Abstract
Chromosomal architecture is known to influence gene expression, yet its role in controlling cell fate remains poorly understood. Reprogramming of somatic cells into pluripotent stem cells by the transcription factors (TFs) Oct4, Sox2, Klf4 and Myc offers an opportunity to address this question but is severely limited by the low proportion of responding cells. We recently developed a highly efficient reprogramming protocol that synchronously converts somatic into pluripotent stem cells. Here, we employ this system to integrate time-resolved changes in genome topology with gene expression, TF binding and chromatin state dynamics. This revealed that TFs drive topological genome reorganization at multiple architectural levels, which often precedes changes in gene expression. Removal of locus-specific topological barriers can explain why pluripotency genes are activated sequentially, instead of simultaneously, during reprogramming. Taken together, our study implicates genome topology as an instructive force for implementing transcriptional programs and cell fate in mammals.
Introduction
Somatic cell reprogramming into pluripotent stem cells (PSCs) represents a widely studied model for dissecting how transcription factors (TFs) regulate gene expression programs to shape cell identity1,2. Chromosomal architecture was recently shown to be cell type-specific and critical for transcriptional regulation3–5, but its importance for cell fate decisions remains poorly understood.
Two major levels of topological organization have been identified in the genome6–8. The first level segregates the genome, at the megabase scale, into two subnuclear compartments. The A compartment corresponds to active chromatin typically associated with a more central nuclear position, while the B compartment represents inactive chromatin enriched at the nuclear periphery/lamina9–14. Compartmentalization is consistent amongst individual cells and a potential driver of genome folding15. A second sub-megabase level consists of topologically associated domains (TADs)16–18 and chromatin loops11, which restrict or facilitate interactions between gene regulatory elements19,20. Importantly, modifying chromatin architecture can lead to gene expression changes19,21–24. Moreover, de novo establishment of TAD structure during zygotic genome activation has been shown to be independent of ongoing transcription, demonstrating that chromatin architecture is not simply a consequence of transcription25–27. Genome topology could therefore be instructive for gene regulation28,29, but whether this reflects a general principle that occurs on a genome-wide scale in space and time is unknown.
Mechanistic studies with mammalian cell reprogramming systems have been hampered by the typically small percentage of responding cells1,30. To overcome this shortcoming, we recently developed a highly efficient and synchronous reprogramming system based on the transient expression of the TF C/EBPα prior to induction of the Yamanaka TFs Oct4, Sox2, Klf4 and Myc (OSKM)31,32. OSKM activates the endogenous core pluripotency TFs sequentially in the order of Oct4, Nanog and Sox2, implying that locus-specific barriers dictate gene activation kinetics33–35. Here, we studied how C/EBPα and OSKM affect genome topology, the epigenome and gene expression during reprogramming. We found that the TFs bind hotspots of topological reorganization at both the compartment and TAD levels. Dynamic reorganization of genome topology frequently preceded gene expression changes at all levels and provided an explanation for the sequential activation of core pluripotency genes during reprogramming. Together, our observations indicate that genome topology has an instructive role in implementing transcriptional programs relevant for cell fate decisions in mammals.
Results
Transcription factors prime the epigenome for reprogramming
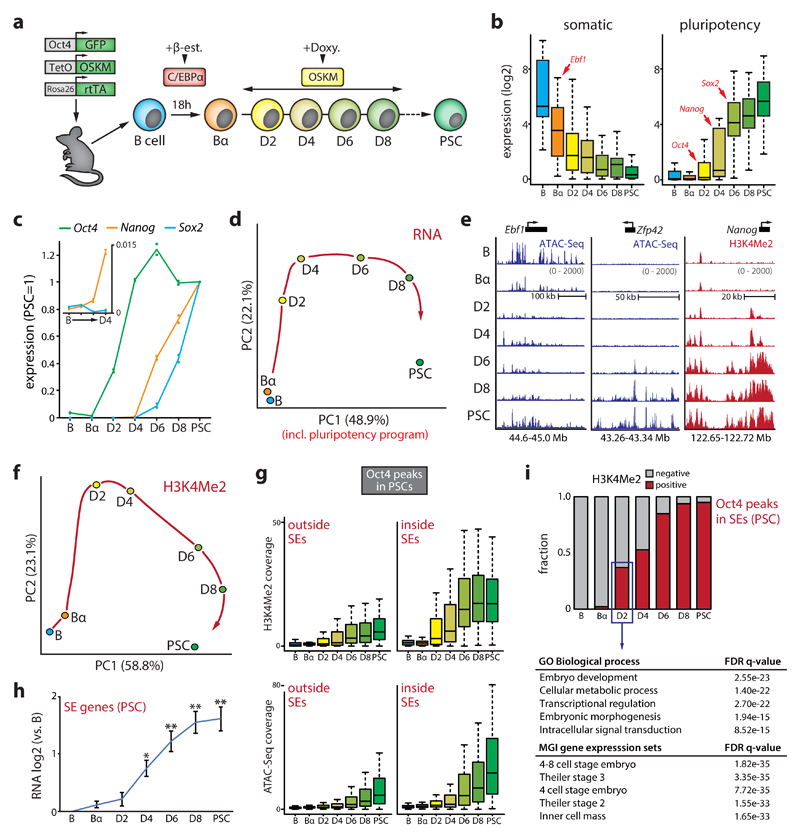
We exposed bone marrow-derived pre-B cells to the myeloid TF C/EBPα to generate ‘Bα cells’. Subsequent activation of OSKM induces reprogramming of nearly 100% of Bα cells into PSC-like cells within 4-8 days31,32. To obtain a high-resolution map of changes in gene expression and chromatin structure we examined 6 different reprogramming stages (B, Bα, D2, D4, D6, and D8) and PSCs (Fig.1a). We profiled the transcriptome by RNA-Seq, active chromatin deposition by H3K4Me2 ChIPmentation36, and chromatin accessibility by ATAC-Seq37 (Supplementary Fig.1). Expression of half of all genes was significantly affected (FDR<0.01) between any two time points, starting with the rapid silencing of the core B cell program initiated by C/EBPα. Pluripotency genes were then activated sequentially, with the core pluripotency factors Oct4, Nanog and Sox2 being activated at D2, D4 and D6, respectively (Fig.1b-c). RT-PCR measurements of primary Nanog and Sox2 transcription confirmed their activation timing (Supplementary Fig.1e).
Figure 1. Dynamics of the transcriptome and epigenome during reprogramming.
(a) Schematic overview of the reprogramming system. C/EBPα-ER in B cells is translocated into the nucleus upon beta-estradiol (β-est.) treatment. After β-est. wash-out, Oct4, Sox2, Klf4 and Myc (OSKM) are induced by doxycycline (doxy.). (b) Box plots of gene expression dynamics (normalized counts) of a set of core B cell (‘somatic’, n=25) and PSC (‘pluripotency’, n=25) identity genes. (c) Average gene expression kinetics of Oct4, Nanog and Sox2 during reprogramming (n=2, relative to the levels in PSCs). Inset shows Nanog expression first appears at D4. (d) Principal component analysis (PCA) of gene expression dynamics (n=16,332 genes) during reprogramming. A red arrow indicates hypothetical trajectory. (e) Representative examples of chromatin opening (measured by ATAC-Seq) and H3K4Me2 deposition (measured by ChIPmentation) at gene regulatory elements controlling B cell (Ebf1) or pluripotency (Zfp42 and Nanog) genes. (f) PCA of H3K4Me2 dynamics during reprogramming (n=26,351 100kb genomic bins). A red arrow indicates hypothetical trajectory. (g) Box plots of dynamics of H3K4Me2 deposition (top) and chromatin accessibility (bottom) at Oct4 binding sites outside (n=31,869) and inside (n=821) PSC superenhancers (SEs). (h) Expression dynamics of genes associated with a SE in PSCs (mean values shown, n=328 genes). Error bars denote 95% CI. (*P<0.01, **P<0.001 versus B cells, unpaired two-tailed t-test). (i) Fraction of H3K4Me2+ Oct4 binding sites in PSC SEs (n=821) during reprogramming; table shows a gene ontology (GO) analysis for SE genes (n=212) associated with early Oct4 recruitment.
Principal component analysis (PCA) revealed a trajectory along which B cells acquire a PSC gene expression program (Fig.1d). Epigenome remodeling showed similar dynamics, with an early loss of chromatin accessibility at gene regulatory elements controlling the B cell program induced by C/EBPα followed by the establishment of active and open chromatin at pluripotency genes by OSKM (Fig.1e, Supplementary Fig.1). OSKM induction led to a genome-wide expansion of active chromatin marked by H3K4Me2, known to be deposited at both primed and active gene regulatory elements38 (Supplementary Fig.1f). The H3K4Me2 landscape more rapidly converged on a pluripotent state than gene expression, suggesting that OSKM primes regulatory elements for subsequent gene activation (Fig.1f). Many regions bound by Oct4 in PSCs39 had already acquired H3K4Me2 by D2 and chromatin opening occurred progressively at Oct4 binding sites (Fig.1g, Supplementary Fig.1g-i). 37% of Oct4 binding sites in predicted PSC superenhancer (SE) elements39 were already H3K4Me2+ by D2, while activation of most associated genes (assigned using in-situ Hi-C data, see Supplementary Materials) occurred 2 days later (Fig.1i-h). These early targeted SEs were linked to genes involved in embryo development (e.g. Oct4, Nanog, Klf9) and metabolism (e.g. Upp1, Uck2); a gene signature strongly associated with 4 to 8 cell stage embryos (Fig.1i).
