SUMMARY
In contrast with numerous studies of glutamate receptor-associated proteins and their involvement in the modulation of excitatory synapses, much less is known about mechanisms controlling postsynaptic GABAA receptor (GABAAR) numbers. Using tandem affinity purification from tagged GABAAR γ2 subunit transgenic mice and proteomic analysis, we isolated several GABAAR-associated proteins including Cleft lip and palate transmembrane protein 1 (Clptm1). Clptm1 interacted with all GABAAR subunits tested and promoted GABAAR trapping in the endoplasmic reticulum. Overexpression of Clptm1 reduced GABAAR-mediated currents in a recombinant system, in cultured hippocampal neurons, and in brain, with no effect on glycine or AMPA receptor-mediated currents. Conversely, knockdown of Clptm1 increased phasic and tonic inhibitory transmission with no effect on excitatory synaptic transmission. Furthermore, altering the expression level of Clptm1 mimicked activity-induced inhibitory synaptic scaling. Thus, in complement to other GABAAR-associated proteins that promote receptor surface expression, Clptm1 limits GABAAR forward trafficking and regulates inhibitory homeostatic plasticity.
Keywords: inhibitory synapse, GABAergic transmission, receptor trafficking, synaptic scaling, homeostasis, tonic inhibition
eTOC BLURB
Ge et al. identify Clptm1 as a GABAAR associated protein that interacts with multiple subunits. They show that Clptm1 traps GABAARs in the ER and Golgi to scale phasic and tonic inhibitory transmission and modulate activity-induced inhibitory homeostasis.
INTRODUCTION
GABAARs mediate the major inhibitory neurotransmission in the mammalian central nervous system. Precise developmental and activity-dependent regulation of GABAAR expression, localization and function is essential in virtually all aspects of brain function. Deficits in GABAAR-mediated transmission contribute to the etiology of many brain disorders including epilepsy (Fritschy, 2008), major depressive disorder (Luscher et al., 2011b), anxiety disorders (Lydiard, 2003), and neurodevelopmental disorders including autism and schizophrenia (Marin, 2012; Nelson and Valakh, 2015).
Native GABAARs are heteropentamers assembled from 19 subunits, α1–6, β1–3, γ1–3, δ, ε, θ, π, and ρ1–3, resulting in various subunit composition across different regions and cell types of the central nervous system. Postsynaptic GABAARs mediating phasic inhibition in the brain are mainly composed of two α1, α2, or α3 subunits together with two β2 or β3 subunits and a single γ2 subunit (Olsen and Sieghart, 2008). The γ2 subunit is essential for postsynaptic clustering of GABAARs (Essrich et al., 1998). In contrast, the α4, α5, α6, and δ subunits are major components of extrasynaptic GABAARs mediating tonic inhibition (Belelli et al., 2009).
The local concentration of GABAARs at postsynaptic sites is a critical determinant of inhibitory synaptic strength and is tightly regulated. Among GABAAR interacting proteins, the best characterized is the scaffolding protein gephyrin. Originally identified as a glycine receptor-associated protein, gephyrin also binds GABAAR α1–3 subunits and localizes to GABA synapses (Tretter et al., 2012; Tyagarajan and Fritschy, 2014). Yet substantial GABAAR clusters and inhibitory transmission remain in the absence of gephyrin (Levi et al., 2004), indicating additional mechanisms controlling postsynaptic GABAAR numbers. Collybistin, neuroligin-2 and lipoma HMGIC fusion partner-like 4 also contribute to the postsynaptic aggregation of surface GABAARs (Poulopoulos et al., 2009; Yamasaki et al., 2017). Most other GABAAR-interacting proteins including GABA receptor-associated protein GABARAP and PLC-related catalytically inactive proteins (PRIPs) function in promoting surface expression of GABAAR by enhancing forward trafficking, inhibiting endocytosis, or enhancing recycling (Kanematsu et al., 2007; Leil et al., 2004; Luscher et al., 2011a; Vithlani et al., 2011). However, overall there is a paucity of studies on GABAAR trafficking mechanisms and interacting proteins, in contrast with the enormous effort and knowledge base on glutamate receptors. Part of this deficit may be due to technical factors, with the lack of free intracellular C-termini limiting utility of two-hybrid screens for GABAARs, and the low abundance, heterogeneity, and detergent sensitivity of GABAergic synaptic complexes hampering biochemical studies.
Here we used a mouse transgenic epitope-tagging tandem affinity purification and mass spectrometry approach to isolate and characterize complexes associated with GABAAR subunit γ2. A similar approach reported during the course of this work also identified a set of proteins associated with GABAAR subunit α2 (Nakamura et al., 2016). We further extensively characterized Clptm1, a multi-pass transmembrane protein that appears to be ubiquitously expressed (Yoshiura et al., 1998) including in brain (Lein et al., 2007). We show that Clptm1 interacts with multiple recombinant GABAAR subunits, regulates phasic and tonic inhibitory transmission in an unusual manner by limiting receptor surface expression, and mimics homeostatic inhibitory synaptic scaling.
RESULTS
Tandem Epitope-tagged GABAARγ2 Transgenic Mice
For tandem affinity purification of GABAAR complexes from brain, we generated Thy1-His6-Flag-YFP-GABAARγ2 (HFY-GABAARγ2) transgenic mice which use the neuron-specific Thy1 promoter to express GABAAR γ2 subunit with multiple epitope tags near its mature N-terminus (Bleckert et al., 2013). The HFY-GABAARγ2 transgene expressed from the Thy1 promoter was neuron specific and broadly expressed in multiple brain regions, including the cortex (Figure 1A) and hippocampus (Figure 1B). To test whether HFY-GABAARγ2 localized properly to inhibitory postsynaptic sites in brain, we did confocal imaging for the YFP signal. HFY-GABAARγ2 localized specifically to inhibitory synapses, as the YFP signal strongly colocalized with the inhibitory postsynaptic scaffold gephyrin and overlapped with the presynaptic synthetic enzyme glutamic acid decarboxylase (GAD) (Figure 1C and 1E). Immunostaining with an antibody against GABAARγ2 which would recognize both native and transgenic receptors revealed overlap between YFP puncta and immunostained GABAARγ2 signals as expected (Figure 1D). However, a small subset of GABAARγ2 puncta did not show overlap with YFP signals. This is due to lack of expression of HFY-GABAARγ2 by a subset of neurons (Figure 1E), although the majority of neurons throughout cortex and hippocampus expressed the transgene. The transgene caused a 31% increase in total GABAARγ2 protein level (Bleckert et al., 2013) which did not affect mouse survival, breeding, or home cage behavior. Indeed, even transgenic expression of GABAARγ2 doubling the total protein level results in no change in many behaviors excepting a difference in ethanol tolerance (Wick et al., 2000).
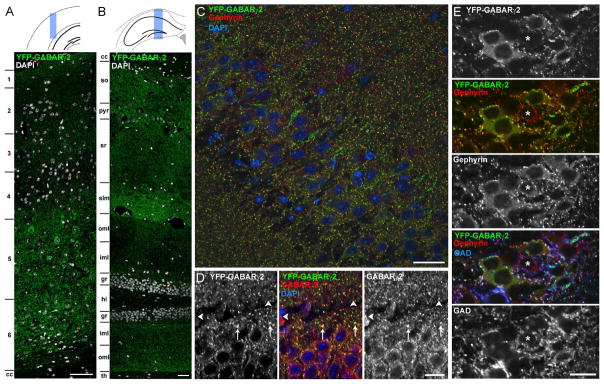
Figure 1. Transgenic HFY-GABAARγ2 Localizes to GABAergic Synapses.
HFY-GABAARγ2 localization was assessed by confocal microscopy of the YFP signal in relation to immunofluorescence for inhibitory synaptic markers in perfused adult mouse brain sections. Panels (A–D) show co-staining for nuclear DAPI. Panels (C–E) are from the hippocampal CA1 region.
(A and B) Composite images through the cortex (A) and hippocampus (B) show widespread expression and punctate distribution of HFY-GABAARγ2. Regions: 1–6, cortical layers; cc, corpus callosum; so, stratum oriens; pyr, stratum pyramidale; sr, stratum radiatum; slm, stratum lacunosum moleculare; oml, outer molecular layer; iml inner molecular layer; gr, granule cell layer; hi, hilus; th, thalamus.
(C) HFY-GABAARγ2 puncta show good colocalization with the postsynaptic inhibitory scaffold gephyrin.
(D) The majority of HFY-GABAARγ2 puncta overlapped with immunostained GABAARγ2 signals (arrows). However, a small subset of GABAARγ2 puncta did not show overlap with HFY-GABAARγ2 YFP signals (arrowheads), presumably from cells lacking transgene expression.
(E) HFY-GABAARγ2 puncta colocalized with gephyrin opposite GAD-labeled terminals, confirming clustering at bona fide GABAergic synapses. The majority of neurons expressed the transgene as detected by the presence of perisomatic HFY-GABAARγ2 clusters but a few neurons did not. The asterisk indicates a neuron lacking detectable transgene expression. Pixel intensity correlation analysis revealed significantly greater colocalization between the original images of HFY-GABAARγ2 and GAD65 or gephyrin than randomized images (p<0.0001, t-test, n=40–41 fields).
Scale bars, 100 μm (A and B), 50 μm (C), and 20 μm (D and E).
Thus, HFY-GABAARγ2 is widely expressed throughout cortex and hippocampus, at low level relative to native GABAARγ2, and localizes properly to inhibitory synapses. The HFY-GABAARγ2 line appears to be a suitable tool to purify and analyze GABAAR-associated proteins.
Purification and Proteomic Analysis of GABAARγ2 Complexes
To purify HFY-GABAARγ2 complexes, we used a procedure previously optimized to purify neuroligin-2 and associated GABAergic postsynaptic proteins including gephyrin and GABAARs from His6-Flag-YFP-neuroligin-2 mouse brain (Kang et al., 2014). In brief, the procedure involves solubilization of complexes with n-dodecyl-β-D-maltoside detergent, initial purification using the His6 tag, and second step purification using anti-GFP antibody. The purified protein complex was then analyzed by liquid chromatography tandem-mass spectrometry (LC-MS/MS). To limit false positive detection, we performed 4 independent experiments using age- and sex-matched HFY-GABAARγ2 mice and wild type mice for comparison. The proteins which passed the detection criteria for selective isolation from HFY-GABAARγ2 mice relative to wild type control included known GABAARγ2-associated proteins enriched in inhibitory postsynaptic sites GABAARα1,α2,α3,α5,β1, β2, and β3 subunits, gephyrin, and neuroligin-2 (Table S1). Furthermore, a gene ontology analysis of the isolated GABAARγ2-associated proteins compared with the entire set of proteins in the mouse database revealed a significant enrichment in the biological processes of chloride transmembrane transport and GABAergic synaptic transmission, and in the cellular components of GABAA receptor complex, postsynaptic membrane, and inhibitory synapse (Figure 2A). This gene ontology characterization along with the successful detection of the known GABAARγ2-associated proteins validates the tandem affinity purification and characterization procedures which may identify novel GABAARγ2-associated proteins.
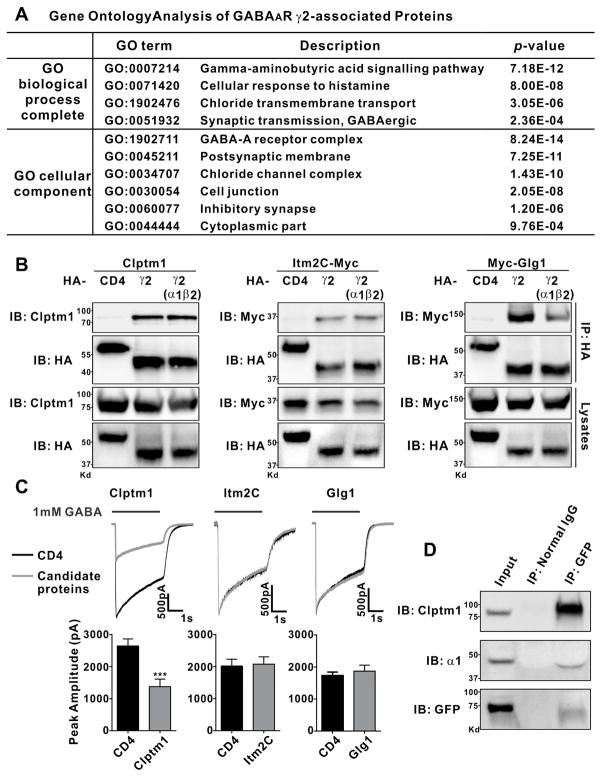
Figure 2. Clptm1, Itm2C, and Glg1 Associate with GABAAR γ2.
(A) For gene ontology analysis, the set of HFY-GABAARγ2-associated proteins listed in Table S1 was compared with the entire mouse database to identify pathways and compartments in which these proteins are significantly enriched using the statistical overrepresentation test (PANTHER with Bonferroni correction for multiple testing). Terms with p value <0.001 are listed here.
(B) HEK293 cells were transfected with Clptm1, Itm2C-Myc, or Myc-Glg1 and HA-tagged GABAAR γ2 alone, GABAAR γ2 with non-tagged α1 and β2, or HA-CD4 as a negative control. Clptm1, Itm2C, and Glg1 were specifically co-immunoprecipitated with GABAAR γ2 but not CD4.
(C) HEK293 cells which stably express GABAARα1/β2/γ2 were transfected with Clptm1, Itm2C, Glg1, or HA-CD4, each together with GFP to detect the transfected cells for recording. GABAAR mediated currents were induced by fast application/removal of GABA (1 mM). Clptm1 significantly reduced GABAAR mediated currents. Clptm1: n=18 cells from 3 independent experiments, Itm2C: n=17 cells from 2 independent experiments, and Glg1: n=21 cells from 2 independent experiments. *** p<0.001, t-test.
(D) Clptm1 was co-immunoprecipitated with GABAAR γ2 and α1 in whole brain homogenate from transgenic HFY-GABAARγ2 mice.
Results are expressed as mean ± SEM.
In addition to expected associated proteins, 39 candidate binding partners were identified for GABAARγ2 (Table S1). To confirm the interaction for selected candidates, we performed co-immunoprecipitation experiments using HA-tagged GABAARγ2 and YFP/CFP-tagged candidate proteins in HEK293 cells. Among the six tested candidate proteins, Clptm1, Integral Membrane Protein 2C (Itm2C), and Golgi Glycoprotein 1 (Glg1) were observed binding to GABAARγ2 compared with HA-CD8 negative control (Figure S1A). Co-immunoprecipitation of Myc-tagged or untagged Clptm1, Itm2C and Glg1 was further confirmed with HA-GABAARγ2 alone and with HA-GABAARγ2 expressed together with non-tagged GABAAR α1 and β2 to form functional receptors (Figure 2B). These data indicate an interaction of these candidate proteins with GABAARγ2 independent of any neuron-specific intermediate factors.
