Abstract
The relationship among diet, human health, and disease is an area of growing interest in biomarker research. Previous studies suggest that the consumption of cranberries (Vaccinium macrocarpon) could beneficially influence urinary and digestive health. The present study sought to determine if daily consumption of sweetened dried cranberries (SDC) changes the urinary proteome and fecal microbiome, as determined in a prospective sample of 10 healthy individuals. Baseline urine and fecal samples were collected from the subjects in the fasted (8–12 h) state. The subjects then consumed one serving (42 g) of SDC daily with lunch for 2 weeks. Urine and fecal samples were collected again the day after 2 weeks of SDC consumption. Orbitrap Q-Exactive mass spectrometry of urinary proteins showed that consumption of SDC resulted in changes to 22 urinary proteins. Multiplex sequencing of 16S ribosomal RNA genes in fecal samples indicated changes in relative abundance of several bacterial taxonomic units after consumption of SDC. There was a shift in the Firmicutes:Bacteroidetes ratio, increases in commensal bacteria, and decreases or the absence of bacteria associated with negative health effects. A decrease in uromodulin in all subjects and an increase in Akkermansia bacteria in most subjects were observed and warrant further investigation. Future larger clinical studies with multiomics and multitissue sampling designs are required to determine the effects of SDC consumption on nutrition and health.
Keywords: : microbiome science, multiomics, nutrigenomics, proteomics, system diagnostics
Introduction
Integrated studies of proteome and microbiome in response to diet are of importance to move omics data to tangible clinical applications. Previously, cranberry polyphenol chemistry has been related to health outcomes (Burleigh et al., 2013; Feliciano et al., 2015; Krueger et al., 2013a; Pierre et al., 2013, 2014). In light of these past studies and the increase in consumption of sweetened dried cranberries (SDC), we determined if the consumption of SDC by healthy human subjects results in detectable changes to the urinary proteome and the fecal microbiome.
Cranberries contain proanthocyanidins (PACs) which improve gut barrier function in mouse models (Anhe et al., 2015; Pierre et al., 2013, 2014). Therefore, discovery and identification of signature shifts in endogenous urinary proteins or the fecal microbiome could inform the development of new biomarkers of gut barrier function. Urine and feces, in particular, have value as diagnostic specimens for tracking dietary interventions for health outcomes because collection is noninvasive. In typical human urine, the excretion of proteins is relatively low, but the sample volume can be large. Urine fluctuates in response to stimulants and contains potential biomarkers (Chen and Kim, 2016; Thomas et al., 2016). Diet may reversibly alter the human fecal microbiome at the species and genera level, although the microbiome is stable at the phylum level in long-term studies (Martinez et al., 2013).
The aims of this prospective study were to determine if daily consumption of SDC alters the urinary proteome and fecal microbiome in 2 weeks and evaluate if these changes might be related to urinary and digestive health.
Materials and Methods
Sweetened dried cranberries
SDC were a blend from two commercial sources (Ocean Spray Cranberries and Mariani) mixed at a 1:1 (w/w) ratio. The blended SDC were repackaged into 42 g individual servings (USDA, 2016) in a food grade processing facility. SDC composition is provided in Table 1 (USDA, 2016). The 4-(dimethylamino)cinnamaldehyde (DMAC) assay was used to quantify PAC content (Feliciano et al., 2012).
Table 1.
Sweetened Dried Cranberry, Nutritional Data
| Nutrient | Unit | Per serving |
|---|---|---|
| Water | G | 6.32 |
| Energy | Kcal | 123 |
| Protein | G | 0.07 |
| Total lipid (fat) | G | 0.44 |
| Carbohydrate, by difference | G | 33.12 |
| Fiber, total dietary | G | 2.1 |
| Sugars, total | G | 29.02 |
| Calcium | Mg | 4 |
| Iron | Mg | 0.16 |
| Magnesium | Mg | 2 |
| Phosphorus | Mg | 3 |
| Potassium | Mg | 20 |
| Sodium | Mg | 2 |
| Zinc | Mg | 0.04 |
| Vitamin C, total ascorbic acid | Mg | 0.1 |
| Thiamin | Mg | 0.005 |
| Riboflavin | Mg | 0.011 |
| Niacin | Mg | 0.219 |
| Vitamin B-6 | Mg | 0.015 |
| Vitamin A, RAE | μg | 1 |
| Vitamin A, IU | IU | 18 |
| Vitamin E (alpha-tocopherol) | mg | 0.84 |
| Vitamin K (phylloquinone) | μg | 3 |
| Fatty acids, total saturated | g | 0.035 |
| Fatty acids, total monounsaturated | g | 0.119 |
| Fatty acids, total polyunsaturated | g | 0.073 |
| Fatty acids, total trans | g | 0.001 |
USDA (2016).
Clinical study design
Healthy human subjects (n = 10, 2 men and 8 women) between the ages of 20 and 41 (mean = 27.5 years ±9.95 standard deviation [SD]) with a body mass index of 20.5–28.7 (mean = 24.07 years ±2.31 SD) were recruited and screened. Volunteers with a medical history of immune-compromising diseases, urinary tract infection (UTI) within the past 6 months, chronic inflammatory bowel disease (IBD), digestive diseases, diabetes, or cranberry allergy were excluded. Subjects who already regularly consumed cranberry products were excluded. Volunteers disclosed prescription and over-the-counter medications, as well as supplements, and these were taken into consideration when we selected the study subjects. One subject maintained a vegetarian diet during the trial period. None of the subjects had notable dietary changes during the trial.
