The structure of the Arabidopsis JMJ14 catalytic domain in complex with H3K4me3 peptide reveals a conserved substrate binding mode shared by both plant and animal KDM5 subfamily histone demethylase.
Abstract
In chromatin, histone methylation affects the epigenetic regulation of multiple processes in animals and plants and is modulated by the activities of histone methyltransferases and histone demethylases. The jumonji domain-containing histone demethylases have diverse functions and can be classified into several subfamilies. In humans, the jumonji domain-containing Lysine (K)-Specific Demethylase 5/Jumonji and ARID Domain Protein (KDM5/JARID) subfamily demethylases are specific for histone 3 lysine 4 trimethylation (H3K4me3) and are important drug targets for cancer treatment. In Arabidopsis thaliana, the KDM5/JARID subfamily H3K4me3 demethylase JUMONJI14 (JMJ14) plays important roles in flowering, gene silencing, and DNA methylation. Here, we report the crystal structures of the JMJ14 catalytic domain in both substrate-free and bound forms. The structures reveal that the jumonji and C5HC2 domains contribute to the specific recognition of the H3R2 and H3Q5 to facilitate H3K4me3 substrate specificity. The critical acidic residues are conserved in plants and animals with the corresponding mutations impairing the enzyme activity of both JMJ14 and human KDM5B, indicating a common substrate recognition mechanism for KDM5 subfamily demethylases shared by plants and animals and further informing efforts to design targeted inhibitors of human KDM5.
INTRODUCTION
Histone modifications represent a pivotal epigenetic regulatory pathway that plays essential roles in various eukaryotic biological processes. Among the diverse histone modifications, lysine methylation is one of the most studied and characterized histone modification types (Kouzarides, 2007). The dynamic regulation of lysine methylation requires specific writer, reader, and eraser proteins to generate, recognize, and remove the modification in both modification-specific and sequence-specific manners. Generally, there are two types of histone lysine methylation erasers: the FAD-dependent KDM1/LSD family demethylases, which are specific to histone 3 lysine 4 dimethylation or monomethylation (H3K4me2/1) and H3K9me2/1, and the α-ketoglutarate- (αKG) and Fe2+-dependent jumonji domain-containing demethylases, which have more diverse specificities, sequences, and functions (Mosammaparast and Shi, 2010). The jumonji family demethylases can be classified into several subfamilies, including the KDM5/JARID subfamily of H3K4me3 site-specific demethylases (Mosammaparast and Shi, 2010).
In humans, the H3K4me3-specific KDM5/JARID subfamily demethylases play an important role in cancer cell proliferation, suppression of tumor-suppressor genes, and promotion of drug tolerance of cancer cells (Rasmussen and Staller, 2014; Taylor-Papadimitriou and Burchell, 2017). Therefore, KDM5/JARID subfamily demethylases are potent drug targets for anticancer drug design, and several inhibitors have been developed to inhibit the activity of human KDM5s as potent anticancer drug candidates (Bavetsias et al., 2016; Horton et al., 2016a, 2016b; Johansson et al., 2016; Labadie et al., 2016; Vinogradova et al., 2016; Taylor-Papadimitriou and Burchell, 2017; Tumber et al., 2017). However, the substrate recognition mechanism for KDM5 demethylases is still unknown, due to the lack of KDM5-H3K4me3 substrate complex structures, which hampers the structure-based design and development of KDM5 targeting drugs, especially KDM5-specific inhibitors.
In plants, histone demethylases show similarities to their animal counterparts but also possess plant-specific features (Lu et al., 2008; Liu et al., 2010). Arabidopsis thaliana JUMONJI14 (AtJMJ14) is a KDM5 subfamily H3K4me3 demethylase that regulates plant flowering time, RNA silencing, and RNA-directed DNA methylation (Lu et al., 2008, 2010; Jeong et al., 2009; Deleris et al., 2010; Searle et al., 2010; Yang et al., 2010; Le Masson et al., 2012; Greenberg et al., 2013). The execution of JMJ14 function requires targeting of the enzyme to certain chromatin loci and subsequent triggering of the demethylation reaction. This process contains two steps: first, the recruitment of the enzyme to the determined chromatin region, and second, the specific capturing of the substrate H3K4me3 by the catalytic domain. JMJ14 is able to use its C-terminal FYR domain to interact with two DNA sequence-specific NAC family transcription factors and is subsequently targeted to certain chromatin regions (Ning et al., 2015; Zhang et al., 2015). However, the molecular mechanism of the specific recognition of H3K4me3 substrate by its catalytic domain is still unknown. Moreover, the mechanism for H3K4me3 substrate specificity of the KDM5 subfamily, either in animals or plants, is still unknown due to the lack of structures of enzyme-H3K4me3 substrate complexes. Here, we report the crystal structure of JMJ14 catalytic domain in both free and substrate-bound forms. The structural analysis indicates that conserved acidic residues in the jumonji and C5HC2 domains are involved in recognition of H3R2 and H3Q5 and play an essential role in the substrate selectivity, which was further confirmed by our in vitro and in vivo enzymatic assays. This specific recognition is conserved not only in plants but also in the animal KDM5 subfamily demethylases, as confirmed by our in vitro activity assay on human KDM5B, suggesting a common KDM5 substrate selection mechanism and providing insight into targeted design of human KDM5-specific inhibitors.
RESULTS
JMJ14 Is an H3K4me3/2 Demethylase
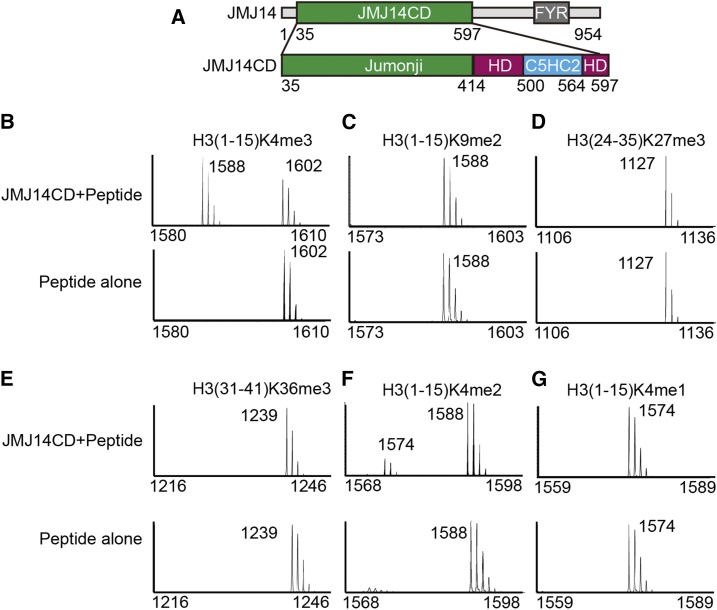
JMJ14 belongs to the KDM5/JARID subfamily and therefore was considered to be an H3K4me3 demethylase similar to other known KDM5/JARID family proteins (Lu et al., 2008). JMJ14 can specifically demethylate H3K4me3/2/1, as observed indirectly by immunostaining-based assays (Jeong et al., 2009; Lu et al., 2010; Yang et al., 2010). In an effort to prevent antibody-introduced bias, we performed a MALDI-TOF mass spectrometry-based histone demethylation assay, which can monitor the demethylation reaction directly based on the molecular weight changes of the substrate peptides. We constructed the JMJ14 N-terminal catalytic domain (residues 35–597, designated JMJ14CD), which contains the jumonji, helical, and C5HC2 zinc finger domains (Figure 1A). The bacterial-expressed JMJ14CD showed unambiguous demethylase activity against H3K4me3 but not H3K9me2, H3K27me3, and H3K36me3 (Figures 1B to 1E). Furthermore, the demethylase assay indicates that JMJ14 can efficiently demethylate both H3K4me3 and H3K4me2 but without prominent activity against H3K4me1 (Figures 1B, 1F, and 1G).
