR. Phox activity of differentiated PLB-985 cells is enhanced, in an agonist specific manner, by the PLA2 activity of Prdx6-PLA2. Eur J Immunol. 2012;42:1609–1617. doi: 10.1002/eji.201142157. [DOI] [PubMed] [Google Scholar]
- 144.Lee I, Dodia C, Chatterjee S, Zagorski J, Mesaros C, Blair IA, Feinstein SI, Jain M, Fisher AB. A novel nontoxic inhibitor of the activation of NADPH oxidase reduces reactive oxygen species production in mouse lung. J Pharmacol Exp Ther. 2013;345:284–296. doi: 10.1124/jpet.112.201079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee I, Dodia C, Chatterjee S, Feinstein SI, Fisher AB. Protection against LPS-induced acute lung injury by a mechanism based inhibitor of NADPH-oxidase (Type 2) Am J Physiol Lung Cell Mol Physiol. 2014;306:635–644. doi: 10.1152/ajplung.00374.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vazquez-Medina JP, Dodia C, Weng L, Mesaros C, Blair I, Feinstein SI, Chatterjee C, Fisher A. The phospholipase A2 activity of peroxiredoxin 6 modulates NADPH oxidase 2 activation via lysophosphatidic acid receptor signaling in the pulmonary endothelium and alveolar macrophages. FASEB J. 2016;30:2885–2898. doi: 10.1096/fj.201500146R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 148.Kwon J, Wang A, Burke DJ, Boudreau HE, Lekstrom KJ, Korzeniowska A, Sugamata R, Kim YS, Yi L, Ersoy I, Jaeger S, Palaniappan K, Ambruso DR, Jackson SH, Leto TL. Peroxiredoxin 6 (Prdx6) supports NADPH oxidase1 (Nox1)-based superoxide generation and cell migration. Free Radic Biol Med. 2016;96:99–115. doi: 10.1016/j.freeradbiomed.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chatterjee S, Fisher AB. Mechanotransduction in the endothelium: role of membrane proteins and reactive oxygen species in sensing, transduction, and transmission of the signal with altered blood flow. Antioxid Redox Signal. 2014;20:899–913. doi: 10.1089/ars.2013.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Browning EA, Chatterjee S, Fisher AB. Stop the flow: a paradigm for cell signaling mediated by reactive oxygen species in the pulmonary endothelium. Annu Rev Physiol. 2012;74:403–424. doi: 10.1146/annurev-physiol-020911-153324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chatterjee S, Fujiwara K, Perez NG, Ushio-Fukai M, Fisher AB. Mechanosignaling in the vasculature: emerging concepts in sensing, transduction and physiological responses. Am J Physiol Heart Circ Physiol. 2015;308:H1451–H1462. doi: 10.1152/ajpheart.00105.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Milovanova T, Chatterjee S, Hawkins BJ, Hong N, Sorokina EM, Debolt K, Moore JS, Madesh M, Fisher AB. Caveolae are an essential component of the pathway for endothelial cell signaling associated with abrupt reduction of shear stress. Biochim Biophys Acta. 2008;1783:1866–1875. doi: 10.1016/j.bbamcr.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang Q, Chatterjee S, Wei Z, Liu WD, Fisher AB. Rac and PI3 kinase mediate endothelial cell-reactive oxygen species generation during nor-moxic lung ischemia. Antioxid Redox Signal. 2008;10:679–689. doi: 10.1089/ars.2007.1521. [DOI] [PubMed] [Google Scholar]
- 154.Chatterjee C, Browning E, Hong N, Debolt KM, Sorokina EM, Liu W, Birmbaum MJ, Fisher AB. Membrane depolarization is the trigger for P13Kinase/Akt activation and leads to the generation of reactive oxygen species. Am J Physiol Heart Circ Physiol. 2011;302:H105–H114. doi: 10.1152/ajpheart.00298.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Browning E, Wang H, Hong N, Yu K, Buerk DG, DeBolt K, Gonder D, Sorokina EM, Patel P, De Leon DD, Feinstein SI, Fisher AB, Chatterjee S. Mechanotransduction drives post ischemic revascularization through K(ATP) channel closure and production of reactive oxygen species. Antioxid Redox Signal. 2014;20:872–886. doi: 10.1089/ars.2012.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wei Z, Manevich Y, Al-Mehdi AB, Chatterjee S, Fisher AB. Ca2+ flux through voltage-gated channels with flow cessation in pulmonary microvascular endothelial cells. Microcirculation. 2004;11:517–526. doi: 10.1080/10739680490476367. [DOI] [PubMed] [Google Scholar]
- 157.Fukai T, Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal. 2011;15:1583–1606. doi: 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fisher AB. Redox signaling across cell membranes. Antioxid Redox Signal. 2009;11:1349–1356. doi: 10.1089/ars.2008.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Frey RS, Ushio-Fukai M, Malik AB. NADPH oxidase-dependent signaling in endothelial cells: role in physiology and pathophysiology. Antioxid Redox Signal. 2009;11:791–810. doi: 10.1089/ars.2008.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bernard K, Hecker L, Luckhardt TR, Cheng G, Thannickal VJ. NADPH oxidases in lung health and disease. Antioxid Redox Signal. 2014;20:2838–2853. doi: 10.1089/ars.2013.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Drummond GR, Selemidis S, Griendling KK, Sobey CG. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat Rev Drug Discov. 2011;10:453–471. doi: 10.1038/nrd3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Benipal B, Feinstein SI, Chatterjee S, Dodia C, Fisher AB. Inhibition of the phospholipase A2 activity of peroxiredoxin 6 prevents lung damage wih exposure to hyperoxia. Redox Biol. 2015;4:321–327. doi: 10.1016/j.redox.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Matot I, Manevich Y, Al-Mehdi AB, Song C, Fisher AB. Fluorescence imaging of lipid peroxidation in isolated rat lungs during nonhypoxic lung ischemia. Free Radic Biol Med. 2003;34:785–790. doi: 10.1016/s0891-5849(02)01435-1. [DOI] [PubMed] [Google Scholar]


