A comparison of open chromatin landscapes reveals commonalities in transcriptional regulation across species and identifies a transcription factor cascade in the Arabidopsis root hair.
Abstract
The transcriptional regulatory structure of plant genomes remains poorly defined relative to animals. It is unclear how many cis-regulatory elements exist, where these elements lie relative to promoters, and how these features are conserved across plant species. We employed the assay for transposase-accessible chromatin (ATAC-seq) in four plant species (Arabidopsis thaliana, Medicago truncatula, Solanum lycopersicum, and Oryza sativa) to delineate open chromatin regions and transcription factor (TF) binding sites across each genome. Despite 10-fold variation in intergenic space among species, the majority of open chromatin regions lie within 3 kb upstream of a transcription start site in all species. We find a common set of four TFs that appear to regulate conserved gene sets in the root tips of all four species, suggesting that TF-gene networks are generally conserved. Comparative ATAC-seq profiling of Arabidopsis root hair and non-hair cell types revealed extensive similarity as well as many cell-type-specific differences. Analyzing TF binding sites in differentially accessible regions identified a MYB-driven regulatory module unique to the hair cell, which appears to control both cell fate regulators and abiotic stress responses. Our analyses revealed common regulatory principles among species and shed light on the mechanisms producing cell-type-specific transcriptomes during development.
INTRODUCTION
The transcription of protein coding genes is controlled by regulatory DNA elements, including both the core promoter and more distal enhancer elements (Lee and Young, 2000). The core promoter is a short DNA region surrounding the transcription start site (TSS), at which RNA polymerase II and general transcription factors are recruited. Enhancer elements act as platforms for recruiting both positive- and negative-acting transcription factors (TFs) and serve to integrate multiple signaling inputs in order to dictate the spatial and temporal control of transcription from the core promoter. As such, enhancer functions are critical for directing transcriptional output during cell differentiation and development, as well as coordinating transcriptional responses to environmental change (Ong and Corces, 2011). Despite their importance, only a small number of bona fide enhancers have been characterized in plants, and we lack a global view of their general distribution and action in plant genomes (Weber et al., 2016).
In large part, our limited knowledge of plant cis-regulatory elements arises from the unique difficulties in identifying these elements. While some enhancers exist near their target core promoter, others can be thousands of base pairs upstream or downstream, or even within the transcribed region of a gene body (Ong and Corces, 2011; Spitz and Furlong, 2012). Furthermore, enhancers generally do not display universal sequence conservation, aside from sharing of individual TF binding sites, which makes them very challenging to locate. By contrast, core promoters can be readily identified through mapping the 5′ ends of transcripts (Morton et al., 2014; Mejía-Guerra et al., 2015). It was recently discovered that many enhancer elements in animal genomes could be identified with relatively high confidence based on a unique combination of flanking histone posttranslational modifications (PTMs), such as an enrichment for H3K27ac and H3K4me1. This characteristic histone PTM signature has led to the annotation of such elements in several animal models and specialized cell types (Heintzman et al., 2009; Bonn et al., 2012). However, the only currently known association between plant cis-regulatory elements and histone PTMs appears to be a modest correlation with H3K27me3 (Zhang et al., 2012b; Zhu et al., 2015). Though encouraging, this mark is not unique to these elements and cannot be used to identify enhancers on its own.
A long-known and general feature of sequence-specific DNA binding proteins is their ability to displace nucleosomes upon DNA binding, leading to an increase in nuclease accessibility around the binding region (Gross and Garrard, 1988; Henikoff, 2008). In particular, DNaseI treatment of nuclei coupled with high-throughput sequencing (DNase-seq) has been used to probe chromatin accessibility. This technology has served as an important tool in identifying regulatory elements throughout animal genomes (Thurman et al., 2012) and more recently in certain plant genomes (Zhang et al., 2012a, 2012b; Pajoro et al., 2014; Sullivan et al., 2014). In addition, a differential micrococcal nuclease sensitivity assay has also been used to probe functional regions of the maize genome, demonstrating the versatility of this approach (Vera et al., 2014; Rodgers-Melnick et al., 2016).
DNase-seq has been used successfully to identify open chromatin regions in different tissues of both rice (Oryza sativa) and Arabidopsis thaliana (Zhang et al., 2012a; Pajoro et al., 2014; Zhu et al., 2015). Over a dozen of the intergenic DNase-hypersensitive sites in Arabidopsis were tested and shown to act as enhancer elements by activating a minimal promoter-reporter cassette, demonstrating that chromatin accessibility is an important factor in enhancer identification (Zhu et al., 2015). Collectively, these DNase-seq studies show that the majority of open chromatin sites exist outside of genes in rice and Arabidopsis, that differences in open chromatin sites can be identified between tissues, and that a large proportion of intergenic open chromatin sites are in fact regulatory, at least in Arabidopsis. Another recent significant advance came from using DNase-seq to examine the changes in Arabidopsis chromatin accessibility and TF occupancy that occur during development and in response to abiotic stress (Sullivan et al., 2014). This work showed that TF-to-TF regulatory network connectivity appears to be similar between Arabidopsis, human, and Caenorhabditis elegans and that such networks were extensively “rewired” in response to stress. This study also showed that many genetic variants linked to complex traits were preferentially located in accessible chromatin regions, portending the potential for harnessing natural variation in regulatory DNA for plant breeding.
We are still left with many open questions regarding the general conservation of transcriptional regulatory landscapes across plant genomes. For example, it remains unclear how many cis-regulatory elements generally exist in plant genomes, where they reside in relation to their target genes, and to what extent these features are conserved across plant genomes. Furthermore, it is not clear how the cis-regulatory elements within a single genome confer cell-type-specific transcriptional activity—and thus cell type identity—during development. In this study, we seek to build on previous work and to address some of these outstanding questions by analyzing chromatin accessibility across multiple, diverse plant species, and between two distinct cell types.
From a methodological perspective, the DNase-seq procedure is relatively labor-intensive and requires a large number of starting nuclei for DNaseI treatment, which can be a major drawback for conducting cell-type-specific profiling investigations. More recently, the assay for transposase-accessible chromatin with sequencing (ATAC-seq) was developed as an alternative approach (Buenrostro et al., 2013). ATAC-seq employs treatment of isolated nuclei with an engineered transposase that simultaneously cleaves DNA and inserts sequencing adapters, such that cleaved fragments originating from open chromatin can be converted into a high-throughput sequencing library by PCR. Sequencing of the resulting library provides readout highly similar to that of DNase-seq, but ATAC-seq requires far fewer nuclei (Buenrostro et al., 2015). The relatively simple procedure for ATAC-seq and its low nuclei input, combined with its recent application in Arabidopsis and rice (Wilkins et al., 2016; Bajic et al., 2018; Lu et al., 2017), has made it widely useful for assaying plant DNA regulatory regions. In this study, we first optimized ATAC-seq for use with crude nuclei and nuclei isolated by INTACT (isolation of nuclei tagged in specific cell types) affinity purification (Deal and Henikoff, 2010). We then applied this method to INTACT-purified root tip nuclei from Arabidopsis, Medicago truncatula, tomato (Solanum lycopersicum), and rice, as well as the root hair and non-hair epidermal cell types of Arabidopsis. The use of diverse plant species of both dicot and monocot lineages allowed us to assay regulatory structure over a broad range of evolutionary distances. Additionally, analysis of the Arabidopsis root hair and non-hair cell types allowed us to identify distinctions in chromatin accessibility that occurred during the differentiation of developmentally linked cell types from a common progenitor stem cell.
In our cross-species comparisons, we discovered that the majority of open chromatin sites in all four species exist outside of transcribed regions. The open sites also tended to cluster within several kilobases upstream of the transcription start sites despite the large differences in intergenic space between the four genomes. When orthologous genes were compared across species, we found that the number and location of open chromatin regions were highly variable, suggesting that regulatory elements are not statically positioned relative to target genes over evolutionary timescales. However, we found evidence that particular gene sets remain under control by common TFs across these species. For instance, we discovered a set of four TFs that appear to be integral for root tip transcriptional regulation of common gene sets in all species. These include ELONGATED HYPOCOTYL5 (HY5) and MYB DOMAIN PROTEIN77 (MYB77), which were previously shown to impact root development in Arabidopsis (Oyama et al., 1997; Shin et al., 2007).
When comparing the two Arabidopsis root epidermal cell types, we found that their open chromatin profiles are qualitatively very similar. However, many quantitative differences between cell types were identified, and these regions often contained binding motifs for TFs that were more highly expressed in one cell type than the other. Further analysis of several such cell-type-enriched TFs led to the discovery of a hair cell transcriptional regulatory module driven by ABA INSENSITIVE5 (ABI5) and MYB33. These factors appear to coregulate a number of additional hair cell-enriched TFs, including MYB44 and MYB77, which in turn regulate many downstream TF genes as well as other genes impacting hair cell fate, physiology, secondary metabolism, and stress responses.
Overall, our work suggests that the cis-regulatory structure of these four plant genomes is strikingly similar and that TF-target gene modules are also generally conserved across species. Furthermore, early differential expression of high-level TFs between the Arabidopsis hair and non-hair cells appears to drive a TF cascade that at least partially explains distinctions between hair and non-hair cell transcriptomes. Our data also highlight the utility of comparative chromatin profiling approaches and will be widely useful for hypothesis generation and testing.
RESULTS AND DISCUSSION
Application of ATAC-Seq in Arabidopsis Root Tips
The ATAC-seq method was introduced in 2013 and has since been widely adopted in many systems (Buenrostro et al., 2013; Mo et al., 2015; Scharer et al., 2016; Lu et al., 2017). This technique utilizes a hyperactive Tn5 transposase that is preloaded with sequencing adapters as a probe for chromatin accessibility. When purified nuclei are treated with the transposase complex, the enzyme freely enters nuclei and cleaves accessible DNA, both around nucleosomes and at nucleosome-depleted regions arising from the binding of TFs to DNA. Upon cleavage of DNA, the transposon integrates sequencing adapters, fragmenting the DNA sample in the process. Regions of higher accessibility will be cleaved by the transposase more frequently and generate more fragments—and ultimately more reads—once the sample is sequenced. Conversely, less accessible regions will have fewer fragments and reads. After PCR amplification of the raw DNA fragments, paired-end sequencing of the ATAC-seq library can reveal nucleosome-depleted regions where TFs are bound.
In this study, we set out to apply ATAC-seq to multiple plant species as well as different cell types from a single species. As such, we first established procedures for using the method with Arabidopsis, starting with root tip nuclei affinity purified by INTACT. We also established a protocol to use nuclei purified by detergent lysis of organelles followed by sucrose sedimentation, with the goal of broadening the application of ATAC-seq to nontransgenic starting tissue. We began with an Arabidopsis INTACT transgenic line constitutively expressing both the nuclear envelope targeting fusion protein (NTF) and biotin ligase (BirA) transgenes. Coexpression of these transgenes results in all the nuclei in the plant becoming biotinylated and, thus, amenable to purification with streptavidin beads (Deal and Henikoff, 2010; Sullivan et al., 2014). Transgenic INTACT plants were grown on vertically oriented nutrient agar plates to facilitate root growth, and total nuclei were isolated from the 1-cm root tip region. These nuclei were further purified either by treatment with 1% (v/v) Triton X-100 and sedimentation through a sucrose cushion (“Crude” purification) or affinity purified using streptavidin-coated magnetic beads (INTACT purification). In both cases 50,000 nuclei from each purification strategy were used as the input for ATAC-seq (Figure 1A). Overall, both Crude and INTACT-purified nuclei yielded very similar results (Figures 1B and 1C; Supplemental Figure 1). One clear difference that emerged was the number of reads that map to organellar DNA between the nuclei preparation methods. While the total reads of Crude nuclei preparations mapped ∼50% to organellar genomes and 50% to the nuclear genome, the total reads of INTACT-purified nuclei consistently mapped over 90% to the nuclear genome (Table 1). The issue of organellar genomes contaminating ATAC-seq reactions is a common one, resulting in a large percentage of organelle-derived reads that must be discarded before further analysis. This issue was also recently shown to be remedied by increasing the purity of nuclei prior to ATAC-seq by use of fluorescence-activated nuclei sorting (Lu et al., 2017). To compare between data sets for the Crude and INTACT preparation strategies, we analyzed the enrichment of ATAC-seq reads using Hotspot peak mapping software (John et al., 2011). Though designed for use with DNase-seq data, Hotspot can also be readily used with ATAC-seq data. The number of enriched regions found with this algorithm did not differ greatly between nuclei preparation types, nor did the SPOT score (a signal specificity measurement representing the proportion of sequenced reads that fall into enriched regions) (Table 1). These results suggest that the data sets are generally comparable regardless of the nuclei purification method.
