Characterization of a novel mutant phenotype in Setaria viridis reveals a role for brassinosteroids in regulating meristem fate during inflorescence development.
Abstract
Inflorescence architecture is a key determinant of yield potential in many crops and is patterned by the organization and developmental fate of axillary meristems. In cereals, flowers and grain are borne from spikelets, which differentiate in the final iteration of axillary meristem branching. In Setaria spp, inflorescence branches terminate in either a spikelet or a sterile bristle, and these structures appear to be paired. In this work, we leverage Setaria viridis to investigate a role for the phytohormones brassinosteroids (BRs) in specifying bristle identity and maintaining spikelet meristem determinacy. We report the molecular identification and characterization of the Bristleless1 (Bsl1) locus in S. viridis, which encodes a rate-limiting enzyme in BR biosynthesis. Loss-of-function bsl1 mutants fail to initiate a bristle identity program, resulting in homeotic conversion of bristles to spikelets. In addition, spikelet meristem determinacy is altered in the mutants, which produce two florets per spikelet instead of one. Both of these phenotypes provide avenues for enhanced grain production in cereal crops. Our results indicate that the spatiotemporal restriction of BR biosynthesis at boundary domains influences meristem fate decisions during inflorescence development. The bsl1 mutants provide insight into the molecular basis underlying morphological variation in inflorescence architecture.
INTRODUCTION
Plant architecture, including the number and placement of lateral organs, is determined by the position and fate of differentiating stem cell populations called meristems (Pautler et al., 2013; Tanaka et al., 2013; Gaillochet and Lohmann, 2015). Meristems that are indeterminate maintain primary meristematic activity, producing daughter cells to grow the plant body, and concomitantly give rise to new axillary meristems (AMs) on their flanks. These AMs can take on one of three possible fates: They too can be indeterminate, they can be determinate and poised to differentiate into a structure such as a leaf or a flower, or they can simply cease development. Much of what we know about inflorescence development comes from model eudicot species (e.g., Arabidopsis thaliana) where an indeterminate inflorescence meristem (IM) gives rise to determinate floral meristems (FMs), which produce flowers. In grass species (family Poaceae), inflorescence architecture is more complex; the IM produces a series of AM types, which finally terminate in short, specialized branches called spikelets, which bear flowers and grain (Bartlett and Thompson, 2014; Zhang and Yuan, 2014). Some grass species (e.g., sorghum [Sorghum bicolor] and Setaria spp) iteratively generate multiple orders of indeterminate AMs called branch meristems (BMs) to produce a highly branched inflorescence. BMs ultimately acquire a spikelet meristem (SM) fate, which specifies spikelet identity, including formation of FMs and subtending glumes. Elucidating the complex molecular framework that modulates the developmental transitions of these meristems is critical to understanding the evolution of various morphologies in grasses, as well as improved crop productivity. For example, the length of a branch, the number of branches, and number of flowers formed all influence grain bearing potential.
A number of regulators of SM identity and determinacy have been identified, yet relatively little is known about how they interact within a larger regulatory framework to control inflorescence architecture in cereals. The maize (Zea mays) branched silkless1 (bd1) gene encodes an AP2-ERF transcription factor that specifies SM identity. In loss-of-function bd1 mutants, SMs take on BM fate and spikelets are replaced by indeterminate branches (Chuck et al., 2002). During specification of SM identity, bd1 mRNA accumulates in a boundary region between the indeterminate meristem and differentiating lateral organ, where it is predicted to suppress ectopic expression of other meristem identity genes in the SM. The orthologous genes in rice (Oryza sativa; FRIZZY PANICLE; Komatsu et al., 2003) and in Brachypodium distachyon (MORE SPIKELETS1; Derbyshire and Byrne, 2013) also function in SM specification, suggesting this role is conserved across grasses. Yet, variation across grass clades is observed in this core morphology, which must result from underlying diversity in the molecular mechanisms that pattern diverse inflorescence architectures. For example, species in the Andropogoneae tribe of panicoid grasses (e.g., maize and sorghum) produce spikelets in pairs, which initiate from a spikelet pair meristem (Kellogg, 2007). Members of the “bristle clade” of grasses (Doust and Kellogg, 2002), including the model grass Setaria viridis and cultivated millet species (Huang et al., 2016), produce sterile, modified branches in their inflorescences called bristles. In these species, after several rounds of branching, BMs differentiate into either a spikelet or a bristle (Supplemental Figure 1), but the nature of this fate decision is unknown. Phenotypic characterization of early inflorescence development in representative “bristle” species suggested that bristle and spikelet are formed as a pair (Doust and Kellogg, 2002), but whether a bristle originates from a modified SM program is unclear.
Meristem maintenance, determinacy, and differentiation are controlled by a complex web of gene regulatory interactions and phytohormone signaling networks. Brassinosteroids (BRs) are polyhydroxylated steroid phytohormones with pleiotropic effects on development (Clouse and Sasse, 1998; Zhu et al., 2013). Spatiotemporal regulation of BR accumulation in Arabidopsis was implicated in formation and maintenance of organ boundaries. Feedback regulation between BRs and boundary identity genes such as CUP-SHAPED COTYLEDON (CUC) and LATERAL ORGAN BOUNDARIES (LOB) excluded growth-promoting BRs from boundaries (Bell et al., 2012; Gendron et al., 2012). Since BRs act locally (Symons et al., 2008), restricted BR synthesis limits growth to subpopulations of cells. Varying levels of BRs can also alter the accumulation or action of other phytohormones such as gibberellic acid (GA) to promote or inhibit cell elongation (Bai et al., 2012; Tong et al., 2014; Best et al., 2016). BRs control several agronomically important traits in rice, including leaf angle and grain size, suggesting that the manipulation of BR homeostasis could be one way to enhance yields (Sakamoto et al., 2006; Vriet et al., 2012; Zhang et al., 2014). In maize, disruption of BR biosynthesis and signaling leads to pistil retention in tassel florets (Hartwig et al., 2011; Best et al., 2016) and altered leaf and auricle morphology (Kir et al., 2015), respectively. The relationship between BR accumulation and growth is not simple. For example, the BR biosynthesis gene, Dwarf4 (Dwf4), had opposite effects on yield in rice when driven by different promoters (Reuzeau et al., 2006; Wu et al., 2008) and the interaction of BR and GA mutants was developmentally context-dependent in maize (Best et al., 2016).
In this work, we demonstrate that BRs contribute to the specification of bristle identity and the maintenance of SM determinacy in S. viridis. We report the molecular identity and characterization of the Bristleless1 (Bsl1) locus in S. viridis, which encodes an inflorescence-expressed paralog of the Cytochrome P450 724B1 (CYP724B1) enzyme required for BR biosynthesis. Loss-of-function bsl1 mutants fail to initiate a bristle identity program, resulting in the homeotic conversion of bristles to spikelets. In addition, SM determinacy is altered in the mutants, which produce two florets per spikelet instead of one. Our results indicate that spatiotemporal regulation of BR biosynthesis during inflorescence development is required for organ fate decisions and provide insight into the molecular basis for morphological variation in inflorescence architecture. By exploiting the molecular genetic tools of the model system S. viridis (Brutnell et al., 2010, 2015; Huang et al., 2016), this study provides opportunities for dissecting the molecular mechanisms by which BRs modulate development and for broadening both fundamental and applied aspects of BR manipulation across panicoid grasses.
RESULTS
bsl Mutant Phenotypes in S. viridis
Screens of ∼3000 N-nitroso-N-methylurea (NMU) mutagenized M2 families of S. viridis (Huang et al., 2017) identified two bsl mutants, both of which showed defects in bristle production (Figures 1A to 1C). One mutant produced few to no bristles while the other displayed a weaker phenotype, producing substantially fewer and shorter bristles than the A10.1 reference line (Supplemental Table 1 and Supplemental Figures 1B and 1C). Both mutants were crossed to the parental A10.1 line, and the resulting progeny of the selfed F1 individuals displayed the expected Mendelian 3:1 ratio for a single locus recessive allele (31:16; P [χ2, 1 df] = 0.15 and 40:10; P [χ2, 1 df] = 0.41], respectively). We crossed the two mutants and they failed to complement, suggesting these mutations are allelic. Biallelic F1 individuals displayed a bsl phenotype and F2 progeny were all bsl. Thus, we named them bsl1-1 and bsl1-2 for strong and weak alleles, respectively.
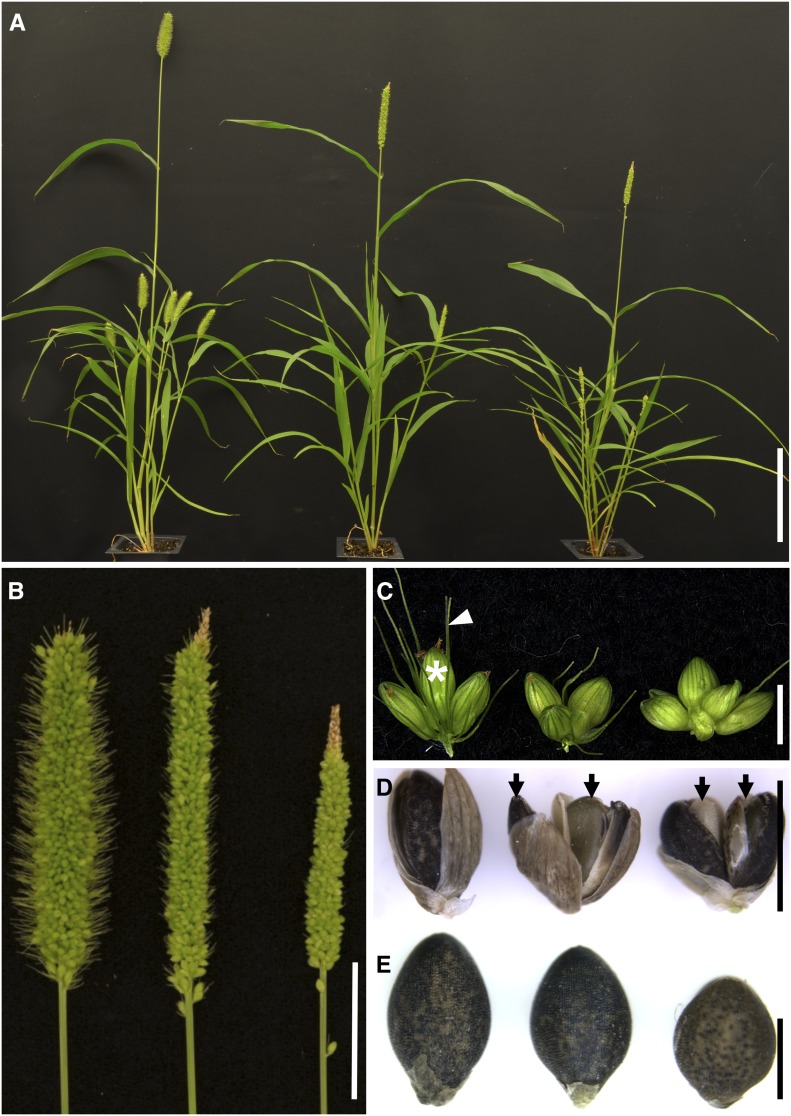
Figure 1.
The bsl Mutants in S. viridis Show Abnormal Inflorescence and Whole-Plant Phenotypes.
(A) Plant morphology of (left to right) wild-type A10.1, bsl1-2, and bsl1-1 mutants. Bar = 10 cm.
(B) Compared with wild-type A10.1 (left), bsl1-1 (right) and bsl1-2 (middle) mutant panicles are skinnier and have reduced bristle phenotypes. Bar = 2 cm.
(C) Spikelet clusters born on primary branches from (left to right) wild-type A10.1, bsl1-2, and bsl1-1 show little to no bristle development. For A10.1, a mature spikelet is marked by a white asterisk and a bristle is marked by a white arrow. Bar = 2 mm.
(D) Representative examples of two seeds (arrows) produced within a single spikelet from bsl1-2 (middle) and bsl1-1 (right), compared with the typical single seed phenotype from wild-type spikelets (left). Bar = 2 mm.
(E) Seed phenotypes of (left to right) A10.1, bsl1-2, and bsl1-1 mutants. Seed size is significantly reduced in the mutants. Bar = 1 mm.
