ABSTRACT
Insulin-like growth factor 1 (IGF1) is a crucial growth factor, that regulates skeletal muscles development during cell growth and repair. Recently, its alternative splicing variant, named IGF1Ec, also named mechano-growth factor (MGF), has gained attentions as a new damage repair factor. However, the structure-function relationships of IGF1Ec have not been fully clarified due to contradictory reports. In this study, we systematically investigated physiologic responses of C2C12 muscle cells to IGF1Ec, IGF1 and MGF E peptide. Our data indicate that while the N-terminal sequence of IGF1Ec, which is homolog in part with IGF1, promotes proliferation; the C-terminal sequence of IGF1Ec, which is identical to MGF E, promotes differentiation and migration of C2C12 cells. Our results suggest that MGF E cannot completely replace all the functions of IGF1Ec on muscle repair and regeneration, and elucidate the relationships between structure and function of IGF1Ec.
KEYWORDS: C2C12 cells, differentiation, IGF1Ec, IGF1, IGF1–24, MGF E, migration, proliferation
Introduction
Alternative splicing is one of the major causes of protein diversity and functional complexity. In response to mechanical activity, insulin-like growth factor 1 (IGF1) undergoes alternative splicing and generates alternative splicing isoforms - 3 in human: IGF1Ea, IGF1Eb and IGF1Ec,1,2 or 2 in rodent: rIGF1Ea and rIGF1Eb. All mRNA splice variants of IGF1 contain exons 3 and 4 that contribute to common mature IGF1 peptide. The differences of these isoforms reside on alternative exons encoding C-terminal peptides called E-peptide. The 3 E-peptide of human IGF1, hEa, hEb and hEc are encoded by exons 4/6, 4/5, and 4/5/6, respectively (Fig. 1A). One distinctive feature of IGF1Ec is that an extra 49 base pair insert was added between exons 4 and 6, which leads to a reading frame shift resulting in a unique 24-amino-acid C-terminus of hEc (MGF E).2 IGF1Ec is also named as mechano-growth factor (MGF), since it was the first to be identified as a mechano sensitive factor.3 In rodent, a human IGF1Ec homolog was identified and named as rIGF1Eb.
Figure 1.
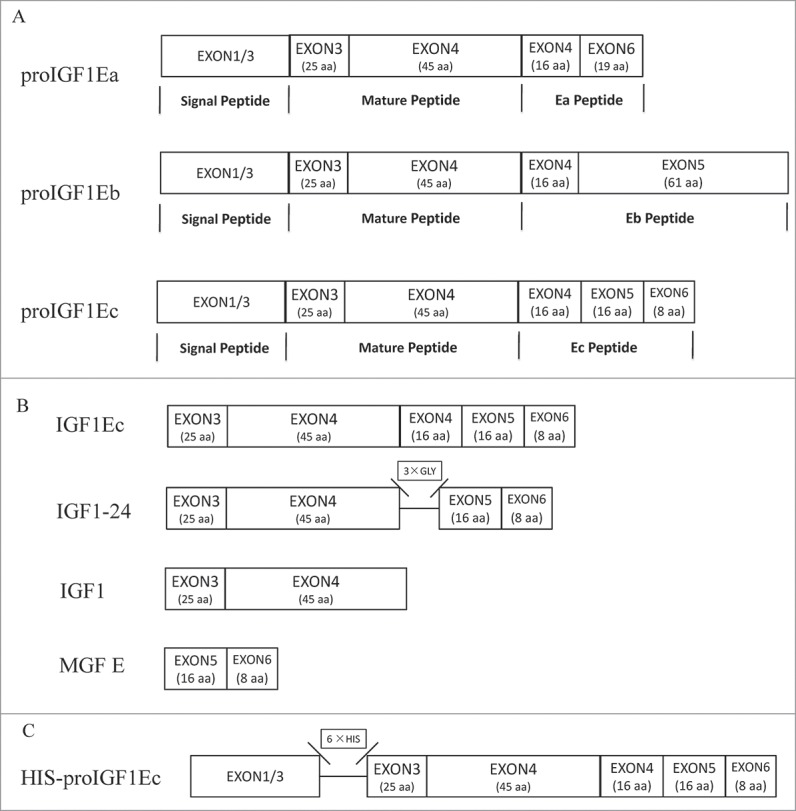
Schematic of cDNA constructs generated for this study. (A) Alternative splicing in IGF1 mRNA. (B) The peptides we used in this research. IGF1Ec contains mature IGF1 and the Ec peptide. IGF1 regards as the mature peptide. MGF E is derived C-terminal 24 amino acid of IGF1Ec. IGF1–24 contains mature IGF1 and MGF E peptide, which is linked by 3 × GLY. (C) A His-tag was inserted between the signal peptide and IGF1Ec immediately after the processing site.
