Abstract
A major goal of modern neuroscience is to understand how ensembles of neurons participate in neural circuits underlying behavior. The recent explosion of genetically-encoded circuit analysis tools has allowed neuroscientists to characterize molecularly-defined neuronal types with unprecedented detail. However, since neurons defined by molecular expression can be functionally heterogeneous, targeting circuit analysis tools to neurons based on their activity is critical to elucidating the neural basis of behavior. Here we review genetic strategies to access activated neurons and characterize their functional properties, molecular profiles, connectivity, and causal roles in sensory-coding, memory, and valence-encoding. We also discuss future possibilities for improving these strategies and using them to screen brain-wide activity patterns underlying adaptive and maladaptive behaviors.
Introduction
A major challenge in cellular and systems neuroscience is to understand how activity in specific neuronal populations produces perception and behavior. Traditional methods including metabolic imaging, pharmacological and physical lesions, and physiological recordings advanced our understanding of how neural activity underlies behavior. However, these techniques are limited in cell-type specificity. Recent studies in the mouse combined molecularly-defined Cre-driver lines (1–3) with Cre-dependent genetic tools to dissect the connectivity (4, 5), physiological response properties (6, 7), and behavioral roles (8–10) of specific neuronal cell-types. However, functionally heterogeneous neurons can belong to the same molecular type and are often spatially intermingled (11–13). Targeting neurons based on activity-dependent gene expression provides an orthogonal approach to dissect the organization and function of activated neural circuits with unprecedented cellular resolution.
When a neuron becomes depolarized, the transient rise of intracellular Ca2+ and downstream second messenger pathways transiently activate the expression of immediate early genes (IEGs) within minutes of neuronal activation (14). Most activity-based genetic strategies rely on the best-characterized IEGs, Fos (15) and Arc (16). Fos is rapidly induced by growth factor stimulation and various patterns of neural activity (15, 17–19) and acts as a transcription factor to regulate expression of many genes (20). Arc is also rapidly induced by neural activity and growth factors; its mRNAs and proteins are enriched in dendrites, which play an important role in synaptic plasticity (21, 22).
The discovery of IEGs enabled functional mapping with cellular resolution throughout the central nervous system (23). Early studies mapped Fos expression patterns that were consistent with known physiological responses to the same stimuli (18, 24). catFISH (compartmental analysis of temporal activity by fluorescent in-situ hybridization) enabled comparison of neuronal populations activated by two experiences ~30 min apart in the same animal (25). While these studies paved the way for mapping experience-driven activity patterns with IEGs, labeling IEGs in fixed tissues precludes examination of the physiological properties of recently activated neurons.
IEG promoters driving effectors for characterizing recently activated neurons
Transgenic animals using IEG promoters to drive fluorescent reporters (Fig. 1A) enabled researchers to characterize the physiological properties of recently activated neurons. For example, the Fos-GFP transgenic mouse, in which the Fos promoter drives a Fos-EGFP fusion protein, enables targeting of GFP+ neurons for electrophysiological recordings hours after an experience (26). In barrel cortex, GFP+ cells delineated an ensemble of synaptically connected neurons that were more likely to be excited during network activity (27) and exhibited larger responses to signals from surrounding whiskers (28). The Arc promoter has also been used to drive expression of fluorescent reporters in transgenic animals (29–31). Arc-GFP knock-in mice allowed tracking of activated populations in primary visual cortex (V1) over time. With repeated stimulus presentations, many GFP+ neurons adapted save a small group of reliable responders (29). Such studies helped elucidate the history of activity preceding IEG expression.
Figure 1.