Clptm1, Itm2C and Glg1 are all transmembrane proteins. Clptm1 is a multi-pass transmembrane protein originally identified from a study of gene mutations in a patient family with cleft lip and palate (Yoshiura et al., 1998) but its biological function has been unknown. Itm2C is a type II transmembrane protein predominantly and widely expressed in the brain (Vidal et al., 2001). Itm2C is also known as BRI3, related to BRI2 implicated in familial British and Danish dementia, and also binds amyloid precursor protein (Matsuda et al., 2009). Glg1, also known as cysteine rich fibroblast growth factor receptor and E-selectin ligand 1, is a type I sialoglycoprotein (Miyaoka et al., 2010; Yang et al., 2010). Supporting a potential role in regulating inhibitory postsynaptic components, we recently isolated Glg1 in association with neuroligin-2 complexes from transgenic mouse brain (Kang et al., 2014). However, another proteomic isolation of GABAAR complexes from Myc-pHluorin-GABAARα2 knockin mice did not yield Clptm1, Itm2C nor Glg1, perhaps related to the use of a different detergent, 2% Triton X-100 (Nakamura et al., 2016) versus 0.5% n-dodecyl-β-D-maltoside in our studies.
We further tested whether these interacting proteins affect GABAAR mediated currents in HEK293 cells stably expressing GABAAR α1, β2, and γ2 subunits. Compared with control cells expressing CD4, expression of Clptm1 but not Itm2C or Glg1 dramatically decreased the peak amplitude of GABAAR mediated currents induced by fast application of 1 mM GABA (Figure 2C). The interaction with GABAAR and reduction of GABA currents reveals a potential specific function of Clptm1.
Clptm1 Binds and Functionally Regulates GABAARs but not Glycine Receptors
To confirm the interaction of Clptm1 as a regulatory protein with GABAARs in vivo, we show that Clptm1 co-immunoprecipitates with HFY-GABAARγ2 from mouse brain (Figure 2D). To test the selectivity of Clptm1 interaction with GABAAR subunits, we expressed HA-tagged γ2, α1, α2, β2, or β3 together with Clptm1 and did co-immunoprecipitation using anti-HA antibody. Clptm1 co-immunoprecipitated with all the tested subunits of GABAAR (Figure 3A), suggesting it may be a rather universal binding partner for different subtypes of GABAAR. In contrast, Clptm1 did not bind glycine receptor (GlyR) subunit α1 which shares similarity with GABAAR in sequence and structure (Grenningloh et al., 1987).
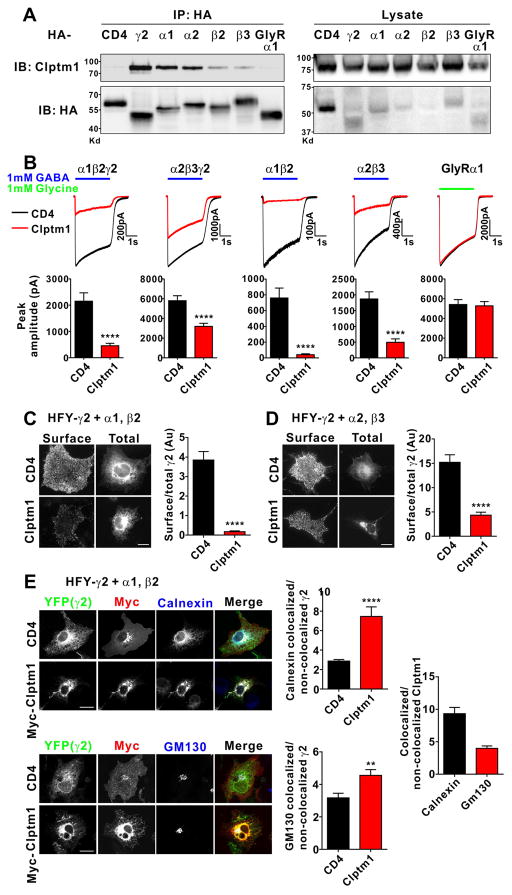
Figure 3. Clptm1 Binds Multiple GABAAR Subunits and Reduces GABAAR Mediated Currents.
(A) HEK293 cells were co-transfected with Clptm1 and HA-tagged GABAAR α1, β2, α2, β3, γ2, GlyR α1 or CD4 as a negative control. Clptm1 was specifically co-immunoprecipitated with GABAAR subunits but not GlyR α1.
(B) HEK293 cells were co-transfected with Clptm1 or HA-CD4 and GABAAR α1/β2/γ2, α2/β3/γ2, α1/β2, α2/β3, or GlyR α1, each together with GFP to detect the transfected cells for recording. GABAAR or GlyR mediated currents were induced by fast application/removal of GABA (1 mM) or glycine (1mM), respectively. Clptm1 significantly reduced GABAAR but not GlyR mediated currents. α1/β2/γ2: n=24 cells from 4 independent experiments, α2/β3/γ2: n=23 cells from 2 independent experiments, α1/β2: n=18 cells from 2 independent experiments, α2/β3: n=18 cells from 2 independent experiments, and GlyRα1: n=22 cells from 3 independent experiments. **** p<0.0001, t-test.
(C and D) COS7 cells were co-transfected with Clptm1 or HA-CD4 and HFY-GABAARγ2 with non-tagged α1/β2 (C) or α2/β3 (D). Surface HFY-GABAARγ2 was immunostained using anti-GFP antibody under non-permeabilized conditions. The surface to total HFY-GABAARγ2 ratio was reduced by Clptm1. Scale bar represents 20 μm. n=30 cells from at least 2 independent experiments. **** p<0.0001, t-test.
(E) COS7 cells co-transfected with Myc-Clptm1 or Myc-CD4 and HFY-GABAARγ2 with non-tagged α1 and β2 were fixed, permeabilized, and immunolabeled for Myc tag and the ER marker calnexin or the Golgi marker GM130. Clptm1 increased ER and Golgi localization of HFY-GABAARγ2 based on quantitation of the average intensity of YFP colocalized with calnexin or GM130 over the average intensity in the rest of the cell not colocalized with calnexin or GM130. Scale bar represents 20 μm. n=29 cells from at least 2 independent experiments. ** p<0.01, **** p<0.0001, t-test.
Results are expressed as mean ± SEM.
The interaction of Clptm1 with all the major GABAAR subunits raised the question of whether Clptm1 modulates GABAAR mediated currents with different subunit composition. We transfected HEK293 cells with the two subunit combinations of GABAAR which are most abundant at inhibitory postsynaptic sites: α1β2γ2 and α2β3γ2. Clptm1 significantly decreased the peak amplitude of GABA currents in the α1β2γ2 transiently transfected cells (Figure 3B) with a more robust effect than in the α1β2γ2 stable cell line (Figure 2C), perhaps due to a stronger interaction of Clptm1 with GABAAR when co-expressed simultaneously. Furthermore, Clptm1 reduced GABAAR α2β3γ2 mediated currents (Figure 3B), suggesting it may be a general modulator of many postsynaptic GABAAR subtypes. To test whether the effect of Clptm1 on the reduction of GABA currents is dependent on the γ2 subunit, we transfected HEK293 cells with α1β2 or α2β3 which also form functional receptors. Clptm1 reduced α1β2 and α2β3 mediated currents similarly (Figure 3B), confirming that γ2 is not the only subunit affected by Clptm1. We also tested functional specificity of Clptm1 using GlyR. In contrast with the reduction of GABA currents, Clptm1 did not change GlyR mediated currents (Figure 2B). Collectively, these results indicate that Clptm1 associates with the major postsynaptic GABAARs through interaction with multiple subunits and functionally modulates their currents. The modulation is specific for GABAAR, as we did not see any Clptm1 interaction with or effect on current amplitude for GlyR.
Clptm1 Reduces Surface Expression and Increases Endoplasmic Reticulum Localization of GABAARs
The reduction of GABA currents in transfected and stable HEK293 cells raised the possibility that Clptm1 may modulate the cell surface expression levels of GABAAR. To test this idea, we transfected COS7 cells with HFY-GABAARγ2 together with α1β2 or α2β3, and labeled cell surface GABAAR using anti-GFP antibody on live cells. The ratio of surface receptor integrated immunofluorescence to total YFP signal was calculated to assess what fraction of GABAAR is present on the cell surface. Co-expression of Clptm1 significantly decreased the surface/total ratio of GABAAR for both α1β2γ2 and α2β3γ2 receptors (Figure 3C and 3D).
This reduction in surface GABAAR mediated by Clptm1 could occur through reduced forward trafficking and exocytosis or through enhanced receptor endocytosis. In cells co-expressing Clptm1 there appeared to be an increase in perinuclear HFY-GABAARγ2 YFP signal, which would be consistent with reduced forward trafficking through increased receptor trapping in the endoplasmic reticulum (ER) and/or Golgi complex. We thus assessed the effect of Clptm1 on colocalization of HFY-GABAARγ2 with the ER marker calnexin and the Golgi marker GM130, in the presence of α1β2 to promote surface expression. Indeed, we observed a greater accumulation of HFY-GABAARγ2 primarily in calnexin-positive ER and to a lesser extent in GM130-positive Golgi when co-expressed with Clptm1 than with CD4 control (Figure 3E). The retention of GABAAR in the ER and Golgi provides a potential mechanism by which Clptm1 decreases surface expression of GABAAR by restricting receptor forward trafficking. The bulk of the retained GABAAR was trapped with Clptm1, indicated by the localization of Clptm1 primarily to the ER with a minority in the Golgi (Figure 3E). This data is consistent with the reported localization of N- or C-terminally tagged Clptm1 exclusively to the ER and Golgi in a large-scale protein localization study (Stadler et al., 2013).
Clptm1 is a multi-pass transmembrane protein (Yoshiura et al., 1998) but its membrane topology is not clear with 5–7 predicted transmembrane domains depending on the prediction software (Figure S2A). To systematically test whether Clptm1 could also be expressed on the cell surface, we added a Myc tag at the N terminus, C terminus, or within each major loop between predicted transmembrane domains. The Myc-Clptm1 constructs were detected with Myc antibody in permeabilized cells. However, we could not detect surface Myc immunoreactivity for any of the expressed Myc-Clptm1 constructs in COS7 cells or in cultured hippocampal neurons (Figure S2B). Altogether, these data indicate that Clptm1 interacts with GABAARs intracellularly, reducing receptor forward trafficking and cell surface expression level.
Overexpression of Clptm1 Induces Multiplicative Down Scaling of Inhibitory Transmission
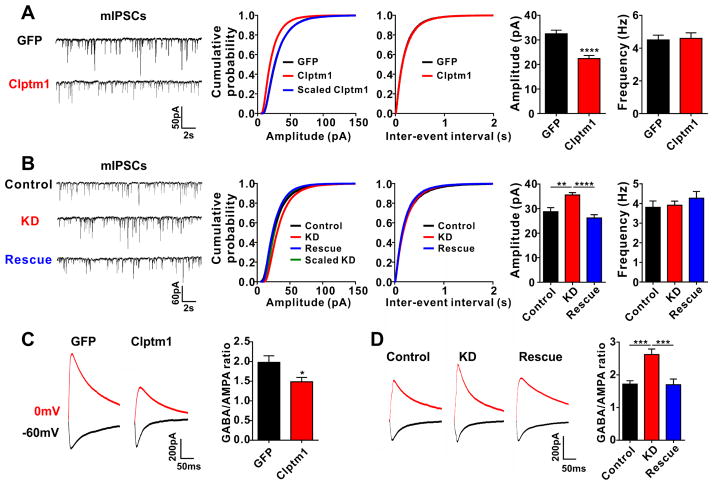
To assess the role of Clptm1 in modulation of native GABAARs, we overexpressed Clptm1 in cultured hippocampal neurons. We first used a low efficiency transfection method to isolate the cell autonomous role of postsynaptic Clptm1, co-expressing GFP to identify transfected cells. Miniature inhibitory postsynaptic currents (mIPSCs) were recorded in the whole cell patch clamp configuration at 9 DIV. Consistent with the reduction of recombinant GABAAR mediated currents in HEK293 cells, the amplitude of mIPSCs was significantly decreased in cultured hippocampal neurons overexpressing Clptm1 relative to control neurons overexpressing CD4 (Figure 4A). To assess whether Clptm1 affects all synapses, we tested whether the relative weights of synapse are preserved corresponding to multiplicative scaling, as occurs during activity-regulated homeostatic synaptic plasticity (Kilman et al., 2002; Kim et al., 2012). Indeed the overexpression of Clptm1 caused a multiplicative scaling down of mIPSC amplitude since the distribution of scaled Clptm1 amplitudes was not significantly different from control (Figure 4A). We also observed a reduction in mIPSC frequency (Figure 4A), which may be due to the reduction in amplitude causing events to fall below the detection threshold. In contrast, the amplitude and frequency of AMPA receptor mediated miniature excitatory postsynaptic currents (mEPSCs) were not altered in neurons overexpressing Clptm1 (Figure 4B). We repeated the Clptm1 overexpression using a high efficiency transfection method and recorded mIPSCs at a more mature stage at 14 DIV. We again found a multiplicative reduction in mIPSC amplitude in cells expressing Clptm1 compared with controls, although no change in frequency (Figure 4C). The specific modulation of inhibitory but not excitatory synaptic transmission indicates that Clptm1, as a GABAAR associated protein, suppresses the function of endogenous postsynaptic GABAARs. The observed multiplicative scaling further suggests that Clptm1 uniformly modulates all postsynaptic GABAARs.
