Plants coordinate their response to above- and belowground competition when growing at high densities; a change in light quality detected by the shoot transmits a signal to adjust root development.
Abstract
Plants in dense vegetation compete for resources and detect competitors through reflection of far-red (FR) light from surrounding plants. This reflection causes a reduced red (R):FR ratio, which is sensed through phytochromes. Low R:FR induces shade avoidance responses of the shoot and also changes the root system architecture, although this has received little attention so far. Here, we investigate the molecular mechanisms through which light detection in the shoot regulates root development in Arabidopsis thaliana. We do so using a combination of microscopy, gene expression, and mutant study approaches in a setup that allows root imaging without exposing the roots to light treatment. We show that low R:FR perception in the shoot decreases the lateral root (LR) density by inhibiting LR emergence. This decrease in LR emergence upon shoot FR enrichment is regulated by phytochrome-dependent accumulation of the transcription factor ELONGATED HYPOCOTYL5 (HY5) in the LR primordia. HY5 regulates LR emergence by decreasing the plasma membrane abundance of PIN-FORMED3 and LIKE-AUX1 3 auxin transporters. Accordingly, FR enrichment reduces the auxin signal in the overlaying cortex cells, and this reduces LR outgrowth. This shoot-to-root communication can help plants coordinate resource partitioning under competition for light in high density fields.
INTRODUCTION
Plants typically grow at high densities in agricultural and natural systems. To compete for the limited light and nutrient resources, they adjust root and shoot architecture. Therefore, it is imperative for plants to sense their neighbors and aboveground they do so through far-red (FR) light that is reflected by leaves of neighboring plants. The resulting FR enrichment of the light is perceived by plants and this typically leads to elongation responses of the stem, petioles, and leaves in shade-intolerant plant species (Pacín et al., 2013; Gommers et al., 2013; Ballaré and Pierik, 2017).
The red (R):FR light ratio is detected by the phytochrome photoreceptors. phyB is the key regulator of shade avoidance in response to low R:FR. It is activated (to Pfr, its active state) by R light and inactivated by FR light (to Pr, its inactive state). In the Pfr state, phyB triggers the phosphorylation and degradation of elongation-promoting PHYTOCHROME INTERACTING FACTORs (PIFs) (Ni et al., 2014; Shin et al., 2016). Upon phyB inactivation by low R:FR, the repression of PIFs is released and stem elongation occurs (Leivar and Monte, 2014). phyB belongs to the class II phytochromes, which all show activation by R light and inactivation by FR. In contrast, phyA, a class I phytochrome shows a unique regulation. phyA is activated by FR light (Rausenberger et al., 2011) and R and white light (WL) promote the degradation of phyA (Li et al., 2011a). However, phyA does remain present at a reduced level and the activation of phyA by FR light (as opposed to deactivation of phyB) means that phyA acts as a negative regulator of hypocotyl elongation under low R:FR conditions (Martínez-García et al., 2014). Low R:FR-mediated activation of PIF4, PIF5, and PIF7 induces shoot elongation (Lorrain et al., 2008; Li et al., 2012; de Wit et al., 2016b), in part through regulation of auxin biosynthesis, transport, and signaling genes (de Wit et al., 2016a). The plant hormone auxin plays an important role in the elongation responses during shade avoidance, and its biosynthesis is rapidly upregulated upon FR perception via TAA1 and YUCCA2, 5, 8, and 9 (Tao et al., 2008; Procko et al., 2014; Kohnen et al., 2016; Müller-Moulé et al., 2016). Auxin is subsequently transported from cell to cell in an active, directed manner. Polar auxin transport through the PIN-FORMED3 (PIN3), PIN4, and PIN7 auxin efflux carriers is required for hypocotyl elongation under low R:FR conditions, and blocking auxin transport through the chemical naphthylphtalamic acid leads to loss of this shade avoidance response (Keuskamp et al., 2010; Kohnen et al., 2016).
The effects of shade on plant aerial tissues are well established (Ballaré and Pierik, 2017). Much less is known about how shade may affect root development (Gundel et al., 2014). Direct application of FR-enriched light to whole seedlings leads to a shorter main root and fewer lateral roots (LRs) (Salisbury et al., 2007); however, it remains unknown to what extent R:FR signaling in the shoot exerts control over root development and through which mechanisms this would occur. Classically, root development is controlled through auxin transport and subsequent signaling (Bhalerao et al., 2002), and auxin is also involved in root growth adjustments in response to stimuli such as gravity (Baster et al., 2013), salt (Galvan-Ampudia et al., 2013), or even blue light (Zhang et al., 2013). It is possible that auxin production and transport associated with shade avoidance responses in the shoot might deplete auxin in the roots, thereby affecting root development. The root system architecture is determined by where and at what frequency LRs emerge. LR development starts at the primary root meristem where a local, regularly oscillating auxin maximum determines a xylem pole pericycle cell to establish and initiate a LR primordium (De Rybel et al., 2010; De Smet et al., 2007). The process of LR development is highly dependent upon the auxin response factors ARF7 and ARF19 (Okushima et al., 2007; Goh et al., 2012; reviewed in detail in Lavenus et al., 2013). After initiation, a lateral root primordium (LRP) undergoes several divisions, which results in a dome-shaped LRP that has to penetrate the endodermis (Vermeer et al., 2014) and then the cortex and epidermis (Péret et al., 2013). This process of LR emergence is highly dependent upon polar auxin transport and occurs through the concerted action of auxin efflux carrier PIN3 and influx carrier LIKE AUX1 3 (LAX3) (Péret et al., 2013). The resulting auxin transport to the LRP tip and the cortex above the developing LRP induces cell wall modifications in the cortex and epidermis, which leads to the separation of these layers (Kumpf et al., 2013; Lee et al., 2013).
Recently, the photoreceptor-targeted transcription factor ELONGATED HYPOCOTYL5 (HY5) was shown to act as a potential shoot-to-root signal transducer. HY5, upon photosynthesis-mediated induction in the cotyledon, can be transported from the shoot to the root, probably via the phloem, and then among others activate NITRATE TRANSPORTER 2:1 in the roots (Chen et al., 2016). hy5 loss-of-function mutants were first identified on the basis of their elongated hypocotyls in WL. HY5 has been demonstrated as a positive driver of photomorphogenesis (Oyama et al., 1997), acting as an integrator of light, hormone, and stress signaling (Cluis et al., 2004; Vandenbussche et al., 2007; Alabadí et al., 2008; Chen et al., 2008; Weller et al., 2009; Toledo-Ortiz et al., 2014; Nawkar et al., 2017). HY5 also functions in regulating root growth, since hy5 mutants display increased LR density and reduced LR gravitropism (Sibout et al., 2006). Interestingly, low R:FR can both induce HY5 gene expression and stabilize HY5 protein (Lee et al., 2007; Pacín et al., 2016), raising the possibility that HY5 may communicate information about the light environment to the root.
Here, we use a setup where the shoot is exposed to low R:FR light conditions via supplemental FR light (WL+FR light), while the root system remains shielded from light. Perception of WL+FR light in the shoot results in a decrease in LR emergence, which does not occur in loss-of-function mutants of certain auxin transporters and transcription factors involved in LR emergence. We show that this root response is dependent on phytochrome (phy) and HY5 function. We propose a regulatory mechanism in which phy-mediated R:FR detection in the shoot induces HY5 accumulation in the LRP, which then inhibits the PIN3- and LAX3-based transport of auxin into the overlaying cells to promote LR emergence.
RESULTS
Supplemental FR Conditions Sensed by the Shoot Decreases LR Density
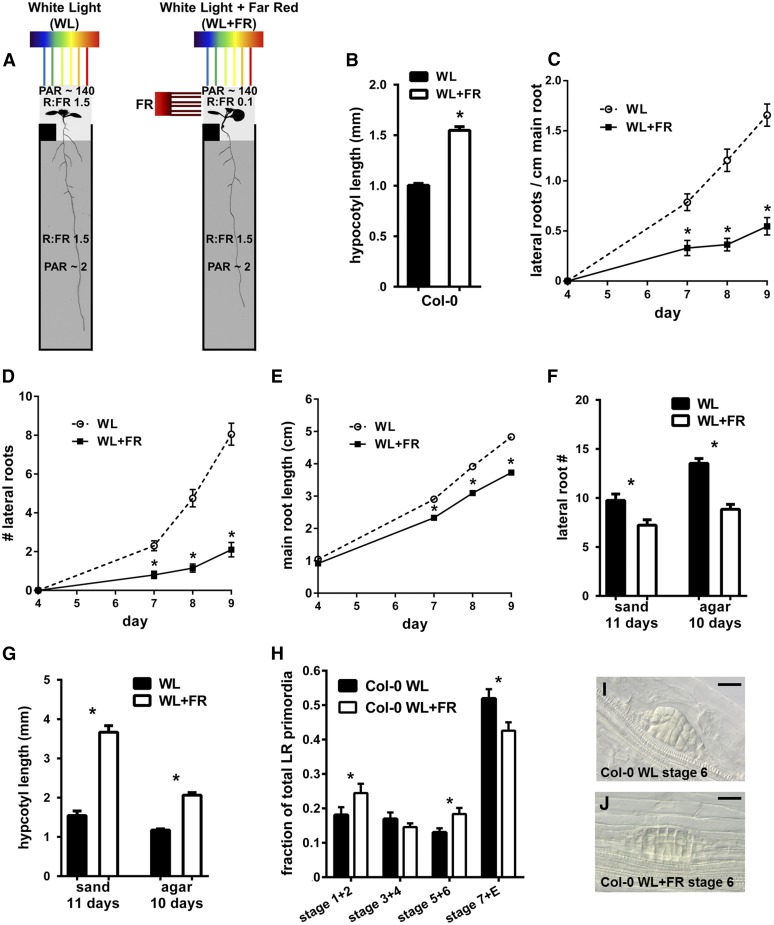
To examine whether supplemental FR irradiance of the shoot would affect root development, we used covers and inserts, for growth of Arabidopsis seedlings on agar-based media, similar to the D-root system (Silva-Navas et al., 2015). Seedlings were grown in either WL or in light supplemented with FR from the side (WL+FR), while blocking as much light as possible with a plate cover and a horizontal, black polycarbonate insert led to a setup with a low R:FR ratio in the shoot compartment (R:FR 0.1, with PAR levels of 130–140 µmol m−2 s−1), while the roots experienced a high R:FR ratio, but in very low light intensity closer to the natural conditions inside soil (Smith, 1982) (R:FR 1.5 and PAR ∼2 µmol m−2 s−1) (Figure 1A). In this setup, we followed wild-type Col-0 root growth during WL or WL+FR conditions for several days. As an indicator for the shoot’s response to WL+FR, we measured hypocotyl length (Figure 1B). In WL+FR conditions, the LR density and LR number was significantly lower than in WL (Figures 1C and 1D), and a decrease in main root length was observed as well (Figure 1E). The LR length also decreased in WL+FR during this experiment, but this response was not as robust as the other aspects of root architecture (Supplemental Figure 1). We found a similar suppression of LR formation, combined with pronounced hypocotyl elongation, by supplemental FR in plants grown on sand in pots (Figures 1F and 1G), indicating that this phenotype is not restricted to plants grown on plates. Since the decrease in LR density will strongly affect the overall root system architecture, we focused on this trait in more detail.