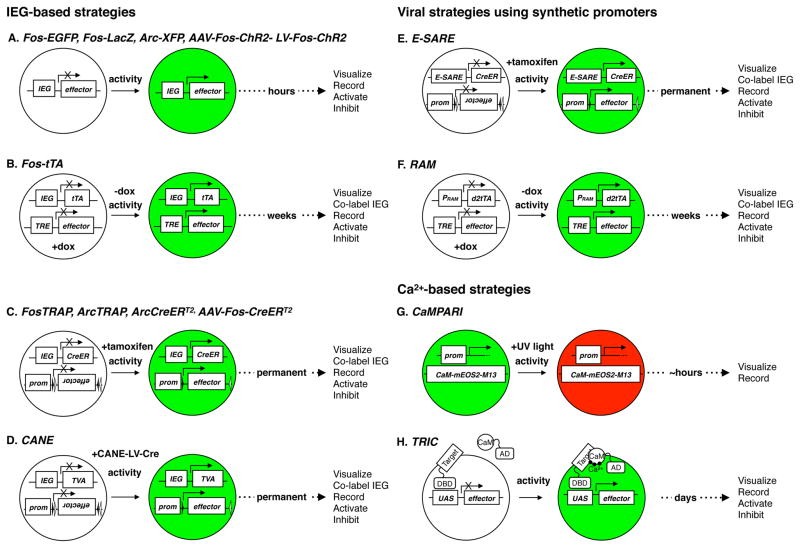
Summary of genetic strategies to visualize or manipulate previously activated neurons. (A) Fos and Arc promoters drive fluorescent proteins or optogenetic tools in both transgenic animals and viruses. Typically, peak labeling occurs a few hours after an experience and effector proteins last less than a day. (B) In Fos-tTA transgenic mice, neural activity in the absence of Dox leads to expression of TRE-conditional effectors including LacZ, chemogenetic tools, and ontogenetic tools for days following an experience. (C) In TRAP and ArcCreERT2 transgenic mice, Fos and Arc promoters drive CreERT2 to achieve permanent expression of Cre-dependent effector proteins in previously activated neurons. (D) In CANE, Fos drives TVA. Injection of EnvA-pseudotyped lentivirus delivers Cre to recently activated neurons, resulting in permanent expression of Cre-dependent effector genes. (E) In E-SARE viruses, a synthetic promoter drives effectors, or CreERT2 to achieve permanent expression of Cre-dependent effector proteins in transiently activated neurons with high S/N. (F) In RAM viruses, a synthetic promoter drives destabilized tTA (d2tTA) which can drive expression of TRE-dependent effectors when activity occurs in the absence of Dox. (G) CaMPARI integrates a green fluorescent Ca2+ indicator with a photoconvertible protein. With the intersection of neural activity and user-delivered UV light, one can visualize recently activated neurons in red until the photoconverted protein is turned over. (H) TRIC uses Ca2+ to bring together DNA-binding (DBD) and activation (AD) domains of a transcription activator to drive effector genes to visualize or manipulate previously activated neurons. TRIC operates on a slower timescale, optimal for measuring slow changes in physiological states.
Fos-LacZ transgenic mice and rats (32, 33) were designed to mark recently activated neurons in fixed tissue. In addition, LacZ product beta-galactosidase converts the prodrug Daun02 into Daunorubicin, which inactivates neurons through apopotosis or blockade of voltage-gated calcium channels (34). This strategy revealed that learned associations between environmental cues and drug reward are stored in small neuronal ensembles in nucleus accumbens (34), medial prefrontal cortex (mPFC) (35), and ventrolateral orbitofrontal cortex (36), and that intermingled ensembles in vmPFC exert opposing effects on food-seeking behavior (37). Only 2–12% of neurons were inactivated in each case, highlighting the specificity of activity-dependent manipulations (38).
The Fos promoter has also been recently used in viruses to drive optogenetic tools to reversibly manipulate the activity of recently activated neurons, revealing the roles of valence-encoding populations in behavior (39, 40). A caveat of transgenic and viral strategies in which IEG promoters directly drive effectors (Fig. 1A) is that effector expression is usually limited to a few hours after experience (Table 1). The next generation IEG-based tools were modular systems designed to drive longer-term effector expression with additional levels of temporal control and have been widely used to investigate the cellular basis of memory.
Table 1.
Summary and comparisons of different genetic strategies
| Method | Construct | Delivery1 | Window2 | Perdurance3 | Strengths | Caveats | Reference |
|---|---|---|---|---|---|---|---|
| IEG-based strategies | |||||||
| Transgenic Mice | |||||||
| Fos-LacZ | Fos>FosLacZ | Tg | < 6hrs | <1d | Daun02 inactivation | Requires fixed tissue to vis. | Smeyne, 1992 |
| Fos-GFP | Fos>FosEGFP | Tg | < 6hrs | <1d | Visualize in living tissue, brain-wide | Impermanent, no manipulation | Barth, 2004 |
| Arc-GFP | Arc>d2EGFP | KI | < 6hrs | <1d | Wang, 2006 | ||
| Arc-dVenus | Arc>dVenus | Tg | < 6hrs | <1d | Eguchi, 2009 | ||
| Arc-GFP | Arc>EGFP | BAC | < 6hrs | <1d | Gong, 2003 | ||
| Fos-tTA | Fos>tTA | Tg | 1–3 days | Weeks | Long-term, brain-wide | Background, long window | Reijmers, 2007 |
| FosTRAP | Fos>CreERT2 | KI | <12 hrs | Permanent | Permanent | ~Long window | Guenthner, 2013 |
| ArcTRAP | Arc>CreERT2 | KI | <12 hrs | Permanent | Background, ~long window | ||
| ArcCreERT2 | Arc>CreERT2 | BAC | ~24h | Permanent | Not knock-in, ~long window | Denny, 2014 | |
| CANE | Fos>Fos-2A-TVA | KI | < 6hrs | Permanent | Permanent together with Cre virus | Limited temporal control; spatially limited | Sakurai, 2016 |
| Transgenic Rats | |||||||
| Fos-LacZ | Fos>FosLacZ | Tg | < 6hrs | <1d | Daun02 inactivation | Requires fixed tissue to vis. | Kasof, 1995 |
| Viruses | |||||||
| LV-Fos-ChR2 | LV-Fos>ChR2 | LV | < 6hrs | <1d | No transgene | Impermanent | Gore, 2015 |
| AAV-Fos-ChR2 | AAV-Fos>ChR2 | AAV | < 6hrs | <1d | No transgene | Impermanent | Ye, 2016 |