IGF1 isoforms play important roles in regulation of muscle growth and regeneration. Under normal condition, the expression level of IGF1Ea is much higher than that of IGF1Ec. However, when muscle cells are subjected to mechano stimuli, injury or exercise, the expression level of IGF1Ec is rapidly upregulated,3-6 suggesting that splicing variants of IGF1 may have differential functions during cells response to stresses. However, the functions among IGF1Ec, hEc and MGF E were highly controversial. It was reported that overexpression of IGF1Ec promoted the proliferation of myoblasts but delayed the myogenic differentiation and fusion with myotubes.7 Interestingly synthetically derived MGF E peptide showed similar functions as IGF1Ec.7 Furthermore, after functional block of IGF1 receptor (IGF1R) using antibody, MGF E peptide can still induce proliferation of C2C12 mouse myoblast, suggesting MGF E peptide-mediated mitogenic activation was independent of IGF1R.7 Based on these results, it was proposed that MGF E peptide could act as an independent factor.7 However, this theory was challenged by some reports showing that rEb, the homolog of hEc in rodent, increased myoblast proliferation and migration, but did not affect myogenic differentiation. And the effects of rEb could be inhibited by pharmacologic IGF1R inhibitor.8 Furthermore, rEb was not able to restore myosin 3 (Myh3) as rodent IGF1 does in IGF1Ec-deficient cells, which suggested that rEb may not replace mature IGF1 to regulate myoblast differentiation.9 In this study, we investigated the regulatory activities and detailed structure-function relationships of IGF1Ec, IGF1 and MGF E (Fig. 1B). We determined the extent of effects of these proteins on the physiological behaviors of C2C12 cells. Our results indicated that the C-terminal and N-terminal domains of IGF1Ec have differential functions in regulation of proliferation, differentiation and migration of C2C12 cells.
Materials and methods
Cell culture
C2C12 muscle cells (Cell Bank of the Chinese Academy of Medical Sciences) were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% Penicillin-Streptomycin (P/S, Hyclone) at 37°C in 5% CO2.
Peptide synthesis
Peptide synthesis was performed as described previously.10 For preparation of GST fusion proteins, pGEX-4T-1 vectors containing coding sequences for IGF1Ec or IGF1–24 were transformed into BL21 (DE3) strain. GST or GST fusion proteins were purified using a glutathione-Sepharose 4B column following the manufacturer's instructions (GE). The native peptides IGF1 and MGF E (without GST tag) are bought from Peprotech and Phoenix Peptide respectively.
Cell proliferation assay
Proliferation was measured using Cell Counting Kit-8 assays (CCK-8, Beyotime). For this plate assay, 2 × 103 C2C12 cells were seeded in 96-well plates. Cells were treated with different concentration of IGF1Ec, IGF1, MGF E and IGF1–24. For IGF1R inhibition, cells were pretreated with PQ401 (Sigma) for 1h before treated peptides as above. After 48h, cell viability was determined and the absorbance of each well was acquired using a microplate reader set (Bio-rad) at 450 nM.
Myogenic differentiation
To induce muscle differentiation, C2C12 muscle cells were growth to near confluence and then switched to DMEM 2% horse serum (Hyclone). Fresh differentiation medium was changed once a day until day 5. IGF1Ec, IGF1, MGF E and IGF1–24 (50 ng/mL) were added to the differentiation media once a day.
Cell migration
Cell migration was tested using a 24-well Transwell (8.0 mm pore size) plate assay (Corning, Costar). When cells were 90% confluent, they were starved for 12 h. C2C12 cells (5 × 104) were seeded in the upper chambers in serum-free media, and IGF1Ec, IGF1, MGF E and IGF1–24 (50 ng/mL) were placed in the bottom chambers. After 6 h migration, non-migrated cells remaining in the upper chamber were removed with a cotton bud and migrated cells were stained with 0.1% crystal violet for 30 mins. The transwell membranes were imaged using light microscopy and cells numbers were analyzed by image J software. For studying effect of inhibitor of IGF1R, cells were pretreated with PQ401 (5 μg/mL) for 1 h before performing migration test.
Transient transfections and detection of IGF1Ec protein
IGF1Ec sequence (Fig. 1C) was inserted in pcDNA 3.1 (Invitrogen) and transfected into cells with Superfect Transfection Reagent (Qiagen). For the detection of IGF1Ec in intra-cellular, lysates from transfected cells were subjected to immunoblotting with an antibody recognizing HIS tag (CWBIO). For the detection of IGF1Ec in cell medium, IGF1Ec peptide is added into serum-free cell medium, and this medium used here was collected from cell supernatant that cultured cells for 24h. At indicated times after adding, media was subjected to immunoblotting with an antibody recognizing GST tag (ProteinTech).
Western blotting
Cells were lysed in ice-cold RIPA buffer with protein inhibitor PMSF for 30 mins and centrifuged (14,000 g) for 10 mins. Protein concentrations were measured using bicinchoninic acid assay (BCA, Beyotime) according to the instructions. 40 μg lysates were run on an 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidenedifluoride (PVDF) membranes (Millipore). Membranes were blocked in 5% skimmed milk for 1 h at room temperature, and processed for protein expression using specific primary antibodies at the indicated dilutions: 1:80 Myogenin (mouse monoclonal, #F5D, Developmental Studies Hybridoma Bank, University of Iowa). Then the membrane was incubated with appropriate secondary antibody (Beyotime) for 1 h. Immunoreactive proteins were visualized via chemiluminescent detection kit (BeyoECL Plus, Millipore).
