ABSTRACT
Kindlins are a small family of 4.1-ezrin-radixin-moesin (FERM)-containing cytoplasmic proteins. Kindlin-3 is expressed in platelets, hematopoietic cells, and endothelial cells. Kindlin-3 promotes integrin activation, clustering and outside-in signaling. Aberrant expression of kindlin-3 was reported in melanoma and breast cancer. Intriguingly, kindlin-3 has been reported to either positively or negatively regulate cancer cell metastasis. In this study, we sought to clarify the expression of kindlin-3 in melanoma cells and its role in melanoma metastasis. Two widely used metastatic mouse and human melanoma cell lines B16-F10 and M10, respectively, were examined and found to lack kindlin-3 mRNA and protein expression. When kindlin-3 was ectopically expressed in these cells, cell migration was markedly reduced. These are attributed to aberrant Rac1 and RhoA activation and overt membrane ruffling. Our data demonstrate for the first time that despite its well established role as a positive regulator of integrin-mediated cell adhesion, aberrant expression of kindlin-3 could lead to imbalanced RhoGTPases signaling that impedes rather than promotes cell migration.
KEYWORDS: cell adhesion, cell migration, cell polarization, cytoskeleton, kindlins, melanoma, metastasis
Introduction
Cell migration is a highly dynamic process that involves cell adhesion, cytoskeletal remodeling, membrane protrusion and rear retraction.1 Transformed cells progress to become life-threatening cancers when they acquire invasive and migratory properties.2 Integrins are heterodimeric cell adhesion receptors that promote cell-cell and cell-extracellular matrix (ECM) interactions.1,3 A large number of cytoplasmic proteins directly associate with the cytoplasmic tails of integrins thereby regulating integrin ligand-binding activity and outside-in signaling.4
The kindlin family of FERM proteins consists of 3 evolutionary conserved members, kindlin-1, kindlin-2 and kindlin-3.5 Kindlin-1 is mainly expressed in epithelial cells while kindlin-2 is expressed in most cell types except hematopoietic cells. The expression of kindlin-3 is restricted to cells of the hematopoietic system and endothelial cells.6,7 The importance of kindlins is underscored by a number of debilitating diseases. Mutations that abrogate kindlin-1 expression lead to skin atrophy and photosensitivity in Kindler's syndrome. A lack of expression of kindlin-3 is the molecular basis of leukocyte adhesion deficiency type (LAD) III in which the leukocytes and platelets of afflicted individuals are defective in adhesion and spreading.8-11 Kindlin-2 is involved in cancer metastasis, myogenesis, and hemostasis.12-15
Kindlin-3 binds to the C-terminal NPxY/F motif of the integrin ß cytoplasmic tail and it serves as a co-activator of talin-induced integrin activation.8,9,16 Kindlin-3 mediates integrin outside-in signaling, cytoskeletal re-modeling and integrin clustering.17-19 Kindlin-3 has also been shown to regulate ß3 and ß1 integrins in platelet adhesion, endothelial tube formation and osteoclast-mediated bone resorption.7,20,21 Recent studies suggest a role of kindlin-3 in cancer progression. A significant elevation of kindlin-3 expression in breast cancer tumors was reported and kindlin-3 overexpression in transfected breast cancer cell line MDA-MB231 promoted tumor growth and lung metastasis.22 However, in another study, kindlin-3 appears to have a tumor suppressor function because silencing of kindlin-3 expression in MDA-MB231, and human melanoma cell lines SKMEL28 and M10 enhanced their metastasic potential.23 A subsequent study by the same group reported a role of kindlin-3 in regulating ß1 integrin-mediated adhesion and migration in the human melanoma cell line M10.24 What accounts for these different observations with regard to the role of kindlin-3 in cancer cell metastasis is unclear. Hence, we sought to clarify the expression of kindlin-3 in melanoma cells and its role in melanoma metastasis in this study.
Materials and methods
General chemicals and reagents
All general chemicals and reagents were purchased from Sigma-Aldrich, St Louis, MO, unless stated otherwise.
Cells and culture conditions
Mouse melanoma cell line B16-F10 was a kind gift from Prof. Nguan-Soon Tan (School of Biological Science, Nanyang Technological University, Singapore). Cells were cultured in DMEM full-medium containing 10% (v/v) heat-inactivated (HI) fetal bovine serum (FBS) and 100 IU/ml penicillin and 100 µg/ml streptomycin. Human melanoma cell line M10 was purchased from American Type Culture Collection (Manassas, VA) and cultured in RPMI1640 full-medium containing supplements aforementioned. Bone marrow derived macrophages were obtained based on previous protocol with modifications.25 Suspension cells were harvested (Day 1) and the medium replaced with one that contained 15% (v/v) L929 supernatant. The medium was discarded and replaced with fresh medium containing L929 supernatant on Day 4. Macrophages were collected on Day 7 using a cell scraper. L929 cells were kindly provided by Prof. K.E. Karjalainen, School of Biological Sciences, Nanyang Technological University, Singapore.
Expression plasmids
The expression plasmid pcDNA3.1(-)zeo-GFP was generated by PCR sub-cloning of EGFP from pEGFP-C1 (kindly provided by Dr. H.Y. Li, Nanyang Technological University, Singapore) using relevant primers into the XbaI and XhoI restriction sites in the MCS of pcDNA3.1(-)zeo (Invitrogen, Life Technologies, Carlsbad, CA). pcDNA3.1(-)zeo-GFP-kindlin-3 was generated using the same strategy except that the EGFP cDNA was sub-cloned into the same restriction enzyme sites found at the 5′ in-frame position of full-length human kindlin-3 in pcDNA3.1(-). Cloning of full-length human kindlin-3 cDNA has been reported previously.18 Both plasmids were verified by sequencing (AITbiotech, Singapore).
Generation of stable B16-F10 cells that over-expressed GFP or GFP-kindlin3
B16-F10 cells were seeded into a 6-well plate and cultured overnight to achieve 60-70% cell confluence at the day of transfection. Expression plasmid pcDNA3.1-GFP or pcDNA3.1-GFP-kindlin-3 (2 μg each) was re-suspended in cell culture medium without HI-FBS and antibiotics in a total volume of 100 µl. Polyfect transfection reagent (Qiagen, Netherlands) (20 µl) was added to the plasmid suspension and incubated for 5-10 min at RT to allow complex formation. Complete cell culture medium (0.6 mL) was added to the plasmid suspension and transferred to cells in the culture dishes. GFP-expressing cells were observed 24 h post-transfection under an epi-fluorescence microscope. Cells stably expressing the constructs were selected in culture medium containing 200 µg/mL of Zeocin (Sigma-Aldrich). When more than ˜50% of cells were observed to be positive for GFP, cells were subjected to single-cell sorting by FACS into wells of 96-well cell culture plates. Cells were maintained in Zeocin-containing medium and surviving clones were expanded and used for subsequent analyses.
Transfection of M10 cells with either GFP or GFP-kindlin-3 expression plasmid
M10 cells (1×106) were transfected with 8 μg of plasmid (either GFP or GFP-kindlin-3 expression plasmid) using the Neon transfection kit on a Digital Bio MP-100 electroporator (Invitrogen). Electroporated cells were grown in RPMI1640 full-medium containing 20% (v/v) HI-FBS for 24 h. Cells were sorted on a FACSAria cell sorter (Becton Dickinson) and grown for 24 h before subsequent analyses.
