Histone deacetylases promote pre-rRNA processing by repressing the expression of key regulators and regulating rRNA methylation through direct binding to rRNA and snoRNAs.
Abstract
Ribosome biogenesis is a fundamental process required for all cellular activities. Histone deacetylases play critical roles in many biological processes including transcriptional repression and rDNA silencing. However, their function in pre-rRNA processing remains poorly understood. Here, we discovered a previously uncharacterized role of Arabidopsis thaliana histone deacetylase HD2C in pre-rRNA processing via both canonical and noncanonical manners. HD2C interacts with another histone deacetylase HD2B and forms homo- and/or hetero-oligomers in the nucleolus. Depletion of HD2C and HD2B induces a ribosome-biogenesis deficient phenotype and aberrant accumulation of 18S pre-rRNA intermediates. Our genome-wide analysis revealed that HD2C binds and represses the expression of key genes involved in ribosome biogenesis. Using RNA immunoprecipitation and sequencing, we further uncovered a noncanonical mechanism of HD2C directly associating with pre-rRNA and small nucleolar RNAs to regulate rRNA methylation. Together, this study reveals a multifaceted role of HD2C in ribosome biogenesis and provides mechanistic insights into how histone deacetylases modulate rRNA maturation at the transcriptional and posttranscriptional levels.
INTRODUCTION
Ribosomes are molecular machines that translate the genetic information from mRNA to protein, a key step for all biological activities. The ribosome is constructed with rRNAs as backbones and ribosomal proteins as fillers (Doudna and Rath, 2002). Three of the four rRNAs (18S, 5.8S, and 25/28S) are transcribed together into a single rRNA precursor (pre-rRNA) by RNA polymerase I (Pol I) in the nucleolus, which are then processed into individual mature rRNAs (Henras et al., 2015). Impaired rRNA transcription and processing are often associated with diseases (e.g., cancers) and other developmental disorders (Byrne, 2009; Higa-Nakamine et al., 2012; Wang et al., 2015; Goudarzi and Lindström, 2016).
Appropriate rRNA production requires precise regulation of both ribosomal DNA (rDNA) transcription and pre-rRNA processing. Of the hundreds to thousands of rRNA genes in yeast and human genomes (Ide et al., 2010), only a subset is actively transcribed (Lawrence and Pikaard, 2004). The rDNA transcription is highly regulated by transcription factors as well as epigenetic modifications such as DNA methylation and histone modifications (Grummt and Pikaard, 2003; Goodfellow and Zomerdijk, 2013). After transcription, pre-RNAs undergo site-specific posttranscriptional modifications mediated by small nucleolar RNAs (snoRNAs) and ribonucleoproteins (Ellis et al., 2010; Bratkovič and Rogelj, 2014). Box C/D and box H/ACA are two major types of snoRNAs responsible for guiding the 2′-hydroxyl ribose methylation (2′-O-methylation) and conversion of uridine to pseudouridine (pseudouridylation) of rRNA, respectively (Bachellerie et al., 2002). These rRNA modifications ensure proper pre-rRNA processing and ribosome biogenesis (Lafontaine, 2015). After assembly with ribosomal proteins, the 35S pre-rRNA is cleaved at specific sites by endonucleases and further trimmed by exonuclease to generate mature rRNAs (18S, 5.8S, and 25/28S) (Henras et al., 2015). The cleavage, trimming, and folding of rRNAs require the assistance of many ribosomal and nonribosomal proteins and snoRNAs (Henras et al., 2015).
Epigenetic factors including DNA methylation, histone modifications, and chromatin remodeling play important roles in rDNA transcription and silencing (Lawrence et al., 2004; Earley et al., 2006; Pontes et al., 2007; Preuss et al., 2008; Pontvianne et al., 2012, 2013). Protein arginine methyltransferases have also been implicated in ribosome biogenesis in both plants and animals (Bachand, 2007; Hang et al., 2014). Histone deacetylases (HDACs) are enzymes that remove acetyl groups from histones and non-histone proteins (Haberland et al., 2009). They are highly conserved in eukaryotes and play pivotal roles in diverse biological processes, including gene expression, genome integrity, development, and diseases (Haberland et al., 2009; Seto and Yoshida, 2014; Bosch-Presegué and Vaquero, 2015). HDACs have been implicated in both rDNA transcription and pre-RNA processing. In mammals, HDAC1 is recruited to specific rDNA loci by Nucleolar Remodeling Complex (NoRC) or transcription factor RUNX2 and represses rDNA transcription (Zhou et al., 2002; Ali et al., 2012). NAD(+)-dependent deacetylase SIRT1 represses rDNA transcription by deacetylating both histones and transcription factors (Salminen and Kaarniranta, 2009; Yang et al., 2013; Voit et al., 2015). SIRT7 facilitates rRNA transcription by stabilizing Pol I at rDNA loci (Ford et al., 2006; Tsai et al., 2012, 2014). A recent study showed that SIRT7 also regulates rRNA maturation through deacetylating a component of U3 snoRNA complex, U3-55k (Chen et al., 2016a).
In the flowering plant Arabidopsis thaliana, RPD3-like histone deacetylase HDA6 is required for the suppression of silenced rDNA loci (Probst et al., 2004; Earley et al., 2006, 2010; Pontes et al., 2007; Pontvianne et al., 2013). Besides the conserved RPD3 and SIR proteins, Arabidopsis has an additional plant-specific HDAC family, termed type-2 HDACs (HD2s), that are phylogenetically conserved in green plants (Bourque et al., 2016). HD2A represses rDNA transcription in nucleolar dominance (Pontes et al., 2007). Overexpression of HD2B attenuates pre-rRNA transcription (Kim et al., 2014). Compared with the large amount of studies on the roles of HDACs in rDNA transcription, little is known about their functions and underlying mechanism in pre-rRNA processing.
In this study, we identify Arabidopsis HD2C as a regulator of ribosome biogenesis. HD2C and HD2B form a complex and are required for normal pre-rRNA processing and ribosome biogenesis. Loss-of-function HD2C and HD2B showed ribosome biogenesis deficiency morphological phenotypes, such as narrow leaves and short roots, and an aberrant accumulation of unprocessed 18S pre-rRNA. Consistent with a functional link between HD2C and pre-rRNA processing, genome-wide profiling of HD2C occupancy combined with transcriptome analysis in hd2b hd2c double mutants revealed that HD2C directly binds and represses the expression of key genes involved in ribosome biogenesis. Intriguingly, we found a direct association of HD2C with pre-rRNA and snoRNAs involved in rRNA methylation. Thus, HD2C exhibits both the canonical role of repressing gene expression as well as the noncanonical action of binding snoRNAs in ribosome biogenesis. This study thus reveals a mechanistic connection between histone deacetylases and pre-rRNA processing in plants.
RESULTS
HD2C Binds Promoters of Genes Involved in Ribosome Biogenesis
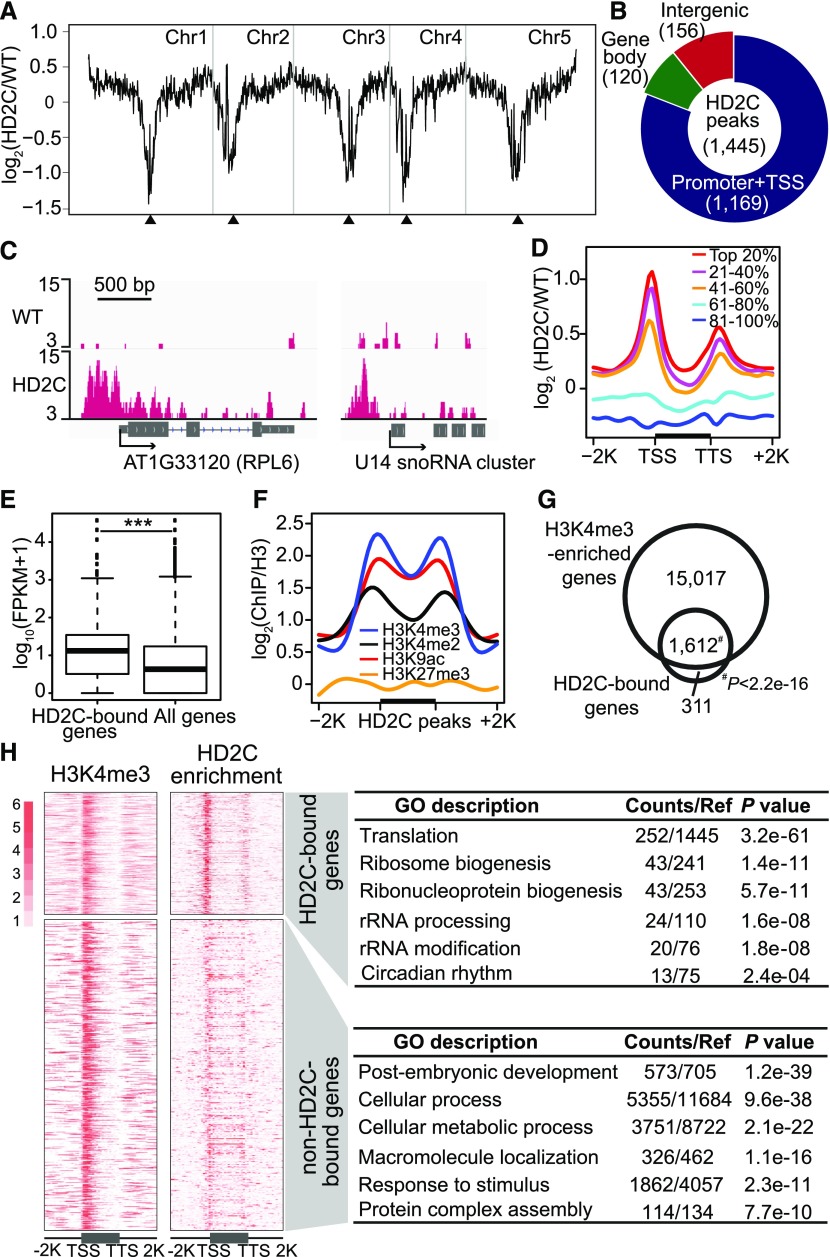
To explore the biological functions of HD2C, we first determined its genome-wide occupancy. We performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) using transgenic Arabidopsis plants expressing FLAG-tagged HD2C driven by its native promoter in hd2c mutant background (pHD2C:HD2C-3×FLAG/hd2c, abbreviated to HD2C-FLAG; Supplemental Figure 1). As a control, the same experiments were performed in parallel with wild-type Col-0. Overall, HD2C is located in gene-rich euchromatin and depleted in transposon-rich heterochromatin (Figure 1A). Further analysis revealed a total of 1445 HD2C binding peaks corresponding to 1923 genes with P < 1e-05 (Supplemental Data Set 1). The majority of the peaks (∼81%) were located in promoter regions (Figures 1B and 1C).
Figure 1.
HD2C Binds Promoters of Active Genes Involved in Ribosome Biogenesis.
(A) Chromosomal view of HD2C binding. The y axis represents the log2 value of HD2C ChIP-seq reads relative to those of untagged wild-type control; Chr1 to Chr5 represent five chromosomes; black triangles indicate positions of centromeres.
(B) Genomic distribution of HD2C binding peaks.
(C) Representative snapshots of HD2C binding to gene promoters.
(D) Metaplots of HD2C ChIP-seq reads over genes. All Arabidopsis genes were divided evenly into five groups based on their expression level in the wild type. Top 20% indicates the 20% genes with highest expression level, and 81 to 100% indicates the 20% genes with lowest expression level. The y axis represents the log2 value of HD2C-FLAG ChIP-seq reads relative to those of untagged wild-type control.
(E) Box plot of expression levels of genes bound by HD2C and all genes in the genome.
(F) Metaplots of histone modification levels over HD2C binding peaks. Black bar in the x axis represents the HD2C binding peaks; y axis represents the levels of histone modifications normalized with H3; −2K and +2K represent 2 kb upstream and downstream of HD2C binding peaks, respectively.
(G) Venn diagram of overlap between HD2C-bound genes and H3K4me3-enriched genes.
(H) Heat maps of HD2C-bound genes and H3K4me3-enriched genes and their functional enrichment. TSS, transcription start site; TTS, transcription terminal site. −2K and +2K represent 2 kb upstream of TSS and 2 kb downstream of transcription terminal site, respectively.