Compared with mature panicles of wild-type A10.1 individuals, bsl1-1 mutant panicles were shorter and had few to no bristles; however, no obvious defects in spikelet or flower development were observed (Figures 1B and 1C). In both bsl1-1 and bsl1-2, we consistently observed a small number of spikelets per mature panicle that produced two seeds instead of the one typically produced from wild-type spikelets (Figure 1D). Bristles were also reduced in bsl1-2 panicles, but to a much lesser extent than in bsl1-1. Interestingly, panicle length was significantly longer in bsl1-2 compared with the wild-type control (Figure 1B); however, bsl1-2 displayed weaker phenotypes for all other traits evaluated (Supplemental Table 1). These phenotypes included skinnier panicles with primary branches clustered tighter to the main rachis (Figure 1B) and significantly smaller seeds borne from bsl1 mutants (Figure 1E). In addition to panicle phenotypes, plant architecture was also altered in bsl1 mutants, which displayed semidwarf phenotypes and increased tiller number compared with wild-type controls (Figure 1A; Supplemental Table 1).
bsl1 Mutants Show Altered Meristem Fate at Two Stages of Inflorescence Development
To investigate how bristle formation was disrupted during development, we used scanning electron microscopy to analyze developmental progression of inflorescence primordia from A10.1 wild-type plants compared with bsl1 mutants. We analyzed images from a developmental series that captured the transition to reproductive development, primary and higher order branch formation, spikelet and bristle differentiation, and floral organ development of wild-type plants (Figure 2; Supplemental Figure 2). At 11 days after sowing (DAS), the shoot apical meristem (SAM) in the wild type had transitioned to an IM and initiation of the first axillary BMs were just visible (Supplemental Figure 2A). Primary, secondary, and tertiary axillary branches developed sequentially from 11 to 15 DAS (Supplemental Figures 2A to 2D). By 16 DAS, the first SMs were initiated at the inflorescence tip and coincided with differentiation of the first bristles. SMs and bristles differentiated from BMs that were morphologically indistinguishable until this transition (Figure 2A). As was previously described, spikelets and bristles differentiated basipetally during development (Doust and Kellogg, 2002). At 17 DAS, AMs that differentiated into bristles developed an indented ring around the meristem tip, which subsequently appeared to sever the tip (Figures 2A and 2B). FM initiation followed spikelet development from 18 to 19 DAS, at which time elongated bristles were observed. By 20 DAS, floral organs had differentiated in the upper floret of each spikelet and the lower floret was evident as a meristematic bulge, which failed to differentiate and later aborted (Figures 2C and 2D).
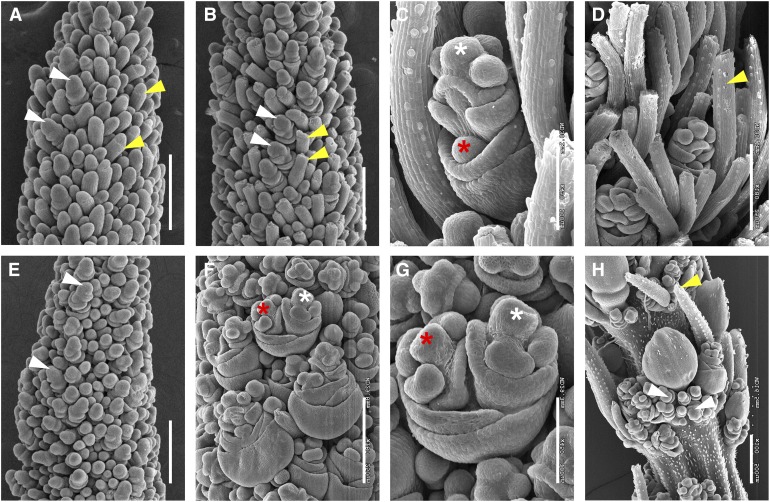
Figure 2.
Morphological Characterization of Inflorescence Development in the bsl1-1 Mutant by Scanning Electron Microscopy Analysis.
(A) and (B) Bristle development was first apparent at 17 DAS in A10.1 wild-type inflorescence primordia (A) and bristle differentiation was obvious by 18 DAS where meristem tips appeared to break off at indentations marked by yellow arrows (B). Differentiating spikelets are marked by white arrows. Bars = 250 μm.
(C) and (D) At 20 DAS, bristles were well developed in the wild-type inflorescences, and within spikelets, an upper floret (white asterisk) developed and a lower floret was visible as a meristematic bulge prior to abortion (red asterisk; [C]). A mature bristle is marked with a yellow arrow (D). Bars = 100 μm in (C) and 250 μm in (D).
(E) In the bsl1-1 mutant inflorescence primordium, bristles were not initiated by 18 DAS, but spikelets appeared to develop normally (white arrows). Bar = 250 μm.
(F) to (H) At 20 DAS, bristle development was highly reduced in the bsl1-1 mutant and upper (white asterisk) and lower (red asterisk) florets were both developed (G). Precocious development of numerous rudimentary spikelets (white arrows) was observed at the base of main spikelets (H). Yellow arrow indicates one of few differentiated bristles formed in the mutant. Bars = 250 μm in (F) and (H) and 100 μm in (G).
The same developmental transitions were investigated in the bsl1-1 mutant. Transition from vegetative to reproductive development was also observed by 11 DAS (Supplemental Figure 2E); however, subsequent developmental progression was delayed after 14 DAS in the mutant. Initiation of AMs and primary branching events appeared unaffected (Supplemental Figures 2F to 2H). By 18 DAS, SMs were formed, but not bristles (Figure 2E). SM development at the base of the inflorescence began at 19 DAS, followed by formation of FMs; however, bristle formation was not observed. Instead, the bsl1-1 mutant formed additional SMs, many of which developed into rudimentary spikelets that appeared to subtend a main spikelet and production of these continued into later stages of development (Figures 2F to 2H). These observations suggest that BMs programmed to form bristles in the wild type took on the fate of SMs in the bsl1-1 mutant. A few bristles were formed in an apparently random distribution, suggesting a threshold of some signal is required for this fate decision. By 20 DAS, floral organs of bsl1-1 spikelets differentiated normally in the upper floret. Remarkably, in many cases, the lower florets did not abort as they had in wild-type controls. Instead, lower florets in bsl1-1 appeared to develop normally, but lateral to the upper floret, likely due to spatial constraints (Figures 2F and 2G). Based on detailed analyses of the scanning electron microscopy images, lower florets appeared to develop from ∼42% of spikelets in bsl1-1 mutants.
Scanning electron microscopy analysis showed that developmental progression of bsl1-2 mutant inflorescences was similar to the wild type until bristles began to differentiate in the wild type (Supplemental Figures 3A and 3F). At this stage, spikelet differentiation was observed in bsl1-2, but bristle formation was apparently delayed (Supplemental Figures 3B and 3G). Like bsl1-1, spikelets appeared to form in place of bristles; however, by 18 DAS, bristles were differentiated in bsl1-2, but fewer compared with the wild type (Supplemental Figures 3C and 3H). Spikelets that developed both upper and lower florets were also observed in bsl1-2 (Supplemental Figures 3D, 3E, 3I, and 3J). As with most other observed phenotypes, spikelet-to-bristle ratio was intermediate in the bsl1-2 mutant compared with the wild type and bsl1-1 (Supplemental Figure 4).
bsl1 Encodes the S. viridis Ortholog of Cytochrome P450 724B1 Involved in BR Biosynthesis
To map the locus responsible for the bsl1 phenotype, we performed bulked segregant analysis (Michelmore et al., 1991; Schneeberger, 2014) using the bsl1-1 allele. DNA from 20 mutant F2 individuals derived from a bsl1-1 × wild-type A10.1 cross was pooled and sequenced to ∼24× coverage (95M reads). Reads were mapped to the A10.1 reference genome (phytozome.jgi.doe.gov; v1.1) and single nucleotide polymorphisms (SNPs) were determined. This revealed a 2-Mb region on chromosome 7 that showed high homozygosity in the bsl1-1 mutant pool (Figure 3A). Within this interval, only three homozygous SNPs resulted in nonsynonymous, start, or stop codon gain or loss, or intron splice site gain or loss. The presence of these SNPs was further validated by whole-genome sequencing of the bsl1-1 mutant genome to 30× coverage (Supplemental Data Set 1). One of these SNPs resulted in a premature stop codon within Sevir.7G140700 (+1860 from the transcriptional start site; Figure 3B), which encodes the syntenic ortholog of Dwarf11 (D11) in rice, the cytochrome P450 724B involved in BR biosynthesis (Tanabe et al., 2005; Sakamoto et al., 2006). The other two nonsynonymous changes in this interval generated missense mutations in Sevir.7G169600 and Sevir.7G173000. We also sequenced the bsl1-2 genome to 30× coverage to determine whether SNPs resided in or around these candidate genes (Supplemental Data Set 1). No SNP variation was associated with Sevir.7G169600 or Sevir.7G173000 and no nonsynonymous changes affected the coding sequence of the D11 ortholog, Sevir.7G140700; however, we did find a homozygous SNP in the second intron of this gene at +1250 from the transcriptional start site (Figure 3B).
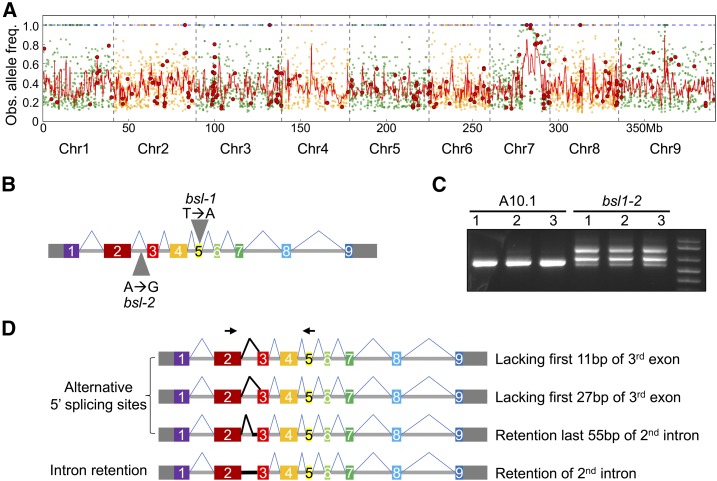
Figure 3.
Mapping of the bsl1 Locus and Analysis of Alternative Transcript Isoforms Produced by the bsl1-2 Allele.
(A) Bulked segregant analysis was performed for the bsl1-1 mutant. Genomic position is plotted on the x axis and observed mutant allele frequency on the y axis. Green and orange dots represent all SNPs of the bsl1-1 mutant genomic pool compared with the A10.1 reference genome and red dots represent nonsynonymous SNPs. The red line is a smoothed curve over a 10-SNP window.
(B) Exon-intron structure of the Bsl1 gene consists of nine exons (solid rectangles) and eight introns (horizontal line). The 5′ and 3′ untranslated regions are shown as gray rectangles. Gray triangles indicate the locations of SNPs responsible for bsl1-1 and bsl1-2 phenotypes.
(C) Gel image of RT-PCR results showing multiple transcript isoforms of Bsl1 in bsl1-2 inflorescence primordia at 15 DAS compared with A10.1.
(D) Diagrammatic image of four transcript isoforms that were detected as alternative splice variants resulting from a SNP in the 2nd intron of bsl1-2. Black arrows indicate the forward and reverse primer sites used for the RT-PCR in (C).
This SNP in bsl1-2 was intriguing since it does not disrupt a canonical splice site and would otherwise have been overlooked in a screen for SNPs with high likelihood of having a phenotypic impact. We examined the Sevir.7G140700 mRNA produced from the hypomorphic bsl1-2 allele by RT-PCR. Total RNA extracted from hand-dissected S. viridis inflorescence primordia at 15 DAS was used to synthesize cDNA for amplification of the bsl1-2 transcript products, which were purified, cloned, and sequenced (Figure 3C). Sequencing results showed that the mRNA produced by bsl1-2 was alternatively spliced compared with the A10.1 control. We observed transcripts with alternative 5′ splice sites and intron retention. Four transcript isoforms unique to the bsl1-2 mutant were identified: (1) retaining the full 2nd intron, (2) an alternative 5′ splice donor site 2 bp after the SNP site, (3) lacking the first 27 bp of the 3rd exon, and (4) lacking the first 11 bp of the 3rd exon (Figure 3D; Supplemental Figure 5). We did not detect any wild-type Bsl1 transcripts in the bsl1-2 mutant and hypothesize that the hypomorphic phenotype observed in bsl1-2 is due to partial functionality of the protein product encoded by the mRNA isoform with the in-frame deletion of the first 27 bp of the 3rd exon. The finding that bsl1-1 and bsl1-2 fail to complement and have SNP variation affecting dramatic changes in the coding capacities of the same gene demonstrate that loss of Sevir.7G140700 function is responsible for the bsl1 mutant phenotypes. We cannot rule out the influence of other SNPs fixed in the mutant populations on seemingly unrelated phenotypes, such as the elongated panicle phenotype displayed by bsl1-2 (Figure 1B), since several rounds of backcrossing were not performed. While the mechanistic basis of the intronic bsl1-2 SNP on splicing is unknown, emerging evidence in animal systems implicates cis-regulatory features in introns as key regulators of splicing (Fu and Ares, 2014).