Figure 1.
Application of ATAC-Seq to Arabidopsis and Comparison with DNase-Seq Data.
(A) Schematic of the INTACT system and strategy for testing ATAC-seq on nuclei with different levels of purity. Upper panel shows the two transgenes used in the INTACT system: the NTF and biotin ligase. Driving expression of both transgenes using constitutive promoters generates biotinylated nuclei in all cell types. Below is a diagram of a constitutive INTACT transgenic plant, showing the 1-cm root tip section used for all nuclei purifications. Root tip nuclei were isolated from transgenic plants and either purified by detergent lysis of organelles followed by sucrose sedimentation (Crude) or purified using streptavidin beads (INTACT). In each case 50,000 purified nuclei were used as input for ATAC-seq.
(B) Genome browser shot of ATAC-seq data along a 170-kb stretch of chromosome 4 from INTACT-purified and Crude nuclei, as well as DNase-seq data from whole root tissue. Gene models are displayed on the bottom track.
(C) Average plots and heat maps of DNase-seq and ATAC-seq signals at the 23,288 ATAC-seq THSs in the INTACT-ATAC-seq data set. The regions in the heat maps are ranked from highest DNase-seq signal (top) to lowest (bottom).
(D) Venn diagram showing the overlap of enriched regions identified in root tip INTACT-ATAC-seq and whole-root DNase-seq data sets.
(E) Genomic distributions of enriched regions identified in DNase-seq, INTACT-ATAC-seq, and Crude-ATAC-seq data sets.
Table 1. ATAC-Seq Reads from Crude and INTACT-Purified Arabidopsis Root Tip Nuclei.
| Experiment | Plastid Mapped Reads (%) | Mitochondrial Mapped Reads (%) | Nuclear Mapped Reads (%) | Total Nuclear Mapped Reads (× 106) | Total Hotspot Enriched Regions Called | SPOT Score |
|---|---|---|---|---|---|---|
| Crude 1 | 25.33 | 22.15 | 52.52 | 40.6 | 43,599 | 0.4339 |
| Crude 2 | 24.40 | 21.03 | 54.58 | 31.0 | 43,043 | 0.4086 |
| Crude 3 | 25.13 | 23.17 | 51.70 | 35.8 | 42,469 | 0.4471 |
| INTACT 1 | 4.62 | 2.44 | 92.94 | 34.6 | 36,463 | 0.4167 |
| INTACT 2 | 3.51 | 2.03 | 94.46 | 34.0 | 41,305 | 0.4004 |
| INTACT 3 | 2.81 | 1.61 | 95.57 | 89.7 | 55,857 | 0.4896 |
ATAC-seq was performed in biological triplicate for both Crude and INTACT-purified nuclei. For each replicate, the table shows the percentage of reads mapping to organelle and nuclear genomes, the total number of enriched regions identified by the peak calling program Hotspot, as well as the SPOT score for each data set. The SPOT score is a measure of specificity describing the proportion of reads that fall in enriched regions, with higher scores indicating higher specificity.
Visualization of the Crude- and INTACT-ATAC-seq data sets in a genome browser revealed that they were highly similar to one another and to DNase-seq data from whole root tissue (Figure 1B). Further evidence of similarity among these data sets was found by examining the normalized read count signal in all data sets (both ATAC-seq and DNase-seq) within the regions called “enriched” in the INTACT-ATAC-seq data set. For this and all subsequent peak calling in this study, we used the findpeaks algorithm in the HOMER package (Heinz et al., 2010), which we found to be more versatile and user-friendly than Hotspot. Using this approach, we identified 23,288 enriched regions in our INTACT-ATAC-seq data. We refer to these peaks, or enriched regions, in the ATAC-seq data as THSs. We examined the signal at these regions in the whole root DNase-seq data set and both Crude- and INTACT-ATAC-seq data sets using heat maps and average plots. These analyses showed that THSs detected in INTACT-ATAC-seq tended to be enriched in both Crude-ATAC-seq and DNase-seq signal (Figure 1C). In addition, the majority of enriched regions (19,516 of 23,288) were found to overlap between the root tip INTACT-ATAC-seq and the whole-root DNase-seq data (Figure 1D), and the signal intensity over DNase-seq or ATAC-seq enriched regions was highly correlated between the data sets (Supplemental Figure 1).
To examine the distribution of hypersensitive sites among data sets, we identified enriched regions in both types of ATAC-seq data sets and the DNase-seq data set and then mapped these regions to genomic features. We found that the distribution of open chromatin regions relative to gene features was nearly indistinguishable among the data sets (Figure 1E). In all cases, the majority of THSs (∼75%) were outside of transcribed regions, with most falling within 2 kb upstream of a TSS and within 1 kb downstream of a transcript termination site (TTS).
Overall, these results show that ATAC-seq can be performed effectively using either Crude or INTACT-purified nuclei and that the data in either case are highly comparable to that of DNase-seq. While the use of crudely purified nuclei should be widely useful for assaying any tissue of choice without a need for transgenics, it comes with the drawback that ∼50% of the obtained reads will be from organellar DNA. The use of INTACT-purified nuclei greatly increases the cost efficiency of the procedure and can also provide access to specific cell types, but requires preestablished transgenic lines.
Comparison of Root Tip Open Chromatin Profiles among Four Species
Having established an efficient procedure for using ATAC-seq on INTACT affinity-purified nuclei, we used this tool to compare the open chromatin landscapes among four different plant species. In addition to the Arabidopsis INTACT line described above, we also generated constitutive INTACT transgenic plants of M. truncatula, rice, and tomato. Seedlings of each species were grown on vertically oriented nutrient plates for 1 week after radicle emergence, and nuclei from the 1-cm root tip regions of each seedling were isolated and purified with streptavidin beads. ATAC-seq was performed in at least two biological replicates for each species, starting with 50,000 purified nuclei in each case. Visualization of the mapped reads across each genome showed notable consistencies in the data for all four species. In all cases, the reads localize to discrete peaks that are distributed across the genome, as expected (Figure 2A). Examination of a syntenic region found in all four genomes suggested at least some degree of consistency in the patterns of transposase accessibility around orthologous genes (Figure 2A).
Figure 2.
ATAC-Seq Profiling of Arabidopsis, M. truncatula, Tomato, and Rice.
(A) Comparison of ATAC-seq data along syntenic regions across the species. The left panel shows a genome browser shot of ATAC-seq data across a syntenic region of all four genomes. ATAC-seq data tracks are shown above the corresponding gene track for each species. The right panel is an enlargement of the region surrounded by a dotted box in the left panel. Orthologous genes are surrounded by black boxes connected by dotted lines between species. Note the apparent similarity in transposase hypersensitivity upstream and downstream of the rightmost orthologs.
(B) Distribution of ATAC-seq THSs relative to genomic features in each species.
(C) Distribution of upstream THSs relative to genes in each species. THSs are binned by distance upstream of the TSS. The number of peaks in each bin is expressed as a percentage of the total upstream THS number in that species.
(D) Number of upstream THSs per gene in each species. Graph shows the percentage of all genes with a given number of upstream THSs.
To specifically identify regions of each genome that were enriched in ATAC-seq signal (THSs), we used the HOMER findpeaks function on each biological replicate experiment. For further analysis, we retained only THS regions that were found in at least two biological replicates of ATAC-seq in each species. These reproducible THSs were then mapped to genomic features in each species in order to examine their distributions. As seen previously for Arabidopsis, the majority of THSs (∼70–80%) were found outside of transcribed regions in all four species (Figure 2B). For this analysis, we classified these extragenic THSs (THSs found anywhere outside of transcribed regions) as proximal upstream (<2 kb upstream of the TSS), proximal downstream (<1 kb downstream of the TTS), or intergenic (>2 kb upstream from a TSS or >1 kb downstream from a TTS). The proportion of THSs in the proximal upstream and intergenic regions varied greatly with genome size and, thus, the amount of intergenic space in the genome. For example, a full 52% of THSs in Arabidopsis—the organism with the smallest genome (∼120 Mb) and highest gene density of the four species—were in the proximal upstream region. This percentage drops as genome size and intergenic space increase, with 37% of the THSs in the proximal upstream region in the rice genome (∼400 Mb), 30% in the M. truncatula genome (∼480 Mb), and a mere 11% in the tomato genome (∼820 Mb). The percentage of total THSs in the proximal downstream region followed a similar pattern, marking 17% of the THSs in Arabidopsis, 12% in rice and M. truncatula, and 6% in tomato. Finally, the proportion of THSs classified as intergenic followed the inverse trend as expected, with 12% of the THSs in intergenic regions for Arabidopsis, 30% for rice and M. truncatula, and 50% for tomato (Figure 2B). Thus, while the overall proportion of extragenic THSs is similar among species, the distance of these sites from genes tends to increase with genome size, which is roughly proportional to the average distance between genes.
Since the majority of THSs were found upstream of the nearest gene for each species, we next classified the regions based on their distance from the nearest TSS. We binned THSs in each genome into twelve distance categories, starting with those >10 kb upstream of the TSS, then into 11 bins of 999 bp moving in toward the TSS, and finally a TSS-proximal bin of 100 to 0 bp upstream of the TSS (Figure 2C). Starting with this TSS-proximal bin, we find that ∼17% of the upstream THSs in Arabidopsis, M. truncatula, and rice are within 100 bp of the TSS, whereas 2.7% of the upstream THSs in tomato are within 100 bp of the TSS. Moving away from the TSS, we find that 91% of the total upstream THSs fall within 2.9 kb of the TSS in Arabidopsis, while this number decreases with genome size, with 84% for rice, 73% for M. truncatula, and 65% for tomato. In the distance bin spanning 9.9 to 3 kb upstream, we find 7% of the total upstream THSs in Arabidopsis, 15% in rice, 23% in M. truncatula, and 32% in tomato. Finally, the THSs that are more than 10 kb away from the TSS accounts for 0.8% of the total upstream THSs in Arabidopsis, 0.9% in rice, 2.3% in M. truncatula, and 3.3% in tomato. Overall, it is clear that in all species the majority of THSs are within 3 kb upstream of a TSS, suggesting that most cis-regulatory elements in these genomes are likely to be proximal to the core promoter. In the species with the largest genomes and intergenic distances (M. truncatula and tomato), THSs tend to be spread over a somewhat wider range upstream of the TSS. However, even in these cases, only a few hundred THSs in total are more than 10 kb away from the nearest gene. It is worth noting that the distribution of THSs in M. truncatula is more similar to that of tomato than rice, despite the genome size being more similar to rice. This suggests that THSs tend to be further away from TSSs in M. truncatula than would be expected based on genome size alone.
As most THSs fall near genes, we next investigated from the opposite perspective: for any given gene, how many THSs were associated with it? In this regard, we find that the Arabidopsis, M. truncatula, and rice genomes are highly similar (Figure 2D). In all three genomes, of the subset of genes that have any upstream THSs, ∼70% of these genes have a single site, ∼20% have two sites, 5 to 7% have three sites, and 2 to 3% have four or more THSs. By contrast, the tomato genome has a different trend. Of the subset of tomato genes with any upstream THSs, only 27% of the genes have a single site, and this proportion gradually decreases with increasing THS number, with 2.7% of the tomato genes in this subset having 10 or more THSs.
Overall, we have found that THSs have similar size and genomic distribution characteristics across all four species (Supplemental Data Set 1). The majority of THSs in all species are found outside of genes, mainly upstream of the TSS, and these sites tend to cluster within 3 kb of the TSS. Furthermore, most genes with an upstream THS in Arabidopsis, M. truncatula, and rice have only one to two THSs, whereas tomato genes tend to have a larger number of upstream THSs. Whether this increase in upstream THSs in tomato is reflective of an increase in the number of regulatory elements per gene based on clade-specific alterations in gene regulation, DNA copy number changes, or simply the greater abundance of transposons and other repeat elements is not entirely clear. Compared with the other species, tomato THSs are much more abundant and tend to be smaller in size than those of the other species, and the tomato ATAC-seq data generally appear to have a lower signal-to-noise ratio (Figure 2A). While it is unclear why the data from tomato are distinct in these ways, it is clear that tomato THSs occupy mostly genic regions of the genome, as expected, and are highly reproducible between biological replicate experiments (Supplemental Figure 2).