Immunofluorescence
Differentiating C2C12 cells were fixed with 4% of paraformaldehyde for 20 mins and rinsed 3 times with phosphate-buffered saline (PBS) at room temperature. Then fixed cells were permeablized with 0.1% Triton-X 100 and stained with antibodies against the differentiation marker myosin heavy chain (MHC) (mouse monoclonal, 1:80, #MF20, Developmental Studies Hybridoma Bank, University of Iowa) according to manufacturer's instructions. Then cells were incubated with Cy3 labeled goat anti-mouse secondary antibody (1:200 dilution; Beyotime) for 1 h. The cells were then washed with PBS and the nuclei were stained with Hoechst (Beyotime) for 5 mins. Images were taken using fluorescence microscope (Olympus BX81). At least 600 nuclei from MHC-positive cells were counted from several random fields. The fusion index was calculated as follows: (MHC-stained myocytes containing > 2 nuclei/total number of nuclei) × 100. All experiments were performed in triplicate.
Statistical analysis
All data are presented as the mean values ± SEM from at least 3 independent experiments. The significance of results was evaluated using one-way ANOVA followed by Dunnett's Multiple Comparison Test. p < 0.05 was considered as significant.
Results
IGF1Ec, but not MGF E, promoted C2C12 cells proliferation
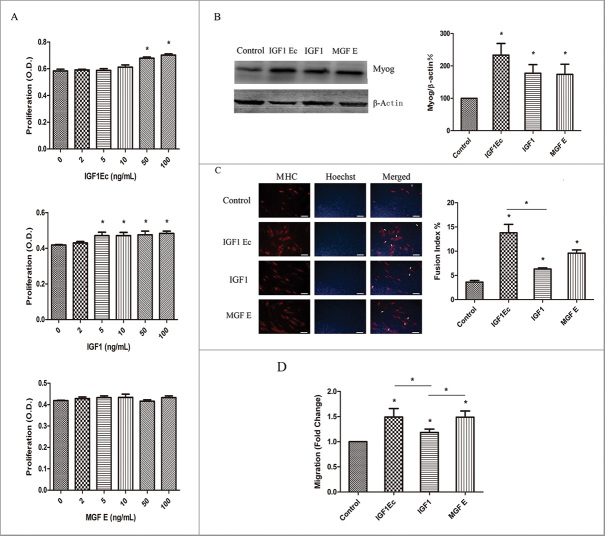
Myogenesis is a complex process, which involves proliferation and then differentiation of muscle progenitor cells into myotubes. To elucidate the structure-function of IGF1Ec in regulation of this process, we first analyzed the effect of different concentrations of IGF1Ec, IGF1 and MGF E on proliferation of C2C12 cells at 48h via CCK-8. Our data indicated that comparing to the non-treated cells, the proliferation of C2C12 cells was statistically significantly increased after being treated with IGF1Ec at 50 ng/mL or 100 ng/mL (Fig. 2A). The cell proliferation was also increased by treating with IGF1 at 5–100 ng/mL (Fig. 2A), ‘but was not affected by MGF E even at 100 ng/mL (Fig. 2A).
Figure 2.
IGF1Ec peptide promoted the proliferation, differentiation, and migration of C2C12 cells. (A) C2C12 cells were treated with different concentrations of IGF1Ec, IGF1 and MGF E. After 48 h, proliferation efficiency was measured via CCK-8. (B) C2C12 cells were treated with 50 ng/mL IGF1Ec, IGF1 and MGF E for 5 d incubation in differentiation medium (DM). The expression of Myog was detected by western blot. (C) C2C12 cells were treated with 50 ng/mL IGF1Ec, IGF1 and MGF E for 5 d incubation in differentiation medium (DM). Skeletal muscle MHC (terminally differentiated state marker) was detected via immunofluorescence (red). Nuclei were visualized using DNA Hoechst staining (blue). Fusion index was defined as the percentage of nuclei belonging to MHC positive cells with 3 or more nuclei. (D) C2C12 cells were seeded in the upper chambers in serum-free media, and IGF1Ec, IGF1 and MGF E (50 ng/mL) were placed in the bottom chambers. After 6h migration, migrated cells were stained with 0.1% crystal violet, imaged and counted. Columns, mean of at least 3 independent experiments; Error bars, SEM. *, p < 0.05. Bars, 100 μm. Arrows indicate multi-nucleated myoblast fusion.
IGF1Ec regulated the expression of myogenic differentiation protein
To determine the structure-function of IGF1Ec in regulation of myogenic differentiation, we determined the effect of IGF1Ec, IGF1 and MGF E (50 ng/mL) on the differentiation of C2C12 cells. Our data showed that all 3 proteins were able to induce the expression of Myog in C2C12 cells (Fig. 2B); and the elongation of the cell body as well as formation of multi-nucleated myoblast fusion, as indicated by myosin heavy chain (MHC) expression (Fig. 2C). And IGF1Ec appeared to be most active among them in stimulating myogenic differentiation.
