Abstract
Cells in our body are constantly exposed to various stresses and threats to their genomic integrity. The tumor suppressor protein p53 plays a critical role in successful defense against these threats by inducing apoptotic cell death or cell cycle arrest. In unstressed conditions, p53 levels and activity must be kept low to prevent lethal activation of apoptotic and senescence pathways. However, upon DNA damage or other stressors, p53 is released from its inhibitory state to induce an array of apoptosis and cell cycle genes. Conversely, inactivation of p53 could promote unrestrained tumor proliferation and failure to appropriately undergo apoptotic cell death, which could, in turn, lead to carcinogenesis. The ubiquitin E3 ligase MDM2 is the most critical inhibitor of p53 that determines the cellular response to various p53-activating agents, including DNA damage. MDM2 activity is controlled by post-translational modifications, especially phosphorylation. However, accumulating evidence suggests that MDM2 is also regulated at the level of protein stability, which is controlled by the ubiquitin-proteasome pathway. Here, we discuss how MDM2 can be regulated in response to DNA damage with particular focus on the regulation of MDM2 protein stability.
DNA damage is one of the major threats to the integrity of the cell. DNA damage can cause cell death or genomic instability, which could lead to tumorigenesis. The p53 protein is considered the “guardian of the genome” due to its role in inducing cell cycle arrest, apoptosis, or senescence upon detection of DNA damage and other stressors (Lane, 1992). In healthy cells or in unstressed conditions, p53 is maintained at low levels by rapid protein turnover to prevent unnecessary activation of apoptotic and senescence pathways. In response to DNA damage, however, p53 protein is stabilized and allowed to accumulate. This induces transcription of an array of genes, such as p21 and BAX, which trigger cell cycle arrest and apoptotic pathways (for review, see Vousden and Prives, 2009). As activation of p53 is crucial to preventing abnormal growth and oncogenesis, in the vast majority of cancers the function of p53 is suppressed genetically (e.g., by mutations) or functionally (e.g., by inhibitors). Several studies have demonstrated that cancer cells can be killed by reactivation of p53 (Tovar et al., 2006; Ventura et al., 2007). Thus, finding a way to restore functional p53 activity is a promising therapeutic option in many cancers.
The murine double minute 2 protein (Mdm2) and its human homolog MDM2 (also referred to as HDM2) have been intensively studied because of their established role as the key negative regulator of p53. Although a number of negative regulators of p53 have been discovered, including more than fifteen E3 ubiquitin ligases that promote rapid protein turnover of p53 through the ubiquitin-proteasomal degradation pathway (for review, see Jain and Barton, 2010), MDM2 is thought to be the most important for two reasons. First, Mdm2 knockout mice die during embryogenesis due to elevated p53 levels. However, this embryonic lethality can be nullified if p53 is co-deleted (Jones et al., 1995; Montes et al., 1995). Second, inhibitors of the p53-MDM2 interaction, such as Nutlin-3a, can restore p53 activity and trigger apoptosis and cell cycle arrest in p53 wild-type cancer cells (Vassilev et al., 2004). MDM2 inhibits p53 in two ways; it physically blocks the transcriptional activity of p53 by binding to the trans-activation domain of p53 as well as promotes p53 protein degradation (Haupt et al., 1997). Importantly, p53 also induces expression of the MDM2 gene (Barak et al., 1993), indicating that MDM2 and p53 form an autoregulatory feedback loop. In unstressed conditions, MDM2 keeps p53 in check. Upon DNA damage or other stressors, however, p53 is released from MDM2 inhibition and allowed to accumulate, inducing gene expression that triggers cell cycle arrest and apoptosis. It should be noted that prior to p53 activation, MDM2 must be inhibited. Although a number of mechanisms that regulate p53 have been discovered, the mechanism that suppresses MDM2 to finally trigger p53 activation in response to cellular stress remains to be fully elucidated. In this review, we will consider the regulation of MDM2 protein stability as a mean to activate p53 following DNA damage.
Structure of MDM2
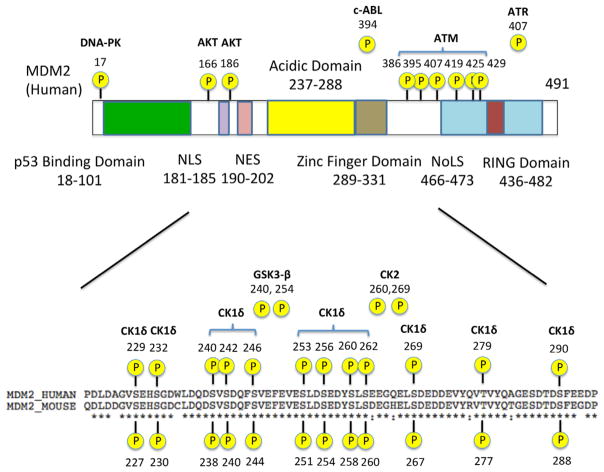
MDM2 consists of several key functional domains, including an N-terminal p53-binding domain, a central acidic domain, a zinc-finger domain, and a C terminal RING domain (Kostic et al., 2006; Priest et al., 2010; Riley and Lozano, 2012) (see Fig. 1). Whereas the Mdm2 gene in mice encodes a 489-amino acid protein, human MDM2 is made of 491 amino acids. Both the N-terminal domain and the central acidic domain are important for p53 recognition, while the C-terminal RING domain is responsible for p53 inhibition (Kussie et al., 1996; Yu et al., 2006). The N-terminal domain of MDM2 binds strongly to the N-terminal transactivation domain of p53 and blocks its transcriptional activity (Yu et al., 2006). Small molecule inhibitors of the p53-MDM2 interaction, such as Nutlin-3a, are designed to target this N-terminal domain of MDM2 (Vassilev et al., 2004). The central acidic domain of MDM2 exhibits only a weak affinity for p53, but many of the post-translational modifications, particularly phosphorylation, of MDM2 that affect its ability to regulate p53 occur in this region (Kulikov et al., 2005; Jin et al., 2006; Lu et al., 2007; Gannon et al., 2012; Wang et al., 2012). Lastly, although the RING domain of MDM2 does not bind to p53 directly, it possesses E3 ligase activity and is thus responsible for ubiquitinating p53 and marking it for degradation. The RING domain is also required for the dimerization of MDM2 with another MDM2 molecule (homodimer) or the structurally similar protein MDMX (also known as MDM4) (heterodimer) (Tanimura et al., 1999). Like other RING-containing E3 ligases, this dimerization is crucial to enabling MDM2’s E3 ligase activity.
Fig. 1.
MDM2 structure. Structural motifs and regions of interest in the MDM2 protein are shown. NLS: Nuclear localization signal; NES: Nuclear export signal; NoLs: Nucleolar localization sequence; RING: Really interesting new gene. Several known phosphorylation sites and their respective kinases are also indicated.