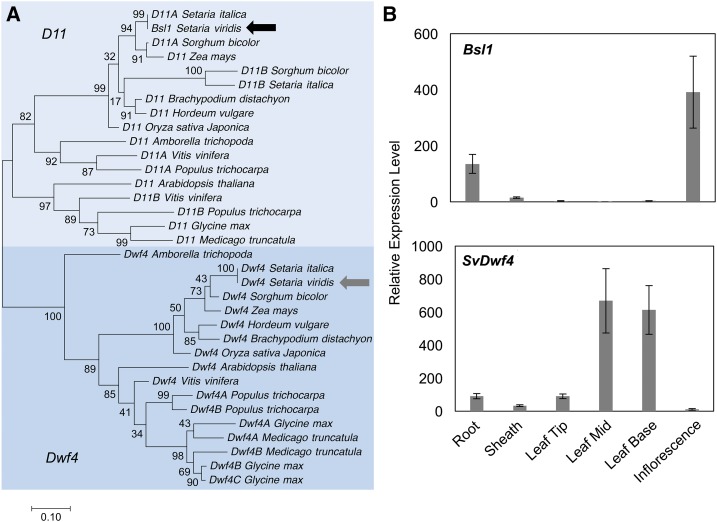
Two distinct clades of cytochrome P450 genes redundantly perform C-22 hydroxylation, the rate-limiting step in BR biosynthesis (Fujita et al., 2006; Sakamoto et al., 2006). In rice, CYP724B1, encoded by the bsl1 ortholog D11, and CYP90B, encoded by OsDwf4, are partially redundant for C-22 hydroxylation and the loss-of-function double mutants in OsDwf4 and D11 exhibited severe dwarfism and malformed, erect leaves (Sakamoto et al., 2006). In Arabidopsis, overexpression of CYP724A1 can complement Atdwf4 mutants (Zhang et al., 2012). A phylogenetic analysis of the OsDwf4 and D11-like genes from S. viridis was performed (Figure 4A; Supplemental Data Set 2). As expected, an ortholog of AtDwf4 and OsDwf4 in S. viridis (SvDwf4; Sevir.9G483600) was identified. We examined expression of these two genes by RT-qPCR analysis and found that SvDwf4 is predominantly expressed in vegetative tissues while Bsl1 is expressed in developing inflorescence primordia as well as in roots (Figure 4B). Therefore, loss of Bsl1 would likely result in an acute loss of C-22 hydroxylation in the inflorescence.
Figure 4.
Phylogeny of Bsl1-Related (D11-Like) and Paralogous Dwf4-Like Genes in the Grasses and Tissue-Specific Expression in S. viridis.
(A) Phylogenetic analysis of D11- and Dwf4-related genes based on coding sequence. Bsl1 and SvDwf4 from S. viridis are noted by black and gray arrows, respectively.
(B) Expression profiles of Bsl1 (upper panel) and SvDwf4 (lower panel) were determined by RT-qPCR. Relative expression was quantified for Bsl1 and SvDwf4 genes in roots, sheaths, and 4th leaves from A10.1 seedlings at 20 DAS and inflorescence primordia at 18 DAS. Error bars indicate standard errors of three biological replicates.
Chemical Inhibition of BR Biosynthesis Suppresses Bristle Formation
While similar phenotypes observed in bsl1 have been described for d11 mutants in rice (e.g., semidwarf stature, small seeds, and clustered panicle branches; Tanabe et al., 2005; Wu et al., 2016), the lack of bristles is a phenotype unique to S. viridis. To test whether disruption of BR biosynthesis is responsible for the bristleless phenotype, we treated A10.1 seedlings with propiconazole (PCZ), an inhibitor of BR biosynthesis (Sekimata et al., 2002; Hartwig et al., 2012). Prior to bristle formation, 250 μM PCZ was applied as a soil drench starting at 13 DAS, and plants were compared with nontreated controls. Treated seedlings showed severe reduction in height, erect leaf angles, and delayed flowering, consistent with phenotypes typical of BR biosynthesis mutants (Clouse and Sasse, 1998) (Figure 5A). Panicles of PCZ-treated seedlings were shorter and developed few bristles, similar to the bsl1-1 mutant phenotype (Figure 5B). Scanning electron microscopy analysis of developing inflorescences from PCZ-treated wild-type plants showed that terminal AMs mostly formed spikelets rather than bristles (Figures 5C and 5D), that there was ectopic development of rudimentary spikelets (Figure 5D), and many spikelets (∼43%) developed two florets instead of one (Figure 5E), very similar to what we observed in bsl1-1 mutants. Thus, chemical disruption of BR biosynthesis recapitulated the bsl1 mutant inflorescence phenotypes. The severe vegetative phenotypes observed in PCZ-treated seedlings (e.g., dwarf and erect leaves) likely result from systematic inhibition of BR biosynthesis, as opposed to the inflorescence-specific loss affected by bsl1.
Figure 5.
Treatment with a BR Inhibitor Suppresses Bristle Formation in S. viridis Panicles.
(A) Compared with untreated wild-type (left) and bsl1-1 mutant (middle) plants, plant height was reduced after treating A10.1 wild-type seedlings with PCZ (right). Bar = 5 cm.
(B) Compared with the untreated control (left), treatment with PCZ suppressed bristle production in A10.1 wild-type panicles (right) and phenocopied untreated bsl1-1 mutant panicles (middle). Bar = 1 cm.
(C) to (E) Scanning electron microscopy analysis of inflorescences from PCZ-treated wild-type plants showed that by 17 DAS (C), there was no sign of bristle differentiation, but spikelets (white arrows) developed normally. At 19 DAS (D), ectopic development of rudimentary spikelets (white arrows) was observed as in bsl1-1 and a few randomly placed bristles (yellow arrow) were observed. Development of both upper (white asterisk) and lower (red asterisk) florets was observed in the PCZ-treated inflorescences (E). Bars = 200 μm in (C) and (D) and 50 μm in (E).
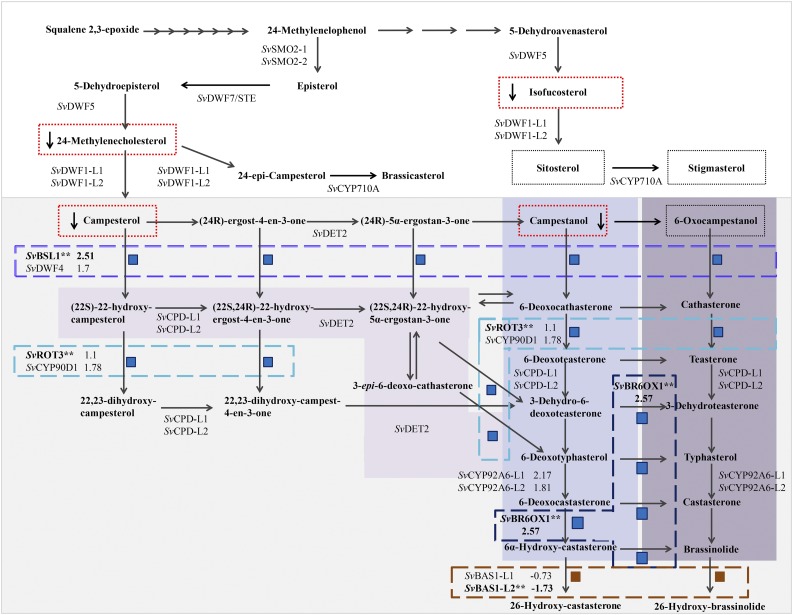
To determine if and to what extent BR metabolism was disrupted in bsl1-1 mutants, we performed a targeted liquid chromatography/mass spectrometry (LC-MS) analysis. Esterification of the 3′ hydroxyl group of BR molecules with picolinic acid was used to provide a cleavable tag for quantitation and detection of BR (Honda et al., 2008). Hand-dissected inflorescence primordia were collected from the bsl1-1 mutant and A10.1 wild-type control seedlings at ∼15 to 17 DAS; during this developmental time window, bristles are initiated and elongating in the wild type (Figure 2) and metabolites were extracted from three biological replicates. We also profiled leaf tissue collected from mutant and wild-type seedlings. Accumulation of sterol and BR metabolites were significantly affected in the bsl1-1 mutant inflorescence samples, but not in leaf tissue. We observed dramatic reductions in sterol and BR intermediates upstream of the proposed step performed by the BSL1 protein including campesterol, campestanol, 24-methylenecholesterol, and isofucosterol (Table 1, Figure 6; adapted from Ohnishi et al., 2012; Vriet et al., 2012; Chung and Choe, 2013). Consistent with the presence of a paralog predominantly expressed in leaves, we did not observe altered accumulation of sterol and BR intermediates in the vegetative tissue (Table 1). Thus, Bsl1 was required for proper BR synthesis in the inflorescence and SvDwf4 likely performs this function in the leaves. Levels of the major structural sterols, sitosterol and stigmasterol, were not significantly changed. An untargeted LC-MS analysis of lipophilic metabolites from the same samples was unable to identify any BR or sterol intermediates as differentially accumulated metabolites. However, one mass feature present at significant levels in bsl1-1 inflorescences, but not in the other samples, matched the exact mass of a pregnane ring-containing molecule, which is predicted as a cleavage product of 3-β-ketone derivatives of BR and sterol precursors 5-dehydroepisterol, 5-dehydroavenasterol, 7-dehydrodesmosterol, and 7-dehydrocholesterol (Supplemental Figure 6).
Table 1. Comparison of Detectable Endogenous BR Intermediates between bsl1-1 Mutants and A10.1 Wild Type in Inflorescence Primordia and Seedling Leaf Tissue.
| Inflorescence |
Seedling |
||||
|---|---|---|---|---|---|
| Chemical | M.W. | A10.1 (WT) | bsl1-1 | A10.1 (WT) | bsl1-1 |
| Isofucosterol | 412.3705 | 29.78 ± 4.53 | 8.85 ± 0.36* | 10.07 ± 2.06 | 9.34 ± 2.52 |
| Sitosterol | 414.3862 | 747.44 ± 122.10 | 577.03 ± 52.15 | 167.22 ± 24.23 | 155.84 ± 44.69 |
| Stigmasterol | 412.3705 | 12.38 ± 2.55 | 13.86 ± 0.46 | 10.52 ± 0.66 | 9.59 ± 2.75 |
| 24-Methylene cholesterol | 398.3549 | 3.06 ± 0.39 | 0.84 ± 0.10* | 1.25 ± 0.34 | 1.29 ± 0.34 |
| Campesterol | 400.3705 | 196.00 ± 33.73 | 122.34 ± 13.70* | 39.44 ± 4.99 | 36.85 ± 11.41 |
| Campestanol | 402.3862 | 8.06 ± 1.52 | 4.23 ± 0.87* | 1.25 ± 0.37 | 1.05 ± 0.40 |
| 6-Oxocampestanol | 416.3654 | 31.51 ± 5.23 | 25.00 ± 2.38 | 7.65 ± 0.83 | 6.65 ± 2.17 |
Values shown are µg/g fresh weight. Asterisk indicates a significant difference in metabolite level between the bsl1-1 mutant and A10.1 wild type (WT) determined by Student’s t test (P < 0.05) of three biological replicates. Inflorescence primordia samples were dissected at 15 to 18 DAS and seedling tissue from 11-DAS seedlings. M.W., molecular weight.
Figure 6.
Proposed Phytosterol and BR Biosynthetic Pathway for S. viridis Shows Where Genes Encoding Enzymes in the Pathway Were Differentially Expressed in bsl1-1 Mutant Inflorescences and Where Changes in Metabolites Were Observed.
BR biosynthesis is highlighted in gray. Subpathways highlighted in purple (from light to dark/left to right) represent the parallel early and late C-6 oxidation pathways and the early C-22 oxidation pathway for BR biosynthesis, respectively. Intermediates that were quantified in wild-type and bsl1-1 mutants are boxed by a dotted line. Those that were significantly decreased in the mutant are boxed in red. Steps catalyzed by enzymes encoded by genes that were differentially expressed in the RNA-seq analyses are boxed by dashed lines (purple = Bsl1; turquoise = SvROT3; dark blue = SvBR6OX1; burnt orange = SvBAS1-L2). Each of these DE genes (asterisks) is associated with two or more metabolic steps, each noted by a blue (upregulated in the mutant) or dark-red (downregulated in the mutant) square. Log2 fold change is indicated for DE genes, their close paralogs, and for SvCYP92A6-L1/2.
Disruption of Bsl1-Mediated BR Biosynthesis Alters Expression of Developmental Transcriptional Regulators and Hormone Signaling Networks
To gain insight into how genes and pathways were perturbed at the molecular level with loss of bsl1 function, we used RNA-seq to profile transcriptional differences in bsl1-1 inflorescence primordia compared with wild-type controls. Inflorescence primordia were hand-dissected at comparable developmental stages in the wild type and mutant based on scanning electron microscopy analyses (Supplemental Figure 2). Wild-type primordia were sampled at ∼15 DAS when bristles were just beginning to differentiate and bsl1-1 mutant primordia, which showed a slight developmental delay, were sampled at ∼16 DAS where terminal AMs were produced and matched the wild type (Supplemental Figure 2). Ten representative primordia were pooled per biological replicate. RNA-seq reads from wild-type and mutant libraries were mapped to the S. viridis reference genome (v1) and annotated gene models used to quantify transcript abundances (Supplemental Data Set 3). Differential expression (DE) analysis identified 689 genes that were significantly altered in expression (corrected P < 0.05) in bsl1-1 inflorescence primordia relative to the wild type (Supplemental Data Set 4). These DE genes were significantly enriched for transcription factors (TFs) (“transcription factor activity” GO:0003700; P = 3.12e−08), and analysis of TF family members showed overrepresentation of AP2-ERF, AUX/IAA, and MYB classes (Supplemental Figure 7). Genes involved in redox activity (“oxidoreductase activity” GO:0016491; P = 7.46e−04), those associated with responses to various stimuli, e.g., phytohormones (“response to hormone” GO:0009725; P = 2.96e−06), and “meristem determinacy” (GO:0010022; P = 7.74e−05) were also overrepresented in the DE gene set (Table 2; Supplemental Figure 8 and Supplemental Data Set 5).