Figure 1.
JMJ14 Is an H3K4me3/2 Demethylase.
(A) A schematic representation of the domain architecture of Arabidopsis JMJ14 (upper panel) and the JMJ14 catalytic domain (lower panel) that was used in this research. CD, catalytic domain; HD, helical domain.
(B) to (G) The MOLDI-TOF-based activity assay for JMJ14CD against histone peptides H3(1-15)K4me3 (B), H3(1-15)K9me2 (C), H3(24-35)K27me3 (D), H3(31-41)K36me3 (E), H3(1-15)K4me2 (F), and H3(1-15)K4me1 (G) with the upper and lower panels showing JM14CD plus peptide and peptide alone, respectively. The molecular weights corresponding to the peaks are labeled. The results show that JMJ14 is predominantly an H3K4me3/2 demethylase.
Structure of JMJ14CD
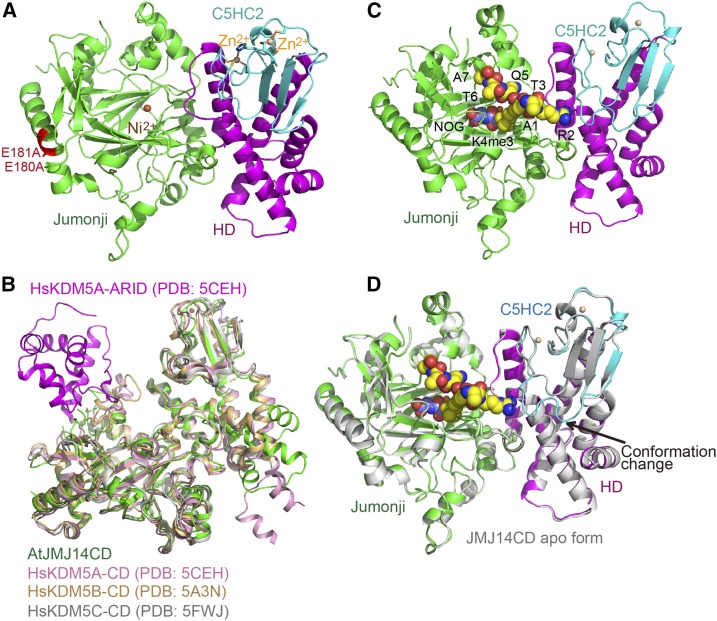
To further investigate the molecular mechanism of H3K4me3 site-specific demethylation by JMJ14, we performed structural studies. As the wild-type protein only produced low-quality crystals, we employed a surface entropy reduction approach to obtain crystals with higher diffraction quality (Goldschmidt et al., 2007). We determined the crystal structure of the substrate-free form JMJ14CD E180A/E181A mutant at 2.3-Å resolution (Figure 2A, Table 1). The mutation sites are located on a solvent-exposed α-helix, which is positioned away from the catalytic center and substrate binding site (Figure 2A). Therefore, we believe that the double mutant does not affect the catalytic function of JMJ14CD and we will not distinguish between this mutant and the wild-type protein in the following discussion.
Figure 2.
Overall Structure of JMJ14CD in Apo- and Substrate-Bound Forms.
(A) Overall structure of JMJCD in a ribbon diagram with the jumonji, helical, and C5HC2 zinc binding domains colored in green, magenta, and cyan, respectively. The nickel and zinc ions were shown in orange and tan balls, respectively. The two surface mutations E180A and E181A are highlighted in red with their side chains shown in stick representation.
(B) Structural comparison of JMJ14 and human KDM5 proteins. The structures of AtJMJ14CD, human KDM5A catalytic domain (HsKDM5A-CD, PDB code: 5CEH), human KDM5B catalytic domain (HsKDM5B-CD, PDB code: 5A3N), and human KDM5C catalytic domain (HsKDM5C-CD, PDB code: 5FWJ) are superimposed together based on their overall structures and are colored in green, pink, orange, and silver, respectively. KDM5A has an ARID domain budding out from the jumonji domain, which is highlighted in magenta.
(C) Overall structure of JMJ14CD-NOG-H3K4me3 complex. The NOG and H3K4me3 are shown in space-filling representation.
(D) A superimposition of JMJ14CD in free form (in silver) and in substrate-bound form (in colored scheme) showing almost identical overall structures. The most significant difference is a loop that is disordered in the structure of the free from but ordered in the substrate bound form. The loop is indicated by an arrow.
Table 1. Data Collection and Refinement Statistics.
| JMJ14CD | JMJ14CD-NOG-H3K4me3 | |
|---|---|---|
| Data collection | ||
| Beamline | SSRF-BL19U1 | SSRF-BL19U1 |
| PDB code | 5YKN | 5YKO |
| Space group | P21 | P212121 |
| Wavelength (Å) | 0.9793 | 0.9793 |
| Cell dimensions | ||
| a, b, c (Å) | 51.1, 69.2, 100.4 | 68.6, 100.1, 109.4 |
| α, β, γ (°) | 90, 104.3, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0–2.3 (2.38–2.30)a | 50.0–2.9 (3.00–2.90) |
| Rmerge | 0.126 (0.952) | 0.161 (0.776) |
| I/σI | 16.0 (1.4) | 13.6 (2.9) |
| Completeness (%) | 99.0 (97.4) | 99.3 (98.6) |
| Redundancy | 3.6 (3.2) | 4.9 (5.0) |
| CC1/2 | 0.567 | 0.810 |
| Refinement | ||
| No. reflections | 30,075 | 17,179 |
| Rwork/Rfree | 0.194/0.222 | 0.204/0.244 |
| No. atoms | 4,022 | 4,031 |
| Protein/peptide | 3,885/– | 3,962/56 |
| Ni2+/Zn2+ | 1/2 | 1/2 |
| Water/NOG | 134/– | – /10 |
| B-factors (Å2) | 56.7 | 45.4 |
| Protein/peptide | 56.8/– | 44.6/95.7 |
| Ni2+/Zn2+ | 103.6/71.6 | 73.9/57.4 |
| Water/NOG | 54.0/– | –/49.5 |
| RMSDsb | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Bond angles (°) | 0.851 | 0.939 |
| Ramachandran plotc | ||
| Favored (%) | 96 | 98 |
| Allowed (%) | 4 | 2 |
| Outlier (%) | 0 | 0 |
Highest resolution shell is shown in parentheses.
