Abstract
The mechanisms by which mitochondrial metabolism supports cancer anabolism are still unclear. Here, we unexpectedly find that genetic and pharmacological inactivation of Pyruvate Dehydrogenase A1 (PDHA1), a subunit of pyruvate dehydrogenase complex (PDC) inhibits prostate cancer development in different mouse and human xenograft tumour models by affecting lipid biosynthesis. Mechanistically, we show that in prostate cancer, PDC localizes in both mitochondria and nucleus. While nuclear PDC controls the expression of Sterol regulatory element-binding transcription factor (SREBF) target genes by mediating histone acetylation, mitochondrial PDC provides cytosolic citrate for lipid synthesis in a coordinated effort to sustain anabolism. In line with these evidence, we find that PDHA1 and the PDC activator, Pyruvate dehydrogenase phosphatase 1 (PDP1), are frequently amplified and overexpressed at both gene and protein level in prostate tumours. Taken together, these findings demonstrate that both mitochondrial and nuclear PDC sustain prostate tumourigenesis by controlling lipid biosynthesis thereby pointing at this complex as a novel target for cancer therapy.
Mitochondrial metabolism is a source of energy and metabolic intermediates that serve various purposes, from redox homeostasis to anabolism or epigenetics1. There is an emerging association between increased mitochondrial metabolism and cancer pathogenesis and progression, but the molecular means underlying this biological process are still unknown1–10. The pyruvate dehydrogenase complex (PDC) is a gatekeeper multi-protein complex that catalyzes the conversion of pyruvate to acetyl coenzyme A (acetyl coA) thereby regulating the mitochondrial activity. This complex includes a major component, PDHA1, that can be dephosphorylated by the Pyruvate dehydrogenase phospatase (PDPs) Pdp1 and Pdp2 and phosphorylated by the pyruvate dehydrogenase kinases (Pdks)11–13. While dephosphorylation of PDHA1 activates the PDC, phosphorylation blocks its activity. In turn, we postulated that by impairing the function of PDHA1 and the PDC we would hamper mitochondrial metabolism and ascertain its consequences in prostate cancer biology.
Results
Pdha1 knockout induces tumour suppression in mice and human prostate tumours
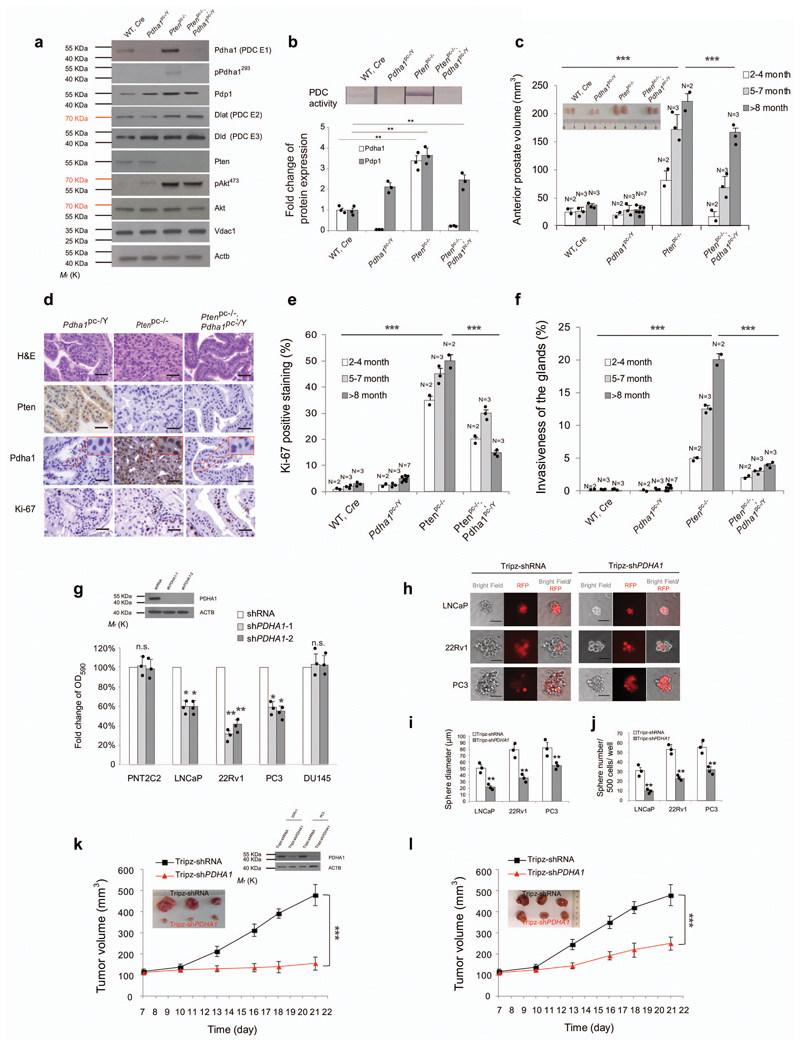
We thus inactivated Pdha1, in Pten-null prostate conditional (Pten pc-/-) mice that develop high-grade intra-epithelial prostate tumours at an early age and invasive prostate cancer at late age14–16. The presence of Cre recombinase, the recombination of the Pten exon 4 and 5 and Pdha1 exon 8, in Ptenpc-/-; Pdha1pc-/Y prostates were verified by genotyping14,17,18 (Supplementary Fig. 1a). Notably, Ptenpc-/- mice develop tumours characterized by increased mRNA and/or protein levels of both Pdha1, Dlat, Dld and Pdp1, but not Pdp2 or Pdks, and an increased PDC activity as compared to normal prostates (Fig. 1a, b, Supplementary Fig. 1b and uncropped blots in Supplementary Fig. 12). Inactivation of Pdha1 in Ptenpc-/- tumours abrogated PDC activity and induced a strong growth inhibition in all the prostate glands of mice of different ages (Fig. 1c). This was associated with a strong reduction in cell proliferation as shown by the decreased Ki-67 staining and number of glands affected by invasive prostate cancer (Fig. 1d-f and Supplementary Fig. 1c). The strong arrest in proliferation in Ptenpc-/-; Pdha1pc-/Y tumours and Pten-/-; Pdha1-/Y MEFs occurred independently of senescence19 and apoptosis, that decreased when compared to controls (Supplementary Fig. 1d-i). Notably, Pdha1 inactivation induced growth arrest exclusively in Pten-/- MEFs, without altering the growth of Ptenwt MEFs (Supplementary Fig. 1g). Moreover, Pdha1 deletion in Pten null tumours and MEFs did not affect the levels of pAkt at Ser 473 and the total amount of mitochondria as indicated by the protein levels of Voltage-dependent anion channel 1 (Vdac1) 5 (Fig. 1a, Supplementary Fig. 1j and uncropped blots in Supplementary Fig. 13).
Figure 1. Pdha1 knockout induces tumour suppression in mice and human prostate tumours.
(a) Western blot analysis of indicated proteins in wild type, Pdha1pc-/Y, Ptenpc-/- and Ptenpc-/-; Pdha1pc-/Y prostates and tumours (n=3, independent prostate samples). Uncropped images are in Supplementary Figure 12. (b) Upper panel, PDC activity measurement in wild type, Pdha1pc-/Y, Ptenpc-/- and Ptenpc-/-; Pdha1pc-/Y prostates and tumours (n=3, independent prostate samples). Lower panel, Quantification of indicated proteins normalized to wild type tissues in indicated prostate tumours in (a) (n=3, independent prostate samples). (c) Comparison of anterior prostate (AP) lobe volumes (mm3, 2 independent lobes per animal are presented) from male of indicated ages and genotypes between wild type, Pdha1pc-/Y, Ptenpc-/- and Ptenpc-/-; Pdha1pc-/Y prostate and tumours (N, the number of mice of indicated ages). Inset is the representative image of anterior prostate lobes in the panel. (d) Representative micrographs in histopathological analysis (haematoxylin/eosin staining and indicated proteins) of anterior prostates (AP) in Pdha1pc-/Y, Ptenpc-/- and Ptenpc-/-; Pdha1pc-/Y prostate tissues from 12 weeks old male mice (n=3) (Scale bar represents 50 μm, insets are regions shown in higher magnification, see also Supplementary Fig. 1c for all the genotypes and images of lower magnification). (e,f) Quantification of the percentage of Ki-67 positive cells (e) and invasive prostate glands (f) in prostates from mice of indicated genotypes at different ages (N, number of mice of indicated ages). (g) Relative cell number quantification by crystal violet staining in the indicated prostate cancer cell lines infected with shPDHA1 or scramble control. Data is normalized to shRNA control. PDHA1 inhibition validated by western blot analysis using two different shPDHA1 was shown in the inset of the panel. Uncropped images are in Supplementary Figure 12. (n=3, independent cell cultures). (h) Spheroid Formation Assays in LNCaP, 22Rv1 and PC3 cells infected with doxycycline-induced Tripz-shPDHA1 or scramble control (n=3, independent cell cultures). Scale bar represents 50 µm. (i,j) Quantification of spheroid diameter (i) and spheroid number per 500 cells (j) in LNCaP, 22Rv1 and PC3 cells infected with doxycycline-induced Tripz-shPDHA1 or scramble Tripz-shRNA control (n=3, independent cell cultures). (k,l) Evaluation of tumour formation in xenotransplantation experiments of 22Rv1 (k) and PC3 (l) cells infected with indicated shPDHA1 or scramble shRNA control. The knockdown of PDHA1 in 22Rv1 and PC3 xenograft tumours is validated by western blot in the inset of panel (k). Uncropped images are in Supplementary Figure 12. (n=6 animals; 12 independent tumour samples). Error bars indicate s.e.m. **P < 0.01; ***P < 0.001. n.s, not significant.
