Abstract
Background
Human gastric cancer (GC) is a leading primary cause of cancer-associated deaths in both males and females worldwide. However, there are few effective diagnostic and therapeutic measures for GC patients due to the complicated underlying mechanisms of GC. Recently, increasing research has indicated that lncRNAs may play a critical role in the progression of GC.
Material/Methods
AI769947, AK054978, DB077273, BG981369, AK054588, and AF131784 expressions were analyzed by qRT-PCR assay in GC tissues and corresponding normal tissues (n=44). BG981369 expression was detected by qRT-PCR assay in GC cells. BG981369 was overexpressed and silenced in AGS and SNU-5 cells. The proliferation ability was detected by MTT and colony formation assays. Cell cycle distribution and cell apoptosis rate were analyzed by flow cytometry. The migration and invasion abilities were measured by Transwell assay. In addition, SOX4 expression was analyzed by qRT-PCR in GC tissues. The correlation between SOX4 and BG981369 was analyzed by Pearson analysis.
Results
The results indicated that lncRNA BG981369 was significantly higher in GC tissues than in normal tissues. Overexpression of BG981369 inhibited the proliferation, migration, and invasion and promoted apoptosis of gastric adenocarcinoma (AGS) cells, and silencing of BG981369 promoted proliferation, migration, and invasion, and inhibited cell apoptosis of SNU-5 cells. Furthermore, we found that SOX4 may act as a downstream mediator of BG981369, suggesting that BG981369 inhibits proliferation, migration, and invasion, and promotes apoptosis by targeting SOX4 in the GC cell lines.
Conclusions
Our results suggest that BG981369 and SOX4 are potentially effective therapeutic targets for GC.
MeSH Keywords: Apoptosis; Cell Migration Assays; Cell Proliferation; RNA, Long Noncoding; SOXC Transcription Factors; Stomach Neoplasms
Background
Gastric cancer (GC), characterized by high incidence and poor prognosis, is among the most common malignant tumors in males and females worldwide [1,2]. According to the Lauren classification, GC can be divided into 2 different types: intestinal (characterized by dysplastic changes) and diffuse types [3]. It was reported that GC is the third leading cause of cancer-related deaths in the males and the fifth most frequent cause in females [4]. According to the cancer statistics of the United State in 2016, it is estimated that more than 26 000 new GC cases and more than 10 000 deaths occurred [1]. Due to enormous progress in diagnostic and therapeutic options, the global incidence of GC is now slowly decreasing [5,6]. However, the prognosis of GC is still extremely poor, especially in advanced GC patients. Due to the lack of effective biomarkers for use in diagnosis, most GC patients are diagnosed at advanced or metastatic stages [7,8]. For these advanced or metastatic GC patients, the main therapeutic measure is chemotherapy, but it has very limited effects and poor prognosis [9–11]. Therefore, it is extremely urgent to determine the exact underlying molecular mechanisms of GC, and to find effective therapeutic targets for patients with GC.
Long non-coding RNAs (lncRNAs), which are more than 200 nucleotides in length and lack protein-coding capacity, are important members of the non-coding RNAs (ncRNAs) family. Although lncRNAs do not code for proteins, they can regulate gene expression by interaction with the corresponding gene at transcriptional and post-transcriptional levels. Emerging evidence shows that lncRNAs are involved in the pathogenesis of various tumors, such as colorectal cancer [12], hepatocellular cancer [13], and lung cancer [14]. Recently, several reports have indicated that many lncRNAs are associated with the progression of GC, including lncRNA H19 [15], lncRNA HOTAIR [16], and lncRNA LINC00152 [17].
In the present study, we first measured the expression levels of 6 lncRNAs: AI769947, AK054978, DB077273, BG981369, AK054588, and AF131784. Our results showed that BG981369 expression was significantly downregulated in GC tissues compared to normal tissues. We then performed qRT-PCR, MTT, apoptosis analysis, wound-healing assay, and Transwell assay in a BG981369-overexpressed cell model and a BG981369 knockdown cell model, showing that knockdown of BG981369 promotes cell proliferation, migration, and invasion, and inhibits cell apoptosis in GC cell lines. In addition, we found the SOX4 expression has a negative correlation with BG981369 expression, and the tumor-suppression effects of BG981369 may be produced through upregulating SOX4 expression.
