ABSTRACT
Mucormycoses are life-threatening infections in immunocompromised patients. This study characterizes the response of human mononuclear cells to different Mucorales and Ascomycota. PBMC, monocytes, and monocyte derived dendritic cells (moDCs) from healthy donors were stimulated with resting and germinated stages of Mucorales and Ascomycota. Cytokine response and expression of activation markers were studied. Both inactivated germ tubes and resting spores of Rhizopus arrhizus and other human pathogenic Mucorales species significantly stimulated mRNA synthesis and secretion of proinflammatory cytokines. Moreover, R. arrhizus spores induced the upregulation of co-stimulatory molecules on moDCs and a specific T-helper cell response. Removal of rodlet hydrophobins by hydrofluoric acid treatment of A. fumigatus conidia resulted in enhanced immunogenicity, whereas the cytokine response of PBMCs to dormant R. arrhizus spores was not influenced by hydrofluoric acid. Scanning electron micrographs of Mucorales spores did not exhibit any morphological correlates of rodlet hydrophobins. Taken together, this study revealed striking differences in the response of human mononuclear cells to resting stages of Ascomycota and Mucorales, which may be explained by absence of an immunoprotective hydrophobin layer in Mucorales spores.
KEYWORDS: hydrophobins, Mucorales, phagocytes, Rhizopus, rodlet, spores
Introduction
Immunocompromised patients are at high risk of acquiring invasive mycoses.1 Besides the most common opportunistic fungal genera Candida and Aspergillus, new emerging opportunistic molds such as Mucorales account for an increasing share of these infections.2,3 Species of the order Mucorales cause life-threatening systemic infections predominantly involving the lung, nasal sinus, and central nervous system. Mucormycoses are often characterized by rapid progression, poor treatment response, and high mortality rates.4,5
The immunocompetent host possesses a vast arsenal of defense strategies to eliminate fungal spores preventing progression to an invasive infection. The mononuclear phagocyte system plays a crucial role in this process.6,7,8 Professional phagocytes destroy fungal pathogens by phagocytosis or release of fungicidal molecules. The presentation of acquired antigens provides T-cells with the required stimulus and serves as an important crosslink between innate and specific immunity.9 Recognition of fungal pathogens by innate immune cells induces the expression and release of a variety of cytokines and chemokines that regulate cell migration and activity, systemic inflammation, and T-cell differentiation.10 This results in a well-orchestrated host response ensuring effective elimination of the pathogen while limiting host damage caused by excessive inflammation.11,12
In mold immunopathology, a key concept is the dependence of the virulence and host defense on the fungal maturational stage.8,13 During their maturation, conidia of Aspergillus fumigatus and other Ascomycota undergo metabolic and morphologic changes.14 These alterations are of major importance for the recognition by the host immune system and its response to the fungus. For example, the loss of the rodlet hydrophobin layer during swelling of Aspergillus conidia is associated with a distinctly stronger stimulation of human innate immune cells.8,15
Though similarities in terms of fungal biology, predisposing factors, and clinical presentation can be observed in Mucorales and Aspergillus infections, recent studies indicate differences in host immunity to these fungi.16,17 However, the morphotype-dependent impact of Mucorales on the human immune system has not yet been conclusively characterized. In this study we therefore sought to expand our knowledge of the interplay between innate human immune cells and various Mucorales species, focusing on the cytokine response of mononuclear phagocytes to different morphotypes.
Results
Dormant Rhizopus spores induce early and strong inflammatory cytokine gene expression in human mononuclear cells
Co-culturing PBMCs, monocytes, and moDCs with ethanol-inactivated germ tubes of Rhizopus arrhizus and Aspergillus fumigatus germ tubes led to significantly increase of proinflammatory cytokine mRNA (Fig. 1A–C). Interestingly, resting spores of R. arrhizus also induced a strong upregulation of IL1B and TNFA transcription compared with unstimulated control cells or those co-cultured with dormant conidia of A. fumigatus. The expression levels of IL1B and TNFA mRNA in PBMCs peaked after 3 h co-culture with resting R. arrhizus spores, and declined subsequently (Fig. 1D). However, significantly upregulated cytokine expression was found compared with unstimulated cells at all studied time points. Elevated IL1B and TNFA gene expression was dependent on the multiplicity of infection (Sup. Fig. 1A), whereas sterile-filtered supernatants of fungal preparations did not cause induction of IL1B and TNFA (Sup Fig. 1B). Upregulated transcriptional activity of IL1B and TNFA was paralleled by markedly elevated secretion of TNFA, IL1B, IL6, IL8, GM-CSF, and MCP-1 in PBMCs co-cultured with inactivated dormant spores and germinated stages of R. arrhizus (Fig. 2). Importantly, stimulation of PBMCs with vital spores resulted in a similarly strong induction of IL1B and TNFA expression (Sup. Fig. 2). Though the peak of TNFA transcription was observed after 6 hours of co-culture, even a 20 min stimulation period resulted in a slight, but significant induction of IL1B and TNFA transcription. Taken together, these observations demonstrate that dormant spores of R. arrhizus are a potent stimulus of an early and strong proinflammatory cytokine response by human mononuclear phagocytes.
Figure 1.