Chromatin state, genome topology and transcription are dynamically coupled
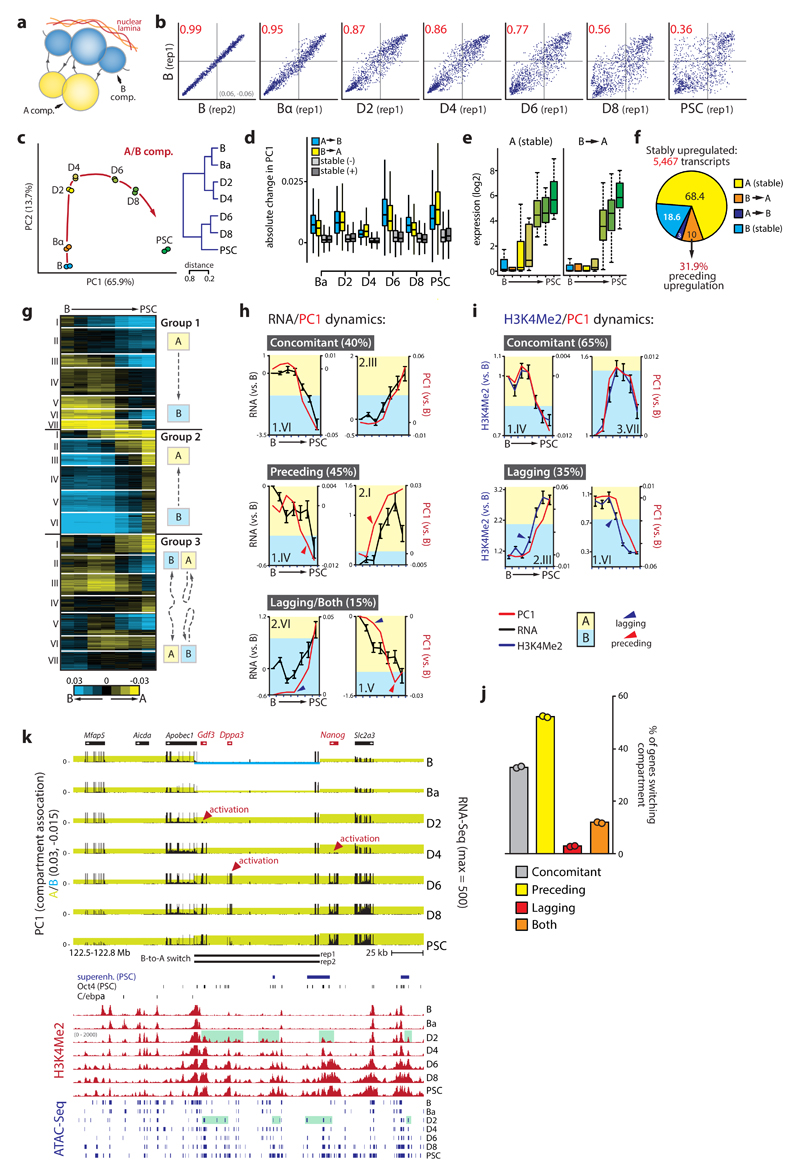
We used in-situ Hi-C11 to map 3D genome organization during cell reprogramming at high resolution and determined genome segmentation into A and B compartments (Supplementary Table 1). Quantitative changes in A/B compartment association (using the PC1 values of a PCA on the Hi-C correlation matrix) during reprogramming were cumulative, widespread and highly reproducible (Pearson R>0.97) (Fig.2a-b, Supplementary Fig.2a-b). Although overall proportions assigned to A and B compartments (40% A - 60% B) remained unchanged throughout reprogramming, compartmentalization strength (as measured by average contact enrichment within and between compartments) was dynamically altered (Supplementary Fig.2c-d). OSKM induction initially (D2-D4) strengthened A-B compartment segregation, followed by substantial loss of compartmentalization due to reduced contact frequencies within the B compartment and increased inter-compartment contacts.
Figure 2. Kinetics of subnuclear compartmentalization, the transcriptome and epigenome.
(a) Schematic representation of chromosome compartments. (b) Scatterplots of PC1 values (n=26,370 100kb bins) showing changes to initial B cell genome compartmentalization for chromosome 13. Pearson correlation coefficient (R2) is indicated in red. (c) Principal component analysis (red arrow indicates hypothetical trajectory) and unsupervised hierarchical clustering (right) of PC1 values (n=26,370 bins). (d) Absolute PC1 changes per timepoint for regions (n=35,348) that switch compartment or do not switch (‘stable’) but increase (-) or decrease (+) in PC1 value. (e) Box plots of normalized transcript counts for key pluripotency genes (n=25) that are stably associated with the A compartment or switch from B to A. (f) Compartment switching at stably upregulated genes. (g) k-means clustering (k=20) of PC1 values for 100kb genomic bins that switch compartment at any timepoint. (h) Examples of individual switching clusters with concomitant mean gene expression and PC1 changes (8/20), clusters with PC1 changes preceding expression changes (9/20), and clusters with expression changes preceding PC1 changes (1/20) or with both phenomena (2/20). (i) Examples of individual switching clusters that show concomitant mean PC1/H3K4Me2 changes (13/20) or H3K4Me2 kinetics preceding PC1 modulation (7/20). (j) Proportion of genes (n=8,218) located in the different categories of switching clusters. (k) Genome browser view of the Gdf3-Dppa3-Nanog locus. Top part shows integrated PC1 (shading denotes A/B compartment status) and RNA-Seq values, with B-to-A switch regions per replicate indicated below. Bottom part depicts superenhancer (SE) location, Oct4 binding, C/EBPα binding, H3K4Me2 dynamics and ATAC-Seq peaks. Green shading indicates priming of Dppa3/Nanog enhancers at D2. Error bars in the figure represent SEM.
Switching of loci between the A/B compartments was frequent, with 20% of the genome changing compartment at any time point during reprogramming. B-to-A and A-to-B switching each occurred in 10% of the genome, with 35% of these regions being involved in multiple switching events (Supplementary Fig.2e). PCA analysis revealed a reprogramming trajectory of genome compartmentalization highly similar to that seen for the transcriptome (Fig.2c, Supplementary Fig.2f). Genes that stably switch compartment after reprogramming tend to change expression accordingly and were enriched for lineage-specific functions: A-to-B switching genes associated with immune system processes, while B-to-A switching genes were enriched for early developmental functions (Supplementary Fig.2g-h). Compartment switching typically occurred in regions with low PC1 values at the edges of A or B compartment domains. At any time point, regions that switched also displayed the most substantial PC1 changes, suggesting that loci with a less pronounced compartment association are more amenable to changing their compartment status (Fig.2d, Supplementary Fig.2i-k).
Our dataset allowed us to monitor genome architecture and to study its interplay with chromatin state and gene expression changes over time. The core transcriptional network that defines B cell identity40 resided primarily (88%) in the A compartment (e.g. Ebf1, Pax5, Foxo1), of which 32% switched to B (Supplementary Table 2, Supplementary Fig.3a). Both switching and non-switching genes were rapidly silenced, but switching genes were repressed to a larger extent. In contrast, 40% of core pluripotency genes41 initially resided in the B compartment of which 90% switched to A (Supplementary Table 2, Supplementary Fig.3b). Pluripotency genes already in the A compartment were activated early (D2-D4, e.g., Oct4), while genes that underwent B-to-A switching were activated late (D6, e.g. Sox2) (Fig.2e). We next divided all genes that change expression between endpoints (>0.5 log2) into stable (non-switching) and compartment-switching groups. Again, downregulated genes that changed compartment from A-to-B (21%) were silenced to a greater extent than non-switching genes in A (Supplementary Fig.3c). Likewise, upregulated genes that switched from B-to-A (16%) were upregulated more substantially than genes already residing in A, albeit with slower kinetics. Interestingly, quantitative changes in compartment association occurred before transcriptional upregulation (Supplementary Fig.3d). To further explore whether compartment switching can precede transcriptional changes we examined four clusters of genes (n=5467 genes) stably upregulated at early, intermediate or late time points (Supplementary Fig.3e). Nearly a third of the genes (n=175/548) that switch from B-to-A in these clusters did so before being upregulated (Fig.2f, Supplementary Fig.3f). Moreover, genes associated with predicted PSC SEs showed a significant increase in A compartment association at D2 prior to transcriptional upregulation at D4 (Supplementary Fig.3g, Fig.1h).
We performed k-means clustering on the PC1 values of the 20% of the genome (n=8218 genes) that switched compartment during reprogramming, identifying 20 clusters with a wide range of switching dynamics that included non-linear and abortive trajectories (Fig.2g). Eight of the 20 clusters displayed concomitant changes in compartmentalization and gene expression (R>0.9, Fig.2h). The remainder, although generally also showing strong correlations between gene expression and PC1 (average R=0.86, range: 0.56-0.97), consisted of clusters with at least one time point at which this correlation was lost (Fig.2h). Genes in these clusters were enriched for metabolic and secretory functions, as well as developmental processes (Supplementary Fig.4a-b). Strikingly, 9 of the 20 clusters showed changes in subnuclear compartment status preceding changes in transcriptional output, involving over half of the genes that switch compartment (e.g. cluster 2.I, Fig.2h and Fig.2j). In only a single cluster compartment modification lagged behind changes in gene expression, while 2 of the 20 clusters displayed both preceding and lagging relationships. We furthermore observed a very strong overall correlation between chromatin state dynamics (gain or loss of H3K4Me2) and genome compartmentalization (average R=0.95, range: 0.93-0.98), with concomitant changes in H3K4Me2 levels and gene expression occurring in 13 of the 20 clusters. However, in 7 of the 20 clusters H3K4Me2 dynamics preceded PC1 changes (Fig.2i), implicating chromatin state as a driver of subnuclear compartmentalization. The extended Nanog locus provides a prime example of modifications to compartmentalization and chromatin state preceding transcriptional changes. It includes a region encompassing Gdf3, Dppa3 and the -45kb Nanog SE39,42, which already switched from B-to-A in Bα cells. OSKM induction strengthened A compartment association of the entire locus, activated Gdf3 expression and primed the Nanog and Dppa3 regulatory elements (H3K4Me2+/ATAC+) at D2 for subsequent gene activation at D4-D6 (Fig.2k).