Material and Methods
Cell lines and tissues
The human gastric cancer cell lines SNU-5, HGC-27, BGC-823, SGC-7901, and AGS, and the non-malignant gastric epithelial cell line GES-1 were all purchased from the American Type Culture Collection (ATCC). All cells were routinely cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin (Biochrom), and 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA) in an atmosphere of 5% CO2 and 95% O2 at 37°C. Gastric cancer tissues (n=44) and corresponding normal tissues (n=44) were obtained from GC patients at the First Affiliated Hospital of Wenzhou Medical University. Informed consent was provided by every GC patient, and ethics approval was obtained from the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University. All tissues were frozen immediately in liquid nitrogen after surgical removal and maintained at −80°C until used.
lncRNA BG981369 overexpression and knockdown experiments
In the overexpression experiment, the cDNA of BG981369 was amplified by primers with 5′BamHI and 3′NotI restriction sites. PCR products were then digested with the 2-restriction endonuclease BamHI and NotI, and subcloned into the pcDNA 3.1 vector. To establish the BG981369 overexpression model, the plasmid was transfected into AGS cells using lipofectamine 2000 (Invitrogen, CA) following the protocol of the manufacturer. In the knockdown experiment, the siRNA targeting human lncRNA BG981369 (si-BG981369) and the negative control (NC) were obtained from Thermo Scientific (Ottawa, ON, Canada). SNU-5 cells were transfected with 20 nmol/L of si-BG981369 or control RNA using lipofectamine 2000 (Invitrogen).
RNA extraction, reverse transcription, and quantitative real-time PCR (qRT-PCR)
Total RNA of tissues or cell lines was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA) and reverse transcription of total RNA was performed using SuperScript III Reverse Transcriptase (Invitrogen) following the instructions of the manufacturer. Quantitative real-time PCR was achieved by using the SYBR-Green PCR Master Mix kit (Takara) in PCR reaction and 7900 HT Fast (Applied Biosystems, CA). The reaction system of qRT-PCR is shown in Table 1. Reaction steps were 95°C for 30 s as the first step in a loop, and 95°C for 5 s and 60°C for 34 s as the second step, for a total of 40 cycles. The sequence of primers used is shown in Table 2. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. The data were analyzed using 2−ΔΔCt calculation [18].
Table 1.
The reaction system of qRT-PCR.
| Reagent | Consumption |
|---|---|
| SYBR® Premix Ex TaqTM II (2×) | 5.0 μl |
| PCR forward primer (10 μM) | 0.4 μl |
| PCR reverse primer (10 μM) | 0.4 μl |
| ROX reference dye (50×) | 0.2 μl |
| DNA template | 1.0 μl |
| ddH2O | 3.0 μl |
| Total | 10.0 μl |
Table 2.
The sequence of corresponding lncRNA primers.
| LncRNAs | Sequence of primers | Product size |
|---|---|---|
| AI769947 | Forward: 5′-gagaccgttacgacacatgc-3′ Reverse: 5′-aggctctgaccagtggaatc-3′ |
208 bp |
| AK054978 | Forward: 5′-gcttcttcccacagcaaaca-3′ Reverse: 5′-gtgtcatgatgcgggtcttc-3′ |
170 bp |
| DB077273 | Forward: 5′-tcgaaccagaaagtccagct-3′ Reverse: 5′-ggtgtcttcagaaggaggct-3′ |
170 bp |
| AK054588 | Forward: 5′-attggtgagatggtggcaga-3′ Reverse: 5′-tcgattttcccctctgctgt-3′ |
163 bp |
| BG981369 | Forward: 5′-taacatggtcgtggcattgg-3′ Reverse: 5′-gtcagttggcacttgtcctg-3′ |
184 bp |
| AF131784 | Forward: 5′-gatgccaagatgctgcctac-3′ Reverse: 5′-tgagacaagcagcttcatgg-3′ |
202 bp |
| GAPDH | Forward: 5′-cacatcgctcagacaccatg-3′ Reverse: 5′-tgacggtgccatggaatttg-3′ |
198 bp |
Cell migration and invasion assay
Transwell chambers (8-μm pore size; BD Biosciences, San Jose, CA), with or without Matrigel coating, were used to detect the invasion or migration ability in treated GC cell lines, respectively. AGS cells with overexpressed BG981369or SNU-5 cells with blocked BG981369 were seeded in serum-free medium in the upper chamber at a concentration of 2×105 cells/ml (migration) or 1×105 cells/ml (invasion), and 600 μl medium containing 15% FBS was added into the lower chamber. After incubation at 37°C for 24 h (migration) or 48 h (invasion), cells remaining on the top layers of the inserts were removed, and the cells on the bottom were fixed and stained with 5% Crystal Violet (Cat # C-3886, Sigma-Aldrich, St. Louis, MO). The cells on the lower surface were counted in at least 6 random fields, and the average number was calculated.