To test the relationship between HD2C binding and gene expression levels, we divided all Arabidopsis genes evenly into five groups based on their expression levels and found that actively expressed genes demonstrated strong enrichment of HD2C (Figure 1D). Consistent with this, the expression levels of HD2C-bound genes were significantly higher than the average expression of all genes (Figure 1E). Furthermore, we noted that HD2C binding sites were co-enriched with active histone modification marks, histone H3 lysine 9 acetylation (H3K9ac) and H3 lysine 4 di- and trimethylation (H3K4me2/3) (Figure 1F). When we overlapped HD2C-bound genes with H3K4me3-enriched genes, we found that 1612 out of 1923 HD2C-bound genes (84%) showed co-enrichment of H3K4me3 (Figure 1G). Interestingly, Gene Ontology (GO) analysis revealed that the 1923 HD2C-bound genes were significantly enriched for translation (P = 3.2e-61), ribosomal biogenesis (P = 1.4e-11), and rRNA processing (P = 1.6e-08; Figure 1H). In contrast, H3K4me3-enriched genes without significant HD2C binding were associated with developmental and metabolic pathways (Figure 1H). Together, these studies showed that HD2C is preferentially associated with promoter of genes involved in ribosome biogenesis pathways.
Depletion of HD2C Induces Genome-Wide H4K16 Hyperacetylation
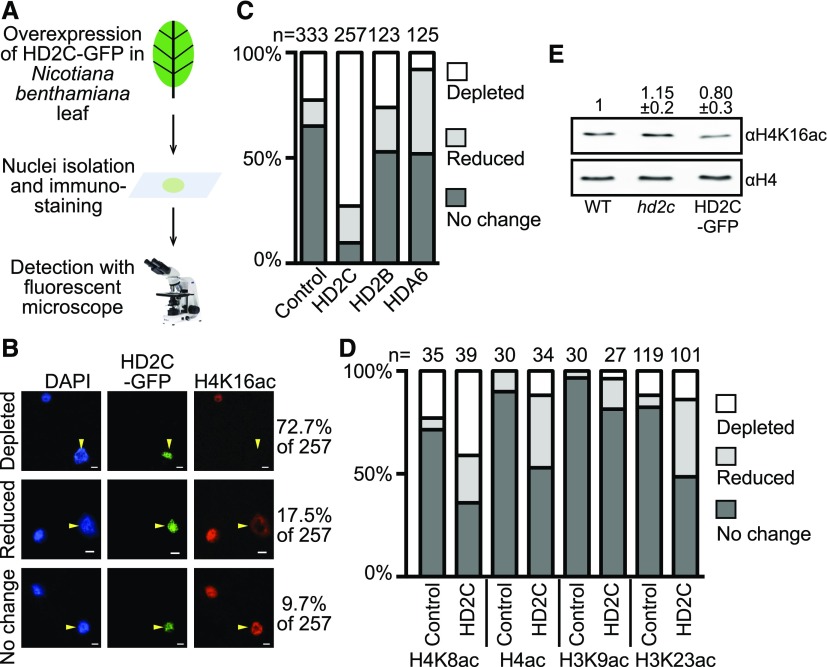
HD2C is predicted to be a histone deacetylase, but its substrate(s) is unknown. To identify the potential histone substrate(s), we overexpressed GFP-tagged HD2C (HD2C-GFP) under the control of a 35S promoter in Nicotiana benthamiana leaves and performed immunostaining with antibodies against various histone acetylation marks (Figure 2A). Of the 257 nuclei overexpressing HD2C-GFP, 72.7% and 17.5% showed depletion or reduction of histone H4 lysine 16 acetylation (H4K16ac), respectively (Figure 2B). To exclude the possibility that the reduction is caused by inefficient recognition of antibodies, we examined the H4K16ac level of nuclei without HD2C-GFP expression (control). Approximately 25% of the control nuclei showed no H4K16ac signals (Figure 2C), much lower than those nuclei with HD2C-GFP. Overexpression of two other putative histone deacetylases (HD2B and HDA6) did not cause significant H4K16ac loss compared with the control (Figure 2C), suggesting that the depletion of H4K16ac is specific to HD2C overexpression. We next examined whether HD2C acts on other histone acetylation marks and found that H4K8ac and H3K23ac also showed a moderate reduction (Figure 2D). There is no noticeable change of H3K9ac when HD2C is overexpressed (Figure 2D).
Figure 2.
Overexpression of HD2C Results in H4K16 Hypoacetylation in N. benthamiana.
(A) Schematic diagram of detecting HD2C-induced histone acetylation change in N. benthamiana.
(B) Immunostaining with H4K16ac antibody in nuclei overexpressing HD2C-GFP. Arrows represent the transfected nuclei expressing HD2C-GFP. Bar = 10 μm.
(C) Quantification of H4K16ac immunostaining performed with HD2C-GFP, HD2B-GFP, and HDA6-GFP. Numbers at the top represent the number of nuclei counted for each experiment.
(D) Quantification of immunostaining performed with HD2C-GFP with other histone acetylation antibodies. Numbers at the top represent the number of nuclei counted for each experiment.
(E) Detection of histone acetylation levels in hd2c mutant and HD2C overexpression (HD2C-GFP) plants by immunoblots. Numbers represent average band intensity of five technical immunoblot repeats (mean ± sd) normalized to H4.
To further test whether HD2C regulates H4K16ac levels in Arabidopsis, we performed immunoblots with H4K16ac antibody in the hd2c mutant. As a further confirmation, we generated HD2C-GFP overexpression transgenic Arabidopsis plants to measure their H4K16ac levels. We found a slight increase of H4K16ac in hd2c and a small decrease of H4K16ac in HD2C-GFP plants (Figure 2E). Taken together, these data suggest that loss-of-function HD2C induces H4K16 hyperacetylation in vivo.
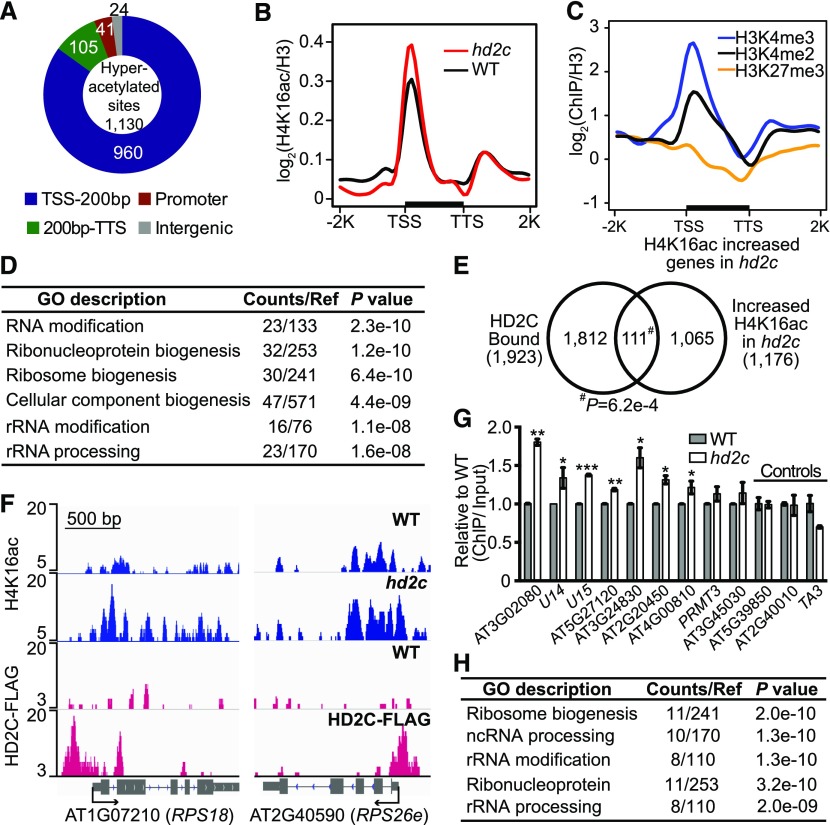
To precisely determine where hyperacetylation occurs, we performed H4K16ac ChIP-seq in an hd2c null mutant. We identified 1130 peaks corresponding to 1176 genes showing increased H4K16ac in hd2c compared with the wild type (P < 1e-05; Supplemental Data Set 2A). Consistent with our previous study showing that H4K16ac marks are abundant at transcription start sites (TSS) and the 200-bp downstream regions (Lu et al., 2015), we found that 960 hyperacetylated peaks (85%) are located around the TSS (Figures 3A and 3B). Genes with increased H4K16ac in hd2c are co-enriched with active histone marks (H3K4me3 and H3K4me2) but not repressive marks (H3K27me3) (Figure 3C), consistent with HD2C-bound genes (Figure 1F). Similar to HD2C-bound genes (Figure 1H), GO analysis of hd2c hyperacetylated genes showed significant enrichment in ribosome biogenesis (P = 6.4e-10), rRNA modification (P = 1.1e-08), and rRNA processing (P = 1.6e-08; Figure 3D).
Figure 3.
Depletion of HD2C Results in H4K16ac Hyperacetylation at Ribosomal Genes in Arabidopsis.
(A) Genomic distribution of H4K16 hyperacetylated peaks in hd2c. TSS-200bp represents transcription start site and its 200 bp downstream region; 200bp-TTS represents the rest gene region and transcription terminal site.
(B) Metaplots of H4K16ac over genes in the wild type and hd2c. TSS, transcription start site; TTS, transcription terminal site. −2K and +2K represent 2 kb upstream of TSS and 2 kb downstream of TTS, respectively. y axis represents the log2 value of H4K16ac ChIP-seq reads normalized with H3 ChIP-seq reads.
(C) Metaplots of histone marks on H4K16ac increased genes in hd2c. y axis represents log2 values ChIP-seq reads of histone marks normalized with those of H3.
(D) GO analysis of genes with increased H4K16ac in hd2c.
(E) Venn diagram of HD2C-bound genes and H4K16ac increased genes in hd2c. P value was calculated with Fisher’s exact test.
(F) Representative snapshots of HD2C occupancy and H4K16ac levels over genes selected from the overlapped group in (E).
(G) Validation of H4K16 hyperacetylation at genes selected from the overlapped group in (E). ChIP-qPCR value was normalized with input and then the wild type was set as 1. Data are represented as mean ± sd with two biological replicates. Two loci (AT5G39850 and AT2G40010) that did not show increased H4K16ac in hd2c and a transposon TA3 were used as negative control. Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001.
(H) GO analysis of genes showed both HD2C binding and increased H4K16ac in hd2c.
To further illustrate the relationship between HD2C binding and increased acetylation, we compared HD2C-bound genes with hyperacetylated genes in hd2c. We found that 111 out of 1923 (∼5.8%) HD2C-bound genes showed increased H4K16ac in hd2c (Fisher’s exact test, P = 6.2e-04; Figures 3E and 3F; Supplemental Data Set 2B). We further validated the H4K16 hyperacetylation at some loci by ChIP-qPCR. Seven of nine targets showed significant increased H4K16ac in hd2c compared with the wild type (Figure 3G). Interestingly, the 111 genes with HD2C binding and H4K16 hyperacetylation in hd2c were enriched in ribosome biogenesis and rRNA modification pathways (Figure 3H; Supplemental Figure 2), further suggesting a function of HD2C in ribosome biogenesis pathways.