Next, we assessed the status of both PDHA1 and PDP1 in human prostate cancers. Bioinformatics analysis revealed that both PDHA1 and PDP1 are frequently amplified at the gene level and over-expressed in human prostate tumours. Interestingly, PDP1 was found amplified (3/3 available datasets) and overexpressed (9/11 datasets) in primary prostate tumours when compared to normal prostate tissues whereas PDHA1 was found amplified (2/3 datasets) and overexpressed (7/13 datasets) (Supplementary Fig. 2a-e) in metastatic when compared to primary tumours. We also stained PDHA1, phospho-PDHA1 and PDP1 on a tissue microarray (TMA) consisting of 128 prostate carcinoma and normal prostate samples. Immunohistochemistry analyses revealed that both PDHA1 and PDP1 are frequently over-expressed in human prostate tumours and their expression increases in tumours with high Gleason score (Supplementary Fig. 3a-c). On the contrary, phospho-PDHA1 staining was found upregulated only in a small portion of cases with a low Gleason score (Supplementary Fig. 3a,d). Interestingly the majority of cases having either high PDP1 or PDHA1 staining were negative for phospho-PDHA1 (Supplementary Fig. 3e). Three out of four prostate cancer cell lines exhibited increased PDC activity compared to non-transformed PNT2C2 prostate cells (Supplementary Fig. 3f). This was associated with increased protein levels of both PDHA1 and PDP1 and enhanced mitochondrial oxygen consumption dependent on both glucose and glutamine utilization (Supplementary Fig. 3f-i and uncropped blots in Supplementary Fig. 14). Taken together these data demonstrate that in prostate tumours and in cancer cell lines the PDC is active.
We next inactivated PDHA1 in prostate cancer cells by means of different PDHA1 shRNAs. In agreement with the results in the mouse model, we found that inactivation of PDC resulted in inhibition of cellular proliferation independently of apoptosis (Fig. 1g and Supplementary Fig. 3j,k and uncropped blots in Supplementary Fig. 12). We also detected, decreased mitochondrial respiration in cells infected with shPDHA1 when compared to control (Supplementary Fig. 3l). Spheres forming ability in LNCaP, 22Rv1, and PC3 was also impaired upon PDHA1 inactivation (Fig. 1h-j, Supplementary Fig. 3m and uncropped blots in Supplementary Fig. 14). 22Rv1 and PC3 prostate cancer cells, infected with shPDHA1 also formed smaller tumours in vivo than controls (Fig. 1k, l and uncropped blots in Supplementary Fig. 12).
Pdha1 inactivation decreases mitochondrial intermediates
Next, we performed metabolomics analysis in WT, Pdha1pc-/Y, Ptenpc-/- and Ptenpc-/-; Pdha1pc-/Y prostates. Ptenpc-/- tumours presented signs of increased TCA cycle as measured by increased, citrate, α-ketoglutarate, acetyl CoA, ATP production and NADH levels (Supplementary Fig. 4a-d, Supplementary Table 4 and Supplementary Note). Increased extracellular lactate levels in Pten null MEFs also suggested increased glycolytic flux in these cells when compared to control (Supplementary Fig. 4e and Supplementary Note). Pdha1 inactivation in Pten null tumours and MEFs resulted in decreased TCA cycle intermediates such as citrate and α-ketoglutarate and affected NADH/NAD+ ratio, ATP and acetyl-CoA levels (Supplementary Fig. 4a-f, Supplementary Table 4 and Supplementary Note). However, glycolysis and lactate production were not consistently affected (Supplementary Fig. 4a, e), in agreement with previous findings in different models20–22. Of note, oxaloacetate levels significantly increased in compound mutant mice, in line with a block in the cycle due to loss of acetyl-CoA production downstream PDC (Supplementary Fig. 4a, d and Supplementary Table 4). To determine whether Pdha1 inactivation induces changes in central carbon metabolism we measured stationary flux5,20 through TCA cycle in prostate epithelial cells derived from transgenic mice of different genotypes, using 13C-stable isotope labeled glucose, glutamine, and palmitate respectively. This analysis showed increased glucose and glutamine incorporation into citrate, fumarate, and malate in Ptenpc-/- cells when compared to normal prostate epithelial cells. Glucose incorporation into citrate in Ptenpc-/-; Pdha1pc-/Y tumor cells, decreased (~70%) when compared to Ptenpc-/- cells. Importantly, PDC inactivation led to compensatory oxidative glutaminolysis and to the production of acetyl-CoA from fatty acid β-oxidation for the reactivation of the TCA (reflected in a 30% increased citrate labeling from palmitate). This compensation, however, did not overcome the decreased citrate synthesis (Supplementary Fig. 4a and Supplementary Table 4). Surprisingly, glutaminolysis compensated the oxidative reactions after α-ketoglutarate to fumarate and malate until aspartate synthesis in TCA cycle. However, glutamine carbon pool was blocked from the reactions for citrate production probably due to the decreased expression of Idh1 in Ptenpc-/-; Pdha1pc-/Y when compared to Ptenpc-/- tumour cells (Supplementary Fig. 5d,g and Supplementary Table 5-7). Finally, the Pyruvate carboxylase activity did not change in response to PDHA1 loss since the M+3 aspartate did not increase in Ptenpc-/-; Pdha1pc-/Y when compared to Ptenpc-/- tumour cells (Supplementary Fig. 4g and and Supplementary Table 5). Taken together, these data demonstrate that PDC inactivation reduces production of TCA intermediates and energetic yield in prostate tumours.
Pdha1 knockdown induces tumour suppression by abrogating lipogenesis
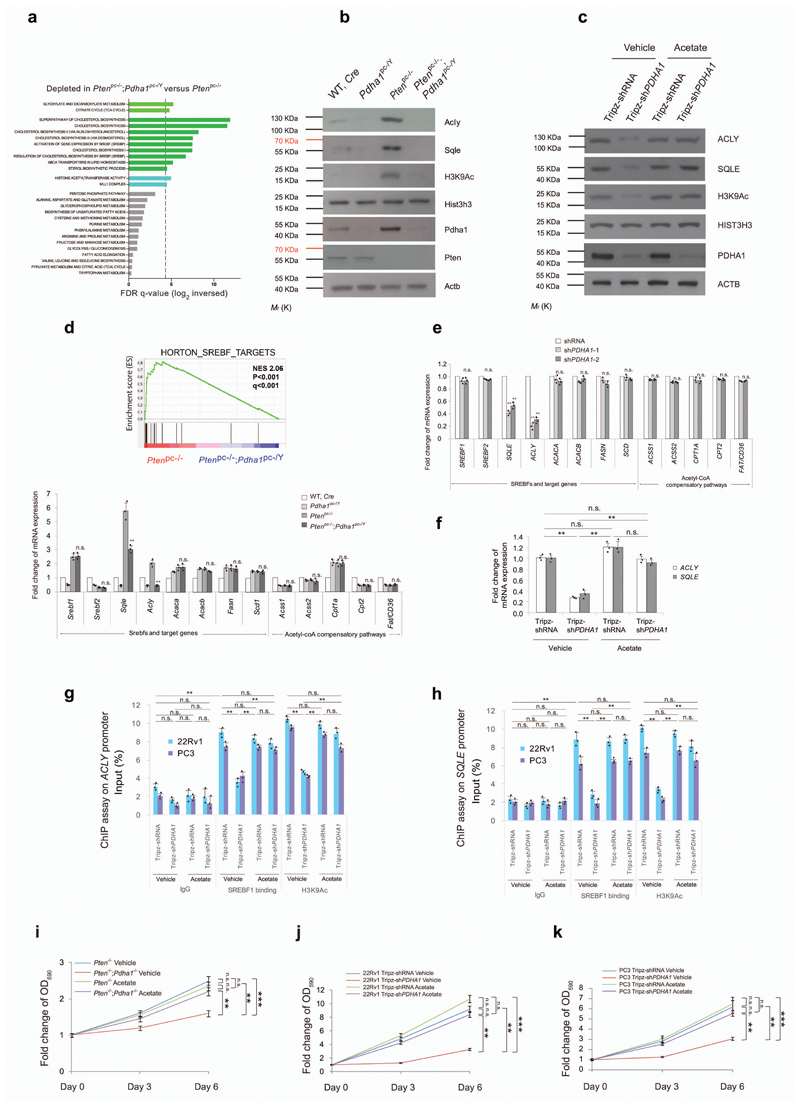
Pdha1 has been long defined as a mitochondrial metabolism regulator. However, recent evidence demonstrates that the PDC controls the nuclear pool of acetyl-CoA thereby promoting histone acetylation and regulating gene expression23,24. In line with this evidence, we detected a strong nuclear localization of PDHA1 in Ptenpc-/- tumours and PDC activity in both the cytosol and nucleus of these tumour cells (see insets in Fig. 1d, Supplementary Figs. 1c and 5a,b and uncropped blots in Supplementary Fig. 14). We also detected Pdha1, Dlat and Dld in the nuclear fractions of Ptenpc-/- tumor cells (Supplementary Fig. 5c and uncropped blots in Supplementary Fig. 14). Transcriptomics followed by GSEA analysis in Ptenpc-/- and Ptenpc-/-; Pdha1pc-/Y tumours demonstrated that fatty acid synthesis, cholesterol biogenesis and genes controlled by the sterol responsive element binding factor (SREBF)25–28 were the most downregulated pathways in Ptenpc-/-; Pdha1pc-/Y tumours (Fig. 2a). Inactivation of Pdha1 in Ptenpc-/- tumours and shPDHA1 human prostate cancer cells decreased Histone H3 Lysine 9 acetylation (H3K9Ac) thereby potentially affecting gene expression (Fig. 2b,c and uncropped blots in Supplementary Fig. 12). Notably, two rate-limiting enzymes engaging fatty acid synthesis and cholesterol biogenesis, ATP Citrate Lyase (Acly)29,30 and Squalene Epoxidase (Sqle) 31,32 were strongly decreased in both Ptenpc-/-; Pdha1pc-/Y tumours and shPDHA1 human prostate cancer cell lines at both protein and gene expression levels (Fig. 2b-f). Downregulation of ACLY in these cells was not associated with the upregulation of Acyl-coenzyme A synthetase short-chain family member 2 (ACSS2)33,34 (Fig. 2d,e). Genes that divert TCA cycle and glutamine intermediates into the lipid metabolisms22,35,36 were also downregulated by PDHA1 inactivation in both mouse and human prostate tumour cells (Supplementary Fig. 5d-f). Importantly, acetate supplementation30 in the culture media of 22Rv1 and PC3 infected with shPDHA1, restored global H3K9Ac, SQLE and ACLY levels (Fig. 2c). Chromatin immunoprecipitation (ChIP) analysis demonstrated that H3K9Ac and the binding of SREBF1 on the promoters of ACLY and SQLE decreased in response to the PDHA1 knockdown in human prostate cancer cells and this was restored by acetate supplementation (Supplementary Fig. 5h, i and Fig. 2g,h). Acetate supplementation restored cell growth in both Pten-/-; Pdha1-/Y MEFs and shPDHA1 22Rv1 and PC3 cells (Fig. 2i-k). Of note, we did not detect any changes in H3K9Ac on the promoter of two cell cycle regulators, E2F Transcription Factor 1 (E2F1) and Cyclin D1 (CCND1), demonstrating the specificity of the epigenetic regulation of PDC in cancer cells (Supplementary Fig. 5j,k). Interestingly, ACLY over-expression in prostate cell lines infected with shPDHA1 rescued the growth arrest induced by loss of PDHA1 only in cells supplemented with citrate (Supplementary Fig. 5l-n and uncropped blots in Supplementary Fig. 14) in agreement with previous data30.
