Abstract
Genetic control of plant diseases has traditionally included the deployment of single immune receptors with nucleotide-binding leucine-rich repeat (NLR) domain architecture. These NLRs recognize corresponding pathogen effector proteins inside plant cells, resulting in effector-triggered immunity (ETI). Although ETI triggers robust resistance, deployment of single NLRs can be rapidly overcome by pathogen populations within a single or a few growing seasons. In order to generate more durable disease resistance against devastating plant pathogens, a multi-tiered strategy that incorporates stacked NLRs combined with other sources of disease resistance is necessary. New genetic and genomic technologies have enabled advancements in identifying conserved pathogen effectors, isolating NLR repertoires from diverse plants, and editing plant genomes to enhance resistance. Significant advancements have also been made in understanding plant immune perception at the receptor level, which has promise for engineering new sources of resistance. Here, we discuss how to utilize recent scientific advancements in a multilayered strategy for developing more durable disease resistance.
INTRODUCTION
The global population is predicted to increase to 9.6 billion by the year 2050 (FAO 2015). To support this rapid rise in population growth, agricultural production must increase without concurrent increases in the levels of land, fertilizer, water, and pesticides utilized. Meeting this enormous challenge will require advancements in all aspects of agricultural production, including decreased losses due to plant diseases (Tilman et al. 2002).
The plant waxy cuticle, cell wall, and preformed antimicrobial compounds are important components inhibiting pathogen colonization. Plants also rely on their innate immune system to actively recognize and respond to invading pathogens. Surface-localized pattern recognition receptors (PRRs) are capable of recognizing conserved microbial patterns, such as bacterial flagellin, fungal chitin, and oomycete glucan, resulting in pattern-triggered immunity (PTI) (Couto and Zipfel 2016). Cell surface receptors have also been identified that recognize non-conserved ligands, including variable fungal effectors (Thomma et al. 2011). Intracellular immune receptors are able to recognize pathogen proteins, called effectors, delivered inside plant cells during infection resulting in effector-triggered immunity (ETI) (Chiang and Coaker 2015; Toruño et al. 2016). These intracellular receptors often possess nucleotide-binding leucine-rich repeat (NLR) domain architecture. In the absence of NLR recognition, effectors inhibit basal defense signaling, enable nutrient acquisition, and affect diverse plant metabolic processes in order to facilitate disease development (Toruño et al. 2016).
Plant resistance (R) proteins often possess NLR domain architecture. NLRs can recognize diverse pathogen effectors, including those from bacteria, fungi, oomycetes, viruses, nematodes, and arthropod pests (Chiang and Coaker 2015). Different plant genomes encode a range of NLRs. For example, Arabidopsis possesses ~151 NLRs while domesticated apple possesses ~737 NLRs (Jones et al. 2016). In addition to the central nucleotide-binding site and C-terminal leucine-rich repeat domains, NLRs can possess an N-terminal coiled-coiled domain (CNLs) or toll/interleukin 1 receptor-like domain (TNLs). NLR effector perception results in a suite of downstream defense responses, including the influx of calcium ions, production of reactive oxygen species, hormonal changes, and transcriptional reprogramming (Chiang and Coaker 2015). A hallmark of ETI is the hypersensitive response (HR), a type of programmed cell death at the penetration site. Localized ETI also induces systemic acquired resistance (SAR), resulting in heightened resistance against subsequent pathogen attack (Spoel and Dong 2012).
NLRs can recognize corresponding pathogen effector proteins directly, indirectly, or in heterologous pairs. The rice CNL Pi-ta directly interacts with and perceives the fungal Magnaporthe oryzae AVR-Pita effector (Jia et al. 2000). Similarly, the flax TNLs L5 and L6 are able to interact with and directly perceive the fungal Melampsora lini rust effector AvrL567 (Dodds et al. 2006). NLRs can also indirectly recognize effector-mediated modification of host proteins. These effector-targeted host proteins could either be bona fide virulence targets (guardees) or decoys of true virulence targets. For example, the Arabidopsis CNL RPS2 senses the Pseudomonas syringae effector protease AvrRpt2 through effector-mediated cleavage of the guardee RIN4 (Axtell and Staskawicz 2003; Mackey et al. 2003). Recently, perception of effectors has been demonstrated through heterologous paired NLR activity (Bernoux et al. 2014). In several cases, the paired NLRs share a promoter and are genetically located in a head-to-head orientation. In paired NLRs, the sensor NLR possesses an additional non-canonical domain which serves as a decoy by mimicking an effector target. Effector binding to the sensor NLR leads to activation of the second signaling NLR (possessing classical domain architecture) resulting in ETI (Bernoux et al. 2014). Two rice CNLs, RGA4 and RGA5, function as a pair to recognize the M. oryzae effectors AVR1-CO39 and AVR-Pia (Césari et al. 2013). The AVR-Pia effector directly interacts with the sensor NLR RGA5 in its RATX1 domain, resulting in the activation of the RGA4 signaling NLR (Césari et al. 2014; Ortiz et al. 2017). Multiple pathogen effectors target similar plant proteins and processes (Mukhtar et al. 2011). Thus, single NLRs or NLR pairs could be capable of recognizing effectors from diverse pathogens.