HD2C Interacts with HD2B and Forms a Homo-Oligomer
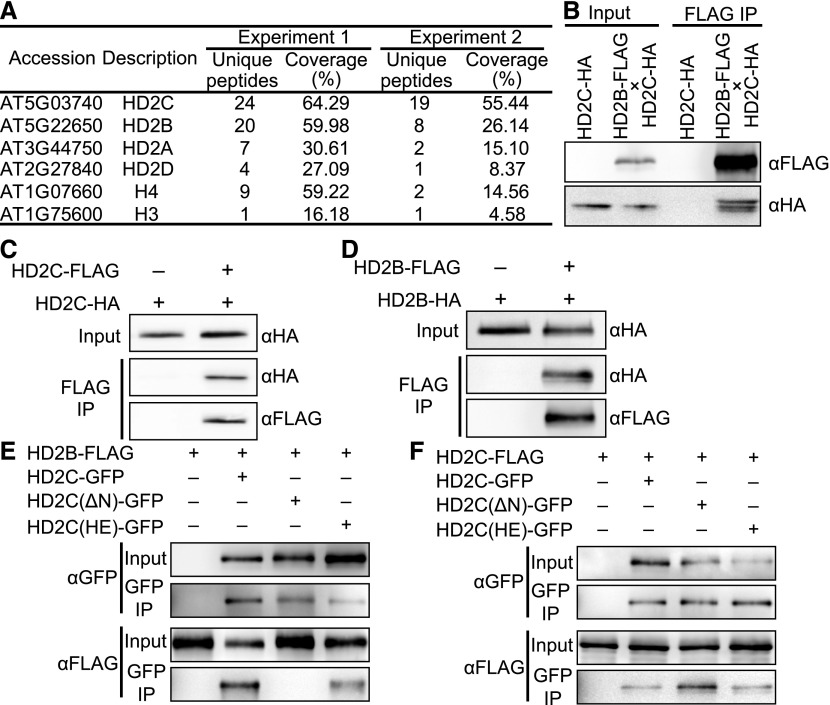
Histone deacetylases often function in collaboration with other proteins (Yang and Seto, 2003). To identify HD2C interacting proteins, we performed immunoprecipitation followed by mass spectrometry analysis (IP-MS) using HD2C-FLAG, the same transgenic lines used for ChIP-seq. The same experiments were performed in Col-0 wild type as a control. We obtained 24 and 19 unique peptides covering 64% and 55% of the HD2C protein sequence in two biologically replicated IP-MS experiments (Figure 4A; Supplemental Data Set 3). Consistent with a previous study, we identified histones (H3 and H4) and HD2A (Buszewicz et al., 2016). Interestingly, we observed copurification of other HD2 members (HD2B and HD2D) (Figure 4A). Both HD2A and HD2B are reported to regulate rRNA transcription (Pontes et al., 2007; Kim et al., 2014). Given that HD2B was the most abundant in our IP-MS, we decided to confirm the HD2C-HD2B interaction. We generated transgenic Arabidopsis expressing FLAG-tagged HD2B driven by its native promoter in the wild-type background (HD2B-FLAG; Supplemental Figure 3A) and crossed with Arabidopsis expressing HA-tagged HD2C driven by the native promoter in hd2c (HD2C-HA). We then performed coimmunoprecipitation (co-IP) in F1 plants and found that HD2C copurified with HD2B (Figure 4B).
Figure 4.
HD2C Interacts with HD2B and Forms Homo-Oligomer.
(A) List of partial proteins copurified with HD2C-FLAG. “Unique peptides” indicates the number of identified peptides that are mapped to only one protein. “Coverage” indicates the percentage of the protein covered by unique peptides identified in this experiment.
(B) Co-IP of HD2C and HD2B using Arabidopsis F1 plants expressing both HD2C-HA and HD2B-FLAG.
(C) Co-IP of HD2C-FLAG and HD2C-HA in N. benthamiana.
(D) Co-IP of HD2B-FLAG and HD2B-HA in N. benthamiana.
(E) Co-IP between GFP tagged HD2C mutants and wild-type HD2B-FLAG in N. benthamiana.
(F) Co-IP between GFP tagged HD2C mutants and wild-type HD2C-FLAG in N. benthamiana.
HD2B was previously shown to form a homopentamer in vitro (Edlich-Muth et al., 2015). Given the high similarity of HD2C and HD2B in amino acid sequences (Supplemental Figure 3B), we hypothesized that HD2C forms a homo-oligomer. To test this, we coinfiltrated HD2C-FLAG and HD2C-HA into N. benthamiana leaves and performed co-IP. HD2C-HA copurified with HD2C-FLAG (Figure 4C), suggesting that HD2C forms a homo-oligomer. Consistent with a previous study (Edlich-Muth et al., 2015), HD2B could also form a homo-oligomer, as shown by our co-IP experiments (Figure 4D). These results suggest that HD2C and HD2B form hetero- and/or homo-oligomer in vivo.
The HD2 proteins share a conserved N-terminal nucleoplasmin-like (NPL) domain (Supplemental Figure 3B) predicted to be a catalytic domain (Bourque et al., 2016). The conserved first five residues (MEFWG), the histidine at position 25 (H25), and the aspartic/glutamic acid at position 69 (D/E69) are critical to the deacetylation activity (indicated with square and asterisks in Supplemental Figure 3B) (Zhou et al., 2004). To test whether these residues are also important for homo- or hetero-oligomerization of HD2 proteins, we generated GFP-tagged HD2C with deletion of the EFWG motif [HD2C(ΔN)-GFP], or double mutations of H25A and E69A [HD2C(HE)-GFP], and then performed co-IP with HD2B-FLAG and HD2C-FLAG in N. benthamiana leaves. The absence of MEFWG disrupted the HD2B-HD2C interaction, but not the HD2C-HD2C interaction (Figures 4E and 4F), suggesting that the MEFWG motif is important for hetero-oligomerization but not homo-oligomerization. In contrast, the H25A E69A mutation of HD2C has no obvious effect on either HD2C-HD2B or HD2C-HD2C interactions (Figures 4E and 4F).
Depletion of HD2C and HD2B Causes Ribosome-Related Developmental Pleiotropy
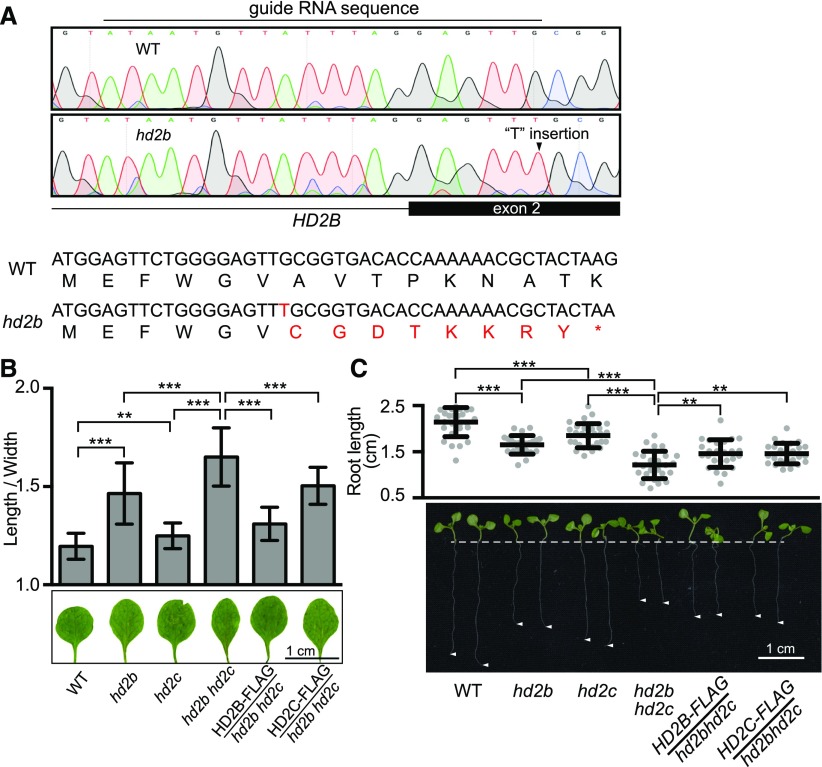
The aforementioned observations prompted us to investigate whether HD2C and HD2B act together in ribosome biogenesis pathways in vivo. To test this idea, we generated an hd2b mutant by CRISPR-Cas9 (Xie et al., 2015) because the available hd2b T-DNA insertion alleles are not null mutants. A single “T” nucleotide insertion between positions 5 and 6 in the second exon of HD2B was created. This insertion resulted in a frame shift and translation termination at position 15 (Figure 5A). In Arabidopsis, aberrant ribosomal processing is associated with multiple abnormalities, with leaf developmental defects being the most prominent (Van Lijsebettens et al., 1994; Ito et al., 2000; Weijers et al., 2001; Byrne, 2009; Abbasi et al., 2010; Hang et al., 2014; Weis et al., 2015). To determine the function of HD2C and HD2B in ribosome biogenesis, we examined the first two true leaves of hd2c and hd2b and noted that both hd2c and hd2b exhibited narrow and pointed leaves compared with the round and regular shape of wild-type leaves (Figure 5B). In addition, the roots of the hd2c and hd2b mutants were significantly shorter than those of wild type (Figure 5C). To test whether HD2C and HD2B act together to regulate the leaf and root development, we generated hd2b hd2c double mutant (Supplemental Figure 4A) and found that hd2b hd2c double mutants displayed an additive effect of narrow leaves and short roots compared with either of the single mutant (Figures 5B and 5C). To further confirm that the phenotypic defects were caused by hd2c and hd2b mutation, we transformed HD2C-FLAG or HD2B-FLAG driven by the respective native promoters into the hd2b hd2c double mutant (Supplemental Figure 4B). As expected, HD2C and HD2B rescued the leaf and root developmental defects of hd2b hd2c to respective single mutant (Figures 5B and 5C), suggesting that HD2C and HD2B are indeed functional and required for normal leaf and root development.
Figure 5.
Depletion of HD2C and HD2B Causes Ribosome-Related Developmental Pleiotropy.
(A) Schematic diagram of loss-of-function hd2b mutant generated with a CRISPR-cas9 system.
(B) Length/width ratio of the first two true leaves of the wild type, hd2b, hd2c, hd2b hd2c double mutant, HD2B-FLAG/hd2b hd2c, and HD2C-FLAG/hd2b hd2c. Data are represented as mean ± sd with at least 30 leaves. Student’s t test, ** P < 0.01 and ***P < 0.001.
(C) Root length of plants in (A). Data are represented as mean ± sd with at least 20 plants. Student’s t test, **P < 0.01 and ***P < 0.001.
HD2C and HD2B Directly Repress Genes Involved in Ribosome Biogenesis
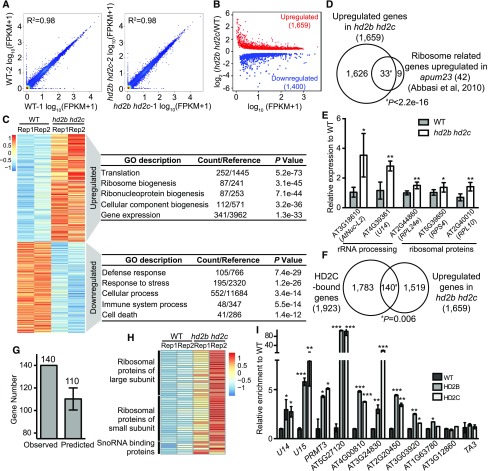
To gain further insights into HD2C and HD2B action, we performed mRNA sequencing in wild type and hd2b hd2c double mutants with two biological replicates (Figure 6A). We identified 1659 significantly upregulated and 1400 downregulated genes (P < 0.05) in hd2b hd2c (Figure 6B; Supplemental Data Set 4). GO analysis revealed that the upregulated genes show significant enrichment in translation (P = 5.2e-73) and ribosome biogenesis (P = 3.1e-45), while downregulated genes are enriched in defense and stress responses (Figure 6C). Interestingly, when we compared our RNA-seq with the transcriptome data of a key rRNA processing regulator APUM23 (Abbasi et al., 2010), we found that 33 out of 42 upregulated ribosome-related genes in apum23 were also significantly upregulated in hd2b hd2c (Figure 6D). We further validated this data by randomly selected 5 out of the 33 genes by RT-qPCR (Figure 6E). Consistent with the gene expression change, apum23 also showed narrow leaves and short roots (Abbasi et al., 2010). These results suggest that HD2C and HD2B play important roles in ribosome biogenesis.
Figure 6.
HD2C and HD2B Directly Repress Expression of Ribosome Biogenesis-Related Genes.
(A) Scatterplots for two RNA-seq biological replicates of the wild type and hd2b hd2c. Expression level was calculated by log10 (FPKM+1).
(B) Scatterplot of differentially expressed genes in hd2b hd2c double mutant. Red dots and blue dots represent upregulated and downregulated genes, respectively. x axis represents the expression level of genes in the wild type by calculating log10 value of FPKM+1. y axis represents the log2 value of gene expression level in hd2b hd2c relative to the wild type. FPKM, fragments per kilobase of transcript per million mapped reads.
(C) Heat maps of differentially expressed genes in two biological replicates of the wild type and hd2b hd2c and their functional enrichment. Color bar indicates the Z-score. Rep1 and Rep2 represent two replicates.
(D) Venn diagram of ribosome biogenesis genes upregulated in apum23 and hd2b hd2c.
(E) RT-qPCR validation of upregulation of overlapped genes in (D). y axis represents the relative expression level to the wild type. Data are represented as mean ± sd from two biological replicates. Student’s t test, *P < 0.05 and **P < 0.01.
(F) Venn diagram of HD2C-bound genes and upregulated genes in hd2b hd2c. P value was calculated with Fisher’s exact test.