In addition to the key domains, MDM2 also contains a nuclear export signal (NES) and a nuclear localization signal (NLS) (Fig. 1). Consequently, MDM2 shuttles between the cytoplasm and the nucleus. Since p53 is primarily in the nucleus, shuttling of MDM2 to the nucleus is essential to its role in inhibiting the transcriptional function of p53. In the nucleus, MDM2 takes p53 away from its transcriptional targets, ubiquitinates p53, and translocates p53 into the cytoplasm for proteasomal degradation (Tao and Levine, 1999), although it has been shown that p53 can be degraded both in the cytoplasm and the nucleus (Stommel and Wahl, 2004). It should be noted that subcellular localization might affect the stability of MDM2 itself. It appears that when forming a homodimer, MDM2 is prone to ubiquitinating itself for degradation (self-ubiquitination) (Tanimura et al., 1999). In contrast, when forming a heterodimer with MDMX, MDM2 is stabilized (Sharp et al., 1999; Gu et al., 2002; Kawai et al., 2007; Tanimura et al., 1999). Since MDMX is a cytoplasmic protein (Migliorini et al., 2002), MDM2 may be more stable in the cytoplasm provided a sufficient amount of MDMX is expressed (note that MDMX expression levels are tissue and cell type specific; Grier et al., 2006). Nevertheless, the mechanism by which cellular stressors such as DNA damage might affect MDM2 stability in different cellular compartments remains elusive.
MDM2 and MDMX cooperate to inhibit p53
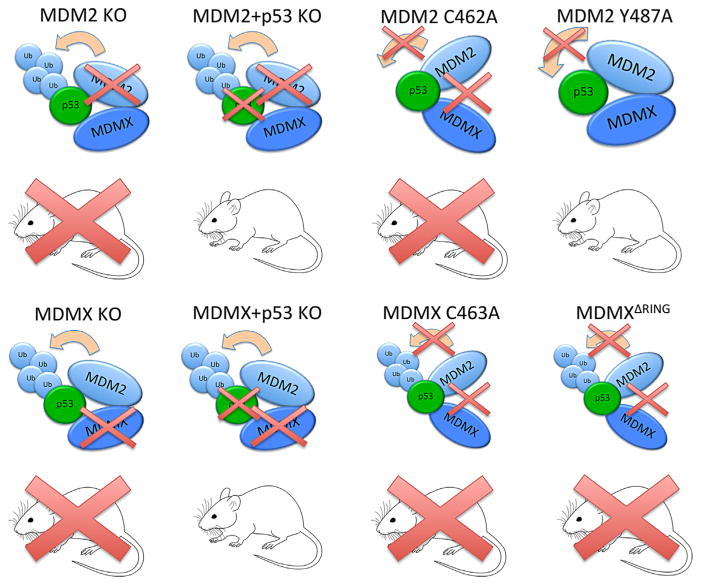
The MDM2 binding partner MDMX is structurally similar to MDM2, consisting of an N-terminal p53-recognition domain and a C-terminal RING domain, which mediates its heterodimerization with MDM2. Unlike MDM2, however, MDMX lacks appreciable E3 ligase activity. Nevertheless, MDMX promotes MDM2-mediated p53 degradation by at least two mechanisms. First, as described above, MDMX stabilizes MDM2 (Sharp et al., 1999; Gu et al., 2002; Kawai et al., 2007; Tanimura et al., 1999). Second, MDM2-MDMX heterodimers have greater E3 ligase activity toward p53 than MDM2 homodimers (Kawai et al., 2007). Because of the lack of intrinsic E3 ligase activity, it was initially thought that MDMX might only play a supporting role for MDM2. However, mouse studies demonstrated that it was not the case (see Fig. 2); the deletion of the Mdmx gene results in embryonic lethality, which can be rescued by co-deletion of p53, reminiscent of Mdm2/ p53 co-knockout mice (Parant et al., 2001). These studies indicate that Mdm2 and Mdmx may be equally important for prevention of lethal p53 activation, at least, during embryogenesis. How can Mdmx suppress p53 without E3 ligase activity? The answer to this question came from subsequent mouse studies. Two studies independently demonstrated that transgenic mice with mutant Mdmx incapable of binding to Mdm2 are embryonic lethal (Huang et al., 2011; Pant et al., 2011). Likewise, mice expressing mutant Mdm2 that is defective in forming a heterodimer with Mdmx but retains the ability to bind to p53 are also embryonic lethal (Itahana et al., 2007). The phenotypes of these mice can be rescued by the concomitant deletion of p53 (Itahana et al., 2007; Huang et al., 2011; Pant et al., 2011). Thus, it is indicated that Mdm2–Mdmx heterodimerization is indispensable for the inhibition of p53 during embryogenesis (Fig. 2). It should be noted, however, that phenotypes of Mdm2 and Mdmx knockout mice are not identical – the deletion of Mdm2 appears to cause more severe phenotypic defects than that of Mdmx. Moreover, cardiomyocyte-specific deletion of Mdm2 results in embryonic lethality, which can be rescued by p53 co-deletion, whereas that of Mdmx results in normal offspring (Grier et al., 2006). Whether MDM2 homodimers or MDM2-MDMX heterodimers play a major role in p53 regulation may be cell-type- or tissue-specific.
Fig. 2.
Summary of Mdm2 and Mdmx knockout mouse studies. Mdm2 knockout (KO) mice die during development but can be rescued if crossed with p53−/− mice (Jones et al., 1995; Montes et al., 1995). Mdmx KO mice also die during development and can be rescued if crossed with p53−/− mice (Parant et al., 2001). Mice with mutant Mdmx that cannot bind to Mdm2 (MdmxC463A and MdmxDRING) also die during development (Huang et al., 2011; Pant et al., 2011). Mice expressing the C462A mutant of Mdm2, which lacks E3 activity and cannot bind to Mdmx, die during embryogenesis (Itahana et al., 2007) while Mdm2Y487A mice, which lack E3 activity but can still bind to Mdmx, remain viable (Tollini et al., 2014).
The E3 ligase activity of MDM2 has long been thought to be essential for the suppression of p53. However, this common notion was recently challenged by an elegant transgenic mouse study. Tollini et al. (2014) created an Mdm2 mutant mouse in which the mutant Mdm2 lacks intrinsic E3 ligase activity but retains the ability to bind to Mdmx. Surprisingly, the resulting mice remain viable (Tollini et al., 2014), indicating that the E3 ligase activity of MDM2 is dispensable for the suppression of lethal p53 activation during embryogenesis. It should be noted, however, that although the mutant mice developed normally, their p53 activation in response to sub-lethal radiation was greater than that of wild-type mice (Tollini et al., 2014). This indicates that the E3 ligase activity of MDM2 is important for p53 regulation but only after cellular stresses such as DNA damage; in the absence of stress, p53 can be inhibited by the MDM2-MDMX heterodimer in an E3 activity-independent manner.
The idea that the MDM2–MDMX heterodimer plays a key role in p53 suppression not only shed light on the novel mechanism of p53 regulation but also opened up a new question regarding how the heterodimer is regulated in response to DNA damage. MDMX is a stable protein, whereas MDM2 is unstable, at least in part, due to its self-ubiquitination. Upon forming the heterodimer complex, MDMX protects MDM2 from degradation. Interestingly, in response to DNA damage, MDM2 promotes MDMX degradation (Kawai et al., 2003; Pan and Chen, 2003). As a result, MDM2 becomes more susceptible to degradation due to not being protected by its binding partner. Indeed, MDM2 undergoes accelerated degradation after DNA damage (Stommel and Wahl, 2004). Whether MDMX protects MDM2 only from self-ubiquitination or from ubiquitination by other E3 ligases and whether DNA-damage induced degradation of MDM2 is mediated by its self-ubiquitination or other E3 ligases remain to be fully elucidated (see below).