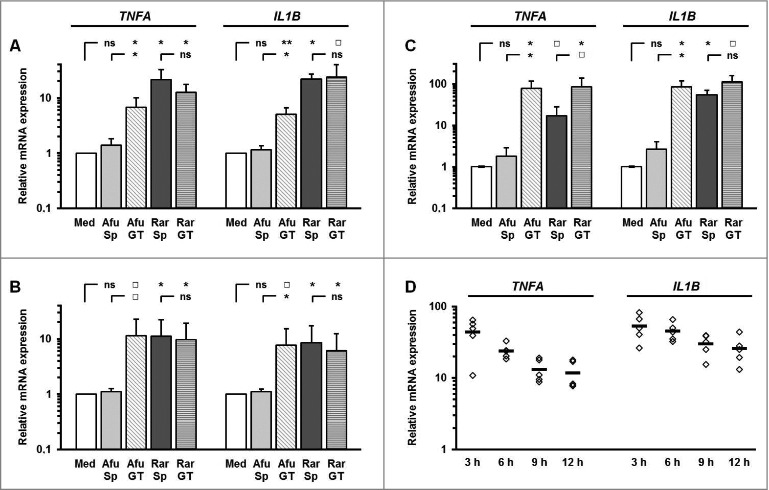
Dormant R. arrhizus spores induce an early and strong upregulation of inflammatory cytokine genes in human mononuclear cells (A) 2 × 106 PBMCs, (B) 1 × 106 monocytes, and 5 × 105 (C) moDCs from healthy donors (n = 5) were co-cultured with an equivalent amount (MOI 1.0) of ethanol-inactivated spores (Sp) and germ-tubes (GT) of R. arrhizus (Rar) and A. fumigatus (Afu) for a period of 6 hours. The expression of important proinflammatory cytokines genes was analyzed by RT-qPCR. Gene expression is shown as relative expression levels compared with unstimulated cells. The errors bars represent the standard deviation. (D) Relative gene expressions of TNFA and IL1B after a 3 to 12 hour co-culture of 2 × 106 PBMCs from 5 healthy donors with an equivalent amount of ethanol-inactivated resting spores of R. arrhizus compared with unstimulated cells. Med. indicates “immune cells stimulated with medium without fungal cells;” horizontal bars indicate the mean values. ns: not significant; p-values: □: 0.05 < p < 0.1; *: 0.01 < p < 0.05; **: 0.001 < p < 0.01.
Figure 2.
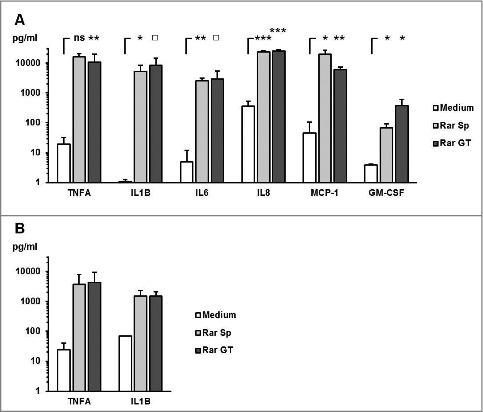
R. arrhizus spores induce the secretion of proinflammatory cytokines (A-B) A total of 2 × 106 PBMCs from healthy donors (n = 4) were co-cultured with 2 × 106 R. arrhizus spores (Sp) and germ tubes (GT) for 9 hours. (A) Cytokine secretion into the culture medium was quantified by bead-based luminex analyses (H-CYTOMAG 60K, Merck-Millipore). ns: not significant; p-values: □: 0.05 <p< 0.1; *: 0.01 < p <0.05; **: 0.001 < p <0.01, ***: p <0.001. (B) TNFA and IL1B concentrations were quantified using ELISA MaxTM deluxe sets (Bio-Legend).
Immunogenicity of dormant spores is observed for various Mucorales species
Further we assessed whether dormant spores of other human pathogenic Mucorales species were capable of stimulating an inflammatory cytokine response in mononuclear cells. All Mucorales species assessed in this study induced an at least 10-fold upregulation of TNFA and IL1B mRNA synthesis in PBMCs after 6 h co-culture (Fig. 3A–B), whereas the presence of Ascomycota, A. fumigatus, and F. solani did not result in significant elevations of proinflammatory cytokine gene expression. A similar pattern was observed for the IL1B release into the culture medium (Fig. 3C). These data suggest that spores of human-pathogenic Mucorales species share common structural or metabolic features triggering a proinflammatory cytokine response by mononuclear cells.
Figure 3.
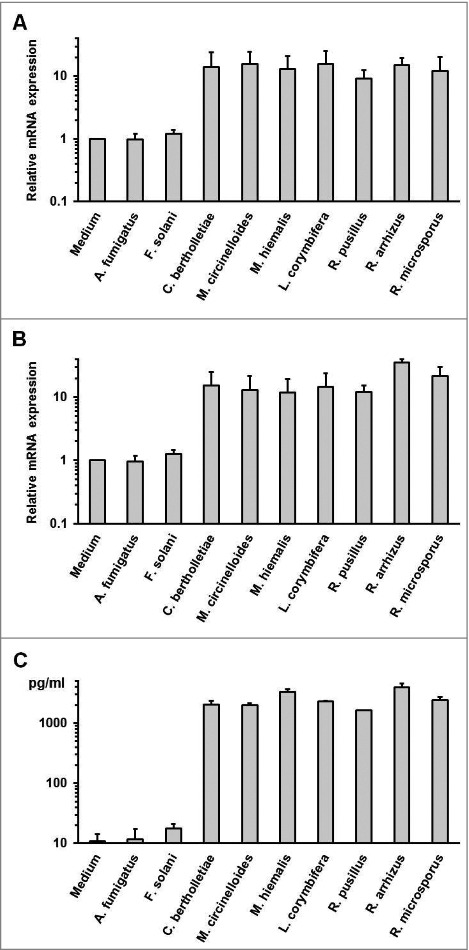
Proinflammatory cytokine response by human PBMCs is caused by exposure to dormant spores of various Mucorales species (A-B) 2 × 106 PBMCs were co-cultured with 2 × 106 ethanol-inactivated resting spores of different opportunistic Ascomycota and Mucorales species for 6 hours. The relative mRNA expression of IL1B (A) and TNFA (B) was assessed by RT-qPCR. (C) IL1B concentrations in the culture medium after a 9 h co-culture of 2 × 106 PBMCS with an equal amount of inactivated dormant Ascomycota and Mucorales spores.