Those selected were enrolled under a protocol approved by the University of Wisconsin Health Sciences Institutional Review Board (IRB 2015-0317). A written and informed consent was obtained from all subjects.
Subjects were directed to complete a daily diet journal for 7 days before the baseline samples. Subjects continued to record dietary intake (including time of consumption) during the 2-week intervention period. Exercise or other daily activity was not recorded.
Baseline urine and fecal samples were collected from the subjects in the fasted (8–12 h) state. The subjects then consumed one serving (42 g) of SDC daily with lunch for 2 weeks. Urine and fecal samples were collected again the day after the 2-week SDC consumption period, as was done previously. A total of 10 paired pre- and post-consumption samples were collected in the present study. Samples were kept in a cooler with frozen gel packs from the time of collection until the time of submission to the laboratory. Urine samples were immediately frozen at −80°C from the time of submission (July and August 2015) until they were defrosted and aliquoted (4–5 mL) for analysis (December 2015). Fecal samples were immediately aliquoted (150 mg) and frozen at −80°C from the time of submission (July and August 2015) until they were defrosted for analysis (December 2015).
Analysis of urinary proteome
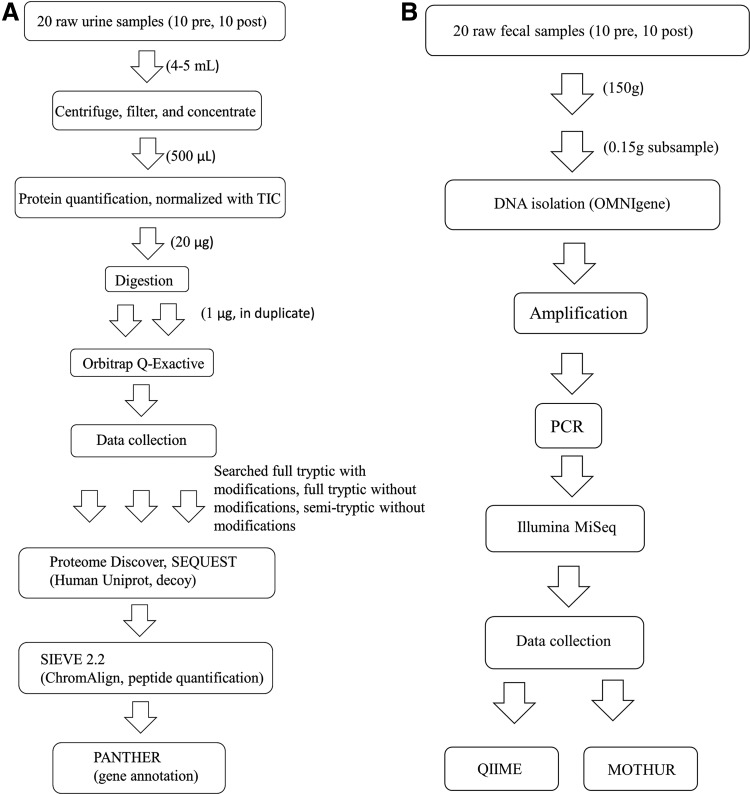
Preparation and proteome analysis of the urine samples were done by the UW School of Pharmacy Analytical Instrumentation Center Mass Spectrometry Facility (Fig. 1A). Using a label-free approach (Jerebtsova and Nekhai, 2014), we used mass spectrometry for characterization of the urinary proteome. The Orbitrap Q-Exactive Mass Spectrometry system combines quadrupole precursor ion selection with high-resolution accurate mass Orbitrap detection to deliver very high resolving power (up to 140,000 full width at half-maximum) to identify, quantify, and confirm compounds in complex samples, which facilitates proteomics, metabolomics, lipidomics, and metabolomics through detection of low-abundance components in complex samples (Michalski et al., 2011).
FIG. 1.
Procedures for proteomic (A) analyses. Procedures for microbiome (B) analyses. TIC, total ion current.
Each subject had a pre- and post-treatment urine sample, and a technical replicate for each sample was run through the Orbitrap Q-Exactive mass spectrometer. All centrifuge steps were spun at 4°C, 20,000 g. Four microliters urine of each sample was concentrated to 500 μL using Amicon Ultra 3 kda molecular weight (MW) spin filters pre-wet with optima liquid chromatograph mass spectrometry (LC-MS) grade water. Samples were buffer exchanged with 3x volume of 50 mM ammonia bicarbonate before protein quantification and digestion. Micro BCA of all 20 samples (pre- and post-) was performed. Urinary proteins were normalized with total ion current. Twenty micrograms protein (determined by Micro BCA) of each sample was digested as per standard digestion protocol.
Digested proteins were cleaned on C18 Zip Tips as per manufacturer's protocol. One microgram of each sample was injected in duplicate on a 115 min increasing acetonitrile (ACN) gradient. Pre- and post-Enolase QC runs to assess mass spec performance were performed, and a 60 min blank run between injections to clean the column and prevent carryover was undertaken. Resulting raw files were searched thrice each (one full tryptic with modifications, one full tryptic without modifications, and one semitryptic without modifications) using Proteome Discoverer 1.4.1.14, SEQUEST HT (Thermo Fisher) against the human UniProt database, including a decoy.
ChromAlign and peptide quantification was performed using SIEVE 2.2 (Thermo Fisher). SIEVE data output includes protein identification for each subject, including the number of peptides, frames, hits, and the pre/post protein ratio calculation. Since our method used a label-free technique, results are relative. Further analysis was conducted with the data from the semitryptic approach using proteins composed of two or more peptides.