Immunoblotting
Proteins separated by SDS-PAGE were electro-transferred onto a PVDF membrane (Millipore, Bedford, MA) according to standard protocol. The PVDF membrane was incubated in TBS-T buffer (150 mM NaCl, 0.1% (v/v) Tween-20, 10 mM Tris, pH 7.0) containing 5% (w/v) non-fat milk for 1 h at RT. The membrane was then incubated in this buffer containing relevant primary antibody: rat anti-human/mouse kindlin-3 mAb clone 918 (1:1000 dilution); rabbit anti-human kindlin-3 pAb (1:2500 dilution, Sigma-Aldrich, cat. no. SAB 4200013); mouse anti-human/mouse actin mAb clone C4 (1:5000 dilution, Becton Dickinson, Franklin Lakes, NJ, cat. no. 612657); mouse anti-human kindlin-3 mAb 3D6 (1: 2500 dilution) (Abcam, Cambridge, UK, cat. no. ab173416). The membrane was washed 3 times and incubated in TBS-T containing 1:5000 dilution of relevant HRP-conjugated secondary antibody. The following antibodies were used: HRP-conjugated goat anti-mouse IgG antibody (Advansta, Menlo Park, CA, cat. no. R-05071-500) or HRP-conjugated goat anti-rabbit IgG antibody (Advansta, cat. no. R-05072-500) or HRP-conjugated goat anti-rat IgG antibody (1:5000 dilution) (GE Healthcare, UK, cat. no. NA935V) for 1 h at RT. The membrane was washed 3 times in TBS-T followed by enhanced chemiluminescence detection using the WesternBright™ ECL kit (Advansta). Data were collected and analyzed on a ChemiDoc™ Touch imaging system (Biorad, Hercules, CA).
Quantitative reverse transcription PCR
Total RNA was extracted from cells using the TRIzol reagent (Invitrogen) as per manufacturer's protocol. cDNA was generated using the Power SYBR Green Cells-to-CT kit (Ambion, Life technologies). Primers for mouse kindlin-3 and GAPDH are as follows: Mouse kindlin-3, F 5′-TACCTGAGGTGCCAGGATGA-3′ and R 5′-GTTTGTTCCCTGAGCCTCCA-3′; Mouse GAPDH, F 5′-GGTGAAGGTCGGTGTGAACG-3′ and R 5′-CTCGCTCCTGGAAGATGGTG-3′. Primers for human GAPDH, actin, kindlin1, kindlin2 and kindlin-3 are as follows: GAPDH, F 5′-GGTGAAGGTCGGAGTCAACG-3′ and R 5′-CTCGCTCCTGGAAGATGGTG-3′; actin, F 5′-GACATGGAGAAAATCTGGCA-3′ and R 5′-TGGGGTGTTGAAGGTCTCAA-3′; kindlin-1, F 5′-AAGCACTTGCGGATATGTACC-3′ and R 5′-TCCTCTTGGATGCCTTGTTC-3′; kindlin-2, F 5′-CTCCTGAATGTTTGGTGTCTCC-3′ and R 5′-CTCATCTTGGCTTCAATTAGACTC-3′; kindlin-3, F 5′- GGCATCGCCAACAACC-3′ and R 5′- CACTGGCGCATGTTGC-3′. The primer sequences for human kindlins were based on that reported previously.23 qPCR was performed on a Bio-Rad C1000 Thermal Cycler CFX96 Real-Time System. Reactions were prepared using the PowerSYBR Green PCR Master Mix (Applied Biosystems, Life technologies) as per manufacturer's protocol. Reaction conditions were 95°C for 10 min followed by 40 cycles of 95°C for 30 sec and 60°C for 1 min. Melt-curves were assessed from 58°C to 95°C with an increment of 0.5°C per 10 sec. Expression levels of transcripts were normalized using 2-ΔΔCt, relative to GAPDH or GAPDH and actin as indicated in figure legends.
Flow cytometry analysis
Cells were washed once in PBS and incubated with 1 μg of APC-conjugated Armenian hamster anti-mouse/rat CD29 (integrin ß1) (clone HMb1-1, cat. no. . 17-0291) or APC-conjugated Armenian Hamster IgG (Isotype Control, clone eBio299Arm, 17-4888) (Affymetrix eBiosciencs, San Diego, CA) in PBS for 1 h at RT. Cells were then washed once in PBS and analyzed on a Fortessa X20 (3 laser) flow cytometer (BD Biosciences). Data were analyzed using the Flowjo software (Tree Star Inc., Ashland, OR). For detecting activated integrin ß1, B16 cells were re-suspended in RPMI 1640 medium containing 1 μg of rat anti-mouse/human CD29 (mAb 9EG7, BD Pharmingen™, cat. no. 550531) or IgG from rat serum (Sigma-Aldrich, cat. no. I8015) and incubated for 30 min at 37°C. Incubation at 37°C for integrin mAb has been reported.26,27 Thereafter, cells were washed once in PBS and labeled with APC-conjugated goat anti-rat secondary antibody (1:400 dilutions) (BD Pharmingen™, cat. no. 551019) for 30 min at 4°C. Flowjo software was used to determine % cells gated positive for mAb 9EG7 staining. The procedure for gating was as follow. Based on the non-specific IgG control for each group of cells, the gating region was first generated such that it contained approximately 5% of the cells with highest intensity. The same gating region was then used to gate cells that were stained with mAb 9EG7 (in this case APC signal).
Cell adhesion assays
Static cell adhesion assays on fibronectin were performed using the method described previously but with modifications.28 Briefly, each well of a 96-well Polysorb microtiter plate (Nunc, Thermofisher scientific, Waltham, MA) was left uncoated or coated with human plasma fibronectin (Sigma) at 2 μg/cm2 in PBS for 2 h at 37°C. Cells (5 × 104) labeled with 2′7′-bis-(2-carboxyethyl)-5-(and-6) carboxyfluorescein (Molecular Probes, Life Technologies, Carlsbad, CA) were seeded into each well and incubated for 30 min at 37°C in a 5% CO2 incubator. Total fluorescence in each well was measured on a fluorescence plate reader (FL600) (Biotek Instruments, Winooski, VT). Wells were washed twice in RPMI1640 medium containing 5% (v/v) HI-FBS and 10 mM HEPES to remove unbound cells. The fluorescence signal of adherent cells was then measured. Percent cell adhesion was calculated based on (fluorescence signal of bound cells/total fluorescence) × 100.
Wound healing and cell random-migration analyses
2D wound healing assays were performed using commercial culture inserts (Ibidi GmbH, Martinsried, Germany). Culture inserts were placed on a non-coated cell culture dish (Nunc A/S, Roskilde, Denmark) using sterile tweezers. 7×104 cells were seeded into each chamber of the culture insert. Cells were incubated for 2 h at 37°C in a 5% CO2 incubator to allow full attachment to the surface. The culture insert was removed and the cells were washed in culture medium to remove non-adherent cells. The culture dish was then filled with an appropriate volume of culture medium. Time-lapse images of the wound were recorded over 20 h at 20 min intervals using a 10X objective (UPLFLN-PH, Olympus) under phase contrast on a motorized Olympus IX83 inverted microscope system equipped with a stage temperature and CO2 incubator, a cooled monochrome digital camera and CellSens Dimension software. The wound area was measured using ImageJ software following the protocol described by Dr Kees Straatman (Advanced Imaging Facilities, University of Leicester, Leicester, UK). Wound closure for each group of cells was calculated (% wound area / total area) and plotted against time (min). The rate of wound closure was calculated based on the change in % wound area/total area between the start of the experiment and the experiment end point/total time taken.