Collectively, these results suggest that there is a relatively small number of regulatory elements per gene in plants. These elements tend to be focused near the promoter rather than at more distal sites, as has been observed in animal, particularly mammalian, genomes (Stadhouders et al., 2012). The assumptions implicit in this argument are that open chromatin sites near a TSS reflect regulatory elements that regulate that TSS and not a more distant one and that upstream elements contribute the majority of regulatory effects. These assumptions appear to be generally validated by many reporter assays showing that an upstream fragment of several kilobases is frequently sufficient to recapitulate native transcription patterns (Medford et al., 1991; Masucci et al., 1996; Ruzicka et al., 2007; Tittarelli et al., 2009; Li et al., 2012), as well as our observation that upstream THSs are the most abundant class of open chromatin sites.
Open Chromatin Features Are Not Directly Conserved among Orthologous Genes
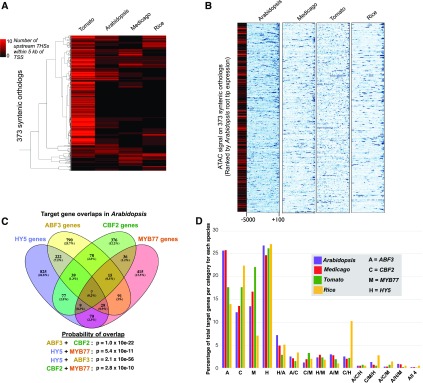
Given that many of the properties of open chromatin regions were shared among Arabidopsis, M. truncatula, rice, and tomato, we next asked whether the numbers and locations of THSs—and thus putative regulatory elements—were conserved among orthologous genes across species. For these analyses, we identified 373 syntenic orthologs (Supplemental Data Set 2) that were found in all four genomes and asked whether members of each ortholog set harbored a similar number of open chromatin regions across the species. Again, using root tip THSs present in at least two biological replicates for each species, we counted the number of THSs within 5 kb upstream of the TSS for each ortholog in each species. We then examined these data for similarities and differences in upstream THS number (Figure 3A). While no clear trend of strong conservation in the number of upstream THSs emerged from this analysis, there was a small subset of orthologs that did have upstream THSs in similar numbers across species. However, this was a very small proportion of the total. As seen in earlier analyses, tomato genes tended to have a larger number of upstream THSs compared with the other species, and most of the 373 orthologs in tomato did have at least one upstream THS. This was not the case in the other three species, where many of the orthologs had no detectable upstream THSs within 5 kb of the TSS. Among the four species, Arabidopsis and M. truncatula showed the greatest similarity in upstream THS number, but even in this case the similarity was minimal despite the relatively closer phylogenetic relationship between these two organisms.
Figure 3.
Characterization of Open Chromatin Regions and Regulatory Elements in Arabidopsis, M. truncatula, Tomato, and Rice.
(A) Heat map showing the number of upstream THSs at each of 373 syntenic orthologs in each species. Each row of the heat map represents a syntenic ortholog, and the number of THSs within 5 kb upstream of the TSS is indicated with a black-to-red color scale for each ortholog in each species. Hierarchical clustering was performed on orthologs using uncentered correlation and average linkage.
(B) Normalized ATAC-seq signals upstream of orthologous genes. Each row of the heat maps represents the upstream region of one of the 373 syntenic orthologs in each species. ATAC-seq signal is shown across each ortholog from +100 to −5000 bp relative to the TSS, where blue is high signal and white is no signal. Heat maps are ordered by transcript level of each Arabidopsis ortholog in the root tip, from highest (top) to lowest (bottom). The leftmost heat map in black-to-red scale indicates the number of upstream THSs from −100 to −5000 bp associated with each of the Arabidopsis orthologs, on the same scale as in (A).
(C) Overlap of predicted target genes for HY5, ABF3, CBF2, and MYB77 in the Arabidopsis root tip. Predicted binding sites for each factor are those THSs that also contain a significant motif occurrence for that factor. Venn diagram shows the numbers of genes with predicted binding sites for each factor alone and in combination with other factors. Significance of target gene set overlap between each TF pair was calculated using a hypergeometric test with a population including all Arabidopsis genes reproducibly associated with an ATAC-seq peak in the root tip (13,714 total genes). For each overlap, we considered all genes cotargeted by the two factors.
(D) Conveying data similar to that in (C), the clustered bar graph shows the percentage of total target genes that fall into a given regulatory category (targeted by a single TF or combination of TFs) in each species.
We next examined the distribution of open chromatin regions across the upstream regions of these 373 orthologous genes relative to their expression level in Arabidopsis, reasoning that there could be patterns of open chromatin similarity based on THS positions, rather than numbers. For this analysis, we examined the normalized ATAC-seq signal across the upstream region of all 373 orthologous genes, from −5000 bp to +100 bp relative to the TSS of each gene (Figure 3B). Orthologs were then ranked within the heat map based on the transcript level of each Arabidopsis ortholog in the root tip (Li et al., 2016a), from highest to lowest expression. For each Arabidopsis ortholog, we also included the upstream THS number to ascertain how this feature might correlate with transcript level for Arabidopsis. While there was some consistency among species in that open chromatin often overlapped with the TSS, we did not observe any clear pattern in transposase hypersensitivity within the upstream regions of these orthologs. K-means clustering of the heat maps similarly did not reveal evidence for conservation of open chromatin patterns among orthologs (Supplemental Figure 3A). An important caveat to this analysis is that many of these syntenic orthologs may not be functional homologs, or “expressologs” (Patel et al., 2012), due to subfunctionalization within gene families. As such, we identified a smaller group (52) of expressologs on which to perform a similar test (Supplemental Data Set 3). While these expressolog genes have both maximally high protein level similarity and expression pattern similarity, including expression in the root, there was also no clear correspondence in upstream THS number among them (Supplemental Figure 3B).
There does not appear to be strong conservation in the number and location of open chromatin sites at orthologous genes across species. Assuming that these genes are still under control of common TFs, this suggests that regulatory elements could be free to migrate, and perhaps split or fuse, while retaining the regulatory parameters of the target gene in question.
One interesting finding from these analyses was that the pattern of upstream THS number does not correlate with expression level, at least for Arabidopsis (Figure 3B). Thus, THSs must not simply represent activating events upstream of the TSS but may also represent binding of repressive factors. Furthermore, we found no correlation between upstream THS number and expression entropy among all genes in the Arabidopsis genome, suggesting a more complex relationship between regulatory element distribution and target gene transcription (Supplemental Figure 3C).
Evidence for Coregulation of Common Gene Sets by Multiple TFs across Species
While there does not appear to be a consistent pattern in the number or placement of open chromatin regions around orthologs or expressologs, we wanted to examine whether it would be possible to find common regulators of specific gene sets among species using a deeper level of analysis. To do this, we first searched for common TF motifs in root tip THSs across the four species. Using the THSs that were found in at least two replicates for each species, we employed the MEME-ChIP motif analysis package (Machanick and Bailey, 2011; Ma et al., 2014) to identify overrepresented motifs of known TFs. We discovered 30 motifs that were both overrepresented and common among all species (Supplemental Data Set 4). We narrowed our list of candidate TFs by considering a variety of factors, including the expression of each TF in the root tip, any known mutant root phenotypes involving those TFs, and whether genome-wide binding information was available for each candidate in Arabidopsis. Ultimately, we selected four TFs for further analysis: HY5, ABSCISIC ACID RESPONSIVE ELEMENTS BINDING FACTOR3 (ABF3), C-REPEAT/DRE BINDING FACTOR2 (CBF2), and MYB77. It is worth noting that among these factors, both HY5 and MYB77 had been previously implicated in root development (Oyama et al., 1997; Zhao et al., 2014). Like HY5 and MYB77, CBF2 and ABF3 have been implicated in stress responses as well as abscisic acid (ABA) signaling (Kang et al., 2002; Knight et al., 2004). Furthermore, overexpression of ABF3 leads to increased tolerance to multiple abiotic stresses in Arabidopsis, rice, cotton (Gossypium hirsutum), and alfalfa (Oh et al., 2005; Abdeen et al., 2010; Wang et al., 2016; Kerr et al., 2017). Given this evidence, we decided to focus on these factors for further study.
We first sought to define the target genes for each of these four TFs in Arabidopsis by combining our chromatin accessibility data with published genome-wide binding data for each factor in Arabidopsis (Table 2). Because an accessible chromatin region (a THS) represents the displacement of nucleosomes by a DNA binding protein, we reasoned that our THS profiles for a given tissue would represent virtually all possible protein binding sites in the epigenomes of root tip cells. Similarly, by using in vitro genomic binding data (DAP-seq) (O’Malley et al., 2016) or ChIP-seq data from a highly heterogeneous tissue, we could identify the spectrum of possible binding sites for that TF, such that the intersection of these data sets would represent the binding sites for that TF in the sample of interest. While there are caveats to this approach, we reasoned that it was more likely to generate false negatives than false positives and would give us a set of high confidence target genes to analyze for each TF. In this regard, ChIP-seq data may be more robust because they represent in vivo binding, while DAP-seq is an in vitro assay and may not capture binding sites that depend on chromatin properties or interactions with other TFs. On the other hand, ChIP-seq data are inherently limited by the cell types present in the sample used.
Table 2. TF Motifs Significantly Enriched in THSs in All Four Species.
| TF | Family | Average Expression in Arabidopsis Root Tip (RPKM) | Percentage of Motif Occurrences in the Arabidopsis Genome That Overlap with a Known Binding Site | Percentage of Motif-Containing THSs in Arabidopsis That Overlap with a Known Binding Site (High-Confidence Binding Sites) |
|---|---|---|---|---|
| AT5G11260 (HY5) | bZIP | 37 | 30.3% (7,156/23,541) | 61.2% (810/1,323) |
| AT4G34000 (ABF3) | bZIP | 1 | 53.3% (3,821/7,164) | 97.1% (1,279/1,316) |
| AT4G25470 (CBF2) | AP2 | 16 | 64.2% (13,144/20,457) | 89.1% (582/653) |
| AT3G50060 (MYB77) | MYB/SANT | 6 | 52.5% (9,147/17,402) | 74.6% (506/678) |
THSs found in at least two replicates for each species were analyzed for overrepresented TF motifs. Four of the 30 TFs that were significantly enriched in THSs of all four species are shown in the table. Significant occurrences of each TF motif were identified across the Arabidopsis genome, and the percentage of these motif occurrences that fall within known binding sites for that factor (based on published ChIP-seq or DAP-seq data sets) are indicated in column 4. The final column indicates the percentage of Arabidopsis root tip THSs that contain a motif for each factor and also overlap with a known binding site for the factor. These are considered high-confidence binding sites (Supplemental Figure 4).
We first tested this approach in Arabidopsis with each of the four TFs of interest. Using THSs from the Arabidopsis root tip that were found in at least two biological replicates, we used the motif identification tool FIMO (Grant et al., 2011) to identify THSs that contained a significant occurrence of the TF motif of interest. The THSs that contained a significant motif match were considered predicted binding sites. We then identified predicted binding sites that also overlapped with a known binding site for that TF (a DAP-seq or ChIP-seq peak), and these were considered high confidence binding sites for that TF in the root tip (Supplemental Figure 4). The predicted binding sites (motif-containing THSs) were themselves very good predictors of the true binding sites for these four TFs (Table 2). For example, of the 1316 Arabidopsis root tip THSs with an occurrence of the ABF3 motif (Mathelier et al., 2014), 1279 (97%) overlapped with an ABF3 ChIP-seq peak from whole 2-d-old seedlings (Song et al., 2016). Similarly, 89% of predicted CBF2 binding sites (Weirauch et al., 2014) overlapped with a CBF2 DAP-seq peak (O’Malley et al., 2016), 74% of predicted MYB77 binding sites (Weirauch et al., 2014) overlapped with a MYB77 DAP-seq peak (O’Malley et al., 2016), and 61% of predicted HY5 binding sites (Mathelier et al., 2014) overlapped with a HY5 DAP-seq peak (O’Malley et al., 2016). In each case, the high confidence binding sites (motif-containing THSs that overlap with a ChIP- or DAP-seq peak) were assigned to their nearest TSS in order to identify the putative target genes for each TF (Supplemental Figure 4).