Figure 1.
Supplemental FR Experienced by the Shoot Leads to Reduced Lateral and Primary Root Growth.
(A) Setup used to illuminate shoot with FR light. The root is shielded by a black cover around the plate and an insert at the shoot-root boundary (black box just below shoot). R:FR ratios of each compartment are indicated, along with the amount of PAR.
(B) Hypocotyl length of 8-d-old seedlings in WL and WL+FR.
(C) LR density (all emerged laterals divided by the total main root length) of Col-0 in WL and WL+FR during 9 d of growth.
(D) Col-0 average LR number.
(E) Col-0 average main root length.
(F) Average number of lateral roots in Col-0 plants grown in sand or on the D-root agar plate system (A), treated with WL or WL+FR.
(G) Hypocotyl length of plants shown in (F).
(H) Distribution of LRP stages (as a fraction of total primordia) of Col-0 seedlings grown for 8 d in WL or WL+FR. Stages were grouped into 1+2, 3+4, 5+6, and 7+ emerged.
(I) and (J) Examples of stage 6 LRP in WL and WL+FR. Bars = 20 µm.
Error bars show se; all experiments n = 15 to 20 seedlings per treatment. Asterisk indicates that difference between means is significant P < 0.05, t test.
To obtain better insight into the processes regulating the decrease in LR density in WL+FR, we quantified LRP stages following the classification of Malamy and Benfey (1997) in 8-d-old seedlings exposed to either WL or WL+FR conditions. A lower fraction of stage 7 and emerged primordia was observed in WL+FR (Figure 1H). Fractions of stage 1+2 and 5+6 were increased in WL+FR, and we observed primordia of stage 6 that apparently failed to penetrate the cortex and epidermis, characterized by a flattened dome shape (Figures 1I and 1J). LRPs cross the endodermis after stage 2 and cross the cortex and epidermis during stage 5+6 (Vilches-Barro and Maizel, 2015); thus, the increase in stage 5+6 primordia, combined with fewer stage 7+ emerged primordia suggested that LR emergence was blocked in WL+FR.
Phytochrome Detection of Supplemental FR in the Shoot Regulates Root Phenotypes
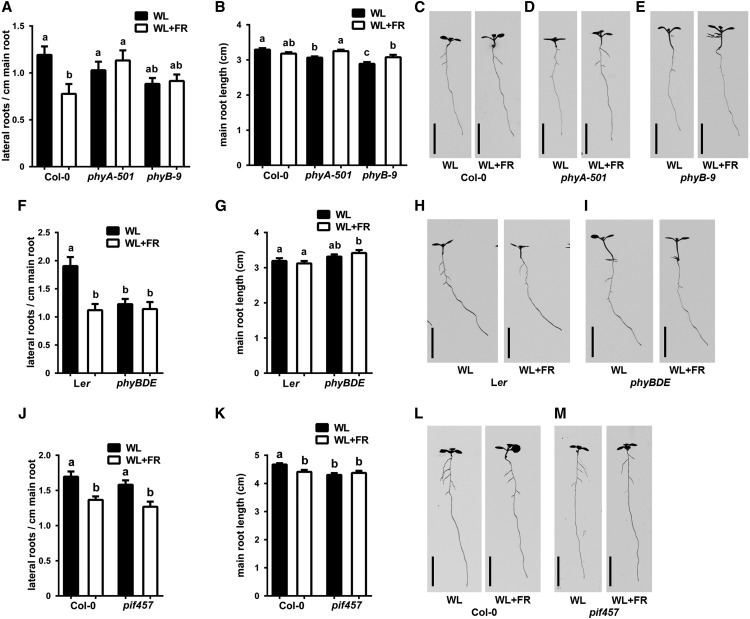
To unravel the mechanism of supplemental FR-induced LR density decrease, we tested mutants of phyB, phyD, phyE, and phyA in our WL+FR light setup. WL+FR light caused a significant decrease in LR density in Col-0, but no effect was observed in either phyA-501 or phyB-9 mutants (Figure 2A). Main root length differed little, but significantly between light treatments and mutants (Figure 2B). In both WL and WL+FR, LR density in phyA-501 was at the level of Col-0 WL, whereas phyB-9 LR density was constitutively low and similar to the level of Col-0 WL+FR. The shoot responses of both mutants were also different: phyA-501 showed a hyper-response to WL+FR, while phyB-9 had a constitutive elongation response in WL and WL+FR (Figures 2C to 2E, Table 1, which includes an overview of hypocotyl lengths for the key experiments in this article). The phyB phyD phyE mutant (phyBDE), in the Ler background, showed a constitutive shoot and LR density phenotype, reminiscent of WL+FR, but was irresponsive to the FR-enriched treatment (Figure 2F), while main root length did not differ significantly between WL and WL+FR. (Figure 2G). PIF4, PIF5, and PIF7 are the principal regulators of hypocotyl elongation downstream of phyB during WL+FR conditions (Hornitschek et al., 2012; Li et al., 2012). Hypocotyls of the pif4 pif5 pif7 mutant (pif457) did not elongate in WL+FR (Figures 2L and 2M, Table 1); however, this mutant did show a decrease in LR density (Figure 2J). Main root length was also still negatively affected in WL+FR (Figure 2K). These results show that phytochromes regulate the decrease in LR density in WL+FR and that hypocotyl elongation during these conditions can be genetically uncoupled from the root response.
Figure 2.
Phytochrome Mutants Do Not Show a Decreased LR Density during WL+FR Experienced by the Shoot, and the Root and Shoot Phenotypes Can Be Uncoupled.
(A) to (E) Col-0, phyA-501, and phyB-9 were grown for 8 d in WL or WL+FR. LR density (A), main root length (B), representative examples at 8 d ([C] to [E]).
(F) to (I) Ler and phyBDE were grown for 8 d in WL or WL+FR. LR density (F), main root length (G), and representative examples ([H] and [I]).
(J) to (M) Col-0 and pif457 were grown for 8 d in WL or WL+FR. LR density (J), main root length (K), and representative examples ([L] and [M]).
Error bars show se; n = 15 to 20 seedlings per treatment. Bars = 1 cm. Letters denote significant difference, P < 0.05, one-way ANOVA.
Table 1. Hypocotyl Response Overview of WL+FR Experiments in This Study.
| Figure 2 | Col-0 |
phyA-501 |
phyB-9 |
Ler |
phyBDE |
Col-0 |
pif457 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 8 hypocotyl | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR |
| Length (mm) | 1.13 | 1.65 | 1.23 | 2.42 | 4.47 | 4.53 | 1.84 | 2.74 | 6.31 | 6.18 | 0.99 | 1.75 | 0.93 | 0.94 |
| % Increase in WL+FR | 45.2 | 96.4 | 1.4 | 49.2 | −2.1 | 76.2 | 0.8 | |||||||
| se | 0.01 | 0.04 | 0.03 | 0.05 | 0.11 | 0.10 | 0.04 | 0.13 | 0.11 | 0.10 | 0.02 | 0.06 | 0.02 | 0.01 |
| Figure 3 | Col-0 | hyh | hy5-2 | hy5 hyh | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 8 hypocotyl | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR | ||||||||||
| Length (mm) | 1.00 | 1.55 | 1.14 | 1.56 | 2.73 | 3.53 | 3.40 | 5.09 | ||||||||||
| % Increase in WL+FR | 54.3 | 36.9 | 29.2 | 49.9 | ||||||||||||||
| se | 0.02 | 0.04 | 0.02 | 0.06 | 0.05 | 0.11 | 0.04 | 0.12 | ||||||||||
| Figure 6 | Col-0a | arf19-1a | arf7-1a | Col-0 | lax3-1 | ida | pin3-3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 7/8 hypocotyl | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR | WL | WL+FR |
| Length (mm) | 1.04 | 1.58 | 1.10 | 1.61 | 1.10 | 1.71 | 1.22 | 1.78 | 1.25 | 1.87 | 1.27 | 1.66 | 1.14 | 1.23 |
| % Increase in WL+FR | 51.9 | 45.7 | 55.6 | 45.9 | 49.6 | 30.7 | 7.9 | |||||||
| se | 0.01 | 0.04 | 0.03 | 0.05 | 0.11 | 0.10 | 0.02 | 0.04 | 0.03 | 0.04 | 0.02 | 0.05 | 0.02 | 0.02 |
FR Light Transmission to the Roots Is Not the Primary Cause of Root Responses to WL+FR
Recently it was shown that light can be transmitted through the shoot to the root in woody tissue, and that this can affect local phyB activity in the roots (Lee et al., 2016). We wanted to investigate the possibility of this light transduction in our WL+FR setup. FR light was applied directly to the root in a compartment that was shielded from the outside WL light, resulting in a low R:FR ratio for the root and a normal R:FR ratio for the shoot (WL+FRroot, Supplemental Figure 2A) with the standard low R:FR treatments (Figure 1) as control. Hypocotyl lengths were increased in WL+FR and not in the WL+FRroot plants (Supplemental Figure 2B). However, the WL+FRroot treatment did show a decrease in main root length and LR emergence, comparable to the normal WL+FR treatment (Supplemental Figures 2C and 2D).