Table 2. Genes Related to Meristem Identity and Determinacy That Were Differentially Expressed in bsl1-1 Mutant Inflorescence Primordia Compared to Wild-Type Controls.
| S. viridis Gene ID | FPKMa |
Log2 FCb | Significantc | Function | Descriptiond | |
|---|---|---|---|---|---|---|
| WT | bsl1-1 | |||||
| Sevir.5G266300 | 15.25 | 29.09 | 0.932 | Up** | Meristem maintenance | WUSCHEL-related homeobox (wox9 a/b) (maize) |
| Sevir.8G062700 | 3.87 | 9.90 | 1.357 | Up** | ATAF2 NAC TF (ATAF2) (Arabidopsis) | |
| Sevir.8G020500 | 0.56 | 2.91 | 2.364 | Up* | narrow sheath1 (ns1) (maize) | |
| Sevir.4G251700 | 2.25 | 0.98 | −1.197 | Down** | AT HOMEOBOX1 (ATH1) (Arabidopsis) | |
| Sevir.2G437800 | 3.81 | 24.69 | 2.698 | Up** | Meristem determinacy | branched silkless1 (bd1) (maize) |
| Sevir.2G209800 | 1.35 | 4.41 | 1.708 | Up* | ramosa1 (ra1) (maize) | |
| Sevir.7G327300 | 1.65 | 3.73 | 1.18 | Up* | BLADE-ON-PETIOLE1/2 (BOP1/2) (Arabidopsis) | |
| Sevir.3G183400 | 89.39 | 55.25 | −0.694 | Down* | ETTIN (ETT) (Arabidopsis) | |
| Sevir.1G183200 | 6.85 | 19.98 | 1.544 | Up** | Floral meristem identity/determinacy | zea centroradialis2 (zcn2) (maize) |
| Sevir.7G234000 | 174.74 | 106.96 | −0.708 | Down* | zea floricaula/leafy1/2 (zfl1/2) (maize) | |
| Sevir.9G196700 | 15.45 | 35.94 | 1.221 | Up** | Differentiation/polarity | yabby9/10 (yab9/10) (maize) |
| Sevir.7G097500 | 17.03 | 34.37 | 1.013 | Up** | LITTLE ZIPPER3 (ZPR3) (Arabidopsis) | |
| Sevir.7G121700 | 1.25 | 3.1 | 1.313 | Up* | Carpel persistence | tasselseed1 (ts1) (maize) |
| Sevir.9G439800 | 3.85 | 10.33 | 1.422 | Up** | tasselseed2 (ts2) (maize) | |
| Sevir.9G508000 | 3.13 | 7.45 | 1.252 | Up** | grassy tillers1 (gt1) (maize) | |
Expression levels indicate mean FPKM values based on RNA-seq data from A10.1 wild-type (WT) and bsl1-1 mutant inflorescence primordia.
Log2 fold change in expression in bsl1-1 mutant inflorescence primordia compared to A10.1 wild type.
Direction of fold change in mutant compared to the wild type and level of significance based on corrected P values from the differential expression analysis (*P < 0.05 and **P < 0.01).
Closest homolog with experimentally defined function in development from maize or Arabidopsis.
Homologs of well-characterized BR biosynthesis genes were significantly upregulated in bsl1-1 mutants compared with wild-type controls (Figure 6; Supplemental Table 2). This is consistent with work in other systems that showed transcripts from BR biosynthesis genes accumulated following loss of BR due to negative feedback regulation (Bancoş et al., 2002; Tanabe et al., 2005; Tanaka et al., 2005). For example, Bsl1 itself was highly increased in bsl1-1 mutants (corrected P = 0.003), as was a gene encoding a BR C-6 oxidase orthologous to brassinosteroid deficient1 (brd1) from maize (Makarevitch et al., 2012) and rice (Hong et al., 2002) (SvBrd1; Sevir.9G171700). The ortholog of the rice ebisu dwarf2 gene, which encodes CYP90D2 (SvROT3/CYP90C1; Sevir.5G137200), a P450 that catalyzes C-23 hydroxylation of BRs (Sakamoto et al., 2012), was significantly upregulated in the bsl1-1 mutant. In addition, a homolog of PHYB ACTIVATION-TAGGED SUPPRESSOR1 (BAS1), which encodes an enzyme that inactivates BRs (Neff et al., 1999), was also significantly reduced in the mutant, suggesting that inactivation was also decreased in response to a lack of BRs.
Based on the morphology of wild-type and bsl1-1 panicles at the sampled developmental stage, we expect that genes expressed at higher levels in the mutant may be involved in promoting SM fate, while those expressed higher in the wild type could be related to specification of bristle identity and differentiation. There were 402 genes (58% of the total DE genes) that were upregulated in bsl1-1 panicles and these were highly enriched for functional processes related to various aspects of hormone biology, including biosynthesis and signaling of auxin, GA, and ethylene. Genes involved in photomorphogenesis (“protein-chromophore linkage” GO:0018298; P = 1.30e−13) were also highly represented, consistent with a role for light signaling in BR-mediated cell elongation (Bai et al., 2012; Supplemental Figure 8 and Supplemental Data Set 5). In addition, a number of key developmental regulators were expressed at higher levels in bsl1-1 mutants, including orthologs of classical genes from maize that specify AM identity and determinacy (e.g., bd1 [Chuck et al., 2002] and ramosa1 [Vollbrecht et al., 2005]), that pattern lateral organ development (e.g., narrow sheath1 [Scanlon et al., 1996] and yabby10 [Juarez et al., 2004]), and those implicated in carpel repression in maize tassels (e.g., grassy tillers1 [Whipple et al., 2011], tasselseed 1 [Acosta et al., 2009], and tasselseed 2 [Irish and Nelson, 1993]) (Table 2). A gene encoding a NAC TF with homology to ATAF2 from Arabidopsis was also expressed at significantly higher levels in bsl1-1. ATAF was shown to maintain BR homeostasis through repression of BAS1, and its expression is feedback inhibited by BRs (Peng et al., 2015).
Among the 287 DE genes that were expressed higher in wild-type panicles, functional enrichment analyses showed that genes involved in nutrient sensing and signaling (“cellular response to nutrient levels”; GO:0031669; P = 8.58e−04), notably nitrogen and sugar related, were overrepresented. Also, genes related to “protein folding” (GO:0006457; P = 7.84e−04) and those responding to osmotic and starvation stress were enriched, suggesting that nutrient remobilization and stress pathways are activated during the loss of meristem activity and development of bristles (Supplemental Data Sets 4 and 5).
Spatiotemporal Expression of Bsl1 Marks Lateral Organ Boundaries during Inflorescence Development and Loss of Function Results in Ectopic SM Identity
We performed in situ hybridization experiments to determine localization of Bsl1 mRNAs at spatiotemporal resolution during S. viridis inflorescence development (Figure 7; Supplemental Figure 9). A10.1 wild-type inflorescence primordia were dissected at sequential developmental stages from 10 to 20 DAS and transcripts were detected using an antisense cDNA probe with specificity to the Bsl1 sequence. Starting at 10 DAS, Bsl1 signal marked primary branches (Supplemental Figure 9A) and persisted during the initial rounds of inflorescence branching (∼12–14 DAS) in a distinct adaxial domain at the base of primary branches (Figures 7A and 7B; Supplemental Figure 9B). At these stages, Bsl1 signal was also detected at the tip of the inflorescence and appeared to mark initiation sites of the first SMs. By 14 DAS, just prior to onset of bristle differentiation, Bsl1 signal was detected at the base of secondary and higher order axillary branches (Figures 7A and 7B). By 15 DAS, expression was no longer detected at the base of primary branches, but accumulated at lateral organ initiation sites and also within newly initiated organ primordia in developing spikelets (Figure 7C; Supplemental Figure 9C). We did not observe Bsl1 signal in newly differentiated bristles at 15 to 16 DAS. As FMs were initiated at 17 DAS, Bsl1 transcripts were detected subtending the differentiating glumes and developing floral organs, including the lower floret (Figure 7D; Supplemental Figure 9D). In bsl1-1 mutants, Bsl1 signal was comparable to the wild type at early stages of development (Supplemental Figure 9E); however, by ∼16 DAS, Bsl1 transcripts became ectopically expressed outside of the defined domain and accumulated at the base of developing spikelets (Figure 7E).
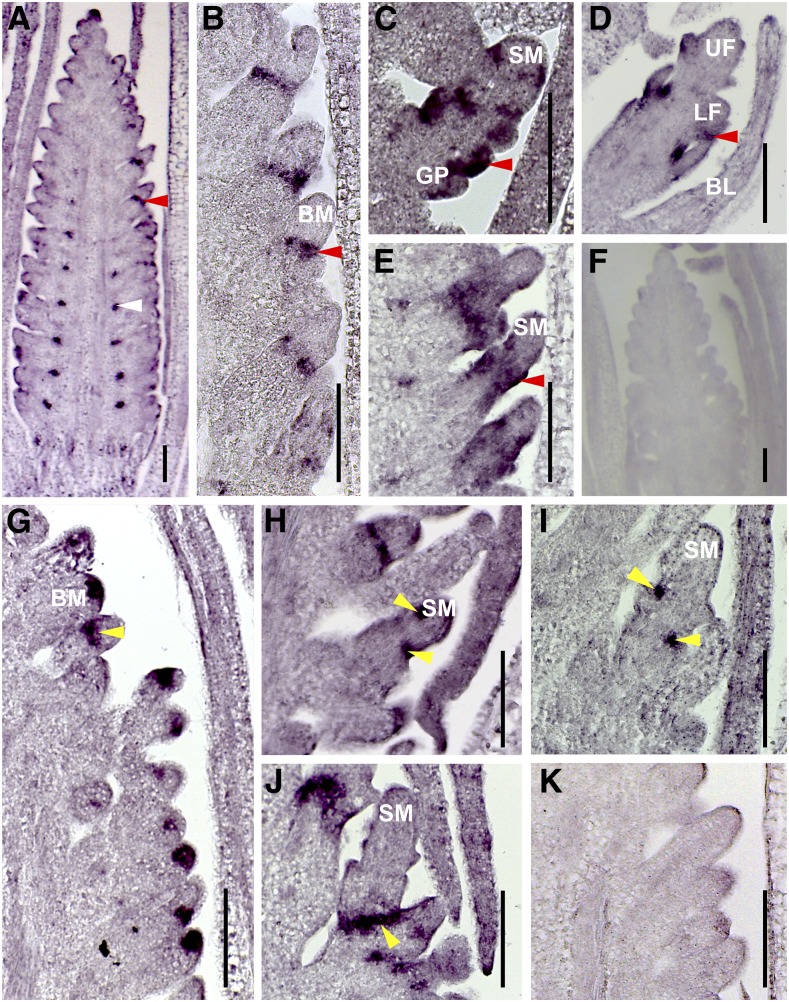
Figure 7.
Localization of Bsl1 and SvBd1 mRNAs in A10.1 Wild-Type and bsl1 Mutant Inflorescence Primordia during Development.
(A) to (E) An antisense Bsl1 probe was used to examine Bsl1 expression during inflorescence development.
(A) and (B) At 14 DAS, Bsl1 mRNAs were localized to the base of primary branches (white arrow; [A]) and lateral organ boundaries (red arrow) of higher order BMs in A10.1 wild-type inflorescences (B).
(C) and (D) Bsl1 signal (red arrows) marked incipient lateral organs in developing spikelets (C) and floral meristems (D) at 15 and 17 DAS, respectively in wild-type inflorescences. Bsl1 signal was not observed in developing bristles.
(E) In bsl1-1 mutants, the Bsl1 signal (red arrow) was expanded and mislocalized to the base of developing spikelet primordia at 16 DAS.
(F) A Bsl1 sense probe showed no signal in A10.1 wild-type inflorescence.
(G) to (J) An antisense SvBd1 probe was used to examine SvBd1 expression during inflorescence development.
(G) At 14 DAS, SvBd1 was expressed in a semicircular domain (yellow arrow) at the base of all AMs prior to spikelet or bristle formation in wild-type inflorescences.
