Abstract
Aim:
Analysis of the properties and chemotherapeutics delivery potential of spheres made of bioengineered spider silks MS1 and MS2.
Materials & methods:
MS1 and MS2 derived from Nephila clavipes dragline silks – MaSp1 and MaSp2, respectively – formed spheres that were compared in terms of physicochemical properties, cytotoxicity and loading/release of chemotherapeutics.
Results:
MS2 spheres were more dispersed, smaller, of solid core, of higher beta-sheet structure content, and of opposite (negative) charge than MS1 spheres. Preloaded MS2 showed greater applicability for mitoxantrone, while postloaded for etoposide delivery compared with MS1 spheres. However, MS1 spheres were a better choice for doxorubicin delivery than MS2.
Conclusion:
Bioengineered silks can be tailored to develop a system with optimal drug loading and release properties.
Keywords: : bioengineered spider silk, biomaterial, biomedicine, cancer therapy, chemotherapeutics, drug carrier, MaSp1, MaSp2, spheres
Conventional chemotherapy suffers from several drawbacks, such as pronounced side effects caused by the general cytotoxicity of anticancer drugs toward healthy tissues. The most effective way to minimize the side effects caused by chemotherapeutics is to accumulate the drug dose within a carrier that enables specific drug delivery to the tumor and allows the amount of the drug administered to a patient to be reduced. To achieve this goal, several conditions should be met, including the drug carrier should target the tumor, and the biomaterial composing the carrier should efficiently encapsulate the drug. Moreover, the drug release process should avoid burst release into the bloodstream before the carrier can reach the tumor, and the effect of the drug on tumor cells should be prolonged.
Bioengineered spider silk possesses extraordinary properties that make it one of the most promising biomaterials for drug delivery. These properties include biocompatibility, biodegradability, simple production (in an Escherichia coli expression system), efficient purification, and facile processing (by self-assembly) under mild, biocompatible conditions into several different morphological forms [1]. Numerous attempts have been made to load and release active compounds from silk-based drug delivery systems, and many of these have been focused on cancer therapy [2]. Among the structural forms that can be prepared by processing silk polymers, such as fibers, films, hydrogels, sponges and scaffolds, the most applicable for use as a systemic drug carrier are spherical particles [1]. Silk particles have been obtained by freezing silk protein in an ethanol solution [3], performing freeze-thaw cycles with liquid nitrogen [4], using a capillary device [5], and mixing the protein with acetone [6], PVA [7] or potassium phosphate [8]. The last method was used to form particles from natural Bombyx mori silk [9] and from bioengineered spider silks derived from the two major spidroins of Araneus diadematus [8,10] and Nephila clavipes [11,12]. Nanoparticles made from natural silkworm silk and bioengineered spider silk have been studied as carriers of model drugs, proteins and polysaccharides [3,4,6,9,11–13].
We have recently showed that MS1 and MS2(9x) bioengineered spider silks derived from the N. clavipes dragline silk proteins (MaSp1 and MaSp2, respectively) can be produced in a bacterial expression system and purified with a high yield using two methods that take advantage of the thermal stability of silk and its resistance to organic acids [12,14]. The resulting bioengineered spider silks were able to form stable and nontoxic spherical particles by mixing them with potassium phosphate [11,12]. Moreover, MS1 protein functionalized with tumor targeting domains formed spheres that were bound and specifically internalized by tumor cells [11]. Compared with functionalized MS1 spheres, particles formed by the blending of functionalized MS1 and MS2 silks at a ratio of 8:2 efficiently killed targeted cancer cells when loaded with a doxorubicin inducing considerably lower nonspecific toxicity [15].
In the present study, we focused on the development of a system for effective drug loading and release. To study the influence of protein sequence on drug loading ability, we employed two bioengineered silk proteins, MS1 and MS2, which are of the same number (15-mers) of the consensus motifs of MaSp1 and MaSp2, respectively. Since, the method of purifying bioengineered spider silk determined the silk sphere properties [12], we purified both silk variants by using the same method. We have made a detailed comparative analysis of the processing conditions and properties of both silk sphere types, including their size and morphology, secondary structure composition, ζ potential and cytotoxicity. Two loading methods, preloading and postloading, were applied to investigate the drug binding potential and the release kinetics. Three commonly used chemotherapeutics (doxorubicin, mitoxantrone and etoposide) were also studied using this system. The differences in amino acid composition between the MS1 and MS2 silks determined the optimal processing conditions, physical properties of the spheres and the loading and release of the drugs.
Materials & methods
Construction of genes encoding bioengineered spider silks
The construction of the pETNX-MS1 plasmid was previously described [14]. The pETNX-MS2 plasmid was based on an expression vector carrying a nine-mer of MS2 protein (pETNX-MS2(9x)) [12]. The 15-mer of the MS2 sequence was obtained by ligation of the 9- and 6-mer of MS2 using a method described previously [12,15]. Enzymes for digestion and ligation were supplied by Fermentas (St. Leon-Rot, Germany). The final construct was sequenced by the University of Adam Mickiewicz Core Facility in Poznan.
Production & purification of bioengineered silk proteins
The pETNX plasmids carrying either the MS1 or MS2 constructs were transformed into BL21 (DE3) E. coli (Life Technologies, CA, USA). Proteins were expressed using a Bioflo 3000 fermenter (New Brunswick Scientific, NJ, USA) under the conditions described previously [14]. Briefly, cells were grown to an OD600 of 10 before the protein production was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG; Life Technologies) to a final concentration of 1 mM. After 4 h of incubation, cells were harvested by centrifugation at 7000 × g for 10 min and stored at -80°C. For protein purification, a thermal extraction method named 80/20 was used as described previously [12]. The protein concentration was determined by spectrophotometry at λ = 280 nm, referring to a molecular weight of 39.25 and 46.72 kDa and extinction coefficient values of 22,350 and 44,700 M-1 cm-1 for the MS1 and MS2 proteins, respectively. Quality and purity of the bioengineered silk proteins were analyzed by electrophoresis, using 4–12% SDS-PAGE gradient gel (Live Technologies). Proteins were stained with a Colloidal Blue Staining Kit (Live Technologies).
MALDI-TOF mass spectrometry
The molecular masses of the purified proteins were confirmed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry performed on a Bruker microflex LT mass spectrometer (Bruker Corp., CA, USA). The MALDI matrix was prepared by dissolving α-cyano-4-hydroxycinnamic acid (GHCA) in a 50% acetonitrile and 0.3% trifluoroacetic acid solution. The protein sample (6 μl, 1 mg/ml) was added to 24 μl of the matrix, and 1 μl of the resulting solution was placed onto a 96 spot target plate and air-dried. MALDI-TOF mass spectra were acquired with 50% laser power and in standard linear positive-ion mode up to 60 kDa.
Formation of bioengineered spider silk spheres
To produce spheres, the bioengineered spider silk proteins were mixed by pipetting with potassium phosphate buffer. To study the influence of protein concentration on sphere formation, 100 μl of the silk protein solution at concentrations of 0.5, 2.5 or 5 mg/ml was mixed with 1 ml of 2 M potassium phosphate, pH 8. To study the influence of salt concentration on sphere formation, 1 ml of potassium phosphate buffer at concentration of 0.5, 1 or 2 M, pH 8 was mixed with 100 μl of silk protein solution at concentration of 0.5 mg/ml. To determine the influence of pH, 1 ml of phosphate at pH of 4, 7 or 10 was mixed with 100 μl of silk protein solution at concentration of 0.5 mg/ml. Phosphate buffers at different pH were obtained by mixing of 1.5 M dibasic and 1.5 M monobasic potassium phosphates (to obtain pH 4 and 7) or 1.5 M tribasic and 1.5 M dibasic potassium phosphates (to obtain pH 10). The spheres were incubated in the phosphate buffer overnight at 4°C, and then dialyzed against ultrapure water using a ZelluTrans regenerated cellulose dialysis tube with the MWCO of 12-14 kDa (Carl Roth, Karlsruhe, Germany). For FTIR analyses, postloading studies, and cytotoxicity studies spheres were prepared by mixing of 2.5 mg/ml silk protein solution with 2 M potassium phosphate, pH 8 at a volumetric ratio of 1:10. The amount of the spheres was determined gravimetrically.