IGF1Ec promoted the in vitro migration of C2C12 cells
Effective extravasation to skeletal muscle is essential for muscle repair. To examine the structure-function of IGF1Ec in regulation of muscle repair, we analyzed the effect of IGF1Ec, IGF1 and MGF E (50 ng/mL) on C2C12 cells transwell migration. Our data showed that IGF1Ec, IGF1 or MGF E treatment increased C2C12 cells migration by 49%, 18% and 48% respectively (Fig. 2D, IGF1Ec, IGF1 or MGF E vs. Control).
Roles of IGF1R in the effects of IGF1Ec on C2C12 cells proliferation and migration
IGF1 signaling pathways play critical roles in proliferation and myogenesis. To examine whether IGF1R mediates the functions of IGF1Ec, we determined the effect of IGF1Ec on the behaviors of C2C12 cells in the presence or absence of PQ401, which is an inhibitor of IGF1R auto-phosphorylation. Our data indicated that cell proliferation induced by IGF1Ec was inhibited by PQ401 in a concentration dependent manner (Fig. 3A), suggesting that IGF1R is involved in IGF1Ec-induced proliferation of C2C12 cells. Similar result was obtained with IGF1 (Fig. 3A), the homolog sequence of IGF1Ec and IGF1 might bind to IGF1R in regulation of proliferation.
Figure 3.
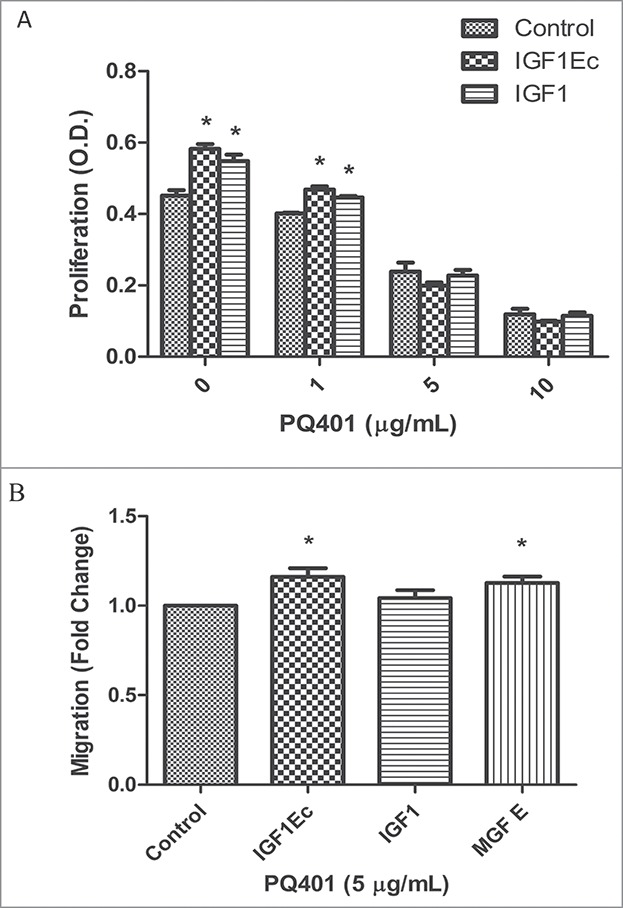
IGF1Ec-increased proliferation in C2C12 cells were IGF1R dependent, while pro-migratory activity partially involved IGF1R. (A) C2C12 cells were pretreated with different concentrations of PQ401 for 1h before treated with IGF1Ec and IGF1 (50 ng/mL). After 48h, cell viability was determined via CCK-8. (B) After pretreated with PQ401 (5 μg/mL), C2C12 cells were seeded in the upper chambers in serum-free medium, and IGF1Ec, IGF1 and MGF E (50 ng/mL) were placed in the bottom chambers. After 6 h migration, the numbers of migrated cells were analyzed as in (B). Columns, mean of at least 3 independent experiments; Error bars, SEM. *, p < 0.05.
In contract to the mediation of IGF1Ec-induced proliferation, the IGF1Ec-induced cell migration was significantly reduced (Fig. 3B vs. 2D), but not completely inhibited by PQ401 (Fig. 3B, IGF1Ec vs. Control), suggesting that other receptors coordinate with IGF1R to mediate IGF1Ec-induced cell migration. As expected, similar results were observed in regulation cell migration by MGF E.
A recombinant protein IGF1–24 has similar functions with IGF1Ec
The above data indicated that IGF1Ec gained its biological activities via its homolog sequences with IGF1 and MGF E. To confirm this observation, we constructed a fusion protein named IGF1–24, which linked the mature IGF1 and MGF E together with 3 Gly (Fig. 1B). Our data showed that IGF1–24 (50 ng/mL) had a similar effect on C2C12 cells proliferation as IGF1Ec (50 ng/mL) had (Fig. 4A); and this effect was IGF1R dependent since it could be inhibited by PQ401 (Fig. 4B). IGF1–24, similar to IGF1Ec, also induced expressions of Myog (Fig. 4C) and MHC (Fig. 4D) as well as formation of large newly fused myofibers with more myonuclei (Fig. 4D) indicating IGF1–24 stimulated myogenic differentiation. In addition, IGF1–24 induced cell migration as IGF1Ec did (Fig. 4E) in an IGF1R partially dependent manner (Fig. 4F). These results demonstrated that IGF1–24 gained activities from the fused IGF1 and MGF E in stimulation of myogenesis.