(G) Relative enrichments of the observed number of overlapped genes between HD2C binding and upregulation in hd2b hd2c compared with the average overlapped genes by 10,000 genome-shuffling experiments.
(H) Heat maps of expression of ribosome biogenesis genes identified from the 140 genes in (F) in the wild type and hd2b hd2c. Color bar indicates the Z-score.
(I) ChIP-qPCR validation of HD2B and HD2C association with ribosome biogenesis genes selected from overlapped genes in (F). Data are represented as mean ± sd from two biological replicates. Student’s t test, *P < 0.05, **P <0 .01, and ***P < 0.001. Transposable element TA3 served as a negative control.
To determine whether HD2C directly represses gene expression, we overlapped HD2C-bound genes with hd2b hd2c upregulated genes and found 140 out of 1659 upregulated genes showed HD2C enrichment (Figure 6F). This is more than expected by chance (Fisher’s exact test, P = 0.006). As a control, we randomly picked 1659 genes in the Arabidopsis genome, shuffled 10,000 times, and found an average of 110 overlapped with HD2C-bound genes (Figure 6G), significantly less than we observed. Within these 140 genes showing HD2C binding and upregulation in hd2b hd2c, we noted genes encoding ribosomal proteins, snoRNA binding proteins, and other known rRNA processing regulators (Figure 6H; Supplemental Table 1). To test whether HD2B binds the same genes as HD2C, we performed HD2B ChIP-qPCR on 10 ribosome-related HD2C targets and found that HD2B was also enriched on nine of them (Figure 6I). Together, these data suggest that HD2C and HD2B regulate ribosome biogenesis by directly repressing key ribosomal proteins and biogenesis factors.
HD2C and HD2B Promote Pre-rRNA Processing
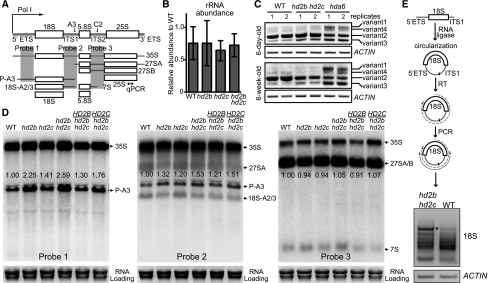
In eukaryotes, two key steps in ribosome biogenesis are pre-rRNA transcription and processing (Woolford and Baserga, 2013; Henras et al., 2015; Weis et al., 2015). To elucidate whether HD2C and HD2B participate in these steps, we first assessed whether HD2C and HD2B affect rRNA transcript levels. With a primer detecting both intermediates and mature rRNA (Figure 7A), we did not observe significant change of rRNA abundance in hd2c, hd2b, or hd2b hd2c mutants (Figure 7B). Previous studies have shown that there are four major pre-rRNA transcript variants and some of them are repressed during development by HDA6 (Pontvianne et al., 2010; Mohannath et al., 2016). To test whether HD2C and HD2B are also involved in repressing pre-rRNA variants, we examined their transcript levels in hd2b hd2c double mutant. Unlike HDA6, we did not observe notable changes of transcript levels of these variants in hd2b hd2c compared with the wild type (Figure 7C), suggesting that HD2C and HD2B are not involved in rRNA silencing. We next investigated the rRNA processing by performing RNA gel blots to detect steady state levels of pre-rRNA intermediates. The three mature rRNAs (18S, 5.8S, and 25S) are transcribed as a single transcript with two internal transcribed spacers (ITS1 and ITS2) and two external transcribed spacers (5′ETS and 3′ETS) (Figure 7A). As used previously (Abbasi et al., 2010; Hang et al., 2014), probe 1 in the 5′ETS was able to detect both 35S and 18S but not 27S and 7S precursors. Probe 2 in ITS1 could distinguish from Probe 1 by recognizing both P-A3 and 18S-A2/3 (Figure 7A). We found modestly increased levels of P-A3 fragment in hd2c or hd2b compared with the wild type (Figure 7D). The aberrant P-A3 accumulation is greater in hd2b hd2c double mutant than any of the single mutant (Figure 7D). Additionally, we found that 7S, the downstream product of 27SA/B, is slightly increased in hd2c, hd2b, and hd2b hd2c mutants as detected by Probe 3 (Figures 7A and 7D). Notably, all these pre-rRNA processing defects were restored to the corresponding single mutant level when introducing either HD2C or HD2B back to hd2b hd2c double mutants (Figure 7D).
Figure 7.
HD2C and HD2B Promote Pre-18S rRNA Processing.
(A) Schematic diagram of rRNA transcription and processing in Arabidopsis. Bidirectional arrow represents the position of the primer used in (B). Probes 1 to 3 represent the positions of the probes used in (D).
(B) RT-qPCR of total rRNA abundance in the wild type, hd2b, hd2c, and hd2b hd2c double mutant. y axis represents the relative transcript level.
(C) RT-PCR detection of four major rRNA transcripts in the wild type, hd2b hd2c, and hda6 in different stages of plants.
(D) RNA gel blots with probes listed in (A). Ethidium bromide staining of mature rRNA as a loading control. Numbers besides the band indicate the relative density of the bands normalized to the loading control.
(E) Circularized RT-PCR for pre-18S rRNA in the wild type and hd2b hd2c. RT was performed with 18S rRNA-specific primer toward 5′ETS. The joint end between 5′ and 3′ of 18S rRNA was amplified in the wild type and hd2b hd2c. The most abundant band in hd2b hd2c compared with the wild type (indicated with an asterisk) was cloned into plasmid and sequenced. ACTIN served as an internal control.
To further confirm the RNA gel blot, we mapped the precise ends of the aberrantly accumulated fragment in hd2b hd2c double mutant by circular RT-PCR. Pre-rRNA intermediates were circularized and reverse transcribed with an 18S specific primer, which were further amplified and cloned into a plasmid for sequencing (Figure 7E). Consistent with the RNA gel blot, we observed overaccumulation of pre-18S rRNA intermediates containing 561 bp of 5′ETS and 191 bp of ITS1 in hd2b hd2c (Figure 7E; Supplemental Figure 5), which is exactly the same as known P-A3 fragment (Sáez-Vasquez et al., 2004; Sikorski et al., 2015). Together, these results suggest that HD2C and HD2B play important roles in proper pre-rRNA processing.
HD2C Is Associated with Pre-rRNA and SnoRNAs
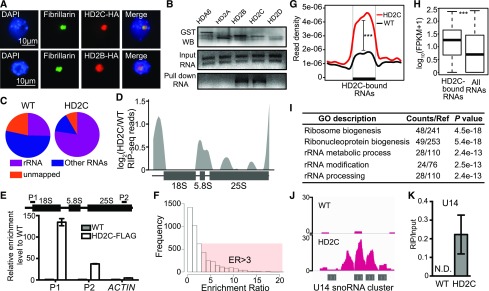
Given the role of HD2C and HD2B in pre-rRNA processing, we performed immunostaining to determine the HD2C and HD2B localization using transgenic Arabidopsis expressing HD2C-HA and HD2B-HA driven by their native promoters. Both HD2C and HD2B located in the nucleolus, as indicated by their colocalization with the nucleolar marker protein Fibrillarin (Figure 8A).
Figure 8.
HD2C Is Associated with Pre-rRNAs and SnoRNAs.
(A) Immunostaining for protein localization of HD2C and HD2B. Fibrillarin was used as a nucleolar marker.
(B) In vitro RNA pull-down assay of HD2C and HD2B. Bands in “GST WB” indicate immunoblots of GST-tagged protein. Bands in “Input RNA” and “Pull down RNA” indicate ethidium bromide staining of rRNA.
(C) Distribution of RIP-seq reads of HD2C and the wild type. rRNA represents the reads mapped to rRNA sequence, and non-rRNA represents reads mapped to other RNAs except rRNA.
(D) HD2C binding peaks on rRNA precursor. y axis represents the log2 value of HD2C RIP-seq normalized with the wild type.
(E) RIP-qPCR confirmation of HD2C binding on rRNA. ACTIN served as a control.
(F) Histogram of frequency distribution of HD2C enrichment ratio on RNAs. ER, enrichment ratio.
(G) Metaplots of HD2C enrichment on HD2C-bound RNAs. y axis represents the read density of RIP-seq. Student’s t test, ***P < 0.001.
(H) Box plots show the abundance of HD2C-bound RNAs and average of all RNAs. y axis represents the log10 value of FPKM+1. Student’s t test, ***P < 0.001.
(I) GO analysis of HD2C-bound RNAs.
(J) Snapshot of HD2C binding peaks on U14 snoRNA cluster RNA transcript.
(K) RIP-qPCR validation of HD2C binding on U14 snoRNA. N.D., not detected.
We next asked whether HD2C and HD2B directly bind rRNAs. We first performed an in vitro pull-down assay with GST-tagged HD2C and HD2B purified from Escherichia coli. HDA6, HD2A, and HD2D were used as controls. Interestingly, only HD2C and HD2B, but not HDA6, HD2A, or HD2D, pulled down rRNAs (Figure 8B). To further confirm the rRNA binding ability of HD2C in vivo, we performed RNA immunoprecipitation followed by sequencing (RIP-seq) in HD2C-FLAG and wild-type plants. We first aligned the total reads to rRNA sequences and found that the majority of HD2C-associated RNA reads (∼78%, compared with ∼25% in the wild type) were indeed aligned to the transcribed regions of rDNA (Figure 8C), suggesting that HD2C binds to nascent pre-rRNA. To map the precise location of HD2C binding, we plotted HD2C RIP-seq reads over rRNAs and found that HD2C preferentially binds 5′ETS of 18S, entire 5.8S, two regions of 25S, and the 3′ end of 25S rRNAs (Figure 8D). This binding pattern was further confirmed by RIP-qPCR (Figure 8E). As a control, we aligned reads to 5S rRNA (transcribed by RNA polymerase III) and did not note significant enrichment of HD2C (Supplemental Figure 6A), suggesting that HD2C specifically binds 18S, 5.8S, and 25S rRNAs.
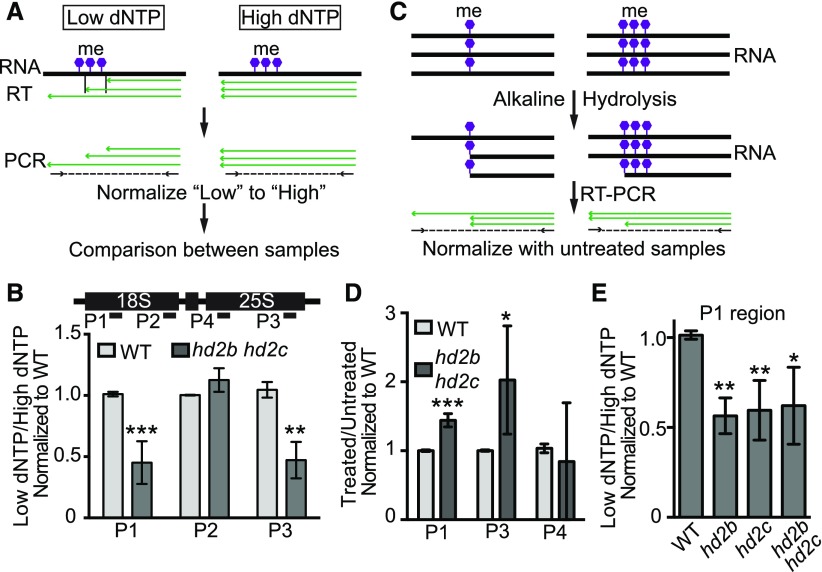
Besides rRNAs, 14% of HD2C RIP-seq reads were mapped to other RNAs including mRNAs and noncoding RNAs (Supplemental Data Set 5A). To identify the HD2C-associated RNAs, we defined an enrichment ratio for each RNA transcript by normalizing the mapped reads of HD2C to the wild type (Lu et al., 2014). With the cutoff enrichment ratio of >3, we identified 1515 RNAs with significant HD2C enrichment (P < 0.05) (Figures 8F and 8G; Supplemental Data Set 5A). Overall, HD2C-bound RNAs were more abundant than the average of total RNAs (Figure 8H). Among the HD2C-associated RNAs, RNAs implicated in ribosome biogenesis, rRNA modification, and rRNA processing were enriched (Figure 8I). Interestingly, we identified a total of 24 snoRNAs comprising both box C/D and box H/ACA snoRNAs that guide rRNA methylation and pseudouridylation, respectively (Bachellerie et al., 2002; Bratkovič and Rogelj, 2014). Strikingly, 21 out of 24 HD2C-bound snoRNAs belong to C/D box class (Supplemental Data Set 5B and Supplemental Figure 6B), suggesting that HD2C may play a role in rRNA methylation. To test this idea, we measured and compared the methylation level of rRNA between the wild type and hd2b hd2c mutant using the feature that reverse transcriptase pauses or stops at the methylated sites in low concentration of deoxynucleotide (dNTP), but not in high dNTP (Dong et al., 2012). The methylation level was calculated by normalizing RT-qPCR value of low dNTP to high dNTP (Figure 9A). We performed RT-qPCR with the primers specific to 18S and 25S rRNA and found a decreased amplification specifically at P1 and P3 regions in hd2b hd2c mutant (Figure 9B), suggesting that certain nucleotide(s) within P1 and P3 regions of 18S and 25S rRNA gain methylation in the absence of HD2C and HD2B.