These data show that genome compartmentalization and chromatin state are dynamically reorganized during cell fate conversion and tightly coupled to global changes in gene expression. In addition, a sizable number of genes are subject to changes in compartmentalization before expression alterations.
Genome partitioning into TADs is largely stable
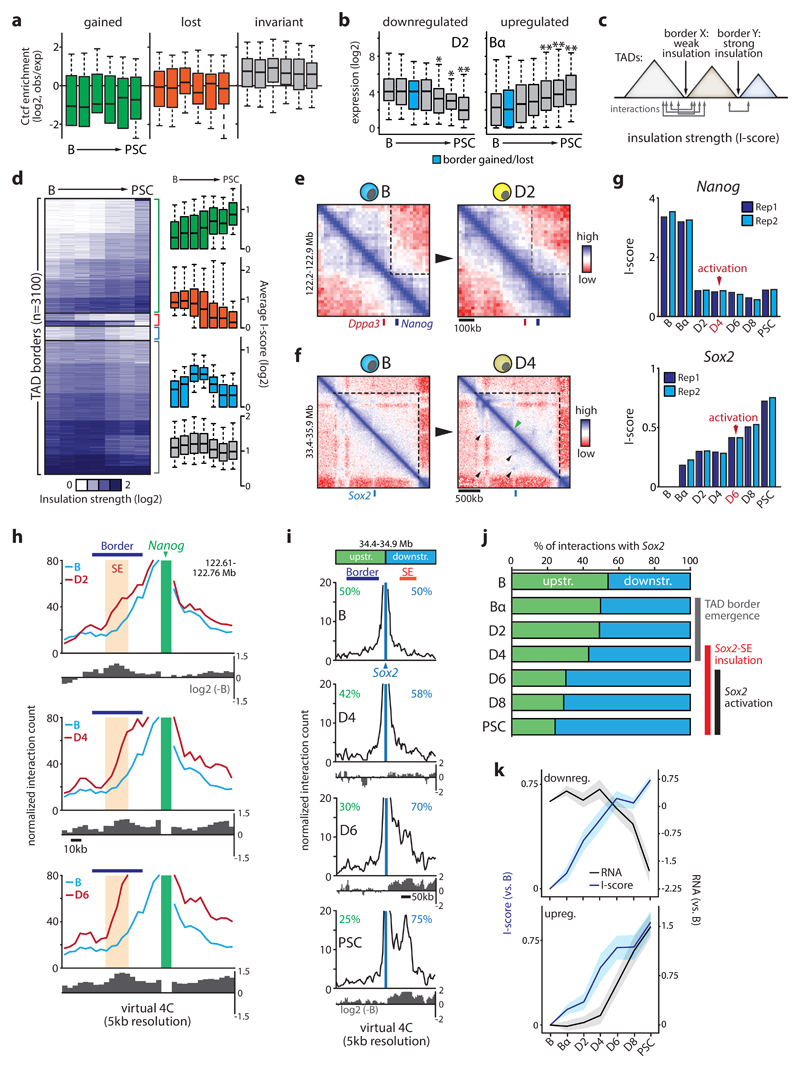
We next used chromosome-wide insulation potential to identify TAD borders and define TADs43, detecting ~2800-3400 borders per time point. Border calls were highly reproducible between biological replicates (Jaccard index > 0.8) and enriched for Ctcf binding sites and transcription start sites (Supplementary Fig.5)17,44. Borders not called in both biological replicates were excluded from all subsequent analyses. Partitioning of the genome into TADs was largely stable during reprogramming, as most TAD borders (>75%) were detected at all stages. Nevertheless, 18% of the 3100 TAD borders were stably acquired (n=431) or lost (=124) during reprogramming, resulting in a net increase in the number of borders and a reduction of average TAD size from 891kb to 741kb (Supplementary Fig.5). Surprisingly, no correlation existed between the stable gain or loss of TAD borders (referred to hereafter as qualitative TAD border changes) and Ctcf binding. In fact, newly acquired TAD borders were relatively depleted for Ctcf binding and Ctcf enrichment levels did not significantly change during reprogramming at borders gained or lost (Fig.3a). However, we did observe specific regions where qualitative TAD changes clearly correlated with Ctcf binding, e.g. at the Sox2 locus where acquisition of a new border and chromatin loop formation (see below) was paralleled by a substantial gain of Ctcf binding sites (Supplementary Fig.5g).
Figure 3. Kinetics of domain insulation during reprogramming.
(a) Ctcf enrichment dynamics (from ChIP-Seq experiments during reprogramming) at TAD borders that are gained (n=431), lost (n=124) or invariant (n=2,185) during reprogramming. (b) Gene expression dynamics at transcriptionally modulated border regions (divided into up or downregulated groups per timepoint) gained or lost D2 or Bα stages (*P<0.05, **P<0.005 versus B cells; unpaired two-tailed t-test). Sample sizes are indicated in Supplementary Fig.5. (c) Cartoon illustrating the concept of the insulation strength score (I-score). (d) k-means clustering (k=20) of I-score. Bar graphs show I-score kinetics for groups that increase (n=1,291), decrease (n=141), transiently increase (n=159) or do not change (n=1,509). (e) Representative in-situ Hi-C contact maps (20kb resolution) of the Dppa3-Nanog border or (f) the internal Sox2 border comparing. Black arrows indicate loops; green arrow indicates border formation. (g) I-score kinetics of the Nanog and Sox2 borders. (h,i) Representative virtual 4C analysis using Nanog (panel h) or Sox2 (panel i) as viewpoints. TAD border and superenhancer (SE) are indicated. Log2 ratio (over B) is shown below each line graph, percentages shown in panel i depict proportions of all interactions with Sox2. (j) Proportion of interactions with Sox2 from the immediate upstream or downstream region (indicated in panel i). Timing of key events involved in Sox2 activation is indicated. (k) Gene expression and I-score kinetics at dynamic border regions where I-score changes precede transcriptional modulation (49%, n=43/88). Line graphs depict I-score and gene expression dynamics for those borders where gene expression is downregulated or upregulated. Error bars/shading represent 95% CI.
The gain or loss of TAD borders did not correlate with overall increased or decreased local gene expression respectively, suggesting that changes in the level of transcription per se are not a main driver of TAD border dynamics (Supplementary Fig.5h). Gene expression changes during reprogramming at dynamic border regions were highly context-dependent, with no apparent correlation between border gain or loss and the direction of transcriptional change (Supplementary Fig.5i). Moreover, these border regions rarely switched compartment (3-9% versus 17% for all borders). Interestingly, at borders that showed transcriptional changes (>0.5 log2 change between endpoints) gene expression was often not significantly altered until after TAD borders were newly acquired or lost (Fig.3b, Supplementary Fig.5j).
Quantitative changes in TAD border strength occur early in reprogramming
Local chromatin insulation by TAD borders can also be approached quantitatively by calculating an insulation strength score (‘I-score’, R2>0.87 between biological replicates) for each border (Fig.3c)43,45. Compared to qualitative border changes (i.e. a gain or loss of border detection), quantitative changes in TAD insulation were more abundant: half of all borders showed a >20% difference in I-score between the first three and last three timepoints of reprogramming (Fig.3d; green, red and blue clusters). Stably acquired or lost borders often had lower average I-scores than invariant TAD borders (Supplementary Fig.6a). Ctcf occupancy correlated with I-score and meta-border plots confirmed that I-score dynamics reflect actual contact maps (Supplementary Fig.6b-c). PCA analysis of I-score kinetics revealed a reprogramming trajectory grossly resembling the transcriptome, PC1 and H3K4Me2 trajectories determined before (Supplementary Fig.6d).
Border regions contained a significant number of genes with cell type-specific functions (e.g. immune system, developmental biology), in addition to the expected housekeeping genes17 (Supplementary Fig.6e-f). Pluripotency genes were often found at or near border regions, including Nanog and Sox2. Both of these loci showed rapid I-score changes that preceded their transcriptional activation (Fig.3e-g). In B and Bα cells Nanog was separated from Dppa3 by a strong border in a region that harbors the -45kb Nanog SE and Gdf3 (Fig.3e, Fig.2k), likely interfering with the reported spatial clustering of these genes and enhancers in PSCs46. I-score was considerably reduced at D2 after OSKM induction (Fig.3g), facilitating interactions between genes and their enhancers required for subsequent transcriptional activation (D4-D6). Consistently, both Hi-C-derived virtual 4C obtained at 5kb resolution and conventional 4C-Seq analyses showed increased (cross-)border contact frequencies of the Nanog promoter as early as D2 (Fig.3h, Supplementary Fig.7a). Within the Sox2 TAD, a new internal border and several chromatin loops appeared between the Bα and D4 stages, before Sox2 activation at D6. High-resolution virtual 4C analysis showed that early border emergence progressively skewed interactions of Sox2 towards its key downstream SE47,48, resulting in the formation of an insulated Sox2-SE subdomain at D6 that is likely critical for Sox2 activation (Fig.3i-j, Supplementary Fig.7b).
To further understand the relationship between I-score changes and local gene expression we analyzed transcriptional changes at the 184 most dynamic borders regions that increase in insulation strength (>75% change in I-score). Gene expression was altered at many of these borders (n=88, >0.5 log2 change between endpoints) during reprogramming, with again no clear bias for activation or repression. At 49% of these borders (n=43/88) I-scores increased before transcriptional changes (Fig.3k), while for the remaining borders a mix of concomitant (n=15), lagging (n=15) and more complex (n=15) kinetics was observed. Likewise, I-score changes also preceded modulation of chromatin state and subnuclear compartmentalization (Supplementary Fig.7). Thus, altered insulation strength at TAD borders is an early reprogramming event linked to transcriptome re-wiring.