| E-SARE | AAV-E-SARE>ERT2CreERT2 -PEST | AAV | < 6hrs | Permanent | High S/N, permanent | Spatially limited | Kawashima, 2013 |
| RAM | AAV-PRAM>mKate2 | AAV | 1–3 days | Weeks | High S/N | Spatially limited, long window | Sorensen, 2016 |
| AAV-Fos-CreERT2 | AAV-Fos>ERT2-CreERT2-PEST | AAV | < 6hrs | Permanent | No transgene, Permanent | Spatially limited | Ye, 2016 |
| Calcium-based strategies | |||||||
| CaMPARI | promoter>CaM-mEOS2-M13 | AAV | > seconds | <1d | Short window | Impermanent | Fosque, 2015 |
| TRIC | promoter>CaM-AD; promoter>GAL4 DBD-CaM target; UAS>effector | Tg (Drosophi la) | Hours | Days | Drives effectors | Long labeling window, background | Gao, 2015 |
Abbreviation: Tg, random transgenic integration; KI, knock-in; BAC, bacterial artificial chromosome-mediated random transgenic integration, LV, lentivirus; AAV, adeno-associated virus.
Refers to the window during which activity of neurons is captured.
Refers to the period during which transiently active neurons can be observed and/or manipulated.
IEG promoters driving tTA allows manipulation of previously activated neurons
A long-standing question in the memory field is whether neurons activated during learning participate in the memory trace. To test this, the Fos-tTA transgenic mouse was developed, in which a Fos promoter fragment drives expression of the doxycycline (Dox)-repressible tetracycline transactivator (tTA). In the absence of Dox, a reporter allele tags activated cells with LacZ (driven by the tetracycline response element, TRE), which perdures for several days (Fig. 1B) (41). Many basolateral amygdala (BLA) neurons tagged during fear conditioning were reactivated (immunopositive for both LacZ and another IEG product Zif) during memory recall, suggesting they were part of a stable fear memory trace.
While LacZ can only be detected in fixed tissue, the flexible design of the Fos-tTA mouse facilitated subsequent studies in which Fos-tTA drove expression of TRE-driven chemogenetic and optogenetic tools to establish causal roles of neurons activated during learning in subsequent memory recall (42, 43). This technique revealed that dentate gyrus (DG) ensembles tagged in a neutral context could serve as a conditioned stimulus during contextual fear conditioning (CFC) (44). While tagged DG ensembles encoding a negative experience could switch their valence when artificially reactivated during a positive experience, BLA neurons could not. Switching valence in DG reduced tag/Fos overlap in BLA, suggesting that functional connectivity had changed (45). These studies indicate that DG encodes stable representations of environmental cues, and that DG→BLA links connect sensory representation to behavioral response. Recently, studies using Fos-tTA to express optogenetic tools in learning-related CA1 neurons revealed their role in activating downstream ensembles in retrosplenial cortex and nucleus accumbens to support contextual fear memory and social memory, respectively (46–48). Of note, silencing CA1 ensembles impaired memory, suggesting that Fos-tTA tagged CFC-related CA1 neurons with high efficiency (46).
Fos-tTA has also been used to map cellular and circuit changes in normal aging and disease states. Aged mice had deficits in freezing to photoactivation of CFC-related neural ensembles in CA1. These deficits were rescued by increasing cellular excitability in CA1, indicating that age-related reduction in neuronal excitability impairs aspects of memory (49). A mouse model of Alzheimer’s Disease (AD) exhibited impairments in long-term episodic memory that could be rescued by reactivating DG neurons that were activated during conditioning, suggesting that memory failure in early AD reflects an impairment in retrieval rather than in encoding (50). Interestingly, photoactivating DG cells tagged during a positive experience rescued depressive behaviors, suggesting a therapeutic strategy (51).
While the Fos-tTA strategy answered fundamental questions about the cellular basis of learning, memory, age-related disease, and maladaptive states, induction of activity-dependent labeling was modest, ranging from ~2–4-fold above home cage levels. Because the slow metabolism of Dox results in a long labeling window on the order of days, background is likely high. Thus, Fos-tTA may only achieve workable signal-to-noise ratio (S/N) in brain regions with very low basal expression of Fos. Also, while reporter expression outlasts Fos protein, Fos-tTA cannot drive permanent effector expression, so manipulations are limited to a few days after the experience (Fig. 1B). These limitations motivated the development of activity-reporter mice that allow permanent labeling of previously activated neurons.