For proteins found to be significantly different, we conducted a batch analysis of gene annotation information through PANTHER version 10: expanded protein families and functions and analysis tools (Mi et al., 2016) and the PANTHER Classification system (Mi et al., 2013).
Analysis of fecal microbiome
The fecal microbiome analysis was performed by the University of Wisconsin-Madison Biotechnology Center (Fig. 1B). DNA was isolated from 0.15 g of fecal matter using the OMNIgene adapted MO BIO PowerFecal DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). DNA concentration was verified using the Qubit® dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA, USA). Samples were prepared as described in the 16S Metagenomic Sequencing Library Preparation Protocol, Part No. 15044223 Rev. B (Illumina, Inc., San Diego, CA, USA) with the following modifications: The 16S ribosomal RNA gene V3/V4 variable region was amplified with nested primers:
forward primer: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTCCTACGGGNGGCWGCAG-3′
reverse primer: 5′-TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGACTACHVGGGTATCTAATCC-3′
Region specific primers were previously described in Klindworth et al., 2013, and were modified to add Illumina adapter overhang nucleotide sequences to the gene-specific sequences. Following initial amplification, library size was verified on an Agilent DNA1000 chip and cleaned using a 1x volume of AxyPrep Mag PCR clean-up beads (Axygen Biosciences, Union City, CA, USA). Illumina dual indexes and sequencing adapters were added using the following primers:
forward primer: 5′-AATGATACGGCGACCACCGAGATCTACAC[55555555]ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′
reverse primer: 5′ CAAGCAGAAGACGGCATACGAGAT[77777777]GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′
Bracketed sequences are equivalent to the Illumina Dual Index adapters D501–D508 and D701–D712. Following PCR, samples were cleaned using a 1x volume of AxyPrep Mag PCR clean-up beads (Axygen Biosciences). Quality and quantity of the finished libraries were assessed using an Agilent DNA1000 chip and Qubit dsDNA HS Assay Kit, respectively. Libraries were standardized to 2 μM and pooled before sequencing. Paired-end 250 bp sequencing was performed using the Illumina MiSeq Sequencer and a MiSeq 600 bp (v3) sequencing cartridge. A technical replicate was run for each sample.
Raw data were analyzed using the standard Illumina Pipeline, version 1.8.2. OTU assignments and diversity plots were created using QIIME analysis pipeline (Caporaso et al., 2010). Additional analysis was done using the program MOTHUR (Schloss et al., 2009).
Statistical analysis
We used a two-tailed binomial test with a p-value of 0.05 to determine if the number of individuals with common proteins or bacteria changing in the same direction was significant. Because there were only 10 subjects, we considered results with a p-value between 0.05 and 0.10 as trending toward significance with a view to potential leads to inform future hypothesis-driven studies with more subjects.
Results
Sweetened dried cranberries
We tested the SDC for composition. By the DMAC assay, we found 0.62 mg/g (A2 equivalents) of soluble PAC and 2.22 mg/g of cranberry PAC (cPAC) equivalents in the SDC product.
Urinary proteome
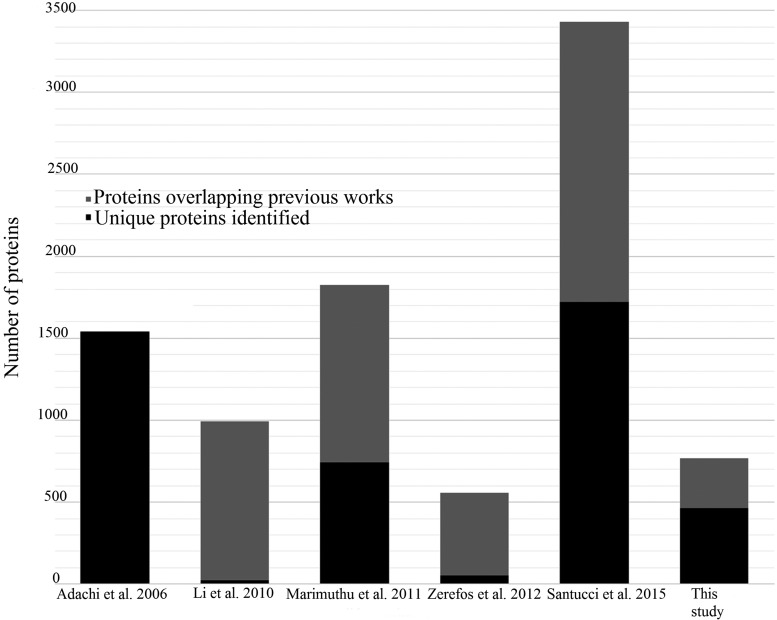
Technical replicate injections had a 71% overlap at the protein level and 68% overlap at the peptide level, which are in line with published results (Addona et al., 2009). We detected 767 proteins with two or more peptides (Supplementary Table S1). To the best of our knowledge, 464 of the 767 proteins have not been previously described in the human urinary proteome (Adachi et al., 2006; Li et al., 2010; Marimuthu et al., 2011; Santucci et al., 2015; Zerefos et al., 2012; Fig. 2). The unique proteins we identified in this study are listed in Supplementary Table S2. Individual variation in both the number of proteins identified and the composition of the proteome was high. Although 64 proteins were found in at least 80% of our subjects, a high percentage of proteins were only identified in individual subjects (420/767, 54.76%).
FIG. 2.
Comparison with previous large urinary proteomic publications. Each bar is the total number of proteins identified in the study; the black portion of each bar represents proteins not previously identified, while the gray portion represents proteins identified in one or more of the previous works.