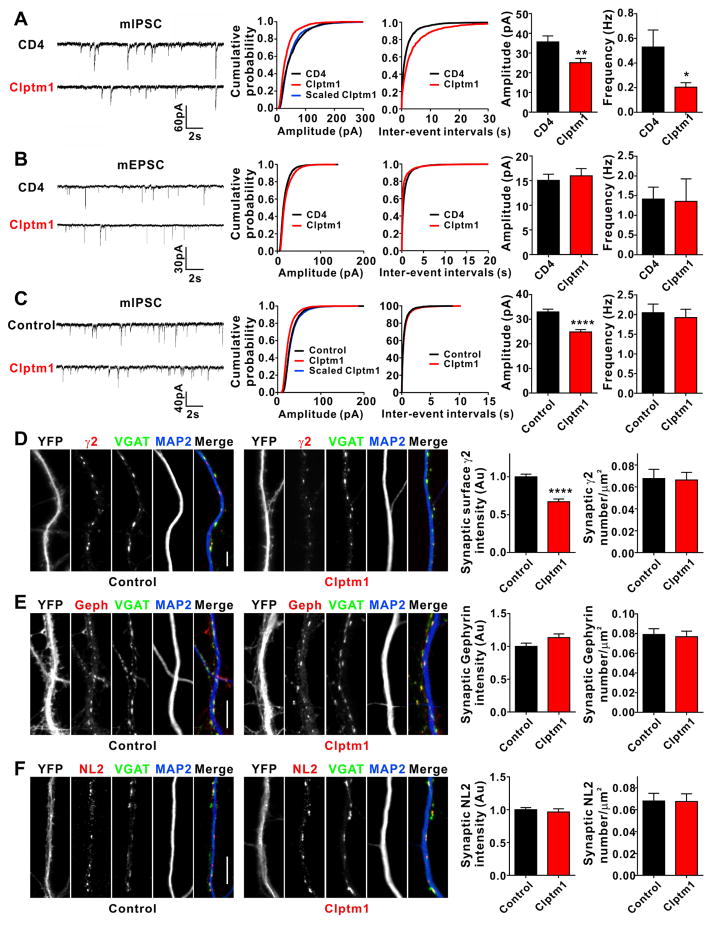
Figure 4. Overexpression of Clptm1 Scales Down Inhibitory Synaptic Strength.
(A and B) Cultured hippocampal neurons were transfected at 7 DIV with Clptm1 or HA-CD4 together with GFP to detect transfected neurons for recording. A low efficiency transfection method was used. mIPSC recordings were performed at 9 DIV and mEPSC recordings at 10 DIV.
(A) Overexpression of Clptm1 significantly reduced mIPSC amplitude compared with control group CD4. mIPSC frequency was also reduced. n=26–28 cells from 3 independent experiments, * p<0.05, ** p<0.01, t-test. The cumulative probability curve of Clptm1 amplitude was scaled by dividing by a factor of 0.70. Kolmogorov-Smirnov (K–S) test showed no significant difference between control and scaled Clptm1 groups.
(B) Overexpression of Clptm1 did not alter mEPSC amplitude or frequency. n=20–21 cells from 3 independent experiments.
(C–F) Cultured hippocampal neurons were transfected with YFP (Control) or YFP-p2a-Clptm1 (Clptm1) at 0 DIV. mIPSC recordings and immunostaining were performed at 14 DIV.
(C) Overexpression of Clptm1 significantly reduced mIPSC amplitude without changing mIPSC frequency. n=40 cells from 6 independent experiments, **** p<0.0001, t-test. The cumulative probability curve of Clptm1 amplitude was scaled by dividing by a factor of 0.78. Kolmogorov-Smirnov (K–S) test showed no significant difference between control and scaled Clptm1 groups.
(D) Neurons were immunostained live using anti-GABAAR γ2 antibody, followed by fixation and immunostaining for VGAT and the dendritic marker MAP2. Overexpression of Clptm1 significantly reduced synaptic surface γ2 intensity without changing postsynaptic γ2 puncta number. Scale bar represents 10 μm. n=30 cells from 2 independent experiments, **** p<0.0001, t-test.
(E–F) Neurons were fixed and immunostained for VGAT, MAP2, and gephyrin (Geph; E) or neuroligin-2 (NL2; F). Overexpression of Clptm1 did not change synaptic intensity or puncta number for either gephyrin or neuroligin-2. Scale bar represents 10 μm. n=30 cells from 2 independent experiments.
Results are expressed as mean ± SEM.
See also Table S2
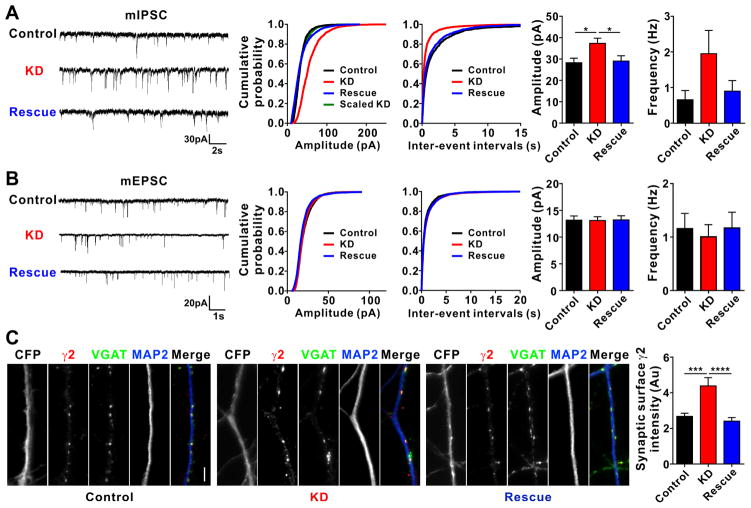
To directly assess whether overexpression of Clptm1 reduces synaptic GABAARs, we immunostained for surface GABAAR γ2 and for vesicular GABA transporter (VGAT), an inhibitory presynaptic marker. Consistent with the reduction in amplitude but not frequency of mIPSCs (Figure 4C), overexpression of Clptm1 reduced the intensity by not the number of postsynaptic surface clusters of GABAAR γ2 (Figure 4D). Overexpression of Clptm1 did not alter the intensity or number of postsynaptic clusters of gephyrin or neuroligin-2 (Figure 4E and 4F), indicating a specific effect on GABAARs.
Knockdown of Clptm1 Induces Multiplicative Up Scaling of Inhibitory Transmission
Based on the above data, we hypothesize that endogenous Clptm1 binds GABAAR in the forward trafficking pathway, blocking GABAAR insertion to inhibitory synapses. In this case, reducing levels of endogenous Clptm1 should result in increased inhibitory synaptic transmission, as Clptm1 is ubiquitously expressed (Yoshiura et al., 1998), including within brain (Lein et al., 2007). To test this idea, we designed a short-hairpin RNA (shRNA) to knock down Clptm1. The knockdown efficacy was confirmed by Western blot in cotransfected HEK293 cells and for native Clptm1 in cultured cortical neurons infected with AAV expressing shClptm1 or its scrambled form shScramble as a control (Figure S3). Knockdown of Clptm1 significantly increased the amplitude of mIPSCs in cultured hippocampal neurons compared with the scrambled control (Figure 5A). The increase in mIPSC amplitude by Clptm1 knockdown was restored back to control level by co-expression of an RNAi-resistant Clptm1* (Figure 5A), suggesting this effect was due to loss of Clptm1 and not off-target effects. Knockdown of Clptm1 caused a multiplicative scaling up of mIPSC amplitude, since the distribution of scaled knockdown amplitudes was not significantly different from control (Figure 5A). As for the overexpression, this multiplicative scaling is consistent with a role for Clptm1 in modulating all postsynaptic GABAARs. Knockdown of Clptm1 had no effect on mEPSCs (Figure 5B), indicating a specific function in regulating inhibitory and not excitatory transmission. Consistent with the effect on mIPSC amplitude, knockdown of Clptm1 resulted in an increase in the intensity of surface synaptic GABAAR γ2 clusters (Figure 5C). These results suggest that endogenous Clptm1 functions to keep GABAARs in check, maintaining inhibitory postsynaptic GABAARs at submaximal level.
Figure 5. Knockdown of Clptm1 Scales Up Inhibitory Synaptic Strength.
Cultured hippocampal neurons were transfected with U6-shScramble-hSyn-CFP and hSyn-YFP as control, U6-shClptm1-hSyn-CFP and hSyn-YFP as knockdown (KD), or U6-shClptm1-hSyn-CFP and hSyn-YFP-p2a-Clptm1* as rescue at 0 DIV. CFP and YFP dual positive neurons were selected for recording or immunostaining. mIPSCs were recorded at 13 DIV, mEPSCs were recorded at 14 DIV, and immunostaining was performed at 14 DIV.
(A) Knockdown of Clptm1 significantly increased mIPSC amplitude compared with the control group, an effect rescued by expressing the shRNA-resistant Clptm1*. n=17–19 cells from 4 independent experiments, p<0.05 one-way ANOVA and * p<0.05 post hoc Holm-Sidak tests. The cumulative probability curve of KD amplitude was scaled by dividing by a factor of 1.65. KS test showed no significant difference between control and scaled KD groups.
(B) Knockdown of Clptm1 did not affect mEPSC amplitude or frequency. n=16–18 cells from 4 independent experiments.
(C) Neurons were immunostained live using anti-GABAAR γ2 antibody, followed by fixation and immunostaining for VGAT and MAP2. Knockdown of Clptm1 significantly increased synaptic surface γ2 intensity, an effect normalized by expressing the shRNA-resistant Clptm1*. Scale bar represents 10 μm. n=30 cells from 3 independent experiments, p<0.0001 one-way ANOVA and *** p<0.001, **** p<0.0001 post hoc Holm-Sidak tests.
Results are expressed as mean ± SEM.
Inhibitory Synaptic Transmission is Regulated by Clptm1 in Vivo
To determine whether Clptm1 regulates inhibitory synaptic transmission in vivo, overexpression or knockdown of Clptm1 was achieved using AAV vectors injected bilaterally into the brain ventricles of newborn mice. Electrophysiological recordings were performed on hippocampal CA1 pyramidal neurons in acute brain slices at postnatal days P14–17. For overexpression, AAV expressing YFP-p2a-Clptm1 was used to mark the transduced cells and the synapsin promoter was used to target neurons. Efficient self-cleavage (Kim et al., 2011) of YFP-p2a-Clptm1 yielding YFP and Clptm1 was confirmed in cultured neurons (data not shown). Consistent with the reduction of mIPSC amplitude in cultured hippocampal neurons, mIPSC amplitude was significantly decreased in CA1 pyramidal neurons expressing YFP-p2a-Clptm1 compared with neurons expressing GFP control (Figure 6A).
Figure 6. Clptm1 Modulates Inhibitory Synaptic Transmission in Vivo.
Neonatal mice were injected with AAV vectors to alter Clptm1 expression and electrophysiological recordings were performed on CA1 neurons expressing the fluorescent protein markers in acute brain slice at P14–17.
(A) Overexpression of YFP-p2a-Clptm1 (Clptm1) significantly reduced mIPSC amplitude compared with control group expressing GFP. n=24–25 cells from 2 mice, **** p<0.0001, t-test. The cumulative probability curve of Clptm1 amplitude was scaled by dividing by a factor of 0.73. K-S test showed no significant difference between control and scaled Clptm1 groups.
(B) Groups were co-injected with AAV-U6-shScramble-hSyn-Tdtomato and AAV-hSyn-GFP as control, AAV-U6-shClptm1-hSyn-Tdtomato and AAV-hSyn-GFP as knockdown (KD), or AAV-U6-shClptm1-hSyn-Tdtomato and AAV-hSyn-YFP-p2a-Clptm1* as rescue. Recordings were collected from Tdtomato and GFP or YFP dual positive CA1 pyramidal neurons. Knockdown of Clptm1 significantly increased mIPSC amplitude compared with the control group, an effect rescued by the shRNA-resistant Clptm1*. n=28 cells from 3–4 mice, p<0.0001 one-way ANOVA and ** p<0.01, **** p<0.0001 post hoc Holm-Sidak tests. The cumulative probability curve of KD amplitude was scaled by dividing by a factor of 1.16. K–S test showed no significant difference between control and scaled KD groups.
(C and D) The GABA/AMPA ratio was recorded in hippocampal CA1 pyramidal neurons by stimulating the Schaffer collateral-commissural fibers and holding the cells at 0 mV for GABAAR currents and −60 mV for AMPAR currents. The GABA/AMPA ratio was significantly reduced in neurons overexpressing Clptm1 (C, n=23–25 cells from 2–3 mice, * p<0.05, t-test). Knockdown of Clptm1 elevated the GABA/AMPA ratio, and co-expressing the shRNA-resistant Clptm1* restored the GABA/AMPA ratio to control level (D, n=23–27 cells from 2–3 mice, p<0.05 one-way ANOVA and *** p<0.001 post hoc Holm-Sidak tests).
Results are expressed as mean ± SEM.
To assess the effects of Clptm1 knockdown in vivo, we co-injected two viral vectors to mediate Tdtomato-labeled knockdown plus GFP/YFP-labeled rescue, or the respective controls. AAV injection was performed in newborn mice and hippocampal CA1 pyramidal cells positive for Tdtomato plus GFP/YFP were recorded in acute brain slices at P14–17. Consistent with the results in cultured hippocampal neurons, the amplitude of mIPSCs was increased in neurons expressing the knockdown vector, and fully restored to control level in the rescue group (Figure 6B). Again, overexpression or knockdown of Clptm1 caused a multiplicative scaling of mIPSC amplitude, since the distribution of scaled overexpression or knockdown amplitudes were not significantly different from the corresponding control (Figure 6A and 6B). No effects were observed on mIPSC frequency, consistent with the proposed role of Clptm1 in controlling postsynaptic GABAAR levels. In addition, the effects of Clptm1 overexpression and knockdown in hippocampal CA1 pyramidal neurons were specific for inhibitory synapses, as no changes were detected in mEPSC amplitude or frequency (Figure S4).
To test whether Clptm1 modifies evoked GABA currents and inhibitory/excitatory ratio, synaptic responses were induced by stimulating Schaffer collateral fibers in the presence of NMDA receptor blocker APV while holding neurons at 0 mV to obtain GABAAR-mediated responses and −60 mV to obtain AMPAR-mediated responses. The ratio between these values, the GABA/AMPA ratio, was significantly reduced in neurons overexpressing Clptm1 compared with control neurons expressing GFP (Figure 6C). Conversely, knockdown of Clptm1 significantly increased the GABA/AMPA ratio, an effect which was fully rescued by the RNAi-resistant Clptm1* (Figure 6D). Taken together, these experiments show that altering the expression level of Clptm1 scales inhibitory synaptic transmission in vivo, with no effect on excitatory transmission thus with a consequent change in inhibition/excitation ratio.