IEG promoters driving Cre allow permanent labeling and manipulation of previously activated neurons
In TRAP (Targeted Recombination in Active Populations) lines, CreERT2 is knocked-in to the Fos or Arc translation start sites, yielding FosTRAP and ArcTRAP (52). When a neuron is activated in the presence of tamoxifen, CreERT2 translocates to the nucleus to recombine floxed alleles, resulting in permanent effector expression (Fig. 1C). Using the active metabolic form of tamoxifen (4-hydroxytamoxifen) limits the drug-active period to <12 hours (52), which is considerably shorter than the labeling window in Fos-tTA. FosTRAP drives reporter expression in sensory cortices and hippocampus following sensory stimulation, but some subcortical regions lack activity-dependent reporter expression. In contrast, ArcTRAP labels subcortical regions, but exhibits tamoxifen-independent labeling in cortico-thalamic projection neurons (52). ArcTRAP has been used to compare neurons across the whole brain that are activated by appetitive and aversive experiences, and allowed identification of molecular signatures of such neurons (40). Though no behavioral deficits were reported (52), it is important to note that CreER replaced the IEG coding sequence and displaced regulatory sequences, essentially creating IEG null alleles. Thus, these experiments were performed in heterozygous Fos and Arc mice. An advantage of BAC transgenics is that the gene of interest remains functional.
A conceptually similar ArcCreERT2 BAC-transgenic mouse (Fig. 1C) was developed to investigate the causal role of hippocampal ensembles in long-term memory. Using these mice to drive inhibitory optogenetic tools revealed the obligatory function of learning-activated hippocampal neurons in long-term memory storage and suggested that ArcCreERT2 labels CA1 ensembles with high efficiency (53). The same genetic strategy revealed that cortical amygdala neurons activated by attractive or aversive odors represent distinct ensembles that can bidirectionally control corresponding innate behaviors (54).
In CANE (Capturing and manipulating Activated Neural Ensembles), 2A-dsTVA is knocked-in to the Fos locus after the Fos open reading frame, such that a destabilized TVA receptor for EnvA (a coat protein for an avian-specific virus) is expressed from the same mRNA as Fos. Companion lentiviruses (LV) pseudotyped with EnvAM21, a mutant with reduced TVA-binding potency, infect dsTVA-expressing neurons to deliver effector proteins. CANE-LV-Cre achieves permanent labeling of previously activated neurons (Fig. 1D). CANE-captured social fear-activated neurons (SFNs) in the ventromedial hypothalamus (VMHvl) bi-directionally controlled social fear. Because SFNs comprise only 3% of VMHvl neurons and specific molecular markers are not available, it would have been difficult to target them without activity-based methods (55).
While these strategies support permanent access to previously activated neurons, the CreERT2-based methods have variable S/N in different parts of the brain in part because, compared with Cre, CreERT2 has lower recombination efficiency even in the presence of tamoxifen. Commensurate with Fos-tTA, salient experiences induced ~2–4-fold increases in labeling in the hippocampus and amygdala of TRAP and ArcCreERT2 mice. CANE may have an improved S/N, but requires virus injections immediately following the experience of interest, which could confound some behaviors. In addition, these strategies may not allow access to all cell-types due to inherent cell-type specificities of endogenous IEGs. Viral strategies based on synthetic promoters feature higher S/N and broader applicability across the cell-type spectrum.
Viral strategies using synthetic promoters improve signal-to-noise
E-SARE is a synthetic promoter in which five repeats of a synaptic-activity responsive element (SARE) (56) that regulates induction of Arc are fused with an Arc minimal promoter. AAV-E-SARE-GFP boasted much greater reporter induction than AAV-Fos-GFP in vivo. E-SARE-labeled populations were further validated by recording their physiological responses. E-SARE driving CreERT2 affords temporal control and permanent labeling of activated neurons (Fig. 1E) (57). RAM (Robust Activity Marking) is based on a synthetic enhancer module in which the AP-1 site and Npas4 binding sites are inserted into an element whose secondary structure is favorable for transcription activation. A small synthetic promoter (PRAM) consists of four synthetic enhancers upstream of a minimal Fos promoter. PRAM reported a higher induction ratio than E-SARE in culture. Higher than Fos-tTA and ArcCreERT2, AAV-PRAM-d2tTA (a destabilized tTA, Fig. 1F) drove ~7-fold induction of TRE-dependent effectors in DG, and a ~37-fold induction in CA3 following CFC. As has been reported for Npas4 (58), these increases were learning-dependent. Although d2tTA had improved fold regulation over tTA, like Fos-tTA, RAM-driven effector expression was highest when animals were off Dox for 3 days (43). A Cre-dependent version of RAM (CRAM) effectively labels GABAergic neurons. Since the enhancer sequences are highly conserved, RAM may also be applicable to other species, as was shown in flies and rats (59).