Figure 9.
HD2C and HD2B Repress RNA Methylation.
(A) Schema diagram of RNA methylation assay using low dNTP method.
(B) RT-qPCR amplification with 18S and 25S rRNA specific primers in the wild type and hd2b hd2c double mutant using method presented in (A). Lower value indicates higher methylation level.
(C) Schema diagram of RNA methylation assay using alkaline hydrolysis method.
(D) RT-qPCR amplification with 18S and 25S rRNA specific primers in the wild type and hd2b hd2c double mutant using method presented in (C). Higher value indicates higher methylation level. Primer positions were shown in (B).
(E) RT-qPCR amplification with 18S rRNA specific primer in the wild type, hd2b and hd2c single mutants, and hd2b hd2c double mutant using method presented in (A). Lower value indicates higher methylation level. Primer position was shown in (B).
All data are represented as mean ± sd with at least four biological replicates. Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001.
To further confirm these results, we performed RT-PCR with RNA treated with alkaline (Figure 9C). The RNA hydrolysis by alkaline is impaired by RNA methylation, which allows us to quantify the relative RNA methylation level (Kiss-László et al., 1996). By normalizing to the untreated RNA, we found higher amplification in P1 and P3 regions in hd2b hd2c (Figure 9D), suggesting an increased RNA methylation level in hd2b hd2c compared with the wild type.
To dissect the respective roles of HD2C and HD2B in RNA methylation, we examined the RNA methylation change in hd2c and hd2b single mutant with low dNTP method. We found similar amplification level between single and double mutants (Figure 9E), suggesting that RNA methylation is similarly affected in hd2c, hd2b, and hd2c hd2b mutants. Taken together, HD2C and HD2B may regulate pre-rRNA processing through snoRNA-mediated rRNA methylation.
DISCUSSION
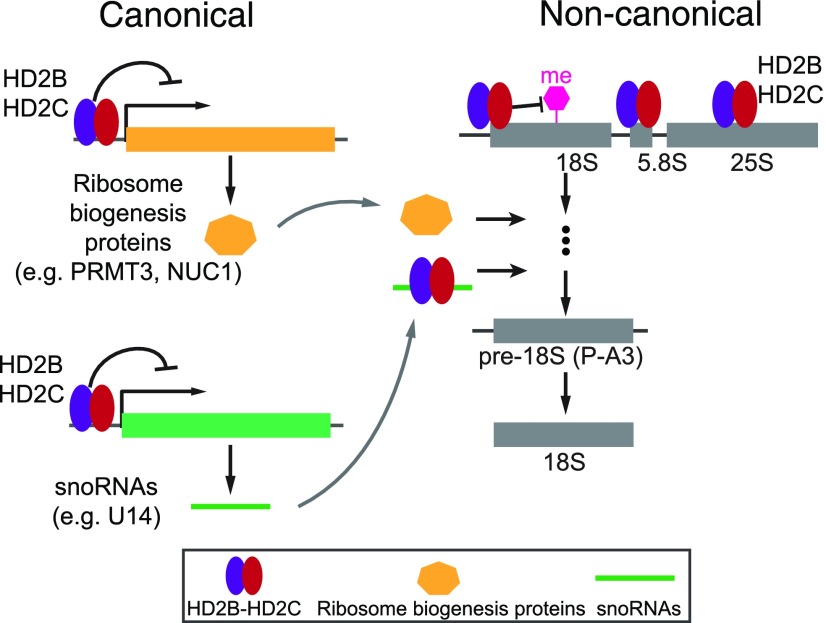
In this study, we provided multiple lines of evidence for a role of Arabidopsis HD2C and HD2B in ribosome biogenesis. HD2C and HD2B form a complex and promote pre-rRNA processing in canonical and noncanonical manners. On the one hand, HD2C and HD2B directly bind and repress the expression of key genes required for ribosome biogenesis, including ribosome biogenesis proteins (e.g., PRMT3 and NUC1) and snoRNAs (e.g., U14). On the other hand, HD2C and HD2B are associated with pre-rRNA and snoRNAs to regulate rRNA modifications (e.g., methylation) and pre-rRNA processing (Figure 10).
Figure 10.
A Working Model for Canonical and Noncanonical Mechanisms of HD2C and HD2B in rRNA Processing.
HD2C and HD2B form complex and drive pre-rRNA processing and ribosome biogenesis via a canonical and/or noncanonical pathway. On the one hand, HD2C and HD2B directly bind and repress the expression of key genes required for ribosome biogenesis, including ribosome biogenesis proteins (e.g., PRMT3 and/or NUC1) and snoRNAs (e.g., U14). On the other hand, HD2C and HD2B are associated with pre-rRNA and snoRNAs to regulate rRNA modifications (e.g., methylation) and pre-rRNA processing.
Canonical Functions of HD2C and HD2B in rRNA Processing
Histone deacetylases play crucial roles in many biological processes, largely through their transcriptional repression activities (Hollender and Liu, 2008; Haberland et al., 2009). Consistent with the canonical gene regulation function, HD2C and HD2B directly bind and repress the expression of many genes involved in ribosome biogenesis (Figure 6). Among them were ribosomal proteins (e.g., RPL11B, RPL11C, RPS7A, RPS16C, and RPS28C), snoRNA binding proteins, and proteins required for pre-rRNA processing. Compared with their human homologs that are implicated in pre-rRNA maturation (Robledo et al., 2008), the function of Arabidopsis ribosomal proteins in pre-rRNA processing is less well understood. SnoRNA binding proteins function together with snoRNAs and play important roles in pre-rRNA processing (Newman et al., 2000; Ellis et al., 2010).
We observed overaccumulation of 18S pre-rRNA intermediate fragment P-A3 in the absence of HD2C and HD2B. Although the precise mechanism is unclear, two possibilities could account for this pattern. First, the immediate downstream product of P-A3, 18S-A2/3, is less abundant in hd2b hd2c compared with the wild type (Figure 7D), suggesting that the processing from P-A3 to its downstream product may be impaired in the mutants. Second, although the steady state level of 27SA/B fragment is unchanged, its downstream product 7S is slightly increased in hd2b hd2c mutants (Figure 7D), which may be the case if 27SA/B is quickly processed into 7S. Given that 27SA/B is generated together with P-A3 from 35S precursor (Figure 7A), the overaccumulation of P-A3 is possibly due to an enhanced processing kinetics at A3 site in hd2b hd2c mutants (Figure 7A). Consistent with this idea, we noted an elevated expression of Arabidopsis protein arginine methyltransferase3 (PRMT3) in hd2b hd2c (Supplemental Data Set 4) accompanied by an overaccumulation of P-A3 (Figure 7D). PRMT3 is implicated in ribosome biogenesis by promoting the 18S pre-rRNA processing (Hang et al., 2014).
NUCLEOLIN1 (NUC1) is another interesting protein that plays an important role in ribosome biogenesis, including pre-rRNA transcription and processing (Sáez-Vasquez et al., 2004; Kojima et al., 2007; Petricka and Nelson, 2007; Pontvianne et al., 2007, 2010). We found that NUC1 is bound by HD2C and upregulated in hd2b hd2c double mutant (Supplemental Data Sets 1 and 4), suggesting that NUC1 is directly regulated by HD2C. Furthermore, our IP-MS showed that NUC1 is copurified with HD2C (Supplemental Data Set 3). More importantly, loss-of-function NUC1 mutation exhibited a similar developmental defective phenotype as hd2c hd2b double mutants (Petricka and Nelson, 2007; Durut et al., 2014). Thus, it is possible that HD2C and HD2B may act through PRMT3 and/or NUC1 to modulate the pre-rRNA processing.
HD2C and HD2B also repress the expression of several snoRNAs including U14 (Figure 6E). In yeast, U14 is required for the generation of 18S pre-rRNA from the upstream precursor (Liang and Fournier, 1995). This function is conserved in Arabidopsis as U14 is a core component of the pre-rRNA processing complex (Qu et al., 2001). Increased U14 level in hd2b hd2c may enable the efficient processing of 35S to its downstream products.
Noncanonical Functions of HD2C and HD2B in Pre-rRNA Processing
Besides the canonical mechanism, we also identified a noncanonical action of HD2C and HD2B in pre-rRNA processing through binding pre-rRNA and snoRNAs, and regulating rRNA methylation (Figures 8 and 9). HD2C is associated with both pre-rRNA and snoRNAs. Notably, snoRNAs are required for posttranscriptional modifications of rRNA, largely through recruiting modifying machineries to specific sites of rRNA (Kiss, 2002; Bratkovič and Rogelj, 2014). The fact that 21 out of 24 HD2C-bound snoRNAs are C/D box type (Supplemental Figure 6B) suggests that HD2C may be involved in rRNA methylation. Consistently, methylation of 18S and 25S rRNA was increased in hd2b hd2c (Figure 9).
The precise mechanism of how HD2C and HD2B inhibit rRNA methylation is unknown. One possibility is that HD2C may act directly through rRNA methyltransferases. We identified two SpoU-family RNA methyltransferases (AT2G19870 and AT4G15520) in HD2C IP-MS (Supplemental Data Set 3). SpoU-family methyltransferases are conserved from prokaryotes to eukaryotes and can methylate both rRNAs and tRNAs (Anantharaman et al., 2002). HD2C may regulate the activity of rRNA methyltransferases. One possibility is that HD2C regulates the recruitment of rRNA methyltransferase. Given that HD2C is capable of binding rRNA, it is possible that HD2C competes with rRNA methyltransferase for rRNA substrates and consequently disables the recruitment of methyltransferase to rRNAs.
We found some of the snoRNAs are bound by HD2C at both the DNA and RNA level (Supplemental Figure 6C). For example, HD2C is associated with both promoter regions of U14 gene as well as its transcription product U14 snoRNA, suggesting that HD2C can regulate pre-rRNA processing at both transcriptional and posttranscriptional levels. Consistent with this idea, mammalian NAD(+)-dependent deacetylase SIRT7 is associated with both rRNA and snoRNAs (Chen et al., 2016a). SIRT7 is also a nucleolar protein and regulates transcription and processing of rRNA by deacetylating U3-55K, a core component of the U3 snoRNP complex (Ford et al., 2006; Chen et al., 2016a). Thus, HD2C displays a multitude of function in regulating pre-rRNA processing. Interestingly, despite the similar amino acid sequence of HD2A and HD2D with HD2C, we did not note that HD2A and HD2D are capable of binding rRNA in vitro. It is possible that HD2A/HD2D and also HDA6 may require posttranslational modifications to bind to rRNA.
Relationship between HD2C and HD2B in Pre-rRNA Processing
Although HD2C and HD2B interact with each other, they may function redundantly in regulating rRNA processing and plant development. The hd2c hd2b double mutants display additively narrow leaves and short roots as well as the abnormal accumulation of P-A3 compared with either hd2c or hd2b single mutant (Figures 5 and 7). One possibility is that HD2C and HD2B may partially mimic the function of each other. These two proteins have the same conserved NPL domain. Our co-IP analyses showed that both of them could form a homo-oligomer. It is possible that HD2C and HD2B may functionally replace each other when one of them is absent. Another possibility is that HD2C and HD2B may have unique and independent functions in ribosome biogenesis. Besides the HD2C-HD2B complex, these two proteins may interact with other proteins involved in ribosome biogenesis. Consistent with this idea, HD2B interacts with ribosomal protein S6 (Kim et al., 2014), which was not copurified with HD2C in our IP-MS. HD2C was also shown to specifically interact with HDA6 that is involved in 5S rDNA transcription (Earley et al., 2006, 2010; Vaillant et al., 2007; Luo et al., 2012). Thus, it will be interesting to decipher the HD2C- and HD2B-interacting partners to further explore their functional link in ribosome biogenesis.