Clptm1 Modulates Inhibitory Homeostatic Synaptic Plasticity
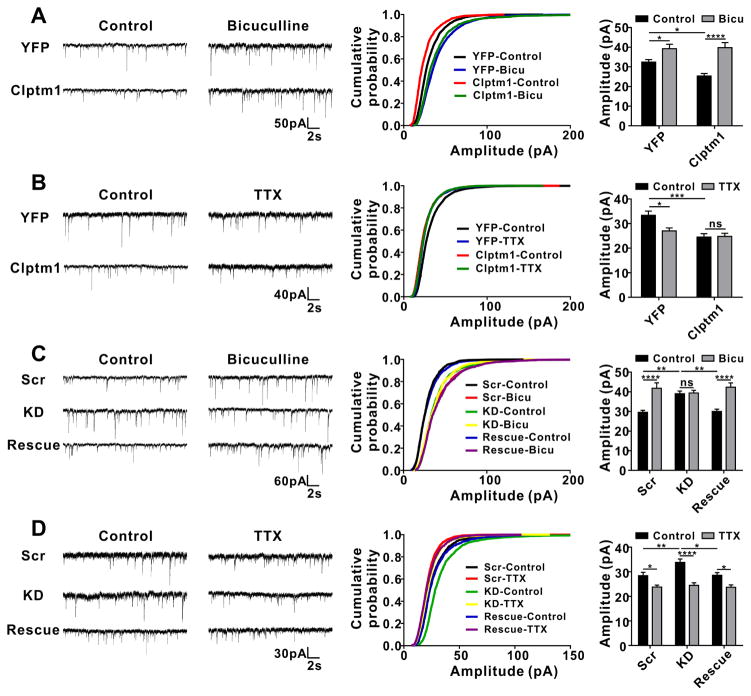
The GABAergic synaptic scaling induced by altering levels of Clptm1 appears similar to that induced by long-lasting alterations in neuronal network activity, a key homeostatic mechanism for maintaining neuron firing within a target range (Kilman et al., 2002; Pribiag et al., 2014; Saliba et al., 2007). To probe the relationship between these phenomena, we tested how altering levels of Clptm1 affects the scaling up of mIPSC amplitude induced by a 24 hour application of GABAAR antagonist bicuculline or scaling down induced by a 48 hour application of voltage-gated Na+ blocker tetrodotoxin (TTX) as demonstrated previously (Pribiag et al., 2014). Consistent with previous studies, mIPSC amplitude was increased following bicuculline treatment in control neurons expressing YFP or scrambled shRNA (Figure 7A and 7C). Knockdown of Clptm1 which elevated mIPSC amplitude in basal conditions, as seen before in cultured neurons (Figure 5A) and brain slices (Figure 6 B), blocked bicuculline-induced scaling up of mIPSC amplitude, an effect which was fully rescued by RNAi-resistant Clptm1* (Figure 7C). In contrast, overexpression of Clptm1 which reduced mIPSC amplitude in basal conditions (Figure 4A and Figure 6A) resulted in a more robust increase in mIPSC amplitude following bicuculline treatment (Figure 7A). Also consistent with previous studies, mIPSC amplitude was reduced following TTX treatment in control neurons expressing YFP or scrambled shRNA (Figure 7B and 7D). Overexpression of Clptm1 blocked TTX-induced scaling down of mIPSCs (Figure 7B) while knockdown of Clptm1 showed a more robust scaling down (Figure 7D). We confirmed that bicuculline- or TTX- induced inhibitory synaptic homeostasis involve multiplicative scaling, as there were no detectable changes in the cumulative distribution of scaled treated groups compared with corresponding control groups, including groups with altered levels of Clptm1 (Figure S5). For all tested conditions, we did not observe any changes in mIPSC frequency (Figure S6). In summary, the synaptic scaling induced by overexpression or knockdown of Clptm1 occluded further homeostatic adjustment in the same direction in response to changes in circuit activity. However, overexpression or knockdown of Clptm1 did not block activity-induced homeostatic adjustment in the opposing direction, indicating alternative pathways.
Figure 7. Changes in Expression Level of Clptm1 Occlude Forms of Inhibitory Synaptic Homeostasis.
Cultured hippocampal neurons were transfected at 0 DIV using nucleofection. Bicuculline (Bicu, 10 μM) or TTX (1 μM) was added into culture medium at 14 DIV. mIPSC recordings were performed 24 hours after bicuculline or 48 hours after TTX application.
(A and B) mIPSC amplitude was significantly increased following bicuculline treatment, and decreased following TTX treatment in neurons expressing YFP. Overexpression of YFP-p2a-Clptm1 (Clptm1), which significantly reduced mIPSC amplitude compared with control group YFP, did not affect bicuculline-induced inhibitory synaptic scaling up (A, n=16–17 cells from 2 independent experiments), but blocked TTX-induced scaling down (B, n=19–24 cells from 4 independent experiments). (A) Interaction: p=0.057, expression level of Clptm1: p=0.095, treatment: p<0.0001, two-way ANOVA and * p<0.05, **** p<0.0001 post hoc Holm-Sidak tests. (B) Interaction: p<0.05, expression level of Clptm1: p<0.001, treatment: p<0.05, two-way ANOVA and * p<0.05, *** p<0.001 post hoc Holm-Sidak tests. There was no significant difference between YFP plus bicuculline and Clptm1 plus bicuculline groups, or between YFP plus TTX and Clptm1 plus TTX groups.
(C and D) The transfection groups were U6-shScramble-hSyn-Tdtomato and hSyn-YFP as control, U6-shClptm1-hSyn-Tdtomato and hSyn-YFP as knockdown (KD), or U6-shClptm1-hSyn-Tdtomato and hSyn-YFP-p2a-Clptm1* as rescue. Tdtomato and YFP dual positive neurons were selected for recording. Knockdown of Clptm1, which significantly elevated mIPSC amplitude compared with the control group, did not affect TTX-induced inhibitory synaptic scaling down (D, n=24–30 cells from 5 independent experiments), but blocked bicuculline-induced scaling up (C, n=19–22 cells from 3 independent experiments). The knockdown effects were fully rescued by expressing the shRNA-resistant Clptm1*. (C) Interaction: p<0.001, expression level of Clptm1: p=0.101, treatment: p<0.0001, two-way ANOVA and ** p<0.01, **** p<0.0001 post hoc Holm-Sidak tests. (D) Interaction: p=0.066, expression level of Clptm1: p<0.01, treatment: p<0.0001, two-way ANOVA and * p<0.05, ** p<0.01, **** p<0.0001 post hoc Holm-Sidak tests.
Results are expressed as mean ± SEM
See also Figure S5, S6 and Table S2.
Clptm1 Suppresses Tonic GABAAR Currents
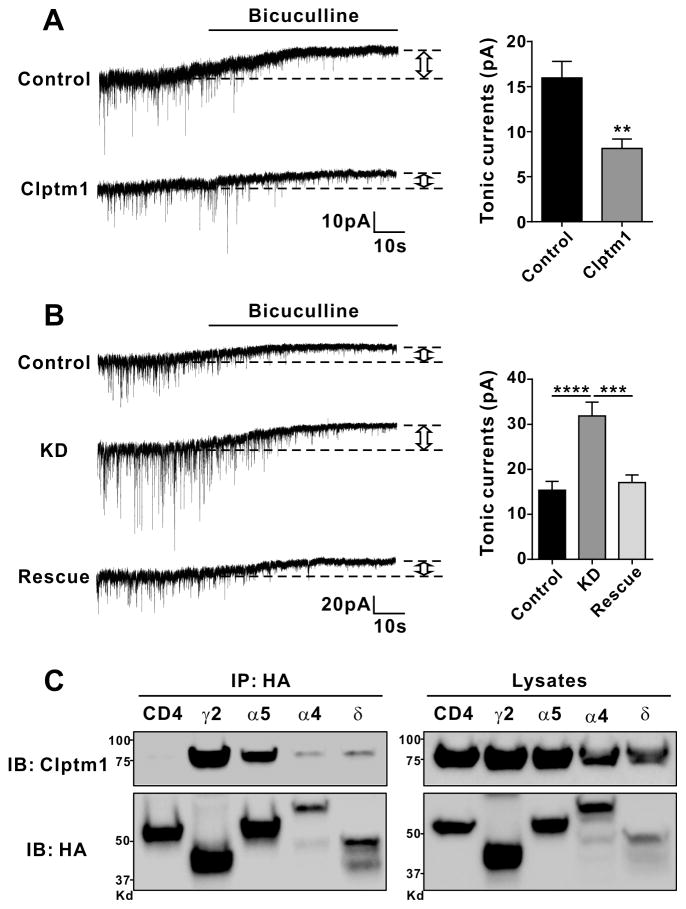
To determine whether Clptm1 controls tonic as well as phasic inhibitory transmission, we overexpressed or knocked down Clptm1 in cultured hippocampal neurons and assessed the tonic current as the change in baseline current upon application of bicuculline. Overexpression of Clptm1 significantly reduced the tonic GABAeric current (Figure 8A). Conversely, knockdown of Cltm1 increased the tonic GABAeric current, which was normalized upon rescue with RNAi-resistant Clptm1* (Figure 8B).
Figure 8. Clptm1 Modulates GABAAR Mediated Tonic Currents.
Cultured hippocampal neurons were transfected at 0 DIV using nucleofection. Recordings were performed at 15 DIV. Bicuculline (10 μM) was added into the recording solution to record tonic currents.
(A) Neurons were transfected with YFP (Control) or YFP-2a-Clptm1 (Clptm1). Overexpression of Clptm1 significantly reduced GABAAR mediated tonic currents. n=16 cells from 3 independent experiments, ** p<0.01, t-test.
(B) The transfection groups were U6-shScramble-hSyn-Tdtomato and hSyn-YFP as control, U6-shClptm1-hSyn-Tdtomato and hSyn-YFP as knockdown (KD), or U6-shClptm1-hSyn-Tdtomato and hSyn-YFP-p2a-Clptm1* as rescue. Knockdown of Clptm1 significantly increased GABAAR mediated tonic currents, an effect normalized by the shRNA-resistant Clptm1*. n=13 cells from 2 independent experiments, p<0.0001 one-way ANOVA and *** p<0.001, **** p<0.0001 post hoc Holm-Sidak tests.
(C) HEK293 cells were co-transfected with Clptm1 and HA-tagged GABAAR γ2, α5, α4, δ or CD4 as a negative control. Clptm1 was co-immunoprecipitated with GABAAR γ2, α5, α4, and δ subunits.
Results are expressed as mean ± SEM
See also Table S2.
Tonic inhibition in hippocampal neurons is mediated by α5-GABAARs and δ-GABAARs (Glykys et al., 2008), the δ subunit often in combination with α4. Thus we tested whether Clptm1 can interact with these subunits by co-expressing Clptm1 with HA-tagged α 5, α4, δ, or γ2 for comparison in HEK293 cells and immunoprecipitated using anti-HA antibody. Clptm1 co-immunoprecipitated with α 5, α4, and δ GABAAR subunits (Figure 8C) which typically function at extrasynaptic sites (Olsen and Sieghart, 2008). Thus, Cptm1 regulates both phasic and tonic currents and interacts with all tested GABAAR subunits.
DISCUSSION
Using a transgenic tandem affinity purification proteomic strategy, we identified Clptm1, Glg1, Itm2C and additional novel candidates as GABAAR-associated proteins. In recombinant systems, Clptm1 co-immunoprecipitated with multiple GABAAR subunits, reduced surface levels of GABAARs and increased their accumulation in the ER and Golgi complex, and reduced GABA currents, with no effect on glycine receptors. In hippocampal cultured neurons and in vivo, overexpression or knockdown of Clptm1 reduced or enhanced, respectively, GABAergic but not glutamatergic transmission. Furthermore, altering levels of Clptm1 occluded some forms of homeostatic activity regulation and affected tonic as well as phasic inhibitory transmission. Together, these data validate Clptm1 as a broad regulator of GABAergic signaling. However, unlike most known GABAAR-associated proteins which promote synaptic accumulation of receptors, Clptm1 reduces GABAAR forward trafficking and limits inhibitory transmission.
CLPTM1 was originally identified by association of a chromosomal translocation with cleft lip and palate (Yoshiura et al., 1998). Altered GABAergic signaling is one among many causes of cleft palate, and it appears that perturbations in either direction from the norm can contribute. For example, administration of either the GABAAR antagonist picrotoxin or GABAAR potentiator lorazepam induces cleft palate (Ding et al., 2004; Jurand and Martin, 1994), as does genetic deletion of GABAAR β3 subunit (Culiat et al., 1995; Homanics et al., 1997). Our findings raise the possibility that CLPTM1 may contribute to cleft palate through altered GABAAR signaling.
One key feature that distinguishes Clptm1 from other GABAAR-associated proteins is its function in limiting surface and synaptic accumulation of receptors. To the best of our knowledge, all other GABAAR-associated proteins with the exception of the rather generic endocytic adapter AP2 function to promote surface and/or synaptic accumulation of GABAARs (reviewed by Luscher et al., 2011a and Vithlani et al., 2011). Mechanisms include facilitation of receptor exit from the ER, translocation from the Golgi to the plasma membrane, surface recycling from endosomes, and confinement at postsynaptic sites. In contrast, Clptm1 increases GABAAR confinement in intracellular compartments including the ER, limiting forward trafficking and acting as a brake on receptor surface expression. This function is consistent with the reported localization of Clptm1 to the ER and Golgi complex (Stadler et al., 2013) and our inability to detect Clptm1 on the cell surface. The function of Clptm1 in reducing surface expression of GABAARs as shown in heterologous cells is also consistent with the effect of Clptm1 knockdown to increase mIPSC amplitude but not frequency in cultured neurons and in vivo.
Thus, Clptm1 may be considered a negative regulator of inhibitory synaptic transmission, one that acts by limiting postsynaptic and extrasynaptic GABAAR accumulation. Clptm1 specifically regulated postsynaptic accumulation of GABAARs without affecting gephyrin or neuroligin-2. Two other negative regulators of inhibitory transmission were recently identified that act by different mechanisms, both involving neuroligin-2. The peptidyl-prolyl isomerase Pin1 binds a phosphorylated neuroligin-2 intracellular domain and reduces its interaction with gephyrin, thus down-regulating GABAergic transmission (Antonelli et al., 2014). The cell surface protein MDGA1 binds the neuroligin-2 extracellular domain and blocks its interaction with neurexin, thus suppressing inhibitory synapse development (Pettem et al., 2013). However, proteins that limit surface expression of selective neurotransmitter receptors are rare. Perhaps most analogous to Clptm1 for GABAARs is Thorase for AMPA receptors. Loss of Thorase increases mEPSC amplitudes and elevates surface expression of GluA1 and GluA2, although by regulating endocytosis rather than forward trafficking (Zhang et al., 2011).