Figure 2. Pdha1 inactivation induces tumour suppression by down-regulating lipogenic genes.
(a) Gene expression profile analysis based on metabolic pathway datasets (GOCC, Gene Ontology Cellular Component; KEGG, Kyoto Encyclopaedia of Genes and Genomes; GOBO, Gene expression-based Outcome; GOMF, Gene Ontology Molecular Function; HumanCyc; Reactome) in Ptenpc-/-; Pdha1pc-/Y tumours versus Ptenpc-/- tumours. Dotted line indicates P=0.05 (n=3). (b) Western blot analysis of indicated proteins in wild type, Pdha1pc-/Y, Ptenpc-/- and Ptenpc-/-; Pdha1pc-/Y prostates and tumours. Uncropped images are in Supplementary Figure 12. (n=3, independent prostate samples). (c) Western blot analysis of indicated proteins in 22Rv1 cells infected with doxycycline-induced Tripz-shPDHA1 or scramble control and treated with 100 μM acetate or vehicle over a 6-day period. Uncropped images are in Supplementary Figure 12. (n=3, independent cell cultures). (d) Upper panel, GSEA of SREBF target genes in Ptenpc-/-; Pdha1pc-/Y versus Ptenpc-/- prostate tumours. Normalized enriched scores (NES) are presented. Data are mean ± standard deviation (s.d.). Lower panel, quantitative real time-PCR analysis of mRNA expression of indicated SREBFs and target genes and genes in acetyl CoA compensatory pathways in mouse prostate and tumours the indicated genotypes (n=3). (e) Quantitative real time-PCR analysis of mRNA expression of indicated SREBFs and target genes and genes in acetyl coA compensatory pathways in 22Rv1 cells infected with shPDHA1 and scramble shRNA control (n=3, independent cell cultures). (f) Quantitative RT-PCR analysis of ACLY and SQLE in 22Rv1 cells infected with doxycycline-induced Tripz-shPDHA1 or scramble control and treated with acetate (100 µM) or vehicle for 6 days (n=3, independent cell cultures). (g,h) Chromatin immunoprecipitation analysis showing the binding of SREBF1 and H3K9Ac on the promoters of ACLY (g) and SQLE (h) in 22Rv1 and PC3 cells infected with doxycycline-induced Tripz-shPDHA1 or scramble control and treated with 100 μM acetate or vehicle over a 6-day period (n=3, independent cell cultures). (i-k) Relative cell number quantification by crystal violet staining of Pten-/- and Pten-/-;Pdha1-/Y MEFs (i) and human cancer cells 22Rv1 and PC3 cells infected with doxycycline-induced Tripz-shPDHA1 or scramble control. (j,k) and treated with acetate (100 µM) or vehicle over a 6-day period (n=3, independent cell cultures). Error bars indicate s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001. n.s, not significant.
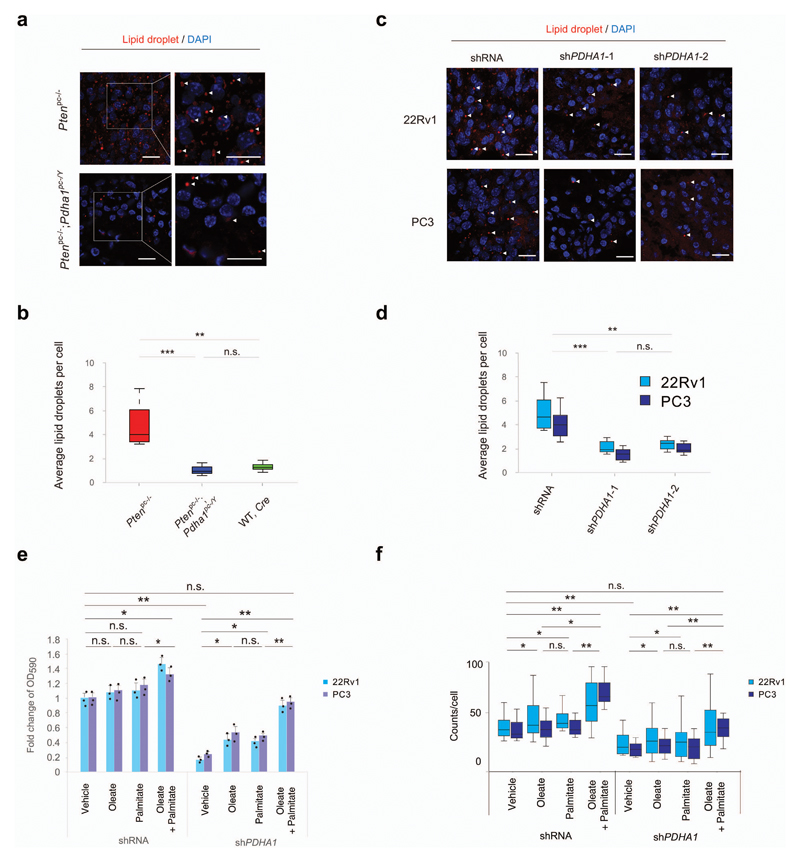
We next performed a direct measurement of lipid species in mouse prostate tumours and human prostate cancer cell lines depleted of PDHA1. Of note, the majority of lipid species, including cholesterol ester were decreased in Ptenpc-/-; Pdha1pc-/Y when compared to Ptenpc-/- tumours and in shPDHA1 22Rv1 and PC3 cells when compared to control cells (Supplementary Fig. 6, Supplementary Table 8-11 and Supplementary Note). 14C tracking experiments in sh-PDHA1 22Rv1 and PC3 cells demonstrated that glucose and glutamine incorporation into lipids and cholesterol (Supplementary Fig. 7a-e and Supplementary Note) were strongly affected when compared to controls whereas glucose and glutamine incorporation into proteins, remained unchanged (Supplementary Fig. 7f,g and Supplementary Note). PDHA1 knockdown did not affect glucose uptake but induced a slight increase of glutamine uptake (Supplementary Fig. 7h, i and Supplementary Note). In agreement with these findings lipid droplets, a known readout of lipid production5,37, were strongly reduced in both mouse and human xenograft tumours upon PDHA1 inactivation (Fig. 3a-d and Supplementary Fig. 7j-o). To assess whether lipid metabolism is affected by PDHA1 inactivation, we cultured sh-PDHA1 22Rv1 and PC3 cells in both the presence and absence of fatty acids and we found that fatty acids fully rescued the growth arrest induced by the PDHA1 knockdown. This was associated with the restoration of lipid droplets in the same cells (Fig. 3e,f and Supplementary Note). Rescue of proliferation upon fatty acid supplementation occurs independently by H3K9 acetylation and reactivation of the expression of ACLY and SQLE (Supplementary Fig. 7p and uncropped blots in Supplementary Fig. 15). This data suggests that fatty acids were directly incorporated into lipids. Collectively, these data demonstrate that PDC activity is required for the proliferation of mouse and human prostate tumour cells and that inactivation of PDC drives tumour growth inhibition by affecting histone acetylation and the expression of genes controlling lipid metabolism.
Figure 3. Pdha1 knockdown induces tumour suppression by abrogating lipogenesis.