Over the last 30 years, research in molecular plant pathology has provided scientists with the knowledge to begin effectively integrating ETI into crop improvement programs. In order to appropriately harness ETI for effective disease resistance, it will first be necessary to identify multiple NLRs recognizing conserved pathogen effectors. A repertoire of NLRs effective against important pathogens can then be combined in single breeding lines. Ideally, these NLR stacks should be deployed in different combinations to maximize their chance for durability. Incorporation of other sources of non-canonical resistance loci should also be used as part of a multi-tiered strategy for durable disease resistance. Other sources of resistance include: quantitative resistance, recessive resistance, susceptibility genes, as well as PRRs into germplasm with stacked NLRs. Figure 1 provides a conceptual overview of how ETI can be incorporated into plant germplasm with the goal of durable disease resistance.
Figure 1. A multilayered strategy for durable disease resistance.
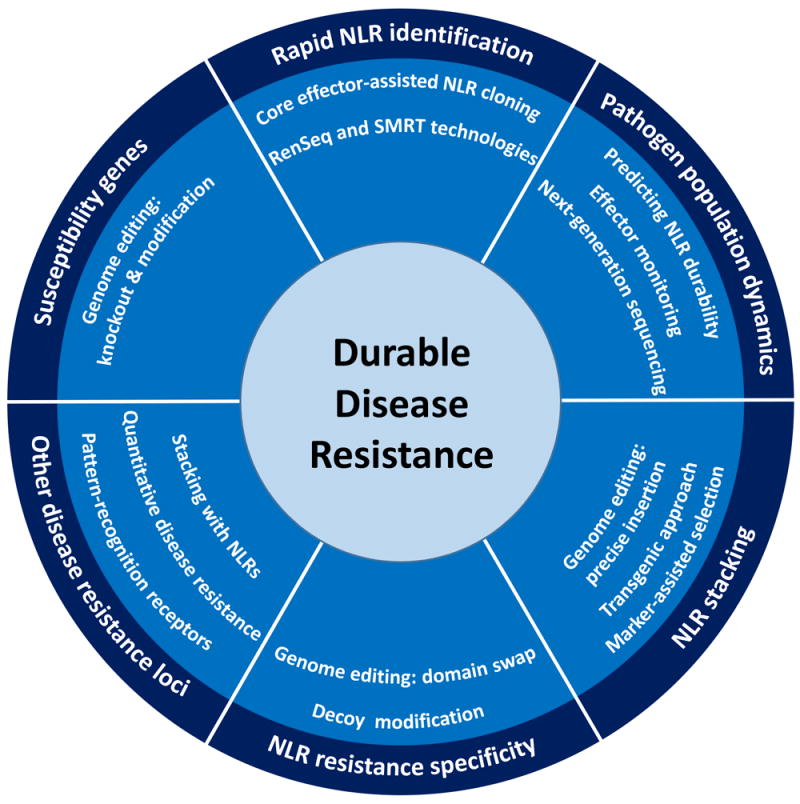
To produce plant germplasm with durable disease resistance, a multilayered strategy is necessary. Plant NLR immune receptors can be identified using RenSeq and their durability predicted by monitoring pathogen population dynamics. Promising NLRs can be stacked in single breeding lines and there is potential for engineering new resistance specificities. As part of a multilayered strategy, NLRs can be combined with other sources of disease resistance loci, including other classes of immune receptors, quantitative sources of resistance, and recessive resistance or susceptibility genes.
RESISTANCE GENE IDENTIFICATION
Effectors as tools to accelerate NLR identification
In order to cause disease, diverse plant pathogens deliver effector proteins directly into host cells. Pathogens possess vast effector repertoires, ranging from 30-40 in bacterial pathogens to hundreds in oomycete pathogens (Toruño et al. 2016). Within a particular species, different pathogen strains possess variable as well as core sets of effectors. These core effectors are defined by their wide distribution across the population of a particular pathogen and their substantial contribution to pathogen virulence (Dangl et al. 2013). Core effectors have been shown to be important virulence components for multiple pathogens, although there are some exceptions (Kombrink et al. 2017). Deploying NLRs recognizing core effectors will require significant evolutionary hurdles for pathogens to overcome and potentially provide durable disease resistance. Therefore, breeding programs should aim to identify and utilize NLRs recognizing these core effectors (Figure 1).
Next-generation sequencing technologies (NGS) have significantly impacted our understanding of plant-microbe interactions and provide an opportunity to rapidly identify effector repertoires from pathogen populations (Baltrus et al. 2011; Studholme et al. 2011; Thakur et al. 2016). The ability to detect effector repertoires from different strains enables the identification of core effectors from a pathogen population. These core effectors can then be used to identify NLRs and to guide NLR deployment. Genome-wide prediction of effectors is possible in multiple pathogens. Oomycete effectors can be predicted based on an N-terminal secretion signal peptide, followed by a conserved translocation motif (RXLR, LXLFLAK) (Haas et al. 2009; Tyler et al. 2006; Whisson et al. 2007) Genome-wide prediction of effectors is also feasible in cyst nematodes (Eves-van den Akker and Birch 2016), bacteria (Alfano 2009) and some pathogenic fungi (Sonah et al. 2016). Evidence of effector selection and contribution to virulence could be integrated using machine learning-based approaches to improve accurate effector identification. Although more and more computational tools are available for the prediction of candidate effectors, there are still some limitations. For example, oomycete pathogens can possess effectors lacking canonical and secretion motifs (Liu et al. 2014). While it is possible to predict effectors based on conserved motifs for some pathogens, it is still challenging to exhaustively identify effectors for many plant pathogens.