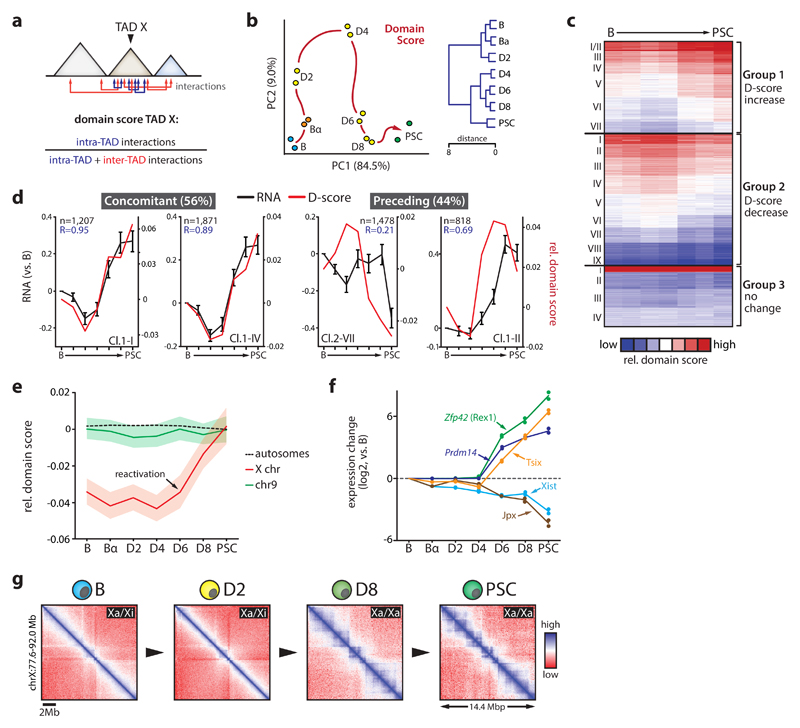
Topological plasticity increases late in reprogramming
TADs differ in their propensity to form contacts with other TADs49,50. To quantify this ‘connectivity’ within a given TAD, we computed a domain score (‘D-score’) defined by the ratio of intra-TAD interactions over all cis interactions (Fig.4a)49. While I-score measures a border’s ability to prevent interactions between two neighboring TADs, D-score quantifies a TAD’s tendency to self-interact. D-scores positively correlated with gene expression and A compartment association (Supplementary Fig.8), as previously noted49,50. Correlations between D-scores, gene expression and compartment association seen at early time points progressively weakened after D4 (Supplementary Fig.8a-b). While TADs explained a greater proportion of expression variability than linear neighborhoods when we estimated the overall impact of TAD structure on gene expression (see Supplemental Materials), this proportion was progressively reduced during reprogramming (Supplementary Fig.8c). Together with the observed decrease in overall A-B compartment segregation (Supplementary Fig.2d) and in line with the previously reported reduced organization of inactive chromatin in PSCs51, these data suggest that at the topological level cells gradually acquire a plastic state characteristic of the pluripotent genome52.
Figure 4. Dynamics of TAD connectivity during reprogramming.
(a) Cartoon depicting domain score (D-score) calculation. Arrows indicate intra or inter TAD interactions. (b) Principal component analysis (left) and unsupervised hierarchical clustering (right) of D-score kinetics (n=2,153 TADs). Red arrow indicates hypothetical trajectory. (c) k-means clustering (k=20) of genome-wide relative D-score (centered on mean). (d) Examples of individual dynamic D-score clusters for which gene expression and D-Score kinetics (mean values presented, number of genes per cluster indicated) are concomitant or where D-score changes precede transcriptional changes. Error bars show SEM. R-values denote Pearson correlation coefficients. (e) Average relative D-score changes for chromosome 9 (n=115 TADs), all autosomes combined (n=1,959 TADs) and the X chromosome (n=106 TADs). Shading denotes 95% CI. (f) Mean gene expression changes (versus B cells, n=2 independent biological replicate reprogramming expriments) of key regulators of X-chromosome re/inactivation during reprogramming. (g) Representative in-situ Hi-C contact maps (50kb resolution) of a 14.5 Mb region on the X chromosome during reprogramming. B-D2 cells carry one inactive X (Xi) and one active X (Xa) chromosome; D8-PSC cells carry two Xa.
Altered TAD connectivity can precede transcriptional changes
PCA analysis of D-score kinetics revealed a reprogramming trajectory for TAD connectivity similar to those for compartmentalization, transcription, active chromatin and I-score (Fig.4b, Supplementary Fig.8d). K-means clustering showed that 79% of TADs exhibited D-score changes (i.e. >20% change between endpoints) (Fig.4c). D-score kinetics correlated closely with compartmentalization (PC1) changes (R>0.84, Supplementary Fig.8e). TADs with the most dynamic connectivity pattern frequently switched compartment and harbored genes enriched for immune cell and stem cell related functions (Supplementary Fig.8f-g). These TADs were highly biased in their compartment association: 88% of TADs that showed a rapid increase in D-scores initially localized to the B compartment, while 83% of the TADs with substantial D-score reductions initially resided in the A compartment (Supplementary Fig.8f).
To assess the correlation between TAD connectivity and gene expression, we compared D-score with intra-TAD gene expression kinetics for the 16 dynamic D-score clusters (Fig.4c). In 9 of 16 clusters D-score changes coincided with gene expression alterations (Fig.4d), in particular for TADs that showed both increased D-scores and intra-TAD expression (R=0.78). However, 7 of 16 clusters showed D-score changes preceding transcriptional changes, with no clusters showing the opposite pattern (Fig.4d). Thus, changes in TAD connectivity frequently precede intra-TAD transcriptional modulation.
X chromosome reactivation evokes TAD reorganization
X chromosome reactivation in PSCs is a classic model for studying the relationship between chromosome structure and gene expression53. The B cells used were derived from female mice carrying one inactive X chromosome, allowing us to study this process using our dataset. While average TAD connectivity for each time point remained similar on autosomes, X chromosome TADs displayed substantial gains in D-scores after D4 (Fig.4e). The observed chromosome-wide D-score increase might be caused by a reactivation of the largely TAD-devoid inactive X chromosome11,54–56. Indeed, after D4 TAD structures were fully re-established and key regulators of X reactivation activated (Zfp42, Prdm14, Tsix), while X chromosome repressors (Xist and Jpx) were downregulated (Fig.4f-g).
Cell type-specific changes in chromatin loops
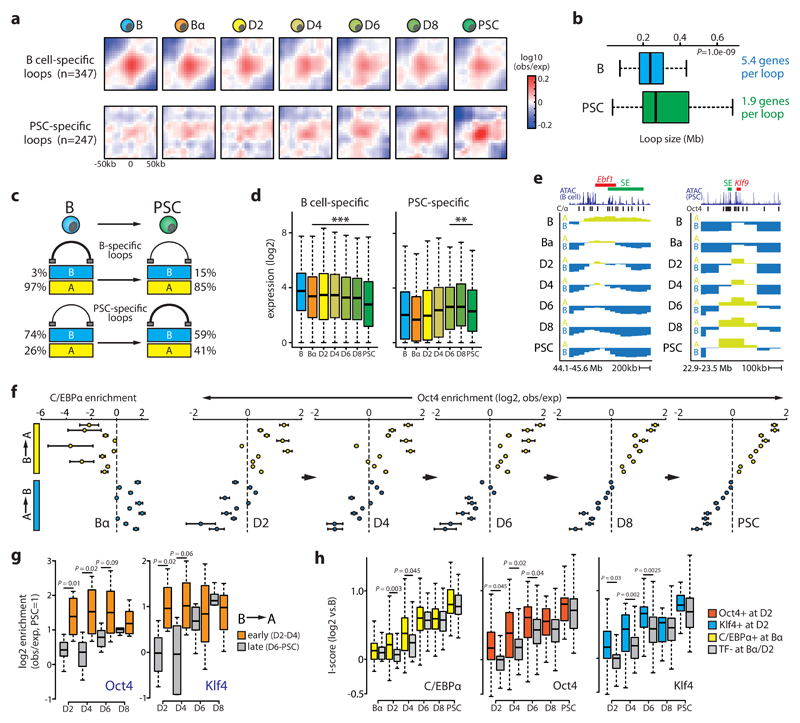
Chromatin loops appear as foci in high-resolution Hi-C maps, representing particularly strong interactions between two distant regions11. We visualized chromatin loop dynamics during reprogramming by performing meta-loop analyses at 5kb resolution of a previously identified set of loops in primary B cells and PSCs49. Similar to the TADs they often demarcate11, chromatin loops in general behave as remarkably stable topological structures during reprogramming (Supplementary Fig.9a). However, cell type-specific loops, representing a minor fraction of all loops (13% for B cells, 5% for PSCs49), showed a dynamic behaviour: while B cell-specific loops lost interaction strength, PSC-specific loops were established de novo from D4 onward (Fig.5a). Intriguingly, the nature of these somatic and pluripotent cell type-specific loops appeared to be different: PSC-specific loops were larger than B cell-specific loops, localized mostly to the B compartment (while virtually all B cell-specific loops localized to A), contained fewer genes that showed lower average gene expression levels and were less enriched for cell type-specific genes (Fig.5b-c, Supplementary Fig.9b). However, in both cases the presence of a loop positively correlated with gene expression changes, indicating that both the formation and loss of cell type-specific loops are dynamically linked to gene regulation (Fig.5d).
Figure 5. Chromatin loop and transcription factor binding dynamics.