Colony formation
Two groups of treated cells (AGS cells transfected with BG981369 or control, and SNU-5 cells transfected with si-BG981369 or si-NC) were collected and plated in 6-well plates at a concentration of 1×103 cells/well and cultured at 37°C in a 5% humidified CO2 atmosphere for 2 weeks. After washing with PBS twice, cells were fixed with 10% formaldehyde and stained with 0.5% crystal violet at room temperature for 15 min. Visible colonies were counted manually.
Cell viability assay (MTT assay)
AGS cells transfected with BG981369 or control, and SNU-5 cells transfected with si-BG981369 or si-NC, were seeded into a 96-well plate at a density of 2×103 cells/well and maintained at 37°C for 24 h. Then, cells were washed 3 times in cold PBS, and 20 μl of MTT solution was added to each well. After incubation with MTT solution (Cat No: M-2128, Sigma, USA) for 30 min at 37°C, 150 μl dimethylsulfoxide (DMSO, Wako, cat. no. 045-24511) was added, and shaken at room temperature for 10 min. The absorbance of each well was determined by a use of a microplate reader at 490 nm.
Cell cycle analysis and apoptosis analysis
The cell cycle distribution of treated AGS and SNU-5 cells was analyzed by flow cytometry. Treated AGS or SNU-5 cells (1×106 cell/well) were seeded into 6-well plates and incubated overnight at 37°C. Cells were collected and fixed in 80% ethanol and maintained overnight at −20°C. Then, cells were washed at least 3 times in cold phosphate-buffered saline (PBS), and 500 μl PI/RNase buffer (BD bioscience, USA) was added for staining at room temperature (30 min in the dark). Cell cycle distribution was determined by use of a FACS-Calibur flow cytometer (Becton Dickinson, USA). For apoptosis analysis, treated AGS and SNU-5 cells were seeded into 6-well plates at a density of 1×106 cell/well and incubated overnight. After washing 3 times with PBS, cells were re-suspended with 0.5 mL binding buffer containing 5 μl phosphatidylethanolamine (PE) and Annexin-V-FITC for 10 min in the dark. Then, flow cytometry was used to analyze the apoptosis rate.
Statistical analysis
All statistical analyses were performed using SPSS 20.0 software (SPSS, Inc., Chicago, IL), and the t test and one-way ANOVA were used, as appropriate. All data are presented as mean ± standard deviation (SD), and P<0.05 was considered to indicate a statistically significant result.
Results
lncRNA BG981369 expression was downregulated in human GC tissues
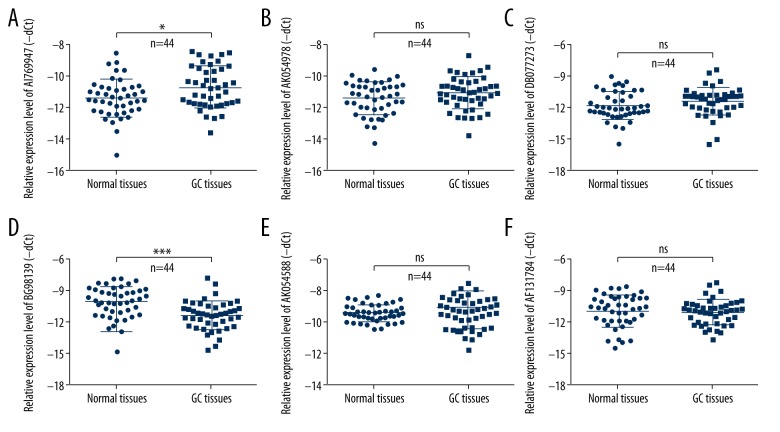
Previously research has analyzed the expression profiles of lncRNAs in human GC tissues relative to normal tissues, and found that expression of more than 100 lncRNAs was altered [19]. To investigate the exact roles of lncRNAs in human GC, we further detected the expression level of 6 different lncRNAs: AI769947, AK054978, DB077273, BG981369, AK054588, and AF131784. Results from qRT-PCR showed that AI769947 expression was higher in the GC tissues than in normal tissues (Figure 1A), and BG981369 expression was significantly lower in the GC tissues than in corresponding normal tissues (Figure 1D). There was no obvious change in the expression levels of AK054978, DB077273, AK054588, and AF131784 in the human GC tissues relative to normal tissues (Figure 1B, 1C, 1E, 1F). There results show that AI769947 and BG981369 may play a role in the progression and development of human GC.