2D cell random-migration assays were performed on fibronectin-coated (2 µg/cm2) coverslip-bottom culture dishes (MatTek Corporation, Ashland, MA). In brief, PBS (200 μl) containing 2 μg of human plasma fibronectin (Sigma-Aldrich) was layered on top of the glass-coverslip of the culture dish and incubated for 3 h at RT. 4×103 cells were seeded onto the coated dish and incubated for 2 h at 37°C in a 5% CO2 incubator to allow attachment onto the coated surface. The culture dish was then filled with appropriate volume of culture medium. Time-lapse images of cell migration were recorded over 20 h at 20 min intervals aforementioned. Cell migration plots were generated using the “Manual Tracking” and “Chemotaxis Tool” plugins for ImageJ as recommended by Ibidi (Ibidi GmbH, Martinsried, Germany).
Membrane dynamics and kymograph analyses were also performed on cells migrating on fibronectin using a 40x objective (LUCPLFLN-PH, Olympus) under phase contrast over 30 min at 6 sec intervals. Kymographs were generated using imageJ, using a modified version of the protocol described by Dr. Kornelis R Straatman (Advanced Imaging Facilities, University of Leicester, Leicester, UK). Briefly, images of single cells were cropped from the field and several areas of interest were marked on the cell image with a line of 5 pixels in width. Images of this area were then generated using the “straighten” function. The images were then lined up to produce a space-time plot using the “montage” function.
The overall brightness and contrast of some video files have been adjusted using ImageJ to improve clarity. 301 frames at 6 sec intervals per image (cell) file were converted to a single AVI video file with a frame speed of 15 frames-per-sec without compression using ImageJ. Video files can be viewed under Supporting Information (S1-S9).
Scanning electron microscopy
Cells were settled onto fibronectin-coated (2 µg/cm2) coverslip glass bottom culture dishes. Cells were washed in PBS at 37°C and fixed in 4% glutaraldehyde for 1 h at 37°C. Dehydration of cells was performed by incubating cells in increasing concentrations of ethanol (50, 60, 70, 80, 90 and 100%, 10 min for each step) before drying with hexamethyldisilazane as previously described.29 The cellular morphology was characterized by scanning electron microscopy (SEM, JEOL 6360, Japan). Cells were sputter-coated with a thin layer of gold and observed under SEM using an accelerating voltage of 5 kV.
Fluorescence microscopy
Cells were washed in PBS once, re-suspended in full DMEM medium and allowed to settle on fibronectin–coated (2 μg/cm2) coverslip-bottom culture dishes (MatTek Corporation). Cells were incubated for 2 h under culture conditions to allow cell spreading on fibronectin. Cells were fixed in 4% (w/v) paraformaldehyde in PBS for 10 min at RT. Cells were then permeabilized with 0.3% (v/v) Triton X-100 in modified cytoskeleton stabilization (CSK) buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 10 mM PIPES, pH 6.8) for 1 min at RT.30 Permeabilized cells were incubated with 1 μg of mouse anti-human paxillin (clone 5H11, Merck Millipore, Damstadt, Germany, cat. no. 05-417) overnight at 4°C. Cells were washed 3 times in PBS followed by incubation in PBS containing Alexa Fluor® 594-phalloidin (0.27 ng/mL) (Thermo Fisher scientific, Waltham, MA, cat. no. A12381) Alexa Fluor® 647-conjugated goat anti-mouse IgG (H+L) (1:400 dilution) (Thermo Fisher scientific, cat. no. A21235), and DAPI (0.1 µg/mL) for 3 h at RT. Cells were washed 3 times in PBS and examined on a LSM 710 confocal laser scanning microscope equipped with a 63×objective (Carl Zeiss, Oberkochen, Germany). Images were processed using the software ZEN2012 and the auto-min/max function was used for optimal setting for contrast and brightness.
Detecting Rac1 and RhoA activities
Rac1 activation was determined using the Rac1 Biochem Kit (Cytoskeleton, Inc., Denver, CO) as per manufacturer's instructions. Briefly, B16 cells were serum starved for 24 h. A total of 3 ×106 cells were then seeded into 3 wells of a 6-well cell culture plate that was each coated with 2 µg/cm2 fibronectin. Cells were incubated under culture conditions for 30 min. Cells were then collected and lysed in buffer provided in the kit. 500 µg of protein lysate and 10 ug of PAK-PBD beads were used for the assay. RhoA activation was determined using the G-LISA Biochem Kit (Cytoskeleton, Inc.) as per manufacturer's instructions. Briefly, B16 cells were serum starved for 24 h. 1 ×106 cells were then seeded into a well of a 6-well plate coated with 2 µg/cm2 fibronectin. Cells were incubated under culture conditions for 1 h. Cells were then collected and lysed followed by RhoA activity measurements.
In vivo lung metastasis study
WT B16-F10 cells or cells expressing either GFP or GFP-kindlin-3 (1×106 in 100 µl PBS) were injected intravenously into 6- to 8-week-old C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME, USA). On day 6 and 8 post-injection, mice were euthanized by CO2 asphyxiation and the lungs were excised to determine the extent of metastasis. This study was carried out in accordance with the protocols ARF-SBS/NIE-A0252 and A0225 approved by the Institutional Animal Care and Use Committee (IACUC), Nanyang Technological University, Singapore. Mice were euthanized by CO2 asphyxiation in this study. The mice were housed and raised under specific-pathogen free (SPF) conditions in individually ventilated cages and were provided with irradiated corn cob, Altromin #1324 maintenance diet and sterile water. Care, treatment and use of mice for scientific purposes comply with the Animals and Birds Act, the Animals and Birds (Care and use of animals for scientific purposes) rules, the National Advisory Committee (Singapore) for laboratory animal researcher's guidelines.
Results
We first examined the expression of kindlin-3 in the highly metastatic mouse B16-F10 melanoma cell line that is commonly used in melanoma studies. The level of kindlin-3 mRNA in B16-F10 cells was minimal compared with that found in mouse bone marrow derived macrophages (BMDMs) based on qRT-PCR analysis (Fig. 1A). In line with this observation, minimal endogenous kindlin-3 protein was detected by immunoblotting using either a mAb specific for kindlin-318 or a polyclonal anti-kindlin-3 antibody (Fig. 1A). There are different observations with regard to the effect of kindlin-3 expression on the migratory property of cancer cells.22,23 Hence we examined the migratory property of B16-F10 cells that overexpressed kindlin-3. Stable B16-F10 cells that either expressed GFP-kindlin-3 or GFP were generated by transfection followed by flow cytometry cell sorting and antibiotic selection. The levels of GFP signal and integrin ß1 expression were determined (Fig. 1B). Expression levels of ß1 integrin were comparable between GFP-kindlin-3, GFP and WT cells. Expression of GFP-kindlin-3 was verified by immunoblotting (Fig. 1C). Flow cytometry analyses using the reporter mAb 9EG731,32 were performed to assess the activity of ß1 integrin in these cells (Fig. 1D). Basal active ß1 integrin was detected in WT and GFP cells. A moderate increase in the population of active ß1 integrin was detected in GFP-kindlin-3 cells. However, there was no marked difference between the 3 groups of cells based on static cell adhesion assay on immobilized fibronectin (Fig. 1E). This could be attributed to the intrinsic property of wild-type B16-F10 cells to adhere avidly to fibronectin without requiring additional stimuli.