A second unusual feature of Clptm1 is its broad association with many GABAAR subunits of both synaptic and extrasynaptic classes. Consistent with its synaptic scaling activity, Clptm1 regulated the main synaptic GABAAR receptor types, α1β2γ2 and α2β3γ2, in a recombinant system. While Clptm1 was isolated by association with HFY-GABAARγ2 and its interaction with the individually expressed γ2 subunit appeared strong, Clptm1 also co-immunoprecipitated with all other GABAAR subunits tested and functionally regulated α1β2 and α2β3 receptors. These findings do not reflect a general lack of specificity as Clptm1 did not co-immunoprecipitate with or functionally modulate glycine receptor α1 and its overexpression and knockdown did not affect excitatory synaptic transmission. Its broad interaction with many GABAAR subunits distinguishes Clptm1 from other GABAAR-associated proteins. For example, gephyrin interacts specifically with α1, α2 and α3 subunits (Tretter et al., 2012), GABARAP with all γ subunits (Nymann-Andersen et al., 2002), and PRIP with β subunits (Terunuma et al., 2004). Consistent with the presence of β subunits in synaptic and extrasynaptic receptors, PRIP regulates phasic and tonic GABAergic transmission (Zhu et al., 2012), like Clptm1.
A third key feature of Clptm1 is its ability to mimic and occlude some forms of activity-regulated inhibitory synaptic scaling. Consistent with a potential role for Clptm1, activity-regulated inhibitory synaptic homeostasis involves changes in the numbers of postsynaptic GABAARs (Kilman et al., 2002; Peng et al., 2010; Rannals and Kapur, 2011). Many molecules have been implicated in controlling excitatory homeostatic synaptic scaling of AMPA receptors, including tumor necrosis factor α (TNFα), brain-derived neurotrophic factor, retinoic acid, fragile-X mental retardation protein, methyl-CpG-binding protein MeCP2, N-cadherin/β-catenin, β3 integrin, stargazin, glutamate receptor-interacting protein 1, protein interacting with C-kinase 1, Homer1a, and activity-regulated cytoskeleton-associated protein Arc/Arg3.1 (reviewed by Fernandes and Carvalho, 2016). However, few signals have been identified that control synaptic scaling of GABAARs. One such signal regulates both excitatory and inhibitory receptors, in opposite directions: elevation of glial-derived TNFα upon TTX treatment both increases surface AMPA receptors and reduces surface GABAARs (Pribiag and Stellwagen, 2013; Stellwagen and Malenka, 2006). Mechanistically, TNFα promotes protein phosphatase 1-regulated endocytosis of the main synaptic GABAARs. The other identified molecular signal involved in GABAergic synaptic scaling, dystroglycan, is selectively required for homeostatic scaling down of inhibitory synaptic strength upon bicuculline treatment (Pribiag et al., 2014). We suggest that Clptm1 may contribute along with signals such as TNFα and dystroglycan to control GABAergic synaptic scaling.
Considering the central role of GABAergic transmission in brain function and dysfunction in epilepsy, major depressive disorder, anxiety disorders, and neurodevelopmental disorders, the identification of novel GABAAR-associated regulatory proteins opens new therapeutic directions. The abilities of Clptm1 to broadly interact with all major synaptic and extrasynaptic GABAAR subunits, to control surface GABAAR levels through limiting forward trafficking, and to multiplicatively scale inhibitory synaptic strength make it an attractive target for intervention.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Dr. Ann Marie Craig (acraig@mail.ubc.ca).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All experimental procedures with animals were conducted following the guidelines of the Canadian Council for Animal Care and the University of British Columbia Animal Care Committee.
Generation of Thy1-HFY-GABAARγ2 transgenic mice was described previously (Bleckert et al., 2013). The epitope tags consist of hexahistidine and FLAG (ASGHHHHHHGMDYKDDDDKGLG) followed by enhanced YFP between amino acids 4 and 5 of the mature GABAARγ2 short isoform coding sequence. The Thy1 vector contains 6.5 kb of the murine thy1.2 gene (Feng et al., 2000). Mice were generated by the Washington University School of Medicine in St. Louis Mouse Genetics Core on a B6/CBA F1 hybrid background and backcrossed at least six generations to C57BL/6J. Genotyping was performed by PCR. Mice were sacrificed between 2 and 4 months of age. Brains from sex- and age-matched transgenic or littermate wild type mice were pooled for tandem affinity purification. Male and female mice were used for imaging and proteomics. Because males and females were pooled for each proteomics experiment, males and females were not analysed separately.
AAV injection was performed on C57BL/6 wild type mice at postnatal day 0. Only male neonates were selected for experimental groups. Electrophysiological recordings were performed at P14–17.
Primary Neuron Culture
Rat hippocampal neurons from embryonic day 18 male and female rats were cultured at low density on poly-L-lysine-coated glass coverslips inverted over a feeder layer of astrocytes in neurobasal medium (Invitrogen) supplemented with B27 (Invitrogen or StemCell). Cortical neurons from embryonic day 18 rats were cultured on poly-L-lysine-coated 12-well plates in the same medium.
Cell Line Culture
HEK293 cells and COS7 cells were cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum. Cell lines were obtained from ATCC and maintained without further authentication.
METHOD DETAILS
Brain Immunofluorescence and Imaging
For confocal imaging, 2–4 month old Thy1-HFY-GABAARγ2 transgenic mice were anaesthetized with 20% urethane and perfused transcardially with 2% paraformaldehyde in PBS. The brains were then collected and post-fixed with 2% paraformaldehyde in PBS for 1 hour, cryoprotected in 20% and then 30% sucrose in PBS, and rapidly frozen. Coronal cryostat sections were cut at 20 μm thickness and mounted on Superfrost Plus slides (Fisher Scientific). The sections were incubated in blocking solution (5% BSA, 5% normal goat serum, and 0.25% Triton X-100 in PBS) for 1 hour at room temperature followed by primary antibodies overnight at 4°C. The following primary antibodies were used: anti-Gephyrin (1:1000; Synaptic Systems); anti-GABAAR γ2 subunit (1:2000; generous gift from Dr. J. M. Fritschy, University of Zurich); anti-GAD65 (1:100; Developmental Studies Hybridoma Bank). For the GAD65 labeling, perfused brains required longer post-fixation which reduced the YFP signal, so anti-GFP (1:500; Invitrogen) was also used. The sections were then washed with PBS and incubated with appropriate Alexa conjugated secondary antibodies (1:500; Invitrogen) for 1 hour at room temperature. For fluorescent labeling of nuclei, 4′,6-diamidino-2-phenylindole (DAPI, 100 ng/ml) was included with the secondary antibodies. The sections were washed with PBS and mounted in Elvanol (Tris-HCl, glycerol, polyvinyl alcohol, 2% 1,4-diazabicyclo[2,2,2]octane). Images were captured using an Olympus Fluoview FV500 confocal on a BX61W microscope with a 60× 1.42 numerical aperture oil-immersion lens and customized filter sets. The colocalization of HFY-GABAARγ2 with GAD65 or gephyrin was quantified using ImageJ colocalization test with Fay randomization method.
Tandem Affinity Purification
All of the protein work was performed at 4°C. Seven brains from sex- and age-matched transgenic or wild type mice between 8 to 16 weeks were collected and pooled in homogenization buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0 with protease inhibitor cocktail (Roche)). The whole brain was first briefly sonicated for two 10 s bursts with a microtip using a Sonic Dismembrator (FB120110, Fisher Scientific) then homogenized using a Dounce homogenizer (099C K54, Thomas Scientific). To get rid of nuclei and cellular debris, the brain homogenate was centrifuged for 10 min at 1500 × g. The supernatant was saved and centrifuged for 1 h at 100,000 × g and the subsequent pellet was resuspended in homogenization buffer and further homogenized using only the Dounce homogenizer, and then lysed with 0.5% (w/v) n-dodecyl-β-D-maltoside (DDM, Sigma) in homogenization buffer rotating slowly for 16 h. By ultracentrifugation at 100,000 × g for 60 min, cytoskeletal debris was removed.
Nickel-nitrilotriacetic acid (Ni-NTA) Agarose (Qiagen) resin was equilibrated with homogenization buffer and incubated with brain lysate for 16 h rotating at 200 rpm on a rotary shaker. The resin was washed twice with 10 mM imidazole in homogenization buffer. Elution of the proteins bound to Ni-NTA resin with 200 mM imidazole in homogenization buffer was repeated three times. Pooled eluate was incubated with biotin conjugated anti-GFP antibody (0.4 μg per mg brain lysate; Rockland) for 1 h, followed by incubation with Streptavidin conjugated Sepharose 4B beads (1 μl per mg brain lysate; US Biological) for 16 h. The immune complexes were washed twice with 0.1% DDM in homogenization buffer, and purified proteins were eluted three times with SDS sample buffer (0.0626 M Tris HCl, 2% SDS, 0.01% bromophenol blue, 100 mM dithiothreitol, 10% glycerol, pH 6.8) and pooled.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
Tandem affinity purified sample from each anti-GFP immunoprecipitation following Ni-NTA elution was run into a 10% SDS-PAGE gel, visualized by colloidal coomassie, split into 6 fractions and then digested out of the gel (Chan et al., 2006). Peptide samples were purified by solid phase extraction on C-18 STAGE tips (Ishihama et al., 2002) and analyzed using a linear-trapping quadrupole Orbitrap mass spectrometer (LTQ-Orbitrap Velos; Thermo Fisher Scientific) on-line coupled to an Agilent 1290 Series HPLC using a nanospray ionization source. The trap column is packed with 5 μm-diameter Aqua C-18 beads (Phenomenex) while the analytical column is packed with 3.0 μm-diameter Reprosil-Pur C-18-AQ beads (Dr. Maisch). Samples were resuspended in buffer A (0.5% aqueous acetic acid) and loaded with the same buffer. A series of gradients of buffer A and buffer B (0.5% acetic acid and 80% acetonitrile in water) were run. The HPLC system included Agilent 1290 series Pump and Autosampler with Thermostat set at 6°C. The sample was loaded on the trap column at 5 μL/min and the analysis was performed at 0.1 μL/min. The LTQ-Orbitrap was set to acquire a full-range scan at 60,000 resolution from 350 to 1600 in the Orbitrap to simultaneously fragment the top ten peptide ions by CID in each cycle in the LTQ (minimum intensity 1000 counts). Parent ions were then excluded from MS/MS for the next 30 seconds. Singly charged ions were excluded in ESI mode as most peptides carry multiple charges. The Orbitrap was continuously recalibrated using lock-mass function.
Protein Identification
The lists of centroided fragment peak were analyzed with Proteome Discoverer v. 1.2 (ThermoFisher Scientific). The search with the Mascot algorithm v. 2.4 was run against the mouse Uniprot database with common contaminants added (50863 total sequences) with the following parameters: peptide mass accuracy 10 ppm; fragment mass accuracy 0.6 Da; trypsin enzyme specificity with one missed cleavage, fixed modifications - carbamidomethyl, variable modifications - methionine oxidation, deamidated N, Q and N-acetyl peptides, ESI-TRAP fragment characteristics. Accurately identified peptides were only confined to those with IonScores higher than 99% confidence. The parameters of Proteome Discoverer were: event detector mass precision 4 ppm, S/N threshold 1; replacement of missing quantitation values with minimum intensity for ratio calculation, use of single peak quantitation channels, use of all peptides for protein quantification.
Selection of GABAARγ2-specific Interactors
From all the proteins identified across 4 independent experiments, those that might be specifically interacting with HFY-GABAARγ2 were selected by applying the following criteria: if a protein was exclusively found in TG sample, it should be detected at least two of four experiments; if a protein was detected in both TG and WT samples, the cumulative PSM ratio TG:WT should be ≥ 6:1.
Overrepresentation Analysis
The possibility that GABAARγ2-associated proteins were enriched in certain cellular pathways or compartments was tested by comparing the set of proteins that meet the selection criteria with the entire mouse database using the statistical overrepresentation test (Geneontology PANTHER (Mi et al., 2013)). GO biological process complete or GO cellular component complete was used as the Annotation Data Set. The Bonferroni correction was applied for multiple testing.
Plasmid Constructs
Clptm1 (Cat#MMM1013-649600; Accession: BC022172), Itm2C (Cat#MMM1013-63801; Accession: BC012952), Glg1 (Cat#MMM4769-99609980; Accession: BC021306), Tfg (Cat#MMM1013-202764274; Accession: BC024638) plasmids were purchased from Open Biosystem. Tmub1 plasmid was a kind gift from Dr. Mitsutoshi Setou (Hamamatsu University, Japan). To add a Myc tag at the N-terminus, Glg1 was subcloned by PCR and insert into modified pEYFP-C1 vector with the signal sequence from TrkC (Takahashi et al., 2011) and the YFP tag replaced with a Myc tag. To add a Myc tag in the C-terminus, Itm2C was subcloned by PCR and inserted into pcDNA4/myc-his vector (Invitrogen). To add an YFP tag, Clptm1, Glg1, and Tmub1 were subloned by PCR and inserted into sp-YFP-C1 vector (Takahashi et al., 2011), Tfg was subloned by PCR and inserted into pEYFP-C1 vector (Clontech), and Itm2C was subcloned by PCR and inserted into pEYFP-N1 vector (Clontech). The construction of Myc-Clstn3-CFP was described previously (Pettem et al., 2013a). Rat GABAAR subunits α1, α2, β2, γ3, γ2 plasmids are kind gifts from Drs. Peter H. Seeburg and Hartmut Lüddens (University of Heidelberg, Germany). Mouse GABAAR subunits α5 plasmid was purchased from Dharmacon (Cat#MMM1013-202798742; Accession: BC062112). Human GlyR α1 was a kind gift from Dr. Yu Tian Wang (University of British Columbia, Canada). HA-tagged CD8, GABAARα1, α2, β2, β3, γ2, α5 and GlyRα1 were made by subcloning the mature coding regions of each subunit into a modified pEYFP-C1 vector with the signal sequence from TrkC and the YFP tag replaced with an HA tag (Pettem et al., 2013b; Takahashi et al., 2011). HA-tagged GABAARα4, δ plasmids were purchased from Sino Biological. The construction of HFY-GABAAR-γ2 was described previously (Dobie and Craig, 2011). For testing surface expression of Clptm1, a Myc tag was added at the mature N-terminus, amino acid 30, 150, 382, 452, 535, or C-terminus of Clptm1. For shRNA interference knockdown, the oligonucleotides that target nucleotides 1641–1659 of mouse Clptm1 (5′-GGCCCTCAACACTTTCATT -3′) were subcloned into pLenLox3.7 variant pLL(syn)CFP (Takahashi et al., 2011) to express CFP and shClptm1 under the human synapsin promoter and U6 promoter, respectively. To make pAAV-U6-shRNA-hSyn-Tdtomato, CFP was replaced by Tdtomato, and U6-shRNA-hSyn-Tdtomato was then cut and inserted into a modified pAAV-MCS vector (Agilent Technologies) with the CMV promoter deleted. We used a scrambled shClptm1 (5′-GTACACTCACTGTCACTCT -3′) as the control shRNA. The RNA-resistant Clptm1 construct Clptm1* was made by site-directed mutagenesis of 1647 C to T. To use YFP as a visualized signal for the expression of Clptm1, the p2a peptide (GSGATNFSLLKQAGDVEENPGP) was used to link YFP with Clptm1 and YFP-p2a-Clptm1* was subcloned into pLenLox3.7 vector under the human synapsin promoter. pAAV-hSynapsin-EGFP-WPRE-bGH (p1696) was a gift from UPenn Vector Core, and GFP was replaced by YFP-p2a-Clptm1* for overexpression or rescue.