Phosphorylation of MDM2 in response to DNA damage
While many of the kinases involved in the DNA damage response phosphorylate p53 directly, several kinases also interact with MDM2 (see Figs. 1 and 3). Phosphorylation of MDM2 by these kinases may directly affect the binding affinity of MDM2 for p53. Alternatively, they could impact the stability or the intrinsic E3 ligase activity of MDM2. While some kinases of MDM2 are not involved in the DNA damage response, here we will only focus on kinases that are linked to the DNA damage response (see Fig. 1 for the phosphorylation sites and the kinases discussed in this section). An entire list of the kinases that phosphorylate MDM2 can be found elsewhere (Riley and Lozano, 2012).
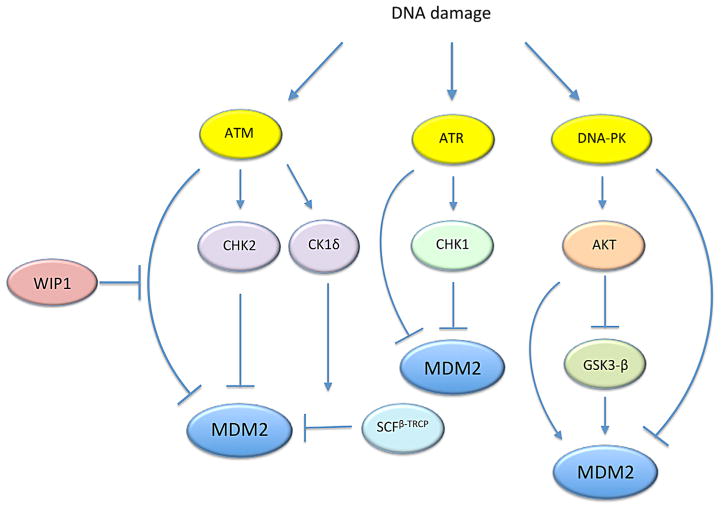
Fig. 3.
MDM2 regulation by the DNA damage kinases. MDM2 is phosphorylated at multiple sites by DNA damage kinases, ATM, ATR, and DNA-PK. These kinases can also phosphorylate MDM2 indirectly via their downstream effectors. CK1δ is phosphorylated by ATM and targets MDM2 for degradation by β-TrCP. See text for more details.
Kinases phosphorylating the central acidic domain
Many of the serine residues within the central acidic domain are targeted by various kinases, such as Glycogen Synthase Kinase 3β (GSK-3β), Casein Kinase 1 (CK1), and CK2 (Fig. 1) (Winter et al., 2004; Allende-Vega et al., 2005; Kulikov et al., 2005). An earlier study noted that the central acidic domain of MDM2 is phosphorylated at multiple serine residues and that the hypo-phosphorylation of these residues impairs the ability of MDM2 to promote p53 degradation (Blattner et al., 2002). Moreover, this hypo-phosphorylation naturally occurs after exposure to ionizing radiation (Blattner et al., 2002), suggesting that dephosphorylation of these residues is part of the DNA damage response. Phosphorylation of the central acidic domain by kinases, such as GSK-3 and CK2, promotes degradation of p53 (Allende-Vega et al., 2005; Kulikov et al., 2005). Conversely, inhibition of GSK-3 or CK2 results in the accumulation of p53 by preventing its MDM2-mediated degradation (Allende-Vega et al., 2005; Kulikov et al., 2005). Of note, GSK-3 is a downstream target of the pro-survival kinase AKT that is activated in response to DNA damage by the DNA-dependent protein kinase (DNA-PK), a serine/threonine kinase. Thus, it is interesting to know how the phosphorylation status of the central acidic domain is coordinately regulated by GSK-3 and other kinases following DNA damage.
Although the central acidic domain is known to be the secondary site of interaction with p53, modulating the phosphorylation status of the acidic domain does not abrogate the ability of MDM2 to bind to p53 (Yu et al., 2006). Thus, it is suggested that phosphorylation of the acidic domain may be linked to the suppression of MDM2 E3 activity or MDM2 degradation. In this regard, it is interesting to note that an E3 ubiquitin ligase complex, SCFβ-TrCP, directly ubiquitinates and targets MDM2 for proteasomal degradation in a manner depending on CK1 phosphorylation of the central acidic domain (Inuzuka et al., 2010). Nonetheless, SCFβ-TrCP promotes MDM2 degradation when the acidic domain is phosphorylated by CK1, which is, at first, contradictory to earlier observations (see discussion below).
ATM and ATR
The ataxia telangiectasia-mutated kinase (ATM) is activated by DNA double strand breaks and has been linked to phosphorylation of both the central acidic and the C-terminal RING domains of MDM2 in response to DNA damage (Cheng et al., 2011) (Fig. 1). Among the multiple ATM phosphorylation sites on MDM2, the most characterized residue is S395 (S394 in mouse Mdm2). Phosphorylation of S395 by ATM was initially noted for its ability to promote p53 activation through impaired p53 nuclear export and reduced p53 degradation (Maya et al., 2001). The phosphatase Wip1 counteracts this action by dephosphorylating S395 (Lu et al., 2007). Moreover, Wip1 can stabilize MDM2 by inhibiting its ubiquitination (Lu et al., 2007). However, a transgenic mouse study has demonstrated that ATM phosphorylation of S394 by itself impacts p53 activation but does not appear to affect MDM2 protein half-life following DNA damage (Gannon et al., 2012). Therefore, MDM2 stabilization by Wip1 may be mediated by dephosphorylating other residues on MDM2 or a binding partner of MDM2, such as MDMX (Zhang et al., 2009).
In addition to S395, ATM phosphorylates five other sites near the C-terminal RING domain of MDM2 after DNA damage (Cheng et al., 2009) (Fig. 1). Mutation of these phosphorylation sites, including S395, to alanine or aspartic acid resulted in enhanced p53 degradation or stability, respectively (Cheng et al., 2009). However, the half-life of these mutant MDM2 proteins remained similar to that of wild-type MDM2 (Cheng et al., 2009), reinforcing the idea that ATM phosphorylation of MDM2 does not directly affect its stability. On the other hand, the mutagenesis study showed that ATM phosphorylation prevents dimerization of the MDM2 RING domain induced by DNA damage (Cheng et al., 2009), which will significantly diminish MDM2’s E3 activity toward p53. In addition, phosphorylation of MDM2 by ATM also reduces the affinity of the central acidic domain for p53, suggesting that ATM phosphorylation may also impact the p53–MDM2 interaction by affecting the conformation of the MDM2 protein (Cheng et al., 2011). Taken together, it is suggested that the ATM phosphorylation primarily impact p53 ubiquitination, but not MDM2 ubiquitination.