Subsets of core effectors have been identified in various pathogens, including bacteria, fungi and oomycetes (Bart et al. 2013; Boevink et al. 2016; Hemetsberger et al. 2015; Stork, Kim, and Mudgett 2015). Using core effectors as probes to screen plant germplasm has emerged as a powerful tool to isolate new NLRs (Vleeshouwers and Oliver 2014) (Figure 1). A rapid phenotype that can be easily screened is the ability of an effector to elicit an HR, a hallmark of the ETI response. In several plant species, there are rapid functional assays based on transient gene expression. Agrobacterium can be used to transiently express effectors in planta as well as facilitate infection with viral based vectors such as Potato virus X (PVX) in the Solanacaea, Cucurbitaceae, and Asteraceae (Vleeshouwers et al. 2011). However, for many crops including monocots, leaves are often not amenable to Agrobacterium-based assays. Alternative bacterial delivery systems and viral-based systems for transient expression are being developed to overcome this problem. For example, the bacterial type III secretion system is an efficient tool to deliver effectors from diverse pathogens into plant cells (Rentel et al. 2008; Sohn et al. 2007). Effector genomics coupled with the development of a high-throughput functional screening system would rapidly allow identification of cognate NLRs that recognize effectors. Wild germplasm and non-host plants also provide a rich source of potential NLRs. The ability to screen diverse plants for core effector recognition has potential to facilitate the identification of useful NLRs from non-hosts (Lee et al. 2014).
NLR identification by coupling resistance gene enrichment and high throughput sequencing
Plant genomes frequently contain hundreds of NLR-encoding genes, many of which are genetically clustered in tandem duplications, making it difficult to assemble NLR loci using Sanger or short read sequencing technology (Marone et al. 2013). Recent advancements now enable NLRs to be rapidly identified from complex plant genomes using resistance gene enrichment sequencing (RenSeq, Table 1). RenSeq is an NLR targeted gene enrichment and sequencing approach (Jupe et al. 2013). RenSeq involves capturing DNA fragments from a genomic or cDNA library using biotinylated RNA oligonucleotides complementary to NLR-encoding genes of a reference genome (Jupe et al. 2013). These DNA fragments are then sequenced using the long-read single-molecule real-time (SMRT) sequencing technology (Witek et al. 2016). The ability of the same template to be sequenced multiple times by SMRT sequencing ensures that the NLR sequence is determined as accurately as possible, resulting in the identification and appropriate gene annotation of many NLRs with potential for pathogen detection. Using this technology, the number of identified NLRs in Solanum tuberosum was increased from 438 to 755 (Jupe et al. 2013, Table 1).
Table 1.
RenSeq-mediated discovery of NLR repertoires and identification of NLRs recognizing specific pathogen effectors.
| Plant | Pathogen | NLRs identified | Reference |
|---|---|---|---|
| Potato | - | 755 | Jupe et al. 2013 |
| Tomato | - | 355 | Andolfo et al. 2014 |
| Wild potato (Potato) | Phytophthora infestans | Rpi-amr3 | Witek et al. 2016 |
| Wheat | Puccinia graminis f. sp tritici | Sr22, Sr45 | Steuernage et al. 2016 |
Wild relatives of crops are a rich source of disease resistance. RenSeq has great potential to identify novel NLRs from wild species that could be deployed into commercial germplasm. NLRs are conserved among plant species and capture based on their sequence homology in conserved regions enables the identification of NLR loci from uncharacterized genomes. A new NLR for resistance to the oomycete pathogen Phytophthora infestans was identified using this RenSeq approach. (Witek et al. 2016). Most commercial potato varieties are susceptible to late blight caused by P. infestans. However, many wild potato relatives show variation for resistance and are a potential source of resistance to P. infestans (Rpi) genes. To accelerate Rpi gene cloning, RenSeq was used to assemble the NLR repertoire from a previously unsequenced wild relative of cultivated potato (Witek et al. 2016). Combined with initial bulk-segregant genetic mapping of the resistance locus, followed by fine mapping, the cognate NLR Rpi-amr3 was identified and a transgenic potato line was shown to be resistant to P. infestans (Witek et al. 2016, Table 1).
RenSeq technology can also be used in combination with mutagenesis (MutRenSeq) in order to facilitate the cloning of new sources of resistance. MutRenSeq is divided into three steps: (i) generate mutants from resistant wild-type plants and identify mutants with loss of disease resistance, (ii) capture and sequence fragments from genomic or cDNA libraries of both resistant wild-type plants and mutants with loss of disease resistance, (iii) compare genes in mutants and wild-type plants to identify the exact mutations responsible for the loss of disease resistance. This strategy was recently used to clone two stem rust resistance genes, Sr22 and Sr45, from hexaploid bread wheat (Steuernage et al. 2016, Table 1). The RenSeq technology provides an efficient strategy to rapidly isolate candidate NLRs to expand the pool of usable NLRs (Figure 1). This approach can also be used to capture other gene families with potential to be applied for crop improvement.
With the above advancements in sequencing and NLR annotation, scientists now possess thousands of potential NLR candidates that can be used in breeding for disease resistance. The current bottleneck in NLR deployment is the ability to rapidly identify multiple NLRs that can recognize effectors for important pathogens from a large pool of potential resistance proteins. Core effectors can be used as probes to screen NLR pools from diverse host species to facilitate more rapid identification of NLR-effector pairs. Core effectors from diverse pathogens can be transiently co-expressed with individual NLRs in the model plant Nicotiana benthamiana, with an HR phenotype indicating effector recognition. The approach proposed here has the potential for a rapid identification and utilization of multiple NLRs from diverse sources. However, advancements still need to be made to facilitate rapid cloning of thousands of NLRs with high sequence similarity. Developing high-throughput pipelines for phenotyping NLRs and corresponding effectors will greatly facilitate the identification of multiple promising NLRs recognizing important pathogens that can be combined in a breeding program (Figure 1).