Next we used a phyb-9 Pro35S:PHYB-GFP complementation line to analyze PHYB-GFP photobodies in the root after 4 to 6 d of growth (Supplemental Figures 2I to 2K) as a proxy for phyB activity (Trupkin et al., 2014). In WL, PHYB-GFP photobodies were clearly visible, indicating that the low light levels in the root compartment already activated phyB (Van Buskirk et al., 2014). WL+FR(shoot) did not result in a difference compared with WL, arguing against direct FR light conductance (Supplemental Figures 2I, 2J, and 2L to 2N). However, direct FR application to the root caused a decrease in the average size and intensity of photobodies, but not in the number of photobodies (Supplemental Figures 2K to 2N).
In addition to FR light, we also applied red light directly to the root R(root), with or without FR(shoot) (Supplemental Figure 2E). Hypocotyl length was not affected by R(root) (Supplemental Figure 2F). Compared with WL, WL+R(root) led to a decrease in LR density, which is in accordance with the previously published result using the D-root system (Silva-Navas et al., 2015) (Supplemental Figures 2G and 2H). However, the combination of WL+FR(shoot)+R(root) led to a further decrease in LR density, showing that the red light applied to the root did not counteract any effect of FR(shoot) and arguing against FR light transmission from the shoot if this would act through the Pr-Pfr photoequilibrium of phyB. Furthermore, PHYB-GFP photobodies in the elongation zone of the root were not changed significantly by applying R(root) (Supplemental Figures 2O to 2T). Together, these additional light treatments on the root indicate that the WL+FR effects are unlikely occurring through direct FR-light transmission to the roots.
HY5 Is Involved in the Root Response to WL+FR Light Experienced by the Shoot
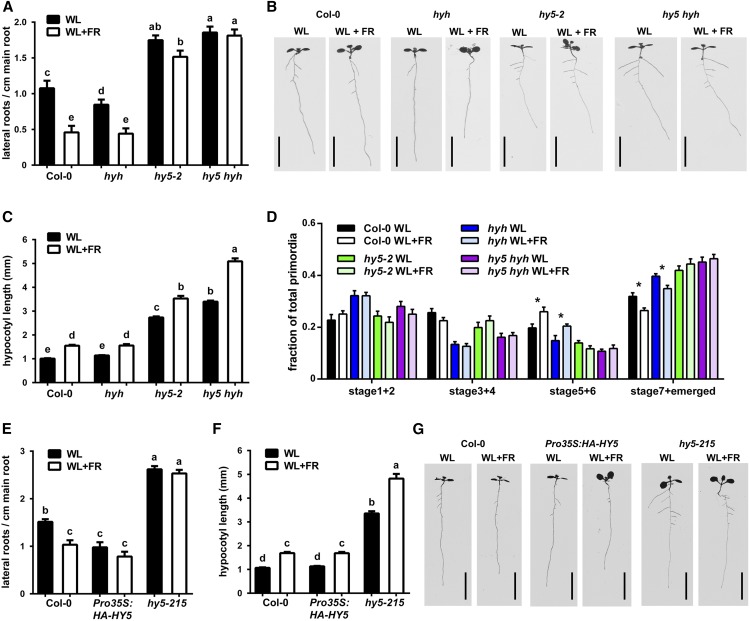
HY5 is a transcription factor involved in a broad range of responses to, among others, light cues (Gangappa and Botto, 2016). Recently a study found that HY5 can be transported from the shoot to the root, through the phloem, when induced in light conditions advantageous for photosynthesis (Chen et al., 2016). We performed a microarray analysis using root RNA of 7-d-old plants in WL and WL+FR. Although fold changes were relatively low, possibly due to the dilution of LRPs in the rest of the root RNA (Supplemental Data Set 1), we did find 409 differentially expressed genes. Among these, there was a significant enrichment of HY5 transcription factor-promoter binding targets (based on Lee et al., 2007 and Zhang et al., 2011) (Supplemental Figure 3A). This prompted us to investigate mutants of HY5 and HY5 HOMOLOG (HYH) in our WL+FR setup. Compared with the wild type, the hy5 hyh double mutant had a higher LR density in control conditions and, contrary to Col-0, this density did not decrease in WL+FR (Figures 3A and 3B). The hyh hy5 mutant had a very long hypocotyl in WL; however, it elongated in WL+FR at a similar rate to Col-0 wild type (Figure 3C, Table 1). The hy5 single mutant also did not show a significant difference in LR density between WL and WL+FR (Figures 3A and 3B). The hyh single mutant had a slightly decreased LR density in control conditions compared with Col-0; however, its LR density was still significantly decreased in WL+FR (Figures 3A and 3B). Next, we analyzed the LRPs from this experiment and found that Col-0 and hyh had an increase in the fraction of stage 5+6 primordia in WL+FR compared with WL (Figure 3D). The hy5-2 and hy5 hyh double mutants did not have any differences in the proportion of LRP stages between WL and WL+FR and generally had a far higher proportion of emerged primordia. This indicated that HY5 represses LR formation and that this transcription factor plays a major role in the FR-enriched LR decrease. This indication was further supported by an experiment using a Pro35S:HA-HY5 line (Li et al., 2011b), which showed a constitutively low LR density in WL and WL+FR (Figures 3E to 3G). We also included another knockout allele of hy5 (hy5-215), which showed a phenotype similar to hy5-2 and had a constitutively high LR density (Figures 3E to 3G).
Figure 3.
HY5 Is Involved in Supplemental FR-Induced LR Reduction.
(A) LR density of 8-d-old Col-0, hyh, hy5-2, and hy5-2 hyh seedlings in WL and WL+FR.
(B) Representative scans of 8-d-old seedlings.
(C) Hypocotyl lengths of experiment shown in (A) and (B).
(D) LRP analysis of seedlings from (A) to (C).
(E) and (F) LR density and hypocotyl length of 8-d-old seedlings, Col-0, hy5-215, and Pro35S:HA-HY5 lines in WL and WL+FR.
(G) Representative scans of 8-d-old seedlings from (E) and (F).
Bars = 1 cm. Letters show statistically significant classes, P < 0.05, one-way ANOVA. Asterisk indicates significant statistical difference, P < 0.05, two-way-ANOVA. Error bars show se; n = 15 to 20 seedlings per treatment.
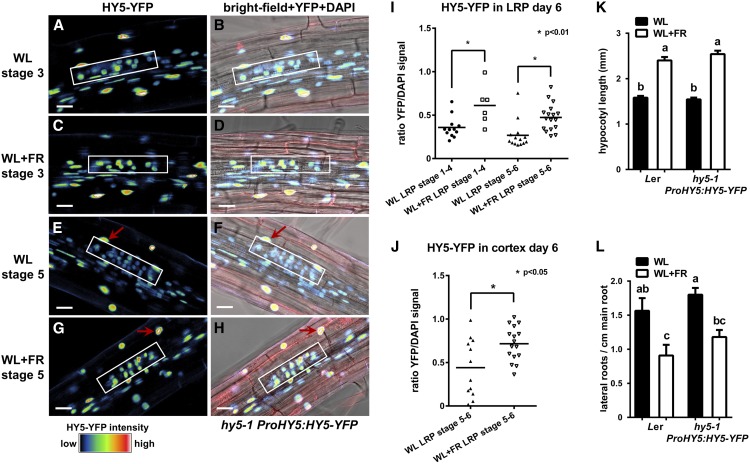
As HY5 is known to be stabilized in the shoot in low R:FR (Pacín et al., 2016), we next investigated changes in HY5 protein level around LRPs with confocal microscopy analysis using the hy5-1 ProHY5:HY5-YFP line (Oravecz et al., 2006). In WL, HY5-YFP appeared to be present less in the LRPs, compared with cortex and epidermis (Figures 4A and 4E). Compared with WL, HY5-YFP in WL+FR was present at higher levels in LRPs of 6-d-old seedlings, both in earlier (stage 1+2+3+4) and later (stage 5+6) stages (Figures 4A to 4I). This agreed well with the repressive role of HY5 in LR formation (Figure 3) (Sibout et al., 2006) because in the wild type the number of emerged LRs in WL+FR is less. Furthermore, the abundance of HY5 observed in the nucleus of the cortex cell overlaying stage 5+6 LRPs was increased in WL+FR as well (Figure 4J). We confirmed that this hy5-1 PropHY5:HY5-YFP complementing line indeed had a wild-type (Ler) response to WL+FR in our standard root phenotyping experiment (Figures 4K and 4L).
Figure 4.
HY5-YFP Is Increased in the LRP and Cortex above LRP during WL+FR.
(A) to (H) Representative confocal microscopy images of LRPs (white box) from 6-d-old hy5-1 ProHY5:HY5-YFP seedlings grown in either WL or WL+FR. Images are of stage 3 ([A] to [D]) and stage 5 ([E] to [H]) LRPs, YFP signal ([A], [C], [E], and [G]), and bright-field plus YFP and 4′,6-diamidino-2-phenylindole (DAPI) ([B], [D], [F], and [H]). Bars = 20 µm.
(I) Quantification of LRP stages 1+2+3+4 and 5+6 of experiment shown in (A) and (H) of the nuclear YFP signal normalized against DAPI staining in the cortex and epidermis.
(J) Quantification of the YFP signal from the nucleus of the cortex cell above the LRP (red arrow in [E] to [H]). Asterisk indicates statistically significant difference, P < 0.01, one-way-ANOVA. Error bars show se; for microscopy, n = 8 seedlings per treatment; for phenotyping, n = 20 seedlings per treatment.
To investigate transcriptional changes in HY5 and HYH, we isolated root and shoot RNA from growth day 4 to day 7 of our experimental setup and performed an RT-qPCR analysis. We observed a significant upregulation of HY5 in WL+FR in the shoot and root on day 4; however, this upregulation mostly disappeared at day 5-7 (Supplemental Figures 3B and 3C). HYH was increased in the shoot, but not in the root (Supplemental Figures 3D and 3E). It was recently shown that HY5 binds to its own promoter to enhance its own expression (Abbas et al., 2014). It is possible that HY5 protein traveling from the shoot to the root enhances expression of HY5 transcripts in the root. We therefore compared HY5 and HYH expression in the hy5-215 mutant background with Col-0 in both WL and WL+FR, using primers designed to anneal to the first exon, to circumvent the hy5-215 mutation, which is located in the last base of the first intron (Oyama et al., 1997). Using these primers, we detected a strong decrease in HY5 expression in the root of hy5-215 compared with Col-0, but not in the hypocotyl or cotyledon (Supplemental Figures 3F to 3K), consistent with functional HY5 regulating its own expression. HYH expression in the root showed a similar dependence on HY5.