Cell Line Culture and Transfection
HEK293 cells and COS7 cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS. Cells were transfected using TransIT-LT1 Transfection Reagent (Mirus) with the recommended protocols as per manufacturer’s instructions. To detect transfected cells for recording, GFP was cotransfected with other plasmids in a ratio of 1:13. Immunoprecipitation, immunocytochemistry, or electrophysiological recording were performed 48 hours after transfection.
Immunoprecipitation and Western Blotting
Transfected HEK293 cells were homogenized in ice-cold RIPA buffer containing 50 mM Tris–HCl, pH 7.4, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 0.5% deoxycholic acid sodium, and a cocktail of protease inhibitors (Roche). After incubation on ice for 30 min, the homogenates were centrifuged at 14,000 rpm at 4°C for 15 min. The supernatants were collected and the total protein concentrations were measured using DC™ Protein Assay Kit (Bio-Rad). The whole brains lysates from HFY-GABAARγ2 transgenic mice at the age of p15 were prepared as described in Tandem Affinity Purification. Protein concentration was determined using DC™ Protein Assay Kit (Bio-Rad).
For immunoprecipitation, 500μg cell lysates were incubated with anti-HA antibody (1 μg; Roche) or anti-GFP antibody (1 μg; Invitrogen) for 4 hours at 4°C. Protein G-sepharose (GE Healthcare) was then added to the mixture and incubated for overnight. The complex was isolated by centrifugation and washed twice with washing buffer (500 mM NaCl, 1% Triton X-100, 50 mM Tris–HCl, pH 7.4) and twice with PBS. The precipitated proteins were eluted from the sepharose beads by heating in 2× sample buffer at 60°C for 5 min, and 25 μg lysate was used as control for total protein expression level.
Proteins eluted from the beads or total lysates were subjected to a sodium dodecyl sulfate-polyacrylamide gel (prepared using TGXTM FastCastTM Acrylamide Kit, 10%; Bio-Rad) electrophoresis and were transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with 5% milk for 1 hour at room temperature, immunoblotted with primary antibodies overnight at 4°C, and then incubated with HRP-conjugated secondary antibodies (1:10000, Southern Biotech) for 1 hour at room temperature. Blots were developed using the Immobilon Western Chemiluminescent HRP Substrate kit (Millipore) and imaged with the Bio-Rad gel imaging system (Bio-Rad). Protein band intensities were quantified with Image Lab software (Bio-Rad).
The following primary antibodies were used: anti-Clptm1 (1:2000; Abcam), anti-Myc (1:500; Santa Cruz), anti-HA (1:1000; Roche), anti-GFP (1:2000; Invitrogen), anti-GABAARα1 (1:1000; Millipore), anti-α-Tubulin (1:10000; Millipore).
Neuron Culture and Transfection
Dissociated hippocampal neuron cultures were prepared from embryonic day 18 rats as previously described (Kaech and Banker, 2006). The hippocampal cells were plated on glass coverslips coated with poly-L-lysine (PLL) at a density of 300,000 per 60 mm dish. The coverslips were inverted over a feeder layer of astrocytes in neurobasal medium (Invitrogen) supplemented with B27 (Invitrogen or StemCell). Cytosine arabinoside (5 μM) was added to neuron culture dishes at 2 days in vitro (DIV) to prevent overgrowth of glial cells. For overexpression studies, hippocampal neurons were transfected with Clptm1 or HA-CD4 and GFP with a ratio of 13:1 at 7 DIV using ProFection® Mammalian Transfection System (Promega), and electrophysiological recordings were performed at 9–10 DIV; or hippocampal neurons were transfected with YFP-p2a-Clptm1 or YFP as control at 0 DIV using nucleofection (AMAXA Biosystems), and electrophysiological recordings or immunostaining were performed at 14 DIV. For knockdown and rescue studies, hippocampal neurons were transfected at 0 DIV with U6-shScramble-hSyn-CFP and hSyn-YFP as control, U6-shClptm1-hSyn-CFP and hSyn-YFP as knockdown, or U6-shClptm1-hSyn-CFP and hSyn-YFP-p2a-Clptm1* as rescue with a ratio of 4:1 using nucleofection (AMAXA Biosystems) and seeded at a density of 1 million per 60 mm dish, and electrophysiological recordings or immunostaining were performed at 13–14 DIV. For homeostasis and tonic GABAAR current studies, hippocampal neurons were transfected with hSyn-YFP, hSyn-YFP-p2a-Clptm1*, U6-shScramble-hSyn-Tdtomato and hSyn-YFP, U6-shClptm1-hSyn-Tdtomato and hSyn-YFP, or U6-shClptm1-hSyn-Tdtomato and hSyn-YFP-p2a-Clptm1* with a ratio of 4:1 at 0 DIV using nucleofection (AMAXA Biosystems) and seeded at a density of 1 million per 60 mm dish, and electrophysiological recordings were performed at 15–16 DIV
For the knockdown efficiency assay, rat cortical neurons were plated on 12-well plates coated with PLL at a density of 1 million per well and cultured in the same medium as hippocampal neurons without addition of cytosine arabinoside. AAV virus was added in the medium at 2 DIV, and cell lysates were harvested at 14 DIV.
Immunocytochemistry
All imaging and analysis was done blind to treatment group. For surface HFY-GABAARγ2 labelling, transfected COS7 cells were live stained with anti-GFP antibody (1:500; Invitrogen) in conditioned media for 30 min at 37 C. Coverslips were washed 3 times with PBS, and then fixed for 12 min with prewarmed parafix (4% paraformaldehyde and 4% sucrose in PBS, pH 7.4), followed by permeabilization with 0.2% Triton X-100 in PBS. The coverslips were blocked with blocking solution (3% bovine serum albumin and 5% normal goat serum in PBS) for 45 min at 37 C, and then incubated with Alexa568-conjugated anti-rabbit secondary antibody (1:500; Invitrogen) for 45 min at 37 C. After washing 6 × 2 min with PBS, coverslips were mounted in Elvanol (Tris-HCl, glycerol, polyvinyl alcohol, and 2% 1, 4-diazabicyclo[2,2,2]octane). Images were acquired on a Zeiss Axioplan2 microscope with an oil immersion objective (63× 1.4 numerical aperture) and cooled CCD camera (Sensys, Photometrics) using MetaMorph software (Molecular Devices) and customized filter sets. For quantification of surface/total HFY-GABAARγ2, the surface anti-GFP immunofluorescene (Alexa568) and YFP integrated intensity were measured for the entire COS7 cell area using ImageJ (NIH) and the ratio was calculated after subtracting off-cell background intensity.
For the ER or Golgi colocalization, transfected COS7 cells were fixed and permeabilized prior to incubation with anti-Calnexin (1:500; Enzo Life Sciences) or anti-GM130 primary antibody (1:500; BD Transduction), followed by the appropriate Alexa 647-conjugated secondary antibody (1:500; Invitrogen). Clptm1 localization was shown by immunostaining Myc-tag using anti-Myc antibody followed by Alexa 568-conjugated secondary antibody (1:500; Invitrogen). Images were obtained on a Zeiss LSM 700 confocal microscope with a 40× 1.4 numerical aperture oil objective and sequential scanning with individual lasers and optimized filters. For quantification, a mask was generated from the anti-Calnexin or anti-GM130 (Alexa647) signal, and average intensity of HFY-GABAARγ2 (YFP) or Myc-Clptm1 (Alexa 568) within and outside of this mask was measured using ImageJ (NIH), and the ratio of HFY- GABAARγ2 or Myc-Clptm1 colocalized with anti-Calnexin or anti-GM130 over non-colocalized was calculated after subtracting off-cell background intensity.
For detecting surface expression of Clptm1, surface Myc-tagged Clptm1 was stained with anti-Myc antibody (Mouse, 1:500; Santa Cruz) in non-permeabilized condition and total Myc-tagged Clptm1 was stained with anti-Myc antibody (Rabbit, 1:2000; Sigma) in permeabilized condition, followed by Alexa488-conjugated anti-mouse and Alexa568-conjugated anti-rabbit secondary antibodies (1:500; Invitrogen). Images were acquired on a Zeiss Axioplan2 microscope with an oil immersion objective (63× 1.4 numerical aperture) and Orca-Flash4.0 CMOS camera (Hamamatsu) using MetaMorph software (Molecular Devices) and customized filter sets.
For detecting surface GABAARγ2 when overexpressing or knocking down Clptm1, cultured hippocampal neurons were live stained with anti-GABAARγ2 antibody (1:500; Synaptic Systems) in conditioned media for 60 min at 37 C. Neurons were then fixed, permeabilized, and incubated with anti-vGAT (1:3000; Synaptic Systems) and anti-Map2 (1:4000; Abcam) antibodies, followed by the appropriate Alexa conjugated secondary antibody (1:500; Invitrogen). For detecting synaptic intensity and puncta number of Gephyrin and Neuroligin2 when overexpressing Clptm1, cultured hippocampal neurons were fixed, permeabilized, and incubated with anti-vGAT (1:3000; Synaptic Systems), anti-Map2 (1:4000; Abcam), and anti-Gephyrin (1:500; Synaptic Systems) or anti-Neuroligin2 (1:1000; Synaptic Systems) antibodies, followed by the appropriate Alexa conjugated secondary antibody (1:500; Invitrogen). Images were acquired on a Zeiss Axioplan2 microscope with an oil immersion objective (63× 1.4 numerical aperture) and Orca-Flash4.0 CMOS camera (Hamamatsu) using MetaMorph software (Molecular Devices) and customized filter sets. Synapses were identified as clusters with pixel overlap between the separately thresholded vGAT and GABAARγ2, Gephyrin, or Neuroligin2 channels. The number of synapses was normalized to the area of Map2 positive dendrites that were randomly selected.
AAV Injection
The packaging of AAV was performed by the University of Pennsylvania Vector Core. On the day of birth (P0), C57BL/6 neonates were anesthetized using isoflurane and bilaterally injected with 0.5 μl of viral vector into each cerebral lateral ventricle through a finely drawn glass micropipette as described previously (Kim et al., 2014). Electrophysiological recording was performed at P14–17.
Electrophysiological Recording
For GABAAR or GlyR mediated currents in HEK293 cells, whole-cell recordings were performed at room temperature (20–22°C) 2 days after transfection. The patch pipettes were pulled from borosilicate glass capillary tubes (World Precision Instruments) using a PP-830 pipette puller (Narishige). The resistance of pipettes was 4–6 MΩ. The electrode pipette solution contained the following (mM): CsCl 140, CaCl2 0.1, MgCl2 2, HEPES 10, BAPTA 10, ATP 4, and QX314 5 (pH 7.2; osmolality 280–290 mosmol/Kg). The extracellular (perfusion or bathing) solution was of the following composition (mM): NaCl 140, KCl 5.4, CaCl2 1.3, MgCl2 2, HEPES 10, Glucose 20 (pH 7.4, osmolality 300–310 mosmol/Kg). Whole-cell currents were recorded at a holding potential of −60 mV. The series resistance in these recordings varied from 6 to 10 MΩ. Rapid application/removal of GABA (1 mM) or glycine (1 mM) was performed using a computer-controlled multi-barrel fast perfusion system (Warner Instruments).
For recoding in cultured hippocampal neurons, whole-cell recordings were made from the cultures at room temperature (20–22°C) at 13–16 DIV or 2–3 days after calcium phosphate transfection. Neurons were continuously perfused (1 ml/min) with the extracellular solution (ECS) containing (in mM): NaCl 140, CaCl2 1.3, KCl 5.4, MgCl2 1, HEPES 25, glucose 33 (pH 7.35; osmolality 300–310 mosmol/Kg). mEPSCs were recorded in the presence of 0.5 μM tetrodotoxin, 10 μM bicuculline methiodide and 100 μM DL-2-Amino-5-phosphonopentanoic acid sodium salt (APV); mIPSCs were recorded in the presence of 0.5 μM tetrodotoxin, 10 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) and 100 μM APV. To record tonic GABAergic currents, hippocampal neurons were recorded in ECS supplemented with 0.5 μM tetrodotoxin, 10 μM CNQX and 100 μM APV. The GABAA receptor competitive antagonist bicuculline (10 μM) was added in perfusion solution after obtaining a stable baseline recording. The amplitude of the tonic current was calculated as the difference between the holding current measured before and after the application of bicuculline. The holding current was calculated from segments containing no synaptic currents. The patch pipettes were pulled from borosilicate glass capillary tubes (World Precision Instruments) using a PP-830 pipette puller (Narishige). The resistance of pipettes was 4–6 MΩ. For mEPSC recording, the patch pipette solution contained (in mM): Cs gluconate 122.5, CsCl 17.5, MgCl2 2, HEPES 10, BAPTA 10, ATP 4, and QX314 5 (pH 7.2; osmolality 280–290 mosmol/Kg). For mIPSC or tonic GABAergic currents recording, the patch pipette solution contained (in mM): CsCl 140, CaCl2 0.1, MgCl2 2, HEPES 10, BAPTA 10, ATP 4, and QX314 5 (pH 7.2; osmolality 280–290 mosmol/Kg). The series resistance in these recordings varied from 6 to 10 MΩ, and recordings where series resistance varied by more than 10% were rejected. No electronic compensation for series resistance was employed. Cells that demonstrated a change in “leak” current of more than 10% were rejected from the analysis.