These viral strategies bypass the need for transgenic mice and thus can be used in other species to access activated neurons and can be more conveniently used in mice (e.g., in combination with other genetic manipulations). However, a caveat viral methods share with CANE is the limited spatial reach of viral transductions. Only transgenic strategies such as TRAP, when combined with transgenic reporters, can identify activated neurons throughout the brain and thus can be used for unbiased mapping of whole-brain activity patterns.
Whole-brain imaging of activated neurons
By combining new imaging technologies with genetic strategies to label activated neurons, it is possible to map the brain-wide projections of activated neurons and screen brain-wide activity patterns following an experience. For example, the brain-clearing method CLARITY and light-sheet microscopy (60) have been used in combination with a viral Fos promoter-driven CreERT2 (Fig. 1C) to map the brain-wide projections of prefrontal cortical neurons activated by appetitive and aversive experiences (40). Similarly, Fos-GFP was used in combination with serial two-photon tomography to screen sex-specific activation patterns during social recognition and sex discrimination (61). Other studies used CLARITY (60), CUBIC (62), and iDISCO+ (63) to clear brains for whole-mount imaging of IEG reporter mice or Fos immunostaining to characterize patterns of neurons activated by specific experiences. Such combinations of powerful technologies will generate new hypotheses about how neuronal activity underlies behavior, and what abnormalities in spatial patterns of activity may be associated with pathological conditions.
Ca2+-based methods
Fluorescent reporters of intracellular Ca2+ concentration such as GCaMP are powerful tools for monitoring real-time neural activity in vivo (64). CaMPARI (calcium-modulated photoactivatable ratiometric integrator) is a GCaMP derivative that labels neurons activated during user-defined epochs (65). Composed of the photoactivatable protein mEOS2 fused to a calmodulin domain and M13 peptide, CaMPARI fluorescence tracks Ca2+ levels and photoconverts from green to red when UV illumination occurs coincidentally with Ca2+ influx (Fig. 1G), allowing subsequent characterization of previously activated neurons by tracking red fluorescence. As proof of principle, V1 neurons with the highest red-to-green fluorescence ratio were specifically tuned to the stimulus presented during photoconverting light pulses. CaMPARI also exhibits experience-specific labeling in zebrafish and Drosophila (65).
A complementary method available in Drosophila called TRIC (Transcriptional Reporter of Intracellular Ca2+) captures brain-wide changes in neural activity over many hours and can drive effector gene expression for subsequent manipulation. In TRIC, calmodulin (CaM) and its target peptide MKII are fused to the transcriptional activation domain and the DNA binding domain of a transcription factor, respectively. When Ca2+ enters the cell, it brings the binding and activation domains together to drive transcription of effector genes (Fig. 1H). TRIC is particularly useful for monitoring and manipulating long-term changes in activity underlying physiological states in Drosophila (66).
These two Ca2+-based strategies support different applications. While CaMPARI does not allow subsequent manipulation of activated neurons, it can label neurons during very brief epochs on the order of seconds. CaMPARI has low background, but the signal is limited by the rate of CaMPARI protein turnover. Being transcription based, TRIC has temporal resolution on the order of hours. However, as TRIC can drive effector expression it can be used to determine causal roles of activated populations in behavior. Though it has not been demonstrated in TRIC, both systems can theoretically be applied to multiple species, serving as alternatives to transgenic IEG-based tools.
Summary and Future Perspectives
Methods that provide genetic access to activated neurons allow users to specifically target behaviorally-relevant neurons in spatially intermingled, functionally heterogeneous populations. Many groups have applied these tools to elucidate the cellular basis of sensory coding, memory, and emotional valence, and to reveal how these processes are disrupted in aging or disease. An exciting new direction is to combine activity-based genetic strategies with whole-brain imaging and sequencing methods for unbiased mapping and profiling of activated populations.
With many options now available, it is critical to consider the advantages and caveats of each genetic strategy (Table 1) and choose the activity reporter best suited to each specific experimental question. Since different IEGs have unique induction profiles in different cell-types (38, 67, 68), it is critical to characterize IEG expression profiles for brain regions of interest before beginning experiments. With IEG promoters driving reporters to visualize activated neurons, the design of the transgene may dictate how faithfully the reporter follows activity. In tTA- and CreERT2-based strategies, transgene expression and drug metabolism will determine labeling window. New viral strategies using synthetic promoters offer higher S/N and experimental flexibility, with the caveat that RAM labeling may be learning-dependent in hippocampus and amygdala. RAM, CaMPARI, and likely E-SARE and TRIC support labeling in other species, and CaMPARI boasts the shortest temporal window for labeling though it does not allow subsequent manipulation of active neurons.