NLR DEPLOYMENT
Stacking multiple NLRs to confer resistance
Durable disease resistance has been a longstanding goal of crop improvement, but it is rarely achieved. Here, we define durable disease resistance as sources of resistance that remain effective over multiple growing seasons under environmental conditions favoring disease. For example, Rps1k in soybean has provided resistance to Phytophthora sojae for 40 years (Sugimoto et al. 2012). The tomato NLR Mi-1.2 confers resistance to root-knot nematodes, potato aphid and whitefly (Nombela et al. 2003; Sugimoto et al. 2012). Mi-1.2 was introgressed into cultivated tomato in 1944 and has conferred durable resistance to root-knot nematodes as the frequency of resistance breaking strains is very low (Smith 1944; Vos et al. 1998). Although there are examples of single immune receptors providing durable resistance, single loci are more likely to be quickly defeated under high disease pressure (Fry 2008). Pathogens are able to rapidly acquire mutations in effectors and lose variable effectors from their genomes. NGS technologies could enable surveillance of pathogen populations and prediction of effector repertoires in real time. The ability to rapidly identify shifts in effector repertoires allows breeders to evaluate current germplasm and predict the utility of future NLR deployment. It is essential to monitor for any breakdown of individual NLRs so that new combinations are continuously assembled for effective disease control (Figure 1).
One useful strategy to overcome single NLR breakdown is to stack multiple NLRs recognizing core effectors in one genotype, ideally in combination with other sources of disease resistance (Figure 1, Table 2). Deploying germplasm with stacked NLRs is more likely to provide durable resistance, since it would be less likely that a pathogen would be able to lose or mutate multiple core effectors simultaneously. Molecular markers tightly linked to NLRs can help minimize linkage drag after backcrossing and facilitate stacking sources of resistance (Miedaner and Korzun 2012; Tiwari et al. 2013) (Figure 1). Wild germplasm represents an important source of resistance, but issues with linkage drag and sexual incompatibility are barriers to their rapid transfer into commercial cultivars through traditional plant breeding approaches. Transgenic approaches can also be employed to effectively stack NLRs from different sources. For example, three Rpi genes have been stacked in potato simultaneously using a transgenic approach, resulting in robust resistance against P. infestans (Zhu et al. 2012, Table 2). This example illustrates the power and utility of combining multiple sources of resistance into a single cultivar to control devastating diseases.
Table 2.
Examples of resistance loci that have been stacked into individual plant genotypes for disease control.
| Plant | Pathogen | Disease | Genetic loci | Reference |
|---|---|---|---|---|
|
| ||||
| Rice | Magnaporthe oryzae | rice blast | pi21 (proline-rich protein) | Fukuoka et al. 2015 |
| Pi34 (unknown) | ||||
| qBR4-2 (unknown) | ||||
| qBR12-1 (unknown) | ||||
|
| ||||
| Rice | Magnaporthe oryzae | rice blast | Pi2 (NLR) | Ellur et al. 2016 |
| Pi54 (NLR) | ||||
|
| ||||
| Rice | Xanthomonas oryzae pv. oryzae | bacterial blight | Xa21 (RLK) | Huang et al. 1997 |
| xa5 (TFIIAγ5) | ||||
| Xa4 (unknown) | ||||
| xa13 (MtN3 sugar transporter) | ||||
|
| ||||
| Potato | Phytophthora infestans | late blight | Rpi-sto1 (NLR) | Zhu et al. 2012 |
| Rpi-vnt1.1 (NLR) | ||||
| Rpi-blb3 (NLR) | ||||
|
| ||||
| Wheat | Puccinia triticina | leaf rust | Lr41 (unknown) | Cox et al. 1994 |
| Lr42 (unknown) | ||||
| Lr43 (unknown) | ||||
|
| ||||
| Wheat | Puccinia triticina Puccinia striiformis f. sp. tritici Blumeria graminis | leaf rust stripe rust powdery mildew | Lr34 (ATP binding cassette transporter) | More et al. 2015 |
| Lr67 (hexose transporter) | Krattiger et al. 2009 | |||
| Dyck 1977 | ||||
Parenthesis in the Genetic loci column indicate gene architecture, NLR = nucleotide-binding leucine-rich repeat, RLK = receptor-like kinase, and TFIIAγ5 = small subunit of the TFIIA transcription factor.
One potential barrier to NLR deployment from diverse sources is that it is unclear if most NLRs can function when transferred between diverse taxa, a term known as broad taxonomic functionality. The maize NLR RxoI controls resistance to Burkholderia andropogonis, the causal agent of bacterial stripe disease (Zhao et al. 2004). When maize RxoI is transferred to rice, it is able to recognize Xanthomonas oryzae pv. oryzicola, the causal agent of bacterial streak disease (Zhao et al. 2005). The Arabidopsis RPS4 and RRS1 NLR pair elicit ETI against effectors from the bacterial pathogens Pseudomonas syringae and Ralstonia solanacearum as well as the fungal pathogen Colletotrichum orbiculare (Narusaka et al. 2009). Using a transgenic approach, RPS4 and RRS1 were successfully transferred into members of the Brassicaceae, Solanaceae, and Cucurbitaceae resulting in recognition of specific bacterial and fungal effectors (Narusaka et al. 2013). These examples demonstrate that NLRs can be effectively transferred from model plants into crop systems.