For recording in hippocampal slices, hippocampal slices were prepared from C57BL/6 mice as previously described (Liu et al., 2004). Briefly, the mouse was decapitated and the brain was rapidly removed and immersed in ice cold artificial cerebrospinal fluid (ACSF) containing (mM): 125 NaCl, 2.5 KCl, 2 CaCl2, 2 MgCl2, 1.25 NaH2PO4, 26 NaCO3, 25 Glucose (osmolality 310–320 mosmol/Kg) which was continuously bubbled with carbogen (95% O2/5% CO2) to adjust the pH to 7.35. The brain was then cut into 400 μm thick coronal slices containing hippocampus with a Leica vibratome (VT1000s, Leica). Freshly cut slices were placed in a recovery chamber with carbogenated ACSF at 31 °C for 60 min. Slices were then maintained at room temperature prior to recording. For whole cell recording, slices were transferred to a recording chamber continuously perfused with carbogenated ACSF (2 ml/min). AAV infected neurons were visualized using infrared differential interference contrast and fluorescence microscopy (Olympus) and only GFP, YFP and Tdtomato positive neurons were chosen for recording. The patch pipettes were pulled from borosilicate glass capillary tubes (World Precision Instruments) using a PP-830 pipette puller (Narishige). The resistance of pipettes was 4–6 MΩ. mEPSC and mIPSC recordings were performed on hippocampal CA1 pyramidal neurons, the same blockers were supplemented in the perfusion ACSF and the same patch pipette solutions were used as in cultured hippocampal neurons. For GABA/AMPA ratio recording, 100 μM APV was added in the perfusion solution to block NMDA receptor mediated currents. The patch pipette solution contained (in mM): Cs gluconate 122.5, CsCl 8, MgCl2 2, HEPES 10, BAPTA 10, ATP 4, and QX314 5 (pH 7.2; osmolality 280–290 mosmol/Kg). Evoked GABAAR and AMPAR mediated currents were recorded on hippocampal CA1 pyramidal neurons by stimulating the Schaffer collateral-commissural fibers and holding the cells at 0 mV and −60 mV, respectively.
All recordings were performed using a MultiClamp 700A amplifier under voltage clamp mode (Molecular Devices), filtered at 2 kHz, and acquired with pCLAMP10 software (Molecular Devices). Peak amplitudes and tonic GABAergic currents were analyzed using Clampfit10 software (Molecular Devices). mIPSCs or mEPSC recorded in a 2 min period of each condition were analyzed using Mini 6.0 software (Synaptosoft). The trigger level for detection of events was set approximately three times higher than the baseline noise and inspection of the raw data was used to eliminate any false event. The cumulative probability distribution plots were generated from randomly selected 50 mIPSC or mEPSC events from each cell in a given condition using GraphPad Prism software. A scaling factor was calculated by linear regression fits of rank ordered amplitudes of treatment group against control group (Kim et al., 2012). The treatment group was then scaled by dividing each event amplitude by the scaling factor. A Kolmogorov-Smirnov (K-S) test was used to test whether there are significant differences between control and scaled treatment group.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis and plotting were performed with Prism 6 (GraphPad Software). Values are expressed as mean ± SEM, and analyzed using a student t-test for comparison between two groups and ANOVA followed by post hoc Holm-Sidak tests for comparisons among multiple groups. Statistical significance is defined as * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. No methods were used to determine whether the data met assumptions of the statistical approach. Each experiment was repeated at least in 2 independent experiments, and n represents cell number, or number of fields for Figure 1. Detailed information can be found in the figure legends.
DATA AND SOFTWARE AVAILABILITY
Electrophysiological recording data is summarized in Table S2.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-HA (3F10) | Roche | Cat#11867431001; RRID: AB_390919 |
| Rabbit monoclonal anti-Clptm1 (EPR8801) | Abcam | Cat#ab174839 |
| Mouse monoclonal anti-Myc (9E10) | Santa Cruz | Cat#sc-40; RRID: AB_627268 |
| Rabbit polyclonal anti-Myc | Sigma | Cat#C3956; RRID: AB_439680 |
| Rabbit polyclonal anti-GFP | Invitrogen | Cat#11122; RRID: AB_221569 |
| Mouse monoclonal anti-α-Tubulin (DM1A) | Millipore | Cat#05-829; RRID: AB_310035 |
| Mouse monoclonal anti-GM130 | BD Transduction Laboratories | Cat#610823; RRID: AB_398142 |
| Rabbit polyclonal anti-Calnexin | Enzo Life Sciences | Cat# ADI-SPA-860; RRID: AB_10616095 |
| Mouse monoclonal anti-gephyrin (mAb7a) | Synaptic Systems | Cat#147011; RRID: AB_887717 |
| Rabbit polyclonal anti-GABAAR γ2 subunit | (Fritschy et al., 1992) | N/A |
| Rabbit polyclonal anti-GABAAR α1 subunit | Millipore | Cat#06-868; RRID: AB_310272 |
| Rabbit polyclonal anti-GABAAR γ2 subunit | Synaptic Systems | Cat#224 003; RRID: AB_2263066 |
| Mouse monoclonal anti-GAD65 | Developmental Studies Hybridoma Bank | Cat#GAD-6; RRID: AB_528264 |
| Guinea pig polyclonal anti-vGAT | Synaptic Systems | Cat#131 004; RRID: AB_887873 |
| Chicken polyclonal anti-Map2 | Abcam | Cat#ab5392; RRID: AB_2138153 |
| Rabbit polyclonal anti-Neuroligin2 | Synaptic Systems | Cat#129 203; RRID: AB_993014 |
| Goat polyclonal anti-GFP biotin conjugated | Rockland | Cat#600-106-215; RRID: AB_218204 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| n-dodecyl-β-D-maltoside (DDM) | Sigma | Cat#D4641 |
| CNQX | Abcam | Cat#ab120044 |
| DL-APV | Abcam | Cat#ab120271 |
| Bicuculline methiodide | Abcam | Cat#ab120109 |
| Tetrodotoxin (TTX) | Abcam | Cat#ab120054 |
| Protein G Sepharose 4 Fast Flow | GE Healthcare | Cat#17061801 |
| Protease Inhibitor Cocktail | Roche | Cat#05892988001 |
| Ni-NTA Agarose | Qiagen | Cat#30230 |
| Streptavidin (Sepharose 4B) | US Biological | Cat#S7973-89E |
| Aqua C-18 beads | Phenomenex | Cat#00G-4299-N0 |
| Reprosil-Pur C-18-AQ beads | Dr. Maisch | Cat#r13.aq |
| C-18 STAGE tips | (Ishihama et al., 2002) | N/A |
| TransIT-LT1 Transfection Reagent | Mirus | Cat#MIR2300 |
| Critical Commercial Assays | ||
| ProFection® Mammalian Transfection System-Calcium Phosphate | Promega | Cat#E1200 |
| Immobilon™ Western chemiluminescent HRP Substrate | MIllipore | Cat#WBKLS0500 |
| DC™ Protein Assay Kit | Bio-Rad | Cat#5000111 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Human: HEK293 cells | ATCC | Cat#CRL-1573 |
| Human: GABAA receptor α1/β2/γ2 stable cell line HEK293 cells | ATCC | Cat#CRL-2029 |
| Monkey: COS7 cells | ATCC | Cat#CRL-1651 |
| Rat: embryonic day 18 cortical primary neuron culture | This paper | N/A |
| Rat: embryonic day 18 hippocampal primary neuron Culture | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Thy1-His6-Flag-YFP-GABAARγ2 (HFY-GABAARγ2) also called Thy1-YFPγ2 Transgenic Mice | (Bleckert et al., 2013) | N/A |
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| Recombinant DNA | ||
| Plasmid: Clptm1 | Open Biosystems | Cat#MMM1013-649600; GenBank: BC022172.1 |
| Plasmid: Itm2C | Open Biosystems | Cat#MMM1013-63801; GenBank: BC012952.1 |
| Plasmid: Glg1 | Open Biosystems | Cat#MMM4769-99609980; GenBank: BC021306.1 |
| Plasmid: Tfg | Open Biosystems | Cat#MMM1013-202764274; GeneBank: BC024638.1 |
| Plasmid: HA-CD4 | (Connor et al., 2016) | N/A |
| Plasmid: HA-CD8 | This paper | N/A |
| Plasmid: Itm2C-Myc | This paper | N/A |
| Plasmid: Myc-Glg1 | This paper | N/A |
| Plasmid: YFP-Clptm1 | This paper | N/A |
| Plasmid: Itm2C-YFP | This paper | N/A |
| Plasmid: YFP-Glg1 | This paper | N/A |
| Plasmid: YFP-Tfg | This paper | N/A |
| Plasmid: Myc-Clstn3-CFP | (Pettem et al., 2013) | N/A |
| Plasmid: YFP-Tmub1 | This paper | N/A |
| Plasmid: GABAARα1 | (Pritchett et al., 1989) | N/A |
| Plasmid: GABAARα2 | (Pritchett et al., 1989) | N/A |
| Plasmid: GABAARβ2 | (Ymer et al., 1989) | N/A |
| Plasmid: GABAARβ3 | (Ymer et al., 1989) | N/A |
| Plasmid: GABAARγ2 | (Pritchett et al., 1989) | N/A |
| Plasmid: GlycineRα1 | (Liu et al., 2010) | N/A |
| Plasmid: GABAARα5 | Dharmacon | Cat#MMM1013-202798742; GenBank: BC062112.1 |
| Plasmid: HFY-GABAARγ2 | (Dobie and Craig, 2011) | N/A |
| Plasmid: HA-GABAARα1 | This paper | N/A |
| Plasmid: HA-GABAARα2 | This paper | N/A |
| Plasmid: HA-GABAARβ2 | This paper | N/A |
| Plasmid: HA-GABAARβ3 | This paper | N/A |
| Plasmid: HA-GABAARγ2 | This paper | N/A |
| Plasmid: HA-GlycineRα1 | This paper | N/A |
| Plasmid: HA- GABAARα5 | This paper | N/A |
| Plasmid: HA- GABAARα4 | Sino Biological | Cat#HG14855-NY |
| Plasmid: HA- GABAARδ | Sino Biological | Cat# HG14909-NY |
| Plasmid: Myc-NT-Clptm1 | This paper | N/A |
| Plasmid: Myc-30-Clptm1 | This paper | N/A |
| Plasmid: Myc-150-Clptm1 | This paper | N/A |
| Plasmid: Myc-382-Clptm1 | This paper | N/A |
| Plasmid: Myc-452-Clptm1 | This paper | N/A |
| Plasmid: Myc-535-Clptm1 | This paper | N/A |
| Plasmid: Myc-CT-Clptm1 | This paper | N/A |
| Plasmid: U6-shScramble-hSyn-CFP | This paper | N/A |
| Plasmid: U6-shClptm1-hSyn-CFP | This paper | N/A |
| Plasmid: Clptm1* | This paper | N/A |
| Plasmid: hSyn-YFP-p2a-Clptm1* | This paper | N/A |
| Plasmid: pAAV-hSyn-YFP-p2a-Clptm1* | This paper | N/A |
| Plasmid: pAAV-U6-shScramble-hSyn-Tdtamato | This paper | N/A |
| Plasmid: pAAV-U6-shClptm1-hSyn-Tdtamato | This paper | N/A |
| Sequence-Based Reagents | ||
| shRNA targeting sequence: Clptm1 Sh2: GGCCCTCAACACTTTCATT | This paper | N/A |
| shRNA scramble sequence: Sh2 scramble: GTACACTCACTGTCACTCT | This paper | N/A |
| Software and Algorithms | ||
| Pclamp 10.5 | Molecular Devices | https://www.moleculardevices.com/systems/conventional-patch-clamp/pclamp-10-software |
| Mini Analysis | Synaptosoft Inc | http://www.synaptosoft.com/MiniAnalysis/ |
| Image J | National Institute of Health | https://imagej.nih.gov/ij/index.html |
| GraphPad Prism | GraphPad Software Inc | http://www.graphpad.com/scientific-software/prism/ |
| Image Lab | Bio-Rad | http://www.bio-rad.com/en-ca/product/image-lab-software |
| Proteome Discoverer v. 1.2 | ThermoFisher Scientific | https://www.thermofisher.com/order/catalog/product/IQLAAEGABSFAKJMAUH |
| Over-representation analysis | Geneontology | http://pantherdb.org/geneListAnalysis.do |
| Uniprot | Uniprot | http://www.uniprot.org/ |
| Tmpred | Omic Tools | https://omictools.com/tmpred-tool |
| Other | ||
Supplementary Material
Table S2. Electrophysiology Data Summary, Related to Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure S4, and Figure S6 (Excel file)
HIGHLIGHTS.
Transgenic proteomic analysis reveals Clptm1 association with many GABAAR subunits
Clptm1 limits GABAAR forward trafficking by trapping receptors in the ER and Golgi
Altering the expression of Clptm1 scales phasic and tonic inhibitory transmission
Clptm1 modulates activity-dependent inhibitory synaptic homeostasis
Acknowledgments
We thank Mia Wallace of the Washington University Mouse Genetics Core and thank Xiling Zhou and Nazarine Fernandes for excellent technical assistance. This work was supported by Canadian Institutes of Health Research FDN 143206 and Canada Research Chair Awards (to A.M.C.), National Institutes of Health EY10699 (to R.O.L.W.) and a Bluma Tischler Postdoctoral Fellowship (to Y.K.). The mass spectrometry infrastructure used here was supported by the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund and the British Columbia Proteomics Network.
Footnotes
AUTHOR CONTRIBUTIONS
Y.K. and A.M.C. designed the screen and initiated the study. Y.K., R.L., R.O.L.W. and A.M.C. generated the mouse line together with the Washington University Mouse Genetics Core. R.A.C. assessed the localization of HFY-GABAARγ2, Y.K. purified the GABAAR complexes, K.M.M. and L.F. performed the proteomic analysis, and Y.G. designed, performed and analyzed all experiments beyond the proteomics screen. Y.G. and A.M.C. wrote the paper with input from all authors.