Resting spores of R. arrhizus induce the upregulation of co-stimulatory molecules on dendritic cells
Apart from orchestrating the host defense against fungal pathogens by release of cytokines, mononuclear phagocytes serve a crucial role in triggering specific immune response with their ability to acquire, process and present fungal antigens to naïve T cells. The upregulation of co-stimulatory molecules and maturation markers on moDCs exposed to dormant spores and germ tubes of A. fumigatus and R. arrhizus was assessed (Fig. 4). Germ tubes of both fungi and resting spores of R. arrhizus stimulated CD83 and CD86 upregulation on moDCs. In contrast, co-culture with dormant conidia of A. fumigatus only led to slightly increased CD83 and CD86 expression (Fig. 4A+B). This suggests that dormant spores of R. arrhizus induced the maturation of moDCs.
Figure 4.
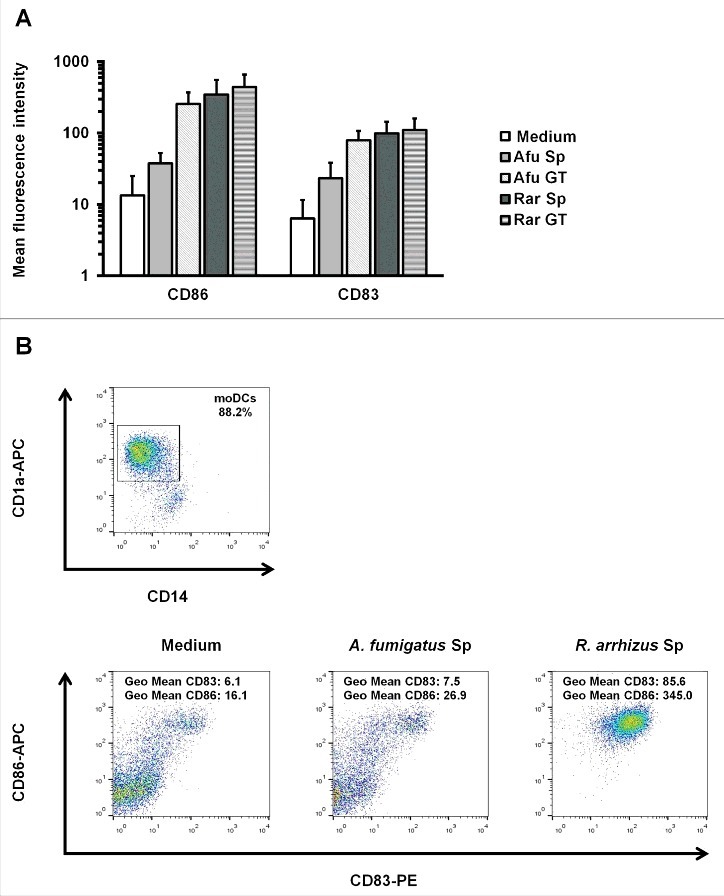
Dormant R. arrhizus spores induce the upregulation of co-stimulatory molecules on moDCs (A) Geometric mean fluorescence values for CD83 and CD86 expression after an 18 h co-culture of 5 × 105 moDCs with 5 × 105 spores (Sp) and germ tubes (GT) of A. fumigatus (Afu) or R. arrhizus (Rar). (B) Gating strategy and CD83/CD86 plots for moDCs from one representative donor. CD1+ CD14− cells were identified among all harvested cells. Within this subset, geometric mean fluorescence for CD83-PE and CD86-APC was quantified.
T-helper cells specifically responding to Rhizopus spores can be detected in healthy subjects
To evaluate whether a specific T-cell response to Mucorales spores can be observed in healthy subjects, a described previously assay19 was used to quantify the frequency of T-helper cells responding to inactivated spores and germ tubes of A. fumigatus and R. arrhizus by CD154 upregulation (Fig. 5). After overnight stimulation with germ tubes of A. fumigatus and R. arrhizus, mean frequencies of 0.29% (+/− 0.15%) and 0.39% (+/− 0.19%) CD154+/CD4+ T-cells were measured, respectively. Stimulation with dormant spores of R. arrhizus resulted in a proportion of 0.35% (+/− 0.18%) specific T-helper cells, whereas only 0.08% (+/− 0.05%) T-cells were CD154 positive in response to resting A. fumigatus spores, which only slightly exceeds the unspecific background frequencies in unstimulated cells.
Figure 5.
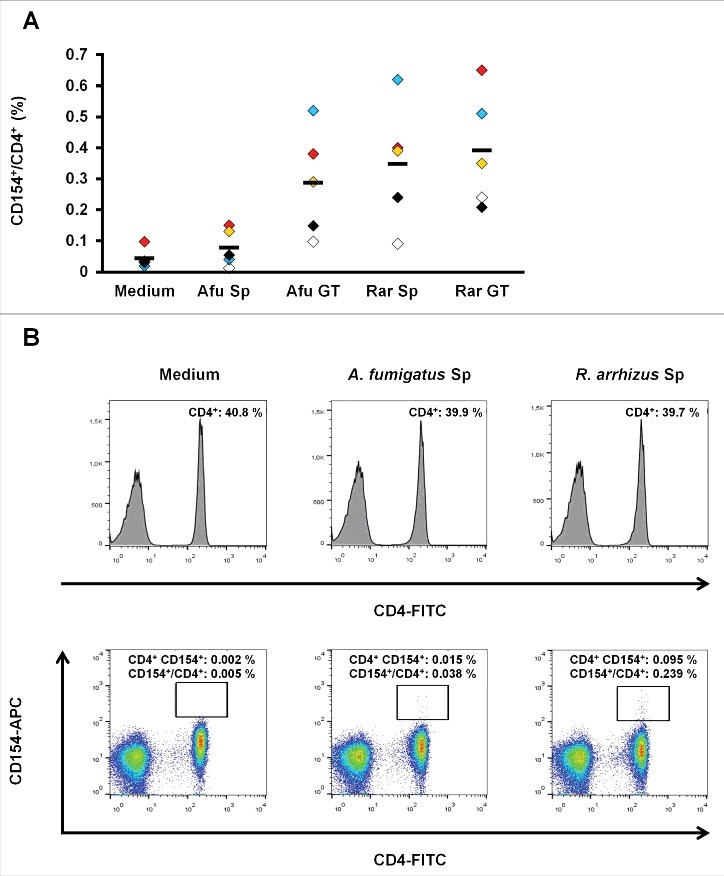
(A) subset of T-helper cells specifically upregulating CD154 after exposure to R. arrhizus spores can be identified in healthy donors (A) Proportion of CD154+/CD4+ T-lymphocytes after an 18 h co-culture of PBMCs with ethanol-inactivated resting spores (Sp) and germ tubes (GT) of A. fumigatus (Afu) or R. arrhizus (Rar). Samples from 5 healthy donors, pre-screened for a proportion of at least 0.2% CD154+/CD4+ T-cells after 18 h stimulation with 5 mg of a commercially available crude mycelial lysate of A. fumigatus (+ 0.1 µg αCD28), have been analyzed. Horizontal bars indicate the mean values. (B) Representative flow cytometry data of one donor (black symbols in Fig. 5A) after stimulation of 1 × 106 PBMCs with 1 × 106 inactivated spores of A. fumigatus or R. arrhizus. 0.1 µg αCD28 antibody were used for co-stimulation. Medium control contains the co-stimulatory antibody, but no fungal cells.