With these lists of target genes for each TF in the Arabidopsis root tip, we looked for gene sets that were regulated by more than one factor, as a means of identifying coregulatory associations between these four TFs. We found extensive cotargeting among these four TFs, with gene sets being targeted by one, two, three, or all four of these TFs to a degree that was far higher than what would be expected by chance (Figure 3C). For example, of the 1271 ABF3 target genes, 297 (23%) are also targeted by HY5 (hypergeometric P = 2.1 × 10−56). Among these 297 genes, 46 are targeted by ABF3, HY5, and CBF2, and seven are targeted by all four TFs. We also asked where the binding sites driving this pattern were located relative to the target genes. To do this, we considered only binding sites within the 5-kb upstream region of a TSS and repeated the target gene assignment and analysis of target gene overlaps between TFs. This subsetting reduced the total number of target genes for each factor by ∼20% but did not substantially alter the percentages of target gene overlap among the four TFs (Supplemental Figure 5A). These results collectively suggest that these four TFs have important roles in root tip gene regulation both individually and in combination and that the majority of their binding sites (∼80%) fall within the 5-kb region upstream of the TSS for target genes. In addition, we find that the binding sites for multiple TFs often occur in the same THS (Supplemental Figure 5B).
We next sought to examine the target genes and proportions of target gene overlaps between the four species to address the conservation of coregulatory relationships among these four TFs. Given that no TF binding data are available for the other three species and knowing that the majority of our predicted binding sites in Arabidopsis corresponded to known binding sites (Table 2; 61–97%), we opted to also use the predicted binding sites for each of the four TFs in M. truncatula, tomato, and rice, with the knowledge that these sets may contain some false positives. For these analyses, we used the Arabidopsis TF motifs, since these have not been directly defined for the other species, with the caveat that the DNA binding specificity of these factors may not be identical among species.
We again used FIMO to identify significant occurrences of each TF motif within the root tip THSs found in at least two biological replicates for each of our four species. We then mapped the predicted binding sites of each TF to the nearest TSS to define target genes for each TF in each species (Supplemental Data Set 5). We then analyzed the overlap of TFs at target genes in each species using four-way Venn diagrams, similar to Figure 3C. To compare regulatory associations across species, we considered each of the 15 categories in every species-specific four-way Venn diagram as a regulatory category. For example, one regulatory category consists of the genes targeted only by ABF3 alone, another would be those targeted only by HY5 and ABF3 at the exclusion of the other two TFs, and so on. For each regulatory category in each species, we calculated the percentage of the total target genes in that category (number of genes in the regulatory category/total number of genes targeted by any of the four TFs) and then compared these percentages between species (Figure 3D). We found remarkably consistent proportions of the target genes in nearly all regulatory categories across all four species. However, notable deviations from this consistency among species were seen in the proportion of rice genes targeted by MYB77 alone and rice genes targeted by CBF2 and HY5 together. In most cases, the proportions of target genes in different regulatory categories were most similar between Arabidopsis and M. truncatula, and these were generally more similar to tomato than to rice, consistent with the evolutionary distances between the species (Vanneste et al., 2014). Commonly overrepresented Gene Ontology (GO) terms among gene sets in particular regulatory categories across species further support the notion of regulatory conservation (Supplemental Figure 5C), although these analyses are limited by the depth of GO annotation in some of these species.
These findings suggest that while neither syntenic orthologous gene sets nor expressolog gene sets tend to share open chromatin patterns, the genes under control of specific TFs or specific combinations of TFs appear to be relatively stable over evolutionary time, at least for the four TFs we examined. One simple explanation for this phenomenon is that the locations of transcriptional regulatory elements are somewhat malleable over time as long as proper transcriptional control is maintained. In this model, these elements would be free to relocate in either direction, and potentially even merge or split. This would maintain proper control over the target gene, but give each ortholog or expressolog a unique chromatin accessibility profile depending on the exact morphology and distribution of the functionally conserved regulatory elements. This idea of modularity is consistent with previous observations that the Drosophila melanogaster even-skipped stripe 2 enhancer can be rearranged and still retain functionality (Ludwig et al., 2000, 2005).
The results also shed light on the interconnectedness of specific TFs in root tip cells and indicate durability of these coregulatory relationships over time. They also generate readily testable hypotheses regarding how HY5, ABF3, MYB77, and CBF2 operate during root development. For example, given that HY5 appears to regulate over 1000 genes in the Arabidopsis root tip (Figure 3C), and that hundreds of these are annotated with GO terms including biological regulation and response to stimulus, we predict that hy5 mutants would have defects in root tip morphology and growth. Indeed, HY5 was previously shown to be involved in the regulation of lateral root growth initiation and gravitropism (Oyama et al., 1997), and we observe that the primary root tips in hy5 mutants also frequently show a bulging and malformed appearance, as well as severe gravitropism defects (Supplemental Figure 6).
Commonalities and Distinctions in the Open Chromatin Landscapes of Arabidopsis Root Epidermal Cell Types
Having examined questions of regulatory conservation between species, we then explored regulatory element and TF relationships between cell types within a single species. In this case, we chose to focus on the root epidermal hair and non-hair cell types in Arabidopsis. Since these two cell types are derived from a common progenitor, they are prime candidates to offer insight into the epigenomic alterations that occur during—and likely drive—cell differentiation. Specifically, we investigated to what extent the open chromatin landscapes would differ between cell types and whether differences in THSs could pinpoint the sites of differential transcriptional regulation. Furthermore, we wanted to understand whether we could use this information to examine the TF-to-TF regulatory connections that underlie the transcriptomic and physiological differences between these cell types.
We used two previously described INTACT transgenic lines as starting material for these experiments: one having biotin-labeled nuclei exclusively in the root hair (H) cells and another with labeled nuclei only in the root epidermal non-hair (NH) cells (Deal and Henikoff, 2010). Nuclei were purified from each fully differentiated cell type by INTACT, and 50,000 nuclei of each type were subjected to ATAC-seq. Visualization of these cell-type-specific data sets in a genome browser, along with the Arabidopsis whole 1-cm root tip ATAC-seq data, showed a high overall degree of similarity among the three data sets (Figure 4A). Comparison of the ATAC-seq signal intensity at common THS regions genome-wide revealed that these two cell types have open chromatin patterns that are highly similar to one another, but distinct from that of the whole root tip (Supplemental Figure 7).
Figure 4.
Characterization of Open Chromatin Regions in the Arabidopsis Root Hair and Non-Hair Cell Types.
(A) Genome browser shot of ATAC-seq data from root hair cell, non-hair cell, and whole root tip representing 50 kb of chromosome 4.
(B) Overlap of THSs found in two biological replicates of each cell type or tissue. Numbers in bold indicate THSs that are only found in a given cell type or tissue (dTHSs).
(C) Average plots and heat maps showing normalized ATAC-seq signals over 7537 root hair cell dTHSs (left panels) and 2574 non-hair cell-enriched dTHSs (right panels). Heat maps are ranked in decreasing order of total ATAC-seq signal in the hair cell panel in each comparison. Data from one biological replicate is shown here and both replicate experiments showed very similar results.
(D) Venn diagram of overlaps between cell-type-enriched gene sets and genes associated with cell-type-enriched dTHSs. Transcriptome data from hair (purple) and non-hair cells (yellow) are from Li et al. (2016a). Genes were considered cell-type-enriched if they had a 2-fold or higher difference between cell types and a read count of 5 RPKM or greater in the cell type with higher expression.
To identify regions of differential accessibility between the cell types and the whole root tip, we considered THS regions that were found in at least two biological replicates of each cell type or tissue. The total number of these reproducible THSs was 32,942 in the whole root tip, 35,552 for the H cells, and 28,912 for the NH cells. The majority of these sites (18,742) were common (overlapping) in all three sample types (Figure 4B) and thus likely represent regulatory sites that are utilized in multiple Arabidopsis root cell types. We also found 6562 THSs that were common to both root epidermal cell types but were not found in the whole root tip, suggesting that these may represent epidermal-specific regulatory elements. In a search for unique THSs in each of the three sample types (those not overlapping with a THS in any other sample), we found 10,455 THSs that were unique to the whole root tip, 7537 unique to the H cells, and 2574 that were unique to the NH cells. We refer to these regions as differential THSs (dTHSs). The dTHSs identified only in the H or NH cell type were of further interest because they may represent regulatory elements that drive the transcriptomic differences between these two epidermal cell types.
To examine the extent of chromatin accessibility differences at these dTHSs, we visualized the accessibility signals from each cell type at both H cell dTHSs and NH cell dTHSs. First, using the 7537 regions identified as H cell dTHSs, we used heat maps and average plots to examine the normalized ATAC-seq read count across these regions in each cell type (Figure 4C, left panel). We then repeated this analysis using the 2574 NH cell dTHSs (Figure 4C, right panel). In each case, it was clear that the regions we identified as dTHSs showed significant differences in chromatin accessibility between the two cell types. However, the differences in chromatin accessibility between cell types were quantitative (varying intensity) rather than qualitative (all-or-nothing). This indicates that, at large, the dTHSs represent sites that are highly accessible in one cell type and less so in the other, rather than being strictly present in one and absent in the other. Therefore, we refer to these sites from this point on as cell-type-enriched dTHSs to convey the notion of quantitative differences between cell types.
To identify the genes that might be impacted by cell-type-enriched dTHSs, we mapped each dTHS to its nearest TSS and considered that to be the target gene. We found that the 7537 H-enriched dTHSs mapped to 6008 genes, while the 2574 NH-enriched dTHSs mapped to 2295 genes. Thus, the majority of genes that are associated with a dTHS are only associated with one such site. This is consistent with our previous finding that most Arabidopsis genes are associated with a single upstream THS (Figure 2D).
We then asked how the set of genes associated with dTHSs overlapped with those whose transcripts that show differential abundance between the two cell types. Using data from a recent comprehensive RNA-seq analysis of flow sorted Arabidopsis root cell types (Li et al., 2016a), we identified sets of transcripts that were more highly expressed in H versus NH cell types. To be considered a cell-type-enriched gene, we required a gene to have a transcript level with twofold or greater difference in abundance between H and NH cell types, as well as at least five reads per kilobase per million mapped reads (RPKM) in the cell type with a higher transcript level. Using this relatively conservative approach, we derived a list of 3282 H cell-enriched genes and 2731 NH cell-enriched genes. We then asked whether the genes associated with cell-type-enriched dTHSs were also cell-type-enriched genes (Figure 4D). Of the 3282 H cell-enriched genes, 743 were associated with an H cell-enriched dTHS, 258 were associated with a NH cell-enriched dTHS, and 108 genes were associated with a dTHS in both cell types. Among the 2731 NH cell-enriched genes, 156 were associated with a NH cell-enriched dTHS, 516 were associated with a H cell-enriched dTHS, and 52 genes showed dTHSs in both cell types. These results suggest that cell-type-enriched expression of a gene is frequently associated with a dTHS in the cell type where the gene is highly expressed but is also often associated with a dTHS in the cell type where that gene is repressed. This highlights the importance of transcriptional activating events in the former case and repressive events in the latter. Interestingly, for a smaller set of cell-type-enriched genes, we observed dTHSs at a given gene in both cell types, indicating regulatory activity at the gene in both cell types.
We next asked what proportion of the transcriptome differences between H and NH cells might be explained based on differential chromatin accessibility. Of the 3282 H cell-enriched genes, 1109 have a dTHS in one or both of the cell types, and among the 2731 NH cell-specific genes, 724 have a dTHS in one or both cell types. Assuming that each dTHS represents a regulatory event contributing to the differential expression of its identified target gene, we could explain differential expression of 33% of the H cell-enriched genes and 27% of the NH cell-enriched genes. The remaining ∼70% of the identified cell-type-enriched genes without clear chromatin accessibility differences may be explained in numerous ways. These genes may not require a change in chromatin accessibility, changes in chromatin accessibility may fall below our limit of detection, or these transcripts may be primarily regulated at the posttranscriptional level rather than at the chromatin accessibility level that we measured.
Another key question relates to the significance of the cell-type-enriched dTHSs that do not map to differentially expressed genes. These could be explained by an inability to detect all differentially expressed genes, perhaps simply due to the stringency of our definition of cell-type-enriched genes. An important biological possibility to consider is that many of these regulatory regions do not in fact regulate the closest gene, but rather act over a distance such that they are orphaned from their true target genes in our analysis. Another possibility is that many of the differential protein binding events represented by these dTHSs are unrelated to transcriptional regulation.
Overall, the accessible chromatin landscapes of the root epidermal H and NH cells appear to be nearly identical in a qualitative sense, but differ significantly at several thousand sites in each cell type. The reasons for the quantitative, rather than all-or-nothing, nature of this phenomenon are not entirely clear. Are the accessibility differences between cell types reflective of unique protein assemblages at the same element in different cell types, or do they instead reflect differences in abundance of the same proteins at an element in different cell types? While these questions certainly warrant further investigation and experimentation, we can gain further insight into the regulatory differences between cell types through deeper examination of the differentially accessible chromatin regions in each.