In addition to phosphorylating MDM2 directly, ATM also regulates MDM2 indirectly by phosphorylating MDMX (Chen et al., 2005). Moreover, upon DNA damage, ATM activates CHK2, which also phosphorylates MDMX (Pereg et al., 2005). Phosphorylation of MDMX by ATM and CHK2 promotes ubiquitination and degradation of MDMX by MDM2 in response to DNA damage (Chen et al., 2005; Pereg et al., 2005), resulting in p53 activation. Notably, creation of an MDMX mutant that cannot be phosphorylated by the DNA damage kinases results in mice that are highly resistant to radiation but also highly tumorigenic (Wang et al., 2009). Combined with previous studies indicating that ATM inhibits MDM2 dimerization and that c-ABL, another downstream target of ATM, can promote MDM2-mediated degradation of MDMX (Waning et al., 2011), these results showcase a critical role of ATM phosphorylation in down-regulating MDM2-MDMX heterodimers to allow for p53 activation.
A related protein, the Rad3-related kinase (ATR), also phosphorylates MDM2 after DNA damage. Phosphorylation of MDM2 by ATR results in a reduction in MDM2-mediated nuclear export of p53 (Shinozaki et al., 2003). ATR has also been shown to phosphorylate the promyelocytic leukemia (PML) tumor-suppressor protein, thereby sequestering MDM2 in the nucleolus (Bernardi et al., 2004). However, RNAi knockdown of ATR does not prevent p53 activation in response to DNA damage (Boehme et al., 2008), suggesting that the ATR-mediated pathway may not be the primary mechanism of MDM2 regulation upon DNA damage. This agrees with the fact that, as discussed earlier, nuclear export of p53 is not necessary for its degradation (Stommel and Wahl, 2004).
AKT and DNA–PK
AKT is somewhat controversial kinase of MDM2 because accumulating evidence suggests that AKT may both promote and inhibit the activity of MDM2. Phosphorylation of MDM2 by AKT has long been known to repress p53 activation. An early study showed that in HER2/neu transformed NIH3T3 cells, AKT phosphorylated MDM2 at S166 and S186, which was thought to facilitate its transport into the nucleus where it can inhibit p53 (Zhou et al., 2001). However, as mentioned earlier, p53 does not need to be taken out of the nucleus for degradation (Stommel and Wahl, 2004). Later on, it was demonstrated that AKT phosphorylation stabilizes MDM2 protein levels by inhibiting its self-ubiquitination (Feng et al., 2004). Furthermore, it was also shown that AKT stabilizes MDMX by phosphorylating S367, which enhances MDM2 stability and, in turn, results in the suppression of p53 activation (Lopez-Pajares et al., 2008). It should be noted that the CHK1 kinase, a downstream target of ATR, also phosphorylates this residue following DNA damage (Jin et al., 2006).
In contrast to the MDM2-stabilizing ability of AKT, there is some evidence to suggest that AKT may also contribute to MDM2 inhibition. For instance, the PI3K inhibitor wortmannin, which would result in the inhibition of AKT, stabilizes MDM2 and prevents DNA damage-induced MDM2 degradation (Stommel and Wahl, 2004). Moreover, Akt1−/− mice exhibit diminished expression of p53-target genes in response to DNA damage (Bozulic et al., 2008), suggesting that AKT may play a role in p53 activation after DNA damage. How can we reconcile the two apparently contradictory results? It is possible that AKT coordinates p53 activation in response to DNA damage by phosphorylating specific substrates other than MDM2. Another possibility, however, is that a role for AKT in the regulation of MDM2 stability may be stimulus-specific. In particular, whereas AKT promotes MDM2 stabilization under normal conditions, the kinase may, directly or indirectly, facilitate MDM2 degradation following DNA damage.
AKT is a downstream target of multiple DNA damage response kinases. In particular, activation of AKT by DNA-PK has been well characterized. Upon DNA damage, AKT is phosphorylated and activated by DNA-PK in the nucleus (Boehme et al., 2008; Bozulic et al., 2008). Once activated, AKT modulates DNA repair and DNA checkpoint response by phosphorylating a number of substrates (Bensimon et al., 2011). Also, Akt1−/− mice and DNA-PKcs−/− mice, in which the catalytic subunit of DNA-PK is deleted, exhibit similar diminished expression of p53-target genes in response to DNA damage (Wang et al., 2000; Bozulic et al., 2008). These results strongly suggest that AKT functions downstream of DNA-PK and has a role in p53 activation following DNA damage (of note, DNA-PK also directly phosphorylates MDM2, which reduces the binding of MDM2 to p53; see Mayo et al., 1997).
It is interesting to note that AKT directly phosphorylates and inhibits GSK-3 in response to various cellular stimuli. As mentioned earlier, GSK-3 is one of the major kinases that phosphorylate the central acidic domain of MDM2, which leads to p53 inhibition by MDM2 (Kulikov et al., 2005; Boehme et al., 2008). In this model, DNA damage induces the activation of AKT through DNA-PK and triggers the inhibition of GSK-3 (Fig. 3). The inhibition of GSK-3 would result in the hypo-phosphorylation of MDM2 in the acidic domain, which in turn leads to p53 stabilization (Boehme et al., 2008). In summary, it appears that in unstressed conditions AKT stabilizes MDM2 by phosphorylating MDM2 at S166 and S188, which suppress its self-ubiquitination. However, following DNA damage, AKT may trigger the hypo-phosphorylation of MDM2 in the central acidic domain, which inhibits MDM2 activity.
Regulation of MDM2 protein stability following DNA damage
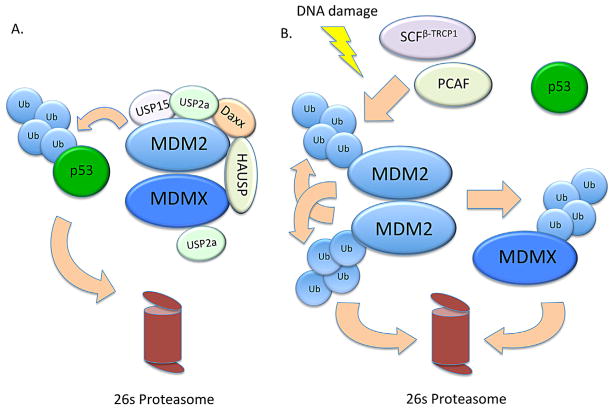
Multiple studies have confirmed that one of the main changes that occurs after DNA damage is a drastic reduction in the protein half-life of MDM2 (Stommel and Wahl, 2004; Itahana et al., 2007; Linares et al., 2007), suggesting that the primary mechanism of MDM2 regulation following DNA damage may be controlled at the level of its protein stability. The stability of MDM2 protein is regulated by the ubiquitin-proteasome system. In brief, ubiquitin is activated by an ubiquitin-activating enzyme (E1), transferred to an ubiquitin E3 ligase through an ubiquitin-conjugating enzyme (E2), and conjugated to a substrate protein by the E3 ligase. Consecutive rounds of ubiquitination (e.g., on K48 of ubiquitin) create an ubiquitin chain covalently bound to a lysine residue on the substrate protein, which will tag the substrate for proteasomal degradation. While MDM2 can serve as an E3 ligase for itself, self-ubiquitination is not the only mode of MDM2 regulation. Several E3 ligases and deubiquitinases have been identified for MDM2 and may play more crucial roles in MDM2 regulation after DNA damage (see Fig. 4).