Figure 4.
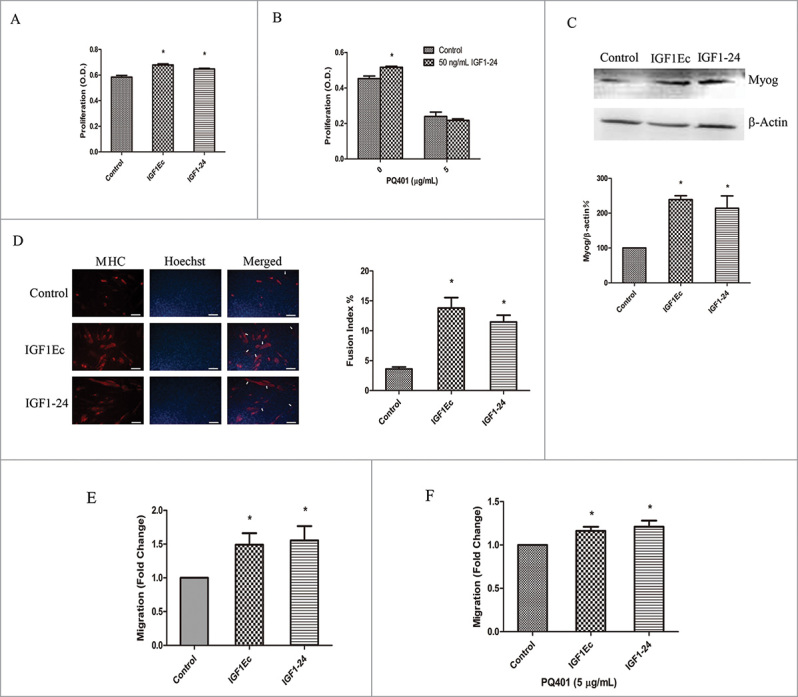
(A)recombinant protein IGF1–24 has similar function with IGF1Ec. (A) C2C12 cells were treated with IGF1Ec and IGF1–24 (50 ng/mL) for 48 h, and proliferation efficiency was measured via CCK-8. (B) After pretreated with PQ401 (5 μg/mL), C2C12 cells were treated and analyzed as in (A). (C) C2C12 cells were treated with IGF1Ec and IGF1–24 (50 ng/mL) for 5 d in DM. The expression of Myog was detected by western blot. (D) C2C12 cells were treated with IGF1Ec and IGF1–24 (50 ng/mL) for 5 d in DM. Skeletal muscle MHC (terminally differentiated state marker) was detected via immunofluorescence (red). Nuclei were visualized using DNA Hoechst staining (blue). Fusion index was defined as the percentage of nuclei belonging to MHC positive cells with 3 or more nuclei. (E) C2C12 cells were seeded into the upper chambers in serum-free media, and 50 ng/mL IGF1Ec and IGF1–24 were placed in the bottom chambers. After 6 h migration, migrated cells were stained with 0.1% crystal violet, imaged and counted. (F) After pretreated with PQ401 (5 μg/mL), C2C12 cells were treated and analyzed as in (E). Columns, mean of at least 3 independent experiments; Error bars, SEM. *, p < 0.05. Bars, 100 μm. Arrows indicate multi-nucleated myoblast fusion.
Existence forms and stability of IGF1Ec
To determine the existence forms of IGF1Ec, we analyzed the intracellular products of overexpressed HIS-proIGF1Ec in cells. Our data showed that only intact IGF1Ec was immunoprecipitated by anti-His monoclonal antibody (mAb) (Fig. 5A). To investigate whether IGF1Ec is stable, we analyzed the products of a GST-tagged IGF1Ec incubated in cell medium for 2 to 24 h. Our data showed that only GST-IGF1Ec was detected by anti-GST mAb even to 24 h, and no sign of instable was observed in the medium (Fig. 5B).
Figure 5.
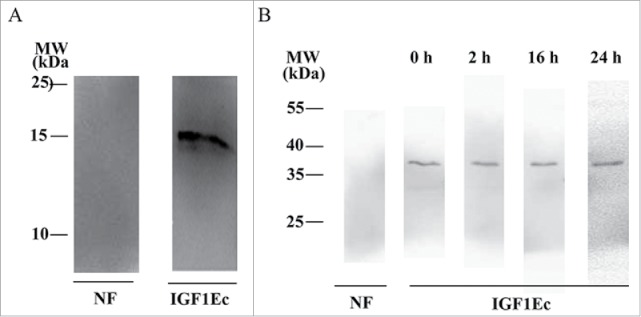
The existing form of IGF1Ec is mostly as full-length pattern. (A) After cells were transfected with HIS-proIGF1Ec construct for 48h, the form of IGF1Ec was immunoprecipitated with anti-His MAb. (B) IGF1Ec peptide was added into cell medium for up to 24 h. The form of IGF1Ec was immunoprecipitated with anti-GST mAb. NF, no factors.