TF Motifs in Cell-Type-Specific THSs Identify Regulators and Their Target Genes
As a means of identifying specific TFs that might be important in specifying the H and NH cell fates, we sought to identify overrepresented motifs in the differentially accessible regions of each cell type. We used each set of cell-type-enriched dTHSs as input for MEME-ChIP analyses (Machanick and Bailey, 2011) and examined the resulting lists of overrepresented motifs. We initially found 219 motifs that were significantly overrepresented relative to genomic background only in H-cell-enriched dTHSs and 12 that were significantly overrepresented only in NH-cell-enriched dTHSs (Supplemental Data Set 6). To narrow our list of candidate TFs to pursue, we vetted these lists of potential cell-type-enriched TFs by considering their transcript levels in each cell type as well as the availability of genome-wide binding data. Based on the available data, we narrowed our search to five transcription factors of interest: four H-cell-enriched TF genes (MYB33, ABI5, NAC083, and At5g04390) and one NH-enriched TF gene (WRKY27) (Table 3).
Table 3. TF Motifs Overrepresented in Cell Type-Enriched dTHSs.
| TF | Family | Cell Specificity | Hair/Non-Hair FPKM Ratio | Percentage of Genomic Motif Occurrences in Known Binding Sites | Percentage of Motif-Containing THSs That Overlap a Known Binding Site (High-Confidence Binding Sites) |
|---|---|---|---|---|---|
| AT5g06100 (MYB33) | MYB | Hair | 2000 | 42.2% (7,038/16,655) | 69.9% (1,473/2,106) |
| AT2g36270 (ABI5) | bZIP | Hair | 3.3 | 26.2% (7,261/27,656) | 57.5% (2,814/4,891) |
| At5g04390 | C2H2 | Hair | 17 | 9.5% (3,850/40,305) | 8.5% (282/3,290) |
| At5g13180 (NAC083) | NAC | Hair | 2 | 51.3% (13,762/26,815) | 48.3% (1,169/2,419) |
| ATgG52830 (WRKY27) | WRKY | Non-hair | 0.35 | 15.3% (4,458/29,126) | 23.6% (1,169/2,419) |
Cell-type-enriched dTHSs were analyzed for overrepresented TF motifs using MEME-ChIP software, and several significantly matching factors are shown in the table. Cell specificity indicates the cell-type-enriched dTHS set from which each factor was exclusively enriched, and hair/non-hair FPKM ratio indicates expression specificity of each factor using RNA-seq data from Li et al. (2016a). Significant occurrences of each TF motif were identified across the Arabidopsis genome, and the percentage of these motif occurrences that fall within known binding sites for that factor (based on published ChIP-seq or DAP-seq data sets) are indicated in column 5. Percentages are calculated by the number of motif occurrences in known binding sites/total number of motif occurrences in the genome. Column 6 indicates the percentage of THSs from the relevant cell type that contain a motif for a factor and also overlap with a known binding site for the factor (high-confidence binding sites).
We next attempted to directly identify the binding sites for each TF by differential ATAC-seq footprinting between the cell types. The logic behind this approach is the same as that for DNase-seq footprinting: that the regions around a TF binding site are hypersensitive to the nuclease or transposase due to nucleosome displacement, but the sites of physical contact between the TF and DNA will be protected from transposon insertion/cutting and thus leave behind a characteristic “footprint” of reduced accessibility on a background of high accessibility (Hesselberth et al., 2009; Vierstra and Stamatoyannopoulos, 2016). We reasoned that we could identify binding sites for each of these cell-type-enriched TFs by comparing the footprint signal at each predicted binding site (a motif occurrence within a THS) between H and NH cells.
For this analysis, we examined the transposase integration patterns around the motifs of each TF in both cell types as well as in purified genomic DNA subjected to ATAC-seq, to control for transposase sequence bias. It was recently reported in Arabidopsis that many TF motifs exhibit conspicuous transposase integration bias on naked DNA (Lu et al., 2017), and our results were in line with these findings for all five TFs of interest here (Supplemental Figure 8). While we observed footprint-like patterns in the motif-containing THSs in our ATAC-seq data, these patterns in each case were also evident on purified genomic DNA. As such, it was not possible to distinguish true binding sites from these data, as any footprint signal arising from TF binding was already obscured by the transposase integration bias. For unknown reasons, many TF motif DNA sequences seem to inherently evoke hyper- and/or hypo-integration by the transposase, and this automatically obscures any potentially informative footprint signal that could be obtained by integration during ATAC-seq on nuclei. Similar technical concerns have also been raised for DNaseI footprinting (Sung et al., 2016). These results suggest that the ATAC-seq footprinting approach may be useful for certain TFs, but these will likely need to be examined on a case-by-case basis. Given this issue and the resulting lack of evidence for footprints of our TFs of interest, we decided to take the approach of defining TF target sites as we did for our studies of root tip TFs.
As described earlier, we defined high confidence binding sites for the five TFs of interest as TF motif-containing THSs in the cell type of interest (predicted binding sites) that also overlapped with an enriched region for the TF in publicly available DAP-seq data (O’Malley et al., 2016) or ChIP-seq data (Supplemental Figure 4). Assigning these high confidence binding sites to their nearest TSS allowed us to define thousands of target genes for these factors in the root epidermal cell types (Table 3; Supplemental Data Set 7). Compared with our analysis of root tip TFs, our capability to predict target sites based on motif occurrences in THSs was much reduced for the four H-cell-enriched and one NH-cell-enriched TFs examined here. For further analyses, we decided to focus on three of the TFs that were more highly expressed in the H cell type and had the largest number of high confidence target genes: ABI5, MYB33, and NAC083.
We first asked how many of the high confidence target genes for these TFs were also preferentially expressed in one cell type or the other. We found that for all three TFs, a large percentage of the total target genes are H cell enriched in their expression (17–21%), while many others are NH cell enriched (6–9%) (Figure 5A). These results are intriguing as they suggest that the activities of these TFs may be generally context dependent. At the same time, however, the majority of the target genes for each TF were not more highly expressed in one cell type compared with the other.
Figure 5.
Targeting of Cell-Type-Enriched Genes by H-Cell-Enriched TFs and Coregulatory Associations among H-Cell-Enriched TFs.
Genome-wide high confidence binding sites for each TF were defined as open chromatin regions in the hair cell that contain a significant motif occurrence for the factor and also overlap with a known enriched region for that factor from DAP-seq or ChIP-seq data. Target genes were defined by assigning each high confidence binding site to the nearest TSS.
(A) Venn diagrams showing high confidence target genes for ABI5, MYB33, and NAC083 and their overlap with cell-type-enriched genes.
(B) Overlap of ABI5, MYB33, and NAC083 high confidence target genes.
(C) GO analysis was performed to illuminate biological functions of genes cotargeted by ABI5 and MYB33. The upper panel shows significantly enriched GO terms for all 288 genes targeted by both ABI5 and MYB33. For each enriched annotation term, the number of genes in the set with that term is shown, followed by the FDR-corrected P value. The lower panel lists significantly enriched GO-terms for the 57 hair-cell-enriched genes cotargeted by ABI5 and MYB33. The seven hair-cell-enriched genes associated with the term regulation of transcription were chosen for further analysis. All annotation terms in the lists are at the biological process level except for the KEGG pathway term “plant hormone signal transduction.”
Each of these H-cell-enriched TFs could activate other H-cell-enriched genes, but what are their functions at regulatory elements near genes that are expressed at low levels in the H cell and high levels in the NH cell? One possibility is that these factors are activators of transcription in the context of H-cell-enriched genes but act as repressors or are neutral toward the target genes that are NH-cell-enriched in their expression. This may reflect context dependency in the sense that the effect on transcription of a target gene may depend on the local milieu of other factors.
We next examined whether ABI5, MYB33, and NAC083 target any of the same genes. Similar to the root tip TFs examined previously, we found that these three TFs also appear to have extensive coregulatory relationships (Figure 5B). For example, 207 target genes were shared between ABI5 and NAC083, 238 were shared between ABI5 and MYB33, and 50 target genes were shared by all three factors. We further analyzed the genes that were cotargeted by ABI5 and MYB33, finding that 57 of the cotargeted genes were H cell enriched. As such, we performed GO analysis on the H-cell-enriched targets as well as the full set of target genes to gain insight into the functions of this coregulatory relationship (Figure 5C). Many of the ABI5/MYB33 target genes were annotated as being involved in responses to ABA as well as water, salt, and cold stress. This is consistent with the known roles of these proteins in ABA signaling (Finkelstein and Lynch, 2000; Reyes and Chua, 2007). Interestingly, seven of the 57 ABI5/MYB33 target genes that were H cell-enriched were also annotated with the term regulation of transcription, suggesting that ABI5 and MYB33 may be at the apex of a transcriptional regulatory cascade in the H cell type.
Identification of a New Regulatory Module in the Root Hair Cell Type
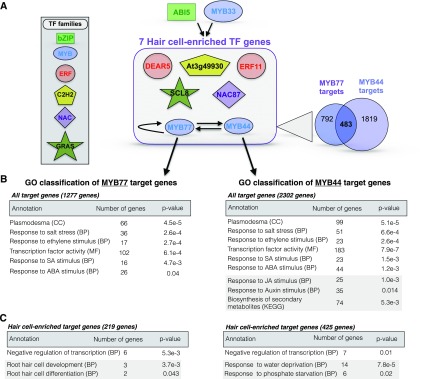
Based on our findings that ABI5 and MYB33 cotarget seven H-cell-enriched TFs, we decided to investigate this potential pathway further. Among the seven TFs putatively coregulated by ABI5 and MYB33 and having H-cell-enriched transcript expression were DEAR5, ERF11, At3g49930, SCL8, NAC087, and two additional MYB factors: MYB44 and MYB77. Aside from MYB77, none of these TFs had been previously reported to produce root-specific phenotypes when mutated. MYB77 was previously shown to interact with auxin response factors (Shin et al., 2007) and to be involved in lateral root development through promotion of auxin-responsive gene expression (Shin et al., 2007). Interestingly, the ABA receptor, PYL8, was shown to physically interact with both MYB77 and MYB44 and to promote auxin-responsive transcription by MYB77 (Zhao et al., 2014). MYB44 has also been implicated in ABA signaling through direct interaction with an additional ABA receptor, PYL9 (Li et al., 2014), as well as repression of jasmonic acid (JA)-responsive transcription (Jung et al., 2010). These factors have additionally been implicated in salicylic acid (SA) and ethylene signaling (Yanhui et al., 2006; Shim et al., 2013). Given that MYB44 and MYB77 are paralogs (Dubos et al., 2010) that appear to integrate multiple hormone response pathways in a partly redundant manner (Jaradat et al., 2013), we decided to identify high confidence target genes (Supplemental Figure 4) for each of them for further study.
We again defined high confidence binding sites as THSs in H cells that contain a significant motif occurrence for the factor and also overlap with a DAP-seq or ChIP-seq enriched region for that factor. Using this approach, we found that MYB44 and MYB77 each target over 1000 genes individually and cotarget 483 genes (Figure 6A). In addition, MYB44 and MYB77 appear to regulate one another, while MYB77 also appears to target itself. This feature of self-reinforcing coregulation could serve as an amplifying and sustaining mechanism to maintain the activity of this module once activated by ABI5, MYB33, and potentially other upstream factors.
Figure 6.
A Transcriptional Regulatory Module in the Root Hair Cell Type.
(A) Diagram of the proposed regulatory module under control of ABI5 and MYB33. As referenced in Figure 5C, ABI5 and MYB33 cotarget seven TFs that are preferentially expressed in the hair cell relative to the non-hair cell type. The family classification of each of the seven TFs is denoted in the figure key. Among the seven hair-cell-specific target TFs are two MYB family members, MYB77 and MYB44. High-confidence binding sites for these two MYB factors were again defined as open chromatin regions in the hair cell that contain a significant motif occurrence for the factor and also overlap with a known enriched region for that factor from DAP-seq or ChIP-seq data. Each high-confidence binding site was then assigned to the nearest TSS to define the target gene for that site. This analysis revealed that MYB44 and MYB77 target each other, and MYB77 targets itself. Both factors target thousands of additional genes, 483 of which are in common (Venn diagram on the lower right of the schematic). Arrows pointing down from MYB77 and MYB44 indicate GO analyses of that factor’s target genes.