Evolution of HD2 Proteins in Plants
HD2C and HD2B first appeared in green algae (Bourque et al., 2016) and are classified into a plant-specific HD2 group. HD2 proteins share a conserved N-terminal NPL domain predicted to be the catalytic domain (Supplemental Figure 3B) (Hollender and Liu, 2008; Bourque et al., 2016). Although the HD2 proteins are specific in plants, the conserved NPL domain has been found in histone chaperon protein nucleoplasmin (NUP) and some of the FK506 binding protein (FKBP) family in fungi, plants, and animals (Frehlick et al., 2007; Edlich-Muth et al., 2015). Like HD2C and HD2B (Figure 4), the NPL domain of NUP and FKBP form a homopentamer (Edlich-Muth et al., 2015). Interestingly, the pentamer of the NPL domain is able to bind histones (Dutta et al., 2001), consistent with our IP-MS results showing that histones copurified with HD2C (Figure 4A). Furthermore, FKBP has conserved histidine and aspartate residues predicted to be critical for the catalytic activity of HD2 proteins (Aravind and Koonin, 1998; Edlich-Muth et al., 2015; Bourque et al., 2016). Besides the domain similarity, some FKBPs show similar cellular localization patterns and functions as HD2 proteins. Like HD2C and HD2B, human FKBP25 and Arabidopsis FKBP53 locate in the nucleolus and play important roles in ribosome biogenesis (Brown et al., 2005; Li and Luan, 2010; Gudavicius et al., 2014). Thus, we postulate that the nucleolus specific HD2 proteins may have evolved from the same ancestor as FKBPs.
Unlike animals that can escape from unfavorable environments, plants have evolved distinct mechanisms to cope with and thrive under such conditions. It is possible that HD2 proteins may have evolved to play important roles in the environmental adaption. Genetic studies showed that HD2 proteins are involved in various stress responses (Luo et al., 2012; Buszewicz et al., 2016; Han et al., 2016). Loss-of-function HD2C is hypersensitive to salt stress (Luo et al., 2012) and plants overexpressing HD2C confer resistance to ABA and drought treatments (Sridha and Wu, 2006). Overexpression of HD2D enhances salt and cold tolerance (Han et al., 2016). We observed upregulation of many ribosome biogenesis genes and downregulation of many stress response genes in hd2b hd2c (Supplemental Data Set 4). Interestingly, the expression levels of all four HD2 genes are downregulated under the ABA and salt stress condition (Luo et al., 2012). Given the positive role of HD2C and HD2B in rRNA maturation, the downregulation of HD2C and HD2B may reduce rRNA synthesis and subsequently slow down the cellular activity and development. It is possible that HD2 proteins may function to balance development and stress tolerance by coordinating rRNA synthesis and fine-tuning the expression of responsive genes.
One distinguishing feature of HD2s is their nucleolar localization. Interestingly, mammalian SIRT7 is located in the nucleolus in normal conditions and is released from the nucleoli upon stresses (Chen et al., 2016a). Like HD2C and HD2B (this study), SIRT7 plays an important role in ribosome biogenesis by regulating rRNAs processing (Chen et al., 2016a). Compared with seven mammalian SIRTs (Haigis and Guarente, 2006), there are only two SIRT proteins in Arabidopsis, rice (Oryza sativa), and maize (Zea mays) (Szućko, 2016). These observations raise an interesting hypothesis that HD2 proteins may have evolved to partially complement the missing SIRT proteins in plants.
Deacetylase Activity of HD2 Proteins
We provided evidence showing that loss-of-function HD2C induces H4K16 hyperacetylation in vivo (Figures 2 and 3). Surprisingly, our ChIP-seq experiments revealed a low overlap between HD2C-bound genes and H4K16 hyperacetylation genes in hd2c. While the precise mechanism is unclear, several possibilities could account for this low overlap. First, we noted that HD2C has additional substrates besides H4K16ac. Thus, HD2C may deacetylate other substrates at the nonoverlapping loci. Second, other HDACs might function redundantly with HD2C to modulate H4K16ac levels; thus, loss of HD2C itself is insufficient to induce H4K16ac hyperacetylation. Third, HD2C may bind to certain loci for other functions (e.g., protein-protein interaction) rather than for deacetylating its targets. Finally, some of the HD2C enriched loci may not be its bona fide targets.
On the other hand, although HD2s are classified into the histone deacetylase family, there is no direct evidence that these proteins are bona fide deacetylases. Current knowledge of HD2 substrates, activity, and functions are largely via genetic studies in hd2 mutant background (Ding et al., 2012; Luo et al., 2012; Buszewicz et al., 2016). Maize extracts enriched with HD2 proteins were used to measure the deacetylase activity (Brosch et al., 1996; Lusser et al., 1997). However, such study is indirect and does not rule out the possible involvement of other HDACs. In this study, we showed that overexpression of HD2C caused reduction of H4K16ac in N. benthamiana and depletion of HD2C induced global H4K16 hyperacetylation in Arabidopsis (Figure 2). Despite numerous attempts, we have been unable to find in vitro conditions to measure deacetylase activity of recombinant HD2C. Thus, it remains to be determined whether HD2C directly removes H4K16ac in vivo. One possibility is that HD2C action on H4K16ac may be mediated by the deacetylation activity of another HDAC(s) that forms complex with HD2C. A previous study using in vitro pull down or overexpressing HD2C in a N. benthamiana transient system reported that HD2C interacts with HDA6 (Luo et al., 2012). However, we did not detect HDA6 in our IP-MS when HD2C is expressed under the native promoter and at the physiologically relevant conditions. Our immunostaining experiment also does not support an active role of HDA6 in modulating H4K16ac levels (Figure 2). Instead, our IP-MS identified one peptide of Histone Deacetylase Complex1 (HDC1) that interacts with HDA6 and HDA19 to promote histone deacetylation (Perrella et al., 2013). Interestingly, human FKBP25 is also associated with Rpd3 like HDAC1 and HDAC2, which may contribute the deacetylation activity of enriched FKBP25 complex in vivo (Yang et al., 2001). Thus, future development of robust in vitro deacetylase activity assay is critical to determine whether HD2C and other HD2s are bona fide deacetylases.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the wild type for all experiments. The T-DNA insertion line of hd2c (SALK_129799) was obtained from the ABRC. Seeds were sown in soil or Murashige and Skoog medium and kept at 4°C for 2 d before transferring to 16-h light using a set of fluorescent lamps (Philips; 40 W) at around 100 μmol m−2 s−1 and 8-h dark at 22°C.
Construction of Vectors and Generation of Transgenic Plants
Genomic DNA of HD2C and HD2B with their 1-kb promoters were amplified and cloned into pENTR/D-TOPO (Thermo Fisher; K240020), using primers shown in Supplemental Table 2. These constructs were recombined into the pEarleyGate302 binary vectors to create epitope-tagged FLAG or HA fusions. For GFP-tagged proteins, cDNA of HD2C, HD2B, and HDA6 were amplified and cloned into pENTR/D-TOPO, then recombined into pEarleyGate103 binary vector. All constructs were then transformed by Agrobacterium tumefaciens-mediated infection into corresponding mutants or wild-type plants. CRISPR-Cas9 construct of HD2B was generated using the system provided by Yinong Yang (Xie et al., 2015). Briefly, two guide RNAs of HD2B were designed with online tool (Lei et al., 2014). The guide RNAs (Supplemental Table 2) were cloned into pRGE32 and transformed into wild-type Arabidopsis Col-0. T1 transgenic plants with HD2B mutations were selected for next generations. Seeds were collected from single branch of T-DNA free T2 plants containing HD2B mutation. T3 plants with HD2B mutations were then crossed back to wild-type Col-0 plants to exclude potential off-target mutations. HD2B homozygous mutants were obtained and used for all experiments.
RNA Extraction, Quantitative RT-PCR, and RNA Gel Blot Analysis
Total RNA was extracted from 6-d-old seedlings with Trizol reagent (Thermo Fisher; 15596026) and treated with DNase I (NEB; M0303S). One microgram of RNA was reverse-transcribed into cDNA with SuperScript III (Thermo Fisher; 18080093) followed by quantitative PCR assay with SYBR Green Master Mix using CFX96 Real-Time System 690 (Bio-Rad). Relative transcript level to ACTIN was calculated with the 2−ΔCT method (Livak and Schmittgen, 2001).
For rRNA methylation assay with low concentration of dNTP, 100 ng DNase I treated total RNA was reverse transcribed with M-MLV (Promega; M1701) in high dNTP (1 mM) and low dNTP (2 μM) condition, respectively. Relative RNA methylation level was calculated by normalizing qPCR values of low dNTP with values of high dNTP. For alkaline hydrolysis assay, 500 ng of RNA was treated in 50 mM Na2CO3 (pH 9) at 90°C for 4 min. RNAs were precipitated with ethanol and then served as a template for RT with rRNA specific primers. For RNA gel blots, seven micrograms of total RNA was separated with 1.5% agarose gel containing 2% formaldehyde and then transferred to a nylon membrane (GE Healthcare; RPN303B). Probe labeling and hybridization were performed with a DIG-High Prime DNA Labeling and Detection Starter Kit (Roche; 11585614910) according to the manufacturer’s instructions. Images were taken using ImageQuant LAS 4000 (GE Healthcare Life Sciences).
Immunostaining
Infiltrated Nicotiana benthamiana leaves were fixed in Tris buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10 mM EDTA) containing 4% paraformaldehyde for 20 min and then washed with Tris buffer three times. Nuclei were isolated from leaves by chopping in nuclei isolation buffer (15 mM Tris-HCl, pH 7.5, 2 mM EDTA, 0.5mM spermine, 80 mM KCl, 20 mM NaCl, and 0.1% Triton X-100). After filtering through Miracloth, the nuclei slurry was diluted three times with sorting buffer (100 mM Tri-HCl, pH 7.5, 50 mM KCl, 2 mM MgCl2, 0.05% Tween20, and 5% sucrose) and then dropped on poly-lysine-coated slides to air-dry. Slides were fixed again with 4% paraformaldehyde in wash buffer (PBS with 0.1% Triton X-100) for 20 min and then washed three additional times with wash buffer. The nuclei were blocked with 1% BSA in wash buffer for 30 min at 37°C and then washed three additional times with wash buffer. The nuclei were then incubated with primary antibodies with 1:200 dilution overnight at 4°C. After washing with PBS three times, the nuclei were blocked and incubated with a secondary antibody with 1:300 dilution for 2 to 4 h at 37°C. The slides were washed with PBS three times before imaging. Images were taken with a Nikon A1R confocal microscope. Acetylation antibodies used here were H3K9ac (Millipore; 07-352), H3K23ac (Millipore; 07-355), H4K8ac (Millipore; 07-328), H4K16ac (Millipore; 07-329), and H4ac (Cell Signaling; 39243).
Library Construction, Sequencing, and Data Analysis
RNA-seq and ChIP-seq libraries were constructed using a TruSeq RNA Library Preparation Kit (Illumina; RS-122-2002) and the Ovation Ultralow DR Multiplex System (NuGEN; 0330), respectively. Libraries were sequenced on a HiSeq 2500 in the UW-Madison Biotechnology Center. For RNA-seq, TopHat (2.0.8b) and Cufflinks (2.1.1) were used for differential expression analysis (Trapnell et al., 2012). The genes showing a P < 0.05 were considered as significantly differentially expressed. Two biological replicates were performed for RNA-seq. For ChIP-seq, reads were aligned to the Arabidopsis reference genome (TAIR10) using Bowtie2 (v2.1.0) with default parameters. Reads that mapped to identical positions in the genome were collapsed into one read. MACS (1.4.2) was used for peak calling with P < 1e-05. BEDTools (2.17.0) and custom PERL scripts were used for further analysis. In HD2C binding profiling, log2 value of normalized HD2C ChIP reads normalized by wild-type reads was calculated and binned in 100-kb intervals. All statistical analysis and figures were done using R (3.2.3). For RIP-seq, RNAs pulled down in HD2C-FLAG and the wild type were transferred to double-strand DNA and then treated as ChIP products for library construction. Reads were aligned to rRNA sequence first with Bowtie2 (v2.1.0). The unmapped reads were then aligned to TAIR10 reference sequence with TopHat (2.0.8b). For enrichment ratio, genes with <10 reads were filtered, then log2 value of normalized HD2C RIP reads divided by wild-type reads was calculated for each transcript. GO analysis was performed using agriGO (http://bioinfo.cau.edu.cn/agriGO/). The numbers of total reads obtained for each sample are listed in Supplemental Table 3.