Discussion
IGF1 is an important factor in promoting cell proliferation, differentiation and survival. In recent years, IGF1Ec, as one of alternative splicing variants of IGF1, has been the focus of intense study. However, there still remains ambiguity in the structure-function relationships of IGF1Ec due to contradictory reports. Therefore, we designed this research to figure out the relatedness of IGF1Ec to IGF1 and MGF E.
In our research, IGF1Ec has been shown to induce the proliferation and differentiation in myoblast cells. And this result is similar to past studies which indicated the positive activities of IGF1Ec by overexpression of IGF1Ec in myoblast cells.7,8 Except proliferation and differentiation, the mobilization of muscle progenitor cells is another important factor for tissue repair.11 As a repair growth factor, IGF1 has been shown to reinforce preferential recruitment of circulating muscle stem cells (SCs) to regenerating tissue.12 Migration is a multiple process, including extension of cellular protrusions, attachment to extracellular matrix, retraction of cell body, and detachment of tail.13 In view of this, we evaluated the effect of IGF1Ec on cell migration in C2C12 cells. Our results have shown that this process was enhanced by IGF1Ec treatment. And compared with IGF1, IGF1Ec has been shown more effective migration enhancement. This result may suggest that alternative splicing of IGF1 might be an import mechanism for cells to respond to tissue damage and participate in the regeneration process since IGF1Ec expression is induced by mechanical stress.6 Through alternative splicing, cells will generate a diverse of biologic factors to enhance cell-extracellular matrix adhesion and align cell motion, which provide an intriguing guide for adaption of mechanical environment. Therefore, considering the promotion effects on myoblast cell proliferation, differentiation and migration, IGF1Ec maybe potentially applied to muscle tissue repair engineering.
In previous researches, MGF E has not only been shown to increase the proliferation of C2C12 cells,7 but also has been shown to have zero effect on the proliferation of hMSCs, C2C12 or primary human skeletal muscle myoblasts cells.14,15 And for its function on differentiation, all of reduced, enhanced and adiaphorous characters have been detected in these cells.7,15,16 While in our observation, the proliferation of C2C12 cells was not affected by MGF E treatment and the differentiation was increased. At the moment, we are not so sure why our results are not able to repeat previous study in C2C12 cells.7 And we are also not certain why not any united and definite functions about MGF E in muscle myoblasts have been made up to now. The reasons for these conflicts always possibly lay on the following aspects. One is the source of MGF E, bought it from different companies or synthesized it from laboratories. Another is the condition of C2C12 cells line. As we all known, different culture condition and cell aging will attribute to variability of phenotype and lineage. As a result, even performing the same treatment, distinct responses will be got from cells. Last, maybe the different experimental conditions and operation by different persons are also conducive to these discrepancies.
Upon IGF1 binding to IGF1R, IGF1R is activated by auto-phosphorylation, which initiates to regulate cell proliferation and differentiation. Through pharmacologic inhibition of IGF1R activity, we showed that IGF1Ec-prompted proliferation was dependent on IGF1R and IGF1Ec-promoted migration was partial dependent on IGF1R. These results indicated that except binding to the canonical IGF1R, IGF1Ec might exert functions via novel receptors or other signal pathways. And the newly function regulation mechanisms of IGF1Ec might be largely related to the structure located in C-terminus of IGF1Ec, especially is its homolog sequence of MGF E.
By comparing the effect of IGF1Ec with IGF1 and MGF E on C2C12 proliferation, we found that both IGF1Ec and IGF1 have promoted roles, while MGF E has not. For this reason, we speculated that the homolog sequence of IGF1 has a more effective enhancement on C2C12 proliferation. And by comparing the effect of IGF1Ec with IGF1 and MGF E on C2C12 differatiation, we found that IGF1Ec was more significantly active than IGF1, and MGF E was also more active than IGF1 in stimulating myogenic differentiation. For this reason, we speculated that the homolog sequence of MGF E has a more effective enhancement on C2C12 differentiation. At last, by comparing the effect of IGF1Ec with IGF1 and MGF E on C2C12 migration, we found that both IGF1Ec and MGF E were more significantly active than IGF1 in stimulating C2C12 migration. For this reason, we speculated that the homolog sequence of MGF E also has a more effective enhancement in C2C12 migration. Therefore, we concidered that IGF1Ec gained its biological activities via its homolog sequences with IGF1 and MGF E.
To confirm this observation, we first constructed a fusion protein named IGF1–24, which linked the mature IGF1 and MGF E together with 3 Gly. We observed quite similar functions of IGF1–24 with IGF1Ec. Then, we detected existence form of IGF1Ec via analyzing the intracellular products of overexpressed HIS-proIGF1Ec in cells. Only intact IGF1Ec has been observed, which is similar to previous findings.17–20 And this result suggested that IGF1Ec was somehow unlike IGF1Ea since mature IGF1, Ea-peptides and IGF1Ea have all been detected to secrete out of the cell.21-23 Previous report implied that cells might first secrete protease-processed IGF1 and then switch to secretion of unprocessed proIGF1Ea at times of limited growth,24 which suggested that full-length peptide of IGF1Ea was more capable to protect cells from terrible environment. Therefore, we speculated that IGF1Ec secreted as unprocessed pattern might have duty to withstand atrocious survival environment and safeguard the cells since it has been reported to be rapidly upregulated after stretch, overload, and injury which will cause severe condition.3-6 Given these observations, we assert that the functions of IGF1Ec are assuredly obtained from its homolog sequence of IGF1 and MGF E, and these 2 parts are both important for the integrity of IGF1Ec functions.