Scanning electron microscopy
A drop of the sphere suspension was dried on a silicon wafer or a glass cover slide attached to a Standard error of the mean (SEM) stub. The samples were sputter coated with Cr for 40 s in Quorum Sputter Coater Q150T ES (Quorum Technologies, UK). Spheres were observed using a JEOL JSM-7001F field emission scanning electron microscope (JEOL. Ltd, Tokyo, Japan) at 5 kV accelerating voltage. Sphere size was analyzed using ImageJ software. At least 100 individual spheres were measured for each sample. Measurements were performed on independent SEM samples at least three-times.
Fourier transform infrared spectroscopy
A drop of silk protein solution (5 mg/ml) or sphere suspension in water was air-dried on a CaF2 crystal. FTIR spectra were obtained using a Bruker Equinox 55/S FTIR spectrometer (Bruker Corp., CA, USA) equipped with a deuterated triglycine sulfate detector and multiple reflection horizontal MIRacle ATR attachment with a diamond crystal. For each measurement, 128 scans were averaged at a resolution of 2 cm-1 within a wavenumber range of 600–4000 cm-1. Spectral analysis and curve fitting were performed using Opus 5.0 software (Bruker Corp.). The second derivative was obtained from the amide I spectra region (range from 1595 to 1705 cm-1) using a third-degree polynomial function. Fourier self-deconvolution of the amide I band was performed using the Lorentzian line slope, half-bandwidth of 25 cm-1 and noise reduction factor of 0.3. The spectra were curve-fitted using mixed Gaussian and Lorentzian peak shapes set in positions determined by the second derivative spectral minima. The secondary structure components were assigned to the amide I band components according to previously published data: 1605–1615 cm-1 for tyrosine side chains, 1616–1637 cmp-1 and 1697–1705 cm-1 for beta sheets, 1638–1655 cm-1 for random coils, 1665–1662 cm-1 for helices, and 1663–1696 cm-1 for turns [16]. The percentage of each secondary structure was assessed by totaling the integrals of the associated peaks and normalizing them to the total amide I integral. The experiment was repeated three-times.
Zeta potential measurements
Zeta potential (ZP) was measured with a Zetasizer Nano XS (Malvern Instruments Ltd., UK). A sphere suspension in water (750 μl) was loaded into a capillary cell, and three measurements were performed at 25°C according to the manufacturer's instructions. The experiment was repeated three-times.
Focused ion beam scanning electron microscopy
Silk particle suspension (10 μl) was applied on a silicon wafer and dried overnight under ambient conditions. The samples were coated with a thin layer (10 nm) of Pt/Pd using a sputter coater (208HR, Cressington Scientific Instruments Inc., PA, USA). Imaging was performed using a Zeiss NVision 40 dual-beam focused ion beam scanning electron microscope (Carl Zeiss SMT Inc., MA, USA). A focused beam of gallium ions (Ga+, 30 keV) was used to cut cross-sections of the silk particles. Specimen were imaged in situ before, during and after FIB milling using the field emission SEM system operating at 2 kV.
Drug loading & release
MS1 and MS2 spheres were loaded with three anticancer drugs: doxorubicin (Medac, Hamburg, Germany), mitoxantrone (Oncotron, Mumbai, India) and etoposide (Ebewe, Unterach, Austria) using either the preloading or postloading method.
The preloading method was applied for loading of mitoxantrone and etoposide to both sphere types. In this method, 50 μl of protein solution at a concentration of 5 mg/ml was initially mixed with 50 μl of a drug solution at a concentration of 2 mg/ml and then mixed with 1000 μl of 2 M potassium phosphate (pH 8). The spheres were incubated overnight at room temperature. Next, samples were centrifuged for 15 min at 10,000 × g, and the amount of drug loaded was determined spectrophotometrically.
The postloading method was applied to load doxorubicin, mitoxantrone and etoposide to both sphere types. The same amount of spheres as was used in the preloading method (250 μg) was suspended in 250 μl of phosphate buffer and mixed with 50 μl of a drug solution at a concentration of 2 mg/ml. The spheres were then incubated overnight with the drugs at room temperature. The spheres were centrifuged for 15 min at 10,000 x g, and the drug concentration in the supernatant was measured spectrophotometrically. The drug absorbance in the supernatants was measured at wavelengths of 508, 610 and 230 nm for doxorubicin, mitoxantrone and etoposide, respectively. Quantification of the drugs was based on standard calibration curves made for each drug. A corresponding buffer (potassium phosphate or PBS) was used as a reference. The drug loading efficiency was calculated using following equation:
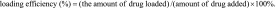 |
For the drug release study, drug-loaded spheres were suspended in 1 ml of phosphate-buffered saline (PBS) at a pH of 4.5, 6.0 or 7.4 and incubated at 37°C with agitation. PBS was collected and replaced at specific time points. The drug concentration in PBS was measured spectrophotometrically as described above. The experiment was repeated three-times.
Cytotoxicity assay
Mouse fibroblast cells (NIH 3T3) were maintained in DMEM medium (Sigma, MO, USA) supplemented with 10% FBS (Sigma) and 80 μg/ml gentamycin (KRKA Nove Mesto, Slovenia). Cells were grown in a humid atmosphere of 5% CO2 at 37°C. For the cytotoxicity assay, cells were seeded onto a 96-well plate (25 × 103 cells/well) and incubated for 24 h. Silk spheres were resuspended in the culture media at appropriate concentrations and then added to the cells. The particles were incubated for 48 h within the cell culture and then the MTT cytotoxicity assay was performed as described previously [11] by using MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; Sigma, MO, USA). The percentage of cell viability was calculated referring to the untreated control sample. The experiment was repeated three-times.
Statistics
The statistical significance of the differences between sphere groups was calculated using analysis of variance. Post hoc tests with the Bonferroni correction were performed. The differences in encapsulation efficiency were tested using an unpaired t-test. The differences between groups were considered significant if the p-value was less than 0.05.
Results
Expression & purification of recombinant spider silk proteins
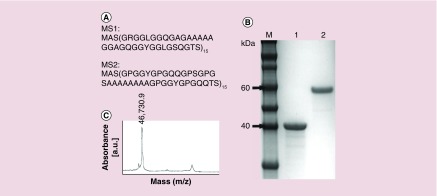
MS1 and MS2 bioengineered spider silk proteins (Figure 1A) were purified by the thermal extraction method (80/20). The average yield was 1.71 and 1.57 mg of silk per gram of bacterial pellet for the MS1 and MS2 proteins, respectively. SDS-PAGE analysis indicated that the proteins were free from impurities and nondegraded (Figure 1 B). SDS-PAGE analysis indicated a higher-than-predicted molecular weight for MS2; however, MALDI-TOF mass spectrometry results confirmed the calculated value (46.72 kDa) (Figure 1C).
Figure 1. . Analysis of MS1 and MS2 proteins.
(A) Amino acid sequences of MS1 and MS2. Both proteins consist of 15 repeats of the monomeric units derived from the consensus motifs of the dragline silk proteins of the N. clavipes spider: MS1 from MaSp1 and MS2 from MaSp2. (B) SDS-PAGE analysis of the purified bioengineered spider silk. Lines: 1 - MS1, 2 - MS2, M-Novex sharp protein molecular mass marker (Live Technologies). (C) MALDI-TOF spectrum of the MS2 protein.