Taken together, these results show that HY5 is necessary for the supplemental FR-induced LR reduction, consistent with the hypothesis that HY5 acts as a shoot-to-root signal during FR enrichment of the shoot.
Auxin Application Rescues LR Density Decrease under WL+FR, but Is Unlikely to Be the Shoot-to-Root Signal in This Context
Since LR formation is highly dependent upon auxin transport (Lavenus et al., 2013), we performed an experiment in which auxin was directly applied to the agar plate in a range of concentrations. Although low doses of indole-3-acetic acid (IAA; 1, 10, and 30 nM) hardly affected hypocotyl elongation (Supplemental Figure 4A), they did rescue LR density in WL+FR to that of WL levels (Supplemental Figure 4B); 10 and 30 nM of IAA had an additional limiting effect on main root growth, but a stimulatory effect on lateral root length (Supplemental Figures 4C to 4H). In a separate experiment with 10 nM of IAA added to the medium, we quantified LRPs and found that the WL+FR-induced accumulation of stage 5-6 primordia was lost and LR emergence was restored to control light levels (Supplemental Figures 4I and 4J). Interestingly, quantification of the Prodr5V2:tdTomato auxin reporter signal from 4-d-old seedlings showed that the auxin response increased in the cotyledons and hypocotyl during the WL+FR treatment (Supplemental Figures 4K and 4L) but that overall auxin signaling in the root was not affected. An overexpression line of the low R:FR-induced auxin biosynthesis gene YUCCA8 (Pro35S:YUC8), a yuc8 yuc9 double knockout line, and a knockout of the auxin biosynthesis gene TAA1 (wei8-1) all showed a LR density reduction in WL+FR, despite the changes in LR density values in WL (Supplemental Figures 5A to 5D). Of these mutant lines, both yuc8 yuc9 and wei8-1 did not have any hypocotyl elongation response to WL+FR (Supplemental Figures 5E to 5H). Additionally, in WL+FR, YUC8 expression did increase in the shoot, but not in the root (Supplemental Figure 5I). These results show that a WL+FR-induced change in auxin biosynthesis in the shoot does not necessarily affect the root response to WL+FR, since both the yuc8 yuc9 and wei8-1 mutants did not respond to WL+FR with an enhanced hypocotyl length, but still had an LR density reduction. Also, the increase in WL+FR-induced auxin signaling in the shoot in Prodr5V2:tdTomato did not affect the overall root auxin signal.
Established Regulators of LR Emergence Are Involved in WL+FR LR Reduction
Using the Pro35S:DII-venusYFP reporter line (DII-vYFP; Brunoud et al., 2012), we were able to follow auxin concentrations more directly and in more detail around the LRP (Figures 5A to 5F). In the LRP itself, no effects of WL+FR on nuclear DII-vYFP were detected; however, in the cortex cells overlaying stage 4 to 6 primordia, an increase in DII-vYFP was observed, indicating a decreased amount of auxin (Figure 5G). The auxin response in the cortex layer above the primordium is of great importance for the penetration of the LR primordium through the root (Vilches-Barro and Maizel, 2015) and the increase in stage 5+6 primordia in WL+FR reported above gave further indication of a reduction in LR emergence.
Figure 5.
In WL+FR, Auxin Is Decreased in the Cortex Overlaying Stage 4-6 LRPs.
(A) to (F) Representative confocal microscopy images of 6-d-old seedlings expressing Pro35S:NLS-DII-vYFP and stained with DAPI. Bars = 20 µm.
(A) and (D) DII-YFP signal of WL-grown (A) or WL+FR-grown (D) seedlings; arrow points to the nucleus of the cortex cell above the LRP.
(B) and (E) DAPI staining image of (A) and (D); arrow denotes measured nucleus.
(C) and (F) Bright-field image merged with YFP and DAPI signal; arrow denotes measured nucleus.
(G) Quantification of DII-YFP signal of cortex nuclei situated above stage 4-6 LRPs, normalized against the DAPI signal. Letters depict significant difference (P < 0.05, Student’s t test). Error bars show se; n = 10 seedlings.
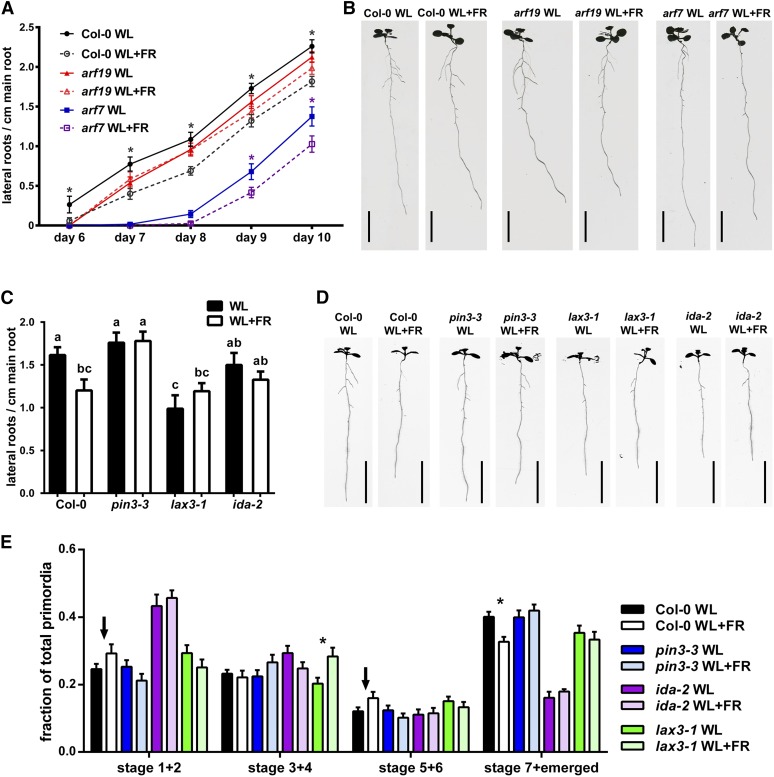
To investigate this notion, we used mutants of genes known to be involved in the process of LR emergence. AUXIN RESPONSE FACTOR7 (ARF7) and ARF19 are transcription factors involved in LR emergence and formation directly downstream of the auxin response (Okushima et al., 2007; Ubeda-Tomás et al., 2008; De Rybel et al., 2010; Goh et al., 2012; Porco et al., 2016). We therefore followed lateral root formation kinetics in the arf7-1 and arf19-1 mutants under WL and WL+FR light conditions. Although arf7-1 forms fewer LRs than Col-0, it is still further inhibited by WL+FR light. Interestingly, arf19-1 is only slightly impaired in LR formation under WL conditions, but does not display any WL+FR-mediated inhibition of LR density at all (Figures 6A and 6B). In addition, we investigated three other lateral root emergence mutants, ida-2, lax3-1, and pin3-3, of which pin3-3 and lax3-1 are auxin transport mutants. In control conditions, lax3-1 had a reduced LR density compared with Col-0 (Figures 6C and 6D), consistent with previous literature (Swarup et al., 2008).
Figure 6.
LR Formation Mutants Show No WL+FR-Induced LR Density Decrease.
(A) Time-course analysis of LR density of Col-0 (dark circles), arf19-1 (red triangles), and arf7-1 (blue squares) in WL or WL+FR. The x axis displays days after germination.
(B) Representative scans of 10-d-old seedlings from (A). Bars = 1 cm.
(C) LR density 8 d after germination of Col-0, pin3-3, lax3-1, and ida-2 grown in either WL or WL+FR with representative scans shown in (D). Bars = 1 cm.
(E) LRP analysis from experiment shown in (C) and (D); the arrows highlight the trend of increasing stage 1+2 and 5+6 in WL+FR Col-0 seedlings. Asterisk indicates statistical significance between WL and WL+FR, P > 0.05, one-way ANOVA. Error bars show se; n = 15 to 20 seedlings per treatment.
Interestingly, pin3-3 and ida-2 did not have a reduced LR density in WL compared with Col-0, which is different from earlier studies (Kumpf et al., 2013; Chen et al., 2015) and might be related to the fact that we kept the roots in darkness rather than in the light. In WL+FR, ida-2, lax3-1, and pin3-3 did not have a reduced LR density compared with WL (Figures 6C and 6D), showing that these LR emergence regulators are involved in the LR root response to WL+FR. These mutants still showed a WL+FR-mediated reduction in main root length, indicating their specificity to the LR response (Supplemental Figure 6H). Furthermore, pin3-3, lax3-1, and ida-2 mutants had no difference between WL and WL+FR in the frequency of stage 1+2 and 5+6 primordia as opposed to the distribution of these stages in Col-0 in WL+FR (Figure 6E). Unexpectedly, lax3-1 accumulated more stage 3+4 LRPs in WL+FR.