(H) and (I) At 15 DAS (H) and 16 DAS (I), SvBd1 was expressed at the boundaries of incipient lateral organs (yellow arrows) in developing spikelets in A10.1 wild-type inflorescences, but not in bristles.
(J) In bsl1-1 mutants, SvBd1 signal was mislocalized to the base of developing spikelets (yellow arrow) at 17 DAS and expression was maintained into later stages of development.
(K) A SvBd1 sense probe showed no signal in A10.1 wild-type inflorescence.
Bars = 100 μm. BL, bristle; GP, glume primordium; UF, upper floret; LF, lower floret.
Scanning electron microscopy analyses suggest that Bsl1-mediated BR accumulation controls differentiation of bristles after the last axillary branching event. Prior to bristle differentiation, it is unclear how this fate decision is made. We did not detect differences in Bsl1 expression among terminal BMs at this stage, suggesting inherent differences between BMs that will become SMs and those that will differentiate into bristles are independent of Bsl1 expression. We next asked whether a loss of meristem activity coincided with bristle differentiation. To test this, we performed in situ hybridizations using a probe designed to the S. viridis ortholog of the maize knotted1 (kn1) gene, a key marker of meristem maintenance (Kerstetter et al., 1997; Bolduc et al., 2012) (Supplemental Figure 10). As expected, SvKn1 (Sevir.9G107600) signal was detected at the tip of every BM prior to bristle differentiation in wild-type and bsl1-1 mutant primordia (Supplemental Figures 10A and 10C). As bristles elongated in the wild type at ∼15 DAS, SvKn1 signal was lost, but was maintained at the tip of every developing SM in the wild type and bsl1-1 mutants (Supplemental Figures 10B and 10D), consistent with acquisition of bristle fate involving a loss of meristematic activity.
Next, we sought to address whether SM identity was preferentially established in certain BMs but not in those that would acquire bristle fate. Our RNA-seq analyses showed that SvBd1 (Sevir.2G437800), encoding the ortholog of a conserved AP2-ERF TF, which specifies SM identity in maize and other grasses (Chuck et al., 2002), was expressed at substantially higher levels in the bsl1-1 mutant compared with the wild type, consistent with increased production of spikelets in place of bristles (Table 2). To test whether the acquisition of spikelet identity is critical for SM versus bristle fate, we performed in situ hybridization with an antisense probe designed to SvBd1. We found that prior to bristle differentiation in 14 DAS wild-type primordia, SvBd1 signal accumulated in a crescent-shaped domain subtending all terminal BMs, similar to its localized expression in maize (Figure 7G). Remarkably, this result indicated that the SM identity program is initiated in all terminal BMs and that bristle identity is established later. This is consistent with a conserved function for SvBd1 in repressing an indeterminate BM program. Furthermore, expression of SvBd1 was transient, gradually diminishing by 16 DAS as spikelets developed, and was completely absent in differentiating bristles (Figures 7H and 7I). In bsl1-1 mutants, however, SvBd1 signal was also localized to a region subtending the developing SM, but expression persisted at the base of developing mutant spikelets later into development (Figure 7J). This persistent SvBd1 signal marked the site of ectopic SM initiation in the bsl1-1 mutants as observed by scanning electron microscopy (Figure 2H).
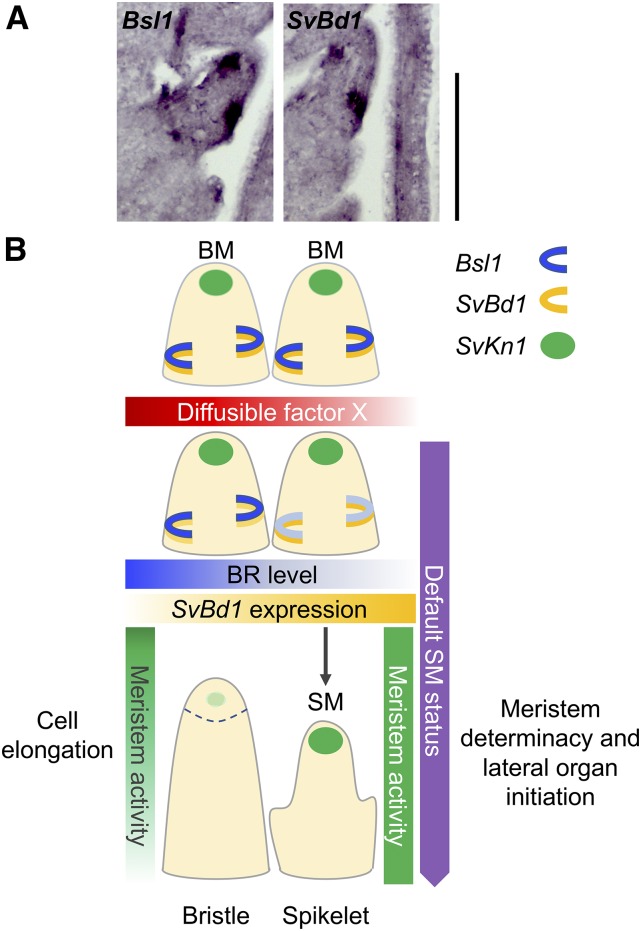
To test the spatiotemporal relationship of Bsl1 and SvBd1, we probed adjacent longitudinal sections of inflorescence primordia at 15 DAS (Figure 8A; Supplemental Figure 11). Bsl1 expression marked a domain directly adjacent to and partially overlapping that of SvBd1. Together, our results suggest that spatiotemporal accumulation of BRs may antagonize the SM identity program by repressing SvBd1 either directly or indirectly.
Figure 8.
A Proposed Model for Bsl1-Dependent BR Control of Spikelet versus Bristle Fate in S. viridis Inflorescence Development.
(A) Adjacent sections from an A10.1 wild-type inflorescence primordium at 15 DAS were probed with Bsl1 (left) and SvBd1 (right) and showed adjacent and partially overlapping domains of expression. Bars = 100 μm.
(B) One proposed model for bristle versus spikelet differentiation in a wild-type S. viridis inflorescence depends on a diffusible factor that enhances spatiotemporal accumulation of BRs. Paired BMs are indistinguishable during early development; SvKn1 (green circle) is expressed in the meristem tip and Bsl1 (blue semicircle) and SvBd1 (yellow semicircle) in adjacent domains at the sites of lateral organ initiation. BMs are poised to become determinate SMs where opposing levels of SvBd1 expression and BRs maintain the boundary. We propose that the presence of a diffusible factor that promotes BR accumulation over a certain threshold would stimulate rapid cell elongation and cessation of meristem activity, leading to formation of a bristle.
DISCUSSION
The bsl1 Mutant in S. viridis Reveals a Role for BRs in Modulating Inflorescence Architecture
In this study, we used S. viridis as a model system to investigate the role of BRs in specifying SM versus bristle fate during inflorescence development. While the functional significance of the bristle structure is unclear, our findings reveal insight into the molecular mechanisms that not only control bristle development, but also the fate decisions that modulate inflorescence architecture and developmental plasticity in grasses. We report evidence for spatiotemporal control of a BR biosynthesis gene in regulating meristem fate to pattern inflorescence form. The highly tractable “bristleless” phenotype in S. viridis provides a robust system for precisely dissecting regulation of BR biosynthesis in space and time, and the mechanisms by which it controls inflorescence development.
The bsl1 mutant phenotype suggests that production of bristles is a trade-off for yield and that through manipulation of this pathway, sterile bristles can be converted to productive spikelets, potentially increasing the number of grains on an inflorescence. Furthermore, the mutant often produces two florets per spikelet instead of one, with some producing seed from both florets, although smaller in size. These phenotypes represent two potential avenues for enhancing grain production in millets, including orphan crops that provide subsistence in developing countries and remain largely unimproved. Likewise, in economically important cereal crops such as maize, rice, and wheat (Triticum aestivum), precision engineering of localized BR biosynthesis and signaling can be harnessed to fine-tune plant development and generate high-yielding crop ideotypes (Vriet et al., 2012). By using S. viridis as a model system, we anticipate that our findings will be readily translated to these crops.
Feedback Regulation of BR Biosynthesis and Accumulation of Pathway Intermediates
BR biosynthesis is achieved by parallel and highly branched pathways. Understanding the regulation of these metabolic pathways will help determine how optimal tissue-specific concentrations of BRs are achieved during development (Fujioka and Yokota, 2003; Chung and Choe, 2013; Vriet et al., 2013). The two rate-limiting steps of BR biosynthesis are C-22 oxidation and C-6 oxidation. C-22 oxidation is catalyzed by the cytochrome P450s CYP724B1/D11/BSL1 and CYP90B1/DWF4 (Ohnishi et al., 2006; Sakamoto et al., 2006; Zhang et al., 2012), while C-6 oxidation is catalyzed by the cytochrome P450 CYP85/BR6OX1 (Shimada et al., 2001; Hong et al., 2002). BR homeostasis is controlled through transcriptional feedback regulation of these biosynthetic genes by known BR signaling TFs, BRASSINAZOLE RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1)/BZR2. The mRNAs encoding BR biosynthesis enzymes, including OsD11 (Tanabe et al., 2005), accumulate during BR deficiency, and decrease with exogenous BRs (Bancoş et al., 2002; Sun et al., 2010; Yu et al., 2011). Bsl1 is the syntenic ortholog of OsD11 (Figure 4A), and conservation of the transcriptionally regulated BR homeostatic mechanism in S. viridis was evidenced by upregulation of Bsl1, SvBr6ox1, and the S. viridis ortholog of ROT3/CYP90C1 in bsl1-1 mutant inflorescences.
BR accumulation was altered in bsl1-1 inflorescences, but not in vegetative tissue, consistent with a tissue-specific role for Bsl1. Interestingly, levels of BR precursors upstream of C-22 hydroxylation were decreased in bsl1-1 inflorescences rather than accumulated as might be predicted for a simple block in a biosynthetic pathway. However, similar trends have been observed in rice (Tanabe et al., 2005; Sakamoto et al., 2006), which suggest some negative feedback reduces precursor levels when C-22 hydroxylation is blocked. Failure to accumulate upstream biosynthetic intermediates has also been shown in the Arabidopsis BR-deficient dwarf5 mutant, defective in the delta-7 sterol reductase (Choe et al., 2000), as well as the dwarf br6-oxidase double mutants, which had lower 24-methylene cholesterol (Kwon et al., 2005). We did not observe downregulation of BR biosynthetic genes that might account for the loss of BR upstream intermediates. Thus, we expect that BR precursors are either shunted via alternative reactions into other intermediates or catabolic products, as has been demonstrated in Arabidopsis dwarf5 (Choe et al., 2000) and rice d2 mutants (Hong et al., 2005).
Our targeted profiling approach used a chemical derivatization of the 3′ hydroxyl group of the steroid A ring to form a sterol 3-β-picolinate ester. Since some BR intermediates contain a 3′ ketone, it is possible that ketone intermediates accumulated in bsl1-1 mutant inflorescences. If a catabolic shunt operates via a ketone intermediate, we would not have observed accumulation of this BR precursor. Interestingly, the unique mass feature identified in our nontargeted assay of bsl1-1 inflorescences is consistent with a C21 pregnane that would result from side chain cleavage (Kolbe et al., 1994) (Supplemental Figure 6). One possibility is that decreased levels of BR precursors (e.g., isofucosterol and 24-methylene cholesterol) resulted from increased catabolism through side chain cleavage. This may be performed by oxidative cleavage similar to the human P450 encoding cholesterol desmolase (Burstein et al., 1975), which also leaves a ketone at the C20 position, as indicated in our hypothetical structure (Supplemental Figure 6). We do not know the structure of the proposed pregnane, and many possible structures are consistent with the exact mass and chemical formula we infer from it.
BR-Mediated Regulation of Plant Development Is Dynamic and Context-Dependent
While core pathways for BR metabolism and signaling have largely been defined, little is known about the molecular mechanisms regulating BR homeostasis and how these in turn modulate plant development. Since BRs are not transported long distances, they act locally to promote growth by enhancing cell elongation and/or expansion (Symons and Reid, 2004; Symons et al., 2008). Therefore, understanding how local BR levels are maintained provides insight into BR-dependent mechanisms for spatiotemporal growth and development. BRs have been implicated in boundary formation between developing organ primordia and the SAM in Arabidopsis (Bell et al., 2012; Gendron et al., 2012). A lower rate of cell division must be maintained in boundary regions. This involves a feedback loop where BZR1 directly represses certain boundary identity genes (e.g., CUC and LOF) and others (e.g., LOB) function to keep BR from accumulating in the boundaries through direct activation of BAS1 (Neff et al., 1999; Bell et al., 2012).