An important consideration in deployment of NLR stacks will be to identify permissive sites in plant genomes for NLR integration. Transgene expression and silencing in later generations is influenced by the genome context surrounding the insertion site (Matzke and Matzke 1998). Insertion of foreign DNA into genes or genetic regions that impact plant growth and production should be avoided. Furthermore, identification and extensive characterization of permissive sites for NLR stacks will promote effective NLR expression, reduce deleterious effects, speed up evaluation and could also accelerate regulatory approval of future transgenic materials (Figure 1). Site-specific recombination technology can be used to precisely insert DNA into a predefined genomic location and facilitate the removal of unwanted DNA such as antibiotic selection markers (Wang et al. 2011). Genome editing technology using CRISPR/Cas9 (Clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9) has promise for facilitating precise insertion at defined genomic locations (Khatodia et al. 2016). For example, it would be ideal to integrate NLRs from wild species into existing NLR loci or clusters in cultivated crop genomes. It may also be feasible to use pathogen-inducible promoters to drive NLR gene expression and boost resistance.
Engineering new NLR-mediated resistance specificities
An elusive goal in molecular plant pathology is the ability to directly engineer novel resistance specificities. Significant effort has focused at the receptor level and defined mutations in the nucleotide binding site of NLRs can render them autoactive through constitutive ATP binding (Chiang and Coaker 2015; Takken and Goverse 2012). However, scientists have been unable to rationally design NLR receptors to recognize diverse pathogen effectors. Some progress has been made in engineering related NLR receptors for enhanced effector recognition. I2 is the tomato ortholog of the potato NLR R3a, which recognizes the P. infestans effector Avr3a (Armstrong et al. 2005; Bos et al. 2006; Ori et al. 1997). I2 is also able to weakly respond to Avr3a and a point mutation in I2’s coiled-coiled domain resulted in enhanced perception of Avr3a (Giannakopoulou et al. 2015). This example illustrates that there is promise for synthetic NLR engineering.
Recently, there has been a significant breakthrough in engineering host proteins guarded by NLRs to generate new resistance specificities. The Arabidopsis RPS5 NLR guards the host kinase PBS1 (Shao et al. 2003). The P. syringae bacterial effector AvrPphB is a protease and cleaves PBS1 at a defined region (Shao et al. 2003). RPS5 detects a conformational change in PBS1 resulting from cleavage (Qi et al. 2013). Effector proteases are common in both bacterial and viral pathogens. Substituting the AvrPphB cleavage site within PBS1 with those from a bacterial or viral protease enabled RPS5 recognition of these proteases upon infection (Kim et al. 2016). This study indicates that decoys can be used to expand the recognition specificity of a plant NLR, with potential for rational engineering of disease resistance (Figure 1). This approach could also be used to engineer resistance against a variety of other pathogens using well-characterized NLRs. Identification of fungal effectors that act as proteases inside host cells would enable this strategy to be applied to other important pathogen classes. Furthermore, it may be possible to fuse guarded proteins with signaling NLRs to engineer new resistance specificities.
COMBINING NLR-MEDIATED RESISTANCE WITH OTHER TYPES OF DISEASE RESISTANCE
Pattern Recognition Receptors (PRRs)
Plants use PRRs on the plasma membrane to recognize conserved microbial features termed pathogen- or microbe-associated molecular patterns (PAMPs/MAMPs) resulting in Pattern-Triggered Immunity (PTI). Recognition of particular MAMPs, like bacterial flagellin, is widely conserved across many plant species (Couto and Zipfel 2016). However, recognition of other MAMPs can be restricted to particular plant species, indicating that transfer of PRRs is a viable strategy for enhancing disease resistance. Although the intensity of PTI responses is usually not as strong as ETI, there is significant overlap in PTI and ETI outputs (Thomma et al. 2011). Since MAMPs are conserved and essential for pathogen viability, PRRs tend to be able to confer resistance to a broad range of pathogens. The rice PRR Xa21 recognizes a conserved sulfated peptide from Xanthomonas (Pruitt et al. 2015; Song et al. 1995). Xa21 has been widely deployed in cultivated rice germplasm and is still effective for disease control. The Arabidopsis Elongation factor Tu receptor (EFR) recognizes elongation factor Tu (EF-Tu), a widely conserved bacterial MAMP (Zipfel et al. 2006). Transfer of Arabidopsis EFR to Nicotiana benthamiana and tomato (Solanum lycopersicum) confers responsiveness to EF-Tu, resulting in resistance against bacterial pathogens from different genera (Lacombe et al. 2010). Xa21 and EFR exhibit broad taxonomic functionality and can be effectively transferred from a monocot to dicot and within the Solanacaea (Holton et al. 2015; Lacombe et al. 2010; Schwessinger et al. 2015). This research suggests that heterologous expression of PRRs could be used to engineer broad-spectrum disease resistance to diverse pathogens, potentially enabling more durable resistance in the field. Additional layers of disease resistance can also be combined with stacks of PRRs and NLRs (Figure 1, Table 2).