The field should continue to explore how labeling by existing methods reflects neuronal activity and to improve labeling specificity. These efforts can direct development of new strategies that fill gaps in the existing toolset. For instance, new tools could specifically access neurons based on subthreshold depolarization of membrane potential (69). Several groups rigorously characterized the physiological response properties of labeled neurons (57, 65). Performing similar experiments using the other genetic strategies will elucidate the nature of labeled populations. For example, pairing chronic Ca2+ imaging with IEG-based reporters will reveal the cellular activity patterns preceding reporter labeling. Redesigning targeting alleles for existing tools could improve the spatial and temporal profiles of reporter expression throughout the brain. Furthermore, new reporters that allow users to genetically access multiple experiences in the same brain could directly compare two different experiences and help subtract away non-task relevant background cells. When characterizing new genetic strategies, side-by-side comparisons with existing technologies are particularly informative for potential users (59, 66). Developing methods that combine the advantages of existing tools (Table 1), such as the temporal precision of Ca2+-based methods with genetic accessibility, will enhance our ability to interrogate the causal relationships between active populations, circuit function, and animal behavior.
Highlights.
IEG promoters driving reporters allow targeting of recently active neurons in vivo.
Fos-tTA mice drive effectors in previously active neurons for several days.
IEG promoters drive CreER for permanent effector expression in once active neurons.
Viruses with synthetic promoters drive effectors in active neurons with high S/N.
New technologies allow whole-brain mapping of active populations.
Acknowledgments
We thank members of the Luo lab and Dr. Adi Mizrahi for helpful comments on the manuscript. Dr. Luo is supported by funds from the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80(6):1368–83. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71(6):995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeNardo LA, Berns DS, DeLoach K, Luo L. Connectivity of mouse somatosensory and prefrontal cortex examined with trans-synaptic tracing. Nature neuroscience. 2015;18(11):1687–97. doi: 10.1038/nn.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyamichi K, Shlomai-Fuchs Y, Shu M, Weissbourd BC, Luo L, Mizrahi A. Dissecting local circuits: parvalbumin interneurons underlie broad feedback control of olfactory bulb output. Neuron. 2013;80(5):1232–45. doi: 10.1016/j.neuron.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A. Cortical interneurons that specialize in disinhibitory control. Nature. 2013;503(7477):521–4. doi: 10.1038/nature12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kvitsiani D, Ranade S, Hangya B, Taniguchi H, Huang JZ, Kepecs A. Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature. 2013;498(7454):363–6. doi: 10.1038/nature12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courtin J, Chaudun F, Rozeske RR, Karalis N, Gonzalez-Campo C, Wurtz H, et al. Prefrontal parvalbumin interneurons shape neuronal activity to drive fear expression. Nature. 2014;505(7481):92–6. doi: 10.1038/nature12755. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura T, Pignatelli M, Suh J, Kohara K, Yoshiki A, Abe K, et al. Island cells control temporal association memory. Science. 2014;343(6173):896–901. doi: 10.1126/science.1244634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparta DR, Hovelso N, Mason AO, Kantak PA, Ung RL, Decot HK, et al. Activation of prefrontal cortical parvalbumin interneurons facilitates extinction of reward-seeking behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34(10):3699–705. doi: 10.1523/JNEUROSCI.0235-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinto L, Dan Y. Cell-Type-Specific Activity in Prefrontal Cortex during Goal-Directed Behavior. Neuron. 2015;87(2):437–50. doi: 10.1016/j.neuron.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagler M, Ozdemir AT, Lagoun S, Malagon-Vina H, Borhegyi Z, Hauer R, et al. Divisions of Identified Parvalbumin-Expressing Basket Cells during Working Memory-Guided Decision Making. Neuron. 2016;91(6):1390–401. doi: 10.1016/j.neuron.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Ohki K, Chung S, Ch’ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433(7026):597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- 14.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4(4):477–85. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311(5985):433–8. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 16.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, et al. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14(2):433–45. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234(4772):80–3. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- 18.Sagar SM, Sharp FR, Curran T. Expression of c-fos protein in brain: metabolic mapping at the cellular level. Science. 1988;240(4857):1328–31. doi: 10.1126/science.3131879. [DOI] [PubMed] [Google Scholar]
- 19.Douglas RM, Dragunow M, Robertson HA. High-frequency discharge of dentate granule cells, but not long-term potentiation, induces c-fos protein. Brain research. 1988;464(3):259–62. doi: 10.1016/0169-328x(88)90033-2. [DOI] [PubMed] [Google Scholar]