Similarly, BR homeostasis is critical to meristem function. In rice, Oryza sativa HOMEOBOX1, a key regulator of meristem maintenance similar to kn1 from maize, controls local BR accumulation by directly targeting and upregulating genes encoding BR catabolism and conjugation enzymes (Tsuda et al., 2014). In roots, BRs promote division of quiescent center (QC) cells at the root stem cell niche (González-García et al., 2011; Heyman et al., 2013). A mechanism that counteracts this was recently described in Arabidopsis; the MYB TF, BRASSINOSTEROIDS AT VASCULAR AND ORGANIZING CENTER (BRAVO), is a cell-specific repressor of cell division in the QC (Vilarrasa-Blasi et al., 2014). BES1 physically interacts with and represses BRAVO, creating a tightly regulated switch that maintains rates of division in the QC. BRAVO expression also requires auxin, which was shown to act in opposition to BRs along a developmental gradient in roots. This balance between hormones underlies the dynamics between stem cell maintenance and differentiation (Chaiwanon and Wang, 2015). These studies provide evidence for BR as a positional cue or morphogen in plant development. In this work, we propose a model wherein BRs fine-tune the decision to maintain SM identity or acquire bristle fate through interactions with additional molecular factor(s) that are asymmetrically localized (Figure 8B).
A Proposed Function for bsl1 in Specifying Bristle Identity in S. viridis
Our morphological and molecular analyses of the bsl1 mutants showed that disruption of BR biosynthesis can result in homeotic conversions within the inflorescence and that spatiotemporal BR accumulation provides a mechanism for fine-tuning meristem fate decisions. The effects of loss of bsl1 function were highly dependent on spatiotemporal context. For instance, progression of the first branching events proceeds as normal in the bsl1 mutants even though in situ hybridization detected Bsl1 signal soon after the floral transition at the base of primary branches (Supplemental Figure 9). At these early stages, this signal is apparently unchanged in bsl1-1 mutants, although primary branches were obviously shorter in the mutant, causing spikelets to be arranged more tightly on the rachis. Perhaps BR accumulation at the base of primary branches helps stimulate modest growth and elongation of adaxial cells. At later stages of inflorescence development, Bsl1 signal is localized to the base of BMs and associated with boundary regions of all lateral organ primordia, yet disruption of bsl1 causes defects in production of bristles but not spikelets. Accumulation of Bsl1 signal at sites of lateral organ outgrowth appears to traverse L1 and inner meristem tissues, consistent with models for BR-mediated growth in Arabidopsis (Savaldi-Goldstein et al., 2007; Vragović et al., 2015).
Perhaps a universal role for localized BR biosynthesis adjacent to boundaries of developing organs is to fine-tune cellular growth response in coordination with other cell-specific growth promoting or repressing factors. As mentioned, BRs promote cell division and expansion in the root QC, organ primordia adjacent to boundary domains, and in localized regions of the SAM; all regions that are required to maintain low cell division and therefore molecular mechanisms are in place to keep this in check, in some cases involving spatiotemporal regulation of TFs or diffusible gradients of other hormones. We propose that spatiotemporal accumulation of BRs is required for promoting bristle differentiation in S. viridis inflorescences. Our observation that SvBd1 is expressed in all terminal meristems suggests that SM identity is the default state and that this program is suppressed in a BR-dependent manner during acquisition of bristle fate. SvBd1 appears to come on transiently in a boundary domain subtending the developing SM to specify its identity, and localized expression of Bsl1 adjacent to this boundary could modulate expression of SvBd1 via BRs.
The question remains, what differentiates SMs at the transition from SM to bristle fate? Bsl1 is expressed in all SMs and spikelet development is, for the most part, normal in the bsl1 mutant. Perhaps during normal inflorescence development, SMs poised to become bristles accumulate higher levels of BR, resulting in local increased cell division and expansion, loss of boundary identity genes, and repression of the SM identity program. In Arabidopsis roots, it was shown that BR-mediated cell elongation leads to exit from the meristem and subsequent decrease in meristem size and growth rate after an initial growth increase (González-García et al., 2011; Chaiwanon and Wang, 2015). A burst of BR-mediated elongation could displace the SM from local signals that maintain its identity until a threshold is reached, e.g., loss of boundary and ultimately loss of meristem. This hypothesis is consistent with the apparent loss of meristem activity that coincides with detachment of the BM tip as bristles differentiate (Figures 2A and 2B). We also showed that SvBd1 expression persists in bsl1-1 mutants, consistent with continued generation of SMs later into development and replacement of bristles with SMs (Figure 2H). This is in line with findings from BR deficient mutants in Arabidopsis, which produced extra floral organs due to ectopic boundary formation (Gendron et al., 2012).
Our model depends on the presence of at least one other factor that promotes spatially restricted BR accumulation and subsequent bristle outgrowth. This could be either a signal that is attenuated by BRs in meristems that become bristles (Figure 8B) or a counteracting signal present in those that become spikelets. This could be auxin, as a number of auxin response genes were misexpressed in bsl1 inflorescences, suggesting that BRs influence auxin signaling during the SM to bristle fate decision. Spatial and dose-dependent interactions between auxin and BR were shown in Arabidopsis (Chaiwanon and Wang, 2015). GA is another candidate for attenuation of the BR signal. In Arabidopsis, BR and GA each enhance the growth response in hypocotyls that is dependent on the other (Bai et al., 2012). Similarly, in rice, BR and GA levels fine-tune each other and an imbalance in either one results in distinct phenotypes (Tong et al., 2014), and in maize, this was dependent on a tissue-specific context (Best et al., 2016).
Numerous studies have identified links between BR synthesis, sensing, and signaling with other hormone and light pathways in Arabidopsis; e.g., some of the first BR mutants were isolated in screens for light-dependent hypocotyl elongation (Clouse et al., 1996; Li et al., 1996; Neff et al., 1999; Luccioni et al., 2002). Consequently, many genes at the interface of these pathways have been annotated based on the experimental context in which they were found. It is likely that modules of genes with analogous functions in various developmental contexts, e.g., grass inflorescence development, have been co-opted to link these pathways where BRs play a role. For example, a recent study described a central growth regulating module that integrates BR, GA, and light signaling (Bai et al., 2012). Our transcriptome analysis identified suites of genes that have previously been associated with light, ethylene, and other hormone signaling pathways (Supplemental Figure 8). Further investigation of these conserved gene modules in a range of developmental- and species-specific contexts should enable their functional dissection.
While our model infers spatiotemporal accumulation of BR via activity of BSL1, the coordinated activities of other BR biosynthesis and inactivation enzymes undoubtedly play integral roles in this program as well. Future work investigating outputs of local BR signaling, perhaps through localization of BZR1, will help resolve the precise mechanisms that integrate BR sensing and signaling with developmental transitions (Chaiwanon and Wang, 2015).
Persistence of the Lower Floret in bsl1 and Analogies to Carpel Retention
Our model is based on a threshold effect of BR that commits a developing SM to bristle identity. Since spikelet to bristle ratio is not random in S. viridis, the accumulation of BR and/or other factors needed to reach this threshold are likely tightly regulated. Mechanistically, such regulation is most parsimoniously described with bristles and spikelets being paired, where a diffusible gradient underlying the two developing SMs would switch on the bristle program (and switch off the SM program) in one (Figure 8B). This paired arrangement of spikelet and bristle could be analogous, in terms of a BR-mediated development switch, to the paired florets that develop within the spikelet. Perhaps at later stages of normal spikelet development, a similar localized action represses floral identity genes in the lower floret. Evidence from maize indicates that there are underlying molecular differences between the upper and lower floret, for example, in regulation of certain MADS box TFs (Thompson et al., 2009).
The retention of the lower floret in spikelets of bsl1 mutants is reminiscent of certain tasselseed (ts) mutants from maize, e.g., ts2, which also fail to abort the lower floret in the ear, leading to disorganized rows (Irish and Nelson, 1993). While most grasses produce perfect flowers that bear both male and female organs, maize produces unisexual male and female flowers on separate structures, the tassel and ear, respectively. Wild-type maize generates staminate flowers on the tassel by selective abortion of pistil primordia and pistillate flowers on the ear by arresting developing stamens (Dellaporta and Calderon-Urrea, 1994; Li and Liu, 2017). Interestingly, these phenotypes are also characteristic of maize BR-deficient mutants. For example, nana plant1 (na1) encoding a homolog of the steroid-5-α-reductase, DET2 (Hartwig et al., 2011), na2 encoding a homolog of the BR biosynthetic gene DWF1, which catalyzes the C-24 reduction step (Best et al., 2016), and Zmbrd1 (Makarevitch et al., 2012), all exhibit persistent pistils in tassel florets, in addition to dwarfism and abnormal leaf morphology (Tao et al., 2004).
GA also affects retention of floral organs in maize. Classic experiments showed that exogenous GA on maize tassels resulted in pistil retention in the tassel florets as well as increased tassel branching (Nickerson, 1959, 1960), but loss of GA resulted in stamen retention in ears (Evans and Poethig, 1995). In addition, pistil retention in tassels of BR-deficient mutants required GA biosynthesis, demonstrating that these two pathways collaborate in specifying floral organ fates (Best et al., 2016). The persistence of the lower floret in BR-deficient bsl1 mutants of S. viridis suggests a conserved role for BRs in grass inflorescence architecture. Furthermore, orthologs of genes controlling pistil retention in maize were misregulated in bsl1-1 mutants. We propose that the homeotic transformation of meristem types in the bsl1 mutants and the mechanism of pistil persistence in maize tassels may share an underlying physiological mechanism. If BRs are critical regulators of differentiation and determinacy in the inflorescence, maize tassel architecture may have evolved from the reuse of this function to reduce organ outgrowth in the pistils of the tassel floret. A careful examination of tassel development in the maize BR mutants, and evaluation of gene expression patterns during BR loss and GA supplementation, may help shed light on links between these two systems.
METHODS
Plant Materials and Growth Conditions
The bsl1-1 and bsl1-2 alleles were isolated from NMU-mutagenized M2 population of Setaria viridis (Huang et al., 2017). S. viridis plants were greenhouse-grown under long-day conditions (28°C/22°C [day/night], 16 h light/8 h dark, 40% relative humidity, 700-square-foot greenhouse outfitted with 11 1000-W metal halide and 10 1000-W high-pressure sodium bulbs). Both mutant alleles were backcrossed to the reference mutagenized line (A10.1) and multiple F1 individuals were used to generate F2 segregating populations. Phenotyping was done using F3 seed grown in the above conditions. Genetic crosses were performed as previously described (Jiang et al., 2013). Plants used for scanning electron microscopy, RNA-seq, in situ hybridization, metabolic profiling, and PCZ treatment were grown in a high-light growth chamber under short-day conditions (31°C/22°C [day/night], 12 h light/12 h dark, 50% relative humidity, chamber outfitted with 6 T8 fluorescent 5-foot bulbs per 10-square-foot canopy, light intensity at 200 µmol m−2 s−1 PAR) at the Danforth Center’s growth facility.
Genome Sequencing and Bulked Segregant Analysis of bsl1 Mutants
For bulked segregant analysis, bsl1-1 plants were backcrossed to the reference line A10.1 as the pollen parent. Self-pollinated F1 individuals generated segregating F2 families. Mutant and wild-type individuals from the F2 generation were identified and the segregation ratio tested with a χ2 test. DNA extracted from 20 F2 mutant individuals was pooled to generate a library for DNA sequencing. Additional DNA libraries were generated from single bsl1-1 and bsl1-2 mutant individuals in the M2 generation and sequenced. All sequencing was performed using the Illumina Hi-Seq 2500 platform. DNA libraries were size selected for inserts of 500 to 600 bp and 100-bp single-end reads were sequenced using standard Illumina protocols. Read mapping and SNP calling were conducted as described (Huang et al., 2017).
Phylogenetic Analysis
The coding sequences of the genes most similar to the rice D11 gene and Arabidopsis Dwf4 gene were obtained from the Phytozome (phytozome.jgi.doe.gov) and Gramene (gramene.org) databases (Supplemental Data Set 2). Sequences were aligned using ClustalW to construct a maximum likelihood tree based on the Tamura-Nei model in MEGA7 (Kumar et al., 2016). Bootstrap support was based on 1000 iterations.
RNA Isolation and RT-qPCR
Root, sheath, and 4th leaf tissues were sampled from each of three individual A10.1 wild-type seedlings at 20 DAS (each representing a biological replicate). Inflorescence primordia sampled from six individual wild-type plants at 18 DAS were pooled for each of three biological replicates. Total RNA was isolated using the Quick-RNA MiniPrep kit (Zymo Research) with in-column DNase I treatment following the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg total RNA by SuperScript III reverse transcriptase (Invitrogen). RT-qPCR was performed using a SYBR Premix Ex Taq II (Tli RNaseH Plus) kit (TaKaRa) in a CFX96 real-time PCR detection system (Bio-Rad). Experimental design included three biological replicates and three technical replicates. Expression levels of Bsl1 and SvDwf4 were analyzed by a standard curve method using equal amounts of total RNA per sample. Standard curves were generated from plasmid DNAs containing the target fragments of Bsl1 and SvDwf4, respectively. Copy number for each point of the plasmid DNA dilution series was calculated. The log base 10 of the copy number was taken for each dilution point and Ct values of dilution points measured by real-time PCR were fitted by linear regression. Ct values were then converted to a relative expression value based on the standard curve. Primers for RT-qPCR are listed in Supplemental Table 3.