The absence of rodlet hydrophobins contributes to the immunogenicity of dormant Mucorales spores
We hypothesized that differences in outer cell wall composition may contribute to the immunogenicity of dormant Mucorales spores. Particularly, the existence or absence of a hydrophobin layer associated with immunoprotective properties in Ascomycota has not been demonstrated in Mucorales yet. Consistent with previous findings,15 treatment of A. fumigatus conidia with 48% hydrofluoric acid for cleavage of phosphodiester bonds in GPI anchors connecting the hydrophobins with other cell wall components led to an upregulation of IL1B, TNFA, and further pro-inflammatory cytokines in PBMCs exposed to these conidia, whereas a slight downregulation of IL10 was observed (Fig. 6A–B, Sup. Fig. 3). Conversely, the immunogenicity of dormant R. arrhizus spores was not enhanced when treated with hydrofluoric acid. Though there was a tendency toward a slightly less pronounced cytokine response to hydrofluoric acid treated R. arrhizus spores, significant pro-inflammatory cytokine gene induction and a specific T-helper cell to these spores by CD154 upregulation were observed (Fig. 6A–B, Sup. Figs. 3 and 4). In a time-course analysis using different durations of hydrofluoric acid treatment, neither shorter nor longer hydrofluoric acid treatment resulted in an increased immunogenicity of dormant Rhizopus spores (Fig. 6C–D).
Figure 6.
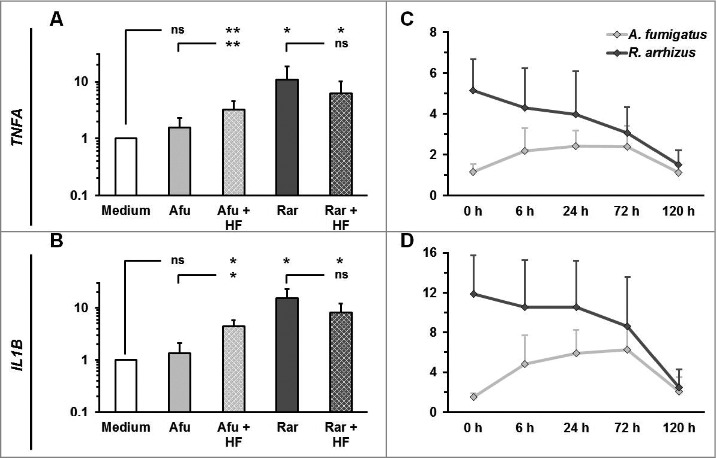
Hydrofluoric acid treatment does not increase the immunogenicity of resting R. arrhizus spores (A-B) Dormant spores of A. fumigatus and R. arrhizus were fixed with 2.5% paraformaldehyde for 12 h, followed 48% hydrofluoric acid treatment of 72 h. After extensive washing with dH2O to remove residual hydrofluoric acid 2 × 106 spores were co-cultured with 2 × 106 PBMCs obtained from healthy donors (n = 4). The expression of TNFA and IL1B was analyzed by RT-qPCR (crosshatched bars). For comparison, the expression of these genes was also analyzed after co-culture of PBMCs with PFA-fixed untreated dormant spores (monochromatic bars). (C-D) Relative TNFA and IL1B mRNA expression levels after a 6 h co-culture of 2 × 106 PBMCs with dormant A. fumigatus and R. arrhizus spores treated with 48% hydrofluoric acid for 0 to 120 hours. ns: not significant; p-values: □: 0.05 <p < 0.1; *: 0.01 < p< 0.05; **: 0.001< p < 0.01.
High-resolution SEM was performed to obtain a direct morphologic image of the cell wall surface morphology of Mucorales. While A. fumigatus conidia exhibited the described previously fibrillary rodlet surface pattern,20 a morphologic correlate to these structures was not observed in resting spores of R. arrhizus, C. bertholletiae, and R. pusillus (Fig. 7A). Treatment of R. arrhizus spores with hydrofluoric acid did not lead to a significantly altered morphology in the outermost cell wall layer, whereas the A. fumigatus spores lost their typical rodlet surface pattern (Fig. 7B).
Figure 7.