HY5 Regulates Root-Cortical Plasma Membrane Abundance of PIN3-GFP and LAX3-YFP
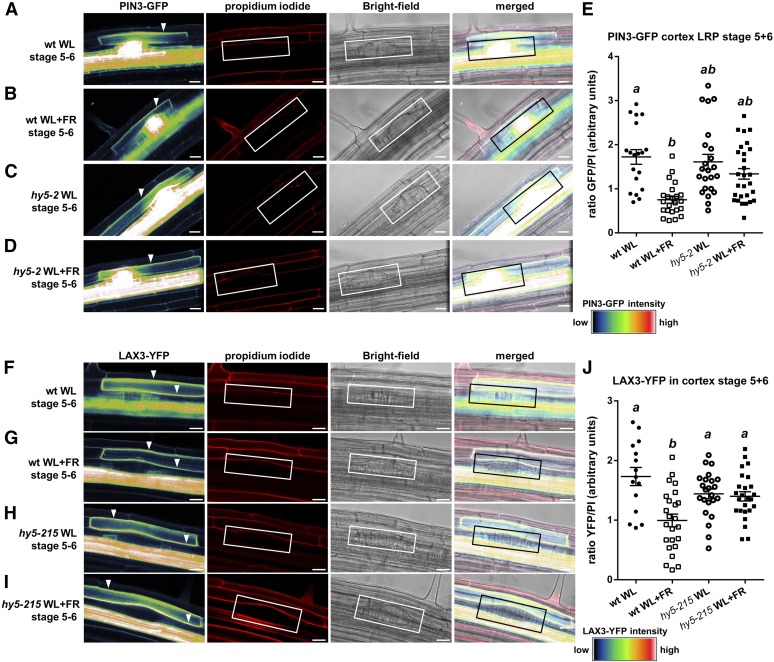
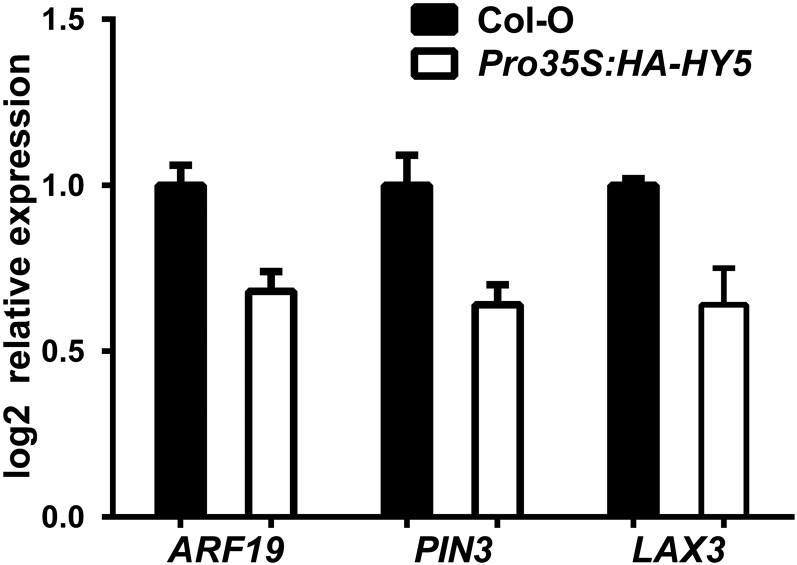
PIN3 and LAX3 regulate LR emergence by cooperatively and sequentially promoting auxin transport to the endodermis, cortex, and epidermis, where this phytohormone is needed for LR emergence (Péret et al., 2013). We subsequently studied their subcellular localization in WL and WL+FR and investigated if they are regulated in a HY5-dependent manner. We used pin3-4 ProPIN3:PIN3-GFP (Zádníková et al., 2010) and lax3-1 ProLAX3:LAX3-YFP (Swarup et al., 2008) lines and crossed them with hy5-2 and hy5-215 respectively. Confocal microscopy images of 6-d-old pin3-4 PIN3-GFP seedlings revealed that the plasma membrane signal in the cortex cell above stage 5+6 LRPs was decreased in WL+FR (Figures 7A, 7B, and 7E). In the hy5-2 background, PIN3-GFP intensity in the aforementioned cortex cells did not differ significantly between WL+FR and WL and was at a similar level as PIN3-GFP in pin3-4 ProPIN3:PIN3-GFP WL (Figures 7C to 7E). In ProLAX3:LAX3-YFP, LAX3-YFP plasma membrane signal in the cortex was also decreased in WL+FR, similar to PIN3-GFP. Furthermore, LAX3-YFP in the hy5-215 background was the same between WL+FR and WL (Figures 7F to 7J). We used the Pro35S:HA-HY5 line from Figures 3E to 3G for an expression analysis, comparing it against Col-0 in WL, and found the expression of ARF19, PIN3, and LAX3 to be reduced in the HY5 overexpression line (Figure 8). However, we observed no significant transcriptional differences for PIN3 and LAX3 between WL and WL+FR (Supplemental Figure 5I). The LAX3 and PIN3 promoters were not found to be bound by HY5 in a ChIP-microarray study (Lee et al., 2007); nevertheless, it is possible that the native HY5 effect is extremely local and we could not detect it due to dilution effects. Together, these results show that HY5 is required for a WL+FR-mediated reduction of the plasma membrane abundance of PIN3 and LAX3 in the cortex, which is a critical area involved in LR emergence (Péret et al., 2012, 2013).
Figure 7.
PIN3-YFP and LAX3-GFP Plasma Membrane Abundance in the Cortex Cell Overlaying the LRP Is Decreased in WL+FR Conditions and Is Regulated through HY5.
(A) to (D) Representative confocal microscopy images of stage 5+6 LRPs from pin3-4 ProPIN3:PIN3-GFP seedlings in Col-0 ([A] and [B]) or hy5-2 ([C] and [D]) background grown for 6 d in WL ([A] and [C]) or WL+FR ([B] and [D]). Left panel: GFP signal (white arrowhead shows plasma membrane signal used for quantification). Middle left panel: Propidium iodide (PI) staining (white box demarcates the LRP). Middle right panel: Bright-field image. Right panel: Merge of previous three panels.
(E) Quantification of the plasma membrane signal of PIN3-GFP from stage 5+6 LRPs as shown in (A) to (D) normalized against the PI staining (wt = pin3-4 ProPIN3:PIN3-GFP and hy5-2 = hy5-2 pin3-4 ProPIN3:PIN3-GFP).
(F) to (I) Representative confocal microscopy images of stage 5+6 LRPs from lax3-1 ProLAX3:LAX3-YFP seedlings in Col-0 ([F] and [G]) or hy5-215 ([H] and [I]) set up and presented similarly as in (A) to (D).
(J) Quantification of the plasma membrane signal of LAX3-YFP from stage 5+6 LRPs as shown in (G) to (I) normalized against the PI staining (wt = lax3-1 ProLAX3:LAX3-YFP and hy5-215 = hy5-215 lax3-1 ProLAX3:LAX3-YFP). Letters depict different statistically significant classes, P < 0.05 one-way ANOVA. Error bars show se; n = 10 to 12 seedlings per treatment. Bars = 20 µm.
Figure 8.
HY5 Overexpression Leads to Lower ARF19, PIN3, and LAX3 Expression Levels.
qPCR experiment using root material of 6-d-old Col-0 and Pro35S:HA-HY5 seedlings. Three biological replicates were performed with 15 seedlings per sample. Relative expression was calculated using the ΔΔCt method. Error bars show the se.
DISCUSSION
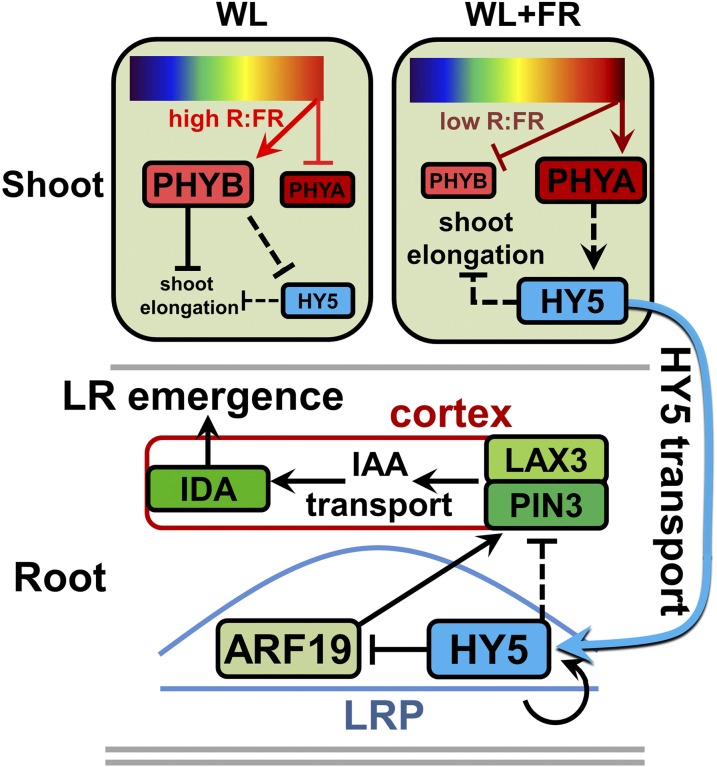
We have shown that supplemental FR enrichment of the shoot leads to a reduction in LR density. This decrease was caused by a reduction in LR emergence, which was HY5 dependent. HY5-YFP was detected at higher levels in the LRP in WL+FR. The plasma membrane abundance of PIN3-GFP and LAX3-YFP was downregulated in WL+FR in the cortex cell above the developing LRP in a HY5-dependent manner, putatively decreasing the auxin concentration in this cortex cell, as measured with DII-YFP. This reduction in auxin can lead to a reduced IDA expression (Kumpf et al., 2013) and a reduction in LR emergence (Figure 9).
Figure 9.
Enhanced Availability of HY5 under FR Enrichment Inhibits Lateral Root Emergence.
Model depicting our hypothesis for the stabilization of HY5 in WL+FR (supplemental FR light, low R:FR ratio) and its subsequent action in the LRP leading to decreased LR emergence. Top part illustrates the shoot. In WL+FR, phyB is converted to the inactive form (Pr) by FR light (dark T-bar), which releases repression of PIFs, leading to shoot elongation. In WL, phyB indirectly promotes HY5 degradation (dashed T-bar), which is relieved in WL+FR, where phyA indirectly promotes HY5 stabilization (dashed arrow). HY5 indirectly represses shoot elongation in both WL and WL+FR (dashed T-bar). The size of the boxes reflects the protein amounts. Bottom part illustrates the root. HY5 is small enough to be transported to the root through the phloem and in the root HY5 can induce its own transcription (circular arrow). HY5 has a negative effect on PIN3 and LAX3 levels in the cortex overlaying the LRP, although it is not clear if this is a direct effect (dashed T-bar). One way of achieving this is to reduce the expression of ARF19 (dark T-bar and arrow). Lower PIN3 and LAX3 abundance leads to reduced auxin concentrations in the overlaying cortex cell (red box, black arrows), which is necessary for IDA induction and cell separation. Ultimately this leads to a reduced LR emergence. Red boxes are the PHYs, green boxes are auxin signaling and transport components, and HY5 is blue.