Figure 1.

Expression analyses of kindlin-3 in mouse B16-F10 melanoma cells. (A) (left panel) Kindlin-3 mRNA expression analyses in BF16-F10 cells and mouse bone marrow derived macrophages (BMDM) by RT-qPCR. Values represent the mean ± SD of technical triplicates. (right panel) Immunoblot analyses of endogenous kindlin-3 expression in BMDM and B16-F10 cells. Actin serves as loading control. Antibodies used were mAb clone 9 and polyclonal antibody (pAb) against kindlin-3. The three faint bands in B16 WT kindlin-3 immunoblot using pAb are most likely non-specific bands. Representative blots of at least 2 independent experiments are shown. (B) Flow cytometry analyses of GFP signal and ß1 integrin expression in WT B16-F10 cells and cells stably expressing GFP or GFP-kindlin-3. (C) Immunoblot analysis of GFP-kindlin-3 expression in the stable cell lines using mAb clone 9. Actin serves as loading control. Representative blots from 2 independent experiments are shown. (D) Flow cytometry analyses of activated ß1 integrin in WT and the 2 stable cell lines using the reporter mAb 9EG7. Based on the non-specific IgG control for each group of cells, the gating region was first generated such that it contained approximately 5% of the cells with the highest intensity. The same gating region was then used to gate cells that were stained with mAb 9EG7. Percent gated 9EG7 positive cells are indicated in each histogram plot. Representative data from 3 independent experiments are shown. (E) Static cell adhesion assay on immobilized fibronectin. Values represent the mean ± SD of technical triplicates. Representative data from 2 independent experiments are shown. For B & D, gating percentages were re-labeled with larger font size for clarity.
We then examined the migratory properties of these cells by performing in vitro wound healing assay (Fig. 2A-C). Whereas gap closure by WT and GFP cells was evident by 20 h, gap closure by GFP-kindlin-3 cells was slower. Cell-tracking analyses were performed on fibronectin (2 μg/cm2)-coated glass coverslips (Fig. 2D). Consistent with the wound healing data, GFP-kindlin-3 cells showed reduced migration over a period of 20 h as compared to WT or GFP cells. Inspection of the lamellipodia of cells on fibronectin under time-lapse imaging revealed lamellipodial waves in WT and GFP cells, whereas ineffective jagged membrane protrusions were observed in GFP-kindlin-3 cells (Fig. 3A). GFP-kindlin-3 cells also showed overt ruffling activities (Supporting Information S1-S9). These data were corroborated by scanning electron microscopy analyses (Fig. 3B). Lamellipodium formation and membrane ruffling are regulated by the Rho GTPases.33 Both Rac1 and RhoA activities were higher in GFP-kindlin-3 cells compared with WT or GFP cells (Fig. 3C & D).
Figure 2.

In vitro 2D-migration assays of WT and stable B16-F10 cells. (A) Wound healing assays were performed over a period of 20 h. Closure of the gap was monitored by live-cell imaging. Representative still images at 0, 10 and 20 h are shown. Scale bar represents 100 μm. (B). Plot of % wound area/total area against time (min). Data points represent the mean ± SD of three independent experiments. (C) The rate of wound closure was calculated and plotted for WT and stable B16-F10 cells. (D) Random migration of cells on fibronectin-coated (2 μg/cm2) coverslip-bottom culture dishes as determined by live cell imaging followed by cell tracking analyses. Twenty cells were tracked in each group over a period of 20 h. Representative data from at least 2 independent experiments are shown. X and Y axes are relabeled with larger fonts for clarity.
Figure 3.

Analyses of membrane ruffling and activities of Rac-1 and RhoA in WT and stable B16-F10 cells. (A) (left panels) Still images of cells on fibronectin. Scale bar represents 10 μm. (right panels) Kymograph analyses of cells on fibronectin. Images of membrane protrusions (3 regions per cell were selected as indicated by dotted lines) at the leading edge of cells were taken and kymographs were generated for a duration of 30 min. Video files of these cells (and additional cells) can be found under Supporting Information (S1-S9). (B) Scanning electron micrographs of WT and stable B16-F10 cells on fibronectin. White arrowheads indicate extensive membrane ruffles in kindlin-3 expressing cells. (C) Rac1 activity in cells on fibronectin was determined by PAK-BD pull-down assay. Representative data from 2 independent experiments are shown. Mean fold differences based on densitometric analyses (image J) of the ratio of active/total Rac1 are shown. WT value is normalized to 1.0. (D) RhoA activity was determined by ELISA-based assay. Representative data from 3 independent experiments are shown. Data points are mean ± SD of triplicates. Student's t-test was used for statistical analysis.
Next, we performed confocal laser scanning microscopy to examine the localization of focal adhesions in these cells by staining for focal adhesion protein paxillin and actin filaments (Fig. 4 & Supporting Information S10). It has been reported that there are 2 populations of B16 melanoma cells, one that is well spread but appears to have a “stationary” morphology and another that is polarized and migratory.34 We therefore examined these 2 groups of cells herein. There was no significant difference in the formation of punctate focal adhesions in polarized WT, GFP and GFP-kindlin-3 cells (Fig. 4A). Comparing the non-polarized cells, GFP-kindlin3 cells showed a rounded morphology with actin filaments around the cell periphery, whereas WT and GFP cells showed actin-rich spindle-like membrane protrusions (Fig. 4B). The morphology of the non-polarized GFP-kindlin3 cells was similar to that seen in MDA-MB-231breast carcinoma cells with overt Rho activation.35
Figure 4.

Immunofluorescence analyses of focal adhesions and actin filaments in WT and stable B16-F10 cells. In both groups (A & B), cells were settled onto fibronectin-coated coverslip-bottom culture dish and incubated for 2 h under culture conditions. Cells were fixed and permeabilized followed by antibody staining for the focal adhesion protein paxillin. Actin filaments were stained with Alexa Fluor® 594-phalloidin and nuclei were stained with DAPI. (A) Polarized cells. (B) Non-polarized cells. GFP and GFP-kindlin-3 were detected in the B16-F10 stable cells. Consistent with previous findings in HUVECs , kindlin-3 did not show punctate localization at mature focal adhesion sites containing paxillin. Scale bar represents 10 μm. Representative images of cells from 2 independent experiments are shown. Images were processed using the auto-min/max function in the ZEN2012 software for optimal setting for contrast and brightness. Also see Supporting Information Fig. S10.
To obtain in vivo data, we performed lung metastasis experiments in syngenic C57BL/6 mice (Fig. 5). Tumor nodules (black) were readily observed in mice on day 6 after intravenous injection of WT or GFP cells, and a high tumor load was observed on day 8. By contrast, tumor formation was comparatively less in the lungs of mice injected with GFP-kindlin3 cells.
Figure 5.