Figure 1.
Expression of 6 different lncRNAs in GC tissues. (A–F) The relative expression level of 6 different lncRNAs – AI769947, AK054978, DB077273, BG981369, AK054588, and AF131784 – was analyzed by qRT-PCR in GC tissues and corresponding normal tissues (n=44). GC – gastric cancer.
lncRNA BG981369 inhibited cell proliferation and altered cell cycle in GC cell lines
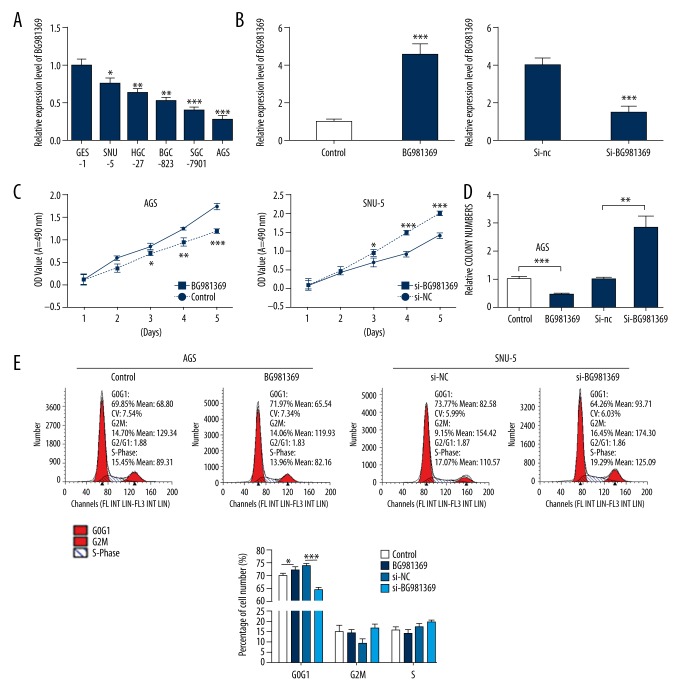
To explore the biological functions of BG981369, we first measured the BG981369 expression in human GC cell lines by qRT-PCR. The results indicated that BG981369 expression was significantly downregulated in all 5 GC cell lines (SNU-5, HGC-27, BGC-823, SGC-7901, and AGS) compared with GES-1 cell lines (Figure 2A). We then established a BG981369-overexpressed AGS cell model and a BG981369 knockdown SNU-5 cell model by transfecting them with BG981369 or si-BG981369, respectively. The efficiency of overexpression and knockdown was verified by qRT-PCR. BG981369 expression was significantly higher in the AGS cells transfected with BG981369 than in cells transfected with control RNA, and the BG981369 expression was obviously downregulated in the SNU-5 cells treated with si-BG981369 compared to si-NC (Figure 2B). MTT assay was performed to examine the roles of BG981369 in cell proliferation, and results showed that overexpression of BG981369 in AGS cells inhibited cell proliferation, and blocking BG981369 in SNU-5 cells promoted cell proliferation (Figure 2C). Results from colony formation assay showed that overexpression of BG981369 in AGS cells significantly decreased the numbers of colonies, and knockdown of BG981369 in SNU-5 cells significantly increased the numbers of colony (Figure 2D). Flow cytometry analysis was used to detect the effects of BG981369 on cell cycle distribution of GC cell lines. Results indicated that overexpression of BG981369 significantly increased the percentage of AGS cells in G0/G1 phase, and knockdown of BG981369 significantly decreased the percentage of cells in G0/G1 phase (Figure 2E). These results suggested that BG981369 acts as a cell proliferation inhibitor in GC cell lines.
Figure 2.
Effects of BG981369 overexpression or knockdown on cell proliferation in GC cell lines. (A) The relative expression of BG981369 in a non-malignant gastric epithelial cell line GES-1, and 5 gastric cancer cell lines – SNU-5, HGC-27, BGC-823, SGC-7901, and AGS – was measured by qRT-PCR. (B) The efficiency of overexpression in AGS cells or knockdown in SNU-5 cells was validated by qRT-PCR. (C, D) MTT assay and colony formation assay were carried out to measure the cell proliferation in AGS cells transfected with control or BG981369, or in SNU-5 cells treated with si-NC or si-BG981369. (E) Flow cytometry analysis was used to determine the cell cycle distribution of BG981369-overexpressed AGS cells and BG981369-blocked SNU-5 cells. GC – gastric cancer.