(a,b) Representative confocal images (a) and quantification of average lipid droplets per cell (Lipidtox, red) (b) in the indicated genotypes in Ptenpc-/-; Pdha1pc-/Y versus Ptenpc-/- prostate tumours. DAPI, blue. (n=3 mice, Scale bar: 20 µm, 1 tumour per mouse, 5 fields acquired). (c,d) Representative confocal images (c) and quantification of average lipid droplets per cell (Lipidtox, red) (d) in 22Rv1 and PC3 shPDHA1 and shRNA xenograft tumours. DAPI, blue. (n=3 mice, Scale bar represents 20 µm, 1 tumour per mouse, 5 fields acquired). (e,f) Relative cell number quantification by crystal violet staining (e) and quantification of lipid droplets by average lipid droplet per cell (f) in 22Rv1 and PC3 cells infected with a shPDHA1 and scramble shRNA control and treated with exogenous fatty acids in fatty acid free media; oleate (25 μM), palmitate (25 μM) or a combination of these two were used(n=3, independent cell cultures). Error bars indicate s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001. n.s, not significant.
Nuclear PDC regulates the expression of lipid biosynthesis genes independently of mitochondrial PDC
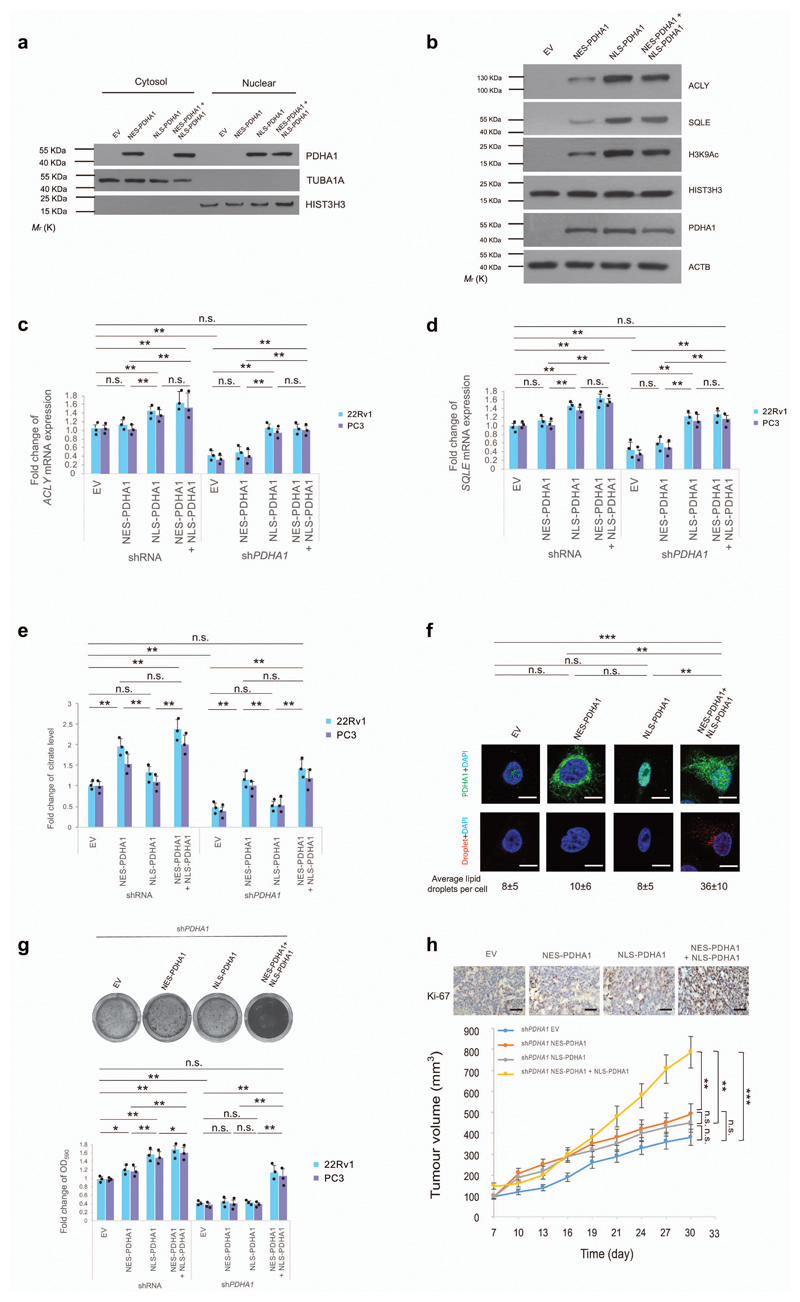
Cellular compartmentalization allows cells to carry out different metabolic reactions at the same time, and pathological protein compartmentalization is associated with cancer38. We, therefore, dissected the role of nuclear and cytosolic PDC in prostate cancer, by expressing nuclear export signal (NES) and/or nuclear localization signal (NLS) fused PDHA1 vectors in prostate cancer cell lines previously infected with (shRNA) and without PDHA1 (shPDHA1) (Fig. 4a, uncropped blots in Supplementary Fig. 13 and Supplementary Fig. 8a). NES-PDHA1 and NLS-PDHA1 constructs were engineered as shPDHA1 resistant vectors. Expressions of these vectors in 22Rv1 cells did not affect SREBF1 maturation and nuclear translocation (Supplementary Fig. 8a). Restoration of NLS-PDHA1 in sh-PDHA1 cells recovered H3K9Ac and the expression of fatty acid synthesis genes (Fig. 4b-d, Supplementary Fig. 8b for the full panel and uncropped blots in Supplementary Fig. 13,15), but did not rescue citrate levels, lipids content and cell growth both in vitro and in vivo (Fig. 4e-h and Supplementary Fig. 8c-g). In contrast, overexpression of both NES-PDHA1 and NLS-PDHA1 restored lipid synthesis genes, citrate levels, lipids content and cell growth (Fig. 4b-h and Supplementary Fig. 8b-g). Interestingly, the selective ACLY inhibitor, SB-204990 partially decreased H3K9 acetylation in prostate cancer cells with high PDC activity and SB-204990 did not further increase the growth arrest in prostate cancer cells infected with a shRNA against PDHA1 (Supplementary Fig. 8h-j and uncropped blots in Supplementary Fig. 15). These data demonstrate that PDC can directly contribute to histone acetylation, independently by the regulation of ACLY30. These experiments reveal that nuclear PDC regulates the expression of lipid synthesis gene in an autonomous manner and both nuclear and cytosolic PDHA1 are needed for the proliferation of prostate cancer cells.
Figure 4. Nuclear PDC regulates the expression of lipid biosynthesis genes independently of mitochondrial PDC.
(a) Western blot analysis of indicated proteins in nuclear and cytoplasmic fractions of shPDHA1 22Rv1 cells infected with NES-PDHA1 and NLS-PDHA1 alone or in combination. Uncropped images are in Supplementary Figure 13. (n=3, independent cell cultures). (b) Western blot analysis of indicated proteins in shPDHA1 22Rv1 cells infected NES-PDHA1 and NLS-PDHA1 alone or in combination (see full panel in Supplementary Fig. 5b). Uncropped images are in Supplementary Figure 13. (n=3, independent cell cultures). (c-e) Quantitative real-time PCR analysis of mRNA expression for ACLY (c) and SQLE (d) and determination of citrate levels (e) in shRNA control and shPDHA1 22Rv1 and PC3 infected with NES-PDHA1, NLS-PDHA1 alone or in combination (n=3, independent cell cultures). (f) Representative confocal images and quantification of lipid droplets (average lipid droplets per cell) in shPDHA1 22Rv1 infected with NES-PDHA1, NLS-PDHA1 alone or in combination (n=3, independent cell cultures, Scale bar represents 10 µm, 5 fields acquired for each group). (g) Upper panel, representative images of crystal violet staining of shPDHA1 22Rv1 infected with infected with NES-PDHA1, NLS-PDHA1 alone or in combination (n=3, independent cell cultures). Lower panel, relative cell number quantification by crystal violet staining in shRNA control and shPDHA1 22Rv1 and PC3 infected with NES-PDHA1, NLS-PDHA1 alone or in combination (n=3, independent cell cultures). (h) Upper panel, representative micrographs in histopathological analysis of Ki-67 of these xenograft tumours. Lower panel, evaluation of tumour formation in xenotransplantation experiments of shPDHA1 22Rv1 infected with NES-PDHA1, NLS-PDHA1 alone or in combination (n=6 animals; 12 injections, Scale bar represents 50 µm, also see Supplementary Fig. 7d for the full panel). Error bars indicate s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001. n.s, not significant.