RMSDs, root mean square deviations.
Ramachandran plot was calculated using the program Molprobity (Chen et al., 2010).
The structure of JMJ14CD is composed of the jumonji, helical, and C5HC2 zinc binding domains (Figure 2A). The three domains are arranged together with the jumonji domain positioned on one side and the helical plus C5HC2 zinc finger domains positioned on the other side. The jumonji domain adopts a typical αKG-dependent oxygenase superfamily fold (Figure 2A). The helical domain is composed of six α-helices to form a helical bundle-like structure (Figure 2A). Although the primary sequence of C5HC2 domain is inside the helical domain (Figure 1A), structurally, it buds out from the helical domain and forms an independent domain that stacks with the helical domain (Figure 2A). Two Zn2+ ions are coordinated within the C5HC2 domain (Figure 2A).
Overall, the JMJ14CD structure resembles reported human KDM5 structures, with root mean square deviations of 1.7, 1.8, and 1.3 Å to the overall structures of human KDM5A, KDM5B, and KDM5C, respectively, upon superimposition (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017) (Figure 2B). A substantial difference is that the human KDM5 proteins have inserted ARID and PHD domains inside the primary sequence of the jumonji domain. However, the ARID and PHD domains protrude out from the jumonji domain in these structures, which leaves the jumonji-helical-C5HC2 catalytic cassette of human KDM5 as a completely independent structural unit (Figure 2B) (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017). The inserted ARID and PHD domains are conserved in some but not all plant KDM5 demethylases (Lu et al., 2008). In our JMJ14CD structure, the region corresponding to human KDM5 ARID-PHD cassette (residues 125–168) has no annotated domain and is disordered in the structure. Nevertheless, the structures of the catalytic domain (including jumonji, helical, and C5HC2 domains) of JMJ14 and human KDM5A/B/C adopt similar folds, indicating a plausible common substrate binding and catalytic mechanism (Figure 2B).
Structure of the JMJ14CD-NOG-H3K4me3 Complex
We further obtained the crystal structure of JMJ14CD in complex with substrates by soaking the JMJ14CD crystals with N-oxalylglycine (NOG), an analog of αKG, and H3(1-10)K4me3 peptide. The structure was determined to 2.9-Å resolution (Figure 2C, Table 1). The substrates bound structure is quite similar to the apo form structure with an root mean square deviation of 0.40 Å for 487 aligned Cαs upon superimposition (Figure 2D). The most significant difference is the region from Glu-508 to Glu-516 of the C5HC2 domain, which is disordered in the apo JMJ14CD structure but forms an ordered conformation in the structure of the complex, which is probably due to the interactions with the peptide (Figure 2D). The peptide can be traced from H3A1 to H3A7 and adopts an extended conformation (Figure 2C; Supplemental Figure 1).
Interaction between JMJ14 and the H3K4me3 Peptide
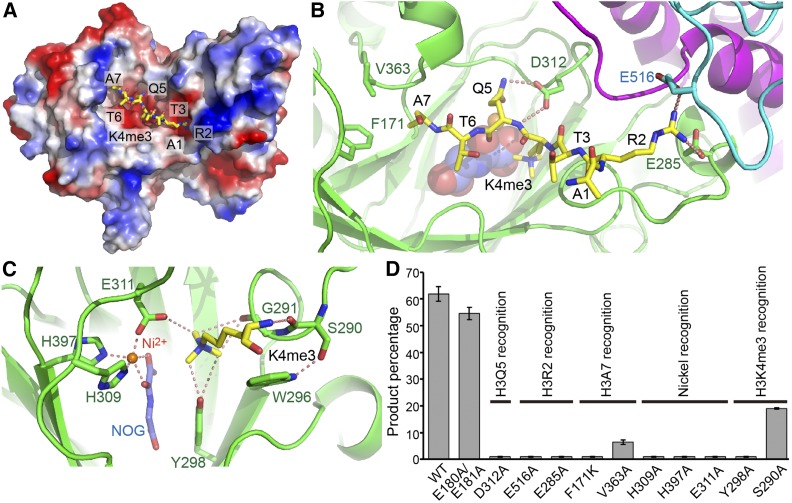
The peptide docks into a negatively-charged surface cleft of JMJ14 (Figure 3A). The intermolecular interactions between JMJ14 and H3K4me3 primarily involve H3R2, H3K4me3, H3Q5, and H3A7, while other residues only contribute to surface contacts but not specific interactions. The guanidino group of H3R2 inserts into a negatively charged pocket at the junction between the jumonji, helical, and C5HC2 domains and makes contacts with all the three domains (Figures 3A and 3B). The guanidino group of H3R2 forms hydrogen bonding and salt bridge interactions with two negatively charged residues, Glu-285 of the jumonji domain and Glu-516 of the C5HC2 domain (Figure 3B). The H3Q5 extends its side chain into the jumonji domain (Figures 3A and 3B). The jumonji domain residue Asp-312 forms two hydrogen bonds with the main chain and side chain amide groups of H3Q5, respectively (Figure 3B). The methyl group of H3A7 is positioned into a shallow hydrophobic pocket formed by Phe-171 and Val-363 of the JMJ14 jumonji domain (Figure 3B). H3A1, H3T3, and H3T6 direct their side chains opposite JMJ14 and have no direct interaction with the protein.
Figure 3.
Interactions between JMJ14 and H3K4me3 Peptide.
(A) An electrostatics surface view of JMJ14CD with the H3K4me3 peptide shown in stick representation. The peptide docks in a negatively charged surface cleft of JMJ14.
(B) The specific recognition of H3R2, H3Q5, and H3A7 by JMJ14. The H3R2 side chain inserts into a binding pocket at the junction formed by jumonji, helical, and C5HC2 domains and forms hydrogen bonding and salt bridge interactions with Glu-285 and Glu-516. The hydrogen bonds are highlighted by dashed red lines. H3Q5 forms two hydrogen bonds with Asp-312. The side chain methyl group of H3A7 inserts into a shallow hydrophobic pocket formed by Phe-171 and Val-363.