Twenty-two proteins were found to significantly change (p < 0.05) between the pre- and postsamples (Table 2). Collectively, they belong to the gene ontology (GO) biological process categories of platelet degranulation (53.96-fold enrichment, p = 9.75 × 10−6) and single multicellular organism process (3.13-fold enrichment, p = 6.41 × 10−4). Seven of these proteins are mapped to GO pathways (Table 3).
Table 2.
Notable Differences in Pre- Versus Post-trial Proteins in Our Study Sample (n = 10 Subjects)
| UniProt KB | Description | Change |
|---|---|---|
| P02768 | ALBU Serum albumin | 9/10 decreased |
| P10909 | CLUS Clusterin | 9/10 decreased |
| P39059 | COFA1 Collagen alpha_1 (XV) chain | 9/10 decreased |
| P01133 | EGF Pro_epidermal growth factor | 9/10 decreased |
| Q16270 | IBP7 Insulin_like growth factor_binding protein 7 | 9/10 decreased |
| P01834 | IGKC Ig kappa chain C region | 9/10 decreased |
| P05154 | IPSP Plasma serine protease inhibitor | 9/10 decreased |
| P01042 | KNG1 Kininogen_1 | 9/10 decreased |
| P10253 | LYAG Lysosomal alpha_glucosidase | 9/10 decreased |
| P10451 | OSTP Osteopontin | 9/10 decreased |
| P98160 | PGBM Basement membrane_specific heparan sulfate proteoglycan core protein | 9/10 decreased |
| Q12907 | LMAN2 Vesicular integral_membrane protein | 10/10 decreased |
| Q96FE7 | P3IP1 Phosphoinositide_3_kinase_interacting protein 1 | 10/10 decreased |
| P07911 | UROM Uromodulin | 10/10 decreased |
| P01009 | A1AT Alpha_1_antitrypsin | 8/9 decreased |
| P05090 | APOD Apolipoprotein D | 8/9 decreased |
| O60494 | CUBN Cubilin | 8/9 decreased |
| P05155 | IC1 Plasma protease C1 inhibitor | 8/9 decreased |
| Q6GTX8 | LAIR1 Leukocyte_associated immunoglobulin_like receptor 1 | 8/9 decreased |
| P35555 | FBN1 Fibrillin_1 | 8/9 increased |
| P55290 | CAD13 Cadherin_13 | 8/8 decreased |
| Q7Z5L0 | VMO1 Vitelline membrane outer layer protein 1 homolog | 8/8 decreased |
Table 3.
Gene Ontology Pathway Analyses for Proteins Observed as Being Different Between Pre- Versus Posttrial
| Pathway accession | Mapped ID | Pathway name |
|---|---|---|
| P00057 | P55290 CAD13 | Wnt signaling pathway |
| P06664 | P01133 EGF | Gonadotropin releasing hormone receptor pathway |
| P00011 | P01042 KNG1 | Blood coagulation |
| P01009 A1AT | ||
| P00034 | P39059 COFA1 | Interin signaling pathway |
| P98160 PGBM | ||
| P00012 | P55290 CAD13 | Cadherin signaling pathway |
| P06959 | P10909 CLUS | CCKR signaling map |
| P00018 | P01133 EGF | EGF receptor signaling pathway |
Of the 25 proteins found in all subjects (Table 4), three proteins, namely the vesicular integral membrane protein, phosphoinositide 3-kinase interacting protein 1, and uromodulin, were observed to significantly decrease in all subjects after the consumption of SDC (p = 0.000977). Eleven proteins were observed to decrease in 9 of 10 subjects (p = 0.010742) after SDC consumption: serum albumin, clusterin, collagen alpha 1 (XV) chain, pro-epidermal growth factor, insulin-like growth factor binding protein, Ig kappa chain C region, plasma serine protease inhibitor, kininogen 1, lysosomal alpha glucosidase, osteopontin, and basement membrane specific heparan sulfate proteoglycan core protein. Fibrillin-1 increased in 8/9 subjects (p = 0.019531) after SDC consumption. Cadherin 13 and vitelline membrane outer layer protein 1 homolog were found to decrease after SDC consumption in the 8 subjects in which it was detected (p = 0.003906).
Table 4.
Urinary Proteins Identified in All Subjects
| UniProt KB | Description |
|---|---|
| P02768 | ALBU_HUMAN Serum albumin |
| P10909 | CLUS_HUMAN Clusterin |
| P39059 | COFA1_HUMAN Collagen alpha_1 (XV) chain |
| P01133 | EGF_HUMAN Pro_epidermal growth factor |
| Q16270 | IBP7_HUMAN Insulin_like growth factor_binding protein 7 |
| P01834 | IGKC_HUMAN Ig kappa chain C region |
| P05154 | IPSP_HUMAN Plasma serine protease inhibitor |
| P01042 | KNG1_HUMAN Kininogen_1 |
| P10253 | LYAG_HUMAN Lysosomal alpha_glucosidase |
| P10451 | OSTP_HUMAN Osteopontin |
| P98160 | PGBM_HUMAN Basement membrane_specific heparan sulfate proteoglycan core protein |
| Q12907 | LMAN2_HUMAN Vesicular integral_membrane protein VIP36 |
| Q96FE7 | P3IP1_HUMAN Phosphoinositide_3_kinase_interacting protein 1 |
| P07911 | UROM_HUMAN Uromodulin |
| P14209 | CD99_HUMAN CD99 antigen |
| Q14624 | ITIH4_HUMAN Inter_alpha_trypsin inhibitor heavy chain H4 |
| P10153 | RNAS2_HUMAN Non_secretory ribonuclease |
| P30530 | UFO_HUMAN Tyrosine_protein kinase receptor UFO |
| P12109 | CO6A1_HUMAN Collagen alpha_1 (VI) chain |
| Q6EMK4 | VASN_HUMAN Vasorin |
| P02461 | CO3A1_HUMAN Collagen alpha_1 (III) chain |
| P41222 | PTGDS_HUMAN Prostaglandin_H2 D_isomerase |
| Q9HCU0 | CD248_HUMAN Endosialin |
| P02671 | FIBA_HUMAN Fibrinogen alpha chain |
| P02751 | FINC_HUMAN Fibronectin |
VIP36, vesicular integral membrane protein.