Nuclear PDC regulates fatty acid synthesis in presence of mitochondrial citrate
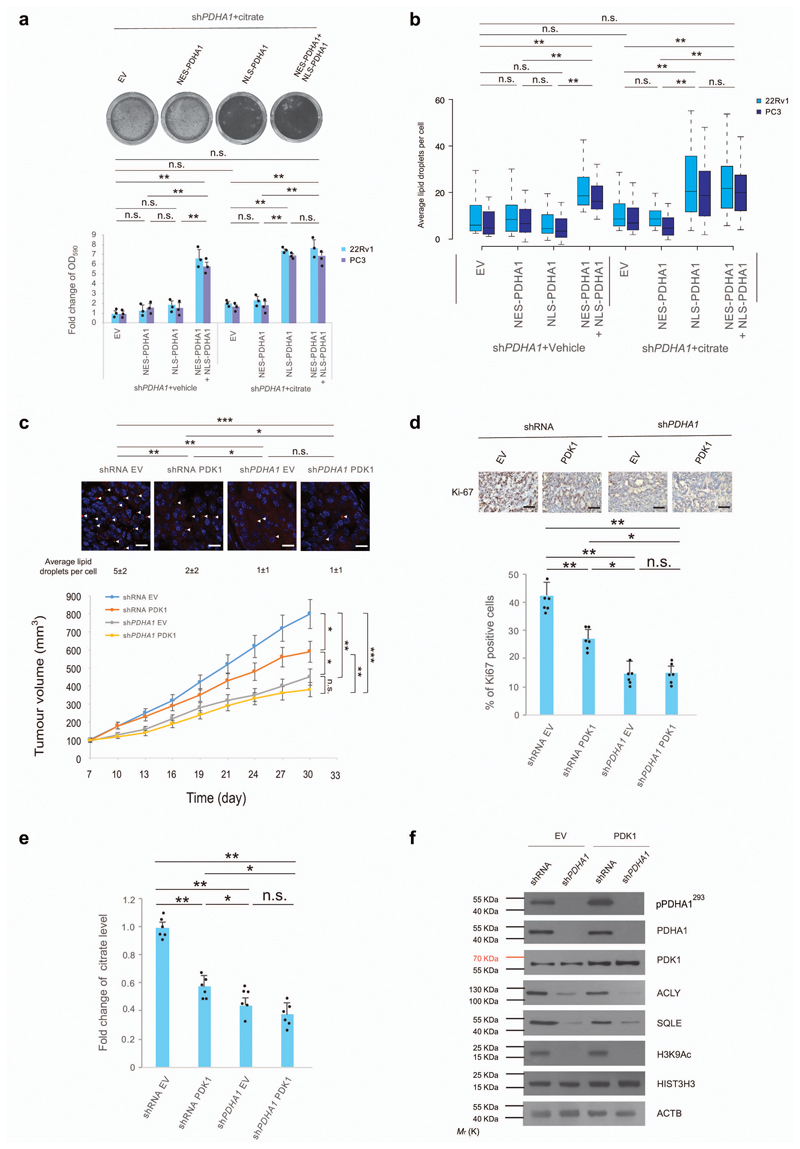
Consistently with these findings, we proved that overexpression of NLS-PDHA1 rescued growth arrest and lipid synthesis in cells cultured in the presence of citrate (Fig. 5a, b and Supplementary Fig. 9a for the whole panel). To validate these findings using an additional system, we overexpressed PDK1 in prostate cancer cells. Previous evidence demonstrates that PDK1 overexpression suppresses mitochondrial metabolism decreasing the intracellular levels of citrate without halting the nuclear function of PDC23,39–41. In line with this evidence overexpression of PDK did not promote phosphorylation of nuclear PDHA1 (Supplementary Fig. 9b and uncropped blots in Supplementary Fig. 15). Overexpression of PDK1 in 22Rv1 cells decreased citrate levels and slightly suppresses cancer cell proliferation both in vitro and in vivo (Fig. 5c-e and Supplementary Fig. 8c-e). However, it did not decrease histone acetylation and expression of fatty acid synthesis genes (Fig. 5f, uncropped blots in Supplementary Fig. 13 and Supplementary Fig. 9b). In contrast, concomitant inactivation of PDHA1 and overexpression of PDK1 decreased citrate level, histone acetylation and lipids content thereby suppressing tumourigenesis of a greater extent than in cells infected with PDK1 alone (Fig. 5c-f and Supplementary Fig. 9c-e). Taken together, these results demonstrate that both mitochondrial and nuclear PDC is required for prostate tumour growth.
Figure 5. Nuclear PDC regulates fatty acid synthesis in presence of mitochondrial citrate.
(a,b) Relative cell number quantification by crystal violet (a, also see full panel in Supplementary Fig. 8a) and quantification by confocal microscopy of average lipid droplets per cell (b) in shPDHA1 22Rv1 and PC3 cells infected with NES-PDHA1, NLS-PDHA1 alone or in combination and treated with citrate (100 µM) or vehicle for 6 days (n=3, independent cell cultures). (c) Upper panel, Representative confocal images, and quantification of average lipid droplets per cell, in xenograft tumours from shRNA control and shPDHA1 22Rv1 cells infected with PDK1 or empty vector. Lower panel, Evaluation of tumour formation in xenotransplantation experiments in shRNA control and shPDHA1 22Rv1 cells infected with PDK1 or empty vector (n=6 animals; 12 injections, 5 fields acquired for each group and Scale Bar represents 20 μm). (d) Upper panel, representative immune-histochemistry micrographs for Ki-67 staining in tumours of the indicated genotypes. (n=6 animals; 12 injections, 5 fields acquired for each group and scale bar represents 50 μm). Lower panel, quantification of the percentage of Ki-67 positive cells in different tumour genotypes (n=6 animals; 12 injections, 5 fields acquired for each group). (e,f) Determination of citrate levels (e) and western blot analysis of indicated proteins (f) in xenograft tumours from shRNA control and shPDHA1 22Rv1 cells infected with PDK1 or empty vector. Uncropped images are in Supplementary Figure 13. (n=6, independent tumour samples). Error bars indicate s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001. n.s, not significant.
Pharmacological inhibition of PDHA1 arrests mouse and human prostate tumours
Given that prostate cancer relies on PDC for proliferation we assessed whether pharmacological inhibition of PDHA1 could also block tumourigenesis in Ptenpc-/- tumours and human xenograft models of prostate cancer. We reasoned that a compound that could affect both the mitochondrial and nuclear function of PDC could be effective in blocking prostate cancer. To this extent, we took advantage of 3-fluoropyruvate (3-FP), a competitive inhibitor of the PDHA142–45. Computer simulations confirmed that 3-FP and pyruvate bind to PDHA1 with similar affinity (Supplementary Fig. 10a, b and Supplementary Note). To assess the specificity of 3-FP for the PDC, cell lysates treated with 3-FP were incubated in presence or absence of pyruvate. 3-FP decreased the activity of PDC and this was reversed by adding pyruvate in a dose dependent manner to the reaction buffer (Supplementary Fig. 10c). 3-FP administered by intraperitoneal injection in Ptenpc-/- mice inhibited the PDC activity in vivo in absence of systemic toxicity and induced a strong decrease of tumour cells proliferation as demonstrated by the reduced tumour size, tumour invasiveness and Ki-67 staining (Supplementary Fig. 10d-j). Importantly, 3-FP inhibited PDC both in the nucleus and the cytosol of prostate tumour cells and decreased H3K9Ac, Acly and Sqle levels in vivo at both protein and mRNA levels (Supplementary Fig. 10k-m and uncropped blots in Supplementary Fig. 15). Metabolomics analysis via LC-MS/MS46 confirmed that 3-FP strongly affected pyruvate metabolism and TCA cycle (Supplementary Fig. 11a). Finally, lipids droplet staining showed a decreased percentage of the number, size and intensity of lipid bodies in Ptenpc-/- tumours treated with 3-FP (Supplementary Fig. 11b-e). 3-FP treatment blocked the proliferation of different human prostate cancer cells and this was associated with a decreased PDC activity (Supplementary Fig. 11f, g). Finally, 3-FP treatment decreased the proliferation of different human prostate cancer xenografts (LNCaP, 22Rv1 and PC3) promoting long-lasting anti-tumour responses in vivo (Supplementary Fig. 11h-j). Taken together these data suggest that pharmacological inactivation of PDC hampers prostate cancer progression.
Discussion
Whether the PDC acts as an oncogene or tumour suppressor in cancer is still under debate19,22,23,39,40,47–49. A previous report shows that reduction in PDC activity through overexpression of PDK1 in melanoma cells promote tumour cell proliferation by enhancing glycolysis and reductive carboxylation19. In this and other models, activation of PDC by dichloroacetate promoted tumour suppression19,47. Our data provide a new angle and demonstrate that the subunits of the PDC such as PDHA1 and PDP1 are amplified and overexpressed at both gene and protein levels and that the PDC is active in prostate cancer. In line with this evidence, we demonstrate that inactivation of PDHA1 hampers prostate cancer progression in vivo in both mouse and human prostate cancer models. These findings are in agreement with recent studies demonstrating that tumor cells, in vivo, increased glucose oxidation via PDC. PDHA1 inactivation in non-small lung cells cancer (NSCLC) xenografts resulted in a decreased tumor formation capacity. An increased TCA cycle via PDC was also observed in poorly perfused tumours areas of patients affected by NSCLC9,10,50. These data, together with our data in prostate cancer, challenged the notion that tumours switch from glucose derived oxidative metabolism to aerobic glycolysis to support their growth6. However, the mechanisms by which tumour cells in vivo become addicted to PDC-mediated mitochondrial metabolisms still remain unknown. As suggested by previous studies this may be explained by the genetic background of the tumours, a difference in tissue requirements or in the composition of the tumour microenvironment10. Intriguingly normal prostate epithelial cells exhibit a “truncated” TCA cycle due to inhibition of aconitase needed for the production and secretion of a high quantity of citrate into the seminal fluid51–53. Therefore, it is not surprising that prostate cancer relies on mitochondrial metabolism more than other tumour types.
Our results reveal that in order to support prostate cancer growth the PDC must be functional in both mitochondria and the nucleus of cancer cells. Mitochondrial PDC function is not sufficient to support lipid biosynthesis in tumour cells since nuclear PDC regulates the transcription of the enzymes that convert cytosolic citrate into acetyl-coA and fatty acids (Supplementary Fig. 11k, l). This compartmentalization allows the mitochondrial production of citrate in the cytosol and the expression of enzymes required for de novo lipid biosynthesis. Prior studies demonstrate that prostate cancer also benefits of enhanced glycolytic metabolism. However, these aggressive glycolytic tumour cells retain an active glucose-derived lipid biosynthesis54, thus in line with the results presented herein.
Our findings also hold important implications for cancer therapy and pave the way for the targeting of the nuclear function of PDC to eradicate prostate cancer. This may be achieved in the future as shown in this paper by using pyruvate analogs or alternatively by developing small molecule inhibitors of PDHA1.