ChIP
H4K16ac ChIP was performed as previously described (Lu et al., 2015). Two grams of leaves were ground into powder in liquid nitrogen and cross-linked in nuclei isolation buffer I (10 mM HEPES, pH 8, 1 M sucrose, 5 mM KCl, 5 mM MgCl2, 5 mM EDTA, 0.6% Triton X-100, 0.4 mM PMSF, and protease inhibitor cocktail tablet [Roche; 14696200]) with 1% formaldehyde for 20 min at room temperature. The homogenate was filtered through two layers of Miracloth (Millipore; 475855) and pelleted by centrifuging at 4000 rpm for 25 min at 4°C. The pellet was washed with nuclei isolation buffer II (0.25 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 1% Triton X-100, 1 mM EDTA, 5 mM β-mercaptoethanol, 0.4 mM PMSF, and protease inhibitor cocktail tablet), then resuspended with nuclear lysis buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS, 0.4 mM PMSF, and protease inhibitor cocktail tablet) and kept on ice for 10 min. The lysate was diluted 10-fold with ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8, 167 mM NaCl, 0.4 mM PMSF, and protease inhibitor cocktail tablet) and sheared by sonication. After centrifugation at 5000 rpm for 10 min, the supernatant was incubated with 5 μg antibody and 40 μL magnetic protein A/G beads (Life Technologies; 10004D) overnight with rotation at 4°C. After sequential washes with low salt buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8), high salt buffer (500 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8), LiCl buffer (0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8), and TE buffer (10 mM Tris-HCl, pH 8, and 1 mM EDTA), the DNA-protein complex was eluted with ChIP elution buffer (1% SDS and 0.1 M NaHCO3) and reverse cross-linked at 65°C for over 6 h. After proteinase K and RNase treatment, DNA was purified by standard phenol-chloroform method for qPCR.
HD2B-FLAG and HD2C-FLAG ChIP were performed as described previously (Chen et al., 2016b). Nuclei were isolated from 2 g of leaves and cross-linked using the same method as described above. After wash with nuclei isolation buffer II, the nuclei were resuspended with IP binding buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM MgCl2, 5% glycerol, 0.1% Nonidet P-40, 1 mM PMSF, and protease inhibitor cocktail tablet) and sheared by sonication. After centrifugation at 5000 rpm for 10 min, the supernatant was incubated with Anti-FLAG M2 magnetic beads (Sigma-Aldrich; M8823) overnight with rotation at 4°C. After wash with IP binding buffer containing 500 mM NaCl, the protein-DNA complex was eluted with ChIP elution buffer and reverse cross-linked at 65°C for 6 h. After proteinase K and RNase treatment, DNA was purified by standard phenol-chloroform method for qPCR analysis or sequencing.
IP-MS Analysis
Affinity purification and mass spectrometry analysis of HD2C-FLAG were performed as previously described (Chen et al., 2016b). Approximately 20 g of leaves from HD2C-FLAG or the wild type (negative control) were ground into powder and homogenized in 80 mL IP binding buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM MgCl2, 5% glycerol, 0.1% Nonidet P-40, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail tablet). After centrifugation at 10,000g for 15 min, the supernatant was incubated with anti-FLAG M2 magnetic beads (Sigma-Aldrich; M8823) with rotation at 4°C for 3 h. The bead-bound complex was washed four times with IP binding buffer at 4°C for 5 min each. Bound protein was released by two 10-min incubations with elution buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM MgCl2, 5% glycerol, 0.5 mM DTT, 1 mM PMSF, and protease inhibitor cocktail tablet) containing 150 ng/μL 3×FLAG peptide (Sigma-Aldrich; F4799) at room temperature. The eluted protein complexes were precipitated with trichloroacetic acid, further digested with Trypsin (Promega; V5111), and analyzed on an Orbitrap mass spectrometer (LTQ Velos; Thermo Fisher Scientific).
HPLC separation employed a 100 × 365-μm fused silica capillary microcolumn packed with 20 cm of 1.7-μm-diameter, 130 Å pore size, C18 beads (Waters BEH), with an emitter tip pulled to ∼1 μm using a laser puller (Sutter Instruments). Peptides were loaded on-column at a flow-rate of 400 nL/min for 30 min and then eluted over 120 min at a flow rate of 300 nL/min with a gradient of 2% to 30% acetonitrile, in 0.1% formic acid. Full-mass profile scans were performed in the FT Orbitrap between 300 and 1500 m/z at a resolution of 60,000, followed by 10 MS/MS HCD scans of the 10 highest intensity parent ions at 42% relative collision energy and 7500 resolution, with a mass range starting at 100 m/z. Dynamic exclusion was enabled with a repeat count of two over the duration of 30 s and an exclusion window of 120 s.
For data analysis, the acquired precursor MS and MS/MS spectra were searched against a Mus musculus protein database (Uniprot reviewed canonical database, containing 16,639 sequences) using SEQUEST, within the Proteome Discoverer 1.3.0.339 software package (Thermo Fisher Scientific). Masses for both precursor and fragment ions were treated as mono-isotopic. Oxidized methionine (+15.995 D) and the Gly-Gly footprint on lysine (+114.043 D) were allowed as dynamic modifications and carbamidomethylated cysteine (+57.021 D) was searched as a static modification. The database search permitted for up to two missed trypsin cleavages and ion masses were matched with a mass tolerance of 10 ppm for precursor masses and 0.1 D for HCD fragments. The output from the SEQUEST search algorithm was validated using the Percolator algorithm. The data were filtered using a 1% false discovery rate (Rohrbough et al., 2006), based on q-values, with a minimum of two peptide matches required for confident protein identification.
Co-IP Analysis
Co-IP was performed with 1.5 g of F1 Arabidopsis plants coexpressing HD2C-HA and HD2B-FLAG or 0.5 g infiltrated N. benthamiana leaves similar to IP-MS mentioned above. Total protein extracts were incubated with 20 μL FLAG magnetic beads for FLAG purification or 4 μg anti-GFP antibody (Roche; 11814460001) for GFP-tagged protein purification. After washing with IP buffer containing 300 mM NaCl four times, beads were boiled in SDS loading buffer.
In Vitro Protein-RNA Pull-Down
Ten micrograms of GST tagged protein was incubated with 20 μL GSH agarose bead slurry (GE Healthcare; 17-0756-01) for 1 h at room temperature. After washing with TBS three times, bead-protein complex was incubated with 5 μg total RNA in TBS containing 80 units/mL RNaseOUT (Thermo Fisher; 10777019) for 1 h at room temperature. After washing with TBS, half of the products were loaded to agarose gel for RNA detection. The other half products were boiled with SDS loading buffer for GST immunoblot.
RIP-Seq
RIP experiments were performed as previous described with slight modifications (Rowley et al., 2013; Xing et al., 2015; Böhmdorfer et al., 2016). One gram of leaves was ground into fine powder in liquid nitrogen and suspended with 15 mL nuclear isolation buffer as used in ChIP containing 8 units/mL RNaseOUT. The homogenate was cross-linked with 0.5% formaldehyde and filtered through two layers of Miracloth. The nuclei were collected by centrifuging at 3000g for 10 min and washed once with nuclei isolation buffer II as used in ChIP containing 8 units/mL RNaseOUT. The nuclei were resuspended in nuclear lysis buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, and 1% SDS) containing 160 units/mL RNaseOUT followed by sonication. After centrifuging at 16,000g for 10 min, the supernatant was diluted 10× with ChIP dilution buffer as used in ChIP containing 160 units/mL RNaseOUT and incubated with anti-FLAG M2 beads (Sigma-Aldrich; M8823) for 3 h with rotation. The bead-protein-RNA complex was washed with ChIP dilution buffer containing 300 mM NaCl and 80 units/mL RNaseOUT. Protein-RNA complex was eluted with elution buffer (100 mM Tris-HCl, pH 8, 10 mM EDTA, and 1% SDS) containing 800 units/mL RNaseOUT for 10 min at room temperature. The elution was repeated at 65°C for another 10 min. The elution was combined and treated with proteinase K for 1 h at 65°C. RNA was extracted with Trizol and treated with DNase I before library construction. The library was constructed with TruSeq RNA Library Preparation Kit (Illumina; RS-122-2002) and sequenced on a HiSeq 2500 in the UW-Madison Biotechnology Center.
Immunoblots
FLAG- and HA-tagged proteins were detected with horseradish peroxidase-conjugated anti-FLAG (Sigma-Aldrich; A8592) and anti-HA antibody (Roche; 12013819001), respectively. The GFP and GST antibodies were purchased from Roche (11814460001) and Thermo Fisher (CAB 4169), respectively. Immunoblots were developed using ECL Plus Western Blotting Detection System (GE Healthcare; RPN2132). For quantification of H4 and H4K16ac, a fluorescent secondary antibody was used (LI-COR; 92532211). Signal was detected with Odyssey infrared imaging system (LI-COR).
Protein Expression and Purification
The full-length cDNAs of HD2A, HD2B, HD2C, HD2D, and HDA6 were cloned into the pGOOD vector with a GST tag at the N terminus and a hexahistidine tag at the C terminus. The plasmid was transformed into Escherichia coli strain BL21 (DE3) RIL (Stratagene). The protein expression was induced by adding IPTG to the cell culture with a final concentration of 0.25 mM at OD600 of 0.8 at 20°C. The protein was purified using HisTrp, Heparin, Q Fastflow, and Superdex G200 columns (GE Healthcare). The purified protein was concentrated to 20 mg/mL and frozen in −80°C for further use.
Accession Numbers
RNA-seq, ChIP-seq, and RIP-seq data were deposited in the Gene Expression Omnibus under accession number GSE107883. The accession numbers for genes are as follows: HD2A (AT3G44750), HD2B (AT5G22650), HD2C (AT5G03740), HD2D (AT2G27840), and HDA6 (AT5G63110).
Supplemental Data
Supplemental Figure 1. Validation of hd2c T-DNA Mutant and HD2C-FLAG Transgenic Plants.
Supplemental Figure 2. List of Ribosome Biogenesis Related Genes with Both HD2C Binding and H4K16 Hyperacetylation in hd2c.
Supplemental Figure 3. Validation of HD2B-FLAG Transgenic Plant and Amino Acid Sequence Alignment of HD2 Protein in Plants.
Supplemental Figure 4. Validation of hd2b hd2c Double Mutant and Rescued Plants.
Supplemental Figure 5. Sequencing Results of Overaccumulated cRT-PCR Products in hd2b hd2c.
Supplemental Figure 6. HD2C Is Associated with Pre-rRNAs and SnoRNAs.
Supplemental Table 1. List of Ribosome Biogenesis Related Genes with Both HD2C Binding and Upregulation in hd2b hd2c.
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Read Numbers of ChIP-seq, RNA-seq, and RIP-seq.
Supplemental Data Set 1. List of HD2C-Bound Genes.
Supplemental Data Set 2. List of Genes with Increased H4K16ac in hd2c.
Supplemental Data Set 3. List of Proteins Identified by IP-MS of HD2C.
Supplemental Data Set 4. Differentially Expressed Genes in the hd2b hd2c Double Mutant.
Supplemental Data Set 5. List of HD2C-Bound RNAs.
Acknowledgments
We thank the UW-Madison Biotech Center for high-throughput sequencing. We also thank Yinong Yang at Pennsylvania State University for providing the CRISPR-Cas9 system. Work in X.Z.’s laboratory was supported by the Alexander von Humboldt Foundation (Alfred Toepfer Faculty Fellow), by an NSF CAREER award (MCB-1552455), and by NIH-NIGMS (R35GM124806). Work in L.M.S.’s laboratory was supported by NIH-NIGMS (R01GM114292).
AUTHOR CONTRIBUTIONS
X.C. and X.Z. designed this study. X.C. performed all experiments except for the mass spectrometry analysis. L.L. performed all bioinformatics analysis. S.Q. provided research materials and experimental supports. M.S. and L.M.S. performed mass spectrometry analysis. X.C. and X.Z. analyzed the data and wrote the manuscript.