Fecal microbiome
A principal coordinate analysis plot of beta-diversity is shown in Figure 3. The pre- versus post-treatment samples did not group together. When we compared the microbial phyla present in the initial samples versus the final samples, the differences were not significant. Six of 10 subjects increased the representation of Bacteroidetes and 7 of 10 subjects decreased the representation of Firmicutes in the total bacteria present. Taken together, 7 of 10 subjects had a decreased Firmicutes:Bacteroidetes ratio. Although not significant, 7 of 10 subjects had increased species diversity in the post sample, and one subject had no change in the total number of species present. Therefore, 8 of 10 subjects had increased or unchanged species diversity, approaching statistical significance (p = 0.0546875).
FIG. 3.
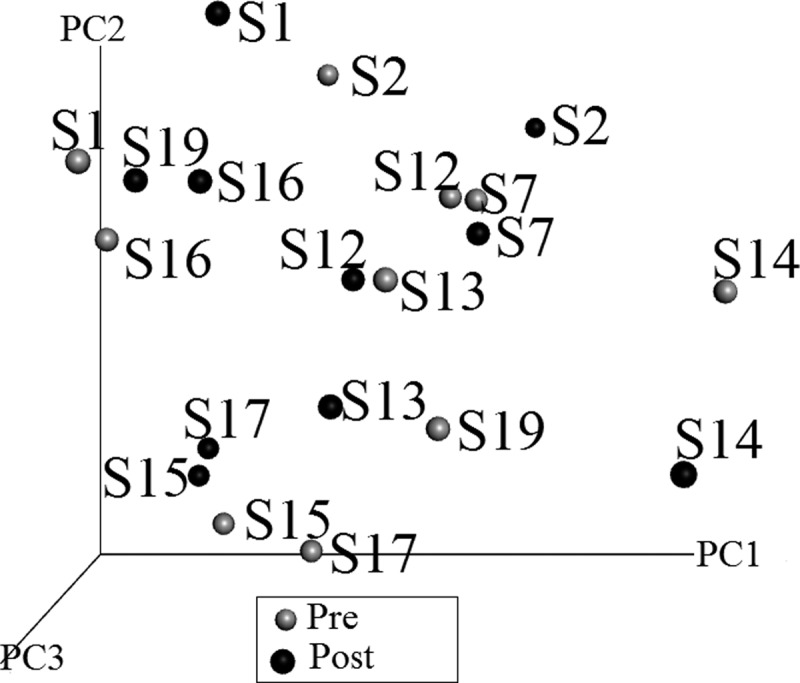
A weighted principal coordinate analysis plot of fecal microbiome beta-diversity. Each dot represents a single subject; gray dots are pretreatment and black dots are posttreatment. A single subject tends to group with itself, rather than with time (pre- vs. posttreatment).
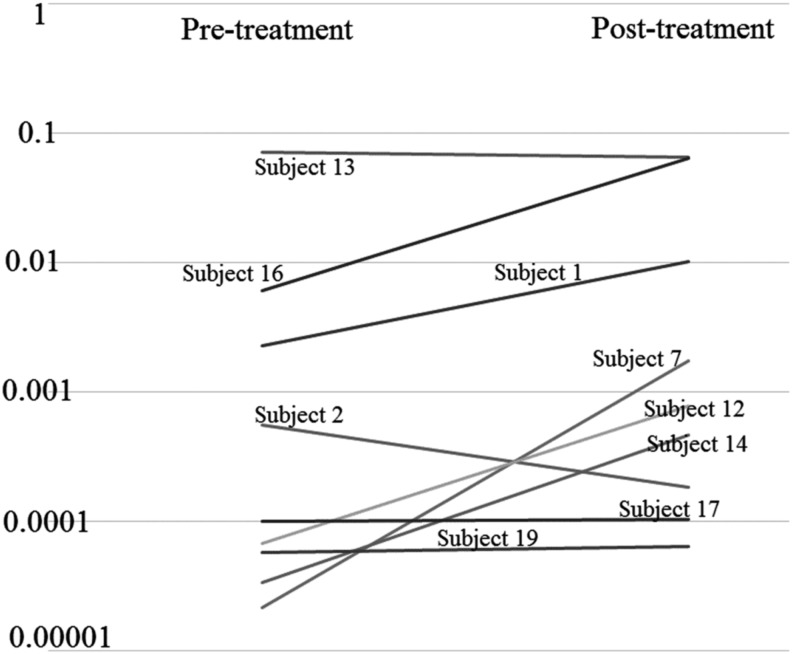
Nine subjects had measurable levels of Akkermansia, and 7 of 9 had a higher relative abundance of Akkermansia in the total bacterial population in the postsample, with a p-value of 0.08984375, trending toward significance (Fig. 4). Akkermansia was not detected in the fecal microbiome of one subject, while another subject had a relatively large decrease. In contrast, five subjects had greater than a one log increase in relative abundance of Akkermansia.