Although HY5 could be controlled by FR light transmission from the shoot into the roots, we did not find evidence in our experiments for a functional consequence of such potential light piping. Direct application of FR light to the root did lead to a reduced root growth similar to shoot applied FR; however, only when FR light was directly applied to the root did we observe changes in root-localized PHYB-GFP photobodies. FR enrichment of the shoot had no such effect on photobodies in the roots. Also, FR light applied to the root did not lead to changes in hypocotyl length. Since transmission of light through tissues should not be direction dependent, FR light application on the root would be expected to stimulate hypocotyl length if FR light transmission occurred at physiologically meaningful levels. Possibly, light transmission through the vasculature in physiologically meaningful quantities likely occurs mostly in mature, woody tissues of adult plants (Sun et al., 2003, 2005; Lee et al., 2016). Therefore, we propose that WL+FR signaling in the shoot initiates a signaling transduction cascade that controls LR formation remotely in the root system with HY5 playing a central role.
HY5 is known to act negatively on auxin signaling (Cluis et al., 2004) and LR development (Sibout et al., 2006). In our system, HYH acted redundantly with HY5, but gauging from the single mutant phenotype, probably plays a minor role. In WL+FR, PIN3-GFP and LAX3-YFP plasma membrane abundance in the cortex was decreased in a HY5-dependent manner. The PIN3 and LAX3 signals in the cortex are thought to be tightly regulated transcriptionally in time (Péret et al., 2013), but PIN3 and LAX3 have not been identified as HY5 targets in ChIP-seq analyses. Indeed, PIN3 does not have a high affinity for the HY5 binding site in its promoter (RRTGACGTVD), while LAX3 has one 2.5 kb upstream of the start codon and both have a few core C/G box motifs (ACGT) ∼2.5 kb upstream of the start codon (Song et al., 2008). The expression change observed in Figure 8 does suggest HY5 can regulate PIN3 and LAX3 expression negatively, either directly or indirectly. It is at the same time likely that HY5 regulates ARF19, whose promoter it can bind (Lee et al., 2007) and whose expression is also lowered by HY5 overexpression. Previously it was shown that ARF19 acts redundantly with ARF7 to control LR emergence (Okushima et al., 2005); however, we found that the inhibition of LR emergence by WL+FR was dependent on ARF19, but not ARF7. It has been shown that ARF7 induces the expression of PIN3, LAX3, and IDA and ARF19 is assumed to induce the same targets (Lavenus et al., 2013). We therefore propose that HY5 negatively affects the transcription of ARF19, thereby affecting PIN3 and LAX3 transcription in a negative manner (Figure 9). It is possible that in the microarray (and qPCR) experiments with Col-0 in WL+FR, the rest of the root material diluted out the changes in the LRPs. A future direction of study would probably benefit from cell type-specific expression analyses.
HY5 was upregulated in WL+FR and HY5 is known to be stabilized in low R:FR (Pacín et al., 2016). HY5-GFP can be transported toward the root, probably through the phloem (Chen et al., 2016). We observed that HY5-YFP increased in the LRPs and overlaying cortex during WL+FR, so HY5 may well be the shoot-to-root mobile factor in this context. Although grafting experiments could test this hypothesis, we found that grafted very young seedlings are not suited for quantitative physiological experiments: The vasculature in the grafting junction is typically perturbed (Marsch-Martínez et al., 2013), grafting recovery takes several days, and grafted plant sizes are variable. It is thus still an open question how and when HY5 is unloaded in the root. According to recent literature, small proteins (<70 kD) can be freely taken up into the phloem of the shoot via the companion cells next to the sieve elements, after which they could then be unloaded at the root tip into the phloem pole pericycle (Ross-Elliott et al., 2017). Since HY5-GFP was shown to be mobile despite the increased size of the HY5-GFP protein compared with HY5 itself, we expect that HY5-YFP (38 kD) can also be mobile via the phloem. HY5 can induce transcription of its own gene in the root (Supplemental Figure 3); thus, an increase in HY5 protein can lead to an increase in HY5 expression (Lee et al., 2007; Binkert et al., 2014; Campos et al., 2016; Zhang et al., 2017). It is possible that HY5 unloaded at the root tip remains in the cells close to where LRPs are initiated and then upregulates their own expression, leading to increased HY5 in the LRP. It is not known if small proteins like HY5 can be unloaded directly at the site of developing LRPs.
The fact that LR density in phyA-501 did not respond to WL+FR is interesting, since recently it has been shown that phyA and phyB both bind the SUPPRESSOR OF PHYTOCHROME A (SPA) proteins (Sheerin et al., 2015; Lu et al., 2015; Zheng et al., 2013). The degradation of HY5 involves ubiquitination by COP1 and its SPA cofactors (Huang et al., 2014). HY5 is stabilized in low R:FR (Pacín et al., 2016), whereas in WL (high R:FR), phyBPfr promotes the nuclear accumulation of SPA1 and COP1 and thereby the degradation of HY5 (Zheng et al., 2013). Contrary to phyB, low R:FR allows active phyApfr in the nucleus (Rausenberger et al., 2011) and phyA negatively influences COP1 activity (Osterlund and Deng, 1998; Osterlund et al., 2000), while phyA interacts with multiple SPAs (Sheerin et al., 2015; Lu et al., 2015). Therefore, we hypothesize that in WL+FR (low R:FR), phyA indirectly enhances the stability of HY5 (summarized in Figure 9).
In several mutant lines such as phyA-501, pif457, pin3-3, lax3-1, and ida-2, we observed the uncoupling of shoot and root responses to WL+FR, indicating that either the shoot or the root alone responded to the WL+FR stimulus. A spectacular genotype is the hyh hy5 double mutant; these seedlings had drastically elongated hypocotyls that still displayed pronounced elongation upon exposure to WL+FR, and a root system with an even higher lateral root density than the wild type in WL, which is not inhibited by WL+FR. This shows that the growth investment in the shade avoidance response of the shoot itself does not have to occur at the expense of lateral root growth.
Conclusions
FR enrichment of the shoot resulted in increased auxin signaling in the shoot; however, we did not observe a coinciding general reduction or increase of auxin signaling in the root. We therefore challenge the hypothesis that auxin is the shoot-to-root signal during WL+FR-induced lateral root growth inhibition. Physiologically relevant light transmission itself is also unlikely causal, as discussed above. We hypothesize that HY5 is the most likely candidate for shoot-to-root communication in response to shoot signaling of WL+FR, consistent with the recent observation that HY5 can move from the shoot to the root (Chen et al., 2016). Future studies should establish the tissue types involved in HY5 transport, its mode of transport and its cell type-specific site of action in modulating root development in response to neighbor-induced phytochrome signaling.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown in a controlled environment growth chamber programmed for a 16-h-light/8-h-dark cycle with a temperature of 20°C and a light level of PAR ∼140 µmol/m2/s (Philips HPI 400 W). Lines harboring point mutations were described previously: phyB-9 (Shinomura et al., 1994), phyBDE (Shalitin et al., 2002), pin3-3 (Friml et al., 2002), hy5-215 (Oyama et al., 1997), pif457 (de Wit et al., 2015), and wei8-1 (Stepanova et al., 2008). T-DNA insertion lines used are as follows: phyA-501 (SALK_014575) (Martínez-García et al., 2014), ida-2 (SALK_133209) (Stenvik et al., 2008), lax3-1 (Swarup et al., 2008), arf7-1 (SALK_040394), arf19-1 (SALK_009879) (Okushima et al., 2005), hy5-2 (SALK_056405C), hyh (WiscDsLox253D10), yuc8-1 (SALK_096110) (Sun et al., 2012), and yuc9-1 (sail_871G01). hy5-1 ProHY5:HY5-YFP (Oravecz et al., 2006), Pro35S:HA-HY5 (Li et al., 2011b), pin3-4 ProPIN3:PIN3-GFP (Zádníková et al., 2010), lax3-1 ProLAX3:LAX3-YFP (Swarup et al., 2008), Pro35S:YUC8 (Hentrich et al., 2013), phyB-9 Pro35S:PHYB-GFP (Hiltbrunner et al., 2005), Pro35S:DII-YFP (Brunoud et al., 2012), and Prodr5V2:tdTom (Liao et al., 2015) were published previously. hy5_SALK (SALK_056405C) and hy5-215 were crossed with pin3-4 ProPIN3:PIN3-GFP and lax3-1 ProLAX3:LAX3-YFP, respectively. The hyh hy5 double mutant was created by crossing hyh (WiscDsLox253D10) and hy5-2 (SALK_056405C); a similar cross was recently published (Zhang et al., 2017). Genotyping on T-DNA insertions was performed with a left border primer on the T-DNA combined with a left and right primer on the genome. The right primer + left border primer yielded the T-DNA PCR fragment. Genotyping on point mutations was performed by sequencing a PCR fragment made with primers flanking the mutation. All primers used for genotyping can be found in Supplemental Table 1.
Plant Growth on Square Plates and Root Quantification
For root phenotyping, seeds were surface sterilized using a 2- to 3-h treatment with chlorine gas. Seeds were sown on 0.5× Murashige and Skoog, 0.1% MES, pH 5.8, 0.8% plant agar plates, at 25 per plate (12.5 × 12.5 × 1.75 cm) on a single line at 9-cm height. The D-root insert was placed in the plate just below the seeds and then the plates were put in darkness at 4°C for 6 d for seed stratification. Plates were put in the growth chamber (16/8 light/dark photoperiod, PAR = 140 µmol m−2 s−1, 21°C) for 2 to 3 h after subjective dawn and allowed to germinate for 24 h, after which either white light treatment continued or the WL+FR treatment started. The root part of the plates was covered with a custom made black paper cover measuring 12.5 × 9 × 1.7 cm. For the WL+FR treatment, plates were placed 20 cm in front of a row of FR LEDs (Phillips GreenPower LED research module far red, 24Vdc/10 W, 730-nm peak), to achieve a R:FR ratio of 0.1 in the shoot part, which was measured inside the plate using a small, flexible R:FR light meter (Skye Spectrosense2, with a custom-made sensor part). For further information on LED lamps, growth lights, and spectra, see Gommers et al. (2017). For the experiments with direct application of red light, Phillips GreenPower deep-red LEDs were used at 20-cm distance and in this case larger black covers were constructed to cover the whole 20 cm from the red light source to the plate. After 4 d of growth, seedlings were transferred in the afternoon to a new plate at five seedlings per plate to ensure homogeneous growth and prevent intermingling of root systems. Square Petri dishes were scanned at 600 dpi using an EPSON V850 photonegative scanner. Time series were analyzed using Smartroot (Lobet et al., 2011), while scans of 8-d-old plants were analyzed using WinRhizo Arabidopsis (http://regent.qc.ca/assets/winrhizo_software.html). After scanning, seedling hypocotyl lengths were determined manually.