(C) The specific recognition of H3K4me3 and the active site conformation. The H3K4me3 main chain forms a hydrogen bond with Ser-290. The methyl groups of the trimethyllysine are specifically recognized by a CH-O hydrogen bonding network. The nickel ion is coordinated by His-309, Glu-311, His-397, and the cofactor analog NOG.
(D) The quantified in vitro demethylation assay of JMJ14CD and its mutants. The percentages of the product peptide are shown as means ± sd (n = 3). The functional annotations of the mutation sites are indicated above the transverse lines. Mutations of important residues involved in substrate binding are shown to significantly impair activity.
The Active Site Conformation and Recognition of Methyllysine
The side chain of H3K4me3 is specifically anchored into the active site of the jumonji domain through extensive interactions. The main chain amide group has a hydrogen bonding interaction with the main chain carbonyl of Ser-290 (Figure 3C). The aliphatic portion of the methyllysine side chain forms hydrophobic contacts with Trp-296, whose conformation is stabilized by a side chain hydrogen bond with Ser-290 (Figure 3C). Similar to what has been observed in the classic model of JMJD2A (Chen et al., 2007; Couture et al., 2007; Ng et al., 2007), Gly-291, Tyr-298, and Glu-311 of JMJ14 each have an oxygen atom to form CH-O hydrogen bonds with the methyl groups of trimethyllysine (Figure 3C), resulting in an extensive hydrogen bond network to coordinate the trimethyllysine in a fixed conformation, confirming the preference of H3K4me3 substrate over H3K4me2 by JMJ14 (Figures 1B and 1F). The Ni2+ ion and NOG are specifically coordinated by surrounding residues, similar to what has been observed for other αKG-dependent histone demethylases such as JMJD2A, UTX, JMJD3, and KDM2A (Chen et al., 2007; Couture et al., 2007; Ng et al., 2007; Sengoku and Yokoyama, 2011; Kruidenier et al., 2012; Cheng et al., 2014) (Figure 3C).
We further performed mass spectrometry-based in vitro enzymatic assays on mutations of critical residues involved in catalysis and substrate recognition of JMJ14 (Figure 3D). The E180A/E181A double mutation, which is necessary for obtaining high-quality crystals, possesses a high activity similar to that of the wild-type protein, consistent with the notion that the mutation site is far from the substrate binding site (Figures 2A and 3D). Mutations of important residues involved in peptide recognition, including D312A for H3Q5 recognition, E285A and E516A for H3R2 recognition, F171K and V363A for H3A7 recognition, and S290A and Y298A for H3K4me3 recognition, all significantly decrease the enzyme activity (Figure 3D). The mutations of residues involving in Fe2+ coordination, including H309A, H397A, and E311A, result in a complete elimination of activity (Figure 3D). It is striking that the V363A mutant version also showed a remarkable decrease in the activity (Figure 3D). Val-363 is involved in the hydrophobic interaction with H3A7 (Figure 3B). The V-to-A mutation can weaken the hydrophobic interaction, but the change looks to be not strong enough to yield such significant decrease. A possible explanation is that the mass spectrum-based in vitro assay is quite sensitive to the mutations. For example, the mutations of D312A, E285A, E516A, and F171K almost completely abolish the activity (Figure 3D), although these residues are only involved in substrate recognition, not in catalysis. Indeed, the V363A mutant still possesses significantly higher activity than D312A, E285A, E516A, and F171K (Figure 3D). Therefore, we conclude that V363A is a weak mutant that has less important role on the substrate recognition than the strong mutants such as D312A, E285A, E516A, and F171K, consistent with our structural observations.
The Substrate Sequence Specificity of KDM5 Subfamily Demethylases
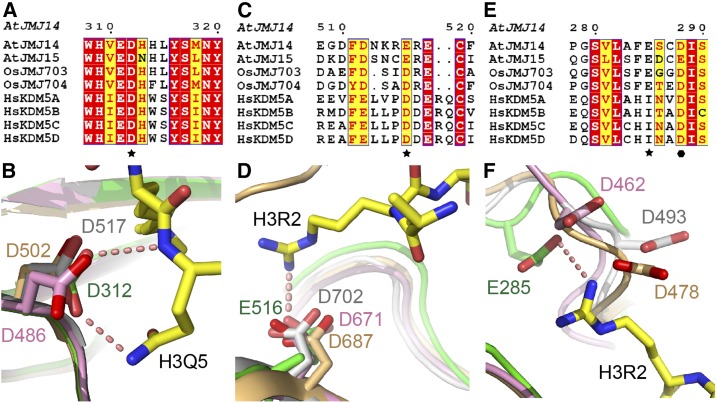
KDM5 subfamily demethylases are solely H3K4me3 specific demethylases (Mosammaparast and Shi, 2010). However, the molecular mechanism for their substrate sequence specificity is still unknown. The structures of human KDM5A/B/C catalytic domains have been reported as enzyme-inhibitor complexes but not substrate complexes (Bavetsias et al., 2016; Horton et al., 2016a, 2016b; Johansson et al., 2016; Labadie et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017). In plants, although the crystal structure of rice (Oryza sativa) JMJ703 jumonji domain in complex with an H3(1-10)K4me3 peptide has been reported, only three peptide residues T3K4me3Q5 were observed and the JMJ703 construct lacked the helical and C5HC2 domains, and hence could not explain the substrate sequence specificity (Chen et al., 2013). In our JMJ14CD-NOG-H3K4me3 complex structure, H3R2 and H3Q5 are specifically recognized by the acidic residues Glu-285, Glu-516, and Asp-312 (Figure 3B), which is the potential determinant for the H3K4me3 specificity for JMJ14. The insertion of H3R2 into the negatively charged pocket could induce the disordered loop of the C5HC2 domain (residues 508–516) to switch to an ordered conformation (Figures 2D and 3A). In addition, Asp-312 can recognize H3Q5 by both main chain and side chain hydrogen bonds (Figure 3B). We compared the primary sequences and three-dimensional structures of KDM5 subfamily demethylases both from animals and plants (Figure 4; Supplemental Figure 2) (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017). The sequence alignment and the structural comparison indicate that the Asp-312 of the jumonji domain and Glu-516 of C5HC2 domain are strictly conserved to be acidic residues and adopt similar conformation not only in plants but also in human KDM5 proteins, ranging from human KDM5A to KDM5D, indicating that the specific recognition of H3R2 by Glu-516 and H3Q5 by Asp-312 are common features of KDM5 family demethylases in plants and animals (Figures 4A to 4D) (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017). By contrast, Glu-285 is conserved only in plant but not human KDM5s (Figure 4E). However, an adjacent Asp residue that is strictly conserved in human KDM5s (highlighted with solid hexagon in Figure 4E) is structurally positioned nearby (Figure 4F) (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017). The flanking regions nearby are of a flexible nature with diversified conformations in different structures (Figure 4F), which is probably due to the absence of the H3K4me3 substrate for the three human KDM5 structures (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017). Indeed, the Asp residues of human KDM5A/B/C adopt different directionalities that cannot be aligned together structurally (Figure 4F) (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017). In addition, the Asp residues were modeled as Ala in their original coordinates, further confirming the flexible nature of this region (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017). Therefore, it is very likely that the binding of H3R2 may induce the conserved Asp residues of human KDM5s to undergo a conformational change and use this Asp to compensate for the function of Glu-285 in JMJ14 that interacts with H3R2.