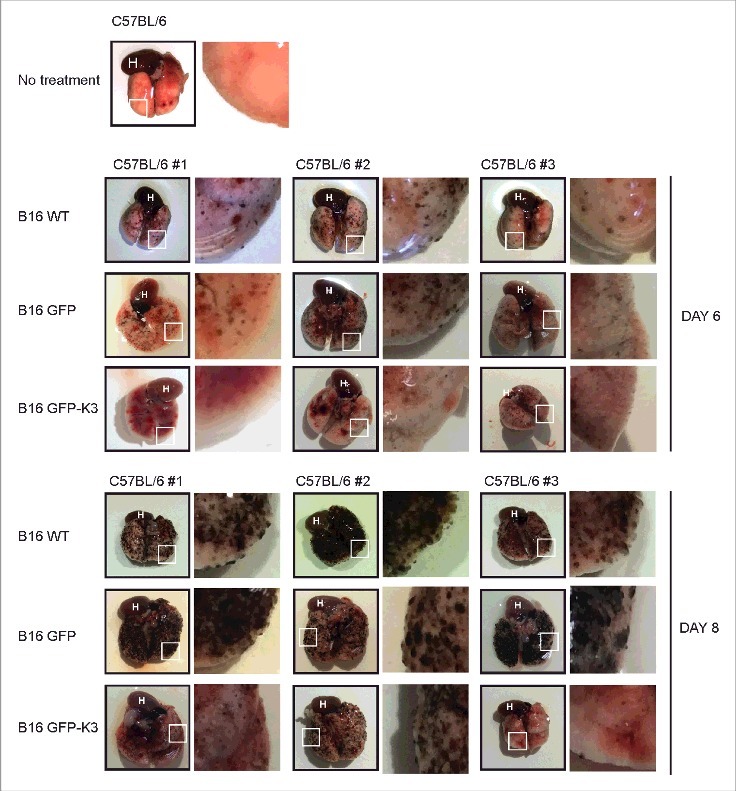
Mouse lung tumor metastasis study. Intravenous injections of WT or B16-F10 cells expressing GFP or GFP-kindlin-3 into C57BL/6J mice were performed and tumor formation in the lungs of mice was examined on day 6 and day 8. Except for the no-treatment group (1 mouse), data from 3 mice per group for different days are shown. The insets show an enlarged view of demarcated sections (white outlined boxes) of lungs. H: heart.
Finally, we asked if overexpressing kindlin-3 in human melanoma cells impedes their migration. We first examined the expression of kindlin-3 in M10 melanoma cells. Kindlin-3 mRNA expression in M10 cells was minimal compared with K562 cells, a human chronic myeloid leukemia cell line that expressed kindlin-3,36 whereas kindlin-1 and kindlin-2 were detected in M10 cells (Fig. 6A). Minimal kindlin-3 mRNA expression in M10 cells was verified in a separate experiment that included the human fibroblast cell line 293T, which is not known to express kindlin-3 (Fig. 6B). Consistent with the above data, we did not detect kindlin-3 protein expression in M10 cells using 2 different anti-kindlin-3 mAbs, namely mAb clone 918 and mAb 3D637 (Fig. 6C and 6D). The human breast cancer line MDA-MB-231 was included in this analysis because it has been shown to be lacking in kindlin-3 expression.37 Given that M10 cells do not express detectable level of kindlin-3, we examined the effect of kindlin-3 overexpression in these cells. M10 cells were transiently transfected with GFP or GFP-kindlin-3 and were sorted by flow cytometry (Fig. 7A). Their migratory properties on fibronectin were determined (Fig. 7B). Consistent with previous data using B16-F10 cells, M10 cells expressing GFP-kindlin3 showed reduced migration on fibronectin compared with WT or GFP cells.
Figure 6.

Analyses of kindlin-3 expression in M10 human melanoma cell line. (A) qRT-PCR analyses of kindlin-1, −2 and −3 mRNA expression levels in M10 cells and the human chronic myeloid leukemia cell line K562. Values are mean ± SD of technical triplicates. A representative experiment from 3 independent experiments is shown. (B) qRT-PCR analyses of kindlin-3 expression in M10, K562 and the human fibroblast cell line 293T. In (A) and (B), GAPDH and actin were used as internal controls. (C) Immunoblot analyses of kindlin-3 protein expression in M10 cells, K562, 293T and the human breast cancer cell line MDA-MB-231 using the anti-kindlin-3 mAb clone 9. Actin serves as loading control. (D) Immunoblot analyses using the anti-kindlin-3 mAb 3D6.
Figure 7.

Analyses of the migratory properties of M10 cells transfected with GFP-kindlin-3. (A) Flow cytometry analyses and sorting of cells transiently transfected with GFP or GFP-kindlin-3 expression plasmid. (B) Sorted cells were subjected to 2D-random migration assays on fibronectin-coated (2 μg/cm2) coverslip-bottom culture dishes. Twenty cells were tracked and migratory profiles were plotted. Representative data from at least 2 independent experiments are shown. X and Y axes are relabeled with larger fonts for clarity.
Discussion
Kindlins not only function as co-activators with talin which positively regulates integrin ligand-binding affinity,16,38 they modulate integrin clustering.18,39 They are also involved in integrin independent processes, including clathrin-mediated endocytosis, Wnt-signaling, and the assembly of erythrocyte membrane-cytoskeleton.15,40,41 All kindlin paralogs are involved in cancer progression but they have different roles. Kindlin-1 is highly expressed in pancreatic cancer cells and enhances their metastatic potential.42 Kindlin-1 expression also correlates with hepatocellular carcinoma tumor size and metastasis.43 However, loss of kindlin-1 expression increases skin tumor susceptibility.44 Kindlin-2 has been shown to promote breast cancer metastasis, but it functions as a tumor suppressor in serous epithelial ovarian cancer.14,45 Kindlin-2 has also been shown to negatively regulate mesenchymal cancer cell invasion.46 Kindlin-3 expression was reported to be up-regulated in late stage malignant breast cancer cells and promoted metastasis,22 and it is involved in chronic myeloid leukemia cell proliferation.36 However, another study reported kindlin-3 having a tumor suppressor function.23
Kindlin-3 was reported to form a complex with EMMPRIN and integrin ß1 in human M10 melanoma cells, suggesting that the protein is endogenously expressed in this cell line.24 In our hands, we did not detect significant kindlin-3 expression at both mRNA and protein levels in either M10 cells or the mouse melanoma cell line B16-F10. What accounts for the difference in observations is not known. Because there are conflicting reports on the function of kindlin-3 in cancer cell metastasis,22,23 we overexpressed kindlin-3 in both B16-F10 and M10 cells and examined their migratory properties. Interestingly, kindlin-3 expression in these cells reduced their cell migration. These observations are not due to clonal effects because, in addition to the B16-F10 stable cells expressing GFP-kindlin-3, we observed a similar migratory defect in M10 cells transiently transfected with GFP-kindlin-3. High fibronectin concentrations (> 50 μg/ml) inhibit cell polarization and migration via RhoA activation.47 In our study, we used a relatively low coating concentration of fibronectin (10 μg/ml). Hence, the migratory defect observed is unlikely a consequence of high fibronectin concentration used.