(B) and (C) The upper tables (B) represent enriched annotation terms for all target genes of the factor, regardless of differential expression between H and NH cells, while the lower tables (C) represent enrichment of terms within target genes that are preferentially expressed in the hair cell relative to the non-hair cell. Annotation term levels are indicated as cellular component (CC), biological process (BP), molecular function (MF), or KEGG pathway (KEGG). For each annotation, the number of target genes associated with that term is shown to the right of the term, followed by the FDR-corrected P value for the term enrichment in the rightmost column. Groups of terms boxed in gray are those that differ between MYB44 and MYB77. The structure of the module suggests that ABI5 and MYB33 drive a cascade of TFs including MYB77 and MYB44, which act to amplify this signal and also further regulate many additional TFs. Additional target genes of MYB77 and MYB44 include hair cell differentiation factors, hormone response genes, secondary metabolic genes, and genes encoding components of important cellular structures such as plasmodesmata.
To gain a deeper understanding of the impact of MYB44 and MYB77 on downstream processes, we performed GO analysis of the target genes for each factor. First considering all target genes, regardless of their expression in the H cell type, we found a variety of overrepresented GO terms for each that were consistent with the known roles of these factors in hormone signaling (Figure 6B). For example, both factors targeted a large number of genes annotated with the terms response to ABA stimulus, response to ethylene stimulus, and response to SA stimulus. Additionally, MYB44 alone targeted many genes with the annotation response to JA stimulus, consistent with its previously reported role as a negative regulator of JA signaling (Jung et al., 2010). Interestingly, the largest overrepresented gene functional category for both factors was transcription factor activity (102 genes for MYB77 and 183 genes for MYB44). This indeed further suggests that these factors initiate a cascade of transcriptional effects. The next-largest overrepresented term was plasmodesma, indicating that production and/or regulation of cell-cell connecting structures are likely controlled by these factors. Plasmodesmata are important for numerous epidermal functions including cell-to-cell movement of TFs such as CPC and TRY (Schellmann et al., 2002; Wada et al., 2002) and transport of other macromolecules and metabolites (Lucas and Lee, 2004).
We also analyzed overrepresented ontology terms in the MYB77 and MYB44 targets that were classified as H-cell-enriched genes. Among the MYB77 target genes in this category were known regulators of H cell fate, while numerous H-cell-enriched MYB44 target genes were annotated as being involved in response to water and phosphate starvation (Figure 6C). The ontology category that was overrepresented in both target lists was negative regulation of transcription (six MYB77 targets and seven MYB44 targets), suggesting that these factors exert additional specific effects on the H cell transcriptome by regulating a subset of potentially repressive TFs.
The fact that MYB77 and MYB44 target a large number of genes that show H-cell-enriched expression suggests that these factors serve as activators of transcription at these targets, and this is supported by published accounts of transcriptional control by these factors (Persak and Pitzschke, 2014). However, both factors also target NH-cell-enriched genes as well as genes without preferential expression between the cell types. This phenomenon was also observed for the H-enriched TFs ABI5, MYB33, and NAC083 (Figure 5), suggesting that certain TFs may generally serve as activators but may also have context-dependent repressive functions. Such a functional switch could occur through direct mechanisms such as structural alteration by alternative splicing or posttranslational modification, functional alteration by partnering with a specific TF or chromatin-modifying complex, or perhaps indirectly by binding to a target site to occlude the binding of other factors necessary for transcriptional activation. The numerous reports of dual function transcription factors in animals and plants support the notion that this may be a general phenomenon (Ikeda et al., 2009; Boyle and Després, 2010; Li et al., 2016b).
Collectively these results suggest that the MYB44/MYB77 module in the H cell specifies a cascade of downstream transcriptional regulation, some of which is positive and some of which is negative. This module likely represents an important hub in controlling H cell fate as well as a variety of physiological functions and environmental responses in this cell type. The fact that MYB77 was also discovered in our analyses of root tip TFs suggests that this factor likely has a broader role in other cell types during early root development, in addition to a role in specification of the H cell versus the NH cell fate. An important next step will be to perform genetic manipulations of these factors (knockout and inducible overexpression, for example), in order to test and elaborate on the specific predictions made by our model.
Summary and Conclusions
In this study, we used ATAC-seq profiling of accessible chromatin to investigate questions regarding the transcriptional regulatory landscape of plant genomes and its conservation across species. We also investigated the similarities and differences in open chromatin landscapes in two root cell types that arise from a common progenitor, allowing us to identify and analyze TFs that act specifically in one cell type versus the other. Overall, we are able to gain several new insights from this work.
In optimization of our ATAC-seq procedures, we found that the assay can be performed effectively on crudely purified nuclei but that this approach is limited by the large proportion of reads arising from organelle genomes (Table 1). This issue is ameliorated by the use of the INTACT system to affinity-purify nuclei for ATAC-seq, which also provides access to individual cell types. Consistent with previous reports, we found that the data derived from ATAC-seq are highly similar to those from DNase-seq (Figure 1). In comparing our root tip ATAC-seq data to DNase-seq data from whole roots, we found that some hypersensitive regions were detected in one assay but not the other. This discrepancy is most likely attributable to differences in starting tissue and laboratory conditions, rather than biological differences in the chromatin regions sensitive to DNaseI versus the hyperactive Tn5 transposase. This interpretation would fit with the large number of differences also observed in THS overlap between Arabidopsis root tip and epidermal cell types.
In a comparison of open chromatin among the root tip epigenomes of Arabidopsis, M. truncatula, tomato, and rice, we found the genomic distribution of THSs in each were highly similar. About 75% of THSs lie outside of transcribed regions, and the majority of these THSs are found within 3 kb upstream of the TSS in all species (Figure 2). Thus, the distance of upstream THSs from the TSS is relatively consistent among species and is not directly proportional to genome size or intergenic space for these representative plant species. Among genes with an upstream THS, 70% of these genes in Arabidopsis, M. truncatula, and rice have a single such feature, 20% have two upstream THSs, and <10% have three or more. By contrast, only 27% of tomato genes with an upstream THS have a single THS, 20% have two, and the proportion with 4 to 10 THSs is 2 to 7 times higher than that for any other species examined. This increase in THS number in tomato could be reflective of an increase in the number of regulatory elements per gene, but is perhaps more likely a result of the greater number of long-terminal repeat retrotransposons near genes in this species (Xu and Du, 2014). In either case, our investigation revealed that open chromatin sites—and by extension transcriptional regulatory elements—in all four species are focused in the TSS-proximal upstream regions and are relatively few in number per gene. This suggests that transcriptional regulatory elements in plants are generally fewer in number and are closer to the genes they regulate than those of animal genomes. For example, the median distance from an enhancer to its target TSSs in Drosophila was found to be 10 kb, and it was estimated that each gene had an average of four enhancers (Kvon et al., 2014). It was also recently reported that in human T cells, the median distance between enhancers and promoters was 130 kb, far greater than the distances we have observed here across plant species (Mumbach et al., 2017).
Analysis of overrepresented TF motifs in THSs across species suggested that many of the same TFs are at play in early root development in all species. Perhaps more surprisingly, coregulation of specific gene sets by multiple TFs seems to be frequently maintained across species (Figure 3). Taken together with the lack of shared open chromatin profiles among orthologous genes and expressologs, these findings suggest that transcriptional regulatory elements may relocate over evolutionary time within a window of several kilobases upstream of the TSS, but regulatory control by specific TFs is relatively stable.
Our comparison of the two Arabidopsis root epidermal cell types, the H and NH cells, revealed that open chromatin profiles were highly similar between cell types. By examining THSs that were exclusive to one cell type, we were able to find several thousand THSs that were quantitatively more accessible in each cell type compared with the other (Figure 4). Mapping of these dTHSs to their nearest genes revealed that in each cell type there were many dTHSs that were near genes expressed more abundantly in that cell type, as well as many near genes with the opposite expression pattern. This suggests that some dTHSs represented transcriptional activating events whereas others were repressive in nature.
Analysis of TF motifs at these dTHSs between cell types identified a suite of TFs that were more highly expressed in H cells and whose motifs were significantly overrepresented in H-cell-enriched dTHSs. Analysis of three of these TFs—ABI5, MYB33, and NAC083—revealed that each factor targets a large number of H-cell-enriched genes as well as a smaller number of NH-cell-enriched genes (Figure 5). These factors also have many overlapping target genes among them, and ABI5 and MYB33 both target seven additional H-cell-enriched TFs. Among these seven H-enriched TFs are two additional MYB factors: MYB77 and MYB44 (Figure 6). Examination of the high confidence target genes of MYB77 and MYB44 revealed that these paralogous factors appeared to regulate each other as well as many other common target genes, including large numbers of other TF genes. Hundreds of the MYB77 and MYB44 target genes were also more highly expressed in the H cell relative to the NH cell, suggesting that these factors set off a broad transcriptional cascade in the H cell type. In addition, they appear to directly regulate many H-cell-enriched genes involved in cell fate specification and water and phosphate acquisition. This type of cooperative action by pairs of MYB paralogs has also been documented recently in Arabidopsis and other species (Millar and Gubler, 2005; Matus et al., 2017; Wang et al., 2017), and the fact that many target genes for each MYB factor are not regulated by the other may reflect a degree of subfunctionalization between the paralogs.
An important question arising from our results is whether classifying a TF as strictly an activator or repressor is generally accurate in most cases. For example, the H-cell-enriched TFs that we examined all have apparent target genes that are highly expressed in the H cell type as well as targets that are expressed at very low levels, if at all, in the H cell type. In fact, these latter genes are often much more highly expressed in the NH cell type. Given that a number of these TFs have been shown to activate transcription in specific cases, this suggests that they promote the transcription of H-cell-enriched targets and either repress or have no effect on NH-cell-enriched target genes. One explanation for this phenomenon is that these TFs have “dual functionality” as activators and repressors, depending on the context (Bauer et al., 2010). However, it is equally possible that these factors do not play a direct role in gene repression. For example, the binding of an activator near a repressed gene may be functionally irrelevant to the regulation of that gene, or it may be that other gene-specific repressors are also bound nearby and override the activity of the activator. This phenomenon will be worth exploring as it may deepen our understanding of the intricacies of transcriptional control.
In this study, we outline a widely applicable approach for combining chromatin accessibility profiling with available genome-wide binding data to construct models of TF regulatory networks. The putative TF regulatory pathways we have illuminated through our comparison across species and cell types provide important hypotheses regarding the evolution of gene regulatory mechanisms in plants and the mechanisms of cell fate specification that are now open to experimental analysis.
METHODS
Plant Materials and Growth Conditions
Plants used in this study were of the Arabidopsis thaliana Col-0 ecotype, the A17 ecotype of Medicago truncatula, the M82 LA3475 cultivar of tomato (Solanum lycopersicum), and the Nipponbare cultivar of rice (Oryza sativa). Transgenic plants of each species for INTACT were produced by transformation with a binary vector carrying both a constitutively expressed biotin ligase and constitutively expressed NTF protein containing a nuclear outer membrane association domain (Ron et al., 2014). The binary vector used for M. truncatula was identical to the tomato vector (Ron et al., 2014) but was constructed in a pB7WG vector containing the phosphinothricin resistance gene for plant selection and it retains the original AtACT2p promoter. The binary vector used for rice is described elsewhere (Reynoso et al., 2017). Transformation of rice was performed at UC Riverside and tomato transformation was performed at the UC Davis plant transformation facility. Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998), and composite transgenic M. truncatula plants were produced according to established procedures (Limpens et al., 2004).
For root tip chromatin studies, constitutive INTACT transgenic plant seeds were surface sterilized and sown on 0.5× Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) with 1% (w/v) sucrose in 150-mm-diameter Petri plates, except for tomato and rice, where full-strength MS medium with 1% (w/v) sucrose and without vitamins was used. Seedlings were grown on vertically oriented plates in controlled growth chambers for 7 d after germination, at which point the 1-cm root tips were harvested and frozen immediately in liquid N2 for subsequent nuclei isolation. The growth temperature and light intensity was 20°C and 200 μmol/m2/s for Arabidopsis and M. truncatula, 23°C and 80 μmol/m2/s for tomato, and 28°C/25°C day/night and 110 μmol/m2/s for rice. Light cycles were 16 h light/8 h dark for all species, and light was produced with a 50:50 mixture of 6500K and 3000K T5 fluorescent bulbs.