Figure 4.
Three Acidic Residues Involved in Sequence-Specific Recognition of H3K4me3 Are Conserved.
(A), (C), and (E) The sequence alignment of two Arabidopsis, two rice, and four human KDM5 demethylases indicates that important residues for recognition of H3R2 and H3Q5 are conserved. Asp-312 (A) and Glu-516 (C) are conserved and are shown to be acidic residues in both plants and animals, while Glu-285 (E) is only conserved in plants. The conserved residues are highlighted with stars. An Asp residue of human KDM5s, which is near JMJ14 Glu-285, is strictly conserved in human KDM5s and is highlighted with a solid hexagon. A fully structure-based sequence alignment of the catalytic domain and the sequences used in the alignment can be found in Supplemental Figure 2.
(B), (D), and (F) A structural view of the conserved acidic residues corresponding to Asp-312 (B), Glu-516 (D), and Glu-285 (F) from the structures of JMJ14CD (in green), human KDM5A (in pink, PDB code: 5CEH), KDM5B (in orange, PDB code: 5A3N), and KDM5C (in silver, PDB code: 5FWJ). It is worth noting that in Figure 4F, the Asp-462 in KDM5A, Asp-478 in KDM5B, and Asp-493 in KDM5C were originally built as an Ala model and were manually modeled to Asp based on their Cα and Cβ positions.
The Three Acidic Residues Are Important for JMJ14 in Vivo Function
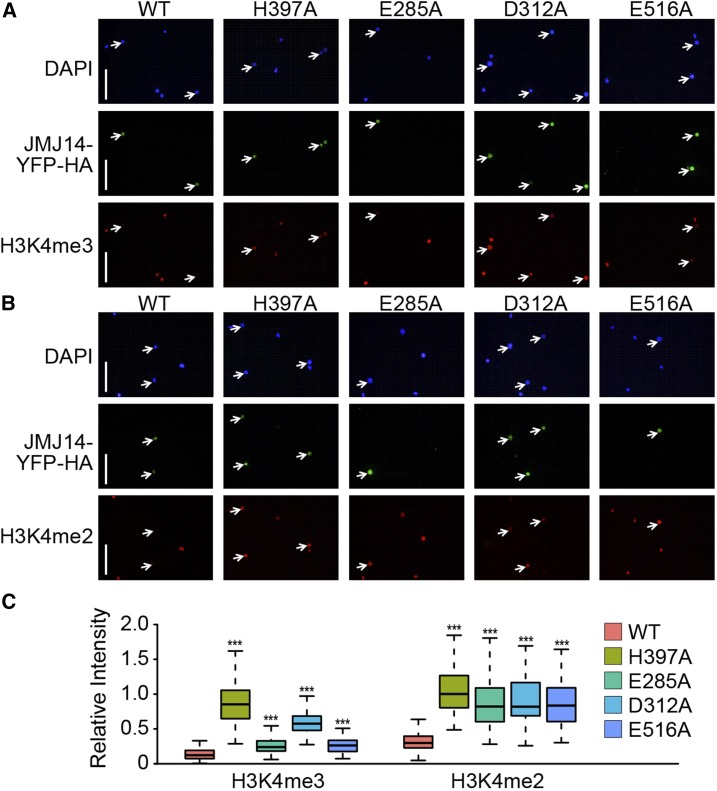
To further confirm the important role of these conserved acidic residues in H3K4me3 substrate recognition, we performed in vivo activity assays in Nicotiana benthamiana leaves (Figure 5). The YFP-HA tagged JMJ14 and its mutants were transformed and overexpressed in N. benthamiana leaves and the remaining H3K4me3 or H3K4me2 levels were detected using the corresponding antibodies (Figures 5A and 5B). The wild-type YFP-HA-JMJ14 can efficiently remove the in vivo H3K4me3 and H3K4me2, confirming JMJ14 is an active H3K4me3/2 demethylase (Figures 5). As a negative control, the H397A mutant, which eliminates the Fe2+ coordination, has a high in vivo H3K4me3 and H3K4me2 levels, indicating a loss of catalytic activity (Figure 5). While the D312A mutant shows a significant loss of activity against H3K4me3, the E285A and E516A mutants retain a certain amount of H3K4me3 demethylase activity (Figure 5A). However, the statistics of the immune fluorescence signal indicates that the E285A and E516A mutants also have a remarkable decrease of H3K4me3 demethylase activity (Figure 5C, left panel). In addition, all three acidic residue mutants, E285A, D312A, and E516A, show significant decrease of activity against H3K4me2 (Figures 5B and 5C), consistent with our in vitro activity assays that all these three residues are important for the activity of JMJ14 (Figure 3D). Because JMJ14 possesses a higher demethylation activity toward H3K4me3 over H3K4me2 and we used an overexpression system in the N. benthamiana leaves (Figures 1B and 1F), any mutations partially affected JMJ14 catalytic activity can have greater effects on H3K4me2 than H3K4me3 in vivo, resulting in more significant decreases of the activity on H3K4me2 than H3K4me3 by mutation of the acidic residues (Figure 5). Together, our in vivo and in vitro activity assays confirmed the importance of the three acidic residues of JMJ14 in substrate recognition.
Figure 5.
The Key Acidic Residues Are Important for in Vivo Sequence-Specific Demethylation by JMJ14.
(A) and (B) Mutations of JMJ14-pEG101 reduced its demethylase activity of H3K4me3 (A) and H3K4me2 (B) in vivo. The wild type and mutations of JMJ14-YFP-HA fusion protein were transiently expressed in N. benthamiana, and nuclei were isolated for immunostaining. Arrows indicate transfected nuclei. Bars = 50 μm.
(C) Quantitative analysis of (A) and (B). Quantification was performed using ImageJ software by calculating the ratio of histone methylation integrated staining density of nuclei overexpressing different variants of JMJ14 to that of the local neighboring wild-type nuclei. More than 50 pairs of nuclei for each variant were observed. Box plots show the median (middle bar) and interquartile range (from the 25th to 75th percentile); whiskers extend to minimum and maximum values within 1.5 times the interquartile range. Mann-Whitney-Wilcoxin test was performed between wild-type JMJ14 and its variants with different mutations (***P < 0.001).