Online Methods
Animals
All mice were maintained under specific-pathogen-free conditions in the animal facilities of the IRB Institute, and experiments were performed according to state guidelines and approved by the local ethics committee. The PtenloxP and Pdha1loxP conditional knockout alleles have been described15,18. Female PtenloxP/loxP; Pdha1loxP/Y mice were crossed with male Probasin-Cre4 (Pb-Cre4) transgenic mice14 for the prostate-specific deletion of Pten and Pdha1. For genotyping, tail DNA was subjected to polymerase chain reaction analysis with the primers listed in Supplementary Table 1. Mice of correct genotypes were randomly chosen and allocated into experimental groups. For the transgenic mice, the experiment was carried on in single blind.
Cell culture and reagents
Human prostate carcinoma cell lines, 22Rv1, LNCaP, PC3, and DU145 were purchased from ATCC and human prostate epithelial PNT2C2 cell line55 were gifted from Prof. Norman James Maitland, University of York and kept in Institute of Oncology Research (IOR) in Bellinzona, Switzerland. The cells were cultured in RPMI medium 1640 (Catalog# 21875034, Thermo Fisher Scientific) with 10% fetal bovine serum (FBS) (Catalog# 10500-064, Thermo Fisher Scientific), 10,000 I.U./mL Penicillin and 10,000 (μg/mL) Streptomycin (Catalog# P4333-20ML, Sigma) under the condition of 37 °C and 5% CO2. Cells were transduced with TRIPZ doxycycline inducible lentiviral construct against human PDHA1 gene (Catalog# V2THS_75677, Dharmacon) and TRIPZ Inducible Lentiviral Empty Vector shRNA Control (Catalog# RHS4750, Dharmacon). Flat bottom ultra low attachment multiple well plates (Catalog# 3473, Corning) were used for sphere formation assay. Cells were transduced with pLKO lentiviral constructs against human PDHA1 gene (Catalog# SHCLNG-NM_000284, Sigma, clone# shPDHA1-1: TRCN0000028582 and shPDHA1-2: TRCN0000028627) and Lentiviral Empty Vector shRNA Control (Catalog# SHC002, Sigma). PDHA1 was PCR-amplified from complementary DNA of a normal human prostate sample. For the overexpression of cytosol and nuclear-localized PDHA1, nuclear export signal (NES) or nuclear localization signal (NLS) was fused PDHA1 as NES-PDHA1 and NLS-PDHA1. NES-PDHA1 and NLS-PDHA1 constructs were engineered as sh-PDHA1 (Catalog# SHCLNG-NM_000284, Sigma, clone# shPDHA1-1: TRCN0000028582) resistant vectors. NES-PDHA1 and NLS-PDHA1 were cloned into pLenti CMV Neo (Addgene) and the empty vector was used as a control. Cells were infected shRNA control or shPDHA1 and overexpressed PDK1 by pLZRS-PDK1-IRES-puro or FG12-eGFP control19 (gifted from Prof. Daniel Peeper, The Netherlands Cancer Institute). Lentiviral and retroviral infections were performed using HEK293T cells and Phoenix cells, respectively, as producers of viral supernatants. Primary MEFs were derived from littermate embryos and obtained by crossing Pdha1loxP/loxP or PtenloxP/loxP animals. Embryos were harvested at 13.5 days post coitum, and individual MEFs were produced and cultured as previously described14–16. For senescence experiments, MEFs were infected with pMSCV hygro-Cre (Plasmid #34565, Addgene) or empty vector control (pMSCV hygro) retrovirus, selected pharmacologically in hygromycin (50 µg/mL, Catalog# H0654-250MG, Sigma) for 48 hr. After selection, cells were seeded and analyzed for SA-β-gal activity (Catalog #9860, Cell Signaling Technology). Cell number quantification with crystal violet was performed as referenced15. The chemicals used for cell and tumour treatments were purchased from Sigma: Doxycycline hyclate (Catalog# D9891-1G, Sigma) and β-Fluoropyruvic acid sodium salt monohydrate (3-Fluoropyruvate, 3-FP, Catalog #F4004, Sigma). SB-204990 (Catalog# 4962, Bio-Techne AG). For the xenograft experiments, 2×106 22Rv1 cells infected Tripz-shPDHA1 or Tripz-shRNA control were injected subcutaneously (s.c.) into the lower flank of the same nude mouse on different sides. The mice started to be fed with Doxycycline (0.2 g/L) water supplemented with 5% sucrose in another day. Tumour formation from each individual injection was monitored every three days until the 22nd day of experimental termination. 2×106 22Rv1 cells infected shRNA control or shPDHA1 and overexpressed PDK1 by pLZRS-PDK1-IRES-puro or FG12-eGFP control19 (gifted from Prof. Daniel Peeper, The Netherlands Cancer Institute) were injected subcutaneously (s.c.) into the lower flank of the same nude mouse on different sides. Tumour formation from each individual injection was monitored every three days until the 30th day of experimental termination. For experiments with xenograft, mice were excluded when the tumour size exceeded the average tumour size at the moment of randomization. The experiments with Xenograft (including tumour measurements) were carried on in single blind. For preclinical treatment on Ptenpc-/- males, 80 mg/Kg of 3-FP were intraperitoneally injected every three days on mice of ages at 8 weeks to 12 weeks. For the xenograft tumours treated with 3-FP or vehicle control, 2×106 LNCaP, PC3, and 22Rv1 cells were injected subcutaneously (s.c.) into the lower flank of the nude mice. The treatment started when tumour size reached 100 mm3 and the treatment continued for a period of two months. When the mice were sacrificed, tumours were disserted and analyzed. Mice were fasted for 6 h prior to tissue harvest (9 am-3 pm) in order to prevent metabolic alterations due to immediate food intake.
Real-time PCR, western blotting, and immunohistochemistry
RNA was extracted using TRIzol® Plus RNA Purification Kit (Catalog# 12183555, Life technologies). 1 μg of total RNA was used for cDNA synthesis using SuperScript® III Platinum® One-Step qRT-PCR Kit (Catalog# 11732-020, Life technologies). Quantitative Real Time PCR (q-RT PCR) was performed as previously described15. Primers employed are detailed in Supplementary Table 2. All qRT-PCR data presented was normalized using β-actin. Tissue and cell lysates were prepared with RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Catalog# 78429, Thermo Scientific)). The following antibodies were used for western blotting: rabbit polyclonal anti-Pten antibody (Catalog # 9552, Cell Signaling Technology), mouse monoclonal anti-PDHA1 antibody (Catalog # 459400, Invitrogen), rabbit polyclonal anti-phospho-PDHE1-A type I (Ser293) antibody (Catalog# ABS204, EMD Millipore), goat polyclonal anti-PDC-E2 Antibody (N-20) (Catalog# sc-16890, Santa Cruz Biotechnology), mouse monoclonal anti-DLD Antibody (E-3) (Catalog# sc-376890, Santa Cruz Biotechnology), rabbit polyclonal anti-PDP1 antibody (Catalog# HPA019081, Sigma), rabbit polyclonal anti-Akt antibody (Catalog# 9272, Cell Signaling Technology), rabbit monoclonal anti-p-Akt Ser473 antibody (Catalog# 4060, Cell Signaling Technology), rabbit monoclonal anti-Acetyl-Histone H3 (Lys9) antibody (Catalog# 9649, Cell Signaling Technology), rabbit monoclonal anti-Histone H3 antibody (Catalog# 4499, Cell Signaling Technology), rabbit polyclonal anti-ACLY antibody (Catalog# 4332, Cell Signaling Technology), goat polyclonal anti-SQLE antibody (Catalog# sc-49754, Santa Cruz Technology), rabbit monoclonal anti-VDAC1 (Catalog# 4661, Cell Signaling Technology), rabbit polyclonal anti-α-Tubulin antibody (Catalog# 2144, Cell Signaling Technology) and mouse monoclonal anti-β-actin (Catalog# A5316, Sigma). For immune-histochemistry (IHC), tissues were fixed in 10% formalin and embedded in paraffin in accordance with standard procedures. Sections were stained with Rabbit polyclonal anti-PTEN antibody (Catalog# 51-2400, Thermo Fisher Scientific), mouse monoclonal anti-PDHA1 (Catalog# 459400, Invitrogen), rabbit polyclonal Anti-PDP1 (Catalog# HPA019081, Sigma), anti-Ki-67 (clone SP6, Lab Vision), or anti-cleaved-caspase-3 (Catalog#9661, Cell Signaling Technology) antibodies. Prostate disease spectrum tissue array, including TNM, clinical stage and pathology grade, 80 cases/80 cores were purchased from US Biomax, Inc, (Catalog# PR8011a) and prostate cancer tissue array, including TNM, clinical stage and pathology grade, 48 cases/48 cores was purchased from US Biomax, Inc (Catalog# PR483b). Ki-67 and gland invasiveness assessment was performed by three prostate pathologists independently, in a blinded manner.
Gene Expression Analysis
Gene expression profiling (GEP) was done using the MouseRef-8 v2.0 Expression BeadChip (Illumina, San Diego, CA, USA), following the manufacturer’s protocol. Arrays were read on an Illumina HiScanSQ system. Data were first extracted with the Illumina GenomeStudio software and then imported in Genomics Suite 6.4 (Partek Incorporated, Saint Louis, MO USA) and quantile normalized. Transcripts with differences in expression were identified by ANOVA. Enrichment analysis was performed using Gene Set Enrichment Analysis (GSEA) 56. Raw data have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO accession no. GSE74245. GSEA was performed on entire gene list ranked according to fold changes observed between Ptenpc-/- and Ptenpc-/-; Pdha1pc-/Y mice. The GS collection assessed includes all GSs smaller than 10 and larger than 500 (8335 out of 22423 GSs retained) compiled according to57 the 24.03.15. GSs yielding significance ((FDR <0.05; nominal p-value <0.005; TAGS ≥50%) were retained and assessed for their role in metabolic processes after visualizing the data as described 58. For the 3 relevant clusters identified, their GSs’ FDR q-values were, together with unaffected metabolic processes log2 transformed and inversed for display in Fig. 2a.