FIG. 4.
Log-scale plot of relative pre- versus posttreatment Akkermansia to total microbial population values in individual subjects. Subject 15 did not have measurable levels of Akkermansia. Seven of nine subjects had relative increases in Akkermansia.
Discussion
Integrated omics studies in multiple tissues and samples offer a system scale perspective on host–environment interactions, including those with food and nutrient intake. In this context, urinary proteomics has been noted as an emergent field of omics science and diagnostic medicine over the past decade (Adachi et al., 2006). Seminal articles in this field (Adachi et al., 2006; Li et al., 2010; Marimuthu et al., 2011; Santucci et al., 2015; Zerefos et al., 2012) have described roughly 4100 urinary proteins.
In the present study, we sought to prospectively determine whether the effects of daily consumption of SDC would lead to changes in the urinary proteome and fecal microbiome in 10 subjects in 2 weeks. Based on our own research (Krueger et al., 2013b) and of others (Liu et al., 2012; Nagaraj and Mann, 2011), we anticipated that the present sample size would offer new insights on proteome and microbiome changes in response to cranberry consumption. We observed 767 proteins in 10 subjects. To the best of our knowledge, 464 of those proteins have not been previously described in the human urinary proteome.
In contrast to a previous report (Nagaraj and Mann, 2011), we did not find a large list of proteins common to all subjects. We observed that 420 of 767 identified proteins were unique to a single subject.
Interestingly, of the 25 proteins found to be common in all 10 subjects in this study, LMAN2 was not previously reported in the publications described above. LMAN2 binds the sugar residues of glycoproteins and the sugar chain in bacteria (Shirakabe et al., 2011) and is involved in protein sorting and segregation (Fiedler et al., 1994). Two proteins, IC1 and LAIR1, found in 9 of 10 subjects, were also not a part of the common core lists. Complement inhibitor 1 is part of the classical primary complement pathway, and defects can result in a primary immune deficiency illness (Grumach and Kirschfink, 2014). Supplementation of complement inhibitor 1 slows the clotting process, binds gram-negative bacteria, and has anti-inflammatory effects (Landsem et al., 2016). LAIR1 is expressed on hematopoietic cells and carries immunoreceptor motifs on its tail (Cao et al., 2015). LAIR1 is considered an immunoinhibitory collagen receptor, and defects in LAIR1 can result in autoimmune disorders, viral illnesses, and cancer (Sun et al., 2014).
Taken together, our report of 464 urinary proteins that have not been previously reported in the literature and the addition of the proteins above to the common core of proteins indicate that there is still much to learn about the urinary proteome.
Compared to a previous study using a dietary supplement consisting of cranberry powder (Krueger et al., 2013b), only one of the eight previously reported proteins was detected in the current study [Ig kappa chain V_III region CLL (P04207)]. In the current study, it was neither found to be common to all subjects nor was it found to be significantly different in the subjects in which it was found. The reasons for these findings are unclear. The previous study was shorter (6 days), and cranberry powder was administered, rather than SDC. Alternatively, with the small sample size of that study (n = 10) and our study (n = 10), the discrepancy may be attributable to individual differences.
Uromodulin is the most abundant urinary protein produced in the kidney and is associated with an increase in innate immune responses in the kidney (Rampoldi et al., 2011). Mouse studies suggest that uromodulin may provide protection to the bladder in a UTI by binding with bacteria (Rampoldi et al., 2011). Uromodulin is also a damage associated molecular pattern that is associated with kidney injury (Anders and Schaefer, 2014) and stimulation of a pro-inflammatory response in the urinary tract (Darisipudi et al., 2012; Garimella and Sarnak, 2017; Rampoldi et al., 2011). Therefore, the function of uromodulin in the urinary tract remains unclear. Our subjects were prescreened for UTIs before beginning the trial. Since we assume that the subjects maintained a healthy urinary tract system throughout the trial, a decrease in all subjects is an important observation regarding the relationship between uromodulin and urinary tract health. The effect of SDC consumption on uromodulin requires more research.
Although it is not clear if microbial dysbiosis is the cause or effect of disease (Round and Mazmanian, 2009), characterization of the microbial community can highlight such dysbiosis. Bacterial diversity is decreased in microbial dysbiosis (Kim et al., 2016; Zhernakova et al., 2016). Seven of our subjects had a richer microbiota after the intervention, and one had no change. The trend we observed may suggest that SDC favor species richness in the gut, perhaps because SDC contain a diversity of microbial substrates such as fiber and polyphenols (Blumberg et al., 2016).
Another measure commonly addressed in discussions of digestive health is the Firmicutes:Bacteroidetes ratio (Ley et al., 2006; Mariat et al., 2009). A reduction in the ratio could indicate an increase in Bacteroidetes, a decrease in Firmicutes, or both. Firmicutes are associated with increased energy efficiency and absorption, a condition that could cause obesity. Seven of our subjects lowered their Firmicutes:Bacteroidetes ratio in the trial. Specifically, 6/10 subjects increased Bacteroidetes and 7/10 subjects decreased Firmicutes. An interesting anomaly occurred with the subject who maintained a vegetarian diet during the trial—the ratio increased. In that subject, Prevotella (a Bacteroidetes) was the largest contributor to the microbiome in the pretrial sample, and Ruminococcus (a Firmicutes) was the largest contributor to the microbiome in the post-intervention sample.