(a) Meta-loop analysis at 5kb resolution of B cell or PSC-specific loops49. Area shown is centered on the respective TF binding sites (+/- 50kb). (b) Boxplot showing median loop size (P=1.0e-09, Wilcoxon rank sum test) and average number of genes per loop for B cell (n=347) or PSC-specific (n=247) loops. (c) Cartoon depicting percentage of B cell or PSC-specific loops within A or B compartments in reprogramming end stages. (d) Boxplot showing gene expression dynamics of genes within B cell (left, n=1874)) or PSC-specific (right, n=469) loops (**P<0.005, ***P<0.001 versus B cells; Wilcoxon rank sum test). (e) Examples of C/EBPα-mediated A-to-B switching (Ebf1 locus) and OSKM-mediated B-to-A switching (Klf9 locus). Superenhancer (SE) location is indicated. (f) C/EBPα and Oct4 binding enrichment (inferred from ChIP-Seq and ATAC-Seq, respectively, see Supplemental Materials) relative to the genome-wide average at the 20 switching clusters shown in Fig.2g. Mean values with 95% CI are shown. (g) Box plots showing Oct4 and Klf4 binding enrichment in clusters (n=10) that switch B-to-A compartment early (D2-D4) or late (D6-PSC). Statistically significance was assessed using an unpaired two-tailed t-test. (h) Insulation strength (I-score) dynamics at hyper-dynamic borders (n=184) bound (n=123 for C/EBPα; n=37 for Oct4; n=22 for Klf4) or not bound (n=61 for C/EBPα; n=147 for Oct4; n=162 for Klf4) by the indicated TFs. Statistically significance was assessed using an unpaired two-tailed t-test).
Transcription factors drive topological genome reorganization
We investigated the impact of C/EBPα and OSKM on genome topology. Approximately 5% of the genome switched compartment during the C/EBPα-induced B-to-Bα transition and 5% during the OSKM-induced Bα-to-D2 transition. Of these early switching regions, only 29% (B-to-Bα) and 36% (Bα-to-D2) represented stable switches (Supplementary Fig.10a). C/EBPα had a largely repressive effect (66% A-to-B switches, e.g. Ebf1), while OSKM operated predominately as an activator (70% B-to-A switches, e.g. Klf9) (Fig.5e, Supplementary Fig.10a). Both C/EBPα and OSKM evoked A-to-B switching and transcriptional silencing of B cell-related loci. At D2, OSKM induced B-to-A switching and activation of known target genes of pluripotency factors involved in developmental processes (Supplementary Fig.10b). However, genes undergoing stable B-to-A switching in Bα cells were only upregulated after OSKM activation, including genes implicated in early embryonic development (e.g. Dppa3, Supplementary Fig.10c). Globally, C/EBPα binding was strongly enriched in the previously identified A-to-B switching clusters and depleted in B-to-A switching clusters (Fig.5f). In contrast, Oct4 and Klf4 binding (as inferred by ATAC-Seq) was concentrated in B-to-A switching regions (Fig.5f, Supplementary Fig.10d). This biased genomic distribution was already apparent at D2 and was stably maintained or reinforced, with early switching clusters (D2-D4) being rapidly targeted by Oct4 and Klf4 and late switching clusters (D6-PSC) becoming more gradually enriched (Fig.5g).
We next examined TF action at TAD borders. Oct4 target sites within ~30% of all border regions were already accessible at D2 (Supplementary Fig.10e). Oct4 or Klf4 recruitment to the most dynamic borders at D2 correlated with accelerated I-score gains as compared to borders bound at later time points (Fig.5h, Supplementary Fig.10f). C/EBPα-bound borders increased their I-scores more rapidly only after OSKM activation at D2 (Fig.5h) and Oct4 enrichment was significantly higher at borders previously bound by C/EBPα (Supplementary Fig.10g), suggesting that C/EBPα primes border regions for subsequent OSKM-induced topological changes. In agreement, Oct4, Klf4 and C/EBPα were frequently recruited to the same dynamic borders early in reprogramming (Supplementary Fig.10h).
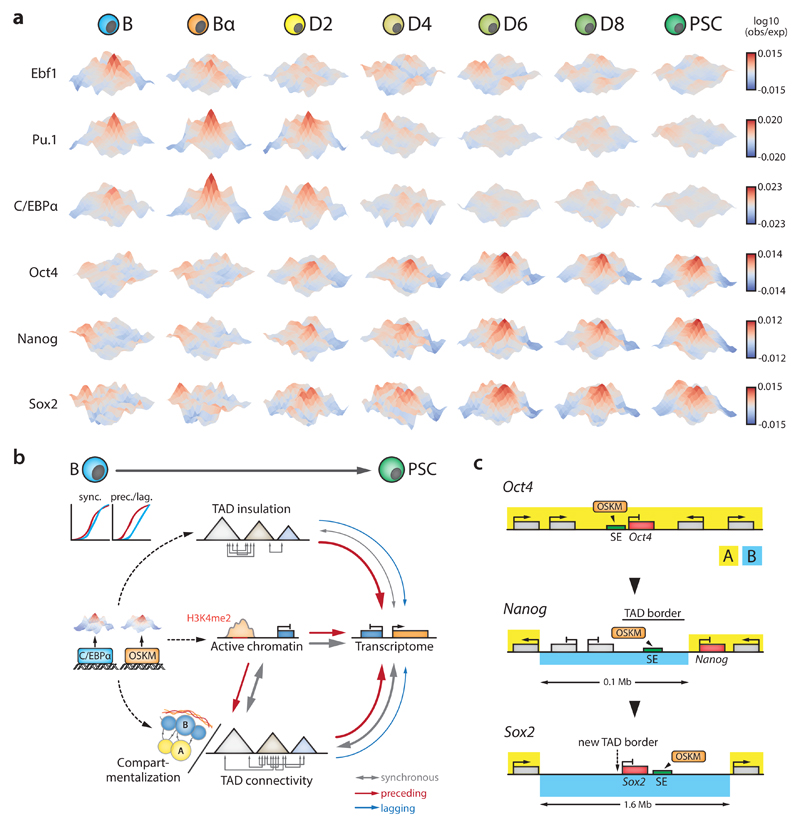
TF-bound sites cluster over large distances14,51,57,58, prompting us to address the dynamics of such 3D crosstalk during reprogramming by measuring inter-TAD spatial connectivity between TF-bound genomic sites at 5 kb resolution (within a 2-10 Mb window, analogous to PE-SCAn51). We observed strong interactions between Ebf1 or Pu.1 binding sites in B cells in agreement with their function as key B cell regulators (Fig.6a). These interaction networks largely disappeared for Ebf1 in Bα and for Pu.1 in D4 cells. Spatial clustering of C/EBPα targets was already present in B cells (Fig.6a), indicating that C/EBPα exploits existing 3D interaction hubs, such as those formed by Pu.1. Alongside interaction hubs mediated by hematopoietic TFs, Oct4 binding sites clustered from D2 onwards to establish 3D crosstalk between PSC-specific regulatory elements, showing that interaction hubs mediated by lineage-specific and pluripotency TFs can coexist (Fig.6a). Moreover, Nanog targeted regions formed interaction hubs as early as D2, before the gene becomes expressed at D4 (Fig.6a), suggesting that late pluripotency factors hitchhike onto an OSKM-mediated interaction hub to lock-in the PSC fate.
Figure 6. Dynamics of 3D crosstalk between transcription factor target sites and model schemes.
(a) 3D interaction meta-plots (5kb resolution) depicting interaction frequencies of sites bound by the indicated TFs during reprogramming. Hubs visualize inter-TAD crosstalk between TF binding sites 2-10 Mb apart. Area shown is centered on the respective TF binding sites (+/- 50kb). (b) Summary scheme depicting the interplay between TF binding, chromatin state, various aspects of genome topology and gene regulation during cell reprogramming. Arrows denote synchronous, preceding or lagging relationships. Arrow thickness indicates prevalence. (c) Activation scenarios for the pluripotency factors Oct4, Nanog and Sox2. Oct4 activation does not seem to require major topological modifications, as the gene and its superenhancer (SE) already reside in the A compartment in B cells and TAD border strength is unaltered. In contrast, Nanog activation is preceded by B-to-A compartment switching of its nearby SE as well as a decrease in TAD border strength that facilitates Nanog-SE interaction. Sox2 activation is preceded by the formation of a new TAD border through chromatin loop formation that progressively insulates the gene and its SE into a smaller subdomain. The complete 1.6 Mb Sox2 region switches from the B to the A compartment, concomitant with activation of the gene at D6.
In summary, C/EBPα and OSKM binding correlates with accelerated topological remodeling of compartmentalization and TAD insulation. In addition, computing inter-TAD 3D crosstalk between TF targets enabled us to visualize the stage-specific formation and disassembly of TF interaction hubs during reprogramming.
Discussion
Our analysis of somatic cell reprogramming (summarized in Supplementary Fig.10i) revealed that the overall dynamics of genome topology, chromatin state and gene expression are closely coupled. Nevertheless, this coupling often occurs in a non-synchronous manner: changes in subnuclear compartmentalization, TAD connectivity and TAD border insulation strength frequently precede transcriptional changes, with the reverse situation occurring only at low frequencies. We propose that transcription factors induce successive changes in chromatin state and genome architecture to enable gene regulatory rewiring during cell reprogramming (Fig.6b). Genome topology as an instructive force that facilitates transcriptional changes may represent a general principle for cell fate decisions.