lncRNA BG981369 promoted cell apoptosis and inhibited cell migration and invasion in GC cell lines
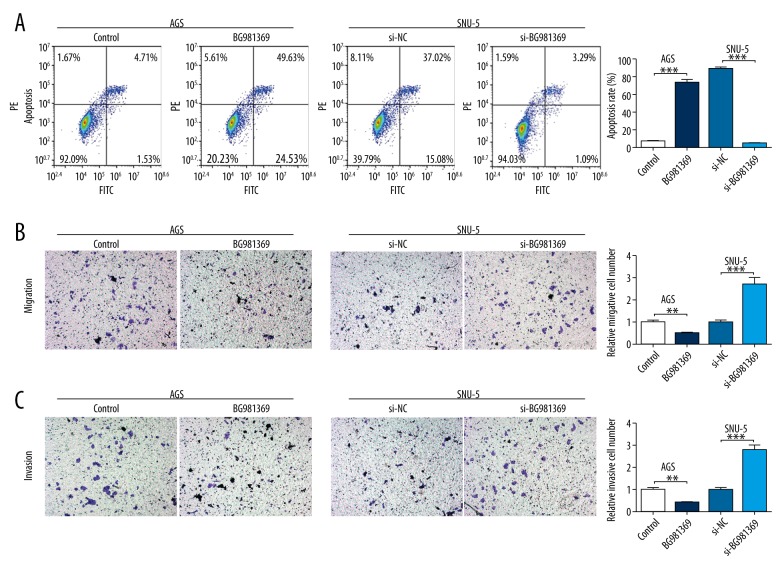
We used Annexin V-FITC/PE double-staining and flow cytometry to measure the effects of BG981369 on cell apoptosis in GC cell lines. Results suggest that cell apoptosis was significantly higher in AGS cells transfected with BG981369 than in those transfected with control RNA, and the cell apoptosis rate was obviously lower in SNU-5 cells treated with si-BG981369 than in cells treated with si-NC (Figure 3A). Transwell assay was used to determine the effects of BG981369 on cell migration and invasion in GC cell lines. Results showed that the migration and invasion abilities were significantly decreased in the BG981369-overexpressed AGS cells, and were significantly increased in the BG981369-blocked SNU-5 cells (Figure 3B, 3C). These results indicate that BG981369 induces cell apoptosis and inhibits cell migration and invasion in GC cell lines.
Figure 3.
Effects of BG981369 overexpression or knockdown on cell migration and invasion in GC cell lines. (A) The cell apoptosis rate of BG981369-overexpressed AGS cells and BG981369-blocked SNU-5 cells was analyzed by flow cytometry. (B) The effects of BG981369 on migration was detected by Transwell assay in BG981369 overexpression or knockdown GC cell lines. (C) The effects of BG981369 on invasion was analyzed by Transwell assay in BG981369 overexpression or knockdown GC cell lines. GC – gastric cancer.
lncRNA BG981369 inhibited cell proliferation, migration, and invasion in a SOX4-dependent manner
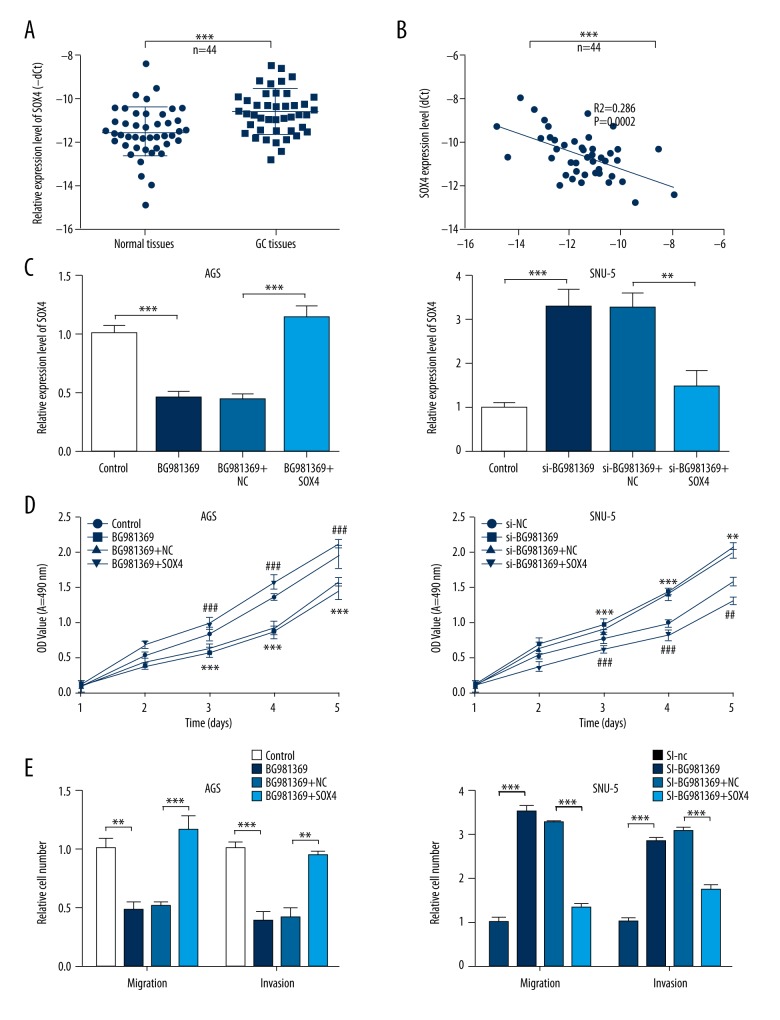
Numerous of studies have demonstrated that the aberrant expression of SOX4 is involved in progression of various cancers [20–23]. To investigate whether SOX4 plays a role in the effects of BG981369 on cell proliferation, migration, and invasion of GC cells, we further analyzed the SOX4 expression in GC tissues. Results from qRT-PCR showed that the relative SOX4 expression was significantly upregulated in GC tissues compared to normal tissues, and SOX4 expression was negatively correlated with BG981369 expression (Figure 4A, 4B). Furthermore, the expression level of SOX4 was significantly decreased in the BG981369-overexpressed AGS cells, and was obviously increased in the BG981369 knockdown SNU-5 cells (Figure 4C). MTT assay results indicated that BG981369 overexpression inhibited the proliferation of AGS cells, BG981369 knockdown