Our findings may appear contrary to the role of kindlin-3 as a positive regulator of integrin-mediated cell adhesion and migration, which is well exemplified in the etiology of LAD III.9,10,48 However, effective cell migration requires coordinated spatio-temporal regulation of cell-substrate adhesion, cytoskeletal remodeling, membrane protrusion, adhesome turnover and rear retraction. Overt and deregulated cell adhesion could impede rather than promote migration. For example, integrin αLß2 (LFA-1) is widely reported to be essential for leukocyte migration but overt activation of integrin αLß2 impedes rather than promotes T cell migration.49
Our data suggest that Rac1 and RhoA activities in GFP-kindlin-3-expressing cells were higher than the basal activities found in either WT cells or cells expressing GFP. Mutual antagonism of Rac1 and RhoA has been demonstrated in migrating cells by several groups.50-52 Conceivably, the loss of this balance as a consequence of deregulated activation of these Rho GTPases could lead to defective cell migration. Although Rac1 is important for lamellipodium formation, there is evidence suggesting RhoA activation precedes that of Rac and Cdc42, and that it plays a role in membrane ruffling.53 High RhoA activity has also been shown to restrict membrane protrusions at the leading edge of migrating leukocytes,54 and it impedes the movement of fibroblasts and lung adenocarcinoma cells.55,56 Expression of constitutively activated Rho also inhibits fibroblast cell line invasion into 3D collagen matrices.57 Hence, the overt activation of RhoA in B16-F10 cells expressing kindlin-3 could have disrupted the balanced activities of RhoGTPases required for effective cell migration. We also observed actin-rich spindle-like membrane protrusions in a population of non-polarized WT or GFP-expressing B16-F10 cells, which is in contrast to the round morphology with radial peripheral actin filaments seen in non-polarized cells expressing GFP-kindlin-3. Interestingly, breast cancer MDA-MB-231 cells treated with the compound strongylophorine-26 that overtly activates Rho showed similar extensive membrane ruffling and non-directional lamellipodia formation, leading to a rounded morphology with radial peripheral filaments.35 Taken together, we rationalized that expression of kindlin-3 in melanoma cells that lack endogenous kindlin-3 could lead to 2 levels of “defects” that impede cell migration. First, cells that are spreading on fibronectin could be poor in polarization. Second, cells that could eventually become polarized exhibit extensive membrane ruffling that impede productive lamellipodia formation. It should be noted that although we observed a population of non-polarized cells at the end of the 2 h incubation on fibronectin by immunofluorescence, many of the non-polarized cells could become polarized with longer incubation time based on live cell imaging (data not shown). The migratory defects of B16-F10 cells ectopically expressing kindlin-3 observed in vitro were verified in vivo using the mouse lung metastasis model, hence it is unlikely that the defect in migration seen in kindlin-3-expressing cells is an aberrant observation based on 2D monolayer culture systems. Our data may suggest that loss of kindlin-3 expression positively regulates cancer cell migration. However, we should be cautious with this interpretation because careful examination and comparison of kindlin-3 expression at both mRNA and protein levels between normal cells and their cancer counterparts are required. We acknowledged that by overexpressing a protein in cells to investigate the function of the protein is not an ideal approach. However, with our existing experimental setup, we have utilized cells that do not express endogenous kindin-3 and employed GFP-empty vector as control.
In conclusion, our study showed the lack of kindlin-3 expression in 2 widely used mouse and human melanoma cell lines. Ectopic expression of kindlin-3 in these cells leads to imbalanced Rho GTPase activation and overt membrane ruffling that impedes rather than promotes cell migration.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Prof. Klaus E. Karjalainen, School of Biological Sciences, Nanyang Technological University, Singapore, for providing the L929 cell line. We thank Prof. Nguan-Soon Tan, School of Biological Sciences, Nanyang Technological University, Singapore, for his advice on the mouse lung metastasis study. We thank Prof. Mark Featherstone, School of Biological Sciences, Nanyang Technological University, Singapore, for proofreading the manuscript.
Funding
This work was supported by grants RG149/14 and M4081320.080 Nanyang Technological University, Singapore. We thank Dr. Lay-Poh Tan, School of Materials Science & Engineering, Nanyang Technological University, Singapore, for her advice on SEM work. W.K. Wee is supported by the program Undergraduate Research Experience on Campus (URECA), Nanyang Technological University, Singapore.
Author contributions
CF, WKW and SMT conceived and designed all the experiments in this study. CF and WKW contributed equally and performed most of the experiments in this study. HC performed the scanning electron microscopy. LTO provided assistance in the mouse husbandry and performed the BMDM experiments. JQ assisted in the qRT-PCR experiments. HFT provided technical assistance in this study. SMT coordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
References
- [1].Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harb Perspect Biol 2011; 3:a005074; PMID:21885598; https://doi.org/ 10.1101/cshperspect.a005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57-70; PMID:10647931; https://doi.org/ 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- [3].Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell 2002; 110:673-87; PMID:12297042; https://doi.org/ 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- [4].Harburger DS, Calderwood DA. Integrin signalling at a glance. J Cell Sci 2009; 122:159-63; PMID:19118207; https://doi.org/ 10.1242/jcs.018093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Meves A, Stremmel C, Gottschalk K, Fassler R. The Kindlin protein family: new members to the club of focal adhesion proteins. Trends Cell Biol 2009; 19:504-13; PMID:19766491; https://doi.org/ 10.1016/j.tcb.2009.07.006 [DOI] [PubMed] [Google Scholar]
- [6].Ussar S, Wang HV, Linder S, Fassler R, Moser M. The Kindlins: subcellular localization and expression during murine development. Exp Cell Res 2006; 312:3142-51; PMID:16876785; https://doi.org/ 10.1016/j.yexcr.2006.06.030 [DOI] [PubMed] [Google Scholar]
- [7].Bialkowska K, Ma YQ, Bledzka K, Sossey-Alaoui K, Izem L, Zhang X, Malinin N, Qin J, Byzova T, Plow EF. The integrin co-activator kindlin-3 is expressed and functional in a non-hematopoietic cell, the endothelial cell. J Biol Chem 2010; 285:18640-9; PMID:20378539; https://doi.org/ 10.1074/jbc.M109.085746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med 2008; 14:325-30; PMID:18278053; https://doi.org/ 10.1038/nm1722 [DOI] [PubMed] [Google Scholar]