Our findings also provide an explanation for the sequential activation of the genes encoding the pluripotency factors Oct4, Nanog and Sox2 in spite of the cells’ continuous exposure to the Yamanaka factors (Fig.6c). The embedding of Oct4 and its enhancers within an A compartment domain, surrounded by genes highly expressed in B cells, may explain its almost immediate activation by OSKM without detectable topological alterations. In contrast, the late activation of Nanog and Sox2 is preceded and accompanied by substantial changes in compartmentalization and TAD structure, indicating that the removal of topological barriers creates new opportunities for gene regulation. That active chromatin dynamics often anticipate changes in subnuclear compartmentalization suggests it plays a major role in mediating switches between the active A and the inactive B compartments (Fig.6b), in line with imaging and local chromatin conformation analyses59,60. Given the strong correlation between compartmentalization and DNA replication timing61, it will be of interest to attempt coupling changes in replication timing with the dynamics of genome topology and gene regulation. A preliminary analysis suggests that replication timing in the starting cell state is not a strong predictor of ordered A/B compartment switching (Supplementary Fig.11). Perturbation experiments aimed at demonstrating causality between specific topological changes and their effects on reprogramming represent a next frontier in dissecting the relationship between genome form, genome function and cell fate.
Previous studies have defined TADs as stable topological structures with little cell type-specificity17,50. At a qualitative level (i.e. present or not), we indeed find a minor portion of TAD borders to be altered during reprogramming. However, there are notable exceptions (e.g. de novo border establishment near Sox2), thus cautioning against the use of TAD definitions from unrelated cell types for the interpretation of gene regulatory processes. However, quantitative aspects of TADs, namely their connectivity and insulation potential, are subject to substantial changes during reprogramming and therefore more cell type-specific in nature.
How do TFs drive 3D genome changes? C/EBPα and Oct4 are selectively enriched in different regions destined to switch compartment. Here, TFs could act by inducing the subnuclear repositioning of specific loci62, for example by initiating modification of local chromatin states. In addition, the TFs rapidly induce insulation strength changes at the most dynamic TAD borders, independent of major changes in compartmentalization or chromatin state. Separate modes of action for TFs at these two topological levels seem plausible, as compartmentalization and TAD organization have been suggested to depend on distinct mechanisms63,64. Mechanistically, intrinsic abilities (e.g. via TF dimerization24) or interactions with canonical architectural proteins65–67 could allow TFs to modify genome topology. Here, inter-TAD hubs of TF target regions could contribute to topological reorganization by TFs, possibly through exploiting architecture previously established by other factors. As early targets (Fig.1), superenhancer regions may provide key platforms for TFs to achieve topological genome remodeling68. The ability of lineage instructive regulators to alter genome topology raises the possibility that in addition to their classical role as transcriptional regulators they possess unappreciated architectural functions at distinct topological layers.
Online Methods
Mice
We crossed ‘reprogrammable mice’ containing a doxycycline-inducible OSKM cassette and the tetracycline transactivator69 with an Oct4-GFP reporter strain70, as previously described31,32. B cells were isolated from 8 to 16 week old female mice (n=6 animals per biological replicate). Mice were housed in standard cages under 12h light–dark cycles and fed ad libitum with a standard chow diet. All experiments were approved by the Ethics Committee of the Barcelona Biomedical Research Park (PRBB) and performed according to Spanish and European legislation.
Cell culture & somatic cell reprogramming
Embryonic stem cells (E14TG2a) and short-term induced PSCs were cultured on gelatinized plates or Mitomycin C inactivated mouse embryonic fibroblasts (MEFs) in N2B27 medium (50% DMEM-F12, 50% Neurobasal medium, N2 (100x), B27 (50x)) supplemented with small-molecule inhibitors PD (1μM, PD0325901) and CHIR (3 μM, CHIR99021), as well as LIF (10 ng ml-1). Reprogramming of primary B cells isolated from the bone marrow of reprogrammable/Oct4-GFP mice was performed as previously described32. Two independent biological replicate reprogramming experiments were used for data generation. Briefly, pre-B cells were infected with C/EBPαER-hCD4 retrovirus, plated at 500 cells cm-2 in gelatinized 12 well plates on Mitomycin C inactivated MEF feeders in RPMI medium. C/EBPα was activated by adding 100 nM β-estradiol (E2) for 18 hours. After E2 washout, the cultures were switched to N2B27 medium supplemented with IL-4 (10 ng ml-1), IL-7 (10 ng ml-1) and IL-15 (2 ng ml-1). OSKM was activated by adding 2 μg ml-1 of doxycycline. Harvesting was done at indicated time points by trypsinization followed by a 20 min pre-plating step to remove feeder cells. All cell lines have been routinely tested for mycoplasma contamination.
RNA isolation, quantitative RT-PCR and RNA-Sequencing (RNA-Seq)
RNA was extracted using the miRNeasy mini kit (Qiagen) and quantified by Nanodrop. cDNA was produced with the High Capacity RNA-to-cDNA kit (Applied Biosystems) and used for qRT-PCR analysis in triplicate reactions with the SYBR Green QPCR Master Mix (Applied Biosystems). Primers are available upon request. Libraries were prepared using the Illumina TruSeq Stranded mRNA Library Preparation Kit followed by paired-end sequencing (2x125bp) on an Illumina HiSeq2500.
Assay for Transposase-Accessible Chromatin with high throughput sequencing (ATAC-Seq)
ATAC-seq was performed as previously described32. 100,000 cells were washed once with 100 μl PBS and resuspended in 50 μl lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.2% IGEPAL CA-630). Cells were centrifuged for 10 min at 500g (4°C), supernatant was removed and nuclei were resuspended in 50 μl transposition reaction mix (25 μl TD buffer, 2.5 μl Tn5 transposase and 22.5 μl nuclease-free water) and incubated at 37°C for 45 min. DNA was isolated using the MinElute DNA Purification Kit (Qiagen). Library amplification was performed by two sequential PCR reactions (8 and 5 cycles, respectively). Library quality was checked on a Bioanalyzer, followed by paired-end sequencing (2x75bp) on an Illumina HiSeq2500.
Chromatin Immunoprecipitation followed by high throughput sequencing (ChIP[m]-Seq)
ChIP-Seq using tagmentation (ChIPmentation) was performed as previously described36 with 100,000 crosslinked cells using 1 μl of H3K4me2 antibody (Abcam, ab32356) per IP. Tagmentation of immobilized H3K4me2-enriched chromatin was performed for 2 min at 37°C in 25 μl transposition reaction mix (12.5 μl TD buffer, 1.0 μl Tn5 transposase and 11.5 μl nuclease-free water). Library amplification was performed as described for ATAC-Seq. Library quality was checked on a Bioanalyzer, followed by sequencing (1x75bp) on an Illumina NextSeq500. Conventional ChIP-Seq was performed as previously described71 with 300,000 crosslinked cells using 5 μl of Ctcf antibody (Millipore, 07-729). Libraries were prepared using the Illumina TruSeq ChIP Library Preparation Kit and sequenced (1x50bp) on an Illumina HiSeq2500.
Chromosome Conformation Capture followed by high-throughput sequencing (4C-Seq)
4C-seq was performed as described previously72,73. Briefly, 0.5-1.0 million crosslinked nuclei were digested with Csp6I followed by ligation under dilute conditions. After decrosslinking and DNA purification, samples were digested overnight with DpnII and once more ligated under dilute conditions. Column-purified DNA was directly used as input for inverse PCR using primers (available upon request) with Illumina adapter sequences as overhangs. Several PCR reactions were pooled, purified and sequenced (1x75bp) on an Illumina HiSeq2500.
Gene Ontology (GO) analysis
GO analyses were performed using the Molecular Signatures Database (MSigDB)74 for gene lists or GREAT75 for peak lists. Only statistically significant (FDR<0.01) terms and pathways were used.
In-situ Hi-C library preparation
In-situ Hi-C was performed as described11 with the following modifications: 1) Two million cells were used as starting material; 2) chromatin was initially digested with 100 U MboI (New England Biolabs) for 2 hours, followed by addition of another 100U (2 hour incubation) and a final 100U before overnight incubation; 3) before fill-in with bio-dATP, nuclei were pelleted and resuspended in fresh 1x NEB2 buffer; 4) ligation was performed overnight at 24°C using 10,000 cohesive end units per reaction; 5) decrosslinked and purified DNA was sonicated to an average size of 300-400 bp using a Bioruptor Pico (Diagenode; 7 cycles of 20 s on and 60 s off); 6) DNA fragment size selection was only performed after final library amplification; 7) library preparation was performed with the NEBNext DNA Library Prep Kit (New England Biolabs) using 3 μl NEBNext adaptor in the ligation step; 8) libraries were amplified for 8-12 cycles using Herculase II Fusion DNA Polymerase (Agilent) and purified/size-selected using Agencourt AMPure XP beads (>200 bp). Hi-C Library quality was assessed by ClaI digest and low-coverage sequencing on an Illumina NextSeq500, after which every technical replicate (n=2) of each biological replicate (n=2) was sequenced at high-coverage on an Illumina HiSeq2500. Data from technical replicates was pooled for downstream analysis. We sequenced >18 billion reads in total to obtain 0.78-1.21 billion valid interactions per timepoint per biological replicate (see Supplementary Table 1 for dataset statistics).
Gene expression analysis using RNA-Seq data
Reads were mapped using STAR76 (-outFilterMultimapNmax 1 -outFilterMismatchNmax 999 -outFilterMismatchNoverLmax 0.06 -sjdbOverhang 100 –outFilterType BySJout -alignSJoverhangMin 8 -alignSJDBoverhangMin 1 -alignIntronMin 20 -alignIntronMax 1000000 -alignMatesGapMax 1000000) and the Ensembl mouse genome annotation (GRCm38.78). Gene expression was quantified using STAR (--quantMode GeneCounts). Sample scaling and statistical analysis were performed using the R package DESeq277 (R 3.1.0 and Bioconductor 3.0) and vsd counts were used for further analysis unless stated otherwise. Standard RPKM values were used as an absolute measure of gene expression. Genes changing significantly at any time point were identified using the nbinomLRT test (FDR<0.01) and for>2-fold change between at least two time points (average of two biological replicates, vsd values). Clustering was performed using the Rpackage Mfuzz (2.26.0).