DECLARATION OF INTEREST
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonelli R, Pizzarelli R, Pedroni A, Fritschy JM, Del Sal G, Cherubini E, Zacchi P. Pin1-dependent signalling negatively affects GABAergic transmission by modulating neuroligin2/gephyrin interaction. Nat Commun. 2014;5:5066. doi: 10.1038/ncomms6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belelli D, Harrison NL, Maguire J, Macdonald RL, Walker MC, Cope DW. Extrasynaptic GABAA receptors: form, pharmacology, and function. J Neurosci. 2009;29:12757–12763. doi: 10.1523/JNEUROSCI.3340-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckert A, Parker ED, Kang Y, Pancaroglu R, Soto F, Lewis R, Craig AM, Wong RO. Spatial relationships between GABAergic and glutamatergic synapses on the dendrites of distinct types of mouse retinal ganglion cells across development. PLoS One. 2013;8:e69612. doi: 10.1371/journal.pone.0069612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan QW, Howes CG, Foster LJ. Quantitative comparison of caste differences in honeybee hemolymph. Mol Cell Proteomics. 2006;5:2252–2262. doi: 10.1074/mcp.M600197-MCP200. [DOI] [PubMed] [Google Scholar]
- Connor SA, Ammendrup-Johnsen I, Chan AW, Kishimoto Y, Murayama C, Kurihara N, Tada A, Ge Y, Lu H, Yan R, et al. Altered Cortical Dynamics and Cognitive Function upon Haploinsufficiency of the Autism-Linked Excitatory Synaptic Suppressor MDGA2. Neuron. 2016;91:1052–1068. doi: 10.1016/j.neuron.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Culiat CT, Stubbs LJ, Woychik RP, Russell LB, Johnson DK, Rinchik EM. Deficiency of the beta 3 subunit of the type A gamma-aminobutyric acid receptor causes cleft palate in mice. Nat Genet. 1995;11:344–346. doi: 10.1038/ng1195-344. [DOI] [PubMed] [Google Scholar]
- Ding R, Tsunekawa N, Obata K. Cleft palate by picrotoxin or 3-MP and palatal shelf elevation in GABA-deficient mice. Neurotoxicol Teratol. 2004;26:587–592. doi: 10.1016/j.ntt.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Dobie FA, Craig AM. Inhibitory synapse dynamics: coordinated presynaptic and postsynaptic mobility and the major contribution of recycled vesicles to new synapse formation. J Neurosci. 2011;31:10481–10493. doi: 10.1523/JNEUROSCI.6023-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essrich C, Lorez M, Benson JA, Fritschy JM, Luscher B. Postsynaptic clustering of major GABAA receptor subtypes requires the [gamma]2 subunit and gephyrin. Nat Neurosci. 1998;1:563–571. doi: 10.1038/2798. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fernandes D, Carvalho AL. Mechanisms of homeostatic plasticity in the excitatory synapse. J Neurochem. 2016;139:973–996. doi: 10.1111/jnc.13687. [DOI] [PubMed] [Google Scholar]
- Fritschy JM. Epilepsy, E/I Balance and GABA(A) Receptor Plasticity. Front Mol Neurosci. 2008;1:5. doi: 10.3389/neuro.02.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy JM, Benke D, Mertens S, Oertel WH, Bachi T, Mohler H. Five subtypes of type A gamma-aminobutyric acid receptors identified in neurons by double and triple immunofluorescence staining with subunit-specific antibodies. PNAS. 1992;89:6726–6730. doi: 10.1073/pnas.89.15.6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glykys J, Mann EO, Mody I. Which GABA(A) receptor subunits are necessary for tonic inhibition in the hippocampus? J Neurosci. 2008;28:1421–1426. doi: 10.1523/JNEUROSCI.4751-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh G, Gundelfinger E, Schmitt B, Betz H, Darlison MG, Barnard EA, Schofield PR, Seeburg PH. Glycine vs GABA receptors. Nature. 1987;330:25–26. doi: 10.1038/330025b0. [DOI] [PubMed] [Google Scholar]
- Homanics GE, DeLorey TM, Firestone LL, Quinlan JJ, Handforth A, Harrison NL, Krasowski MD, Rick CEM, Korpi ER, Makela R, et al. Mice devoid of gamma -aminobutyrate type A receptor beta 3 subunit have epilepsy, cleft palate, and hypersensitive behavior. PNAS. 1997;94:4143–4148. doi: 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Andersen JS, Mann M. Microcolumns with self-assembled particle frits for proteomics. J Chromatography A. 2002;979:233–239. doi: 10.1016/s0021-9673(02)01402-4. [DOI] [PubMed] [Google Scholar]
- Jurand A, Martin LV. Cleft palate and open eyelids inducing activity of lorazepam and the effect of flumazenil, the benzodiazepine antagonist. Pharmacol Toxicol. 1994;74:228–235. doi: 10.1111/j.1600-0773.1994.tb01103.x. [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protocols. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kanematsu T, Mizokami A, Watanabe K, Hirata M. Regulation of GABA(A)-receptor surface expression with special reference to the involvement of GABARAP (GABA(A) receptor-associated protein) and PRIP (phospholipase C-related, but catalytically inactive protein) J Pharmacol Sci. 2007;104:285–292. doi: 10.1254/jphs.cp0070063. [DOI] [PubMed] [Google Scholar]
- Kang Y, Ge Y, Cassidy RM, Lam V, Luo L, Moon KM, Lewis R, Molday RS, Wong RO, Foster LJ, et al. A combined transgenic proteomic analysis and regulated trafficking of neuroligin-2. J Biol Chem. 2014;289:29350–29364. doi: 10.1074/jbc.M114.549279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilman V, van Rossum MC, Turrigiano GG. Activity deprivation reduces miniature IPSC amplitude by decreasing the number of postsynaptic GABA(A) receptors clustered at neocortical synapses. J Neurosci. 2002;22:1328–1337. doi: 10.1523/JNEUROSCI.22-04-01328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Tsien RW, Alger BE. An improved test for detecting multiplicative homeostatic synaptic scaling. PLoS One. 2012;7:e37364. doi: 10.1371/journal.pone.0037364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Grunke SD, Levites Y, Golde TE, Jankowsky JL. Intracerebroventricular viral injection of the neonatal mouse brain for persistent and widespread neuronal transduction. JoVE. 2014:51863. doi: 10.3791/51863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leil TA, Chen ZW, Chang CS, Olsen RW. GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. J Neurosci. 2004;24:11429–11438. doi: 10.1523/JNEUROSCI.3355-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Levi S, Logan SM, Tovar KR, Craig AM. Gephyrin is critical for glycine receptor clustering but not for the formation of functional GABAergic synapses in hippocampal neurons. J Neurosci. 2004;24:207–217. doi: 10.1523/JNEUROSCI.1661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wong TP, Pozza MF, Lingenhoehl K, Wang Y, Sheng M, Auberson YP, Wang YT. Role of NMDA receptor subtypes in governing the direction of hippocampal synaptic plasticity. Science. 2004;304:1021–1024. doi: 10.1126/science.1096615. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu DC, Wang YT. Allosteric potentiation of glycine receptor chloride currents by glutamate. Nat Neurosci. 2010;13:1225–1232. doi: 10.1038/nn.2633. [DOI] [PubMed] [Google Scholar]
- Luscher B, Fuchs T, Kilpatrick CL. GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron. 2011a;70:385–409. doi: 10.1016/j.neuron.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Shen Q, Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry. 2011b;16:383–406. doi: 10.1038/mp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydiard RB. The role of GABA in anxiety disorders. J Clin Psychiatry. 2003;64(Suppl 3):21–27. [PubMed] [Google Scholar]
- Marin O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Matsuda Y, D’Adamio L. BRI3 inhibits amyloid precursor protein processing in a mechanistically distinct manner from its homologue dementia gene BRI2. J Biol Chem. 2009;284:15815–15825. doi: 10.1074/jbc.M109.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protocols. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Tanaka M, Imamura T, Takada S, Miyajima A. A novel regulatory mechanism for Fgf18 signaling involving cysteine-rich FGF receptor (Cfr) and delta-like protein (Dlk) Development. 2010;137:159–167. doi: 10.1242/dev.041574. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Morrow DH, Modgil A, Huyghe D, Deeb TZ, Lumb MJ, Davies PA, Moss SJ. Proteomic Characterization of Inhibitory Synapses Using a Novel pHluorin-tagged gamma-Aminobutyric Acid Receptor, Type A (GABAA), alpha2 Subunit Knock-in Mouse. J Biol Chem. 2016;291:12394–12407. doi: 10.1074/jbc.M116.724443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, Valakh V. Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 2015;87:684–698. doi: 10.1016/j.neuron.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymann-Andersen J, Wang H, Chen L, Kittler JT, Moss SJ, Olsen RW. Subunit specificity and interaction domain between GABA(A) receptor-associated protein (GABARAP) and GABA(A) receptors. J Neurochem. 2002;80:815–823. doi: 10.1046/j.0022-3042.2002.00762.x. [DOI] [PubMed] [Google Scholar]
- Olsen RW, Sieghart W. International Union of Pharmacology. LXX. Subtypes of gamma-aminobutyric acid(A) receptors: classification on the basis of subunit composition, pharmacology, and function. Update. Pharmacol Rev. 2008;60:243–260. doi: 10.1124/pr.108.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Zeng SY, Song HL, Li MY, Yamada MK, Yu X. Postsynaptic spiking homeostatically induces cell-autonomous regulation of inhibitory inputs via retrograde signaling. J Neurosci. 2010;30:16220–16231. doi: 10.1523/JNEUROSCI.3085-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettem KL, Yokomaku D, Luo L, Linhoff MW, Prasad T, Connor SA, Siddiqui TJ, Kawabe H, Chen F, Zhang L, et al. The specific alpha-neurexin interactor calsyntenin-3 promotes excitatory and inhibitory synapse development. Neuron. 2013;80:113–128. doi: 10.1016/j.neuron.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettem KL, Yokomaku D, Takahashi H, Ge Y, Craig AM. Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development. J Cell Biol. 2013;200:321–336. doi: 10.1083/jcb.201206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, Zhang M, Paarmann I, Fuchs C, Harvey K, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63:628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- Pribiag H, Peng H, Shah WA, Stellwagen D, Carbonetto S. Dystroglycan mediates homeostatic synaptic plasticity at GABAergic synapses. Proc Natl Acad Sci U S A. 2014;111:6810–6815. doi: 10.1073/pnas.1321774111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribiag H, Stellwagen D. TNF-alpha downregulates inhibitory neurotransmission through protein phosphatase 1-dependent trafficking of GABA(A) receptors. J Neurosci. 2013;33:15879–15893. doi: 10.1523/JNEUROSCI.0530-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchett DB, Sontheimer H, Shivers BD, Ymer S, Kettenmann H, Schofield PR, Seeburg PH. Importance of a novel GABAA receptor subunit for benzodiazepine pharmacology. Nature. 1989;338:582–585. doi: 10.1038/338582a0. [DOI] [PubMed] [Google Scholar]
- Rannals MD, Kapur J. Homeostatic strengthening of inhibitory synapses is mediated by the accumulation of GABA(A) receptors. J Neurosci. 2011;31:17701–17712. doi: 10.1523/JNEUROSCI.4476-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba RS, Michels G, Jacob TC, Pangalos MN, Moss SJ. Activity-dependent ubiquitination of GABA(A) receptors regulates their accumulation at synaptic sites. J Neurosci. 2007;27:13341–13351. doi: 10.1523/JNEUROSCI.3277-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler C, Rexhepaj E, Singan VR, Murphy RF, Pepperkok R, Uhlen M, Simpson JC, Lundberg E. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat Methods. 2013;10:315–323. doi: 10.1038/nmeth.2377. [DOI] [PubMed] [Google Scholar]
- Stellwagen D, Malenka RC. Synaptic scaling mediated by glial TNF-alpha. Nature. 2006;440:1054–1059. doi: 10.1038/nature04671. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Arstikaitis P, Prasad T, Bartlett TE, Wang YT, Murphy TH, Craig AM. Postsynaptic TrkC and presynaptic PTPsigma function as a bidirectional excitatory synaptic organizing complex. Neuron. 2011;69:287–303. doi: 10.1016/j.neuron.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terunuma M, Jang IS, Ha SH, Kittler JT, Kanematsu T, Jovanovic JN, Nakayama KI, Akaike N, Ryu SH, Moss SJ, et al. GABAA receptor phospho-dependent modulation is regulated by phospholipase C-related inactive protein type 1, a novel protein phosphatase 1 anchoring protein. J Neurosci. 2004;24:7074–7084. doi: 10.1523/JNEUROSCI.1323-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter V, Mukherjee J, Maric HM, Schindelin H, Sieghart W, Moss SJ. Gephyrin, the enigmatic organizer at GABAergic synapses. Front Cell Neurosci. 2012;6:23. doi: 10.3389/fncel.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagarajan SK, Fritschy JM. Gephyrin: a master regulator of neuronal function? Nat Rev Neurosci. 2014;15:141–156. doi: 10.1038/nrn3670. [DOI] [PubMed] [Google Scholar]
- Vidal R, Calero M, Revesz T, Plant G, Ghiso J, Frangione B. Sequence, genomic structure and tissue expression of Human BRI3, a member of the BRI gene family. Gene. 2001;266:95–102. doi: 10.1016/s0378-1119(01)00374-2. [DOI] [PubMed] [Google Scholar]
- Vithlani M, Terunuma M, Moss SJ. The dynamic modulation of GABA(A) receptor trafficking and its role in regulating the plasticity of inhibitory synapses. Physiol Rev. 2011;91:1009–1022. doi: 10.1152/physrev.00015.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick MJ, Radcliffe RA, Bowers BJ, Mascia MP, Luscher B, Harris RA, Wehner JM. Behavioural changes produced by transgenic overexpression of gamma2L and gamma2S subunits of the GABAA receptor. Eur J Neurosci. 2000;12:2634–2638. doi: 10.1046/j.1460-9568.2000.00160.x. [DOI] [PubMed] [Google Scholar]
- Yamasaki T, Hoyos-Ramirez E, Martenson JS, Morimoto-Tomita M, Tomita S. GARLH Family Proteins Stabilize GABAA Receptors at Synapses. Neuron. 2017;93:1138–1152. e1136. doi: 10.1016/j.neuron.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Mendoza-Londono R, Lu H, Tao J, Li K, Keller B, Jiang MM, Shah R, Chen Y, Bertin TK, et al. E-selectin ligand-1 regulates growth plate homeostasis in mice by inhibiting the intracellular processing and secretion of mature TGF-beta. J Clin Invest. 2010;120:2474–2485. doi: 10.1172/JCI42150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ymer S, Schofield PR, Draguhn A, Werner P, Kohler M, Seeburg PH. GABAA receptor beta subunit heterogeneity: functional expression of cloned cDNAs. EMBO J. 1989;8:1665–1670. doi: 10.1002/j.1460-2075.1989.tb03557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura K, Machida J, Daack-Hirsch S, Patil SR, Ashworth LK, Hecht JT, Murray JC. Characterization of a novel gene disrupted by a balanced chromosomal translocation t(2;19)(q11.2;q13.3) in a family with cleft lip and palate. Genomics. 1998;54:231–240. doi: 10.1006/geno.1998.5577. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang Y, Chi Z, Keuss MJ, Pai YM, Kang HC, Shin JH, Bugayenko A, Wang H, Xiong Y, et al. The AAA+ ATPase Thorase regulates AMPA receptor-dependent synaptic plasticity and behavior. Cell. 2011;145:284–299. doi: 10.1016/j.cell.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Yoshida S, Migita K, Yamada J, Mori F, Tomiyama M, Wakabayashi K, Kanematsu T, Hirata M, Kaneko S, et al. Dysfunction of extrasynaptic GABAergic transmission in phospholipase C-related, but catalytically inactive protein 1 knockout mice is associated with an epilepsy phenotype. J Pharmacol Exp Ther. 2012;340:520–528. doi: 10.1124/jpet.111.182386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Electrophysiology Data Summary, Related to Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure S4, and Figure S6 (Excel file)