The Conserved Acidic Residues Are Functionally Important in Human KDM5B
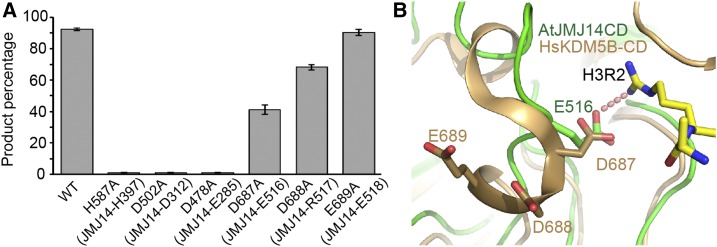
To explore whether the three conserved acidic residues also have equally important functions in animal KDM5/JARID subfamily demethylases, we performed in vitro activity assays toward human KDM5B and the corresponding mutants. The catalytic portion of human KDM5B (residues 1–750, also known as JARID1B), including the jumonji, PHD, ARID, helical, and C5HC2 domains, was expressed and purified as described previously (Xiang et al., 2007). The wild-type KDM5B shows high H3K4me3 demethylation activity (Figure 6A). The H587A mutation, which disrupts Fe2+ binding, eliminates the activity, as expected (Figure 6A). The D478A and D502A mutations, which correspond to E285A and D312A of JMJ14 in H3R2 and H3Q5 recognition, respectively, significantly decreased the demethylation activity to a background level (Figure 6A), suggesting that these two residues are important in human KDM5B substrate recognition as in JMJ14. D687A, which corresponds to E516A of JMJ14, can partially decrease the activity to a level that is half of the wild-type KDM5B, in contrast to that E516A of JMJ14, which almost totally disrupts the in vitro activity, suggesting a difference between JMJ14 and human KDM5 (Figures 3D and 6A). We noticed that there are two additional conserved acidic residues Asp-688 and Glu-689 near Asp-687 of KDM5B (Figure 4C). Further demethylation assays indicated that D688A can moderately decrease the activity, while the E689A has no effect and shows an activity comparable to the wild-type KDM5B (Figure 6A). We analyzed the KDM5B structure and found that Asp-688 is near the potential H3R2 binding site and might form a continuous negatively charged surface with Asp-687 upon conformational change to potentially enhance the binding to H3R2, while Glu-689 is extended toward the opposite side and away from the potential H3R2 binding site (Figure 6B). Therefore, the two adjacent acidic residues Asp-687 and Asp-688 may be jointly involved in the H3R2 recognition, serving the role of Glu-516 of JMJ14. The less important role of KDM5B Asp-687 than JMJ14 Glu-516 suggests a plausible different mechanism for the H3R2 recognition between plants and animals. In general, our in vitro demethylation assays demonstrate that the key acidic residues of JMJ14 involving sequence-specific H3K4me3 recognition are structurally conserved in human KDM5B, further supporting the model of the molecular interaction between human KDM5B and the H3K4me3 substrate.
Figure 6.
The Conserved Acidic Residues Are Functionally Important in Human KDM5B.
(A) The quantified in vitro demethylation assay of human KDM5B and its mutants. The percentages of the product peptide are shown as means ± sd (n = 3). The corresponding residues of JMJ14 of the KDM5B mutation sites are indicated in the parentheses.
(B) The structure of AtJMJ14CD is superimposed with human KDM5B catalytic domain (HsKDM5B-CD, PDB code: 5A3N) with JMJ14 colored in green and KDM5B in orange. Asp-688 is near the potential H3R2 binding site, while Glu-689 is far away and adopts an opposite directionality.
Therefore, we conclude that the sequence specificity of KDM5 family H3K4me3 demethylases relies on the conserved acidic residues of JMJ14 and human KDM5 demethylases to specifically recognize the RXKme3Q (X stands for any residue) motif of the substrate. The RXKme3Q motif is unique for the H3K4me3 position both in plant and animal histone proteins. Therefore, this motif itself is sufficient to obtain the histone substrate specificity for KDM5 subfamily demethylases.
DISCUSSION
We performed structural and functional studies on the plant KDM5 subfamily H3K4me3 demethylase JMJ14. JMJ14 can recognize H3R2 and H3Q5 by several acidic residues of the jumonji and C5HC2 domains, with these acidic residues important and conserved in plants and animals, suggesting a common sequence-specific demethylation mechanism shared by plant and animal KDM5 subfamily demethylases. Human KDM5s have ARID and PHD domains inserted inside the sequence of the jumonji domain, but these domains structurally bud out from the jumonji domain (Johansson et al., 2016; Vinogradova et al., 2016; Tumber et al., 2017) (Figure 2B). Although these insertion domains are missing in Arabidopsis JMJ14, the jumonji-helical-C5HC2 catalytic cassette and conserved substrate recognition acidic residues are shared by both animal and plant KDM5 proteins, revealing a common substrate recognition mechanism. Most available human KDM5-targeted inhibitors mimic and block binding of the cofactor αKG, which also inhibits other histone demethylases, especially those of the KDM4 subfamily (Taylor-Papadimitriou and Burchell, 2017). Therefore, there is an urgent need to develop KDM5-specific inhibitors. The structure-based H3K4me3 substrate-mimicking inhibitors could possess higher selectivity and have the capacity to be more specific and potent drugs because different subfamilies of jumonji domain-containing demethylases share a common cofactor αKG but have different specific histone substrates. Our studies not only revealed the molecular mechanism of H3K4me3 substrate specificity of the KDM5 subfamily demethylases, but also shed light on the design of structure-based KDM5 targeting inhibitors and on anticancer drug design.
METHODS
Protein Expression and Purification
The catalytic domain of the Arabidopsis thaliana JMJ14 (residues 35–597) was cloned into a self-modified pET-Sumo vector to fuse a hexahistidine tag plus a yeast sumo tag to the target protein. The plasmid was transformed into Escherichia coli strain BL21(DE3) Codon Plus (Stratagene). The protein expression was induced by adding IPTG to cell culture with a final concentration of 0.2 mM at 16°C when OD600 reached 0.6. The recombinant expressed protein was purified using HisTrap, Heparin, and Superdex G200 columns (GE Healthcare). All the mutations were generated using a PCR-based method and expressed and purified using the same protocol as wild-type protein. The peptides and chemicals were purchased from GL Biochem and Sigma-Aldrich, respectively.
Human KDM5B was expressed and purified using the same protocol as described previously (Xiang et al., 2007). In brief, the catalytic portion (residues 1–750) was cloned into a pET28a vector. The plasmid was transformed into E. coli strain Rosetta 2 (DE3). The protein expression was induced by IPTG with concentration of 0.2 mM at 16°C when the OD600 of cell culture reached 0.6. The protein was purified using a HisTrap column (GE Healthcare). The mutations were generated using a PCR-based method and expressed and purified using the same protocol as wild-type protein.