Plant Growth on Sand
Seedlings were stratified on soil for 4 d. Plants were grown in control light conditions (as described above) for 4 d and then transferred to 70-mL pots containing sand and were kept covered to prevent dehydration. Pots were initially watered with 12 mL nutrient solution per pot [composition: 2.6 mM KNO3, 2.0 mM Ca(NO3)2, 0.6 mM KH2PO4, 0.9 mM MgSO4, 6.6 mM MnSO4, 2.8 mM ZnSO4, 0.5 mM CuSO4, 66 mM H3BO3, 0.8 mM Na2MoO4, and 134 mM Fe-EDTA, pH 5.8; based on Millenaar et al., 2005]. At day 3 and day 6, 2 mL of tap water was added per pot. When the plants were transferred to sand, they were put in a WL+FR compartment (R:FR 0.1) or a control WL compartment. After 7 d of treatment, plants were carefully removed from the pots, the roots carefully washed and scanned at 600 dpi using an Epson V850 photonegative scanner. Lateral roots were counted manually from the images and hypocotyl lengths were determined by manual measurement.
DIC and Epifluorescence Microscopy and LRP Analysis
Seedlings used for LRP analysis were fixed and cleared according to a previously published protocol (Malamy and Benfey, 1997). The seedlings were mounted in 50% glycerol and analyzed using a Zeiss Axioskop2 DIC (differential interference contrast) microscope (40× Plan-NEOFLUAR DIC objective) with a Lumenera Infinity 1 camera. Primordia were counted manually. To image Prodr5V2:tdTom, we used a Leica MZ16FA stereofluorescence microscope with a Planapo 2.0× objective, a Leica DFC420 C camera, and a dsRed filter.
Confocal Microscopy and Analysis
For confocal microscopy, seeds were sown at 14 per plate and were not transferred to another plate during their 6 d of growth. The remaining conditions were as described for the root analysis. Whole seedling roots were mounted on glass slides in water with 1 µg/mL propidium iodide or incubated for 1 min in 1 µg/mL DAPI in water and then washed in water before mounting. For PIN3-GFP, DII-YFP, LAX3-YFP, and HY5-YFP analysis, a Zeiss axioplan LSM5 Pascal microscope was used with an excitation laser of 488 nm and a 500- to 530-nm band-pass filter for GFP and a 505- to 560-nm band-pass for YFP. Images were acquired using a 40× NA1.2 water immersion objective. Within experiments, pinhole, gain, laser power, and detector offset were the same. Single slice images were taken medially through the LRP. For PHYB-GFP, confocal microscopy was performed with a Zeiss Observer Z1 LSM7 confocal imaging system, with a 488-nm excitation laser and a 505- to 560-nm band-pass filter. Slice thickness was always 70 µm and confocal z-stack images were made of 18 to 20 slices of the elongation zone using a 63× NA1.4 oil immersion objective. Within experiments, pinhole, gain, laser power, and detector offset were the same. All images were analyzed using ICY (http://icy.bioimageanalysis.org/). Z-stacks were projected using the built-in projection plug-in at max and PHYB-GFP photobodies were semiautomatically analyzed using the Spotdetector plug-in (Olivo-Marin, 2002).
RNA Extraction and qPCR
For all expression analyses, plants were not transferred at day 4 but were sown 25, 19, 16, and 14 in a row for 4, 5, 6, and 7 d of growth, respectively. The other growth conditions were as described above. The Qiagen plant RNeasy kit was used for RNA extraction. First-strand cDNA was made using the Thermo Scientific RevertAid H Minus Reverse Transcriptase, RiboLock RNase inhibitor, and Invitrogen random hexamer primers. RNA input into the cDNA reaction was kept equal within experiments. Primers were designed preferably across introns and for 100- to 150-bp fragments with an annealing temperature of ∼60°C with primer3plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Primers were tested for efficiency using generic Col-0 cDNA at a concentration range of 2.5→40 ng of cDNA per 5 μL reaction. qPCR reagents used were Bio-Rad SYBR-Green Mastermix on 384-well plates in a Life Technologies ViiA7 real-time PCR system. All CT values were normalized against two validated housekeeping genes: ADENINE PHOSPHORIBOSYL TRANSFERASE1 (APT1) and PROTEIN PHOSPHATASE 2A SUBUNIT A3 (PP2AA3). The ΔΔCT method was used to calculate relative expression values. Primer sequences are provided in Supplemental Table 1.
Microarray Analysis
Material for the microarray analysis was obtained by extracting RNA from ∼25 7-d-old whole-root systems of plants exposed to WL or WL+FR. The other growth conditions were as described above. From this RNA, cDNA from three independent replicate samples was hybridized with a Aragene 1.0 microarray. Raw CEL files were processed using R with Bioconductor and oligo (documentation: http://wiki.bits.vib.be/index.php/Analyze_your_own_microarray_data_in_R/Bioconductor#Open_CEL_files_from_newer_Affymetrix_Arrays_.28HTA.2C_Gene_ST. . ..29_using_oligo). However, background noise correction was omitted due to excessively high medium expression intensities making this method unusable. Differentially expressed genes were selected based on an false discovery rate of P < 0.05.
Statistical Analyses
All statistical significance calculations (except for microarray data) were made using Graphpad Prism software. One-way or two-way ANOVAs were performed with a post-hoc Tukey test at a significance level of P < 0.05. All root phenotype experiments had an n of 15 to 20.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: APT1, AT1G27450; ARF7, AT5G20730; ARF19, AT1G19220; HY5, AT5G11260; HYH, AT3G17609; IDA, AT1G68765; LAX3, AT1G77690; PhyA, AT1G09570; PhyB, AT2G18790; PhyD, AT4G16250; PhyE, AT4G18130; PIF4, AT2G43010; PIF5, AT3G59060; PIF7, AT5G61270; PIN3, AT1G70940; PP2AA3, AT1G13320; TAA1, AT1G70560; YUC8, AT4G28720; and YUC9, AT1G04180.
Supplemental Data
Supplemental Figure 1. Additional LR length data of Figures 1 and 2.
Supplemental Figure 2. Direct red and far-red light applied to the root leads to reduced LR density and PHYB-GFP photobody differences, but not to changes in hypocotyl length.
Supplemental Figure 3. Transcriptional responses in the root and shoot to WL+FR light experienced by the shoot.
Supplemental Figure 4. Auxin application can rescue the WL+FR-induced decrease in LR emergence.
Supplemental Figure 5. Auxin biosynthesis mutants have a reduced LR density in WL+FR.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. Microarray data complementary to Supplemental Figure 3A.
Acknowledgments
We thank Juan Carlos del Pozo for providing details and resources of the D-root system and Christian Fankhauser, Dolf Weijers, Rongfeng Huang, and Roman Ulm for sharing Arabidopsis seeds. We thank Koen Bensink for his help with the qPCR on Pro35S:HA-HY5. This research was funded by the Netherlands Organisation for Scientific Research, open competition grant 823.01.013 and Vidi grant 864.12.003 to R.P., a scholarship of government sponsorship for overseas study, Taiwan, admission number 0991167-2-UK-004 to C.K., and EMBO long-term fellowship EMBO ALTF 407-2015 to S.H.