For studies of the Arabidopsis root hair and non-hair cell types, previously described INTACT transgenic lines were used (Deal and Henikoff, 2010). These lines are in the Col-0 background and carry a constitutively expressed biotin ligase gene (ACT2p:BirA) and a transgene conferring cell-type-specific expression of the NTF gene (from the GLABRA2 promoter in non-hair cells or the ACTIN DEPOLYMERIZING FACTOR8 promoter in root hair cells). Plants were grown vertically on plates as described above for 7 d, at which point 1.25-cm segments from within the fully differentiated cell zone were harvested and flash frozen in liquid N2. This segment of the root contains only fully differentiated cells and excludes the root tip below and any lateral roots above.
Nuclei Isolation
For comparison of ATAC-seq using crude and INTACT-purified Arabidopsis nuclei, a constitutive INTACT line was used (ACT2p:BirA/UBQ10p:NTF) (Sullivan et al., 2014), and nuclei were isolated as described previously (Bajic et al., 2018). In short, after growth and harvesting as described above, 1 to 3 g of root tips was ground to a powder in liquid N2 in a mortar and pestle and then resuspended in 10 mL of NPB (20 mM MOPS, pH 7, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, and 1× Roche Complete protease inhibitors) with further grinding. This suspension was then filtered through a 70 μM cell strainer and centrifuged at 1200g for 10 min at 4°C. After decanting, the nuclei pellet was resuspended in 1 mL of NPB and split into two 0.5-mL fractions in new tubes. Nuclei from one fraction were purified by INTACT using streptavidin-coated magnetic beads as previously described (Bajic et al., 2018) and kept on ice prior to counting and subsequent transposase integration reaction. Nuclei from the other fraction were purified by nonionic detergent lysis of organelles and sucrose sedimentation, as previously described (Bajic et al., 2018). Briefly, these nuclei in 0.5 mL of NPB were pelleted at 1200g for 10 min at 4°C, decanted, and resuspended thoroughly in 1 mL of cold EB2 (0.25 M sucrose, 10 mM Tris, pH 8, 10 mM MgCl2, 1% Triton X-100, and 1× Roche Complete protease inhibitors). Nuclei were then pelleted at 1200g for 10 min at 4°C, decanted, and resuspended in 300 μL of EB3 (1.7 M sucrose, 10 mM Tris, pH 8, 2 mM MgCl2, 0.15% Triton X-100, and 1× Roche Complete protease inhibitors). This suspension was then layered gently on top of 300 μL of fresh EB3 in a 1.5-mL tube and centrifuged at 16,000g for 10 min at 4°C. Pelleted nuclei were then resuspended in 1 mL of cold NPB and kept on ice prior to counting and transposase integration.
For INTACT purification of total nuclei from root tips of M. truncatula, tomato, and rice, as well as purification of Arabidopsis root hair and non-hair cell nuclei, 1 to 3 g of starting tissue was used. In all cases, nuclei were purified by INTACT and nuclei yields were quantified as described previously (Bajic et al., 2018).
ATAC-Seq
Freshly purified nuclei to be used for ATAC-seq were kept on ice prior to the transposase integration reaction and never frozen. Transposase integration reactions and sequencing library preparations were then performed as previously described (Bajic et al., 2018). In brief, 50,000 purified nuclei or 50 ng of Arabidopsis leaf genomic DNA was used in each 50 μL transposase integration reaction for 30 min at 37°C using Nextera reagents (Illumina; FC-121-1030). DNA fragments were purified using the Minelute PCR purification kit (Qiagen), eluted in 11 μL of elution buffer, and the entirety of each sample was then amplified using High Fidelity PCR Mix (NEB) and custom bar-coded primers for 9 to 12 total PCR cycles. These amplified ATAC-seq libraries were purified using AMPure XP beads (Beckman Coulter), quantified by qPCR with the NEBNext Library Quantification Kit (NEB), and analyzed on a Bioanalyzer High Sensitivity DNA Chip (Agilent) prior to pooling and sequencing.
High-Throughput Sequencing
Sequencing was performed using the Illumina NextSeq 500 or HiSeq 2000 instrument at the Georgia Genomics Facility at the University of Georgia. Sequencing reads were either single-end 50-nucleotide or paired-end 36-nucleotide and all libraries that were to be directly compared were pooled and sequenced on the same flow cell.
Sequence Read Mapping, Processing, and Visualization
Sequencing reads were mapped to their corresponding genome of origin using Bowtie2 software (Langmead and Salzberg, 2012) with default parameters. Genome builds used in this study were Arabidopsis version TAIR10, M. truncatula version Mt4.0, Tomato version SL2.4, and Rice version IRGSP 1.0.30. Mapped reads in .sam format were converted to .bam format and sorted using Samtools 0.1.19 (Li et al., 2009). Mapped reads were then filtered using Samtools to retain only those reads with a mapping quality score of 2 or higher (Samtools “view” command with option “-q 2” to set mapping quality cutoff). Arabidopsis ATAC-seq reads were further filtered with Samtools to remove those mapping to either the chloroplast or mitochondrial genomes, and root hair and non-hair cell data sets were also subsampled such that the experiments within a biological replicate had the same number of mapped reads prior to further analysis. For normalization and visualization, the filtered, sorted .bam files were converted to bigwig format using the “bamcoverage” script in deepTools 2.0 (Ramírez et al., 2016) with a bin size of 1 bp and RPKM normalization. Use of the term normalization in this article refers to this process. Heat maps and average plots displaying ATAC-seq data were also generated using the “computeMatrix” and “plotHeatmap” functions in the deepTools package. Genome browser images were made using the Integrative Genomics Viewer (IGV) 2.3.68 (Thorvaldsdóttir et al., 2013) with bigwig files processed as described above.
Identification of Orthologous Genes among Species
Orthologous genes among species were selected exclusively from syntenic regions of the four genomes. Syntenic orthologs were identified using a combination of CoGe SynFind (https://genomevolution.org/CoGe/SynFind.pl) with default parameters, and CoGe SynMap (https://genomevolution.org/coge/SynMap.pl) with the QuotaAlign feature selected and a minimum of six aligned pairs required (Lyons and Freeling, 2008; Lyons et al., 2008).
Peak Calling to Detect THSs
Peak calling on ATAC-seq data was performed using the “Findpeaks” function of the HOMER package (Heinz et al., 2010). The parameters “-region” and “-minDist 150” were used to allow identification of variable length peaks and to set a minimum distance of 150 bp between peaks before they are merged into a single peak, respectively. We refer to the peaks called in this way as transposase hypersensitive sites or THSs.
Genomic Distribution of THSs
For each genome, the distribution of THSs relative to genomic features was assessed using the PAVIS web tool (Huang et al., 2013) with “upstream” regions set as the 2000 bp upstream of the annotated transcription start site and “downstream” regions set as 1000 bp downstream of the transcription termination site.
TF Motif Analyses
ATAC-seq THSs that were found in two replicates of each sample were used for motif analysis. The regions were adjusted to the same size (500 bp for root tip THSs or 300 bp for cell-type-specific dTHSs). The MEME-ChIP pipeline (Machanick and Bailey, 2011) was run on the repeat-masked fasta files representing each THS set to identify overrepresented motifs, using default parameters. For further analysis, we used the motifs derived from the DREME, MEME, and CentriMo programs that were significant matches (E value < 0.05) to known motifs. Known motifs from both Cis-BP (Weirauch et al., 2014) and the DAP-seq database (O’Malley et al., 2016) were used in all motif searches.
Assignment of THSs to Genes
For each ATAC-seq data set, the THSs were assigned to genes using the “TSS” function of the PeakAnnotator 1.4 program (Salmon-Divon et al., 2010). This program assigns each peak/THS to the closest TSS, whether upstream or downstream, and reports the distance from the peak center to the TSS based on the genome annotations described above.
ATAC-Seq Footprinting
To examine motif-centered footprints for TFs of interest, we used the “dnase_average_profile.py” script in the pyDNase package (Piper et al., 2013). The script was used in ATAC-seq mode [“-A” parameter] with otherwise default parameters.
Defining High-Confidence Target Sites for Transcription Factors
We used FIMO (Grant et al., 2011) to identify motif occurrences for TFs of interest, and significant motif occurrences were considered to be those with a P value < 0.0001. Genome-wide high confidence binding sites for a given transcription factor were defined as transposase hypersensitive sites in a given cell type or tissue that also contain a significant motif occurrence for the factor and also overlap with a known enriched region for that factor from DAP-seq or ChIP-seq data (see also Supplemental Figure 2 for a schematic diagram of this process).
GO Analysis
GO analyses using only Arabidopsis genes were performed using the GeneCodis 3.0 program (Nogales-Cadenas et al., 2009; Tabas-Madrid et al., 2012). Hypergeometric tests were used with P value correction using the false discovery rate (FDR) method. AgriGO was used for comparative GO analysis of gene lists among species, using default parameters (Du et al., 2010; Tian et al., 2017).
Accession Numbers
The raw and processed ATAC-seq data described here have been deposited in the NCBI Gene Expression Omnibus database under record number GSE101482. The characteristics of each data set (individual accession number, read numbers, mapping characteristics, and THS statistics) are included in Supplemental Data Set 8. For comparison to our ATAC-seq data from root tips, we used a published DNase-seq data set from 7-d-old whole Arabidopsis roots (SRX391990), which was generated from the same INTACT transgenic line used in our experiments (Sullivan et al., 2014). Publicly available ChIP-seq and DAP-seq data sets were also used to identify genomic binding sites for transcription factors of interest. These include ABF3 (AT4G34000; SRX1720080) and MYB44 (AT5G67300; SRX1720040) (Song et al., 2016), HY5 (AT5G11260; SRX1412757), CBF2 (AT4G25470; SRX1412036), MYB77 (AT3G50060; SRX1412453), ABI5 (AT2G36270; SRX670505), MYB33 (AT5G06100; SRX1412418), NAC083 (AT5G13180; SRX1412546), MYB77 (AT3G50060; SRX1412453), WRKY27 (AT5G52830; SRX1412681), and At5g04390 (SRX1412214) (O’Malley et al., 2016). Raw reads from these files were mapped and processed as described above for ATAC-seq data, including peak calling with the HOMER package. Published RNA-seq data from Arabidopsis root hair and non-hair cells (Li et al., 2016a) were used to define transcripts that were specifically enriched in the root hair cell relative to the non-hair cell (hair-cell-enriched genes), and vice versa (non-hair-enriched genes). We defined cell-type-enriched genes as those whose transcripts were at least 2-fold more abundant in one cell type than the other and had an abundance of at least five RPKM in the cell type with higher expression.
Supplemental Data
Supplemental Figure 1. Comparison of read counts at enriched regions in DNase-seq versus ATAC-seq and Crude-ATAC-seq versus INTACT-ATAC-seq.
Supplemental Figure 2. Analysis of reproducibility in tomato ATAC-seq data
Supplemental Figure 3. Analysis of ATAC-seq signals at orthologous genes.
Supplemental Figure 4. Defining high-confidence binding sites and target genes for each TF.
Supplemental Figure 5. Overlaps of root tip transcription factor target genes.
Supplemental Figure 6. Wild-type and hy5-1 root tip morphology and gravitropism phenotypes.
Supplemental Figure 7. Comparison of ATAC-seq read counts between data sets.
Supplemental Figure 8. Footprinting at motifs of cell-type-enriched TFs in genomic DNA and cell-type-specific ATAC-seq data sets.
Supplemental Data Set 1. Characteristics of THSs in Arabidopsis, M. truncatula, rice, and tomato.
Supplemental Data Set 2. Syntenic orthologous genes in all four species.
Supplemental Data Set 3. Expressolog gene sets in four species.
Supplemental Data Set 4. Motifs common to THSs in all species.
Supplemental Data Set 5. Predicted target genes for ABF3, CBF2, HY5, and MYB77 in all four species.
Supplemental Data Set 6. Motifs overrepresented in cell-type-enriched differential transposase hypersensitive sites.
Supplemental Data Set 7. Binding sites and target genes for cell-type-enriched TFs.
Supplemental Data Set 8. ATAC-seq data set characteristics.
Acknowledgments
We thank Paja Sijacic and Shannon Torres for constructive criticism of the manuscript. This work was supported by funding from the National Science Foundation (Plant Genome Research Program Grant IOS-123843) to J.B.-S., N.S., S.M.B., and R.B.D. D.A.W. was supported in part by funding from the Elise Taylor Stocking Memorial Fellowship, and K.K. was supported in part by the Finnish Cultural Foundation.