Morphology & size of MS1 & MS2 spheres
The silk spheres were formed upon mixing the soluble bioengineered spider silk proteins with potassium phosphate buffer. Particle formation was observed as an immediate increase of the turbidity of the mixed solution. The size and morphology of the spheres were dependent on the type of the protein and particle preparation conditions (Figures 2 & 3).
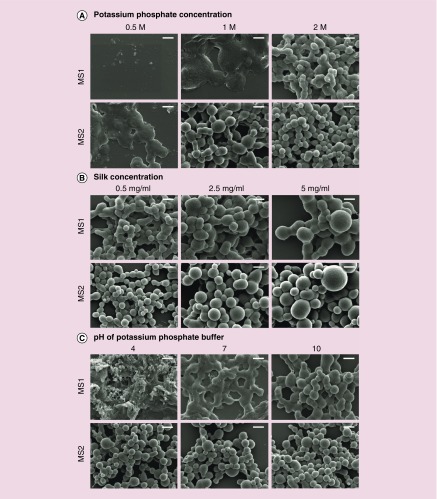
Figure 2. . Scanning electron microscope images of the MS1 and MS2 spider silk spheres.
(A) Particles formed using an initial silk concentration of 0.5 mg/ml and potassium phosphate buffer (pH 8) at different concentrations: 0.5, 1 or 2 M. (B) MS1 and MS2 particles produced at different initial silk concentrations: 0.5, 2.5 or 5 mg/ml, mixed with 2 M potassium phosphate (pH 8). (C) Particles produced using an initial silk concentration of 0.5 mg/ml and 1.5 M potassium phosphate buffer at pH 4, 7 or 10. Scale bars: 1 μm. Representative scanning electron microscope images from three independent experiments are shown.
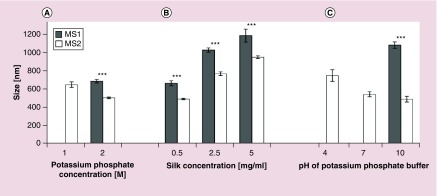
Figure 3. . The average size of MS1 and MS2 silk particles as determined from scanning electron microscope images.
(A) Particles formed at an initial silk concentration of 0.5 mg/ml using potassium phosphate buffer (pH 8) at different concentrations: 1 and 2 M. (B) Particles produced at different initial silk solution concentrations: 0.5, 2.5 and 5 mg/ml, mixed with 2 M potassium phosphate (pH 8). (C) Particles produced using an initial silk concentration of 0.5 mg/ml and 1.5 M potassium phosphate buffer at pH 4, 7 or 10. Data are presented as the mean of three independent experiments. Error bars show the standard deviations.
***p < 0.001.
The concentration of potassium phosphate was critical to sphere formation as no particles were observed at a potassium phosphate concentration of 0.5 M for the MS1 protein, whereas the MS2 protein formed nonspherical aggregates under the same conditions (Figure 2A). Similar aggregates of the MS1 protein were observed at 1 M potassium phosphate. Spherical particles of the MS1 protein were formed at the highest concentration of 2 M, while MS2 spheres were formed at a potassium phosphate concentration of 1 M. In general, as the phosphate concentration increased, both particle types improved in terms of their spherical shape and monodispersity, and the size of the MS2 particles decreased (Figures 2A & 3).
A 2 M solution of potassium phosphate was used to form the spheres at different silk concentrations. For both the MS1 and MS2 proteins, sphere size increased together with the initial protein concentration (Figures 2B & 3). The mean size of the MS1 particles prepared at the lowest (0.5 mg/ml) initial silk concentration was 0.667 ± 0.137 μm and increased to 1.167 ± 0.341 μm at the silk concentration of 5 mg/ml. The MS2 particle mean size was slightly smaller at each silk concentration and ranged from 0.49 ± 0.13 μm (at 0.5 mg/ml silk) to 0.95 ± 0.26 μm (at 5 mg/ml) (Figures 2B & 3).
Phosphate buffer adjusted to pH values of 4, 7 and 10 at a concentration of 1.5 M were used to form spheres at an initial silk concentration of 0.5 mg/ml. At pH 4, amorphous aggregates were observed, and no particles were found for the MS1 protein. The morphology of the MS1 particles improved at pH of 10. The pH value did not influence the morphology of the MS2 spheres (Figures 2C & 3).
SEM images (Figure 2) indicated a difference in the morphology between the MS1 and MS2 spheres. MS1 spheres had a tendency to aggregate, whereas MS2 particles were dispersed and presented a well-defined spherical shape with a smooth surface. Moreover, the difference in the average size of the spheres formed by MS1 and MS2 was significant for spheres prepared at the same processing conditions; furthermore, the MS2 spheres were smaller (Figure 3).
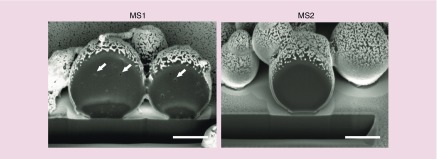
Focused ion beam cross-sections of the particles revealed pores inside of the sphere cores of the MS1 variants. MS2 spheres showed a solid core, and no pores were observed (Figure 4).
Figure 4. . Representative images of FIB-milled MS1 and MS2 particles observed using an FESEM microscope.
Spheres were produced using an initial silk concentration of 5 mg/ml and 2 M potassium phosphate (pH 8). Pores inside of MS1 particles are marked with white arrows. Scale bar: 1 μm.
Secondary structure analysis of MS1 & MS2 spheres
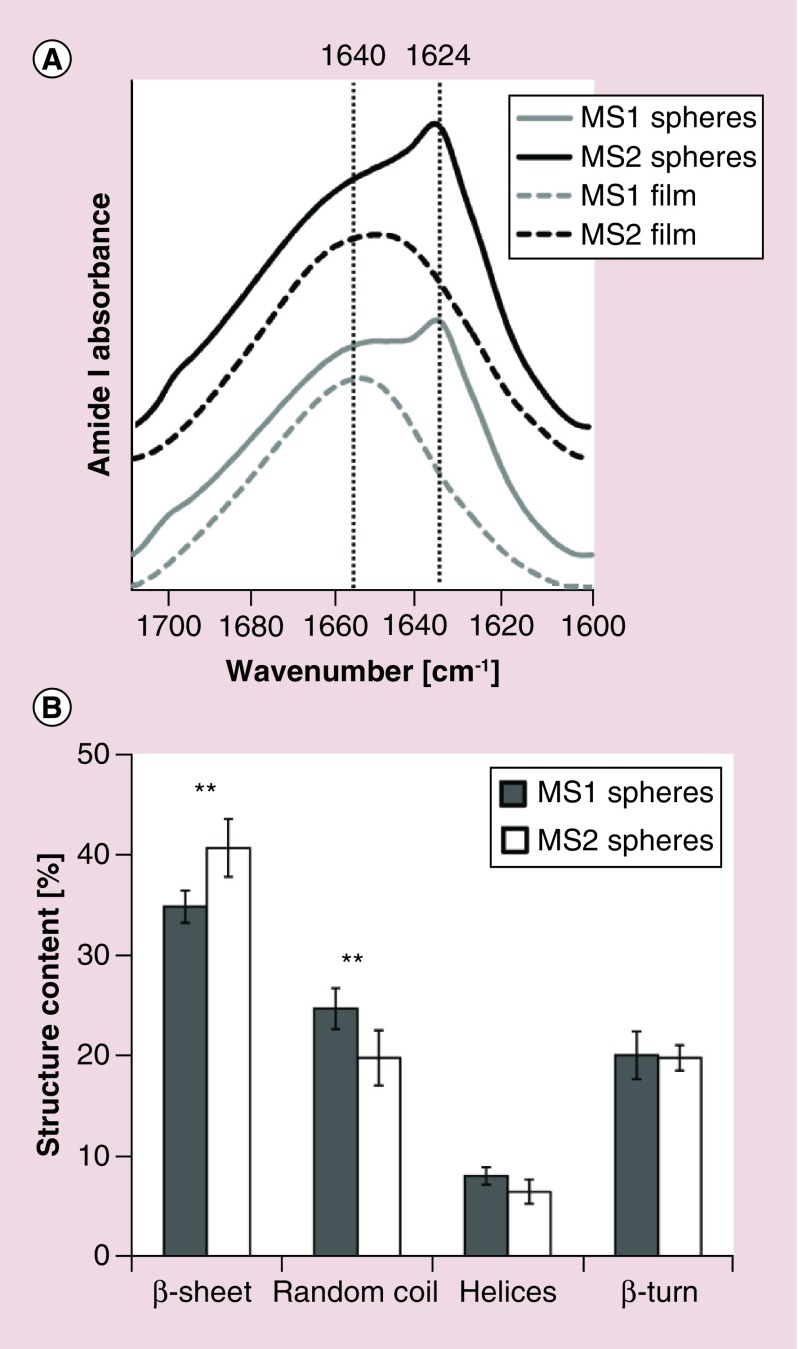
The secondary structure composition of the silk spheres and films formed by air-drying silk protein solutions were estimated using amide I region analysis of the Fourier deconvolved infrared spectra. The original spectra of both particle types showed an increase in absorption at 1623 cm-1 (Figure 5A, solid lines) compared with the spectra of the MS1 and MS2 films (dashed lines), indicating an increase in the beta-sheet structure content. The percentage of secondary structures in the silk particles was deduced from 11 peaks contributing to the amide I band (1600, 1612, 1623, 1634, 1645, 1653, 1660, 1667, 1676, 1686 and 1697 cm-1) that were assigned to a particular secondary structure or a side chain absorbance according to the literature data [16]. The secondary structure content was significantly different between the two types of silk spheres with respect to beta sheet and random coil content, indicating that the MS2 particles were more crystalline than the MS1 particles. The percentages of helices and beta turns were similar in both analyzed particle types (Figure 5B).