promoted proliferation of SNU-5 cells, and SOX4 blocked these effects of BG981369 on GC cell lines (Figure 4D). In addition, the inhibiting effects of BG981369 overexpression in AGS cells, and the promoting effects of BG981369 in SNU-5 cells were also blocked by the application of SOX4 (Figure 4E), suggesting that BG981369 is an inhibitor of cell proliferation, migration, and invasion via the SOX4 signaling pathway.
Figure 4.
The roles of SOX4 in the BG981369 effects on cell proliferation, migration, and invasion in GC cell lines. (A) The relative SOX4 expression was analyzed by qRT-PCR in GC tissues and normal tissues. (B) The expression correlation between SOX4 and BG981369. (C) The expression level of SOX4 was measured by qRT-PCR in AGS cells treated with control, BG981369, BG981369+NC, or BG981369+SOX4, and in SNU-5 cells treated with si-NC, si-BG981369, si-BG981369+NC, or si-BG981369+SOX4. (D) MTT assay was used to detect the cell proliferation in AGS cells treated with control, BG981369, BG981369+NC, or BG981369+SOX4, and in SNU-5 cells treated with si-NC, si-BG981369, si-BG981369+NC, or si-BG981369+SOX4. (E) Transwell assay was performed to determine the cell migration and invasion in AGS cells treated with control, BG981369, BG981369+NC, or BG981369+SOX4, and in SNU-5 cells treated with si-NC, si-BG981369, si-BG981369+NC, or si-BG981369+si-SOX4. GC – gastric cancer, SOX4 – SRY-related high-mobility group box 4.
Discussion
Although the incidence of human gastric cancer has clearly dropped in recent decades, it remains the fourth most common type of cancer, and the 5-year survival rate of GC patients remains only around 30% [24,25]. Since many factors are associated with the progression of GC, clinical treatment of this disease is extremely difficult and the mortality rate is relatively higher [26,27]. Currently, the treatments of GC are mainly surgical removal combined with radiotherapy and chemotherapy; however, the effects of these treatments on GC are disappointing, especially in advanced or metastatic GC [28]. Defining the underlying mechanisms of GC will contribute to discovery of effective new therapeutic targets.
Emerging studies have identified numerous genes involved in the pathogenesis of human GC, and among these, lncRNAs are important [29,30]. Emerging evidence demonstrated that lncRNAs participate in the biological processes of various cancer cells, including cell proliferation, development, apoptosis, and metastases [31,32].