- 20.Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55(5):917–24. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- 21.Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52(3):445–59. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, et al. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20(11):3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annual review of neuroscience. 1991;14:421–51. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- 24.Hunt SP, Pini A, Evan G. Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature. 1987;328(6131):632–4. doi: 10.1038/328632a0. [DOI] [PubMed] [Google Scholar]
- 25.Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nature neuroscience. 1999;2(12):1120–4. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- **26.Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24(29):6466–75. doi: 10.1523/JNEUROSCI.4737-03.2004. Describes the Fos-GFP transgenic mouse. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yassin L, Benedetti BL, Jouhanneau JS, Wen JA, Poulet JF, Barth AL. An embedded subnetwork of highly active neurons in the neocortex. Neuron. 2010;68(6):1043–50. doi: 10.1016/j.neuron.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jouhanneau JS, Ferrarese L, Estebanez L, Audette NJ, Brecht M, Barth AL, et al. Cortical fosGFP expression reveals broad receptive field excitatory neurons targeted by POm. Neuron. 2014;84(5):1065–78. doi: 10.1016/j.neuron.2014.10.014. [DOI] [PubMed] [Google Scholar]
- **29.Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, et al. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126(2):389–402. doi: 10.1016/j.cell.2006.06.038. Describes the Arc-GFP knock-in mouse. [DOI] [PubMed] [Google Scholar]
- **30.Eguchi M, Yamaguchi S. In vivo and in vitro visualization of gene expression dynamics over extensive areas of the brain. NeuroImage. 2009;44(4):1274–83. doi: 10.1016/j.neuroimage.2008.10.046. Describes the Arc-dVenus transgenic mouse. [DOI] [PubMed] [Google Scholar]
- **31.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–25. doi: 10.1038/nature02033. Describes GENSAT atlas which contains the Arc-GFP BAC transgenic mouse. [DOI] [PubMed] [Google Scholar]
- **32.Smeyne RJ, Schilling K, Robertson L, Luk D, Oberdick J, Curran T, et al. fos-lacZ transgenic mice: mapping sites of gene induction in the central nervous system. Neuron. 1992;8(1):13–23. doi: 10.1016/0896-6273(92)90105-m. Describes the Fos-LacZ transgenic mouse. [DOI] [PubMed] [Google Scholar]
- **33.Kasof GM, Mandelzys A, Maika SD, Hammer RE, Curran T, Morgan JI. Kainic acid-induced neuronal death is associated with DNA damage and a unique immediate-early gene response in c-fos-lacZ transgenic rats. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15(6):4238–49. doi: 10.1523/JNEUROSCI.15-06-04238.1995. Describes the Fos-LacZ transgenic rat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Koya E, Golden SA, Harvey BK, Guez-Barber DH, Berkow A, Simmons DE, et al. Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization. Nature neuroscience. 2009;12(8):1069–73. doi: 10.1038/nn.2364. Describes the Daun02 inactivation method. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bossert JM, Stern AL, Theberge FR, Cifani C, Koya E, Hope BT, et al. Ventral medial prefrontal cortex neuronal ensembles mediate context-induced relapse to heroin. Nature neuroscience. 2011;14(4):420–2. doi: 10.1038/nn.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fanous S, Goldart EM, Theberge FR, Bossert JM, Shaham Y, Hope BT. Role of orbitofrontal cortex neuronal ensembles in the expression of incubation of heroin craving. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(34):11600–9. doi: 10.1523/JNEUROSCI.1914-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren BL, Mendoza MP, Cruz FC, Leao RM, Caprioli D, Rubio FJ, et al. Distinct Fos-Expressing Neuronal Ensembles in the Ventromedial Prefrontal Cortex Mediate Food Reward and Extinction Memories. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36(25):6691–703. doi: 10.1523/JNEUROSCI.0140-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruz FC, Koya E, Guez-Barber DH, Bossert JM, Lupica CR, Shaham Y, et al. New technologies for examining the role of neuronal ensembles in drug addiction and fear. Nature reviews Neuroscience. 2013;14(11):743–54. doi: 10.1038/nrn3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Gore F, Schwartz EC, Brangers BC, Aladi S, Stujenske JM, Likhtik E, et al. Neural Representations of Unconditioned Stimuli in Basolateral Amygdala Mediate Innate and Learned Responses. Cell. 2015;162(1):134–45. doi: 10.1016/j.cell.2015.06.027. Describes LV-Fos-ChR2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, et al. Wiring and Molecular Features of Prefrontal Ensembles Representing Distinct Experiences. Cell. 2016;165(7):1776–88. doi: 10.1016/j.cell.2016.05.010. Describes AAV-Fos-ChR2 and AAV-Fos-CreERT2 and first application of CLARITY to genetic strategies to access activated populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **41.Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317(5842):1230–3. doi: 10.1126/science.1143839. Describes the Fos-tTA transgenic mouse. [DOI] [PubMed] [Google Scholar]
- 42.Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, et al. Generation of a synthetic memory trace. Science. 2012;335(6075):1513–6. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *43.Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484(7394):381–5. doi: 10.1038/nature11028. First demonstration that DG ensembles activated during contextual fear conditioning are sufficient to drive freezing during recall. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez S, Liu X, Lin PA, Suh J, Pignatelli M, Redondo RL, et al. Creating a false memory in the hippocampus. Science. 2013;341(6144):387–91. doi: 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- 45.Redondo RL, Kim J, Arons AL, Ramirez S, Liu X, Tonegawa S. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature. 2014;513(7518):426–30. doi: 10.1038/nature13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *46.Tanaka KZ, Pevzner A, Hamidi AB, Nakazawa Y, Graham J, Wiltgen BJ. Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron. 2014;84(2):347–54. doi: 10.1016/j.neuron.2014.09.037. Demonstrated that hippocampal neurons activated during fear conditioning are required for memory recall. [DOI] [PubMed] [Google Scholar]