Figure 5. . Fourier transform IR spectra and secondary structure composition of the MS1 and MS2 spheres.
(A) FTIR absorbance spectra in the amide I region of MS1 and MS2 silk particles (solid lines) and films formed by drying MS1 and MS2 proteins on a CaF2 crystal (dashed lines). (B) Secondary structure content of both types of particles after the Fourier self-deconvolution of the amide I region of the FTIR spectra is shown. Data are presented as the mean of three independent experiments. Error bars show the standard deviations.
**p < 0.01.
FTIR: Fourier transform IR.
Drug loading potential of MS1 & MS2 spheres
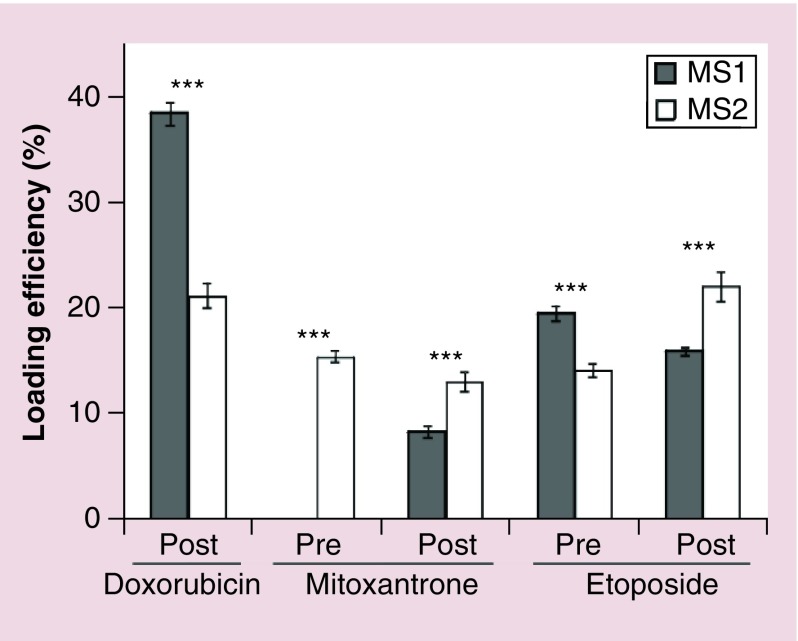
The incorporation of anticancer drugs was tested using spheres prepared by two methods: preloading and postloading. The postloading of doxorubicin into MS1 spheres showed a nearly two-fold higher loading efficiency than its loading into MS2 spheres (38 and 21%, respectively). The preloading of mitoxantrone resulted in no detectable encapsulation of the drug in MS1 spheres and a 15% loading efficiency in MS2 spheres. The postloading of mitoxantrone resulted in a significantly higher incorporation of the drug in MS2 spheres than in MS1 particles. Etoposide was loaded in an efficiency range from 14 to 22% using both methods for both variants of spheres. For MS1 particles, the loading of etoposide was higher using the preloading method, while for MS2, the postloading method was superior (Figure 6). The loading efficiencies differed significantly between the two silk types.
Figure 6. . The loading efficiency of three anticancer drugs (doxorubicin, mitoxantrone and etoposide) into MS1 and MS2 spheres prepared using the preloading method (pre) and postloading method (post).
In the preloading method, a drug was added to the protein solution, and then spheres were formed by mixing the resulting solution with 2 M potassium phosphate (pH 8). In the postloading method, the drug was added to a suspension of particles formed by mixing silk protein at an initial concentration of 2.5 mg/ml with 2 M potassium phosphate (pH 8). The mean of three independent experiments is shown. Error bars show the standard deviation.
***p < 0.001.
Loading doxorubicin into both types of spheres significantly influenced their ζ potentials (Table 1). In contrast to the negatively charged MS2 spheres, doxorubicin-loaded MS2 spheres had positive ζ potential, in other words, after doxorubicin loading, the charge of the MS2 spheres was reversed from negative to positive. Moreover, the ζ potential of MS1 spheres postloaded with doxorubicin was significantly more positive than the ZP of unloaded MS1 spheres. Postloaded and preloaded mitoxantrone significantly increased the ζ potential only of MS1 spheres, whereas etoposide did not significantly modify the ZP of either sphere type loaded by either methods.
Table 1. . ζ potential of MS1 and MS2 silk spheres with and without drug (doxorubicin, mitoxantrone and etoposide).
| ζ potential (mV ± SD) | ||||
|---|---|---|---|---|
| Spheres | MS1 | MS2 | ||
| Without drug | 4.5 | ±2.7 | -13.7 | ±3.0 |
| Drug preloading | ||||
| Mioxantrone | 9.0 | ±1.1* | -13.7 | ±1.1 |
| Etoposide | 5.7 | ±0.7 | -14.6 | ±3.3 |
| Drug postloading | ||||
| Doxorubicin | 14.5 | ±0.8*** | 16.2 | ±0.8*** |
| Mioxantrone | 10.1 | ±2.1** | -9.7 | ±2.1 |
| Etoposide | 4.1 | ±2.1 | -9.5 | ±0.9 |
Spheres were loaded using the pre- and postloading methods. ζ potential was measured immediately after loading. The means and standard deviations of three independent experiments are shown.