A previous study revealed the expression profiles of lncRNAs in human GC tissues, and expression of a total of 135 lncRNAs was found to be altered more than 2-fold in GC tissues compared to normal tissues [19]. Among them, the expression levels of FER1L4, uc001lsz, BG491697, AF131784, AF131784, AK054588, AF147447, HMlincRNA1600, and BG981369 were the most reduced, and the expression levels of H19, HMlincRNA717, AI769947, BQ213083, AK054978, and DB077273 were the most increased [19]. In our study, we first measured the expression of 6 lncRNAs – AI769947, AK054978, DB077273, BG981369, AK054588, and AF131784 – by qRT-PCR, and results indicated that AI769947 expression was significantly higher, whereas BG981369 expression was significantly lower in GC tissues than in corresponding normal tissues. The 4 other lncRNAs showed no significant variation between GC tissues and normal tissues. To investigate the biological roles of BG981369 in human GC, we established a BG981369-overexpressed cell model by transfecting AGS cells with reconstructed plasmid containing BG981369, and we also established a BG981369-silenced cell model by treating SNU-5 cells with si-BG981369. After verifying the efficiency with qRT-PCR, these 2 cell models were used to explore the effects of BG981369 on cell proliferation, cell cycle distribution, apoptosis, migration, and invasion. Results from MTT analysis and colony formation assay showed that BG981369 overexpression inhibited cell proliferation, migration, and invasion, and promoted cell apoptosis, whereas BG981369 knockdown promoted cell proliferation, migration, and invasion, and inhibited cell apoptosis. These results suggest that BG981369 is a tumor suppresser in the progression of GC.
SRY-related high-mobility group box 4 (SOX4), characterized by a highly conserved high-mobility group box domain, is an important transcription factor [33,34]. Studies have demonstrated that SOX4 is involved in numerous biological processes, including central nervous system development and differentiation [35–37]. In addition, increasing evidence shows that SOX4 is involved in the progression of various cancers, such as lung cancer, endometrial cancer, and breast cancer [38–40]. However, whether SOX4 participates in the progression of GC remains unclear.
To explore whether SOX4 participates in the pathogenesis of human GC, we first measured the SOX4 expression by qRT-PCR, and found that SOX4 expression was significantly increased in the GC tissues compared to normal tissues. Furthermore, the expression level of SOX4 showed a negative correlation with BG981369 expression in GC cell lines. In addition, MTT and Transwell assays results indicated that SOX4 blocked the effects of tumor inhibition of BG981369 overexpression, and silenced SOX4 reversed the effects of tumor promotion of BG981369 knockdown in GC cell lines. These results suggest that SOX4 acts as a downstream mediator of BG981369 in GC.
Conclusions
The present research demonstrates that lncRNA BG981369 could be an important tumor growth inhibitor by targeting SOX4 in GC cell lines, suggesting that BG981369 and SOX4 may be potential therapeutic targets for GC.
Footnotes
Conflict of interests
None.
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Polkowski W, van Sandick JW, Offerhaus GJ, et al. Prognostic value of Lauren classification and c-erbB-2 oncogene overexpression in adenocarcinoma of the esophagus and gastroesophageal junction. Ann Surg Oncol. 1999;6:290–97. doi: 10.1007/s10434-999-0290-2. [DOI] [PubMed] [Google Scholar]
- 4.Ohtsu A, Shah MA, Van Cutsem E, et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: A randomized, double-blind, placebo-controlled phase III study. J Clin Oncol. 2011;29:3968–76. doi: 10.1200/JCO.2011.36.2236. [DOI] [PubMed] [Google Scholar]
- 5.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niccolai E, Taddei A, Prisco D, Amedei A. Gastric cancer and the epoch of immunotherapy approaches. World J Gastroenterol. 2015;21:5778–93. doi: 10.3748/wjg.v21.i19.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol. 2016;22:2403–14. doi: 10.3748/wjg.v22.i8.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasechnikov V, Chukov S, Fedorov E, et al. Gastric cancer: Prevention, screening and early diagnosis. World J Gastroenterol. 2014;20:13842–62. doi: 10.3748/wjg.v20.i38.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner AD, Grothe W, Haerting J, et al. Chemotherapy in advanced gastric cancer: A systematic review and meta-analysis based on aggregate data. J Clin Oncol. 2006;24:2903–9. doi: 10.1200/JCO.2005.05.0245. [DOI] [PubMed] [Google Scholar]
- 10.Yamanaka T, Matsumoto S, Teramukai S, et al. The baseline ratio of neutrophils to lymphocytes is associated with patient prognosis in advanced gastric cancer. Oncology. 2007;73:215–20. doi: 10.1159/000127412. [DOI] [PubMed] [Google Scholar]
- 11.Miao RL, Wu AW. Towards personalized perioperative treatment for advanced gastric cancer. World J Gastroenterol. 2014;20:11586–94. doi: 10.3748/wjg.v20.i33.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385–88. [PubMed] [Google Scholar]