Fig. 4.
Regulation of MDM2 and p53 before and after DNA damage. (A) In unstressed cells, MDM2 forms a stable heterodimer with MDMX and ubiquitinates p53 for proteasomal degradation. The deubiquitinating enzymes USP2a and HAUSP can bind MDM2 and MDMX and further increase their stability. MDM2’s affinity for HAUSP is enhanced upon binding to the protein Daxx. (B) After DNA damage, MDM2 stability is down-regulated via activation of E3 ligases such as PCAF and β-TrCP. Phosphorylation of MDM2 by the DNA damage kinases ATM and Chk2 cause it to direct its E3 ligase activity toward MDMX and promote its degradation. Degradation of MDMX in turn causes MDM2 to form homodimers which are unstable and prone to self-ubiquitination. Incidentally, upon DNA damage, the deubiquitinase USP2a is transcriptionally repressed, while HAUSP loses its affinity for MDM2, further contributing to MDM2 destabilization.
Ubiquitination of MDM2
It was initially thought that the primary mechanism of MDM2 regulation was through self-ubiquitination (Stommel and Wahl, 2004). After DNA damage, the protein half-life of wild-type MDM2 decreases dramatically, while the half-life of MDM2 with a C462A mutation in its RING domain that abrogates its E3 ligase activity remains unchanged (Stommel and Wahl, 2004). However, recent studies have found evidence suggesting that MDM2 degradation following DNA damage does not solely depend on its own E3 activity (Itahana et al., 2007; Linares et al., 2007; He et al., 2014). Although the results are conflicting, it may be attributed to differences in methodology and cell types. While the earlier study ectopically overexpressed the C464A mutant in a p53 wild-type cancer cell line (U2OS) (Stommel and Wahl, 2004), the later studies analyzed embryonic fibroblasts derived from C462A Mdm2 knock-in mice (Itahana et al., 2007; He et al., 2014). Itahana et al. (2007) used a murine knock-in model to show that in unstressed conditions the C462A mutant has nearly the same protein half-life as wild-type MDM2. They also showed that irradiation accelerates the degradation of both the C462A and the wild-type MDM2, although the C462A mutant degrades at a slightly slower rate. This indicates that while MDM2’s self-ubiquitinating activity does contribute to its degradation after DNA damage, there are other E3 ligases that target MDM2 for ubiquitin-mediated degradation in response to DNA damage.
Several ubiquitin E3 ligases targeting MDM2 for degradation have been discovered. One example is the p300–CBP-associated factor (PCAF). PCAF is a histone acetyltransferase (HAT) and a transcription co-activator for p53, which promotes gene expression induced by p53. In addition to its HAT activity, PCAF has intrinsic ubiquitin E3 ligase activity and promotes ubiquitination of MDM2 for proteasomal degradation (Linares et al., 2007). Knockdown of PCAF by siRNA stabilizes MDM2 in unstressed conditions and prevents p53 induction after DNA damage. On the other hand, ectopic expression of wild-type PCAF promotes MDM2 degradation. Moreover, a dominant negative form of PCAF, which can bind to MDM2 but lacks E3 ligase activity, can prolong the protein half-life of both wild-type MDM2 and the C464A RING mutant MDM2 following DNA damage (Linares et al., 2007). These observations suggest that PCAF plays a critical role not only in the suppression of MDM2 levels under unstressed conditions but also in MDM2 destabilization induced by DNA damage. It should be noted that MDM2 can also ubiquitinate PCAF for degradation, which indirectly diminishes p53’s transcriptional activity regulated by PCAF’s HAT activity (Jin et al., 2004). Thus, MDM2 and PCAF form an interesting trans-ubiquitination network that control p53 activity.
Another ubiquitin E3 ligase shown to target MDM2 for proteasomal degradation is SCFβ-TrCP (Inuzuka et al., 2010). SCF is a multi-protein ubiquitin ligase complex containing the proteins Skp1, Cullin, and Rbx1/Roc1 whose substrate specificity is determined by an associated F-box protein (reviewed in Ang and Harper, 2005). After noticing that depletion of Cullin leads to an increase in MDM2 levels, Inuzuka et al. (2010) demonstrated that β-TrCP is the F-box protein that directs the SCF complex to MDM2. Unlike PCAF, β-TrCP requires MDM2 to be pre-phosphorylated by the serine/ threonine kinase CK1δ before it can mark it for degradation. CK1δ is known to phosphorylate MDM2 at multiple sites within its acidic domain (Fig. 1) as well as near the N-terminal p53 binding domain and between the acidic domain and RING finger domain. It should be noted that the interaction between MDM2 and β-TrCP is mediated by phosphorylation of multiple serine residues rather than just by one key residue (Inuzuka et al., 2010). In particular, successive phosphorylation of the serine residues by CK1δ increases the binding affinity of MDM2 for β-TrCP (Inuzuka et al., 2010). RNAi knockdown of either β-TrCP (either isoform 1 or 2) or CK1δ can attenuate MDM2 degradation after DNA damage (Inuzuka et al., 2010). Although β-TrCP facilitates MDM2 degradation in the G1/S transition of cell cycle, it also promotes MDM2 destruction in response to DNA damage (Inuzuka et al., 2010). After DNA damage, phosphorylation of CK1δ by ATM triggers the translocation of CK1δ to the nucleus where MDM2 is phosphorylated by CK1δ and targeted by β-TrCP for proteasomal degradation (Wang et al., 2012). Interestingly, although DNA damage enhances the interaction between MDM2 and β-TrCP, siRNA knockdown of β-TrCP does not abrogate initial p53 activation but rather attenuates p53 levels at later time points following DNA damage (Inuzuka et al., 2010). Thus, it is suggested that the role of β-TrCP may not be to trigger p53 activation itself, but rather to suppress MDM2 levels during the recovery phase after DNA damage to maintain sustained p53 activation.
As mentioned earlier, the phosphorylation-dependency of β-TrCP-mediated MDM2 ubiquitination is somewhat counterintuitive, given that degradation of MDM2 by β-TrCP requires phosphorylation of the central acidic domain, whereas earlier studies show that this region is often hypo-phosphorylated after DNA damage (Blattner et al., 2002; Winter et al., 2004). In fact, phosphorylation of the acidic domain by GSK-3 has been implicated in stabilization, rather than destabilization, of MDM2. It is especially noteworthy that GSK-3 and CK1δ share at least one of the phosphorylation sites within the central acidic domain and, moreover, that GSK-3 phosphorylation can be enhanced by prior phosphorylation on a priming site by CK1δ (Winter et al., 2004; Kulikov et al., 2005). Whereas GSK-3 primarily phosphorylates two sites within the acidic domain, CK1δ phosphorylates many sites including serine residues outside of the acidic domain (Inuzuka et al., 2010) (Fig. 1). Thus, phosphorylation of such “CK1δ-specific” residues may be crucial in creating this difference and may play a key role in the phosphorylation-dependent ubiquitination of MDM2 by β-TrCP. Alternatively, it may be that DNA damage first induces hypo-phosphorylation in the acidic domain and that CK1δ re-phosphorylates this region afterwards. This would agree with the observation that β-TrCP-mediated MDM2 degradation does not seem to impact the acute p53 activation phase (Inuzuka et al., 2010).