Chromatin immunoprecipitation
ChIP assays were performed with approximately 6 × 106 cells per experiment. Cells were subject to hypotonic lysis and treated with micrococcal nuclease to recover mono- to tri-nucleosomes. Nuclei were lysed by brief sonication and dialysed into N-ChIP buffer (10 mM Tris pH 7.6, 1 mM EDTA, 0.1% SDS, 0.1% Na-Deoxycholate, 1% Triton X-100) for 2 h at 4 °C. Soluble material was incubated overnight at 4 °C after addition of 0.5–1 μg of antibody bound to 25 μl protein A Dynal magnetic beads (Catalog# 10006D, Invitrogen), with 5% kept as input DNA. Magnetic beads were washed, chromatin was eluted and ChIP DNA was dissolved in 10 mM Tris pH 8 for quantitative PCR reactions (see later). Three separate ChIP experiments were performed on replicate biological samples. The data shown are the average qRT–PCR values (n =3). Primers are listed below. All qPCR was performed using an Applied Biosystems StepOnePlus system and Power SYBR Green PCR master mix. ChIP samples were diluted 1:100 in H2O and 5 μl was used per reaction. ChIP-qPCR signals were calculated as per cent input. Primers used in ChIP were listed in Supplementary Table 3.
Subcellular fractionation
Nuclear and cytoplasmic fractionation was performed by centrifugation technique as described previously59. Nuclear and cytoplasmic extracts were made using NE-PER Nuclear and Cytoplasmic Extraction Reagent Kit (Catalog# 78833, Pierce Biotechnology). Total cell lysate was prepared in radioimmunoprecipitation assay buffer (RIPA; 25 mmol/L Tris (pH 7.4), 150 mmol/L KCl, 5 mmol/L EDTA, 1% NP40, 0.5% sodium deoxycholate, and 0.1% SDS) with protease inhibitor cocktail (Catalog# 88688, Thermo Scientific). Proteins at the same amount were separated by 10% to 15% SDS-PAGE and transferred onto polyvinylidene difluoride membranes. For each fraction, nuclear protein histone 3 (Catalog# 4499, Cell Signaling Technology) and cytoplasmic protein α-Tubulin (Catalog# 2144, Cell Signaling Technology) were used to show clear isolation between nucleus and cytoplasm during fractionation.
Targeted mass spectrometry analysis
For mouse prostate tissues, 500 µL of 80% LC-MS grade methanol was added to each approximately 15 mg sample and incubated at -80°C for 15 min. Tissue samples collected were centrifuged at 18, 470g for 5 min in a cold room to pellet cell debris and proteins. Supernatants were saved. Pellets were resuspended in 500 µl 80% methanol by vortexing and subsequently centrifuged as before. Supernatants were centrifuged one final time at 18, 470g for 10 min at 4°C. Metabolite extractions were dried to a pellet by SpeedVac with no heat. Samples were re-suspended using 20 µL LC-MS grade water and 10 µL was injected and analyzed using a 5500 QTRAP hybrid triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) through selected reaction monitoring (SRM). Two hundred and sixty-one endogenous water-soluble metabolites were targeted for steady-state analyses of samples. Some metabolites were targeted in both the positive and negative ion mode through a positive/negative polarity switching for a total of 296 SRM transitions. The ESI voltage was +4900V in the positive ion mode and -4500V in the negative ion mode. The dwell time was 3ms per SRM transition and the total cycle time was ~1.56 sec. Approximately 10-12 data points were acquired per detected metabolite. Samples were delivered to the MS through normal-phase chromatography using a 4.6mm i.d ×10 cm Amide XBridge HILIC column (Waters Corp.) at 350 µl min-1. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 35% B from 0-3.5 min; 35% B to 2% B from 3.5-11.5 min; 2% B was held from 11.5-16.5 min; 2% B to 85% B from 16.5-17.5 min; 85% B was held for 7 min to re-equilibrate the column. Buffer A was comprised of 20mM ammonium hydroxide/20mM ammonium acetate (pH 9.0) in 95:5 water/acetonitrile. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v2.1 software (AB/SCIEX). Metabolomics data analysis was done in part using Metaboanalyst 2.0 software (www.metaboanalyst.ca).
Isotope Labeling and stationary Profiling
Fresh transgenic mouse tumour tissues were mechanically dissociated, enzymatically digested and filtered to obtain a single-cell suspension as described60. Single cells were stained with fluorescein isothiocyanate (FITC)-anti-CD34 (Catalog# 560238, BD Biosciences) for stroma cells, FITC-anti-Ter119 (Catalog# 557915, BD Biosciences) for erythrocytes, FITC-anti-CD31(Catalog# 561813, BD Biosciences) for endothelial cells, and FITC-anti-CD45 (Catalog# 553079, BD Bioscience) for leukocytes and incubated 20 min on ice. All antibodies (BD Biosciences) were used at 1:300; cells were then loaded into MS column with Anti-FITC MicroBeads (Catalog# 130-048-701, Miltenyi Biotec) for MACS separation, and unstained epithelial cells were collected in the negative fraction. For steady state metabolomic analysis5, prostate epithelial cells derived from transgenic mouse tumours were plated to ~80% confluence on 10 cm dishes in biological quadruplicate. cells were plated in RPMI medium (Catalog# 11875093, Thermo Scientific) was supplemented with 10% dialyzed serum and devoid of glucose or glutamine and supplemented with one of the two carbon-13-labelled substrates [U-13C6]-glucose and [U-13C5] glutamine (Catalog# CLM-1396-1 and CLM-1822-H-0.25, Cambridge Isotope Labs) where the remaining substrates were unlabelled (glucose 11 mM and glutamine 2 mM). Fatty acid oxidation studies20 were conducted using [U-13C16] palmitate (Catalog# CLM-6059-1, Cambridge Isotope Labs) noncovalently bound to fatty acid-free BSA. [U-13C16] palmitate-BSA was added to culture medium at 5% of the final volume (50 mM final concentration) with 1 mM carnitine in medium formulated with delipidated FBS (Catalog# 12676011, Thermo scientific). Additionally, fresh media containing carbon-13-labelled substrates was exchanged 2 h prior to metabolite extraction for steady state analyses. Considering primary epithelial cells isolated from mouse prostate tissue do not attach and fit for long term culture, we labeled these epithelial cells for 6 h and we did not see the difference compared to the results from labeling for 24 h. After 6 h of incubation with labelled substrates, metabolite extraction of medium was performed by adding 1mL cold (−80 °C) 80% methanol (Catalog#34966-1L and 14263-1L, Sigma), incubated at −80 °C for 30 min followed by centrifugation at 10,000g for 10 min at 4 °C. The resultant supernatant was lyophilized by speedvac and stored at − 80 °C until analysis. Dried metabolite pellets were re-suspended in 20 μL LC–MS grade water, 5 μL was injected onto a Prominence UFLC and separated using a 4.6 mm i.d. Å~ 100 mm Amide XBridge HILIC column at 360 μL per minute starting from 85% buffer B (100% ACN) to 0% B over 16 min. Buffer A: 20 mM NH4OH/20 mM CH3COONH4 (pH = 9.0) in 95:5 water/ACN.287 selected reaction monitoring (SRM) transitions were captured using positive/ negative polarity switching by targeted LC-MS/MS using a 5500 QTRAP hybrid triple quadrupole mass spectrometer. As for metabolomics, the quantity of the metabolite fraction analyzed was adjusted to the corresponding protein concentration from a sample processed in parallel. The analysis was performed for each of the three substrates on three independent tumours in biological triplicate.
Lipidomics analysis on transgenic mouse prostate tumours
~ 5 mg of solid tissue was snap frozen in liquid nitrogen (−196°C) as close to the time of resection as possible. Chloroform: methanol (2:1 ratio) was added to a final volume 20 times the volume of the biological sample (100 µL in 2 mL of solvent mixture) in a 12 mL glass vial. The mixture was agitated for 30 min in an orbital shaker at room temperature. 0.2 volume parts of water (400 µL for 2 mL) was added and the mixture was vortexed for 1 min. The mixture was placed still for 10 min and centrifuge at low speed (1000 g) to separate into three phases. The upper aqueous phase was kept (optional) to analyze small organic polar molecules. The middle layer contains protein, DNA, and polar large molecules. The lower phase containing non-polar lipids was collected and evaporated under vacuum using a SpeedVac rotary evaporator or under a nitrogen stream. 10 µL of sample was injected onto LC-MS/MS using a hybrid QExactive Plus Orbitrap mass spectrometer in DDA mode using positive/negative ion polarity switching (Top 8 in both modes). Using a 100 mm x 2.0 mm C18 column at 260 µL/min with a 1100 quaternary HPLC, lipids were eluted over 20 min. from 32% B buffer (90% IPA/10% ACN/10 mM NH4HCO2/0.1% FA) to 97% B. A buffer consisted of 59.9% ACN/40% water/10 mM NH4HCO2/0.1% FA. Lipid molecules were identified and quantified using LipidSearch 4.1.9 software61.
Statistics
For each independent in vitro experiment, at least three technical replicates were used (exceptions: in western blot analysis technical replicates are presented, in targeted metabolomics three technical replicates were used. For data mining analysis, ANOVA test was used for multi-component comparisons and Student T test for paired-comparisons. In the in vitro experiments, data groups were assessed for normal distribution and Student T test was applied for paired-comparison. Data represent mean ± s.e.m. of pooled experiments unless otherwise stated. n values represent the number of experimental samples and all the experiments were repeated at least three times. For in vivo experiments, the equal variance could not be assumed and a non-parametric Mann-Whitney test was used. The confidence level used for all the statistical analyses was of 0.95 (alpha value = 0.05). Two-tail statistical analysis was applied for experimental design without predicted result, and one-tail for validation experiments (*P < 0.05; **P < 0.01; ***P < 0.001).