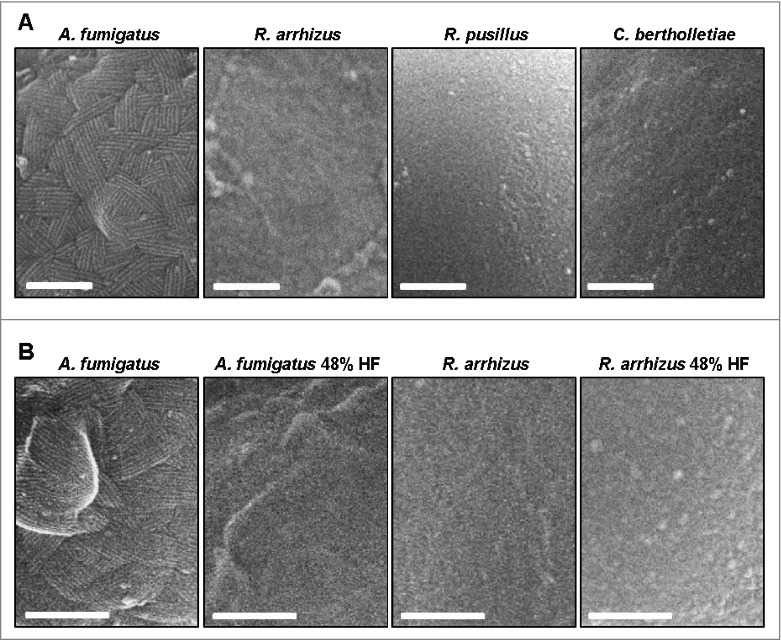
Mucorales do not possess a morphologic correlate of A. fumigatus rodlet hydrophobins (A) Scanning electron micrographs of resting spores of A. fumigatus ATTC 46645 (shows approximately 10 nm wide rodlets), R. arrhizus CBS 110.17, R. pusillus CBS 245.58, and C. bertholletiae CBS 187.84. Scale bar = 200 nm. (B) Scanning electron micrographs of native and hydrofluoric acid treated (48%, 72 h) resting spores of A. fumigatus ATTC 46645 and R. arrhizus CBS 110.17. Scale bar = 200 nm.
In accordance with these findings, pBLAST analyses of the A. fumigatus RodA and RodB sequences against the Joint Genome Institute MycoCosm database (634 organisms including 19 Mucoromycotina) did not reveal any homologues of these proteins in Mucorales, while significant homologies to proteins in numerous Ascomycota species were detected (Table S1). Taken together, the immunological data, morphological findings, and in silico data do not provide any evidence for the existence of immunoprotective rodlet hydrophobins in the cell wall of the studied Mucorales spores.
Discussion
Opportunistic infections caused by molds are a major threat for patients with impaired innate or specific immunity. Mucorales have emerged as increasingly important pathogens in these patients,3 but the immunopathology of mucormycoses is poorly understood. Here we provide evidence that human mononuclear phagocytes exposed to both resting spores and mature stages of R. arrhizus and other opportunistic Mucorales species respond with an early and strong proinflammatory cytokine expression and release.
The data presented here are in accordance with previous findings demonstrating that co-culture of human monocytes with heat-inactivated Rhizopus spores results in a robust induction of IL6 and TNFA secretion.16 Our data further support previously published results demonstrating that dormant conidia of A. fumigatus and other airborne fungal spores induce little inflammatory response.21,22 Though different immune cell subsets, effector-target ratios, and methods for inactivating fungal cells were used in our study and the cited work,16 we share the conclusion of an inflammatory immune response caused by resting Mucorales spores and corroborate these data via direct comparison of different Mucorales species and morphotypes. The observed immunogenicity of resting Mucorales spores contrasts the findings in Ascomycota.
Apart from orchestrating the host defense against invading fungi via the release of cytokines, mononuclear phagocytes serve a crucial role in triggering specific immune response with their ability to acquire, process, and present fungal antigens to naïve T cells. For this reason, we compared the upregulation of co-stimulatory molecules and maturation markers on moDCs exposed to dormant spores and germ tubes of A. fumigatus and R. arrhizus. Confirming previous data, T-cell response is limited to the germinated stages of A. fumigatus,23 whereas a significant proportion of T-cells specifically respond to resting spores of R. arrhizus by upregulating CD154 (Fig. 4B).
Immunogenicity of Mucorales spores may be explained by differences in outer cell wall composition. The cell wall of dormant spores of A. fumigatus is covered by a pigmented rodlet hydrophobin layer.15 These conidial rodlet hydrophobins form highly insoluble complexes in the outermost cell wall layer facilitating aerosolic dispersion of airborne fungal spores and their growth at air-liquid-interfaces.24,25,26 Several studies highlight an additional role of rodlet proteins in mediating a stage-specific immune response to fungal spores. During maturation, conidia of A. fumigatus and other Ascomycota swell and lose their hydrophobin layer. The structures within the cell wall are composed primarily of ß-glucans, galactomannan and chitin14 are exposed to the innate immune cells, leading to an inflammatory response of human and murine phagocytes.22
Paris and colleagues showed that RodAp, the main rodlet hydrophobin of A. fumigatus spores, mediates resistance to host alveolar macrophages. Spores of a rodletless mutant (ΔrodA-47) were significantly more sensitive to killing by macrophages than wild-type conidia.19 Aimanianda and colleagues described that surface hydrophobins prevent immune recognition of spores of A. fumigatus and other Ascomycota species due to the resistance of rodlet proteins to lysosomal proteolytic degradation resulting in a lack of antigenic peptides.15 In another study, this group found that RodA hydrophobins mask the immunogenic cell wall components β1,3 glucan and α-mannose of Aspergillus and Fusarium impairing Dectin-1 and Dectin-2 mediated recognition by macrophages, leading to reduced cytokine secretion. In a corneal murine infection model, ΔRodA Aspergillus spores induced significantly stronger cytokine release and neutrophil recruitment to the site of infection than wild-type conidia reducing fungal survival in infected mice.27 On the other hand, rodlet-mediated masking of the host response is assumed to provide a protective mechanism against tissue damage caused by an excessive inflammatory response to non-invasive fungal stages.28,29
Conidial hydrophobins have been identified in several Ascomycota and Basidiomycota species.26,29 The existence of rodlet hydrophobins in airborne human-pathogenic mold fungi has not been demonstrated in other phyla. In this study we have provided first evidence of the absence of a morphologic correlate of rodlet hydrophobins in dormant Mucorales spores using high resolution SEM, supported by in silico data underlining the absence of RodAp and RodBp homologues in Mucorales. The largely unaltered immunogenicity of R. arrhizus spores after hydrofluoric acid treatment further supports the absence of immunoprotective hydrophobins. The trend toward reduced immunogenicity of spores treated with hydrofluoric acid, especially after treatment of prolonged periods, may be attributable to potential off-target effects of hydrofluoric acid including the depletion of immunogenic proteins or polysaccharids in the fungal cell wall. Our data, however, clearly demonstrate, that both hydrofluoric acid treated and untreated R. arrhizus spores induce a robust pro-inflammatory cytokine response and specific T-cell response. Most importantly, in contrast to A. fumigatus, no evidence for an enhanced immunogenicity of R. arrhizus spores by hydrofluoric acid treatment was obtained. Hence, the findings of this study suggest that the missing coverage of immunogenic carbohydrate and protein structures in an immunoprotective rodlet hydrophobin layer in the fungal cell wall may contribute to the immunogenicity of resting Mucorales spores.