******p < 0.05; p < 0.002; p < 0.001.
Drug release from MS1 & MS2 spheres
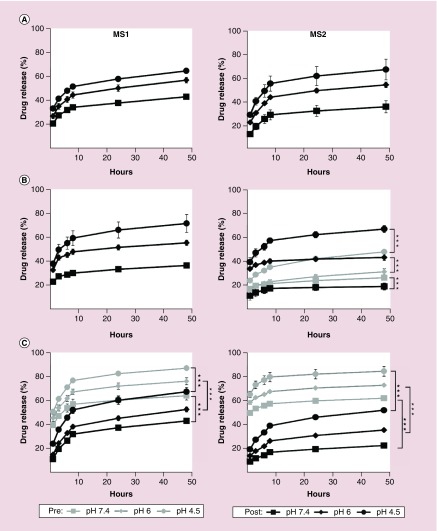
Release profiles of the incorporated drugs were studied over 48 h at three different pH values: 7.4, 6 and 4.5. All drugs were released in a pH-dependent manner from both types of spheres (Figure 7).
Figure 7. . Release of anticancer drugs incorporated into MS1 and MS2 particles over 48 h at 37°C in phosphate-buffered saline adjusted to pH 7.4, 6 or 4.5. The drugs were incorporated by the preloading method (Pre) or the postloading method (Post) as described in the materials and methods section.
The release of (A) doxorubicin, (B) mitoxantrone, and (C) etoposide is demonstrated. Data are expressed as the mean of three independent experiments with standard deviation error bars.
***p < 0.001.
After 48 h, similar release of doxorubicin from MS1 and MS2 spheres was observed at pH 4.5 and 6; however, at pH 7.4, the amount of drug released from the MS1 particles was higher than the amount released from the MS2 particles (Figure 7A).
Postloaded mitoxantrone was released more quickly from MS1 spheres than from MS2 spheres (Figure 7B). For the MS2 spheres at pH 6 and 4.5, the release of preloaded mitoxantrone was lower than that of postloaded mitoxantrone.
Preloaded etoposide showed burst release from both types of the spheres and in all release buffers, although this profile was more evident for MS2 spheres (Figure 7C). Etoposide loaded into MS1 and MS2 spheres using the postloading method showed a lower release profile than the same drug loaded using the other method; meanwhile, MS1 spheres released etoposide faster than MS2 spheres.
Cytotoxicity of the MS1 & MS2 spheres
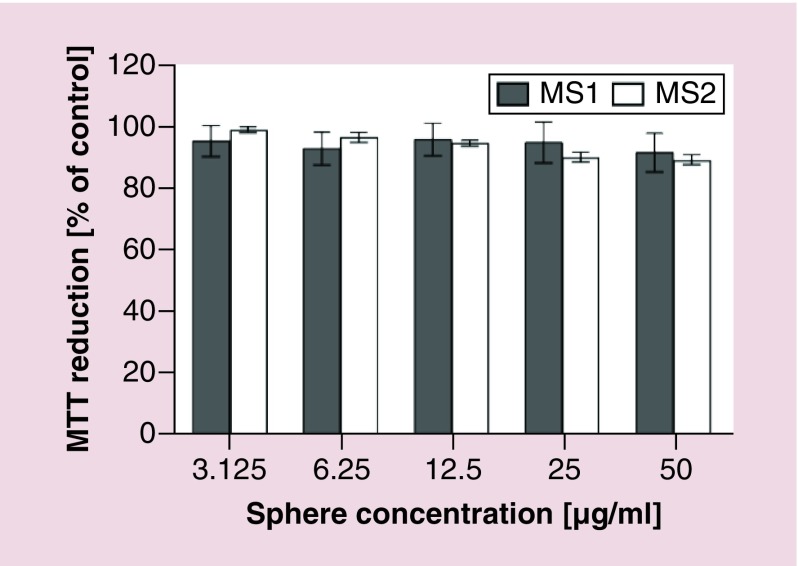
NIH/3T3 cell viability was assessed using the MTT assay. Both MS1 and MS2 particles did not show a cytotoxic effect within the range of concentrations tested (Figure 8).
Figure 8. . Mitochondrial activity of NIH/3T3 cells assessed using MTT assay after a 48-h incubation with the silk particles.
The % MTT reduction was calculated in reference to nontreated control cells. Data represent the mean of three independent experiments, while error bars show the standard deviation.
Discussion
In nature, the amino acid composition of spidroins underlies the diversity of silks and their properties. In the case of bioengineered spider silks, their amino acid composition is a key factor determining the physical properties of silk polymers [10,17]. Thus, the behavior of MS1 and MS2 proteins during particle formation and differences in sphere properties are directly related to their amino acid composition. Additionally, our previous findings indicated that the purification method plays an important role in the final properties of spheres made of bioengineered spider silk [12].
The natural analogs of the studied proteins, in other words, the MaSp1 and MaSp2 spidroins of N. clavipes, share a simple amino acid composition based on alanine and glycine [17]. These two main components are organized in separate blocks: alanine-rich domains of four to seven residues and glycine-rich regions, mostly GGY, GGL and GGQ motifs. Both blocks occur repeatedly and alternately in the protein sequence. At the N- and C-termini, the nonrepetitive, very conservative domains occur [17]. In contrast to the MaSp1 protein, MaSp2 contains proline residues that form GPGGX motifs within a glycine-rich region of the sequence [18]. Proline provides rigidity to the protein chain and acts as a disruptor of the secondary structures by imposing certain torsion angles of the polypeptide chain [19]. The GPGGX motifs form beta-turns that, when combined, can assemble into a beta-spiral structure [20]. The presence of these residues in the MaSp2-derived sequence of the MS2 protein could be one of the sources of the differences in the properties of the silk spheres.
In this work, MS1 and MS2 silk proteins formed solid spherical particles under salting-out conditions provided by mixing the dissolved protein with kosmotropic phosphate ions. Increasing the phosphate concentration above a critical level (which was different for the two studied proteins) induced a phase separation between the solvent-rich and protein-rich phases, followed by precipitation of the silk proteins and formation of hydrophobic interactions between silk protein chains. The interacting proteins underwent structural rearrangements that led to beta-sheet structure formation, stabilizing the silk spheres [8]. This salting-out model was previously proposed by Slotta et al. for the bioengineered spider silk eADF4(C16), which was based on the spidroin of A. diadematus: ADF4 [8]. Our experiments that showed the process of salting out at higher potassium phosphate concentrations suggested that MS2 protein polymerizes more rapidly than MS1 and that the structures formed by this protein have a lower tendency to aggregate. MS2 particles formed smooth, tightly packed spherical particles. As indicated by their FTIR spectra, the beta-sheet content (crystallinity) increased after sphere assembly for both the MS1 and MS2 silks; however, for MS2 spheres, this increase was significantly higher. Previous findings indicated that the number and length of polyalanine blocks influences the crystallinity of silk-derived bioengineered proteins [21]. Moreover, in the case of a polyalanine sequence combined with polyethylene glycol, it was indicated that the longer the polyalanine segment, the greater the tendency to self-assemble into beta-sheet aggregates [22]. Given the equal amount of polyalanine motifs (15 for both proteins), the higher crystallinity of the MS2 spheres can be explained by the greater number (8 vs 5) of alanine residues in one repeat. The high beta-sheet content and presumed presence of beta-spirals in MS2 spheres could lead to the tight packing of the protein matrix of the particles [21]. In contrast, MS1 spheres could present a looser packaging of the protein chains within the core of the particles, which could explain the 30% greater size of these particles and the pores observed inside of the MS1 particles in cross-sectional images. Moreover, the greater number of alanine residues in MS2 protein could cause faster kinetics of beta-sheet formation; thus, a lower potassium phosphate concentration was needed to form MS2 particles. MS1 protein, which does not contain proline, showed the ability to form spherical particles, indicating that proline content may influence but does not determine the ability of a protein to assemble into spheres. The study based on the modification of amino acid sequence (addition/substitution/deletion of single amino acid) is reasonable next step in the silk science. It could determine the critical amino acid residues in the silk sequence that determines the property of silk. However, it was not within the scope of this study.