Yet another Cullin family E3 ligase, the anaphase-promoting complex/cyclosome (APC/C), has also been shown to ubiquitinate MDM2 via a scaffold subunit of the APC complex, APC2 (He et al., 2014). Similar to SCF, APC/C is a RING-type ubiquitin ligase complex. However, whereas SCF complexes are active at all stages of the cell cycle, the activity of APC/C is specific to mitosis and the G1 phase (reviewed in Ang and Harper, 2005). Depletion of APC2 results in an accumulation of MDM2, which delays and diminishes p53 activation in response to DNA damage. Incidentally, APC2 mRNA is down-regulated in ovarian, lung, and renal cancers (He et al., 2014), suggesting that increased MDM2 protein levels in these cancer types may be attributed, at least in part, to attenuation of APC2-mediated MDM2 protein turnover.
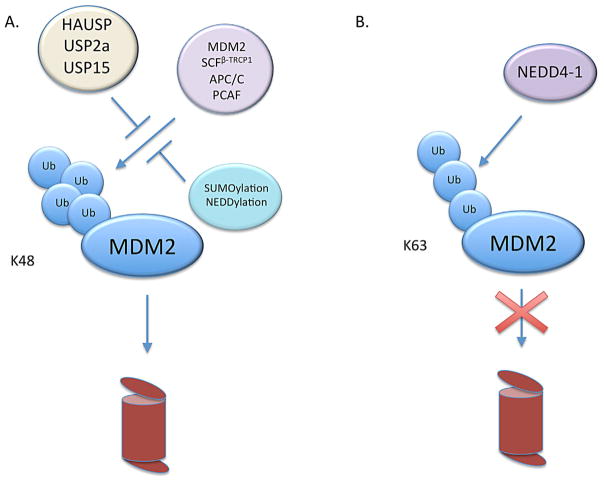
Finally, while other E3 ligases generally destabilize MDM2, NEDD4-1 has been identified as an E3 ligase that rather stabilizes MDM2. NEDD4-1 promotes K63-mediated ubiquitination, instead of K48-mediated ubiquitination, on MDM2 (Xu et al., 2015). Since K63-conjugated polyubiquitination is not linked to proteasomal degradation, it prevents degradation of the target protein by competing with the formation of K48-mediated ubiquitination by other E3 ligases (Xu et al., 2015) (Fig. 5). Therefore, K63 ubiquitination of MDM2 by NEDD4-1 results in the stabilization of MDM2 and, consequently, the suppression of p53 activity (Xu et al., 2015). In the absence of NEDD4-1, MDM2 degrades more rapidly and basal p53 levels are slightly higher than that of control cells (Xu et al., 2015). Moreover, without NEDD4-1, p53 accumulates faster, inducing cell cycle arrest at the G1 phase (Xu et al., 2015). Nevertheless, the presence or absence of NEDD4-1 does not impact DNA damage-induced MDM2 degradation (Xu et al., 2015), suggesting the involvement of another E3 ligase(s) in triggering MDM2 degradation following DNA damage. Unlike the other E3 ligases described above, NEDD4-1 may primarily act to stabilize MDM2 and suppress p53 in the absence of cellular stress.
Fig. 5.
Ubiquitination and ubiquitin-like modifications of MDM2 (A) The E3 ligases SCFβ-TRCP, APC/C, PCAF, and MDM2 itself can attach K48 ubiquitin chains to MDM2, which mark it for proteasomal degradation. Ubiquitination of MDM2 is inhibited by the deubiquitinating enzymes HAUSP, USP2a, and USP15 as well as by the process of SUMOylation and NEDDylation. (B) NEDD4-1 attaches a K63 ubiquitin chain to MDM2, which does not mark it for degradation but does prevent the attachment of K48 ubiquitin chains and enhances MDM2 stability.
SUMOylation of MDM2
Whereas ubiquitination of lysine residues in MDM2 seems to be a major mechanism of MDM2 destabilization, other posttranslational modifications on these lysine residues may also regulate MDM2 stability. One such modification is SUMOylation. Similar to ubiquitination, SUMOylation involves the addition of a small ubiquitin-like modifier (SUMO) protein to a lysine residue on a target protein via series of ligases. The SUMO protein is first activated by an E1 enzyme such as Aos1-Uba2 then conjugated to a E2 enzyme (Ubc9) before being attached to its target protein by a E3 ligase (Buschmann et al., 2001). Once attached, the SUMO protein prevents ubiquitin conjugation in a competitive manner. SUMOylation at K446 in the Ring domain stabilizes MDM2 by preventing its self-ubiquitination (Buschmann et al., 2000). Consequently, this SUMOylation enhances the ability of MDM2 to ubiquitinate p53 without affecting the interaction between MDM2 and p53 (Buschmann et al., 2000). Interestingly, MDM2 SUMOylation is diminished following DNA damage (Buschmann et al., 2000), allowing MDM2 degradation. It is interesting to know whether SUMOylation at K446 also protects MDM2 from other ubiquitin E3 ligases.
Since the identification of MDM2 SUMOylation at K446, multiple SUMOylation sites in MDM2 as well as SUMO E3 ligases were found (Miyauchi et al., 2002). Interestingly, the tumor suppressor protein ARF has been shown to promote MDM2 SUMOylation which is inhibited by MDMX (Xirodimas et al., 2002; Ghosh et al., 2005). ARF binds to MDM2 and inhibits its RING activity (Honda and Yasuda, 1999). It can also sequester MDM2 in the nucleolus, possibly via an interaction within the nucleolar localization sequence within the RING domain (Tao and Levine, 1999; Weber et al., 1999). ARF also promotes MDM2-mediated degradation of MDMX (Pan and Chen, 2003). Given the roles of ARF and MDMX in MDM2 regulation, it is likely that ARF-mediated SUMOylation is inhibitory to the function of MDM2. Thus, SUMOylation of MDM2 appears to have multiple roles.
NEDDylation of MDM2
In addition to SUMO and ubiquitin, lysine residues in MDM2 can be modified by another small ubiquitin-like molecule called NEDD8. Similar to the SUMO protein, NEDD8 also competes with ubiquitin for lysine modification. While ectopic expression of NEDD8 stabilizes MDM2 protein in a dose-dependent manner, deNEDDylation of MDM2 by the NEDD8-specific protease NEDP1 destabilizes MDM2 protein (Watson et al., 2010). Interestingly, NEDP1 levels increase after DNA damage (Watson et al., 2010). However, since NEDP1 induction does not take place until p53 activation (Watson et al., 2010), it is unlikely that NEDP1 is responsible for accelerated MDM2 degradation after DNA damage, which precedes p53 activation. NEDP1 may instead play a role in suppressing MDM2 levels in the later phase of DNA damage response to maintain p53 levels.