Quantification of Endogenous BRs and Intermediates
Shoot tissue (leaf and stem) was harvested from each of three wild-type and bsl1-1 seedlings at 11 DAS (three biological replicates per genotype). Inflorescence primordia were hand-dissected from wild-type and bsl1-1 seedlings at 15 to 17 DAS and pooled for individual biological replicates. For each tissue type and genotype, three biological replicates were analyzed. Metabolite measurements were conducted using a modified protocol for sterol profiling (Honda et al., 2008). An extraction solvent of 8:5:3 (v/v/v) of chloroform to methanol to hexanes was added to ground tissue with a 5 μL spike-in of deuterated d7-campesterol (0.1 mg/mL) standard. Samples were vortexed for 20 min, centrifuged for 5 min (17,300g) and then dried by speed vacuum for 2 h at 37°C. Sterols were labeled with a picolinic ester using a mixture consisting of 100 mg 2 methyl-6-nitrobenzoicanhydride, 30 mg 4-dimethylaminopyridine, 80 mg picolinic acid, and 1.5 mL tetrahydrofuran with 20 mL trimethylamine added last. Samples were vortexed and incubated for 30 min at room temperature and then dried in a speed vacuum for 2 h at 37°C. Dried sample was resuspended in 150 μL acetonitrile. Half of the sample was used for targeted BR measurements and the other half for untargeted metabolite measurements and run at the Bindley Metabolite Profiling Facility at Purdue University. Targeted BR measurements were run on an Atlantis T3 column (Waters) and Agilent 1200 series HPLC system and injected into a 6460 Triple Quad MS/MS (Agilent Technologies) in positive ESI mode. Identification and quantification of BRs were performed using Agilent MassHunter Qualitative Analysis software (Agilent Technologies). Untargeted measurements were run on an Atlantis T3 column and Agilent 1200 series HPLC system and injected into a 6545 Q-TOF MS (Agilent Technologies) in positive ESI mode. Agilent MassHunter Mass Profiler Professional software (v13.11) was used to match mass features to metabolite data with a 10 ppm mass error and 0.35 min retention time window. Statistical analyses were conducted if a mass appeared in two of six seedling or inflorescence samples with a P value < 0.01 and fold change > 2. Significant masses were run through the METLIN MS/MS metabolite database (Smith et al., 2005).
RNA-Seq Library Construction, Sequencing, and Analysis
RNA-seq libraries were generated from pools of inflorescence primordia hand-dissected from wild-type and bsl1-1 mutant seedlings for three and two biological replicates, respectively. Wild-type primordia were sampled at ∼15 DAS. Since bsl1-1 mutants are morphologically comparable to the wild type at ∼16 DAS, they were sown a day earlier in a controlled high-light growth chamber and collected alongside the wild type. Total RNA (1 µg) was extracted (PicoPure RNA isolation kit; Thermo Fisher Scientific), and libraries were generated using the NEBNext Ultra Directional RNA Library Prep Kit (Illumina), size-selected for 200-bp inserts, and quantified on an Agilent bioanalyzer using a DNA 1000 chip. We generated an average of 40 M 100-bp, single-end reads using the Illumina HiSeq 2500 platform at the University of Illinois at Urbana-Champaign W.M. Keck Center. Read mapping and differential expression analysis were performed using the Tuxedo suite of open-source tools (Trapnell et al., 2013). Reads were mapped to the S. viridis A10.1 reference genome (phytozome.jgi.doe.gov; v1.1) using TopHat2 (v2.1.0) and an a priori set of 35,214 gene models. Gene-level expression values were represented by fragments per kilobase exon per million reads mapped (FPKM) and a consensus FPKM was determined for each gene based on its representation across biological replicates. Differential expression was determined using Cuffdiff (v 2.2.1) and a corrected P value of <0.05. Gene Ontology enrichment analysis was performed using the GOStats package in R (Falcon and Gentleman, 2007) based on Setaria italica GO annotations available at Phytozome. P values were calculated using a hypergeometric test. Functional annotations from Ensembl BioMart, TAIR10, Phytozome, and MaizeGDB were based on homologs determined through Ensembl Compara gene trees (gramene.org).
Scanning Electron Microscopy Analysis
For scanning electron microscopy analysis of bsl1-1, bsl1-2, and PCZ-treated and untreated wild-type inflorescences, samples were fixed and dehydrated as described (Hodge and Kellogg, 2014). Samples were critical point dried using a Tousimis Samdri-780a and imaged by a Hitachi S2600 scanning electron microscope at Washington University’s Central Institute of the Deaf. Scanning electron microscopy images were also used to quantify spikelet number and the frequency of two florets per spikelet across samples. For spikelet number, we counted and averaged spikelet number across images (120× magnification) representing equivalent mid sections of mutant and wild-type inflorescences at the time of floral differentiation. To estimate the approximate percent frequency of two florets per spikelet, images were taken in blocks along the length of representative bsl1-1 and PCZ-treated inflorescences at 19 to 20 DAS.
In Situ Hybridization
In situ hybridization was performed as previously described (Kouchi and Hata, 1993). The fixation, embedding, and sectioning of immature S. viridis inflorescences were performed according to the protocol described by Jackson (1991) with adaptations. Developing inflorescences covered by nonemerging leaves were dissected from plants and fixed at 4°C in PFA solution (1.85% [w/v] paraformaldehyde, 5% [v/v] acetic acid, and 63% [v/v] ethanol) for at least 1 d. The fixed materials were dehydrated in an ethanol gradient series (70, 85, 95, and 100% ethanol). Then, the dehydrated material was cleared with 50% histo-clearII/50% ethanol once and 100% histo-clearII three times, followed by paraffin wax infiltration for 3 d and embedding in wax on the fourth day. Slides with microtome sections (10 mm thick) of wild-type and bsl1-1 mutant developing inflorescences were deparaffinized and treated with 5 μg/mL proteinase K at 37°C for 20 min followed by refixation in PFA solution (4% [w/v] paraformaldehyde, 5% [v/v] acetic acid, and 50% [v/v] ethanol) for 10 min. Digoxigenin-UTP-labeled Bsl1, Svbd1, and SvKn1 sense and antisense probes were synthesized by in vitro transcription of 423-, 423-, and 358-bp fragments of Bsl1, Svbd1, and SvKn1 cDNA by DIG RNA labeling kit (SP6/T7; Roche), respectively. The probes were used to detect Bsl1, Svbd1, and SvKn1 transcripts in sections containing the inflorescence meristem. The hybridization was performed at 50°C overnight in 50% formamide buffer [0.5 ng/mL probe, 1 mg/mL tRNA, 0.1 mg/mL poly(A), 30 mM DTT, 0.3 M NaCl, 10 mM Tris-HCl, 1 mM EDTA, 10% dextran sulfate, and 1× Denhardt’s solution]. After being washed with 4× SSC (0.15 M NaCl in 0.015 M sodium citrate), the slides were treated with 20 μg/mL RNaseA at 37°C for 30 min, washed with RNase buffer (0.5 M NaCl, 0.01 M Tris-HCl) three times, 0.5× SSC twice, and 1× buffer1 (0.1 M Tris and 0.15 M NaCl) once, and then incubated in Roche blocking solution for 30 min. Digoxigenin signal was visualized by nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche) overnight, following the manufacturer’s instructions. Slides were observed under a Leica ICC50 HD microscope.
Accession Numbers
RNA-seq data (raw sequence reads and processed data files) have been deposited in the NCBI Gene Expression Omnibus under accession number GSE100423. Whole-genome sequence data for bsl1-1 and bsl1-2 were previously deposited in the NCBI Sequence Read Archive and are available under accession numbers SRX2110281 and SRX2112339, respectively.
Supplemental Data
Supplemental Figure 1. Meristem transitions during S. viridis inflorescence development and panicle morphologies showing reduced bristle production in bsl1 mutants.
Supplemental Figure 2. Scanning electron microscopy analysis of early developmental transitions in wild-type A10.1 and bsl1-1 mutant inflorescence primordia.
Supplemental Figure 3. Morphological characterization of inflorescence development in the bsl1-2 mutant by scanning electron microscopy analysis.
Supplemental Figure 4. Morphological analysis of spikelet-to-bristle ratios in bsl1 mutants compared with the wild type.
Supplemental Figure 5. Sequences of alternative transcript isoforms in bsl1-2 mutants.
Supplemental Figure 6. Possible substrates and pathway for catabolism of BR intermediates to produce a putative C21 pregnane in bsl1 inflorescences.
Supplemental Figure 7. Distribution of TF family members that were differentially expressed in bsl1-1 inflorescence primordia.
Supplemental Figure 8. Differentially expressed genes related to hormone and light signaling pathways based on homology to genes from Arabidopsis.
Supplemental Figure 9. Additional RNA in situ hybridization sections of Bsl1 during S. viridis inflorescence development.
Supplemental Figure 10. RNA in situ hybridization of SvKn1 in A10.1 wild-type and bsl1-1 inflorescence primordia.
Supplemental Figure 11. RNA in situ hybridization on adjacent sections probed with Bsl1 and SvBd1.
Supplemental Table 1. Phenotypic measurements of bsl1 mutant plants.
Supplemental Table 2. Annotation of genes related to sterol and BR biosynthesis and signaling in S. viridis and related expression information.
Supplemental Table 3. Table of primers used in this study.
Supplemental Data Set 1. High-confidence SNP calls for bsl1-1 and bsl1-2 mutants.
Supplemental Data Set 2. Alignment of coding sequences of D11 and Dwf4 genes by ClustalW.
Supplemental Data Set 3. Transcript abundances and raw read counts for all annotated S. viridis genes (v1) in the wild type compared with bsl1-1 mutant inflorescence primordia.
Supplemental Data Set 4. Differentially expressed genes in bsl1-1 mutant inflorescences and functional annotations.
Supplemental Data Set 5. Overrepresentation of functional classes among differentially expressed genes based on Gene Ontology term enrichment.