Despite of the clinical similarities of mucormycoses and invasive mycoses caused by Ascomycota species, this study highlights the existence of considerable differences in the immunopathology and cell wall architecture of these fungi. Further research is required to gain a comprehensive view of the immunogenic structures and antigens of Mucorales spores as well as the receptors and signaling pathways mediating the inflammatory response to these spores. This may open the field of new prophylactic and therapeutic strategies in mucormycosis targeting the early developmental stages of Mucorales.
Materials & methods
Preparation and culture of leukocyte subsets
Whole blood specimens were collected from healthy volunteers after obtaining written informed consent. The study was conducted in full compliance with the Declaration of Helsinki and approved by the Ethics Committee of the University of Wuerzburg. Peripheral blood mononuclear cells (PBMCs) were purified by Ficoll gradient centrifugation. Monocytes were isolated using MACS CD14 positive selection (Miltenyi Biotec, #130–050–201). To generate monocyte-derived dendritic cells (moDCs), monocytes were incubated with IL4 (Miltenyi Biotec, #130–095–373) and GM-CSF (Sanofi, Leukine® sargramostim) for a period of 6 d. The purified leukocyte subsets were resuspended at a concentration of 1 × 106 cells/ml in RPMI 1640 (Gibco / Life Technologies, #72400–021) supplemented with 10% heat-inactivated fetal bovine serum (FCS, Sigma-Aldrich, #F-7524) and 100 µg/ml gentamicin (Merck Serono, Refobacin®). Cells were cultured at 37 °C and 5% CO2.
Fungal strains and preparation of fungal cells
The following fungal strains were used in this study: Rhizopus arrhizus var. arrhizus (CBS 110.17), Rhizopus microsporus (CBS 53680), Rhizomucor pusillus (CBS 245.58), Lichtheimia corymbifera (CBS 271.65), Mucor circinelloides (CBS 192.98), Mucor hiemalis (CBS 200.28), Cunninghamella bertholletiae (CBS 187.84), Fusarium solani (CBS 181.29), and Aspergillus fumigatus (ATTC 46645).
Dormant spores of these isolates were prepared from mature colonies grown on beer wort agar and passed through a 40 µm cell strainer to remove residual mycelium. A total of 1 × 108 spores were incubated in 20 ml RPMI 1640 under constant shaking at 200 rpm at RT. Germ tubes were obtained after 12–14 hours, and hyphae after 16–18 hours. To prevent further maturation during co-culture with immune cells, the fungi were inactivated by incubation in 96% ethanol for 30 min at RT. Subsequently, fungal morphotypes were washed 5 times with dH2O and resuspended at a concentration of 2 × 107 cells/ml in RPMI 1640. Successful inactivation was confirmed by incubation of 10 µl of the spore solution on beer wort agar plates for 7 d.
Chemical removal of conidial hydrophobins by hydrofluoric acid treatment
As described previously,15 dormant spores were fixed with 2.5% paraformaldehyde (Carl Roth, #0335) for 12 h, followed by 48% hydrofluoric acid (Merck, #100334) treatment of 72 h at 4 °C. After extensive washing with dH2O to remove residual hydrofluoric acid, spores were resuspended in RPMI 1640 at a concentration of 2 × 107 cells/ml.
Gene expression analysis
Total RNA from human immune cells was purified using the RNeasy® Mini Kit (Qiagen, #74106) according to the manufacturer's protocol. RNA was eluted in 30 µl of RNase-free water and the concentration was quantified with the NanoDrop spectrophotometer (Peqlab). RNA was determined by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, # 4368814). Quantitative PCR was performed on a StepOneTM plus instrument (Applied Biosystems) using the iTaqTM Universal SBYR® Green Supermix (Biorad, #1725122). An initial denaturation step (90 °C, 15 s) was followed by 40 cycles of repeated denaturation (90 °C, 3 sec) and extension (60°C, 30 sec). The following primer sequences were used: ALAS1 5′-GGCAGCACAGATGAATCAGA-3′ and 5′-CCTCCATCGGTTTTCACACT-3′, IL1B 5′-GGACAAGCTGAGGAAGATGC-3′ and 5′-TCGTTATCCCATGTGTCGAA-3′, and TNFA 5′-TGCTTGTTC CTCAGCCTCTT-3′ and 5′-TGGGCTACAGGCTTGTCACT-3′. Primers were confirmed not to cross-anneal nucleic acid sequences of the studied fungal species.
Analysis of cytokine release
ELISA MaxTM deluxe sets (Bio-Legend, #437004 and #430204) were used to quantify the release of IL1B and TNFA into the culture medium. Bead-based multiplex cytokine assays were conducted using a magnetic Milliplex Human Cytokine Panel (Merck Millipore, HCYTOMAG-60K) according to the manufacturer's instructions. Culture supernatants were pre-diluted 1:2 in fresh RPMI 1640.
Flow cytometry
For staining of surface markers moDCs were harvested 120 hours after seeding, washed with HBSS (Sigma-Aldrich, #H6648) and resuspended in RPMI 1640 + 10% FCS at a concentration of 1 × 106 cells per ml. 300 µl (3 × 105 cells) were plated in each well of a 48-well plate and co-incubated with ethanol-inactivated fungal spores or germ tubes for 18 h. The cells were then transferred to FACS tubes, washed and stained in 100 µl HBSS containing CD1a-FITC, CD14-PerCP, CD83-PE, and CD86-APC antibodies (Miltenyi Biotec, #130–097–903, #130–094–969, #130–094,876; BD Biosciences, #556855). Analysis was performed on a FACSCalibur flow cytometer (BD Biosciences). CD1a+ CD14− cells were considered as moDCs and geometric mean fluorescence intensity of CD83-PE and CD86-APC was determined in the CD1a+ CD14− subset.