- 13.Li C, Chen J, Zhang K, et al. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36:423–34. doi: 10.1159/000430109. [DOI] [PubMed] [Google Scholar]
- 14.Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35:5375–80. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–29. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du M, Wang W, Jin H, et al. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 2015;6:31255–62. doi: 10.18632/oncotarget.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang Q, Ge J, Shao Y, et al. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441–47. doi: 10.1007/s13277-014-1709-3. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego, Calif) 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Song H, Sun W, Ye G, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. doi: 10.1186/1479-5876-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan H, Yang H, Wei H, et al. Overexpression of SOX4 promotes cell migration and invasion of renal cell carcinoma by inducing epithelial-mesenchymal transition. Int J Oncol. 2017;51(1):336–46. doi: 10.3892/ijo.2017.4010. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Liu X, Zhang G, Zhang L. [Role of SOX4 on DDP resistance in non-small cell lung cancer cell of A549]. Zhongguo Fei Ai Za Zhi. 2017;20:298–302. doi: 10.3779/j.issn.1009-3419.2017.05.12. [in Chinese] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao L, Arsenault M, Ng ET, et al. SOX4 regulates gonad morphogenesis and promotes male germ cell differentiation in mice. Dev Biol. 2017;423:46–56. doi: 10.1016/j.ydbio.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Cui L, Huang J, et al. SOX4 promotes progression in OLP-associated squamous cell carcinoma. J Cancer. 2016;7:1534–40. doi: 10.7150/jca.15689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Y, DePinho RA, Ernst M, Vousden K. Cancer research: Past, present and future. Nat Rev Cancer. 2011;11:749–54. doi: 10.1038/nrc3138. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Yong H, Zhu H, et al. Abnormal amphiregulin expression correlates with gastric cancer prognosis. Oncotarget. 2016;7:76684–92. doi: 10.18632/oncotarget.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khanderia E, Markar SR, Acharya A, et al. The influence of gastric cancer screening on the stage at diagnosis and survival: A meta-analysis of comparative studies in the far east. J Clin Gastroenterol. 2016;50:190–97. doi: 10.1097/MCG.0000000000000466. [DOI] [PubMed] [Google Scholar]
- 27.Iacovelli R, Pietrantonio F, Farcomeni A, et al. Chemotherapy or targeted therapy as second-line treatment of advanced gastric cancer. A systematic review and meta-analysis of published studies. PLoS One. 2014;9:e108940. doi: 10.1371/journal.pone.0108940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson M, Okines AF, Starling N. Current and future therapies for advanced gastric cancer. Clin Colorectal Cancer. 2015;14:239–50. doi: 10.1016/j.clcc.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 29.Wan X, Ding X, Chen S, et al. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumour Biol. 2015;36:521–32. doi: 10.1007/s13277-015-3136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wadhwa R, Song S, Lee JS, et al. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–55. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun M, Nie FQ, Wang ZX, De W. Involvement of lncRNA dysregulation in gastric cancer. Histol Histopathol. 2016;31:33–39. doi: 10.14670/HH-11-655. [DOI] [PubMed] [Google Scholar]
- 32.Guo X, Xia J, Deng K. Long non-coding RNAs: emerging players in gastric cancer. Tumour Biol. 2014;35:10591–600. doi: 10.1007/s13277-014-2548-y. [DOI] [PubMed] [Google Scholar]
- 33.Shen H, Blijlevens M, Yang N, et al. Sox4 expression confers bladder cancer stem cell properties and predicts for poor patient outcome. Int J Biol Sci. 2015;11:1363–75. doi: 10.7150/ijbs.13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jafarnejad SM, Ardekani GS, Ghaffari M, Li G. Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci. 2013;70:2677–96. doi: 10.1007/s00018-012-1187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: Extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–70. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 36.Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Brain Res Mol Brain Res. 2000;79:180–91. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- 37.Tiwari N, Tiwari VK, Waldmeier L, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–83. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 38.Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42:425–28. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang YW, Liu JC, Deatherage DE, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–46. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Liang Q, Lei Y, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]