Falony et al. (2016) included only the genus Bifidobacterium of the phylum Actinobacteria as part of the global core bacteria. We found both the genera Bifidobacterium and Eggerthella in all our samples. Actinobacteria produce bioactive metabolites, including antibacterials, antifungals, antivirals, and immunomodifiers. Although it was not significant, we found an increased representation in the total bacterial community in both genera. Based on the functions of the phylum, SDC may be stimulating immune function in the gut.
Our previous work (Pierre et al., 2013) demonstrated that when cPACs are added to enteral nutrition in mice, mucin production in the gut increases. Akkermansia are mucin degrading bacteria. Increased mucin production could support an increased population of Akkermansia. The presence of Akkermansia mediates symptoms associated with metabolic syndrome (Anhe et al., 2015, 2016; Li et al., 2016; Roopchand et al., 2015; Schneeberger et al., 2015). Of our 10 subjects, 9 subjects had detectable levels of Akkermansia in both the pretrial state and post-intervention. Of those, seven had an increased relative abundance of Akkermansia in the total bacterial composition.
Conclusions
The addition of SDC to the diet of 10 subjects daily for 2 weeks influenced the composition of the urinary proteome and fecal microbiome. Twenty-two proteins were found to have differences between pre- and post-treatment, including uromodulin. With the function of uromodulin in the urinary system unclear, but understood to be implicated in urinary health, the decrease found in all subjects was intriguing. Utilizing a labeled isotope approach to target and quantify uromodulin in relation to SDC will be a focus in upcoming studies. The present study also added to the number of proteins found in healthy human urine by about 10%. Targeting the genera of interest, including Akkermansia to obtain more quantitative results, is also warranted in the future.
Supplementary Material
Abbreviations Used
- cPAC
cranberry PAC
- DMAC
4-(dimethylamino)cinnamaldehyde
- GO
gene ontology
- IBD
chronic inflammatory bowel disease
- IRB
Institutional Review Board
- PAC
proanthocyanidin
- SD
standard deviation
- SDC
Sweetened Dried Cranberries
- UTI
urinary tract infection
Acknowledgments
The authors thank the Cranberry Institute for funding this research and the NIH in form of the High-end, Shared Instrumentation grant (1S10RR029531−01) to the Analytical Instrumentation Center. The views expressed in this article are solely of the authors. We acknowledge Madison Cox, Bacteriology Department at University of Wisconsin-Madison; Molly Pellitteri Hahn, UW School of Pharmacy Analytical Instrumentation Center Mass Spectrometry Facility; Marie Adams Hasenstein, Biotechnology Center, Gene Expression Center at University of Wisconsin-Madison; and Michael Polewski, Animal Sciences Department at University of Wisconsin-Madison, for technical assistance in this project.
Author Disclosure Statement
The present research was funded by the Cranberry Institute. The funders had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
References
- Adachi J, Kumar C, Zhang Y, Olsen JV, and Mann M. (2006). The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol 7, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addona TA, Abbatiello SE, Schilling B, et al. (2009). Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders HJ, and Schaefer L. (2014). Beyond tissue injury-damage-associated molecular patterns, toll-like receptors, and inflammasomes also drive regeneration and fibrosis. J Am Soc Nephrol 25, 1387–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anhe FF, Pilon G, Roy D, et al. (2016). Triggering Akkermansia with dietary polyphenols: A new weapon to combat the metabolic syndrome? Gut Microbes 7, 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anhe FF, Roy D, Pilon G, et al. (2015). A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 64, 872–883 [DOI] [PubMed] [Google Scholar]
- Blumberg JB, Basu A, Krueger CG, et al. (2016). Impact of cranberries on gut microbiota and cardiometabolic health: Proceedings of the Cranberry Health Research Conference 2015. Adv Nutr 7, 759S–770S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh AE, Benck SM, McAchran SE, et al. (2013). Consumption of sweetened, dried cranberries may reduce urinary tract infection incidence in susceptible women—a modified observational study. Nutr J 12, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Fu A, Yang S, et al. (2015). Leukocyte-associated immunoglobulin-like receptor-1 expressed in epithelial ovarian cancer cells and involved in cell proliferation and invasion. Biochem Biophys Res Commun 458, 399–404 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, and Kim J. (2016). Urinary proteomics and metabolomics studies to monitor bladder health and urological diseases. BMC Urol 16, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darisipudi MN, Thomasova D, Mulay SR, et al. (2012). Uromodulin triggers IL-1beta-dependent innate immunity via the NLRP3 inflammasome. J Am Soc Nephrol 23, 1783–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G, Joossens M, Viera-Silva S, et al. (2016). Population-level analysis of gut microbiome variation. Science 352, 560–565 [DOI] [PubMed] [Google Scholar]
- Feliciano RP, Heintz JA, Krueger CG, Vestling MM, and Reed JD. (2015). Fluorescent labeling of cranberry proanthocyanidins with 5-([4,6-dichlorotriazin-2-yl]amino) fluorescein (DTAF). Food Chem 166, 337–345 [DOI] [PubMed] [Google Scholar]
- Feliciano RP, Shea MP, Shanmuganayagam D, et al. (2012). Comparison of isolated cranberry (Vaccinium macrocarpon Ait.) proanthocyanidins to catechin and procyanidins A2 and B2 for use as standards in the 4-(dimethylamino)cinnamaldehyde assay. J Agric Food Chem 60, 4578–4585 [DOI] [PubMed] [Google Scholar]