Data availability
The mouse gene expression datasets are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74245
For cellular, molecular and metabolic assays, the lipidomics analysis on prostate cancer cell line, fatty acid rescue assay and computer simulations of 3-FP and PDC docking, the methods are available in Supplementary Note.
Supplementary Material
Acknowledgements
The authors all the members of the IOR/IRB animal core facility for technical assistance and the animal work. We thank Min Yuan and Susanne B. Breitkopf for help with metabolomics analysis. The mass spectrometry work was partially supported by NIH grants P01-CA120964 (J.M.A.) and P30-CA006516 (J.M.A.). Cholesterol measurement in prostate cancer cell lines were supported by AIRC Investigator Grant (IG 2016 Id. 19104) and CARIPARO Pediatric research Grant. The work of A.M. was funded by Fondazione Umberto Veronesi and Associazione Italiana Ricerca sul Cancro and Fondazione Cassa di Risparmio di Firenze (grant 19515 to PC and AM) and AIRC (grant 8797 to PC). We thank Eugenio Ragazzi in Department of Pharmaceutical and Pharmacological Sciences, University of Padova for the multivariate statistical analysis. The work of A.C. is supported by the department of education of the Basque Government (IKERTALDE IT1106-16), the BBVA foundation, the MINECO (SAF2016-79381-R (FEDER/EU); Severo Ochoa Excellence Accreditation SEV-2016-0644) and the European Research Council (Starting Grant 336343, PoC 754627). The participation of A.C. as part of CIBERONC was co-funded with FEDER funds. This work was supported by Swiss national science foundation (SNF) grant Ambizione (PZ00P3_136612/1), European Research Council starting grant (ERCsg 261342), the European Society for Medical Oncology (ESMO) translational research award to A.A., the Dr. Josef Steiner Cancer Research Award and the Swiss Bridge Award to A.A., and Fondazione IBSA fellowship award to J.C.
Footnotes
URLs
Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/; MetaboAnalyst, http://www.metaboanalyst.ca/; UCSC Genome Browser on Human, https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr8%3A124998506-125022283&hgsid=641911343_mdAzqHRm9nypyskAklqb2aiYGn1X
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author Contributions
J.C. and A.A. originally conceived the concept, designed the experiments, interpreted the data and wrote the paper. J.C., I.G., D.B., A.R. and A. Alajati performed experiments and analyzed the data. J.C. crossed and generated transgenic mouse model. G.C., E.P. and J.C. made experiments on xenograft mouse model. A. Alajati., S.P., M.L., M.S. and J.D.B. performed immune-histochemical experiments and analysis. R.D. and J.C. established and carried out fluorescence microscopy. J.M.A. and J.C. conducted metabolomics and lipidomics analysis on mouse prostate tumours. S. C., M. A., M.M. and J.C. performed cholesterol measurements in prostate cancer cell lines. A.M. and P.C. performed carbon-14 tracing experiments and interpreted data. I.M. and A.R. carried out mitochondrial OCR measurements. A.R., N.D. and F.B. performed gene expression profile on mouse tumours and GSEA analysis of metabolic pathways on mouse tumours. R.G.E. provided bioinformatics analysis on PDHA1 and PDP1 amplification and overexpression in human prostate cancer datasets. A.C. provided support on metabolic analysis, interpretation of results and discussion.
Competing interests
The authors declare no competing financial interests.
References
- 1.Zong WX, Rabinowitz JD, White E. Mitochondria and Cancer. Mol Cell. 2016;61:667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeBleu VS, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marin-Valencia I, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vazquez F, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viale A, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn CS, Metallo CM. Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab. 2015;3:1. doi: 10.1186/s40170-015-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vyas S, Zaganjor E, Haigis MC. Mitochondria and Cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hensley CT, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson SM, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wieland OH. The mammalian pyruvate dehydrogenase complex: structure and regulation. Rev Physiol Biochem Pharmacol. 1983;96:123–170. doi: 10.1007/BFb0031008. [DOI] [PubMed] [Google Scholar]
- 12.Kolobova E, Tuganova A, Boulatnikov I, Popov KM. Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem J. 2001;358:69–77. doi: 10.1042/0264-6021:3580069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roche TE, et al. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- 14.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 18.Johnson MT, et al. Inactivation of the murine pyruvate dehydrogenase (Pdha1) gene and its effect on early embryonic development. Mol Genet Metab. 2001;74:293–302. doi: 10.1006/mgme.2001.3249. [DOI] [PubMed] [Google Scholar]
- 19.Kaplon J, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 20.Vacanti NM, et al. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol Cell. 2014;56:425–435. doi: 10.1016/j.molcel.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56:414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajagopalan KN, et al. Metabolic plasticity maintains proliferation in pyruvate dehydrogenase deficient cells. Cancer Metab. 2015;3:7. doi: 10.1186/s40170-015-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutendra G, et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 24.Nagaraj R, et al. Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation. Cell. 2017;168:210–223 e211. doi: 10.1016/j.cell.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briggs MR, Yokoyama C, Wang X, Brown MS, Goldstein JL. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem. 1993;268:14490–14496. [PubMed] [Google Scholar]
- 26.Wang X, et al. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J Biol Chem. 1993;268:14497–14504. [PubMed] [Google Scholar]
- 27.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han J, et al. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature. 2015;524:243–246. doi: 10.1038/nature14557. [DOI] [PubMed] [Google Scholar]
- 29.Hatzivassiliou G, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helms MW, et al. Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease. Br J Cancer. 2008;99:774–780. doi: 10.1038/sj.bjc.6604556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevenson J, Luu W, Kristiana I, Brown AJ. Squalene mono-oxygenase, a key enzyme in cholesterol synthesis, is stabilized by unsaturated fatty acids. Biochem J. 2014;461:435–442. doi: 10.1042/BJ20131404. [DOI] [PubMed] [Google Scholar]
- 33.Zhao S, et al. ATP-Citrate Lyase Controls a Glucose-to-Acetate Metabolic Switch. Cell Rep. 2016;17:1037–1052. doi: 10.1016/j.celrep.2016.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulusu V, et al. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 2017;18:647–658. doi: 10.1016/j.celrep.2016.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguzzi A, Altmeyer M. Phase Separation: Linking Cellular Compartmentalization to Disease. Trends Cell Biol. 2016;26:547–558. doi: 10.1016/j.tcb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Dupuy F, et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015;22:577–589. doi: 10.1016/j.cmet.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Mager J, Blank I. Synthesis of fluoropyruvic acid and some of its biological properties. Nature. 1954;173:126–127. doi: 10.1038/173126a0. [DOI] [PubMed] [Google Scholar]
- 43.Avi-Dor Y, Mager J. The effect of fluoropyruvate on the respiration of animal-tissue preparations. Biochem J. 1956;63:613–618. doi: 10.1042/bj0630613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chari-Bitron A, Avi-Dor Y. Effect of fluoropyruvate on the swelling, phosphorylative activity and respiration of guinea-pig liver mitochondria. Biochem J. 1959;71:572–578. doi: 10.1042/bj0710572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du J, et al. Inhibition of mitochondrial pyruvate transport by zaprinast causes massive accumulation of aspartate at the expense of glutamate in the retina. J Biol Chem. 2013;288:36129–36140. doi: 10.1074/jbc.M113.507285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hitosugi T, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531:110–113. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonnet S, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 50.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costello LC, Franklin RB. Aconitase activity, citrate oxidation, and zinc inhibition in rat ventral prostate. Enzyme. 1981;26:281–287. doi: 10.1159/000459195. [DOI] [PubMed] [Google Scholar]
- 52.Costello LC, Liu Y, Franklin RB, Kennedy MC. Zinc inhibition of mitochondrial aconitase and its importance in citrate metabolism of prostate epithelial cells. J Biol Chem. 1997;272:28875–28881. doi: 10.1074/jbc.272.46.28875. [DOI] [PubMed] [Google Scholar]
- 53.Costello LC, Liu Y, Zou J, Franklin RB. The pyruvate dehydrogenase E1 alpha gene is testosterone and prolactin regulated in prostate epithelial cells. Endocr Res. 2000;26:23–39. doi: 10.1080/07435800009040143. [DOI] [PubMed] [Google Scholar]
- 54.Torrano V, et al. The metabolic co-regulator PGC1alpha suppresses prostate cancer metastasis. Nat Cell Biol. 2016;18:645–656. doi: 10.1038/ncb3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delaleu N, Nguyen CQ, Tekle KM, Jonsson R, Peck AB. Transcriptional landscapes of emerging autoimmunity: transient aberrations in the targeted tissue's extracellular milieu precede immune responses in Sjogren's syndrome. Arthritis Res Ther. 2013;15:R174. doi: 10.1186/ar4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J, et al. TIP30 induces apoptosis under oxidative stress through stabilization of p53 messenger RNA in human hepatocellular carcinoma. Cancer Res. 2008;68:4133–4141. doi: 10.1158/0008-5472.CAN-08-0432. [DOI] [PubMed] [Google Scholar]
- 60.Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat Protoc. 2010;5:702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Breitkopf SB, et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics. 2017;13 doi: 10.1007/s11306-016-1157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mouse gene expression datasets are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74245
For cellular, molecular and metabolic assays, the lipidomics analysis on prostate cancer cell line, fatty acid rescue assay and computer simulations of 3-FP and PDC docking, the methods are available in Supplementary Note.