Crystallization, Data Collection, and Structure Determination
The crystallization was conducted using the sitting drop vapor diffusion method at 20°C. The wild-type protein only produced crystals that diffracted ∼5-Å resolution. Several surface entropy reduction mutations were introduced to improve the diffraction quality (Goldschmidt et al., 2007). Fortunately, an E180A/E181A mutant of JMJ14CD yielded crystals with good diffraction quality in 0.2 M Na2SO4, 20% PEG3350, and 0.1 M bis-tris propane, pH 6.5. For obtaining the substrate bound crystals, the apo-form crystals were soaked in reservoir solution supplemented with 2 mM NOG and 2 mM H3(1-10)K4me3 peptide for 12 h. Before data collection, all the crystals were soaked in the reservoir solution supplemented with 15% glycerol and flash cooled in liquid nitrogen. All the diffraction data were collected at beamline BL19U1 of the National Center for Protein Sciences Shanghai at Shanghai Synchrotron Radiation Facility and were processed using the program HKL3000 (Otwinowski and Minor, 1997). A summary of statistics of the data collection is listed in Table 1.
The structure of the apo-form JMJ14CD was solved using the molecular replacement method with the structure of rice JMJ703 jumonji domain as a search model (PDB code: 4IGQ) with the program Phenix (Adams et al., 2010; Chen et al., 2013). The manual model building and structure refinement were carried our using the program Coot and Phenix, respectively (Adams et al., 2010; Emsley et al., 2010). The geometry of the structure model was monitored using the program Molprobity (Chen et al., 2010). The structure of JMJ14CD-NOG-H3(1-10)K4me3 complex was solved using the molecular replacement method and refined using the same protocol as apo JMJ14CD structure. A summary of the structure refinement statistics is listed in Table 1. The molecular graphics were generated using the program Pymol (DeLano Scientific). The sequences were aligned using the program T-coffee with manual adjustment to make sure the sequence alignment fit the 3D alignment of the structures and further illustrated using ESPript (Di Tommaso et al., 2011; Robert and Gouet, 2014)
In Vitro Histone Demethylation Assay
The purified protein (10 μM) was incubated with histone peptides (80 μM) in a reaction buffer of 80 μM Fe(NH4)2(SO4)2, 2 mM ascorbic acid, 1 mM αKG, 50 mM Tris-HCl, pH 7.3, 150 mM NaCl, and 50 mM Arg/Glu at 25°C. For JMJ14 and its mutants, the reaction system often yielded precipitates and 5% glycerol was added into the reaction system to prevent protein precipitation. Two hours later, the reaction was stopped by heating to 98°C for 3 min. The reaction mixture was further desalted by ZipTip (Millipore). The MALDI-TOF analysis was conducted according to published protocols (Sengoku and Yokoyama, 2011). In brief, the eluted peptides were spotted on the MALDI plate and cocrystallized with 10 mg/mL α-cyano-4-hydroxycinnamic acid in 60% acetonitrile/0.1% TFA. Then, the samples were analyzed with a 5800 MALDI-TOF/TOF mass spectrometer (ABsciex). For quantification, peptide mass spectra were recorded in reflector mode from triplicate reactions and per spectrum was acquired from a total of 1250 laser shots. The resulting mass spectra were processed by the Data Explorer software.
Transgene
JMJ14-pEG101 and JMJ14E285A-pEG101 have been described previously (Earley et al., 2006; Lu et al., 2010). The three mutant versions of JMJ14 were reconstructed from JMJ14-pEG101 using a PCR-based method as described before (Gibson, 2011). The constructs were transformed into Agrobacterium tumefaciens EHA105 strains. For transient expression, agrobacteria carrying different version of JMJ14 were infiltrated into Nicotiana benthamiana leaves. N. benthamiana was grown at 23°C under a 16-h-light/8-h-dark photoperiod in a white light of ∼100 to 120 μE m−2 s−1.
In Vivo Histone Demethylation Assay
Half of each leaf of 4-week-old N. benthamiana was infiltrated with Agrobacterium EHA105 strains to express different versions of JMJ14 with YFP and HA tag. The other halves of leaves without infiltration were used as controls. At 48 h after injection, the YFP signal of leaves were observed with a fluorescence microscope and seven to eight leaves with similar expression levels were pooled together for the following steps. The whole leaves were cut into ∼0.5 × 1-cm pieces and fixed in cold 4% paraformaldehyde in Buffer A (10 mM Tris, pH 7.5, at 25°C, 100 mM NaCl, and 10 mM EDTA) for 20 min. Then, the leaves were washed twice using ice-cold Buffer A for 10 min each with gentle shake. After sopping up water with filter paper, nuclei were released by chopping leaves in LB01 buffer (15 mM Tris-HCl, pH 7.5, 2 mM EDTA, 0.5 mM spermine, 80 mM KCl, 20 mM NaCl, and 0.1% Triton X-100) until very fine and filtered through a 35-μm nylon mesh (BD Falcon; 352235). The flow-through was transferred to a 1.5-mL microcentrifuge tube and centrifuged at 1500g for 5 min. Then the pellet was washed several times with nuclear isolation buffer (35% glycerol, 300 mM sucrose, 20 mM Tris, pH 8.0, 5 mM MgCl2, 5 mM KCl, and 0.2% Triton X-100) (Hetzel et al., 2016) to remove the chloroplasts. The pellet was resuspended with LB01 buffer and 1:4 diluted in sorting buffer (100 mM Tris, pH 7.5, 50 mM KCl, 2 mM MgCl2, 0.05% Tween-20, and 5% sucrose) before being spotted onto microscopy slides and air dried for ∼2 h at room temperature.
After postfixation with 4% paraformaldehyde in PBS buffer (10 mM sodium phosphate, pH 7.0, and 143 mM NaCl), one drop Image-iT FX signal enhancer was added to each slide, and slides were kept at room temperature for 30 min. Immunolabeling was performed using histone methylation-specific antibodies (H3K4me3, Millipore 07-473, 1:100; H3K4me2, Millipore 07-030, 1:1500). The modified histones were revealed by Alexa Fluor 555-conjugated goat anti-rabbit (Invitrogen A31630, 1:500). Transfected cells were revealed by monitoring the YFP signal.
After staining, the slides were mounted in Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (Vector Laboratory H-1200) and then photographed with an Olympus BX51 fluorescence microscope. More than 50 pairs of transfected nuclei versus nontransfected nuclei in the same field of view were observed. Quantification was performed using ImageJ software by calculating the ratio of histone methylation integrated staining density of nuclei overexpressing different versions of JMJ14 to that of the local neighboring wild-type nuclei. Box plot and the Mann-Whitney-Wilcoxin test were performed by R.