- Fiedler K, Parton R, Kellner R, Etzold T, and Simons K. (1994). VIP36, a novel componenet of glycolipid rafts and exocytic carrier vesicles in epithelial cells. EMBO J 13, 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garimella PS, and Sarnak MJ. (2017). Uromodulin in kidney health and disease. Curr Opin Nephrol Hypertens 26, 136–142 [DOI] [PubMed] [Google Scholar]
- Grumach AS, and Kirschfink M. (2014). Are complement deficiencies really rare? Overview on prevalence, clinical importance and modern diagnostic approach. Mol Immunol 61, 110–117 [DOI] [PubMed] [Google Scholar]
- Jerebtsova M, and Nekhai S. (2014). Quantitative mass spectrometry of urinary biomarkers. J Integr OMICS 4, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim DB, and Park JY. (2016). Changes of mouse gut microbiota diversity and composition by modulating dietary protein and carbohydrate contents: A pilot study. Prev Nutr Food Sci 21, 57–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger CG, Meudt JJ, Howell AB, Khoo C, and Shanmuganayagam D. (2013b). Consumption of cranberry powder shifts urinary protein profile in healthy human subjects. FASEB J 27, 637.32 [Google Scholar]
- Krueger CG, Reed JD, Feliciano RP, and Howell AB. (2013a). Quantifying and characterizing proanthocyanidins in cranberries in relation to urinary tract health. Anal Bioanal Chem 405, 4385–4395 [DOI] [PubMed] [Google Scholar]
- Landsem A, Fure H, Mollnes TE, Nielsen EW, and Brekke OL. (2016). C1-inhibitor efficiently delays clot development in normal human whole blood and inhibits Escherichia coli-induced coagulation measured by thromboelastometry. Thromb Res 143, 63–70 [DOI] [PubMed] [Google Scholar]
- Ley R, Turnbaugh P, Klein S, and Gordon J. (2006). Microbial ecology: Human gut microbes associated with obesity. Nature 444, 1022–1023 [DOI] [PubMed] [Google Scholar]
- Li J, Lin S, Vanhoutte PM, Woo CW, and Xu A. (2016). Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe-/- mice. Circulation 133, 2434–2446 [DOI] [PubMed] [Google Scholar]
- Li QR, Fan KX, Li RX, et al. (2010). A comprehensive and non-prefractionation on the protein level approach for the human urinary proteome: Touching phosphorylation in urine. Rapid Commun Mass Spectrom 24, 823–832 [DOI] [PubMed] [Google Scholar]
- Liu X, Shao C, Wei L, et al. (2012). An individual urinary proteome analysis in normal human beings to define the minimal sample number to represent the normal urinary proteome. Proteome Sci 10, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariat D, Firmesse O, Levenez F, et al. (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimuthu A, O'Meally RN, Chaerkady R, et al. (2011). A comprehensive map of the human urinary proteome. J Proteome Res 10, 2734–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Muller CE, and Walter J. (2013). Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS One 8, e69621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, and Thomas PD. (2013). Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8, 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, and Thomas PD. (2016). PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res 44, D336–D342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski A, Damoc E, Hauschild JP, et al. (2011). Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics 10, M111.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, and Mann M. (2011). Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res 10, 637–645 [DOI] [PubMed] [Google Scholar]
- Pierre JF, Heneghan AF, Feliciano RP, et al. (2013). Cranberry proanthocyanidins improve the gut mucous layer morphology and function in mice receiving elemental enteral nutrition. JPEN J Parenter Enteral Nutr 37, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre JF, Heneghan AF, Feliciano RP, et al. (2014). Cranberry proanthocyanidins improve intestinal sIgA during elemental enteral nutrition. JPEN J Parenter Enteral Nutr 38, 107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampoldi L, Scolari F, Amoroso A, Ghiggeri G, and Devuyst O. (2011). The rediscovery of uromodulin (Tamm-Horsfall protein): From tubulointerstitial nephropathy to chronic kidney disease. Kidney Int 80, 338–347 [DOI] [PubMed] [Google Scholar]
- Roopchand D, Carmody R, Kuhn P, et al. (2015). Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes 64, 2847–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, and Mazmanian SK. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci L, Candiano G, Petretto A, et al. (2015). From hundreds to thousands: Widening the normal human Urinome (1). J Proteomics 112, 53–62 [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger M, Everard A, Gomez-Valades AG, et al. (2015). Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep 5, 16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe K, Hattori S, Seiki M, Koyasu S, and Okada Y. (2011). VIP36 protein is a target of ectodomain shedding and regulates phagocytosis in macrophage Raw 264.7 cells. J Biol Chem 286, 43154–43163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Jiao Y, Wei W, et al. (2014). Comparison of LAIR-1 genetic pathways in murine vs human internal organs. Gene 552, 140–145 [DOI] [PubMed] [Google Scholar]
- Thomas S, Hao L, Ricke WA, and Li L. (2016). Biomarker discovery in mass spectrometry-based urinary proteomics. Proteomics Clin Appl 10, 358–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA. (2016). 09079, Cranberries, dried, sweetened. In: National Nutrient Database for Standard Reference, 28 ed. USDA, Agricultural Research Service, Nutrient Data Laboratory; http://www.ars.usda.gov/ba/bhnrc/ndl [Google Scholar]
- Zerefos PG, Aivaliotis M, Baumann M, and Vlahou A. (2012). Analysis of the urine proteome via a combination of multi-dimensional approaches. Proteomics 12, 391–400 [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Kurilshikov A, Bonder M, et al. (2016). Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.