CD154-positive T-cells were detected after an 18 h co-culture of 1 × 106 PBMCs with 5 µg of an A. fumigatus mycelial lysate (Miltenyi Biotec, #130–098–170) or ethanol-inactivated fungal spores or germ tubes and 0.1 µg CD28 co-stimulatory antibody. Cells were stained using the Inside Stain Kit (Miltenyi Biotec, #130–090–477) as well as CD4-FITC and CD154-APC antibodies (Miltenyi Biotec, #130–092–358 and #130–092–290). Lymphocytes were identified by FSC/SSC properties. The frequency of CD154 positive cells among CD4 positive cells was determined.
Scanning electron microscopy (SEM)
Freeze-drying was performed as described previously [18]. The glutaraldehyde (Sigma-Aldrich, #G5882) fixed samples mounted on small silicon chips were washed with dH2O. The chips were blotted with filter paper to remove most of the water before freezing by nitrogen cooled propane. The frozen samples were cryo-transferred to a Baf 300 freeze-etching device (Bal-Tec) and partially freeze-dried for 35 min at −90 °C. The samples were then rotary coated by electron beam evaporation with 2 nm of platinum and kept cold during liquid-nitrogen transfer to the cryo-stage of the SEM. Specimens were investigated at a temperature of −100 °C, using a Gatan cryo-holder 626 in a Hitachi S-5200, in-lens field emission SEM at an accelerating voltage of 10 kV using the secondary electron signal.
Statistical analysis
Unless otherwise indicated, cells from 5 different donors were analyzed. Significance testing was defined p < 0.05 by using the paired 2-sided t-test.
Disclosure of potential conflicts of interests
AJU has received support for travel to meetings from Astellas and Basilea. He is a consultant and on the speakers' bureaus of Astellas, Gilead, MSD, and Pfizer. He has also received support for travel and accommodation from Astellas, Boehringer Ingelheim, Gilead, MSD, and Pfizer for activities unrelated to this study. His institution has received grants from Astellas, Gilead, MSD, and Pfizer. JL declares that he has no conflicts of interest, his institution received a grant from Pfizer. SW, VT, PW, JE, AMWG, MD, and HE declare that they have no conflicts of interest to disclose.
Meetings where the information has previously been presented
Parts of this study have been presented at the European Congress of Clinical Microbiology and Infectious Diseases 2015 (Copenhagen, Denmark), the Spring Meeting of Antifungal Chemotherapy of the Paul-Ehrlich-Society 2015 (Bonn, Germany), and Trends for Medical Mycology 2015 (Lisbon, Portugal).
Supplementary Material
Acknowledgments
We thank the Institute for Hygiene and Microbiology (IHM), University of Würzburg for provision of fungal strains and for the opportunity to use their BSL II facilities for culturing and handling of Mucorales isolates.
Funding
This work was supported by the Interdisciplinary Center for Clinical Research (IZKF) Wuerzburg (grant number Z-3/56 to SW) and the DFG Transregio / SFB 124 (project A2, to HE and JL).
References
- [1].Pappas PG. Opportunistic fungi: a view to the future. Am J Med Sci 2010; 340(3):253-7; PMID:20823702; https://doi.org/ 10.1097/MAJ.0b013e3181e99c88 [DOI] [PubMed] [Google Scholar]
- [2].Kontoyiannis DP, Wessel VC, Bodey GP, Rolston KV. Zygomycosis in the 1990s in a tertiary-care cancer center. Clin Infect Dis 2000; 30(6):851-6; https://doi.org/ 10.1086/313803 [DOI] [PubMed] [Google Scholar]
- [3].Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL, Sein M, Sein T, Chiou CC, Chu JH, et al.. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis 2005; 41:634-53; PMID:16080086; https://doi.org/ 10.1086/432579 [DOI] [PubMed] [Google Scholar]
- [4].Eucker J, Sezer O, Graf B, Possinger K. Mucormycoses. Mycoses 2001; 44(7-8):253-60; PMID:11714058; https://doi.org/ 10.1111/j.1439-0507.2001.00656.x [DOI] [PubMed] [Google Scholar]
- [5].Lanternier F, Dannaoui E, Morizot G, Elie C, Garcia-Hermoso D, Huerre M, Bitar D, Dromer F, Lortholary O, French Mycosis Study Group . A global analysis of mucormycosis in France: the RetroZygo Study (2005-2007). Clin Infect Dis 2012; 54 Suppl 1:S35-43; PMID:22247443; https://doi.org/ 10.1093/cid/cir880 [DOI] [PubMed] [Google Scholar]
- [6].Ribes JA, Vanover-Sams CL, Baker DJ. Zygomycetes in human disease. Clin Microbiol Rev 2000; 13(2):236-301; PMID:10756000; https://doi.org/ 10.1128/CMR.13.2.236-301.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Romani L. In: Calderone R, Cihlar R, eds. Fungal pathogenesis. Principles and clinical applications. New York: Marcel Dekker Inc., 2002: 401-32 [Google Scholar]
- [8].Lass-Flörl C, Roilides E, Löffler J, Wilflingseder D, Romani L. Minireview: host defence in invasive aspergillosis. Mycoses 2013; 56(4):403-13; PMID:23406508; https://doi.org/ 10.1111/myc.12052 [DOI] [PubMed] [Google Scholar]
- [9].Romani L. Immunity to fungal infections. Nat Rev Immunol 2004; 4(1):1-23; PMID:14661066; https://doi.org/ 10.1038/nri1255 [DOI] [PubMed] [Google Scholar]
- [10].Antachopoulos C, Roilides E. Cytokines and fungal infections. Br J Haematol 2005; 129(5):583-96; PMID:15916680; https://doi.org/ 10.1111/j.1365-2141.2005.05498.x [DOI] [PubMed] [Google Scholar]