Accession Numbers
The Arabidopsis Genome Initiative locus identifier (www.arabidopsis.org/) used in this study is AT4G20400 (JMJ14). Coordinates and structure factors have been deposited in the RCSB Protein Data Bank with the accession codes 5YKN (JMJ14CD) and 5YKO (JMJ14CD-NOG-H3K4em3 complex). Sequences for the homologs aligned in Figure 4A and Supplemental Figure 2 can be found in Uniprot with accession codes of AtJMJ14 (Q8GUI6), AtJMJ15 (O64752), OsJMJ703 (Q53WJ1), OsJMJ706 (Q8W3G5), HsKDM5A (P29375), HsKDM5B (Q9UGL1), HsKDM5C (P41229), and HsKDM5D (Q9BY66).
Supplemental Data
Supplemental Figure 1. Electron density map of the H3K4me3 peptide.
Supplemental Figure 2. Structure-based sequence alignment of KDM5 subfamily demethylases.
Acknowledgments
We thank the staff of beamline BL19U1 of the National Center for Protein Sciences Shanghai at the Shanghai Synchrotron Radiation Facility for assistance during data collection, Degui Chen for sharing human KDM5B expression vector, and the proteomic core facility of Shanghai Center for Plant Stress Biology for assistance with the mass spectrometry experiment. This work was supported by the National Key R&D Program of China (2016YFA0503200 to J.D.; 2016YFD0100904 to X. Cao), by the National Natural Science Foundation of China (31622032 to J.D.), by the Chinese Academy of Sciences (to J.D. and XDPB0403 to X. Cao), and by the State Key Laboratory of Plant Genomics.
AUTHOR CONTRIBUTIONS
Z.Y., Q.Q., W.C., B.J., X. Chen, H.H., K.H., X.D., and S.L. performed the experiments. W.A.T. helped design the in vitro activity assay. X. Cao and J.D. conceived the study, designed the experiments, analyzed the data, and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Adams P.D., et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavetsias V., et al. (2016). 8-Substituted pyrido[3,4-d]pyrimidin-4(3H)-one derivatives as potent, cell permeable, KDM4 (JMJD2) and KDM5 (JARID1) histone lysine demethylase inhibitors. J. Med. Chem. 59: 1388–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Chen X., Wang Q., Zhang F., Lou Z., Zhang Q., Zhou D.X. (2013). Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet. 9: e1003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., et al. (2007). Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl. Acad. Sci. USA 104: 10818–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Cheung P., Kuo A.J., Yukl E.T., Wilmot C.M., Gozani O., Patel D.J. (2014). A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev. 28: 1758–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J.F., Collazo E., Ortiz-Tello P.A., Brunzelle J.S., Trievel R.C. (2007). Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 14: 689–695. [DOI] [PubMed] [Google Scholar]
- Deleris A., Greenberg M.V., Ausin I., Law R.W., Moissiard G., Schubert D., Jacobsen S.E. (2010). Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep. 11: 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.M., Taly J.F., Notredame C. (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39: W13–W17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G. (2011). Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt L., Cooper D.R., Derewenda Z.S., Eisenberg D. (2007). Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 16: 1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M.V., Deleris A., Hale C.J., Liu A., Feng S., Jacobsen S.E. (2013). Interplay between active chromatin marks and RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 9: e1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel J., Duttke S.H., Benner C., Chory J. (2016). Nascent RNA sequencing reveals distinct features in plant transcription. Proc. Natl. Acad. Sci. USA 113: 12316–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J.R., Engstrom A., Zoeller E.L., Liu X., Shanks J.R., Zhang X., Johns M.A., Vertino P.M., Fu H., Cheng X. (2016a). Characterization of a linked Jumonji domain of the KDM5/JARID1 family of Histone H3 Lysine 4 demethylases. J. Biol. Chem. 291: 2631–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J.R., et al. (2016b). Structural basis for KDM5A histone lysine demethylase inhibition by diverse compounds. Cell Chem. Biol. 23: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J.H., Song H.R., Ko J.H., Jeong Y.M., Kwon Y.E., Seol J.H., Amasino R.M., Noh B., Noh Y.S. (2009). Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS One 4: e8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C., et al. (2016). Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat. Chem. Biol. 12: 539–545. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Kruidenier L., et al. (2012). A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488: 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labadie S.S., et al. (2016). Design and evaluation of 1,7-naphthyridones as novel KDM5 inhibitors. Bioorg. Med. Chem. Lett. 26: 4492–4496. [DOI] [PubMed] [Google Scholar]
- Le Masson I., Jauvion V., Bouteiller N., Rivard M., Elmayan T., Vaucheret H. (2012). Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell 24: 3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420. [DOI] [PubMed] [Google Scholar]
- Lu F., Cui X., Zhang S., Liu C., Cao X. (2010). JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 20: 387–390. [DOI] [PubMed] [Google Scholar]
- Lu F., Li G., Cui X., Liu C., Wang X.J., Cao X. (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 50: 886–896. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N., Shi Y. (2010). Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79: 155–179. [DOI] [PubMed] [Google Scholar]
- Ng S.S., et al. (2007). Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448: 87–91. [DOI] [PubMed] [Google Scholar]
- Ning Y.Q., Ma Z.Y., Huang H.W., Mo H., Zhao T.T., Li L., Cai T., Chen S., Ma L., He X.J. (2015). Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 43: 1469–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Rasmussen P.B., Staller P. (2014). The KDM5 family of histone demethylases as targets in oncology drug discovery. Epigenomics 6: 277–286. [DOI] [PubMed] [Google Scholar]
- Robert X., Gouet P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42: W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I.R., Pontes O., Melnyk C.W., Smith L.M., Baulcombe D.C. (2010). JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 24: 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T., Yokoyama S. (2011). Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev. 25: 2266–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Papadimitriou J., Burchell J. (2017). JARID1/KDM5 demethylases as cancer targets? Expert Opin. Ther. Targets 21: 5–7. [DOI] [PubMed] [Google Scholar]
- Tumber A., et al. (2017). Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem. Biol. 24: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova M., et al. (2016). An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12: 531–538. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Zhu Z., Han G., Ye X., Xu B., Peng Z., Ma Y., Yu Y., Lin H., Chen A.P., Chen C.D. (2007). JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. USA 104: 19226–19231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Jiang D., Jiang J., He Y. (2010). A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 62: 663–673. [DOI] [PubMed] [Google Scholar]
- Zhang S., et al. (2015). C-terminal domains of a histone demethylase interact with a pair of transcription factors and mediate specific chromatin association. Cell Discov. pii: 15003. [DOI] [PMC free article] [PubMed] [Google Scholar]