AUTHOR CONTRIBUTIONS
R. Pierik and D.K. conceived the project. K.v.G., R. Pierik, and C.K. designed the experiments. K.v.G., C.K., D.K., and R. Paalman performed all experiments. K.v.G., R. Pierik, C.K., and S.H. wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Abbas N., Maurya J.P., Senapati D., Gangappa S.N., Chattopadhyay S. (2014). Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26: 1036–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D., Gallego-Bartolomé J., Orlando L., García-Cárcel L., Rubio V., Martínez C., Frigerio M., Iglesias-Pedraz J.M., Espinosa A., Deng X.W., Blázquez M.A. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53: 324–335. [DOI] [PubMed] [Google Scholar]
- Ballaré C.L., Pierik R. (2017). The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. 40: 2530–2543. [DOI] [PubMed] [Google Scholar]
- Baster P., Robert S., Kleine-Vehn J., Vanneste S., Kania U., Grunewald W., De Rybel B., Beeckman T., Friml J. (2013). SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 32: 260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R.P., Eklöf J., Ljung K., Marchant A., Bennett M., Sandberg G. (2002). Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29: 325–332. [DOI] [PubMed] [Google Scholar]
- Binkert M., Kozma-Bognár L., Terecskei K., De Veylder L., Nagy F., Ulm R. (2014). UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J., Vernoux T. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106. [DOI] [PubMed] [Google Scholar]
- Campos M.L., Yoshida Y., Major I.T., de Oliveira Ferreira D., Weraduwage S.M., Froehlich J.E., Johnson B.F., Kramer D.M., Jander G., Sharkey T.D., Howe G.A. (2016). Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang J., Neff M.M., Hong S.-W., Zhang H., Deng X.-W., Xiong L. (2008). Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. USA 105: 4495–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2015). A coherent transcriptional feed-forward motif model for mediating auxin-sensitive PIN3 expression during lateral root development. Nat. Commun. 6: 8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yao Q., Gao X., Jiang C., Harberd N.P., Fu X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26: 640–646. [DOI] [PubMed] [Google Scholar]
- Cluis C.P., Mouchel C.F., Hardtke C.S. (2004). The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 38: 332–347. [DOI] [PubMed] [Google Scholar]
- De Rybel B., et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20: 1697–1706. [DOI] [PubMed] [Google Scholar]
- De Smet I., et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690. [DOI] [PubMed] [Google Scholar]
- de Wit M., Galvão V.C., Fankhauser C. (2016a). Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 67: 513–537. [DOI] [PubMed] [Google Scholar]
- de Wit M., Keuskamp D.H., Bongers F.J., Hornitschek P., Gommers C.M.M., Reinen E., Martínez-Cerón C., Fankhauser C., Pierik R. (2016b). Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr. Biol. 26: 3320–3326. [DOI] [PubMed] [Google Scholar]
- de Wit M., Ljung K., Fankhauser C. (2015). Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 208: 198–209. [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809. [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia C.S.S., Julkowska M.M.M., Darwish E., Gandullo J., Korver R.A., Brunoud G., Haring M.A., Munnik T., Vernoux T., Testerink C. (2013). Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 23: 2044–2050. [DOI] [PubMed] [Google Scholar]
- Gangappa S.N., Botto J.F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9: 1353–1365. [DOI] [PubMed] [Google Scholar]
- Goh T., Joi S., Mimura T., Fukaki H. (2012). The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893. [DOI] [PubMed] [Google Scholar]
- Gommers C.M.M., Keuskamp D.H., Buti S., van Veen H., Koevoets I.T., Reinen E., Voesenek L.A.C.J., Pierik R. (2017). Molecular profiles of contrasting shade response strategies in wild plants: Differential control of immunity and shoot elongation. Plant Cell 29: 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers C.M.M., Visser E.J.W., St Onge K.R., Voesenek L.A., Pierik R. (2013). Shade tolerance: when growing tall is not an option. Trends Plant Sci. 18: 65–71. [DOI] [PubMed] [Google Scholar]
- Gundel P.E., Pierik R., Mommer L., Ballaré C.L. (2014). Competing neighbors: light perception and root function. Oecologia 176: 1–10. [DOI] [PubMed] [Google Scholar]
- Hentrich M., Böttcher C., Düchting P., Cheng Y., Zhao Y., Berkowitz O., Masle J., Medina J., Pollmann S. (2013). The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 74: 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130. [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Deng X.W. (2014). Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 21: 96–103. [DOI] [PubMed] [Google Scholar]
- Keuskamp D.H., Pollmann S., Voesenek L.A.C.J., Peeters A.J.M., Pierik R. (2010). Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. USA 107: 22740–22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen M.V., Schmid-Siegert E., Trevisan M., Petrolati L.A., Sénéchal F., Müller-Moulé P., Maloof J., Xenarios I., Fankhauser C. (2016). Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf R.P., Shi C.-L., Larrieu A., Stø I.M., Butenko M.A., Péret B., Riiser E.S., Bennett M.J., Aalen R.B. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 110: 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J., Goh T., Roberts I., Guyomarc’h S., Lucas M., De Smet I., Fukaki H., Beeckman T., Bennett M., Laplaze L. (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18: 450–458. [DOI] [PubMed] [Google Scholar]
- Lee H.-J.H., et al. (2016). Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci. Signal. 9: ra106. [DOI] [PubMed] [Google Scholar]
- Lee H.W., Kim M.J., Kim N.Y., Lee S.H., Kim J. (2013). LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 73: 212–224. [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011a). Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang L., Yu Y., Quan R., Zhang Z., Zhang H., Huang R. (2011b). The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 68: 88–99. [DOI] [PubMed] [Google Scholar]
- Liao C.-Y., Smet W., Brunoud G., Yoshida S., Vernoux T., Weijers D. (2015). Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12: 207–210, 2, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G., Pagès L., Draye X. (2011). A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 157: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Lu X.-D., Zhou C.-M., Xu P.-B., Luo Q., Lian H.-L., Yang H.-Q. (2015). Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8: 467–478. [DOI] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Marsch-Martínez N., Franken J., Gonzalez-Aguilera K.L., de Folter S., Angenent G., Alvarez-Buylla E.R. (2013). An efficient flat-surface collar-free grafting method for Arabidopsis thaliana seedlings. Plant Methods 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Gallemí M., Molina-Contreras M.J., Llorente B., Bevilaqua M.R.R., Quail P.H. (2014). The shade avoidance syndrome in Arabidopsis: the antagonistic role of phytochrome a and B differentiates vegetation proximity and canopy shade. PLoS One 9: e109275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar F.F., Cox M.C., van Berkel Y.E., Welschen R.A., Pierik R., Voesenek L.A., Peeters A.J. (2005). Ethylene-induced differential growth of petioles in Arabidopsis. Analyzing natural variation, response kinetics, and regulation. Plant Physiol. 137: 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Moulé P., Nozue K., Pytlak M.L., Palmer C.M., Covington M.F., Wallace A.D., Harmer S.L., Maloof J.N. (2016). YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 4: e2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawkar G.M., Kang C.H., Maibam P., Park J.H., Jung Y.J., Chae H.B., Chi Y.H., Jung I.J., Kim W.Y., Yun D.-J., Lee S.Y. (2017). HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: 2084–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.-L., Tepperman J.M., Stanley D.J., Maltby D.A., Gross J.D., Burlingame A.L., Wang Z.-Y., Quail P.H. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo-Marin J.C. (2002). Extraction of spots in biological images using multiscale products. Pattern Recognit. 35: 1989–1996. [Google Scholar]
- Oravecz A., Baumann A., Máté Z., Brzezinska A., Molinier J., Oakeley E.J., Adám E., Schäfer E., Nagy F., Ulm R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Deng X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16: 201–208. [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacín M., Semmoloni M., Legris M., Finlayson S.A., Casal J.J. (2016). Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol. 211: 967–979. [DOI] [PubMed] [Google Scholar]
- Pacín M., Legris M., Casal J.J. (2013). COP1 re-accumulates in the nucleus under shade. Plant J. 75: 631–641. [DOI] [PubMed] [Google Scholar]
- Péret B., et al. (2012). AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B., et al. (2013). Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol. Syst. Biol. 9: 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco S., et al. (2016). Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 143: 3340–3349. [DOI] [PubMed] [Google Scholar]
- Procko C., Crenshaw C.M., Ljung K., Noel J.P., Chory J. (2014). Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 165: 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825. [DOI] [PubMed] [Google Scholar]
- Ross-Elliott T.J., et al. (2017). Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. eLife 6: e24125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury F.J., Hall A., Grierson C.S., Halliday K.J. (2007). Phytochrome coordinates Arabidopsis shoot and root development. Plant J. 50: 429–438. [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yang H., Mockler T.C., Maymon M., Guo H., Whitelam G.C., Lin C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767. [DOI] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.-D., Huq E., Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin A.-Y., Han Y.-J., Baek A., Ahn T., Kim S.Y., Nguyen T.S., Son M., Lee K.W., Shen Y., Song P.-S., Kim J.-I. (2016). Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7: 11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T., Nagatani A., Chory J., Furuya M. (1994). The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol. 104: 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R., Sukumar P., Hettiarachchi C., Holm M., Muday G.K., Hardtke C.S. (2006). Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2: e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Navas J., Moreno-Risueno M.A., Manzano C., Pallero-Baena M., Navarro-Neila S., Téllez-Robledo B., Garcia-Mina J.M., Baigorri R., Gallego F.J., del Pozo J.C. (2015). D-Root: a system for cultivating plants with the roots in darkness or under different light conditions. Plant J. 84: 244–255. [DOI] [PubMed] [Google Scholar]
- Smith H. (1982). Light quality, photoperception, and plant strategy. Annu. Rev. Plant Physiol. 33: 481–518. [Google Scholar]
- Song Y.H., et al. (2008). DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins. Plant Physiol. 146: 1862–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G.-E., Tandstad N.M., Guo Y., Shi C.-L., Kristiansen W., Holmgren A., Clark S.E., Aalen R.B., Butenko M.A. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.-Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Yoda K., Suzuki H. (2005). Internal axial light conduction in the stems and roots of herbaceous plants. J. Exp. Bot. 56: 191–203. [DOI] [PubMed] [Google Scholar]
- Sun Q., Yoda K., Suzuki M., Suzuki H. (2003). Vascular tissue in the stem and roots of woody plants can conduct light. J. Exp. Bot. 54: 1627–1635. [DOI] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954. [DOI] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Johansson H., Lee K.P., Bou-Torrent J., Stewart K., Steel G., Rodríguez-Concepción M., Halliday K.J. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupkin S.A., Legris M., Buchovsky A.S., Tolava Rivero M.B., Casal J.J. (2014). Phytochrome B nuclear bodies respond to the low red to far-red ratio and to the reduced irradiance of canopy shade in Arabidopsis. Plant Physiol. 165: 1698–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Swarup R., Coates J., Swarup K., Laplaze L., Beemster G.T.S., Hedden P., Bhalerao R., Bennett M.J. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10: 625–628. [DOI] [PubMed] [Google Scholar]
- Van Buskirk E.K., Reddy A.K., Nagatani A., Chen M. (2014). Photobody localization of phytochrome B is tightly correlated with prolonged and light-dependent inhibition of hypocotyl elongation in the dark. Plant Physiol. 165: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Habricot Y., Condiff A.S., Maldiney R., Van der Straeten D., Ahmad M. (2007). HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 49: 428–441. [DOI] [PubMed] [Google Scholar]
- Vermeer J.E.M., von Wangenheim D., Barberon M., Lee Y., Stelzer E.H.K., Maizel A., Geldner N. (2014). A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183. [DOI] [PubMed] [Google Scholar]
- Vilches-Barro A., Maizel A. (2015). Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 23: 31–38. [DOI] [PubMed] [Google Scholar]
- Weller J.L., Hecht V., Vander Schoor J.K., Davidson S.E., Ross J.J. (2009). Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 21: 800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádníková P., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617. [DOI] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang K.X., Xu H.H., Yuan T.T., Zhang L., Lu Y.T. (2013). Blue-light-induced PIN3 polarization for root negative phototropic response in Arabidopsis. Plant J. 76: 308–321. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li C., Zhang J., Wang J., Yang J., Lv Y., Yang N., Liu J., Wang X., Palfalvi G., Wang G., Zheng L. (2017). Dissection of HY5/HYH expression in Arabidopsis reveals a root-autonomous HY5-mediated photomorphogenic pathway. PLoS One 12: e0180449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., et al. (2013). Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]