Acknowledgments
We thank members of the Eveland, Brutnell, and Kellogg labs at the Donald Danforth Plant Science Center (DDPSC) for helpful discussions and advice related to the project. We also thank Tom Brutnell for providing the NMU-mutagenized S. viridis population for screening and for critical review of the manuscript. We thank Lynsey Kovar for computational support, Luke Dell for assistance in seed propagation and plant care, and Kevin Reilly and his team at the DDPSC for growth chamber maintenance. We acknowledge Bruce Cooper and Amber Jannasch of the Metabolite Profiling Facility at Purdue University and SungHua Choe of Seoul National University for metabolite analyses and deuterated campesterol standards. This work was supported by DDPSC start-up funds to A.L.E., grants from the National Science Foundation (IOS-1238202 to A.L.E. and IOS-1444503 to B.P.D.), and a USDA-NIFA fellowship to N.B.B. (NIFA 2017-67011-26077). The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
AUTHOR CONTRIBUTIONS
J.Y., B.P.D., and A.L.E. designed the research. J.Y., S.T., and H.J. performed genetics experiments. J.Y. and S.T. performed developmental and molecular experiments. N.B.B. performed metabolite experiments. P.H. and J.Y. performed bioinformatics analyses. J.Y., N.B.B., P.H., B.P.D., and A.L.E. analyzed the data. J.Y. and A.L.E. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Acosta I.F., Laparra H., Romero S.P., Schmelz E., Hamberg M., Mottinger J.P., Moreno M.A., Dellaporta S.L. (2009). tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323: 262–265. [DOI] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancoş S., Nomura T., Sato T., Molnár G., Bishop G.J., Koncz C., Yokota T., Nagy F., Szekeres M. (2002). Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130: 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett M.E., Thompson B. (2014). Meristem identity and phyllotaxis in inflorescence development. Front. Plant Sci. 5: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E.M., Lin W.C., Husbands A.Y., Yu L., Jaganatha V., Jablonska B., Mangeon A., Neff M.M., Girke T., Springer P.S. (2012). Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 109: 21146–21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best N.B., Hartwig T., Budka J., Fujioka S., Johal G., Schulz B., Dilkes B.P. (2016). nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis gene DWARF1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiol. 171: 2633–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M.K., Morohashi K., O’Connor D., Grotewold E., Hake S. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26: 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell T.P., Bennetzen J.L., Vogel J.P. (2015). Brachypodium distachyon and Setaria viridis: Model genetic systems for the grasses. Annu. Rev. Plant Biol. 66: 465–485. [DOI] [PubMed] [Google Scholar]
- Brutnell T.P., Wang L., Swartwood K., Goldschmidt A., Jackson D., Zhu X.G., Kellogg E., Van Eck J. (2010). Setaria viridis: a model for C4 photosynthesis. Plant Cell 22: 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein S., Middleditch B.S., Gut M. (1975). Mass spectrometric study of the enzymatic conversion of cholesterol to (22R)-22-hydroxycholesterol, (20R,22R)-20,22-dihydroxycholesterol, and pregnenolone, and of (22R)-22-hydroxycholesterol to the lgycol and pregnenolone in bovine adrenocortical preparations. Mode of oxygen incorporation. J. Biol. Chem. 250: 9028–9037. [PubMed] [Google Scholar]
- Chaiwanon J., Wang Z.Y. (2015). Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Tanaka A., Noguchi T., Fujioka S., Takatsuto S., Ross A.S., Tax F.E., Yoshida S., Feldmann K.A. (2000). Lesions in the sterol delta reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 21: 431–443. [DOI] [PubMed] [Google Scholar]
- Chuck G., Muszynski M., Kellogg E., Hake S., Schmidt R.J. (2002). The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241. [DOI] [PubMed] [Google Scholar]
- Chung Y., Choe S. (2013). The regulation of brassinosteroid biosynthesis in Arabidopsis. Crit. Rev. Plant Sci. 32: 396–410. [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D., Sasse J.M. (1998). BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451. [DOI] [PubMed] [Google Scholar]
- Dellaporta S.L., Calderon-Urrea A. (1994). The sex determination process in maize. Science 266: 1501–1505. [DOI] [PubMed] [Google Scholar]
- Derbyshire P., Byrne M.E. (2013). MORE SPIKELETS1 is required for spikelet fate in the inflorescence of Brachypodium. Plant Physiol. 161: 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust A.N., Kellogg E.A. (2002). Inflorescence diversification in the panicoid “bristle grass” clade (Paniceae, Poaceae): evidence from molecular phylogenies and developmental morphology. Am. J. Bot. 89: 1203–1222. [DOI] [PubMed] [Google Scholar]
- Evans M.M., Poethig R.S. (1995). Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol. 108: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S., Gentleman R. (2007). Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258. [DOI] [PubMed] [Google Scholar]
- Fu X.D., Ares M. Jr (2014). Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S., Yokota T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54: 137–164. [DOI] [PubMed] [Google Scholar]
- Fujita S., Ohnishi T., Watanabe B., Yokota T., Takatsuto S., Fujioka S., Yoshida S., Sakata K., Mizutani M. (2006). Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J. 45: 765–774. [DOI] [PubMed] [Google Scholar]
- Gaillochet C., Lohmann J.U. (2015). The never-ending story: from pluripotency to plant developmental plasticity. Development 142: 2237–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron J.M., Liu J.S., Fan M., Bai M.Y., Wenkel S., Springer P.S., Barton M.K., Wang Z.Y. (2012). Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859. [DOI] [PubMed] [Google Scholar]
- Hartwig T., Chuck G.S., Fujioka S., Klempien A., Weizbauer R., Potluri D.P., Choe S., Johal G.S., Schulz B. (2011). Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci. USA 108: 19814–19819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T., Corvalan C., Best N.B., Budka J.S., Zhu J.Y., Choe S., Schulz B. (2012). Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS One 7: e36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J., Cools T., Vandenbussche F., Heyndrickx K.S., Van Leene J., Vercauteren I., Vanderauwera S., Vandepoele K., De Jaeger G., Van Der Straeten D., De Veylder L. (2013). ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863. [DOI] [PubMed] [Google Scholar]
- Hodge J.G., Kellogg E.A. (2014). Patterns of inflorescence development of three prairie grasses (Andropogoneae, Poaceae). Int. J. Plant Sci. 175: 963–974. [Google Scholar]
- Honda A., Yamashita K., Miyazaki H., Shirai M., Ikegami T., Xu G., Numazawa M., Hara T., Matsuzaki Y. (2008). Highly sensitive analysis of sterol profiles in human serum by LC-ESI-MS/MS. J. Lipid Res. 49: 2063–2073. [DOI] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Fujioka S., Takatsuto S., Yoshida S., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M. (2005). The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17: 2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32: 495–508. [DOI] [PubMed] [Google Scholar]
- Huang P., Jiang H., Zhu C., Barry K., Jenkins J., Sandor L., Schmutz J., Box M.S., Kellogg E.A., Brutnell T.P. (2017). Sparse panicle1 is required for inflorescence development in Setaria viridis and maize. Nat. Plants 3: 17054. [DOI] [PubMed] [Google Scholar]
- Huang P., Shyu C., Coelho C.P., Cao Y., Brutnell T.P. (2016). Setaria viridis as a model system to advance millet genetics and genomics. Front. Plant Sci. 7: 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish E.E., Nelson T.M. (1993). Development of Tassel Seed-2 inflorescences in maize. Am. J. Bot. 80: 292–299. [Google Scholar]
- Jackson D.P. (1991). In-situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, Gurr S.J., McPherson M.J., Bowles D.J., eds (Oxford, UK: Oxford University Press; ), pp. 163–174. [Google Scholar]
- Jiang H., Barbier H., Brutnell T. (2013). Methods for performing crosses in Setaria viridis, a new model system for the grasses. J. Vis. Exp. 80: 10.3791/50527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M.T., Twigg R.W., Timmermans M.C. (2004). Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544. [DOI] [PubMed] [Google Scholar]
- Kellogg E.A. (2007). Floral displays: genetic control of grass inflorescences. Curr. Opin. Plant Biol. 10: 26–31. [DOI] [PubMed] [Google Scholar]
- Kerstetter R.A., Laudencia-Chingcuanco D., Smith L.G., Hake S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054. [DOI] [PubMed] [Google Scholar]
- Kir G., Ye H., Nelissen H., Neelakandan A.K., Kusnandar A.S., Luo A., Inzé D., Sylvester A.W., Yin Y., Becraft P.W. (2015). RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 169: 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe A., Schneider B., Porzel A., Voigt B., Krauss G., Adam G. (1994). Pregnane-type metabolites of brassinosteroids in cell suspension cultures of Ornithopus sativus. Phytochemistry 36: 671–673. [Google Scholar]
- Komatsu M., Chujo A., Nagato Y., Shimamoto K., Kyozuka J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130: 3841–3850. [DOI] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238: 106–119. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Fujioka S., Jeon J.H., Kim H.B., Takatsuto S., Yoshida S., An C.S., Choe S. (2005). A double mutant for the CYP85A1 and CYP85A2 genes of Arabidopsis exhibits a brassinosteroid dwarf phenotype. J. Plant Biol. 48: 237–244. [Google Scholar]
- Li J., Nagpal P., Vitart V., McMorris T.C., Chory J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401. [DOI] [PubMed] [Google Scholar]
- Li Q., Liu B. (2017). Genetic regulation of maize flower development and sex determination. Planta 245: 1–14. [DOI] [PubMed] [Google Scholar]
- Luccioni L.G., Oliverio K.A., Yanovsky M.J., Boccalandro H.E., Casal J.J. (2002). Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 128: 173–181. [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Thompson A., Muehlbauer G.J., Springer N.M. (2012). Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS One 7: e30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R.W., Paran I., Kesseli R.V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Nguyen S.M., Malancharuvil E.J., Fujioka S., Noguchi T., Seto H., Tsubuki M., Honda T., Takatsuto S., Yoshida S., Chory J. (1999). BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 15316–15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson N.H. (1959). Sustained treatment with gibberellic acid of five different kinds of maize. Ann. Mo. Bot. Gard. 46: 19–37. [Google Scholar]
- Nickerson N.H. (1960). Studies involving sustained treatment of maize with gibberellic acid II: Responses of plants carrying certain tassel-modifying genes. Ann. Mo. Bot. Gard. 47: 243–261. [Google Scholar]
- Ohnishi T., Godza B., Watanabe B., Fujioka S., Hategan L., Ide K., Shibata K., Yokota T., Szekeres M., Mizutani M. (2012). CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 287: 31551–31560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Watanabe B., Sakata K., Mizutani M. (2006). CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci. Biotechnol. Biochem. 70: 2071–2080. [DOI] [PubMed] [Google Scholar]
- Pautler M., Tanaka W., Hirano H.Y., Jackson D. (2013). Grass meristems I: shoot apical meristem maintenance, axillary meristem determinacy and the floral transition. Plant Cell Physiol. 54: 302–312. [DOI] [PubMed] [Google Scholar]
- Peng H., Zhao J., Neff M.M. (2015). ATAF2 integrates Arabidopsis brassinosteroid inactivation and seedling photomorphogenesis. Development 142: 4129–4138. [DOI] [PubMed] [Google Scholar]
- Reuzeau C., Frankard V., Hatzfeld Y., Sanz A., Van Camp W., Lejeune P., De Wilde C., Lievens K., de Wolf J., Vranken E., Peerbolte R. (2006). Traitmill™: a functional genomics platform for the phenotypic analysis of cereals. Plant Genet. Resour. 4: 20–24. [Google Scholar]
- Sakamoto T., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24: 105–109. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Ohnishi T., Fujioka S., Watanabe B., Mizutani M. (2012). Rice CYP90D2 and CYP90D3 catalyze C-23 hydroxylation of brassinosteroids in vitro. Plant Physiol. Biochem. 58: 220–226. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202. [DOI] [PubMed] [Google Scholar]
- Scanlon M.J., Schneeberger R.G., Freeling M. (1996). The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development 122: 1683–1691. [DOI] [PubMed] [Google Scholar]
- Schneeberger K. (2014). Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 15: 662–676. [DOI] [PubMed] [Google Scholar]
- Sekimata K., Han S.Y., Yoneyama K., Takeuchi Y., Yoshida S., Asami T. (2002). A specific and potent inhibitor of brassinosteroid biosynthesis possessing a dioxolane ring. J. Agric. Food Chem. 50: 3486–3490. [DOI] [PubMed] [Google Scholar]
- Shimada Y., Fujioka S., Miyauchi N., Kushiro M., Takatsuto S., Nomura T., Yokota T., Kamiya Y., Bishop G.J., Yoshida S. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126: 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.A., O’Maille G., Want E.J., Qin C., Trauger S.A., Brandon T.R., Custodio D.E., Abagyan R., Siuzdak G. (2005). METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27: 747–751. [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G.M., Reid J.B. (2004). Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 135: 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G.M., Ross J.J., Jager C.E., Reid J.B. (2008). Brassinosteroid transport. J. Exp. Bot. 59: 17–24. [DOI] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., Yoshimura A., Kitano H., Matsuoka M., Fujisawa Y., Kato H., Iwasaki Y. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Asami T., Yoshida S., Nakamura Y., Matsuo T., Okamoto S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138: 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W., Pautler M., Jackson D., Hirano H.Y. (2013). Grass meristems II: inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 54: 313–324. [DOI] [PubMed] [Google Scholar]
- Tao Y., Zheng J., Xu Z., Zhang X., Zhang K., Wang G. (2004). Functional analysis of ZmDWF1, a maize homolog of the Arabidopsis brassinosteroids biosynthetic DWF1/DIM gene. Plant Sci. 167: 743–751. [Google Scholar]
- Thompson B.E., Bartling L., Whipple C., Hall D.H., Sakai H., Schmidt R., Hake S. (2009). bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Xiao Y., Liu D., Gao S., Liu L., Yin Y., Jin Y., Qian Q., Chu C. (2014). Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26: 4376–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Kurata N., Ohyanagi H., Hake S. (2014). Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26: 3488–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J., González-García M.P., Frigola D., Fàbregas N., Alexiou K.G., López-Bigas N., Rivas S., Jauneau A., Lohmann J.U., Benfey P.N., Ibañes M., Caño-Delgado A.I. (2014). Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev. Cell 30: 36–47. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Springer P.S., Goh L., Buckler E.St. IV, Martienssen R. (2005). Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126. [DOI] [PubMed] [Google Scholar]
- Vragović K., Sela A., Friedlander-Shani L., Fridman Y., Hacham Y., Holland N., Bartom E., Mockler T.C., Savaldi-Goldstein S. (2015). Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc. Natl. Acad. Sci. USA 112: 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriet C., Russinova E., Reuzeau C. (2012). Boosting crop yields with plant steroids. Plant Cell 24: 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriet C., Russinova E., Reuzeau C. (2013). From squalene to brassinolide: the steroid metabolic and signaling pathways across the plant kingdom. Mol. Plant 6: 1738–1757. [DOI] [PubMed] [Google Scholar]
- Whipple C.J., Kebrom T.H., Weber A.L., Yang F., Hall D., Meeley R., Schmidt R., Doebley J., Brutnell T.P., Jackson D.P. (2011). grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 108: E506–E512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Fu Y., Zhao S., Gu P., Zhu Z., Sun C., Tan L. (2016). CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 14: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Zhang C., Bai M.Y., Chong K. (2014). Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 33: 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Yuan Z. (2014). Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 65: 553–578. [DOI] [PubMed] [Google Scholar]
- Zhang R., Xia X., Lindsey K., da Rocha P.S. (2012). Functional complementation of dwf4 mutants of Arabidopsis by overexpression of CYP724A1. J. Plant Physiol. 169: 421–428. [DOI] [PubMed] [Google Scholar]
- Zhu J.Y., Sae-Seaw J., Wang Z.Y. (2013). Brassinosteroid signalling. Development 140: 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]