Chromatin accessibility analysis using ATAC-Seq data
Reads were mapped to the UCSC mouse genome build (mm10) using Bowtie278 with standard settings. Reads mapping to multiple locations in the genome were removed using SAMtools79; PCR duplicates were filtered using Picard. Bam files were parsed to HOMER80 for downstream analyses and browser visualization. Peaks in ATAC-Seq signal were identified using findPeaks (-region -localSize 50000 -size 250 -minDist 500 -fragLength 0, FDR<0.001).
ChIPmentation/ChIP-Seq data analysis
Reads were mapped and filtered as described for ATAC-Seq. H3K4me2 enriched regions were identified using HOMER findpeaks (findPeaks -region -size 1000 -minDist 2500, using a mock IgG experiment as background signal). H3K4me2 coverage per 100kb genomic bin was computed using BEDTools81 and normalized for differences in sequencing depth (normalized coverage = coverage / (number of unique mapped reads in dataset / 1e6)). Ctcf peaks were identified using MACS282 with callpeak --nolambda --nomodel -g mm --extsize 100 -q 0.01.
4C-Seq data analysis
The sequence of the 4C-Seq reading primer was trimmed from the 5’ of reads using the demultiplex.py script from the R package fourCseq83 (allowing 4 mismatches). Reads in which this sequence could not be found were discarded. Reads were mapped using STAR and processed using fourCseq to filter out reads not located at the end of a valid fragment and to count reads per fragment. Signal tracks were made after smoothing RPKM counts per fragment with a running mean over three fragments.
In-situ Hi-C data processing and normalization
We processed Hi-C data using an in-house pipeline based on TADbit84. First, quality of the reads was checked using FastQC to discard problematic samples and detect systematic artifacts. Trimmomatic85 with the recommended parameters for paired end reads was used to remove adapter sequences and poor quality reads (ILLUMINACLIP:TruSeq3-PE.fa:2:30:12:1:true; LEADING:3; TRAILING:3; MAXINFO:targetLength:0.999; and MINLEN:36).
For mapping, a fragment-based strategy as implemented in TADbit was used, which is similar to previously published protocols86. Briefly, each side of the sequenced read was mapped in full length to the reference genome (mm10, Dec 2011 GRCm38). After this step, if a read was not uniquely mapped, we assumed the read was chimeric due to ligation of several DNA fragments. We next searched for ligation sites, discarding those reads in which no ligation site was found. Remaining reads were split as often as ligation sites were found. Individual split read fragments were then mapped independently. These steps were repeated for each read in the input FASTQ files. Multiple fragments from a single uniquely mapped read will result in as many contact as possible pairs can be made between the fragments. For example, if a single read was mapped through three fragments, a total of three contacts (all-versus-all) was represented in the final contact matrix. We used the TADbit filtering module to remove non-informative contacts and to create contact matrices. The different categories of filtered reads applied are:
self-circle: reads coming from a single restriction enzyme (RE) fragment and point to the outside.
dangling-end: reads coming from a single RE fragment and point to the inside.
error: reads coming from a single RE fragment and point in the same direction.
extra dangling-end: reads coming from different RE fragments but are close enough and point to the inside. The distance threshold used was left to 500 bp (default), which is between percentile 95 and 99 of average fragment lengths.
duplicated: the combination of the start positions and directions of the reads was repeated, pointing at a PCR artifact. This filter only removed extra copies of the original pair.
random breaks: start position of one of the reads was too far from RE cutting site, possibly due to non-canonical enzymatic activity or random physical breaks. Threshold was set to 750 bp (default), > percentile 99.9.
From the resulting contact matrices, low quality bins (those presenting low contacts numbers) were removed as implemented in TADbit’s “filter_columns” routine. A single round of ICE normalization87 - also known as “vanilla” normalization11 - was performed. That is, each cell in the Hi-C matrix was divided by the product of the interactions in its columns and the interactions in its row. Finally, all matrices were corrected to achieve an average content of one interaction per cell.
Identification of subnuclear compartments and topologically associated domains (TADs)
To segment the genome into A/B compartments, normalized Hi-C matrices at 100kb resolution were corrected for decay as previously published, grouping diagonals when signal-to-noise was below 0.0511. Corrected matrices were the split into chromosomal matrices and transformed into correlation matrices using the Pearson product-moment correlation. The first component of a PCA (PC1) on each of these matrices was used as a quantitative measure of compartmentalization and H3K4Me2 ChIPmentation data was used to assign negative and positive PC1 categories to the correct compartments. If necessary, the sign of the PC1 (which is randomly assigned) was inverted so that positive PC1 values corresponded to A compartment regions and vice versa for the B compartment.
Normalized contact matrices at 50kb resolution were used to define TADs, using a previously described method with default parameters43,54. First, for each bin, an insulation index was obtained based on the number of contacts between bins on each side of a given bin. Differences in insulation index between both sides of the bin were computed and borders were called searching for minima within the insulation index. The insulation score of each border was determined as previously described43, using the difference in the delta vector between the local maximum to the left and local minimum to the right of the boundary bin. This procedure resulted in a set of borders for each time point and replicate. To obtain a set of consensus borders along the time course, we proceeded in two steps: (a) merging borders of replicates and overlapping merged borders (that is, for each pair of replicates, we expand borders one bin on each side and kept only those borders present in both replicates as merged borders), and (b) we further expanded two extra bins (100kb) on each side and determined the overlap to get a consensus set of borders common to any pair of time points.
Domain scores were obtained by averaging cells over parts of the Hi-C matrix. In nature, this metric is sensitive to outlier cells with a lot of counts and is less sensitive to missing data. For this analysis (and for the meta-loop analysis below) we thus used a more stringent strategy to remove low-coverage bins by fitting a logistic function to the distribution of the sum of interactions in each bin:
Where f is the logistic function optimized by the variables a, b and c. N is the number of bins in the matrix, and x the number of interactions in a given bin. This fit was implemented by weighting bins with higher values of interactions, as we considered bins with lower counts artifacts. We set the weight function as dependent on the bin index, in the context of bins sorted by their sum of interactions:
With i representing the index of the bin and W the weight applied to the fitting. Once the logistic function was fitted, we used it to define a threshold. We removed bins with fewer counts than x when f(x) was equal to zero. The resulting filtered matrices were ICE normalized (1 round, see above). Finally, domain scores were calculated using matrices binned at 50kb by dividing the sum of intra-TAD contacts by the sum of all contacts involving the TAD.
Expression variability explained by TADs
To estimate expression variability, we fitted a hierarchical regression model per gene expression values for each timepoint, including three levels of organization: the gene itself, the local neighborhood (the 50 kb TSS bin) and the TAD. We used the variance associated with each level and the total variance of the model to assess the proportion of variability explained by each factor. In order to test if topology was playing a role beyond linear proximity of genes, we repeated the estimation replacing actual TADs by a fixed segmentation of the genome in domains with the same size as the average TAD (i.e. “fake” TADs, constructed by placing a border at fixed distances that correspond to the average size of TADs). Model estimation was performed using the lme4 R package.
Inter and intra-compartment strength measurements
We followed a previously reported strategy to measure overall interaction strengths within and between A and B compartments63. Briefly, we based our analysis on the 100kb bins showing the most extreme PC1 values, discretizing them by percentiles and taking the bottom 20% as B compartment and the top 20% as A compartment. We classified each bin in the genome according to PC1 percentiles and gathered contacts between each category, computing the log2 enrichment over the expected counts by distance decay. Finally, we summarize each type of interaction (A-A, B-B and A-B/B-A) by taking the median values of the log2 contact enrichment.
Meta-analysis of borders, loops and interactions between transcription factor binding sites
To assess whether particular parts of the Hi-C interaction matrices had common structural features, we performed meta-analyses by merging individual sub-matrices into an average meta-matrix in a similar fashion as previously published51. Three types of meta-analysis were performed. First, we studied TAD border dynamics at 50kb resolution by extracting interaction counts 1.25Mb up and downstream of the TAD border. Extracted matrices were averaged for each group of clustered TAD borders, including those that increase, decrease or do not change in insulation score during reprogramming. Second, using 5kb resolution contact maps, we investigated the dynamics of a previously identified set of chromatin loops in primary B cells and PSCs49 by extracting interaction counts 50 kb up and downstream of loop anchor regions. Meta-loop matrices were then calculated by averaging individually subtracted loop matrices into a single one per group. Third, we studied whether two regions bound by a given transcription factor (TF) are likely to find each other more than expected within a genomic distance ranging from 2 to 10Mb. All sub-matrices at 5kb resolution between pairs of TF binding sites and 50kb up and downstream of a TF peak were extracted and averaged into a single meta-matrix. For Oct4, Nanog and Sox2 meta-analyses we used those TF binding sites that overlapped with an ATAC-Seq peak (see above) at the D2 stage. All meta-analyses were performed using the observed/expected Hi-C matrices, which were filtered, ICE normalized and corrected for decay. For visualization proposes, the resulting meta-analysis matrices were smoothed using a Gaussian filter of sigma=1.
Virtual 4C analysis and promoter-superenhancer linking
For the generation of v4C profiles we first chose a bait region (e.g. Sox2) and (optionally) a window size around the bait (final viewpoint window was centered on the bait). We then extracted the observed Hi-C matrix at 5 kb resolution for that specific region. Rows overlapping the bait were subsetted after which we summed up all bait rows to get the number of observed contacts per bin (column). Aiming to reduce noise, we performed a moving average smoothing (5 bins) to obtain v4C profiles. Count numbers per bin were normalized for differences in sequencing depth between time points. For visualization purposes, we removed all data overlapping the bait extended with one bin per side.