- [9].Moser M, Bauer M, Schmid S, Ruppert R, Schmidt S, Sixt M, Wang HV, Sperandio M, Fässler R. Kindlin-3 is required for b2 integrin-mediated leukocyte adhesion to endothelial cells. Nat Med 2009; 15:300-5; PMID:19234461; https://doi.org/ 10.1038/nm.1921 [DOI] [PubMed] [Google Scholar]
- [10].McDowall A, Svensson L, Stanley P, Patzak I, Chakravarty P, Howarth K, Sabnis H, Briones M, Hogg N. Two mutations in the KINDLIN3 gene of a new leukocyte adhesion deficiency III patient reveal distinct effects on leukocyte function in vitro. Blood 2010; 115:4834-42; PMID:20357244; https://doi.org/ 10.1182/blood-2009-08-238709 [DOI] [PubMed] [Google Scholar]
- [11].Svensson L, Howarth K, McDowall A, Patzak I, Evans R, Ussar S, Moser M, Metin A, Fried M, Tomlinson I, et al.. Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat Med 2009; 15:306-12; PMID:19234463; https://doi.org/ 10.1038/nm.1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhan J, Zhu X, Guo Y, Wang Y, Qiang G, Niu M, Hu J, Du J, Li Z, Cui J, et al.. Opposite role of Kindlin-1 and Kindlin-2 in lung cancers. PloS one 2012; 7:e50313; PMID:23209705; https://doi.org/ 10.1371/journal.pone.0050313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yu Y, Qi L, Wu J, Wang Y, Fang W, Zhang H. Kindlin 2 regulates myogenic related factor myogenin via a canonical Wnt signaling in myogenic differentiation. PloS One 2013; 8:e63490; PMID:23717433; https://doi.org/ 10.1371/journal.pone.0063490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yu Y, Wu J, Guan L, Qi L, Tang Y, Ma B, Zhan J, Wang Y, Fang W, Zhang H. Kindlin 2 promotes breast cancer invasion via epigenetic silencing of the microRNA200 gene family. Int J Cancer 2013; 133:1368-79; PMID:23483548; https://doi.org/ 10.1002/ijc.28151 [DOI] [PubMed] [Google Scholar]
- [15].Pluskota E, Ma Y, Bledzka KM, Bialkowska K, Soloviev DA, Szpak D, Podrez EA, Fox PL, Hazen SL, Dowling JJ, et al.. Kindlin-2 regulates hemostasis by controlling endothelial cell-surface expression of ADP/AMP catabolic enzymes via a clathrin-dependent mechanism. Blood 2013; 122:2491-9; PMID:23896409; https://doi.org/ 10.1182/blood-2013-04-497669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science 2009; 324:895-9; PMID:19443776; https://doi.org/ 10.1126/science.1163865 [DOI] [PubMed] [Google Scholar]
- [17].Feigelson SW, Grabovsky V, Manevich-Mendelson E, Pasvolsky R, Shulman Z, Shinder V, Klein E, Etzioni A, Aker M, Alon R. Kindlin-3 is required for the stabilization of TCR-stimulated LFA-1:ICAM-1 bonds critical for lymphocyte arrest and spreading on dendritic cells. Blood 2011; 117:7042-52; PMID:21536861; https://doi.org/ 10.1182/blood-2010-12-322859 [DOI] [PubMed] [Google Scholar]
- [18].Feng C, Li YF, Yau YH, Lee HS, Tang XY, Xue ZH, Zhou YC, Lim WM, Cornvik TC, Ruedl C, et al.. Kindlin-3 mediates integrin aLb2 outside-in signaling, and it interacts with scaffold protein receptor for activated-C kinase 1 (RACK1). J Biol Chem 2012; 287:10714-26; PMID:22334666; https://doi.org/ 10.1074/jbc.M111.299594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xue ZH, Feng C, Liu WL, Tan SM. A role of kindlin-3 in integrin αMß2 outside-in signaling and the Syk-Vav1-Rac1/Cdc42 signaling axis. PloS One 2013; 8:e56911; PMID:23437269; https://doi.org/ 10.1371/journal.pone.0056911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kasirer-Friede A, Kang J, Kahner B, Ye F, Ginsberg MH, Shattil SJ. ADAP interactions with talin and kindlin promote platelet integrin alphaIIbbeta3 activation and stable fibrinogen binding. Blood 2014; 123:3156-65; PMID:24523237; https://doi.org/ 10.1182/blood-2013-08-520627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schmidt S, Nakchbandi I, Ruppert R, Kawelke N, Hess MW, Pfaller K, Jurdic P, Fässler R, Moser M. Kindlin-3-mediated signaling from multiple integrin classes is required for osteoclast-mediated bone resorption. J Cell Biol 2011; 192:883-97; PMID:21357746; https://doi.org/ 10.1083/jcb.201007141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sossey-Alaoui K, Pluskota E, Davuluri G, Bialkowska K, Das M, Szpak D, Lindner DJ, Downs-Kelly E, Thompson CL, Plow EF. Kindlin-3 enhances breast cancer progression and metastasis by activating Twist-mediated angiogenesis. Faseb J 2014; 28:2260-71; PMID:24469992; https://doi.org/ 10.1096/fj.13-244004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Djaafri I, Khayati F, Menashi S, Tost J, Podgorniak MP, Sadoux A, Daunay A, Teixeira L, Soulier J, Idbaih A, et al.. A novel tumor suppressor function of Kindlin-3 in solid cancer. Oncotarget 2014; 5:8970-85; PMID:25344860; https://doi.org/ 10.18632/oncotarget.2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Delyon J, Khayati F, Djaafri I, Podgorniak MP, Sadoux A, Setterblad N, Boutalbi Z, Maouche K, Maskos U, Menashi S, et al.. EMMPRIN regulates beta1 integrin-mediated adhesion through Kindlin-3 in human melanoma cells. Exp Dermatol 2015; 24:443-8; PMID:25807898; https://doi.org/ 10.1111/exd.12693 [DOI] [PubMed] [Google Scholar]
- [25].Ying W, Cheruku PS, Bazer FW, Safe SH, Zhou B. Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp 2013; PMID:23851980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tan SM, Robinson MK, Drbal K, van Kooyk Y, Shaw JM, Law SK. The N-terminal region and the mid-region complex of the integrin b2 subunit. J Biol Chem 2001; 276:36370-6; PMID:11477072; https://doi.org/ 10.1074/jbc.M102392200 [DOI] [PubMed] [Google Scholar]
- [27].Tan SM, Walters SE, Mathew EC, Robinson MK, Drbal K, Shaw JM, Law SK. Defining the repeating elements in the cysteine-rich region (CRR) of the CD18 integrin ß2 subunit. FEBS Lett 2001; 505:27-30; PMID:11557036; https://doi.org/ 10.1016/S0014-5793(01)02778-8 [DOI] [PubMed] [Google Scholar]
- [28].Tang XY, Li YF, Tan SM. Intercellular adhesion molecule-3 binding of integrin aLb2 requires both extension and opening of the integrin headpiece. J Immunol 2008; 180:4793-804; PMID:18354203; https://doi.org/ 10.4049/jimmunol.180.7.4793 [DOI] [PubMed] [Google Scholar]
- [29].Braet F, De Zanger R, Wisse E. Drying cells for SEM, AFM and TEM by hexamethyldisilazane: a study on hepatic endothelial cells. J Microsc 1997; 186:84-7; PMID:9159923; https://doi.org/ 10.1046/j.1365-2818.1997.1940755.x [DOI] [PubMed] [Google Scholar]
- [30].Tang RH, Law SK, Tan SM. Selective recruitment of src family kinase Hck by leukocyte integrin αMß2 but not αLß2 or αXß2. FEBS Lett 2006; 580:4435-42; PMID:16854414; https://doi.org/ 10.1016/j.febslet.2006.06.099 [DOI] [PubMed] [Google Scholar]