As expected from the calculated isoelectric points of the MS1 and MS2 proteins, which were equal to 10.83 and 5.27, respectively, in ddH2O the MS1 particles showed a positive ZP value, whereas the MS2 particles were negative. The ζ potential value of the MS1 particles indicated their low colloidal stability. Thus, weak repulsion between particles might contribute to the aggregation behavior of the MS1 spheres [23].
Since, amino acid composition determines the properties of bioengineered silk, it is hard to compare our spheres to particles prepared from different silks. The most relevant data concern particles composed of eADF3 and eADF4(C16), which are derived from the spidroins of A. diadematus – ADF3 and ADF4, respectively [10]. Spherical aggregates of eADF3 disassembled upon the replacement of potassium phosphate buffer with distilled water. Moreover, it was proposed that eADF3 particles have hydrophilic side chains that project out from the spheres and form dangling ends, which can mediate interactions with neighboring aggregates [10]. The eADF4(C16) spheres were dense packed with ‘smooth’ surfaces, in other words, presumably without the dangling ends, and were chemically stable [10]. Based on these characteristics, eADF3 spheres are more closely related to MS1 particles, while eADF4(C16) are more closely related to MS2 spheres. Another similarity is that the polyalanine segment is longer in eADF4(C16) and MS2 than in eADF3 and MS1 (8 and 8 vs 6 and 5 residues, respectively). However, based on the presence of proline in the ADF3 and ADF4 proteins, it was proposed that they are both the MaSp2-type proteins [24]. By contrast, Huemmerich et al. proposed that based on the overall hydrophobic nature of spidroins, we can distinguish two types of proteins: MaSp1 and ADF4, which display relatively high hydrophobicity and have approximately 1% charged residues, and MaSp2 and ADF3, which are more hydrophilic and have less than 1% charged residues [24]. Although, in nature ADF3 and ADF4 correspond more closely to MaSp2 and MaSp1, respectively, their bioengineered variants behaved oppositely in terms of the properties of the spherical particles. However, it needs to be mentioned that bioengineered MS1, MS2 and eADF4(C16) comprised corresponding repetitive motifs, while eADF3 was based on a repetitive motif followed by the nonrepetitive region NR3 [10]. For fiber formation, the presence of N- and C-termini was substantial; the fibers made of bioengineered silks containing N- and C-domains revealed significantly different mechanical properties from fibers made of the corresponding ‘plain’ bioengineered silks [25].
To date, bioengineered spider silks have been shown to form vehicles for the delivery of small-molecule drugs or macromolecules, such as proteins, polysaccharides or nucleic acids [13,26–31]. The proposed mechanism for drug release from biodegradable biopolymers usually combines mechanisms such as drug diffusion through pores in a polymer matrix, degradation of the biopolymer and swelling. The initial burst release is caused by a washing out of the fraction of drug weakly bound to the particle surface or between the particle clusters, while the subsequent gradual release is caused by the equilibrium-based diffusion of the drug from the biopolymer matrix to the particle surface and then to the release medium [9,26]. Blüm and Scheibel showed that release kinetics can also be modified by crosslinking the protein molecules that form eADF4(C16) spheres [29]; however, the use of crosslinkers increases the risk of carrier toxicity. The enhanced drug loading and better release kinetics may also be achieved by functionalization of silk by blending with other compounds, chemical conjugation and genetic engineering. These modifications may render a control over desired properties for different purposes, as previously [32–35]. However, after such modifications, the silk-assembly property and biocompatibility may be an issue of concern. Most of the previous studies have focused on model drugs. Here, we employed three chemotherapeutics and two loading methods to compare the loading capacity and the release kinetics from the MS1 and MS2 spheres. Loading was tested by preparing the particles using two methods: preloading, where the drug was incorporated during the sphere formation process in a high molarity phosphate environment, and diffusion-driven postloading, which was conducted under aqueous conditions. The influence of the pH on the drug release kinetics was studied at pH 7.4, 6 and 4.5, which corresponded to the pH values that occur in biological fluids (blood plasma), endosomes and lysosomes, respectively.
In this study, we found that the drug loading efficiency depended on the loading method and differed significantly between the two protein types. During the preloading of doxorubicin, the drug co-precipitated with the silk protein, rendering the spectrometric measurement of unbound drug unreliable (data not shown). Thus, only the postloading efficiency of doxorubicin was explored. The results from the diffusion-driven postloading method indicated that the positively charged MS1 spheres had greater potential to incorporate the positively charged doxorubicin than the negatively charged MS2 spheres. These results suggest that electrostatic attraction between the drug and the silk, which was previously suggested to be a major driving force for drug loading [26], plays a minor role in the case of doxorubicin. Here, two factors could contribute to better loading efficiency of doxorubicin in the MS1 spheres. First, the solid, condensed protein core of the MS2 particles could pose an obstacle to the diffusion-based loading of doxorubicin. The drug could be electrostatically attracted to the MS2 sphere surface, but it could fail to penetrate into the core, which would make it susceptible to being washed out by the release buffer. Indeed, the loading of doxorubicin significantly modified the ζ potential value of the silk spheres: it increased the positive surface charge of the MS1 spheres and inverted the negative ζ potential of the MS2 spheres, making them positive. This corroborated the presence of doxorubicin on the surfaces of both sphere types. On the other hand, the looser packing of the MS1 spheres and their inner pores could enable the penetration of drug molecules through the sphere. The second factor could be associated with the difference in hydrophobicity between the two silk proteins. The grand average of hydropathicity (GRAVY) value of the MS1 protein indicates that it is more hydrophobic than MS2 (-0.145 vs -0.535, respectively) and suggests that it can form stronger interactions with hydrophobic drug molecules.
The release kinetics of doxorubicin were accelerated at low pH values, as reported previously [6,11]. The drug properties could influence the release process. It has been shown that low pH promotes the transition of a drug to its soluble form, which increases its release rate from a carrier [36]. Moreover, the influx of protons at low pH may compete with the hydrogen bonding between doxorubicin molecules and silk, weakening the drug-silk interactions and increasing the rate of drug release. The most important observation concerns the doxorubicin release profile at pH of 7.4; because of its lower release rate than the MS1 spheres, the MS2 spheres are more suitable for carrying this drug through blood plasma.
In contrast to doxorubicin, mitoxantrone was incorporated with a higher efficiency in MS2 particles than in MS1 particles using both loading methods. The ζ potential value of the MS2 spheres did not increase significantly after loading; thus, the drug did not accumulate considerably on the surface of the particles and probably penetrated into the sphere cores. Postloaded drug exhibited a slower release from the MS2 spheres, especially at pH 6.0 and 7.4, than from the MS1 spheres, indicating its higher affinity to the MS2 protein. Since, mitoxantrone is more hydrophilic than doxorubicin (the theoretical logP values are 1 [experimental -3.1] and 1.3 for mitoxantrone and doxorubicin, respectively) and carries two positive charges, a charge-mediated interaction of mitoxantrone with negatively charged MS2 can be assumed. This could partially explain the stronger affinity of mitoxantrone for the negatively charged MS2 protein than for positively charged MS1. Moreover, the faster drug release at pH 4.5 from the MS2 spheres could be partially explained by enhancing the repulsive interactions between mitoxantrone and MS2 (the Ip of MS2 is 5.27; thus, at a pH of 4.5, it should be protonated). Interestingly, no drug was loaded into the MS1 spheres when using the preloading method. Although, the ζ potential value of the MS1 spheres increased significantly, indicating some accumulation of the drug on the surface of the spheres, the spectrophotometric reading was below the detection level. During preloading at pH 8, electrostatic repulsion between the positively charged drugs and cationic MS1 protein could occur. Moreover, mitoxantrone forms dimers in a pH-dependent process, and they are favored in a basic environment [37]. The higher dissociation of mitoxantrone dimers induced by anionic surfactants than that induced by cationic surfactant resulted in a higher encapsulation efficiency of the monomer form of mitoxantrone [37]. A similar effect could occur in the presence of the negatively charged MS2 protein. The charge-mediated interactions between mitoxantrone and MS2 protein could recruit some fraction of the mitoxantrone molecules and hence prevent their dimerization under the preloading conditions, resulting in 15% loading efficiency. In contrast to the postloaded drug, preloaded mitoxantrone was slowly released from MS2 spheres, indicating deeper penetration of the drug into the sphere structure.