Quantitative disease resistance
Plant breeders have incorporated quantitative disease resistance (QDR) loci into elite breeding lines for decades (French et al. 2016). As the name implies, QDR is partial resistance leading to a reduction in disease. QDR loci that have been cloned to date encode a range of proteins such as wall associated kinases, a putative ABC transporter, and a serine hydroxymethyl transferase (Chauhan et al. 2015; Huard-Chauveau et al. 2013; Liu et al. 2012). Some cloned QDR loci are canonical immune receptors such as NLRs and PRRs, with a quantitative effect on disease resistance (French et al. 2016). QDR loci can also be influenced by the environment and epistatic effects. Thus, it is important to pyramid QDRs in appropriate genetic backgrounds to maximize their potential for a reduction in disease. In several cases, pyramiding QDR loci resulted in strong disease resistance (Das and Rao 2015; Ellur et al. 2016; Fukuoka et al. 2015; Yasuda et al. 2015, Table 2). Four QDR loci against the blast fungus M. oryzae were pyramided using marker-assisted selection in rice (Fukuoka et al. 2015, Table 2). Importantly, each locus controlled a different aspect of resistance against M. oryzae, including a loss of function mutation in a negative immune regulator, a gene of unknown function, and an NLR expression polymorphism (French et al. 2016). There is significant promise for pyramiding QDRs targeting different stages of infection with canonical immune receptors with the goal of durable resistance (Figure 1).
Genome editing and susceptibility loci
Within the last decade, significant advances in genome editing technologies have enabled targeted modification of genetic loci. Xanthomonas transcription activator-like effectors (TALEs) are delivered into host cells during infection and act as transcription factors to drive the expression of host genes to promote bacterial virulence in susceptible genetic backgrounds. In 2009, two seminal papers were published that deciphered the TALE code, elucidating that the effector’s central repeat region determined DNA binding specificity (Boch et al. 2009; Moscou and Bogdanove 2009). Deciphering the TALE code enabled scientists to rapidly predict conserved TALE binding sites, revealing that multiple plant susceptibility (S) loci encode SWEET sugar transporters (Streubel et al. 2013). S loci facilitate pathogen growth and virulence, and they are attractive targets for disease control (van Schie and Takken 2014). Furthermore, TALEs have promising applications in engineering resistance (Schornack et al. 2013). For example, synthetic TALEs under the control of a pathogen inducible promoter can be designed to induce the transcription of defense genes, immune receptors, or defense activating transcription factors.
Genome editing uses sequence specific nucleases to alter DNA sequences in a given genome. Genome editing mediated by CRISPR/Cas9 is easy to use and has revolutionized the ability to perform efficient plant genome editing (Paul III and Qi 2016). Cas9 induces double-stranded DNA breaks at precise genetic locations mediated by an RNA-guide (Doudna and Charpentier 2014). These double-stranded breaks are then repaired by endogenous plant DNA repair machinery and can result in mutations or deletions (Doudna and Charpentier 2014). Multiple disease targets have been edited using CRISPR/Cas9, with the current focus on genes encoding S loci or negative immune regulators. Synthetic TALE nucleases and CRISPR/Cas9-mediated genome editing were used to engineer resistance against plant SWEET sugar transporters, which are TALE targets (Jiang et al. 2013; Li et al. 2012). Recessive resistance to potyviruses is frequently controlled by mutations in the cap-binding protein eIF4E, which is a component of the host translation initiation complex (van Schie and Takken 2014). Plant genomes possess multiple eIF4E isoforms and mutation of isoforms required for viral replication has not impacted plant growth (van Schie and Takken 2014). Cas9-mediated genome editing has been used to engineer resistance to multiple potyviruses in cucumber (Chandrasekaran et al. 2016). Recently, Cas9-mediated genome editing with RNA guides targeting geminiviruses resulted in disease resistance against these DNA viruses (Ji et al. 2015). Genome editing also has promise in the future for targeted gene insertion and fine-tuning gene expression, especially with the discovery of more precise and efficient nucleases (Paul III and Qi 2016).
CONCLUDING POINTS
Since the discovery of H. Flor’s gene-for-gene concept in 1942 (Flor 1971), there has been significant progress in understanding the genetic and molecular basis of ETI. Many NLR effector pairs have been identified and modes of NLR recognition have been elucidated. Furthermore, advances in DNA sequencing and genome editing enable rapid identification of effectors and NLRs which can be used to accelerate NLR deployment and help predict the durability of resistance. To achieve more durable disease resistance, it will be necessary to combine different types of resistance acting at different stages in pathogen infection in various combinations. A greater foundational understanding of NLR signaling, pathogen biology, and effector targets is also necessary to effectively engineer new resistance specificities. Advancements are needed in high throughput phenotyping to identify and facilitate the deployment of new sources of resistance to stay ahead of pathogen evolution. We are currently at a point where scientists can begin to use this information to strategically implement more rational disease control.
BOXED TERMS.
Effector – Secreted pathogen proteins that enhance virulence and alter plant physiology in susceptible plant genotypes.
NLR – Nucleotide-binding leucine-rich repeat. An immune receptor present inside plant cells that recognizes a corresponding pathogen effector protein.
ETI – Effector-triggered immunity. Robust resistance caused by recognition of pathogen effectors, often mediated by NLRs.
HR – Hypersensitive Response. Programmed cell death at the site of infection. A hallmark of NLR activation and ETI.
PAMPs/MAMPs – Pathogen Associated Molecular Patterns/Microbe Associated Molecular Patterns. Conserved microbial features that can be perceived by the plant and elicit defense responses.
PRR – Pattern recognition receptor. Surface localized plant receptors that can recognize specific PAMPs/MAMPs.
PTI – Pattern-triggered immunity. Resistance caused by recognition of pathogen PAMPs/MAMPs, often mediated by PRRs.
R gene – Resistance gene. A plant gene that confers pathogen resistance. R genes can be mediated by NLRs as well as other mechanisms.
QDR – Quantitative disease resistance. Intermediate, or quantitative, disease resistance mediated by one or more loci.