- *47.Cowansage KK, Shuman T, Dillingham BC, Chang A, Golshani P, Mayford M. Direct reactivation of a coherent neocortical memory of context. Neuron. 2014;84(2):432–41. doi: 10.1016/j.neuron.2014.09.022. Used catFISH to demonstrate that natural fear recall and artificial stimulation of Fos-tTA tagged cortical neurons tagged during fear conditioning recruit highly overlapping populations in downstream structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okuyama T, Kitamura T, Roy DS, Itohara S, Tonegawa S. Ventral CA1 neurons store social memory. Science. 2016;353(6307):1536–41. doi: 10.1126/science.aaf7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, et al. A shared neural ensemble links distinct contextual memories encoded close in time. Nature. 2016;534(7605):115–8. doi: 10.1038/nature17955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *50.Roy DS, Arons A, Mitchell TI, Pignatelli M, Ryan TJ, Tonegawa S. Memory retrieval by activating engram cells in mouse models of early Alzheimer’s disease. Nature. 2016;531(7595):508–12. doi: 10.1038/nature17172. Describes an elegant control experiment in which Fos-tTA drives both ChR2 and diptheria toxin (DTA). Reactivating tagged neurons promoted behavioral improvement in a model of AD. Improvements were then abolished when tagged neurons were killed with DTA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez S, Liu X, MacDonald CJ, Moffa A, Zhou J, Redondo RL, et al. Activating positive memory engrams suppresses depression-like behaviour. Nature. 2015;522(7556):335–9. doi: 10.1038/nature14514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **52.Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78(5):773–84. doi: 10.1016/j.neuron.2013.03.025. Describes ArcTRAP and FosTRAP knock-in mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **53.Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, et al. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83(1):189–201. doi: 10.1016/j.neuron.2014.05.018. Describes ArcCreERT2 transgenic mouse. First demonstration that hippocampal neurons activated during fear conditioning are required for long term memory recall. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Root CM, Denny CA, Hen R, Axel R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature. 2014;515(7526):269–73. doi: 10.1038/nature13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **55.Sakurai K, Zhao S, Takatoh J, Rodriguez E, Lu J, Leavitt AD, et al. Capturing and Manipulating Activated Neuronal Ensembles with CANE Delineates a Hypothalamic Social-Fear Circuit. Neuron. 2016;92(4):739–53. doi: 10.1016/j.neuron.2016.10.015. Describes CANE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, et al. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(1):316–21. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **57.Kawashima T, Kitamura K, Suzuki K, Nonaka M, Kamijo S, Takemoto-Kimura S, et al. Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nature methods. 2013;10(9):889–95. doi: 10.1038/nmeth.2559. Describes E-SARE. [DOI] [PubMed] [Google Scholar]
- 58.Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334(6063):1669–75. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **59.Sorensen AT, Cooper YA, Baratta MV, Weng FJ, Zhang Y, Ramamoorthi K, et al. A robust activity marking system for exploring active neuronal ensembles. eLife. 2016:5. doi: 10.7554/eLife.13918. Describes RAM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nature protocols. 2014;9(7):1682–97. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *61.Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell reports. 2015;10(2):292–305. doi: 10.1016/j.celrep.2014.12.014. Application of serial two-photon tomography to genetic strategies to access active neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *62.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157(3):726–39. doi: 10.1016/j.cell.2014.03.042. First application of CUBIC to genetic strategies to access activated neurons. [DOI] [PubMed] [Google Scholar]
- 63.Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell. 2016;165(7):1789–802. doi: 10.1016/j.cell.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature methods. 2009;6(12):875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **65.Fosque BF, Sun Y, Dana H, Yang CT, Ohyama T, Tadross MR, et al. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347(6223):755–60. doi: 10.1126/science.1260922. Describes CaMPARI. [DOI] [PubMed] [Google Scholar]
- **66.Gao XJ, Riabinina O, Li J, Potter CJ, Clandinin TR, Luo L. A transcriptional reporter of intracellular Ca(2+) in Drosophila. Nature neuroscience. 2015;18(6):917–25. doi: 10.1038/nn.4016. Describes TRIC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157(5):1216–29. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun X, Lin Y. Npas4: Linking Neuronal Activity to Memory. Trends in neurosciences. 2016;39(4):264–75. doi: 10.1016/j.tins.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154(4):904–13. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]