Deubiquitination of MDM2
In addition to competitive inhibition from SUMOylation and NEDDylation, ubiquitination of MDM2 can be directly inhibited by deubiquitinating enzymes. One of these proteins is HAUSP (also known as USP7). HAUSP was originally reported as an MDM2 antagonist because overexpression of HAUSP promotes stability of p53 (Li et al., 2002). However, it was later shown that depletion of HAUSP promotes MDM2 destabilization and leads to p53 activation (Cummins and Vogelstein, 2004; Li et al., 2004). Importantly, HAUSP can deubiquitinate MDM2, MDMX, and p53. Upon DNA damage, both MDM2 and MDMX reduces their affinity for HAUSP, leading to enhanced ubiquitination of the two proteins, while p53 remains associated with HAUSP (Meulmeester et al., 2005). Thus, it may be that HAUSP switches its binding partner upon DNA damage. One mechanism through which this may occur is via its interaction with the death domain-associated protein 6 (Daxx). Daxx promotes the binding of MDM2 to HAUSP and thus increases MDM2 stability (Tang et al., 2006). After DNA damage, Daxx is phosphorylated by ATM, which triggers its dissociation from MDM2 and leads to MDM2 degradation as well as p53 activation (Tang et al., 2013).
In addition to HAUSP, MDM2 has also been reported to be deubiquitinated by the protein USP2a (Stevenson et al., 2007). Unlike HAUSP, USP2a does not deubiquitinate p53. Ectopic overexpression of USP2a increases MDM2 levels and p53 ubiquitination, while siRNA knockdown of USP2a leads to an increase in p53 expression and a slight decrease in MDM2 half-life (Stevenson et al., 2007). USP2a has also been shown to act as a deubiquitinating enzyme for MDMX (Allende-Vega et al., 2010), suggesting that in addition to directly stabilizing MDM2, USP2a can also promote MDM2 stabilization by preventing degradation of MDMX. Interestingly, USP2a is down-regulated after treatment with cisplatin at both mRNA and protein levels (Allende-Vega et al., 2010). It remains elusive whether this down-regulation of USP2a could fully account for accelerated MDM2 degradation following DNA damage.
Recently, USP15, identified as a negative regulator of T-cell activation, was shown to be a deubiquitinase for MDM2 (Zou et al., 2014). USP15 is abundantly expressed in melanoma and colorectal cancer cell lines where it stabilizes MDM2 by directly removing ubiquitin chains from MDM2 (Zou et al., 2014). Knocking down USP15 in these cell lines results in MDM2 degradation and p53-dependent apoptosis (Zou et al., 2014). Whether USP15 also modulates MDMX stability, how the activity of USP15 is regulated, and whether DNA damage impacts USP15 activity toward MDM2 remain to be elucidated.
Concluding remarks
Although MDM2 was identified more than 15 years ago, much about it remains unknown. A number of posttranslational modifications and potential binding partners indicate that MDM2 is regulated via several different pathways that have yet to be fully elucidated. Phosphorylation can modulate MDM2 stability by modulating its affinity for E3 ligases or indirectly by affecting the MDM2–MDMX interaction. However, phosphorylation by DNA damage response kinases, particularly ATM and ATR, does not seem to play a direct role in MDM2 stability. Nonetheless, these kinases do have effects on p53 activation.
In recent years, more and more evidence suggests that MDM2 regulation of p53 is tied to its interaction with the related protein MDMX. It is the disruption of the MDM2-MDMX heterodimer, but not the lack of MDM2 E3 ligase activity, that causes embryonic lethality in a p53-dependent manner—the loss of MDM2 ligase activity only attenuates the cellular response to DNA damage (Tollini et al., 2014). Although several E3 ligases that regulate MDM2 degradation were identified, their E3 ligase activity toward MDM2 was analyzed without any consideration of whether MDM2 forms homodimers or MDM2-MDMX heterodimers (or exists as monomers). In light of these recent discoveries, it is time to deliberately investigate the action of the E3 ligases on MDM2-MDMX heterodimers. As MDMX protects MDM2 from self-ubiquitination, does it also protect MDM2 from other E3 ligases? Is monomeric MDM2 more susceptible to degradation than MDM2 in (homo or hetero) dimers? Under unstressed conditions, MDMX is much more stable than MDM2. However, given the similarities between MDM2 and MDMX, can E3 ligases specific for MDM2 also act on MDMX or possibly the MDM2-MDMX heterodimer? Finally, although multiple E3 ligases that regulate MDM2 stability were discovered, the mechanism of accelerated MDM2 degradation following DNA damage still remains elusive. These questions should become interesting topics of research in the coming years. Currently, from the pharmacological perspective, inhibitors of the p53-MDM2 interaction, such as Nutlin-3a, seem to be a promising approach to restoring p53 function in cancer. However, the emergence of primary and acquired resistance to this class of compounds has already been demonstrated (Long et al., 2010). Fully elucidating the molecular mechanism of how MDM2 is regulated, particularly at the level of its protein stability, may allow us to circumvent this resistance and help us identify a novel therapeutic target in cancer treatment.
Acknowledgments
Contract grant sponsor: NCI;
Contract grant number: R00 CA140948.
Contract grant sponsor: American Cancer Society;
Contract grant number: IRG-82-003-30.
Contract grant sponsor: NCI;
Contract grant number: P30 CA23108.
We would like to thank the members of Kurokawa laboratory for valuable discussions. This work was supported by an NCI Career Development Award R00 CA140948 (to M.K.), American Cancer Society Institutional Research Grant #IRG-82-003-30 (to M.K.) and a cancer center core grant P30 CA23108.
Footnotes
Conflicts of interest: None.