We took advantage of this approach to link promoters to SEs. For each SE, we set a window of 2 Mb around the SE bait and extracted the corresponding Hi-C matrix at 5 kb resolution, removing low count and/or mappability bins. Using the full inter-chromosomal matrix, we computed an expected Hi-C matrix, averaging all pairs of loci at the same distance per chromosome. After merging the two replicates, we generated virtual 4C profiles for each SE with the observed and corresponding expected number of counts. These profiles allowed us to rank nearby promoters according to their contact enrichment (observed/expected), designating the two highest-ranking genes as putative SE targets. Using this method, we detected a larger number of genes associated with the superenhancer subset (372 versus the 210 assigned by GREAT), which included half of the genes identified using GREAT. GO analyses and gene expression analyses on the GREAT gene set or this extended target gene set were similar, although the Hi-C based gene set showed stronger enrichments for GO terms associated with embryonic development. Analyses on the Hi-C based gene set were used in Fig.1.
Integration of B cell replication timing data
We partitioned the genome in 100 Kb bins, labeling the compartment (A, B or 0) for each time point and biological replicate. Then, we identified the bins with more than one compartment type (i.e. switching bins). For each bin, the residence time in A or B was the number of consecutive time points in A or B previous to a switch. The results presented are the grand sum per compartment, residence time and biological replicate over all switching bins.
Statistics and Reproducibility
In-situ Hi-C data used throughout the paper was generated by analyzing two independent B-to-iPS replicate reprogramming experiments. Representative data was only shown if results were similar for both independent biological replicate experiments. All box plots depict the first and third quartiles as the lower and upper bounds of the box, with a thicker band inside the box showing the median value and whiskers representing the 1.5x interquartile range. Wilcoxon rank sum tests were performed using the wilcox.test() function in R in a two-sided manner. T-tests were performed using the t.test() function in R in an unpaired and two-sided fashion with (n-2) degrees of freedom.
Data availability
All data generated has been deposited in the Gene Expression Omnibus (GEO) under GSE96611. Accession number of published datasets used: Ctcf ChIP-Seq in pre-B cells: SRR39783788; Ctcf ChIP-Seq in induced PSCs: GSE7647849; Oct4 and Nanog ChIP-Seq in PSCs: GSE4428639; Klf4 ChIP-Seq in PSCs: GSE1143189; C/EBPα and Pu.1 ChIP-Seq in Bα cells: GSE7121532; Ebf1 V5-ChIP-Seq in pro-B cells: GSE5359590. CH12 Repli-chip data was obtained from ENCODE (Biosample ENCBS789HDO).
Supplementary Material
Acknowledgements
We thank D. Higgs, J. Hughes, J. Davies and Z. Duan for advice on Hi-C technology; C. Schmidl for ChIPmentation advice; C. van Oevelen for help with Ctcf ChIP-Seq.; C. Segura for mouse colony management and T. Tian for bone marrow collection; the CRG Genomics Core Facility and the CRG-CNAG Sequencing Unit for sequencing and Graf laboratory members for discussions. This work was supported by the European Research Council Synergy Grant (4D-Genome) and Ministerio de Educacion y Ciencia, SAF.2012-37167. R.S. was supported by an EMBO Long-term Fellowship (ALTF 1201-2014) and a Marie Curie Individual Fellowship (H2020-MSCA-IF-2014).
Footnotes
Author Contributions
R.S. and T.G. conceived the study and wrote the manuscript with input from all co-authors. R.S. performed molecular biology, RNA-Seq, ChIP-Seq, ChIPmentation, ATAC-Seq, 4C-Seq and in-situ Hi-C experiments. R.S, E.V., F.S, J.Q., A.G., S.C. and M.A.M-R. performed bioinformatic analyses. R.S., E.V., F.S. and M.A.M-R. integrated and visualized data. B.D.S. performed reprogramming experiments with help from R.S. and C.B. R.S., F.L.D. and Y.C. optimized and implemented in-situ Hi-C technology. J.H. performed high-throughput sequencing. F.L.D., G.F., M.B. and M.A.M-R. provided valuable advice and T.G. supervised the research.
Competing Financial Interests
The authors declare no competing financial interests.
URLs
FastQC quality control tool; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Picard tool; http://broadinstitute.github.io/picard
MACS2 tool; https://github.com/taoliu/MACS/
References
- 1.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–39. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–71. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 4.Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14:762–75. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–21. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denker A, de Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30:1357–82. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon JR, Gorkin DU, Ren B. Chromatin Domains: The Unit of Chromosome Organization. Mol Cell. 2016;62:668–80. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–9. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boettiger AN, et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–22. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353:598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci U S A. 2015;112:4672–7. doi: 10.1073/pnas.1504783112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens TJ, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017 doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Lupianez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Symmons O, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrey G, et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 22.Montavon T, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–45. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Symmons O, et al. The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Dev Cell. 2016;39:529–543. doi: 10.1016/j.devcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng W, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–44. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell. 2017;169:216–228 e19. doi: 10.1016/j.cell.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Ke Y, et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell. 2017;170:367–381 e20. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 27.Du Z, et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 28.Apostolou E, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghavi-Helm Y, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 30.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Stefano B, et al. C/EBPalpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature. 2014;506:235–9. doi: 10.1038/nature12885. [DOI] [PubMed] [Google Scholar]
- 32.Di Stefano B, et al. C/EBPalpha creates elite cells for iPSC reprogramming by upregulating Klf4 and increasing the levels of Lsd1 and Brd4. Nat Cell Biol. 2016;18:371–81. doi: 10.1038/ncb3326. [DOI] [PubMed] [Google Scholar]
- 33.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–22. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bar-Nur O, et al. Small molecules facilitate rapid and synchronous iPSC generation. Nat Methods. 2014;11:1170–6. doi: 10.1038/nmeth.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruetz T, Kaji K. Routes to induced pluripotent stem cells. Curr Opin Genet Dev. 2014;28:38–42. doi: 10.1016/j.gde.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods. 2015;12:963–5. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–54. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–25. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Martello G, Smith A. The nature of embryonic stem cells. Annu Rev Cell Dev Biol. 2014;30:647–75. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- 42.Blinka S, Reimer MH, Jr, Pulakanti K, Rao S. Super-Enhancers at the Nanog Locus Differentially Regulate Neighboring Pluripotency-Associated Genes. Cell Rep. 2016;17:19–28. doi: 10.1016/j.celrep.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crane E, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–4. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–46. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Bortle K, et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes Dev. 2008;22:575–80. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, et al. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014;9:e114485. doi: 10.1371/journal.pone.0114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou HY, et al. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014;28:2699–711. doi: 10.1101/gad.248526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krijger PH, et al. Cell-of-Origin-Specific 3D Genome Structure Acquired during Somatic Cell Reprogramming. Cell Stem Cell. 2016;18:597–610. doi: 10.1016/j.stem.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–6. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Wit E, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–31. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 52.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–16. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasque V, Plath K. X chromosome reactivation in reprogramming and in development. Curr Opin Cell Biol. 2015;37:75–83. doi: 10.1016/j.ceb.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giorgetti L, et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–9. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng X, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16:152. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minajigi A, et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349 doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Z, et al. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. Elife. 2014;3:e04236. doi: 10.7554/eLife.04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Therizols P, et al. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–42. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wijchers PJ, et al. Cause and Consequence of Tethering a SubTAD to Different Nuclear Compartments. Mol Cell. 2016;61:461–73. doi: 10.1016/j.molcel.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–5. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zullo JM, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–87. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 63.Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering C, Mirny L, Spitz F. Two independent modes of chromosome organization are revealed by cohesin removal. bioRxiv. 2016 doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nora EP, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944 e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van den Berg DL, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–81. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–32. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei Z, et al. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 68.Beagrie RA, et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017 doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat Methods. 2010;7:56–9. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boiani M, Eckardt S, Scholer HR, McLaughlin KJ. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 2002;16:1209–19. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Oevelen C, et al. C/EBPalpha Activates Pre-existing and De Novo Macrophage Enhancers during Induced Pre-B Cell Transdifferentiation and Myelopoiesis. Stem Cell Reports. 2015;5:232–47. doi: 10.1016/j.stemcr.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stadhouders R, et al. Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat Protoc. 2013;8:509–24. doi: 10.1038/nprot.2013.018. [DOI] [PubMed] [Google Scholar]
- 73.Brouwer RW, van den Hout MC, van I WF, Soler E, Stadhouders R. Unbiased Interrogation of 3D Genome Topology Using Chromosome Conformation Capture Coupled to High-Throughput Sequencing (4C-Seq) Methods Mol Biol. 2017;1507:199–220. doi: 10.1007/978-1-4939-6518-2_15. [DOI] [PubMed] [Google Scholar]
- 74.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klein FA, et al. FourCSeq: analysis of 4C sequencing data. Bioinformatics. 2015;31:3085–91. doi: 10.1093/bioinformatics/btv335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Serra F, et al. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput Biol. 2017;13:e1005665. doi: 10.1371/journal.pcbi.1005665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ay F, et al. Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics. 2015;16:121. doi: 10.1186/s12864-015-1236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ribeiro de Almeida C, et al. The DNA-binding protein CTCF limits proximal Vkappa recombination and restricts kappa enhancer interactions to the immunoglobulin kappa light chain locus. Immunity. 2011;35:501–13. doi: 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 89.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 90.Schwickert TA, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15:283–93. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated has been deposited in the Gene Expression Omnibus (GEO) under GSE96611. Accession number of published datasets used: Ctcf ChIP-Seq in pre-B cells: SRR39783788; Ctcf ChIP-Seq in induced PSCs: GSE7647849; Oct4 and Nanog ChIP-Seq in PSCs: GSE4428639; Klf4 ChIP-Seq in PSCs: GSE1143189; C/EBPα and Pu.1 ChIP-Seq in Bα cells: GSE7121532; Ebf1 V5-ChIP-Seq in pro-B cells: GSE5359590. CH12 Repli-chip data was obtained from ENCODE (Biosample ENCBS789HDO).