Etoposide was incorporated in a range from 14 to 22% for both particle types and using both loading methods. Etoposide is a neutral compound with a logP value of 0.6. Under the postloading conditions, its affinity for the more hydrophilic MS2 protein was significantly higher than for MS1; however, in the preloading environment, the loading efficiency of the MS1 spheres was significantly higher than the loading efficiency of the MS2 spheres. The release experiments showed that the postloading method resulted in a slower release profile of etoposide, whereas preloaded drug showed a burst release. This suggests that the aqueous postloading conditions are more favorable for establishing stable silk-etoposide interactions. Preloaded etoposide probably accumulated on the surface of the spheres, and the interaction of the drug with silk protein was weak, which was also suggested by the burst release of the preloaded drug within the first few hours.
Conclusion
Our results indicated that spheres made of bioengineered spider silks can be utilized as anticancer drug carriers. The structure, charge and hydrophobicity of both the drug and silk played a crucial role in the drug loading efficiency and release kinetics. MS2 particles presented favorable properties in terms of morphology and colloidal stability. The MS2 particles loaded using the preloading method showed greater applicability for mitoxantrone delivery. Moreover, because they showed the highest postloading efficiency and the slowest release, MS2 particles were best for etoposide delivery. However, because of the more than two-fold higher loading efficiency and slow kinetics of release, MS1 spheres were a better choice for doxorubicin delivery than MS2 spheres.
The silk biomaterial is biocompatible, biodegradable and mechanically durable. The sphere formation process occurs under ambient and aqueous conditions that enable control over particle size and simultaneous loading of anticancer drugs. Moreover, the postloading method of drugs can be used to stably incorporate chemotherapeutics into silk spheres. The bioengineering of the silk sequence provides a unique opportunity to control its material properties, such as charge, hydrophobicity and secondary structure, on the molecular level [17]. Moreover, the addition of a functional domain can provide specificity to the drug delivery system [11,38]. Owing to the use of genetic engineering, the properties of bioengineered silk can be tailored to develop the delivery system with optimal drug loading and release, which is of great importance for the delivery of chemotherapeutics in cancer therapies and beyond.
Future perspective
Spider silk has been considered as a material with a great potential in biomedical applications. The bioengineered spider silk technology can be further developed in order to obtain an optimal drug delivery system for cancer therapies. Recent approaches in bioengineered silk technology encompass functionalization of silk molecules by adding sequences encoding the desired function, for example, ligands that target tumor. This combined with the possibility of controlling silk carrier properties by modification of the silk sequence, by applying desired purification procedure and by blending two different bioengineered spider silk proteins, makes such hybrid particles a very promising strategy for smart delivery of therapeutics for cancer treatment. Moreover, silk-based drug delivery is not limited to the treatment of cancer, but it has much broader application for treating other diseases, not yet investigated. However, there are still some issues that need to be addressed in the near future in order to implement this technology. The study based on the modification of amino acid sequence (addition/substitution/deletion of single amino acid) is a reasonable next step in the silk science. It could determine the critical amino acid residues in the silk sequence that determine the property of silk. The delivery systems have to be easily prepared using an adaptable and up-scalable technique. Thus, for the future drug delivery purposes, the optimization and standardization of the particle preparation are highly needed. The targeted-delivery concept based on the silk spheres and feasibility of the system has already been verified in the in vitro preliminary studies. Future work should, therefore, focus on in vivo studies to verify the potential benefits of these biomaterials. Further, in vivo research concerning toxicity, immunogenicity, biodistribution and anticancer efficacy of silk spheres is of a great importance.
Summary points.
The difference in amino acid composition of silks determined the processing conditions and physical properties of the silk spheres and the loading/release of the drugs.
MS2 spheres were formed at lower concentration of kosmotropic ions, possessed higher beta-sheet structure content, presented a better-defined spherical shape with a smooth surface and a solid core (no pores were observed) in comparison with MS1 particles.
The MS2 particles were more dispersed and smaller than MS1 particles.
MS2 particles were less hydrophobic and of opposite charge (negative) comparing with MS1 spheres what determined the loading and release of drugs.
Negatively charged MS2 particles loaded by the preloading method showed greater applicability for the delivery of the positively charged mitoxantrone comparing with MS1 spheres.
The post loaded MS2 particles were superior for neutral etoposide delivery compared with MS1 spheres.
The more hydrophobic MS1 spheres were a better choice for delivery of the most hydrophobic drug (doxorubicin) in comparison to MS2.
By associating the properties of silk spheres and drugs the optimal drug delivery system can be developed.
Acknowledgements
We would like to thank R Abbott-Beauregard for technical assistance.
Footnotes
Financial & competing interests disclosure
A Florczak and K Jastrzebska were supported by the International PhD Projects Programme of Foundation for Polish Science operated within the Innovative Economy Operational Programme (IE OP) 2007–2013 within European Regional Development Fund. The project was supported by grant from The National Science Centre, Poland (2014/15/B/NZ7/00903). We also thank the National Institutes of Health for support (P41EB00252, R01AR068048). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Hardy JG, Römer LM, Scheibel TR. Polymeric materials based on silk proteins. Polymer. 2008;49(20):4309–4327. [Google Scholar]
- 2.Jastrzebska K, Kucharczyk K, Florczak A, Dondajewska E, Mackiewicz A, Dams-Kozlowska H. Silk as an innovative biomaterial for cancer therapy. Rep. Pract. Oncol. Radiother. 2015;20(2):87–98. doi: 10.1016/j.rpor.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Illustrates the types of silk-based biomaterials that can be used to for cancer treatment and to study the biology of cancer.
- 3.Chen M, Shao Z, Chen X. Paclitaxel-loaded silk fibroin nanospheres. J. Biomed. Mat. Res. A. 2012;100(1):203–210. doi: 10.1002/jbm.a.33265. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Wenk E, Matsumoto A, Meinel L, Li C, Kaplan DL. Silk microspheres for encapsulation and controlled release. J. Control Release. 2007;117(3):360–370. doi: 10.1016/j.jconrel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 5.Mitropoulos AN, Perotto G, Kim S, Marelli B, Kaplan DL, Omenetto FG. Synthesis of silk fibroin micro- and submicron spheres using a co-flow capillary device. Adv. Mater. 2014;26(7):1105–1110. doi: 10.1002/adma.201304244. [DOI] [PubMed] [Google Scholar]
- 6.Seib FP, Jones GT, Rnjak-Kovacina J, Lin Y, Kaplan DL. pH-dependent anticancer drug release from silk nanoparticles. Adv. Health. Mat. 2013;2(12):1606–1611. doi: 10.1002/adhm.201300034. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates the pH-dependent release kinetics of doxorubicin from silk spheres as a cancer treatment.
- 7.Wang X, Yucel T, Lu Q, Hu X, Kaplan DL. Silk nanospheres and microspheres from silk/PVA blend films for drug delivery. Biomaterials. 2010;31(6):1025–1035. doi: 10.1016/j.biomaterials.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slotta UK, Rammensee S, Gorb S, Scheibel T. An engineered spider silk protein forms microspheres. Angew. Chem. 2008;47(24):4592–4594. doi: 10.1002/anie.200800683. [DOI] [PubMed] [Google Scholar]; •• First report describing a simple new method of silk sphere formation.