S loci – Susceptibility loci. Loci that enhance pathogen susceptibility. Mutation of S loci results in enhanced disease resistance.
RenSeq - Resistance gene enrichment sequencing. An NLR targeted gene enrichment and sequencing approach.
Genome editing – A type of genetic modification where DNA is modified (deleted, inserted, or replaced) using engineering nucleases.
Acknowledgments
G.C. and M.X. were supported by the National Institutes of Health Grant RO1GM092772. G.C. was supported by the USDA-NIFA Grant 2015-67013-23082 awarded to G.C. M.X. was supported by the NSFC Grant 31672008.
Contributor Information
Meixiang Zhang, Department of Plant Pathology, University of California, Davis, California 95616, USA; Department of Plant Pathology, Nanjing Agricultural University, Nanjing 210095, China.
Gitta Coaker, Department of Plant Pathology, University of California, Davis, California 95616, USA.
LITERATURE CITED
- Alfano JR. Roadmap for future research on plant pathogen effectors. Mol Plant Pathol. 2009;10:805–813. doi: 10.1111/j.1364-3703.2009.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo G, Jupe F, Witek K, Etherington GJ, Ercolano MR, Jones JD. Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol. 2014;14:120. doi: 10.1186/1471-2229-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong MR, Whisson SC, Pritchard L, Bos JIB, Venter E, Anna O, Avrova AO, Rehmany AP, Bohme U, Brooks K, Cherevach I, Hamlin N, White B, Frasers A, Lord A, Quail MA, Churcher C, Hall N, Berriman M, Huang S, Kamoun S, Beynon JL, Birch PRJ. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci U S A. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011;7:e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Cohn M, Kassen A, McCallum EJ, Shybut M, Petriello A, Krasileva K, Dahlbeck D, Medina C, Alicai T, Kumar L, Moreira LM, Neto JR, Verdier V, Santana MA, Kositcharoenkul N, Vanderschuren H, Gruissem W, Bernal A, Staskawicz BJ. High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proc Natl Acad Sci U S A. 2012;109:E1972–E1979. doi: 10.1073/pnas.1208003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Boevink PC, Wang XD, McLellan H, He Q, Naqvi S, Armstrong MR, Zhang W, Hein I, Gilroy EM, Tian ZD, Birch PRJ. A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat Commun. 2016;7:10311. doi: 10.1038/ncomms10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JIB, Kanneganti TD, Young C, Cakir C, Huitema E, Win J, Armstrong MR, Birch PRJ, Kamoun S. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 2006;48:165–176. doi: 10.1111/j.1365-313X.2006.02866.x. [DOI] [PubMed] [Google Scholar]
- Césari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, Shimamoto K, Dodds P, Terauchi R, Kroj T. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014;33:1941–1959. doi: 10.15252/embj.201487923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Césari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, Morel JB, Fournier E, Tharreau D, Terauchi R, Kroj T. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell. 2013;25:1463–1481. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. 2016;17:1140–1153. doi: 10.1111/mpp.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan H, Boni R, Bucher R, Kuhn B, Buchmann G, Sucher J, Selter LL, Hensel G, Kumlehn J, Bigler L, Glauser G, Wicker T, Krattinger SG, Keller B. The wheat resistance gene Lr34 results in the constitutive induction of multiple defense pathways in transgenic barley. Plant J. 2015;84:202–215. doi: 10.1111/tpj.13001. [DOI] [PubMed] [Google Scholar]
- Chiang YH, Coaker G. Effector triggered immunity: NLR immune perception and downstream defense responses. Arabidopsis Book. 2015;13:e0183. [Google Scholar]
- Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- Cox TS, Raupp WJ, Gill BS. Leaf rust-resistance genes Lr41, Lr42, and Lr43 transferred from Triticum tauschii to common wheat. Crop Sci. 1994;34:339–343. [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Rao GJN. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front Plant Sci. 2015;6:698. doi: 10.3389/fpls.2015.00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CIA, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Dyck PL. Genetics of leaf rust reaction in three introductions of common wheat. Can J Plant Sci. 1977;59:329–332. [Google Scholar]
- Ellur RK, Khanna A, Yadav A, Pathania S, Rajashekara H, Singh VK, Krishnan SG, Bhowmick PK, Nagarajan M, Vinod KK, Prakash G, Mondal KK, Singh NK, Prabhu KV, Singh AK. Improvement of Basmati rice varieties for resistance to blast and bacterial blight diseases using marker assisted backcross breeding. Plant Sci. 2016;242:330–341. doi: 10.1016/j.plantsci.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Eves-van den Akker S, Birch PRJ. Opening the effector protein toolbox for plant-parasitic cyst nematode interactions. Mol Plant. 2016;9:1451–1453. doi: 10.1016/j.molp.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO WI. The State of Food Insecurity in the World 2015. Meeting the 2015 international hunger targets: taking stock of uneven progress; FAO, Rome. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- French E, Kim BS, Iyer-Pascuzzi AS. Mechanisms of quantitative disease resistance in plants. Sem Cell Dev Biol. 2016;56:201–208. doi: 10.1016/j.semcdb.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Fry W. Phytophthora infestans: the plant (and R gene) destroyer. Mol Plant Pathol. 2008;9:727–727. doi: 10.1111/j.1364-3703.2007.