- [11].Bonifazi P, Zelante T, D'Angelo C, De Luca A, Moretti S, Bozza S, Perruccio K, Iannitti RG, Giovannini G, Volpi C, et al.. Balancing inflammation and tolerance in vivo through dendritic cells by the commensal Candida albicans. Mucosal Immunol 2009; 2(4):362-74; PMID:19421183; https://doi.org/ 10.1038/mi.2009.17 [DOI] [PubMed] [Google Scholar]
- [12].Romani L. Immunity to fungal infections. Nat Rev Immunol 2011; 11(4):275-88; PMID:21394104; https://doi.org/ 10.1038/nri2939 [DOI] [PubMed] [Google Scholar]
- [13].Park SJ, Mehrad B. Innate immunity to Aspergillus species. Clin Microbiol Rev 2009; 22(4):535-51; PMID:19822887; https://doi.org/ 10.1128/CMR.00014-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Latge JP. Tasting the fungal cell wall. Cell Microbiol 2010; 12(7):863-72; PMID:20482553; https://doi.org/ 10.1111/j.1462-5822.2010.01474.x [DOI] [PubMed] [Google Scholar]
- [15].Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, et al.. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 2009; 460(7259):1117-21; PMID:19713928; https://doi.org/ 10.1038/nature08264 [DOI] [PubMed] [Google Scholar]
- [16].Warris A, Netea MG, Verweij PE, Gaustad P, Kullberg BJ, Weemaes CM, Abrahamsen TG. Cytokine responses and regulation of interferon-gamma release by human mononuclear cells to Aspergillus fumigatus and other filamentous fungi. Med Mycol 2005; 43(7):613-21; PMID:16396246; https://doi.org/ 10.1080/13693780500088333 [DOI] [PubMed] [Google Scholar]
- [17].Roilides E, Kontoyiannis DP, Walsh TJ. Host defenses against zygomycetes. Clin Infect Dis 2012; 54 Suppl 1:S61-6; PMID:22247447; https://doi.org/ 10.1093/cid/cir869 [DOI] [PubMed] [Google Scholar]
- [18].Walther P. High-resolution cryo-SEM allows direct identification of F-actin at the inner nuclear membrane of Xenopus oocytes by virtue of its structural features. J Microsc 2008; 232(2):379-85; PMID:19017237; https://doi.org/ 10.1111/j.1365-2818.2008.02109.x [DOI] [PubMed] [Google Scholar]
- [19].Bacher P, Kniemeyer O, Teutschbein J, Thön M, Vödisch M, Wartenberg D, Scharf DH, Koester-Eiserfunke N, Schütte M, Dübel S, et al.. Identification of immunogenic antigens from Aspergillus fumigatus by direct multiparameter characterization of specific conventional and regulatory CD4+ T cells. J Immunol 2004; 193(7):3332-43; https://doi.org/ 10.4049/jimmunol.1400776 [DOI] [PubMed] [Google Scholar]
- [20].Paris S, Debeaupuis JP, Crameri R, Carey M, Charlès F, Prévost MC, Schmitt C, Philippe B, Latgé JP. Conidial hydrophobins of Aspergillus fumigatus. Appl Environ Microbiol 2003; 69(3):1581-8; PMID:12620846; https://doi.org/ 10.1128/AEM.69.3.1581-1588.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hohl TM, Van Epps HL, Rivera A, Morgan LA, Chen PL, Feldmesser M, Pamer EG. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog 2005; 1(3):e30; PMID:16304610; https://doi.org/ 10.1371/journal.ppat.0010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gersuk GM, Underhill DM, Zhu L, Marr KA. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J Immunol 2006; 176(6):3717-24; PMID:16517740; https://doi.org/ 10.4049/jimmunol.176.6.3717 [DOI] [PubMed] [Google Scholar]
- [23].Rivera A, Van Epps HL, Hohl TM, Rizzuto G, Pamer EG. Distinct CD4+-T-cell responses to live and heat-inactivated Aspergillus fumigatus conidia. Infect Immun 2005; 73(11):7170-79; PMID:16239511; https://doi.org/ 10.1128/IAI.73.11.7170-7179.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Beever RE, Dempsey GP. Function of rodlets on the surface of fungal spores. Nature 1978; 272(5654):608-10; PMID:148008; https://doi.org/ 10.1038/272608a0 [DOI] [PubMed] [Google Scholar]
- [25].Wösten HA. Hydrophobins: multipurpose proteins. Annu Rev Microbiol 2001; 55:625-46; PMID:11544369; https://doi.org/ 10.1146/annurev.micro.55.1.625 [DOI] [PubMed] [Google Scholar]
- [26].Linder MB, Szilvay GR, Nakari-Setälä T, Penttilä ME. Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev 2005; 29(5):877-96; PMID:16219510; https://doi.org/ 10.1016/j.femsre.2005.01.004 [DOI] [PubMed] [Google Scholar]
- [27].Carrion Sde J, Jr Leal SM, Ghannoum MA, Aimanianda V, Latgé JP, Pearlman E. The RodA hydrophobin on Aspergillus fumigatus spores masks dectin-1- and dectin-2-dependent responses and enhances fungal survival in vivo. J Immunol 2013; 191(5):2581-8; PMID:23926321; https://doi.org/ 10.4049/jimmunol.1300748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Slesiona S, Gressler M, Mihlan M, Zaehle C, Schaller M, Barz D, Hube B, Jacobsen ID, Brock M. Persistence versus escape: Aspergillus terreus and Aspergillus fumigatus employ different strategies during interactions with macrophages. PLoS One 2012; 7(2):e31223; PMID:22319619; https://doi.org/ 10.1371/journal.pone.0031223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bayry J, Aimanianda V, Guijarro JI, Sunde M, Latgé JP. Hydrophobins - unique fungal proteins. PLoS Pathog 2012; 8(5):e1002700; PMID:22693445; https://doi.org/ 10.1371/journal.ppat.1002700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