Literature Cited
- Allende-Vega N, Dias S, Milne D, Meek D. Phosphorylation of the acidic domain of Mdm2 by protein kinaseCK2. Mol Cell Biochem. 2005;274:85–90. doi: 10.1007/s11010-005-3074-4. [DOI] [PubMed] [Google Scholar]
- Allende-Vega N, Sparks A, Lane DP, Saville MK. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. 2010;29:432–441. doi: 10.1038/onc.2009.330. [DOI] [PubMed] [Google Scholar]
- Ang XL, Harper JW. SCF-mediated protein degradation and cell cycle control. Oncogene. 2005;24:2860–2870. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. Mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A, Aebersold R, Shiloh Y. Beyond ATM: The protein kinase landscape of the DNA damage response. FEBS Lett. 2011 doi: 10.1016/j.febslet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- Blattner C, Hay T, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol. 2002;22:6170–6182. doi: 10.1128/MCB.22.17.6170-6182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme Ka, Kulikov R, Blattner C. P53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc Natl Acad Sci USA. 2008;105:7785–7790. doi: 10.1073/pnas.0703423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBα/Akt1 Acts Downstream of DNA-PK in the DNA Double-Strand Break Response and Promotes Survival. Mol Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- Buschmann T, Fuchs SY, Fuchs SY, Lee CG, Lee CG, Pan ZQ, Pan ZQ, Ronai Z, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- Buschmann T, Lerner D, Lee CG, Ronai Z. The Mdm-2 Amino Terminus Is Required for Mdm2 Binding and SUMO-1 Conjugation by the E2 SUMO-1 Conjugating Enzyme Ubc9. J Biol Chem. 2001;276:40389–40395. doi: 10.1074/jbc.M103786200. [DOI] [PubMed] [Google Scholar]
- Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24:3411–3422. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28:3857–3867. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Cross B, Li B, Chen L, Li Z, Chen J. Regulation of MDM2 E3 Ligase Activity by Phosphorylation after DNA Damage. Mol Cell Biol. 2011 doi: 10.1128/MCB.05553-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004 [PubMed] [Google Scholar]
- Feng J, Tamaskovic R, Yang Z, Brazil DP, Merlo A, Hess D, Hemmings BA. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J Biol Chem. 2004;279:35510–35517. doi: 10.1074/jbc.M404936200. [DOI] [PubMed] [Google Scholar]
- Gannon HS, Woda BA, Jones SN. ATM Phosphorylation of Mdm2 Ser394 Regulates the Amplitude and Duration of the DNA Damage Response in Mice. Cancer Cell. 2012;21:668–679. doi: 10.1016/j.ccr.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Weghorst K, Berberich SJ. MdmX inhibits ARF mediated Mdm2 sumoylation. Cell Cycle. 2005;4:604–608. [PubMed] [Google Scholar]
- Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26:192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, Parant J, Lozano G, Yuan ZM. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem. 2002;277:19251–19254. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- He Y, Tollini L, Kim TH, Itahana Y, Zhang Y. The anaphase-promoting complex/ cyclosome is an E3 ubiquitin ligase for Mdm2. Cell Cycle. 2014;13:2101–2109. doi: 10.4161/cc.29106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, Stommel JM, Gygi S, Lahav G, Asara J, Xiao Z-XJ, Kaelin WG, Harper JW, Wei W. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Jain AK, Barton MC. Making sense of ubiquitin ligases that regulate p53. Cancer Biol Ther. 2010 doi: 10.4161/cbt.10.7.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Dai M-S, Lu SZ, Xu Y, Luo Z, Zhao Y, Lu H. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 2006;25:1207–1218. doi: 10.1038/sj.emboj.7601010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zeng SX, Lee H, Lu H. MDM2 Mediates p300/CREB-binding Protein-associated Factor Ubiquitination and Degradation. J Biol Chem. 2004;279:20035–20043. doi: 10.1074/jbc.M309916200. [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 2007;67:6026–6030. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution Structure of the Hdm2 C2H2C4 RING, a Domain Critical for Ubiquitination of p53. J Mol Biol. 2006;363:433–450. doi: 10.1016/j.jmb.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Kulikov R, Boehme KA, Blattner C. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundanc. Mol Cell Biol. 2005;25:7170–7180. doi: 10.1128/MCB.25.16.7170-7180.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, Latreille D, Cuvier O, Lacroix M, Le Cam L, Coux O, Benkirane M. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol. 2007;9:331–338. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H, Wang S, Malek SN. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acute myelogenous leukemia. Blood. 2010;116:71–80. doi: 10.1182/blood-2010-01-261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V, Kim MM, Yuan ZM. Phosphorylation of MDMX mediated by Akt leads to stabilization and induces 14-3-3 binding. J Biol Chem. 2008;283:13707–13713. doi: 10.1074/jbc.M710030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase Acts as a Gatekeeper in the p53-Mdm2 Autoregulatory Loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Martinez Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, Kastan MB, Katzir E, Oren M. ATM-dependent phosphorylation of Mdm2 on serine 395: Role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG. ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle. 2005;4:1166–1170. doi: 10.4161/cc.4.9.1981. [DOI] [PubMed] [Google Scholar]
- Migliorini D, Danovi D, Colombo E, Carbone R, Pelicci PG, Marine JC. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J Biol Chem. 2002;277:7318–7323. doi: 10.1074/jbc.M108795200. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Xiong S, Iwakuma T, Quintás-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci USA. 2011;108:11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- Priest C, Prives C, Poyurovsky MV. Deconstructing nucleotide binding activity of the Mdm2 RING domain. Nucleic Acids Res. 2010;38:7587–7598. doi: 10.1093/nar/gkq669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MF, Lozano G. The Many Faces of MDM2 Binding Partners. Genes Cancer. 2012;3:226–239. doi: 10.1177/1947601912455322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- Shinozaki T, Nota A, Taya Y, Okamoto K. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene. 2003;22:8870–8880. doi: 10.1038/sj.onc.1207176. [DOI] [PubMed] [Google Scholar]
- Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Agrawal T, Cheng Q, Qu L, Brewer MD, Chen J, Yang X. Phosphorylation of Daxx by ATM Contributes to DNA Damage-Induced p53 Activation. PLoS ONE. 2013:8. doi: 10.1371/journal.pone.0055813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Qu L-K, Zhang J, Wang W, Michaelson JS, Degenhardt YY, El-Deiry WS, Yang X. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleocytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollini La, Jin A, Park J, Zhang Y. Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer Cell. 2014;26:235–247. doi: 10.1016/j.ccr.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, Myklebost O, Heimbrook DC, Vassilev LT. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc Natl Acad Sci USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wang S, Guo M, Ouyang H, Li X, Cordon-Cardo C, Kurimasa A, Chen DJ, Fuks Z, Ling CC, Li GC. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc Natl Acad Sci USA. 2000;97:1584–1588. doi: 10.1073/pnas.97.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased Radioresistance and Accelerated B Cell Lymphomas in Mice with Mdmx Mutations that Prevent Modifications by DNA-Damage-Activated Kinases. Cancer Cell. 2009;16:33–43. doi: 10.1016/j.ccr.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Inuzuka H, Zhong J, Fukushima H, Wan L, Liu P, Wei W. DNA damage-induced activation of ATM promotes beta-TRCP-mediated Mdm2 ubiquitination and destruction. Oncotarget. 2012;3:1026–1035. doi: 10.18632/oncotarget.640. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/22976441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waning DL, Lehman JA, Batuello CN, Mayo LD. C-Abl phosphorylation of Mdm2 facilitates Mdm2-Mdmx complex formation. J Biol Chem. 2011;286:216–222. doi: 10.1074/jbc.M110.183012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, Li BK, Roche O, Blanch A, Ohh M, Irwin MS. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene. 2010;29:297–304. doi: 10.1038/onc.2009.314. [DOI] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Chisholm J, Desterro JMS, Lane DP, Hay RT. P14ARF promotes accumulation of SUMO-1 conjugated (H) Mdm2. FEBS Lett. 2002;528:207–211. doi: 10.1016/s0014-5793(02)03310-0. [DOI] [PubMed] [Google Scholar]
- Xu C, Fan CD, Wang X. Regulation of Mdm2 protein stability and the p53 response by NEDD 4–1 E3 ligase. Oncogene. 2015;34:281–289. doi: 10.1038/onc.2013.557. [DOI] [PubMed] [Google Scholar]
- Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci USA. 2006;103:1227–1232. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lin H, Guo H, Yang J, Jones SN, Jochemsen A, Lu X. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res. 2009;69:7960–7968. doi: 10.1158/0008-5472.CAN-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, Nakaya M, Zhou X, Cheng X, Yang P, Lozano G, Zhu C, Watowich SS, Ullrich SE, Sun S-C. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol. 2014;15:562–570. doi: 10.1038/ni.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]