References
- Abbasi N., Kim H.B., Park N.I., Kim H.S., Kim Y.K., Park Y.I., Choi S.B. (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64: 960–976. [DOI] [PubMed] [Google Scholar]
- Ali S.A., Dobson J.R., Lian J.B., Stein J.L., van Wijnen A.J., Zaidi S.K., Stein G.S. (2012). A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation. J. Cell Sci. 125: 2732–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V., Koonin E.V., Aravind L. (2002). SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 4: 71–75. [PubMed] [Google Scholar]
- Aravind L., Koonin E.V. (1998). Second family of histone deacetylases. Science 280: 1167a. [Google Scholar]
- Bachand F. (2007). Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot. Cell 6: 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J.P., Cavaillé J., Hüttenhofer A. (2002). The expanding snoRNA world. Biochimie 84: 775–790. [DOI] [PubMed] [Google Scholar]
- Böhmdorfer G., Sethuraman S., Rowley M.J., Krzyszton M., Rothi M.H., Bouzit L., Wierzbicki A.T. (2016). Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. eLife 5: e19092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegué L., Vaquero A. (2015). Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 282: 1745–1767. [DOI] [PubMed] [Google Scholar]
- Bourque S., et al. (2016). The evolution of HD2 proteins in green plants. Trends Plant Sci. 21: 1008–1016. [DOI] [PubMed] [Google Scholar]
- Bratkovič T., Rogelj B. (2014). The many faces of small nucleolar RNAs. Biochim. Biophys. Acta 1839: 438–443. [DOI] [PubMed] [Google Scholar]
- Brosch G., Lusser A., Goralik-Schramel M., Loidl P. (1996). Purification and characterization of a high molecular weight histone deacetylase complex (HD2) of maize embryos. Biochemistry 35: 15907–15914. [DOI] [PubMed] [Google Scholar]
- Brown J.W., Shaw P.J., Shaw P., Marshall D.F. (2005). Arabidopsis nucleolar protein database (AtNoPDB). Nucleic Acids Res. 33: D633–D636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszewicz D., et al. (2016). HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 39: 2108–2122. [DOI] [PubMed] [Google Scholar]
- Byrne M.E. (2009). A role for the ribosome in development. Trends Plant Sci. 14: 512–519. [DOI] [PubMed] [Google Scholar]
- Chen S., Blank M.F., Iyer A., Huang B., Wang L., Grummt I., Voit R. (2016a). SIRT7-dependent deacetylation of the U3-55k protein controls pre-rRNA processing. Nat. Commun. 7: 10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Lu L., Mayer K.S., Scalf M., Qian S., Lomax A., Smith L.M., Zhong X. (2016b). POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5: e17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Bellizzi Mdel.R., Ning Y., Meyers B.C., Wang G.L. (2012). HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24: 3783–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z.W., Shao P., Diao L.T., Zhou H., Yu C.H., Qu L.H. (2012). RTL-P: a sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 40: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., Rath V.L. (2002). Structure and function of the eukaryotic ribosome: the next frontier. Cell 109: 153–156. [DOI] [PubMed] [Google Scholar]
- Durut N., et al. (2014). A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis. Plant Cell 26: 1330–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S., Akey I.V., Dingwall C., Hartman K.L., Laue T., Nolte R.T., Head J.F., Akey C.W. (2001). The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8: 841–853. [DOI] [PubMed] [Google Scholar]
- Earley K., Lawrence R.J., Pontes O., Reuther R., Enciso A.J., Silva M., Neves N., Gross M., Viegas W., Pikaard C.S. (2006). Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20: 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Pontvianne F., Wierzbicki A.T., Blevins T., Tucker S., Costa-Nunes P., Pontes O., Pikaard C.S. (2010). Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 24: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlich-Muth C., Artero J.B., Callow P., Przewloka M.R., Watson A.A., Zhang W., Glover D.M., Debski J., Dadlez M., Round A.R., Forsyth V.T., Laue E.D. (2015). The pentameric nucleoplasmin fold is present in Drosophila FKBP39 and a large number of chromatin-related proteins. J. Mol. Biol. 427: 1949–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.C., Brown D.D., Brown J.W. (2010). The small nucleolar ribonucleoprotein (snoRNP) database. RNA 16: 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L. (2006). Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frehlick L.J., Eirín-López J.M., Ausió J. (2007). New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. BioEssays 29: 49–59. [DOI] [PubMed] [Google Scholar]
- Goodfellow S.J., Zomerdijk J.C. (2013). Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell. Biochem. 61: 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi K.M., Lindström M.S. (2016). Role of ribosomal protein mutations in tumor development (Review). Int. J. Oncol. 48: 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Pikaard C.S. (2003). Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4: 641–649. [DOI] [PubMed] [Google Scholar]
- Gudavicius G., Dilworth D., Serpa J.J., Sessler N., Petrotchenko E.V., Borchers C.H., Nelson C.J. (2014). The prolyl isomerase, FKBP25, interacts with RNA-engaged nucleolin and the pre-60S ribosomal subunit. RNA 20: 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Montgomery R.L., Olson E.N. (2009). The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis M.C., Guarente L.P. (2006). Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20: 2913–2921. [DOI] [PubMed] [Google Scholar]
- Han Z., Yu H., Zhao Z., Hunter D., Luo X., Duan J., Tian L. (2016). AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front. Plant Sci. 7: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang R., Liu C., Ahmad A., Zhang Y., Lu F., Cao X. (2014). Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc. Natl. Acad. Sci. USA 111: 16190–16195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E. (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa-Nakamine S., Suzuki T., Uechi T., Chakraborty A., Nakajima Y., Nakamura M., Hirano N., Suzuki T., Kenmochi N. (2012). Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 40: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C., Liu Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50: 875–885. [DOI] [PubMed] [Google Scholar]
- Ide S., Miyazaki T., Maki H., Kobayashi T. (2010). Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696. [DOI] [PubMed] [Google Scholar]
- Ito T., Kim G.T., Shinozaki K. (2000). Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 22: 257–264. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Kim S., Shin Y.J., Hur Y.S., Kim W.Y., Lee M.S., Cheon C.I., Verma D.P. (2014). Ribosomal protein S6, a target of rapamycin, is involved in the regulation of rRNA genes by possible epigenetic changes in Arabidopsis. J. Biol. Chem. 289: 3901–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2002). Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T. (1996). Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kojima H., Suzuki T., Kato T., Enomoto K., Sato S., Kato T., Tabata S., Sáez-Vasquez J., Echeverría M., Nakagawa T., Ishiguro S., Nakamura K. (2007). Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 49: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L. (2015). Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22: 11–19. [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Pikaard C.S. (2004). Chromatin turn ons and turn offs of ribosomal RNA genes. Cell Cycle 3: 880–883. [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609. [DOI] [PubMed] [Google Scholar]
- Lei Y., Lu L., Liu H.Y., Li S., Xing F., Chen L.L. (2014). CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7: 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li H., Luan S. (2010). AtFKBP53 is a histone chaperone required for repression of ribosomal RNA gene expression in Arabidopsis. Cell Res. 20: 357–366. [DOI] [PubMed] [Google Scholar]
- Liang W.Q., Fournier M.J. (1995). U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 9: 2433–2443. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lu L., Chen X., Sanders D., Qian S., Zhong X. (2015). High-resolution mapping of H4K16 and H3K23 acetylation reveals conserved and unique distribution patterns in Arabidopsis and rice. Epigenetics 10: 1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Guan X., Schmidt C.A., Matera A.G. (2014). RIP-seq analysis of eukaryotic Sm proteins identifies three major categories of Sm-containing ribonucleoproteins. Genome Biol. 15: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Wang Y.Y., Liu X., Yang S., Lu Q., Cui Y., Wu K. (2012). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63: 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A., Brosch G., Loidl A., Haas H., Loidl P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277: 88–91. [DOI] [PubMed] [Google Scholar]
- Mohannath G., Pontvianne F., Pikaard C.S. (2016). Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon. Proc. Natl. Acad. Sci. USA 113: 13426–13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D.R., Kuhn J.F., Shanab G.M., Maxwell E.S. (2000). Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6: 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella G., Lopez-Vernaza M.A., Carr C., Sani E., Gosselé V., Verduyn C., Kellermeier F., Hannah M.A., Amtmann A. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25: 3491–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J.J., Nelson T.M. (2007). Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 144: 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Lawrence R.J., Silva M., Preuss S., Costa-Nunes P., Earley K., Neves N., Viegas W., Pikaard C.S. (2007). Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS One 2: e1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Matía I., Douet J., Tourmente S., Medina F.J., Echeverria M., Sáez-Vásquez J. (2007). Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell 18: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Blevins T., Chandrasekhara C., Feng W., Stroud H., Jacobsen S.E., Michaels S.D., Pikaard C.S. (2012). Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 26: 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Blevins T., Chandrasekhara C., Mozgová I., Hassel C., Pontes O.M., Tucker S., Mokros P., Muchová V., Fajkus J., Pikaard C.S. (2013). Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 27: 1545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., et al. (2010). Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss S.B., Costa-Nunes P., Tucker S., Pontes O., Lawrence R.J., Mosher R., Kasschau K.D., Carrington J.C., Baulcombe D.C., Viegas W., Pikaard C.S. (2008). Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol. Cell 32: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., Vaucheret H., Mittelsten Scheid O. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L.H., Meng Q., Zhou H., Chen Y.Q. (2001). Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 29: 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo S., Idol R.A., Crimmins D.L., Ladenson J.H., Mason P.J., Bessler M. (2008). The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14: 1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbough J.G., Breci L., Merchant N., Miller S., Haynes P.A. (2006). Verification of single-peptide protein identifications by the application of complementary database search algorithms. J. Biomol. Tech. 17: 327–332. [PMC free article] [PubMed] [Google Scholar]
- Rowley M.J., Böhmdorfer G., Wierzbicki A.T. (2013). Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods 63: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vasquez J., Caparros-Ruiz D., Barneche F., Echeverría M. (2004). A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 24: 7284–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Kaarniranta K. (2009). SIRT1 regulates the ribosomal DNA locus: epigenetic candles twinkle longevity in the Christmas tree. Biochem. Biophys. Res. Commun. 378: 6–9. [DOI] [PubMed] [Google Scholar]
- Seto E., Yoshida M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski P.J., Zuber H., Philippe L., Sement F.M., Canaday J., Kufel J., Gagliardi D., Lange H. (2015). Distinct 18S rRNA precursors are targets of the exosome complex, the exoribonuclease RRP6L2 and the terminal nucleotidyltransferase TRL in Arabidopsis thaliana. Plant J. 83: 991–1004. [DOI] [PubMed] [Google Scholar]
- Sridha S., Wu K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46: 124–133. [DOI] [PubMed] [Google Scholar]
- Szućko I. (2016). Sirtuins: not only animal proteins. Acta Physiol. Plant. 38: 237. [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.C., Greco T.M., Cristea I.M. (2014). Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol. Cell. Proteomics 13: 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.C., Greco T.M., Boonmee A., Miteva Y., Cristea I.M. (2012). Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol. Cell. Proteomics 11: 60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I., Tutois S., Cuvillier C., Schubert I., Tourmente S. (2007). Regulation of Arabidopsis thaliana 5S rRNA Genes. Plant Cell Physiol. 48: 745–752. [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M., Vanderhaeghen R., De Block M., Bauw G., Villarroel R., Van Montagu M. (1994). An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 13: 3378–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R., Seiler J., Grummt I. (2015). Cooperative action of Cdk1/cyclin B and SIRT1 is required for mitotic repression of rRNA synthesis. PLoS Genet. 11: e1005246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Nag S., Zhang X., Wang M.H., Wang H., Zhou J., Zhang R. (2015). Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med. Res. Rev. 35: 225–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Franke-van Dijk M., Vencken R.J., Quint A., Hooykaas P., Offringa R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128: 4289–4299. [DOI] [PubMed] [Google Scholar]
- Weis B.L., Kovacevic J., Missbach S., Schleiff E. (2015). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20: 729–740. [DOI] [PubMed] [Google Scholar]
- Woolford J.L. Jr., Baserga S.J. (2013). Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Minkenberg B., Yang Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 112: 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Wang Y., Hamilton M., Ben-Hur A., Reddy A.S. (2015). Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 27: 3294–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Song T., Chen L., Kabra N., Zheng H., Koomen J., Seto E., Chen J. (2013). Regulation of SirT1-nucleomethylin binding by rRNA coordinates ribosome biogenesis with nutrient availability. Mol. Cell. Biol. 33: 3835–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.M., Yao Y.L., Seto E. (2001). The FK506-binding protein 25 functionally associates with histone deacetylases and with transcription factor YY1. EMBO J. 20: 4814–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.J., Seto E. (2003). Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr. Opin. Genet. Dev. 13: 143–153. [DOI] [PubMed] [Google Scholar]
- Zhou C., Labbe H., Sridha S., Wang L., Tian L., Latoszek-Green M., Yang Z., Brown D., Miki B., Wu K. (2004). Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 38: 715–724. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Santoro R., Grummt I. (2002). The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21: 4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]