AUTHOR CONTRIBUTIONS
R.B.D., S.M.B., N.S., J.B.-S., K.A.M., M.B., K.K., M.R., G.P., and D.A.W. designed the research project. K.A.M. performed all experiments on Arabidopsis root tips as well as hair and non-hair cells. M.B. performed all experiments on M. truncatula root tips. K.K., D.A.W., and K.Z. performed all experiments on tomato root tips, and M.R. and G.P. performed all experiments on rice root tips. M.W. performed all analyses of syntenic regions and identification of orthologous genes among species. K.B., M.D., and C.Q. analyzed ATAC-seq data sets with Hotspot software and also contributed expertise in other analyses. R.B.D., K.A.M., and M.B. analyzed the data. R.B.D. drafted the manuscript with subsequent input and editing from all authors.
References
- Abdeen A., Schnell J., Miki B. (2010). Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genomics 11: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic M., Maher K.A., Deal R.B. (2018). Identification of open chromatin regions in plant genomes using ATAC-seq. Methods Mol. Biol. 1675: 183–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D.C., Buske F.A., Bailey T.L. (2010). Dual-functioning transcription factors in the developmental gene network of Drosophila melanogaster. BMC Bioinformatics 11: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczyński B., Riddell A., Furlong E.E. (2012). Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44: 148–156. [DOI] [PubMed] [Google Scholar]
- Boyle P., Després C. (2010). Dual-function transcription factors and their entourage: unique and unifying themes governing two pathogenesis-related genes. Plant Signal. Behav. 5: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. (2015). ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109: 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Deal R.B., Henikoff S. (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18: 1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., Noble W.S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D.S., Garrard W.T. (1988). Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57: 159–197. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., et al. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. (2008). Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9: 15–26. [DOI] [PubMed] [Google Scholar]
- Hesselberth J.R., Chen X., Zhang Z., Sabo P.J., Sandstrom R., Reynolds A.P., Thurman R.E., Neph S., Kuehn M.S., Noble W.S., Fields S., Stamatoyannopoulos J.A. (2009). Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6: 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Loganantharaj R., Schroeder B., Fargo D., Li L. (2013). PAVIS: a tool for peak annotation and visualization. Bioinformatics 29: 3097–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaradat M.R., Feurtado J.A., Huang D., Lu Y., Cutler A.J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A. (2011). Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43: 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Shim J.S., Seo J.S., Lee H.Y., Kim C.H., Choi Y.D., Cheong J.J. (2010). Non-specific phytohormonal induction of AtMYB44 and suppression of jasmonate-responsive gene activation in Arabidopsis thaliana. Mol. Cells 29: 71–76. [DOI] [PubMed] [Google Scholar]
- Kang J.Y., Choi H.I., Im M.Y., Kim S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr T.C., Abdel-Mageed H., Aleman L., Lee J., Payton P., Cryer D., Allen R.D. (2017). Ectopic expression of two AREB/ABF orthologs increases drought tolerance in cotton (Gossypium hirsutum). Plant Cell Environ., 10.1111/pce.12906. [DOI] [PubMed] [Google Scholar]
- Knight H., Zarka D.G., Okamoto H., Thomashow M.F., Knight M.R. (2004). Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol. 135: 1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon E.Z., Kazmar T., Stampfel G., Yáñez-Cuna J.O., Pagani M., Schernhuber K., Dickson B.J., Stark A. (2014). Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91–95. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Young R.A. (2000). Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34: 77–137. [DOI] [PubMed] [Google Scholar]
- Li D., Li Y., Zhang L., Wang X., Zhao Z., Tao Z., Wang J., Wang J., Lin M., Li X., Yang Y. (2014). Arabidopsis ABA receptor RCAR1/PYL9 interacts with an R2R3-type MYB transcription factor, AtMYB44. Int. J. Mol. Sci. 15: 8473–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Farmer A.D., Lindquist I.E., Dukowic-Schulze S., Mudge J., Li T., Retzel E.F., Chen C. (2012). Characterization of a set of novel meiotically-active promoters in Arabidopsis. BMC Plant Biol. 12: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yamada M., Han X., Ohler U., Benfey P.N. (2016a). High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev. Cell 39: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wu X.Y., Li H., Song J.H., Liu J.Y. (2016b). A dual-function transcription factor, AtYY1, is a novel negative regulator of the Arabidopsis ABA response network. Mol. Plant 9: 650–661. [DOI] [PubMed] [Google Scholar]
- Limpens E., Ramos J., Franken C., Raz V., Compaan B., Franssen H., Bisseling T., Geurts R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55: 983–992. [DOI] [PubMed] [Google Scholar]
- Lu Z., Hofmeister B.T., Vollmers C., DuBois R.M., Schmitz R.J. (2017). Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 45: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W.J., Lee J.Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5: 712–726. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Bergman C., Patel N.H., Kreitman M. (2000). Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Palsson A., Alekseeva E., Bergman C.M., Nathan J., Kreitman M. (2005). Functional evolution of a cis-regulatory module. PLoS Biol. 3: e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E., Freeling M. (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53: 661–673. [DOI] [PubMed] [Google Scholar]
- Lyons E., Pedersen B., Kane J., Freeling M. (2008). The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the rosids. Trop. Plant Biol. 1: 181–190. [Google Scholar]
- Ma W., Noble W.S., Bailey T.L. (2014). Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat. Protoc. 9: 1428–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T.L. (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260. [DOI] [PubMed] [Google Scholar]
- Mathelier A., et al. (2014). JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42: D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus J.T., et al. (2017). A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared to the berry color locus. Plant J. 91: 221–236. [DOI] [PubMed] [Google Scholar]
- Medford J.I., Elmer J.S., Klee H.J. (1991). Molecular cloning and characterization of genes expressed in shoot apical meristems. Plant Cell 3: 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía-Guerra M.K., Li W., Galeano N.F., Vidal M., Gray J., Doseff A.I., Grotewold E. (2015). Core promoter plasticity between maize tissues and genotypes contrasts with predominance of sharp transcription initiation sites. Plant Cell 27: 3309–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.A., Gubler F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A., et al. (2015). Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86: 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton T., Petricka J., Corcoran D.L., Li S., Winter C.M., Carda A., Benfey P.N., Ohler U., Megraw M. (2014). Paired-end analysis of transcription start sites in Arabidopsis reveals plant-specific promoter signatures. Plant Cell 26: 2746–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach M.R., et al. (2017). Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49: 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and biological assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nogales-Cadenas R., Carmona-Saez P., Vazquez M., Vicente C., Yang X., Tirado F., Carazo J.M., Pascual-Montano A. (2009). GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 37: W317–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. [DOI] [PubMed] [Google Scholar]
- Oh S.J., Song S.I., Kim Y.S., Jang H.J., Kim S.Y., Kim M., Kim Y.K., Nahm B.H., Kim J.K. (2005). Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 138: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C.T., Corces V.G. (2011). Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12: 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A., et al. (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R.V., Nahal H.K., Breit R., Provart N.J. (2012). BAR expressolog identification: expression profile similarity ranking of homologous genes in plant species. Plant J. 71: 1038–1050. [DOI] [PubMed] [Google Scholar]
- Persak H., Pitzschke A. (2014). Dominant repression by Arabidopsis transcription factor MYB44 causes oxidative damage and hypersensitivity to abiotic stress. Int. J. Mol. Sci. 15: 2517–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J., Elze M.C., Cauchy P., Cockerill P.N., Bonifer C., Ott S. (2013). Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 41: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.L., Chua N.H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49: 592–606. [DOI] [PubMed] [Google Scholar]
- Reynoso M., Pauluzzi G., Kajala K., Cabanlit S., Velasco J., Bazin J., Deal R., Sinha N., Brady S.M., Bailey-Serres J. (2017). Nuclear transcriptomes at high resolution using retooled INTACT. Plant Physiol., 10.1104/pp.17.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E., Vera D.L., Bass H.W., Buckler E.S. (2016). Open chromatin reveals the functional maize genome. Proc. Natl. Acad. Sci. USA 113: E3177–E3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., et al. (2014). Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 166: 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka D.R., Kandasamy M.K., McKinney E.C., Burgos-Rivera B., Meagher R.B. (2007). The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52: 460–472. [DOI] [PubMed] [Google Scholar]
- Salmon-Divon M., Dvinge H., Tammoja K., Bertone P. (2010). PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer C.D., Blalock E.L., Barwick B.G., Haines R.R., Wei C., Sanz I., Boss J.M. (2016). ATAC-seq on biobanked specimens defines a unique chromatin accessibility structure in naïve SLE B cells. Sci. Rep. 6: 27030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.S., Jung C., Lee S., Min K., Lee Y.W., Choi Y., Lee J.S., Song J.T., Kim J.K., Choi Y.D. (2013). AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 73: 483–495. [DOI] [PubMed] [Google Scholar]
- Shin R., Burch A.Y., Huppert K.A., Tiwari S.B., Murphy A.S., Guilfoyle T.J., Schachtman D.P. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Huang S.C., Wise A., Castanon R., Nery J.R., Chen H., Watanabe M., Thomas J., Bar-Joseph Z., Ecker J.R. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354: aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Furlong E.E. (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13: 613–626. [DOI] [PubMed] [Google Scholar]
- Stadhouders R., van den Heuvel A., Kolovos P., Jorna R., Leslie K., Grosveld F., Soler E. (2012). Transcription regulation by distal enhancers: who’s in the loop? Transcription 3: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A.M., et al. (2014). Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030. [DOI] [PubMed] [Google Scholar]
- Sung M.H., Baek S., Hager G.L. (2016). Genome-wide footprinting: ready for prime time? Nat. Methods 13: 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas-Madrid D., Nogales-Cadenas R., Pascual-Montano A. (2012). GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 40: W478–W483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R.E., et al. (2012). The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45: W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittarelli A., Santiago M., Morales A., Meisel L.A., Silva H. (2009). Isolation and functional characterization of cold-regulated promoters, by digitally identifying peach fruit cold-induced genes from a large EST dataset. BMC Plant Biol. 9: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K., Baele G., Maere S., Van de Peer Y. (2014). Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 24: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera D.L., Madzima T.F., Labonne J.D., Alam M.P., Hoffman G.G., Girimurugan S.B., Zhang J., McGinnis K.M., Dennis J.H., Bass H.W. (2014). Differential nuclease sensitivity profiling of chromatin reveals biochemical footprints coupled to gene expression and functional DNA elements in maize. Plant Cell 26: 3883–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J., Stamatoyannopoulos J.A. (2016). Genomic footprinting. Nat. Methods 13: 213–221. [DOI] [PubMed] [Google Scholar]
- Wada T., Kurata T., Tominaga R., Koshino-Kimura Y., Tachibana T., Goto K., Marks M.D., Shimura Y., Okada K. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419. [DOI] [PubMed] [Google Scholar]
- Wang N., Xu H., Jiang S., Zhang Z., Lu N., Qiu H., Qu C., Wang Y., Wu S., Chen X. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90: 276–292. [DOI] [PubMed] [Google Scholar]
- Wang Z., Su G., Li M., Ke Q., Kim S.Y., Li H., Huang J., Xu B., Deng X.P., Kwak S.S. (2016). Overexpressing Arabidopsis ABF3 increases tolerance to multiple abiotic stresses and reduces leaf size in alfalfa. Plant Physiol. Biochem. 109: 199–208. [DOI] [PubMed] [Google Scholar]
- Weber B., Zicola J., Oka R., Stam M. (2016). Plant enhancers: a call for discovery. Trends Plant Sci. 21: 974–987. [DOI] [PubMed] [Google Scholar]
- Weirauch M.T., et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O., Hafemeister C., Plessis A., Holloway-Phillips M.M., Pham G.M., Nicotra A.B., Gregorio G.B., Jagadish S.V., Septiningsih E.M., Bonneau R., Purugganan M. (2016). EGRINs (Environmental Gene Regulatory Influence Networks) in rice that function in the response to water deficit, high temperature, and agricultural environments. Plant Cell 28: 2365–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Du J. (2014). Young but not relatively old retrotransposons are preferentially located in gene-rich euchromatic regions in tomato (Solanum lycopersicum) plants. Plant J. 80: 582–591. [DOI] [PubMed] [Google Scholar]
- Yanhui C., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60: 107–124. [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang T., Wu Y., Jiang J. (2012a). Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24: 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wu Y., Schnable J.C., Zeng Z., Freeling M., Crawford G.E., Jiang J. (2012b). High-resolution mapping of open chromatin in the rice genome. Genome Res. 22: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Xing L., Wang X., Hou Y.J., Gao J., Wang P., Duan C.G., Zhu X., Zhu J.K. (2014). The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7: ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Zhang W., Zhang T., Liu B., Jiang J. (2015). Genome-wide prediction and validation of intergenic enhancers in Arabidopsis using open chromatin signatures. Plant Cell 27: 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]