Conclusion
IGF1Ec, as one of alternative splicing variants of IGF1, plays an active role in muscle cell proliferation, differentiation and migration. Its function is profited from 2 parts. One is the homolog sequence of IGF1, which mostly contributes to cell proliferation. Another is the homolog sequence of MGF E, which gives IGF1Ec extra functions in inducing cell differentiation and migration than IGF1. In actuality, the effects as cribbed to MGF E will not availably reflect actions of IGF1Ec. Different from IGF1Ea, IGF1Ec exists mostly in full-length form without being processed to form mature IGF1 and MGF E peptide in cells. And it's quite stable in cell medium. Combination of the function of IGF1 and MGF E makes IGF1Ec may be adequate to participate in muscle repair.
After damage to the myofibers, muscle tissue will undergo a cute reconstruction and regeneration process, involved coordinated activation, proliferation, migration and differentiation of muscle progenitor cells to generate new myofibers. IGF1Ec is rapidly generated in response to injury, and IGF1Ec-treated cells exhibit accelerated proliferation, migration, expressed Myog and MHC, commit to terminal differentiation, and form new myofibers. All these data point out that IGF1Ec will be a good muscle regeneration factor, mediating cell transfer to damage tissue, giving rise to myogenesis differentiation and repairing the damaged tissue.
Funding Statement
This study is supported by Natural Science Foundation of China under Grant 31670952 and 30970701; Fundamental Research Funds for the Central Universities under Grant CDJXS102300 and “111” Project under Grant B06023, China.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Okazaki R, Durham SK, Riggs BL, Conover CA. Transforming growth factor-beta and forskolin increase all classes of insulin-like growth factor-I transcripts in normal human osteoblast-like cells. Biochem Biophys Res Commun 1995; 207:963-70; PMID:7864902; https://doi.org/ 10.1006/bbrc.1995.1279 [DOI] [PubMed] [Google Scholar]
- [2].Chew SL, Lavender P, Clark AJ, Ross RJ. An alternatively spliced human insulin-like growth factor-I transcript with hepatic tissue expression that diverts away from the mitogenic IBE1 peptide. Endocrinology 1995; 136:1939-44 [DOI] [PubMed] [Google Scholar]
- [3].Goldspink G. Research on mechano growth factor: its potential for optimising physical training as well as misuse in doping. Br J Sports Med 2005; 39:787-8; PMID:16244184; https://doi.org/ 10.1136/bjsm.2004.015826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang S, Alnaqeeb M, Simpson H, Goldspink G. Cloning and characterization of an IGF1 isoform expressed in skeletal muscle subjected to stretch. J Muscle Res Cell Motil 1996; 17:487-95; PMID:8884603; https://doi.org/ 10.1007/BF00123364 [DOI] [PubMed] [Google Scholar]
- [5].Yang H, Alnaqeeb M, Simpson H, Goldspink G. Changes in muscle fibre type, muscle mass and IGF-I gene expression in rabbit skeletal muscle subjected to stretch. J Anat 1997; 190:613-22; PMID:9183683; https://doi.org/ 10.1046/j.1469-7580.1997.19040613.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McKoy G, Ashley W, Mander J, Yang SY, Williams N, Russell B, Goldspink G. Expression of insulin growth factor-1 splice variants and structural genes in rabbit skeletal muscle induced by stretch and stimulation. J Physiol 1999; 516:583-92; PMID:10087355; https://doi.org/ 10.1111/j.1469-7793.1999.0583v.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang SY, Goldspink G. Different roles of the IGF-I Ec peptide (MGF) and mature IGF-I in myoblast proliferation and differentiation. FEBS Lett 2002; 522:156-60; PMID:12095637; https://doi.org/ 10.1016/S0014-5793(02)02918-6 [DOI] [PubMed] [Google Scholar]
- [8].Brisson BK, Barton ER. Insulin-Like Growth Factor-I E-Peptide activity is dependent on the IGF-I receptor. PLoS One 2012; 7:e45588; PMID:23029120; https://doi.org/ 10.1371/journal.pone.0045588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matheny RW Jr, Nindl BC. Loss of IGF-IEa or IGF-IEb impairs myogenic differentiation. Endocrinology 2011; 152:1923-34; PMID:21406500; https://doi.org/ 10.1210/en.2010-1279 [DOI] [PubMed] [Google Scholar]