- [31].Lenter M, Uhlig H, Hamann A, Jeno P, Imhof B, Vestweber D. A monoclonal antibody against an activation epitope on mouse integrin chain β 1 blocks adhesion of lymphocytes to the endothelial integrin α 6 β 1. Proc Natl Acad Sci U S A 1993; 90:9051-5; PMID:7692444; https://doi.org/ 10.1073/pnas.90.19.9051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bazzoni G, Shih DT, Buck CA, Hemler ME. Monoclonal antibody 9EG7 defines a novel β 1 integrin epitope induced by soluble ligand and manganese, but inhibited by calcium. J Biol Chem 1995; 270:25570-7; PMID:7592728; https://doi.org/ 10.1074/jbc.270.43.25570 [DOI] [PubMed] [Google Scholar]
- [33].Lessey EC, Guilluy C, Burridge K. From mechanical force to RhoA activation. Biochemistry 2012; 51:7420-32; PMID:22931484; https://doi.org/ 10.1021/bi300758e [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ballestrem C, Wehrle-Haller B, Imhof BA. Actin dynamics in living mammalian cells. J Cell Sci 1998; 111:1649-58; PMID:9601095 [DOI] [PubMed] [Google Scholar]
- [35].McHardy LM, Warabi K, Andersen RJ, Roskelley CD, Roberge M. Strongylophorine-26, a Rho-dependent inhibitor of tumor cell invasion that reduces actin stress fibers and induces nonpolarized lamellipodial extensions. Mol Cancer Ther 2005; 4:772-8; PMID:15897241; https://doi.org/ 10.1158/1535-7163.MCT-04-0310 [DOI] [PubMed] [Google Scholar]
- [36].Qu J, Ero R, Feng C, Ong LT, Tan HF, Lee HS, Ismail MH, Bu WT, Nama S, Sampath P, et al.. Kindlin-3 interacts with the ribosome and regulates c-Myc expression required for proliferation of chronic myeloid leukemia cells. Sci Rep 2015; 5:18491; PMID:26677948; https://doi.org/ 10.1038/srep18491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhao Y, Malinin NL, Meller J, Ma Y, West XZ, Bledzka K, Qin J, Podrez EA, Byzova TV. Regulation of cell adhesion and migration by Kindlin-3 cleavage by calpain. J Biol Chem 2012; 287:40012-20; PMID:23012377; https://doi.org/ 10.1074/jbc.M112.380469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bouaouina M, Calderwood DA. Kindlins. Curr Biol 2011; 21:R99-101; PMID:21300280; https://doi.org/ 10.1016/j.cub.2010.12.002 [DOI] [PubMed] [Google Scholar]
- [39].Ye F, Petrich BG, Anekal P, Lefort CT, Kasirer-Friede A, Shattil SJ, Ruppert R, Moser M, Fässler R, Ginsberg MH. The mechanism of kindlin-mediated activation of integrin αIIbß3. Curr Biol 2013; 23:2288-95; PMID:24210614; https://doi.org/ 10.1016/j.cub.2013.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yu Y, Wu J, Wang Y, Zhao T, Ma B, Liu Y, Fang W, Zhu WG, Zhang H. Kindlin 2 forms a transcriptional complex with b-catenin and TCF4 to enhance Wnt signalling. EMBO Rep 2012; 13:750-8; PMID:22699938; https://doi.org/ 10.1038/embor.2012.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fässler R, Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 2008; 134:353-64; PMID:18662549; https://doi.org/ 10.1016/j.cell.2008.05.033 [DOI] [PubMed] [Google Scholar]
- [42].Mahawithitwong P, Ohuchida K, Ikenaga N, Fujita H, Zhao M, Kozono S, Shindo K, Ohtsuka T, Aishima S, Mizumoto K, et al.. Kindlin-1 expression is involved in migration and invasion of pancreatic cancer. Int J Oncol 2013; 42:1360-6; PMID:23440354 [DOI] [PubMed] [Google Scholar]
- [43].Ma HX, Shu QH, Pan JJ, Liu D, Xu GL, Li JS, Ma JL, Jia WD, Yv JH, Ge YS. Expression of Kindlin-1 in human hepatocellular carcinoma and its prognostic significance. Tumour Biol 2015; 36:4235-41; PMID:25592379; https://doi.org/ 10.1007/s13277-015-3060-8 [DOI] [PubMed] [Google Scholar]
- [44].Rognoni E, Widmaier M, Jakobson M, Ruppert R, Ussar S, Katsougkri D, Böttcher RT, Lai-Cheong JE, Rifkin DB, McGrath JA, et al.. Kindlin-1 controls Wnt and TGF-β availability to regulate cutaneous stem cell proliferation. Nat Med 2014; 20:350-9; PMID:24681597; https://doi.org/ 10.1038/nm.3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ren C, Du J, Xi C, Yu Y, Hu A, Zhan J, Guo H, Fang W, Liu C, Zhang H. Kindlin-2 inhibits serous epithelial ovarian cancer peritoneal dissemination and predicts patient outcomes. Biochem Biophys Res Commun 2014; 446:187-94; PMID:24583125; https://doi.org/ 10.1016/j.bbrc.2014.02.087 [DOI] [PubMed] [Google Scholar]
- [46].Shi X, Wu C. A suppressive role of mitogen inducible gene-2 in mesenchymal cancer cell invasion. Mol Cancer Res 2008; 6:715-24; PMID:18505917; https://doi.org/ 10.1158/1541-7786.MCR-07-2026 [DOI] [PubMed] [Google Scholar]
- [47].Cox EA, Sastry SK, Huttenlocher A. Integrin-mediated adhesion regulates cell polarity and membrane protrusion through the Rho family of GTPases. Mol Biol Cell 2001; 12:265-77; PMID:11179414; https://doi.org/ 10.1091/mbc.12.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Manevich-Mendelson E, Feigelson SW, Pasvolsky R, Aker M, Grabovsky V, Shulman Z, Kilic SS, Rosenthal-Allieri MA, Ben-Dor S, Mory A, et al.. Loss of kindlin-3 in LAD-III eliminates LFA-1 but not VLA-4 adhesiveness developed under shear flow conditions. Blood 2009; 114:2344-53; PMID:19617577; https://doi.org/ 10.1182/blood-2009-04-218636 [DOI] [PubMed] [Google Scholar]
- [49].Vararattanavech A, Chng CP, Parthasarathy K, Tang XY, Torres J, Tan SM. A transmembrane polar interaction is involved in the functional regulation of integrin αLß2. J Mol Biol 2010; 398:569-83; PMID:20338181; https://doi.org/ 10.1016/j.jmb.2010.03.027 [DOI] [PubMed] [Google Scholar]
- [50].Rottner K, Hall A, Small JV. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr Biol 1999; 9:640-8; PMID:10375527; https://doi.org/ 10.1016/S0960-9822(99)80286-3 [DOI] [PubMed] [Google Scholar]
- [51].Williams MJ, Habayeb MS, Hultmark D. Reciprocal regulation of Rac1 and Rho1 in Drosophila circulating immune surveillance cells. J Cell Sci 2007; 120:502-11; PMID:17227793; https://doi.org/ 10.1242/jcs.03341 [DOI] [PubMed] [Google Scholar]
- [52].D'Avino PP, Savoian MS, Glover DM. Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J Cell Biol 2004; 166:61-71; PMID:15240570; https://doi.org/ 10.1083/jcb.200402157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].O'Connor K, Chen M. Dynamic functions of RhoA in tumor cell migration and invasion. Small GTPases 2013; 4:141-7; PMID:24025634; https://doi.org/ 10.4161/sgtp.25131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Worthylake RA, Burridge K. RhoA and ROCK promote migration by limiting membrane protrusions. J Biol Chem 2003; 278:13578-84; PMID:12574166; https://doi.org/ 10.1074/jbc.M211584200 [DOI] [PubMed] [Google Scholar]
- [55].Gu J, Sumida Y, Sanzen N, Sekiguchi K. Laminin-10/11 and fibronectin differentially regulate integrin-dependent Rho and Rac activation via p130(Cas)-CrkII-DOCK180 pathway. J Biol Chem 2001; 276:27090-7; PMID:11369773; https://doi.org/ 10.1074/jbc.M102284200 [DOI] [PubMed] [Google Scholar]
- [56].Arthur WT, Burridge K. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol Biol Cell 2001; 12:2711-20; PMID:11553710; https://doi.org/ 10.1091/mbc.12.9.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Banyard J, Anand-Apte B, Symons M, Zetter BR. Motility and invasion are differentially modulated by Rho family GTPases. Oncogene 2000; 19:580-91; PMID:10698528; https://doi.org/ 10.1038/sj.onc.1203338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