- 9.Lammel AS, Hu X, Park SH, Kaplan DL, Scheibel TR. Controlling silk fibroin particle features for drug delivery. Biomaterials. 2010;31(16):4583–4591. doi: 10.1016/j.biomaterials.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rammensee S, Slotta U, Scheibel T, Bausch AR. Assembly mechanism of recombinant spider silk proteins. Proc. Natl Acad. Sci. USA. 2008;105(18):6590–6595. doi: 10.1073/pnas.0709246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florczak A, Mackiewicz A, Dams-Kozlowska H. Functionalized spider silk spheres as drug carriers for targeted cancer therapy. Biomacromolecules. 2014;15(8):2971–2981. doi: 10.1021/bm500591p. [DOI] [PubMed] [Google Scholar]; •• In vitro study on functionalization of bioengineered silk with tumor targeting peptides.
- 12.Jastrzebska K, Felcyn E, Kozak M, et al. The method of purifying bioengineered spider silk determines the silk sphere properties. Sci. Rep. 2016;6:28106. doi: 10.1038/srep28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofer M, Winter G, Myschik J. Recombinant spider silk particles for controlled delivery of protein drugs. Biomaterials. 2012;33(5):1554–1562. doi: 10.1016/j.biomaterials.2011.10.053. [DOI] [PubMed] [Google Scholar]
- 14.Dams-Kozlowska H, Majer A, Tomasiewicz P, Lozinska J, Kaplan DL, Mackiewicz A. Purification and cytotoxicity of tag-free bioengineered spider silk proteins. J. Biomed. Mat. Res. A. 2013;101(2):456–464. doi: 10.1002/jbm.a.34353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florczak A, Jastrzebska K, Mackiewicz A, Dams-Kozlowska H. Blending two bioengineered spider silks to develop cancer targeting spheres. J. Mat. Chem. B. 2017;16(5):3000–3011. doi: 10.1039/c7tb00233e. [DOI] [PubMed] [Google Scholar]
- 16.Hu X, Kaplan DL, Cebe P. Determining beta-sheet crystallinity in fibrous proteinsby thermal analysis and infrared spectroscopy. Macromolecules. 2006;39(18):6161–6170. [Google Scholar]
- 17.Tokareva O, Jacobsen M, Buehler M, Wong J, Kaplan DL. Structure-function-property-design interplay in biopolymers: spider silk. Acta Biomaterial. 2014;10(4):1612–1626. doi: 10.1016/j.actbio.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This is an essential review describing spider silk properties.
- 18.Hinman MB, Lewis RV. Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J. Biol Chem. 1992;267(27):19320–19324. [PubMed] [Google Scholar]
- 19.Deber CM, Brodsky B, Rath A. Proline residues in proteins. Encycloped. Life Sci. 2010 [Google Scholar]
- 20.Lewis RV. Spider silk: ancient ideas for new biomaterials. Chem. Rev. 2006;106(9):3762–3774. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- 21.Rabotyagova OS, Cebe P, Kaplan DL. Role of polyalanine domains in beta-sheet formation in spider silk block copolymers. Macromol. Biosci. 2010;10(1):49–59. doi: 10.1002/mabi.200900203. [DOI] [PubMed] [Google Scholar]
- 22.Rathore O, Sogah DY. Self-assembly of beta-sheets into nanostructures by poly(alanine) segments incorporated in multiblock copolymers inspired by spider silk. J. am. Chem. Soc. 2001;123(22):5231–5239. doi: 10.1021/ja004030d. [DOI] [PubMed] [Google Scholar]
- 23.Neubauer MP, Blüm C, Agostini E, Engert J, Scheibel T, Fery A. Micromechanical characterization of spider silk particles. Biomater. Sci. 2013;1(11):1160–1165. doi: 10.1039/c3bm60108k. [DOI] [PubMed] [Google Scholar]
- 24.Huemmerich D, Scheibel T, Vollrath F, Cohen S, Gat U, Ittah S. Novel assembly properties of recombinant spider dragline silk proteins. Curr. Biol. 2004;14(22):2070–2074. doi: 10.1016/j.cub.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Heidebrecht A, Eisoldt L, Diehl J, et al. Biomimetic fibers made of recombinant spidroins with the same toughness as natural spider silk. Adv. Mat. 2015;27(13):2189–2194. doi: 10.1002/adma.201404234. [DOI] [PubMed] [Google Scholar]
- 26.Lammel A, Schwab M, Hofer M, Winter G, Scheibel T. Recombinant spider silk particles as drug delivery vehicles. Biomaterials. 2011;32(8):2233–2240. doi: 10.1016/j.biomaterials.2010.11.060. [DOI] [PubMed] [Google Scholar]
- 27.Schierling MB, Doblhofer E, Scheibel T. Cellular uptake of drug loaded spider silk particles. Biomater. Sci. 2016;4(10):1515–1523. doi: 10.1039/c6bm00435k. [DOI] [PubMed] [Google Scholar]
- 28.Numata K, Hamasaki J, Subramanian B, Kaplan DL. Gene delivery mediated by recombinant silk proteins containing cationic and cell binding motifs. J. Control Release. 2010;146(1):136–143. doi: 10.1016/j.jconrel.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blüm C, Scheibel T. Control of drug loading and release properties of spider silk sub-microparticles. BioNanoScience. 2012;2(2):67–74. [Google Scholar]
- 30.Doblhofer E, Scheibel T. Engineering of recombinant spider silk proteins allows defined uptake and release of substances. J. Pharmaceut. Sci. 2015;104(3):988–994. doi: 10.1002/jps.24300. [DOI] [PubMed] [Google Scholar]; • A basic model of drug release kinetics from a silk spheres is presented.
- 31.Kozlowska AK, Florczak A, Smialek M, et al. Functionalized bioengineered spider silk spheres improve nuclease resistance and activity of oligonucleotide therapeutics providing a strategy for cancer treatment. Acta Biomaterial. 2017;59:221–233. doi: 10.1016/j.actbio.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardy JG, Torres-Rendon JG, Leal-Egana A, et al. Biomineralization of engineered spider silk protein-based composite materials for bone tissue engineering. Materials. 2016;9(7) doi: 10.3390/ma9070560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manchineella S, Thrivikraman G, Basu B, Govindaraju T. Surface-functionalized silk fibroin films as a platform to guide neuron-like differentiation of human mesenchymal stem cells. ACS Appl. Mat. Interf. 2016;8(35):22849–22859. doi: 10.1021/acsami.6b06403. [DOI] [PubMed] [Google Scholar]
- 34.Manchineella S, Thrivikraman G, Khanum KK, Ramamurthy PC, Basu B, Govindaraju T. Pigmented silk nanofibrous composite for skeletal muscle tissue engineering. Adv. Health. Mat. 2016;5(10):1222–1232. doi: 10.1002/adhm.201501066. [DOI] [PubMed] [Google Scholar]
- 35.Spieß K, Wohlrab S, Scheibel T. Structural characterization and functionalization of engineered spider silk films. Soft Matter. 2010;6(17):4168–4174. [Google Scholar]
- 36.Sanson C, Schatz C, Le Meins JF, et al. A simple method to achieve high doxorubicin loading in biodegradable polymersomes. J. Control Release. 2010;147(3):428–435. doi: 10.1016/j.jconrel.2010.07.123. [DOI] [PubMed] [Google Scholar]
- 37.Enache M, Volanschi E. Spectral characterization of self-association of antitumor drug mitoxantrone. Revue Roumaine de Chimie. 2010;55(4):255–262. [Google Scholar]
- 38.Numata K, Mieszawska-Czajkowska AJ, Kvenvold LA, Kaplan DL. Silk-based nanocomplexes with tumor-homing peptides for tumor-specific gene delivery. Macromol. Biosci. 2012;12(1):75–82. doi: 10.1002/mabi.201100274. [DOI] [PMC free article] [PubMed] [Google Scholar]