- [10].Feng J, Wan R, Yi Q, He L, Yang L, Tang L. Examination of alternate codon bias solutions for expression and purification of recombinant mechano growth factor in Escherichia coli. Biotechnol Appl Biochem 2014; 62:690-8; PMID:25345350; https://doi.org/ 10.1002/bab.1312 [DOI] [PubMed] [Google Scholar]
- [11].Galvez BG, Sampaolesi M, Brunelli S, Covarello D, Gavina M, Rossi B, Constantin G, Torrente Y, Cossu G. Complete repair of dystrophic skeletal muscle by mesoangioblasts with enhanced migration ability. J Cell Biol 2006; 174:231-43; PMID:16831885; https://doi.org/ 10.1083/jcb.200512085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Musarò A, Giacinti C, Borsellino G, Dobrowolny G, Pelosi L, Cairns L, Ottolenghi S, Cossu G, Bernardi G, Battistini L, Molinaro M, Rosenthal N. Stem cell-mediated muscle regeneration is enhanced by local isoform of insulin-like growth factor 1. Proc Natl Acad Sci U S A 2004; 101:1206-10; PMID:14745025; https://doi.org/ 10.1073/pnas.0303792101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell 1996; 84:359-69; PMID:8608589; https://doi.org/ 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- [14].Collins JM, Goldspink PH, Russell B. Migration and proliferation of human mesenchymal stem cells is stimulated by different regions of the mechano-growth factor prohormone. J Mol Cell Cardiol 2010; 49:1042-5; PMID:20875825; https://doi.org/ 10.1016/j.yjmcc.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fornaro M, Hinken AC, Needle S, Hu E, Trendelenburg AU, Mayer A, Rosenstiel A, Chang C, Meier V, Billin AN, et al.. Mechano-growth factor peptide, the COOH terminus of unprocessed insulin-like growth factor 1, has no apparent effect on myoblasts or primary muscle stem cells. Am J Physiol Endocrinol Metab 2014; 306:E150-156; PMID:24253050; https://doi.org/ 10.1152/ajpendo.00408.2013 [DOI] [PubMed] [Google Scholar]
- [16].Barton ER. Viral expression of insulin-like growth factor-I isoforms promotes different responses in skeletal muscle. J Appl Physiol 2006; 100:1778-84; PMID:16439513; https://doi.org/ 10.1152/japplphysiol.01405.2005 [DOI] [PubMed] [Google Scholar]
- [17].Dluzniewska J, Sarnowska A, Beresewicz M, Johnson I, Srai SK, Ramesh B, Goldspink G, Go´ recki DC, Zabłocka B. A strong neuroprotective effect of the autonomous C-terminal peptide of IGF1Ec (MGF) in brain ischemia. FASEB J 2005; 19:1896-8; PMID:16144956 [DOI] [PubMed] [Google Scholar]
- [18].Philippou A, Stavropoulou A, Sourla A, Pissimissis N, Halapas A, Maridaki M, Koutsilieris M. Characterization of a rabbit antihuman mechano growth factor (MGF) polyclonal antibody against the last 24 amino acids of the E domain. In Vivo 2008; 22:27-35; PMID:18396778 [PubMed] [Google Scholar]
- [19].Philippou A, Papageorgiou E, Bogdanis G, Halapas A, Sourla A, Maridaki M, Pissimissis N, Koutsilieris M. Expression of IGF1 isoforms after exercise-induced muscle damage in humans: characterization of the MGF E peptide actions in vitro. In Vivo 2009; 23:567-75; PMID:19567392 [PubMed] [Google Scholar]
- [20].Stavropoulou A, Halapas A, Sourla A, Philippou A, Papageorgiou E, Papalois A, Koutsilieris M. IGF1 expression in infarcted myocardium and MGF E peptide actions in rat cardiomyocytes in vitro. Mol Med 2009; 15:127-35; PMID:19295919; https://doi.org/ 10.2119/molmed.2009.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Conover CA, Baker BK, Bale LK, Clarkson JT, Liu F, Hintz RL. Human hepatoma cells synthesize and secrete insulin-like growth factor Ia prohormone under growth hormone control. Regul Pept 1993; 48:1-8; PMID:8265808; https://doi.org/ 10.1016/0167-0115(93)90330-B [DOI] [PubMed] [Google Scholar]
- [22].Duguay SJ. Post-translational processing of insulin-like growth factors. Horm Metab Res 1999; 31:43-49; PMID:10226780; https://doi.org/ 10.1055/s-2007-978697 [DOI] [PubMed] [Google Scholar]
- [23].Duguay SJ, Milewski WM, Young BD, Nakayama K, Steiner DF. Processing of wild-type and mutant proinsulin-like growth factor-IA by subtilisin-related proprotein convertases. J Biol Chem 1997; 272:6663-70; PMID:9045697; https://doi.org/ 10.1074/jbc.272.10.6663 [DOI] [PubMed] [Google Scholar]
- [24].Wilson HE, Westwood M, White A, Clayton PE. Monoclonal antibodies to the carboxy-terminal Ea sequence of pro-insulin-like growth factor-IA (proIGF-IA) recognize proIGF-IA secreted by IM9 B-lymphocytes. Growth Horm IGF Res 2001; 11:10-7; PMID:11437469; https://doi.org/ 10.1054/ghir.2000.0182 [DOI] [PubMed] [Google Scholar]