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka S, Saka N, Mizukami Y, Koga H, Yamanouchi U, Yoshioka Y, Hayashi N, Ebana K, Mizobuchi R, Yano M. Gene pyramiding enhances durable blast disease resistance in rice. Sci Rep. 2015;5:7773. doi: 10.1038/srep07773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulou A, Steele JF, Segretin ME, Bozkurt TO, Zhou J, Robatzek S, Banfield MJ, Pais M, Kamoun S. Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol Plant Microbe Interact. 2015;28:1316–1329. doi: 10.1094/MPMI-07-15-0147-R. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Orto-Alalibo TT, Bozkurt TO, Ah-Fong AMV, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JIB, Bulone V, Cai GH, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, Fischbach MA, Fugelstad J, Gilroy EM, Gnerre S, Green PJ, Grenville-Briggs LJ, Griffith J, Grunwald NJ, Horn K, Horner NR, Hu CH, Huitema E, Jeong DH, Jones AME, Jones JDG, Jones RW, Karlsson EK, Kunjeti SG, Lamour K, Liu ZY, Ma LJ, MacLean D, Chibucos MC, McDonald H, McWalters J, Meijer HJG, Morgan W, Morris PF, Munro CA, O’Neill K, Ospina-Giraldo M, Pinzon A, Pritchard L, Ramsahoye B, Ren QH, Restrepo S, Roy S, Sadanandom A, Savidor A, Schornack S, Schwartz DC, Schumann UD, Schwessinger B, Seyer L, Sharpe T, Silvar C, Song J, Studholme DJ, Sykes S, Thines M, van de Vondervoort PJI, Phuntumart V, Wawra S, Weide R, Win J, Young C, Zhou SG, Fry W, Meyers BC, van West P, Ristaino J, Govers F, Birch PRJ, Whisson SC, Judelson HS, Nusbaum C. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- Hemetsberger C, Mueller AN, Matei A, Herrberger C, Hensel G, Kumlehn J, Mishra B, Sharma R, Thines M, Huckelhoven R, Doehlemann G. The fungal core effector Pep1 is conserved across smuts of dicots and monocots. New Phytol. 2015;206:1116–1126. doi: 10.1111/nph.13304. [DOI] [PubMed] [Google Scholar]
- Holton N, Nekrasov V, Ronald PC, Zipfel C. The phylogenetically-related pattern recognition receptors EFR and XA21 recruit similar immune signaling components in monocots and dicots. PLoS Pathog. 2015;11:e1004602. doi: 10.1371/journal.ppat.1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Angeles ER, Domingo J, Magpantay G, Singh S, Zhang G, Kumaravadivel N, Bennett J, Khush GS. Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor Appl Genet. 1997;95:313–320. [Google Scholar]
- Huard-Chauveau C, Perchepied L, Debieu M, Rivas S, Kroj T, Kars I, Bergelson J, Roux F, Roby D. An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet. 2013;9:e1003766. doi: 10.1371/journal.pgen.1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhang H, Zhang Y, Wang Y, Gao C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat plants. 2015;1:15144. doi: 10.1038/nplants.2015.144. [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Vance RE, Dangl JL. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354 doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- Jupe F, Witek K, Verweij W, Sliwka J, Pritchard L, Etherington GJ, Maclean D, Cock PJ, Leggett RM, Bryan GJ, Cardle L, Hein I, Jones JDG. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 2013;76:530–544. doi: 10.1111/tpj.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatodia S, Bhatotia K, Passricha N, Khurana SM, Tuteja N. The CRISPR/Cas genome-editing tool: application in improvement of crops. Front Plant Sci. 2016;7:506. doi: 10.3389/fpls.2016.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Qi D, Ashfield T, Helm M, Innes RW. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science. 2016;351:684–687. doi: 10.1126/science.aad3436. [DOI] [PubMed] [Google Scholar]
- Kombrink A, Rovenich H, Shi-Kunne X, Rojas-Padilla E, van den Berg GC, Domazakis E, de Jonge R, Valkenburg DJ, Sánchez-Vallet A, Seidl MF, Thomma BP. Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol Plant Pathol. 2017;18:596–608. doi: 10.1111/mpp.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden Hl, Bossolini E, Selter LL, Keller B. A putative ABC transporter conferse durable resistance to multiple fungal pathogens in wheat. Science. 2009;323(5919):1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B, Jones JD, Zipfel C. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol. 2010;28:365–369. doi: 10.1038/nbt.1613. [DOI] [PubMed] [Google Scholar]
- Lee HA, Kim SY, Oh SK, Yeom SI, Kim SB, Kim MS, Kamoun S, Choi D. Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans. New Phytol. 2014;203:926–938. doi: 10.1111/nph.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, Hill J, Baum TJ, Cianzio S, Whitham SA, Korkin D, Mitchum MG, Meksem K. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature. 2012;492:256–260. doi: 10.1038/nature11651. [DOI] [PubMed] [Google Scholar]
- Liu T, Song T, Zhang X, Yuan H, Su L, Li W, Xu J, Liu S, Chen L, Chen T, Zhang M, Gu L, Zhang B, Dou D. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun. 2014;5:4686. doi: 10.1038/ncomms5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Marone D, Russo MA, Laido G, De Leonardis AM, Mastrangelo AM. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci. 2013;14:7302–7326. doi: 10.3390/ijms14047302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke AJ, Matzke MA. Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol. 1998;1:142–8. doi: 10.1016/s1369-5266(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Miedaner T, Korzun V. Marker-assisted selection for disease resistance in wheat and barley breeding. Phytopathology. 2012;102:560–566. doi: 10.1094/PHYTO-05-11-0157. [DOI] [PubMed] [Google Scholar]
- Moore JW, Hererra-Foessel SH, Lan C, Schnippenkoetter W, Ayliffe M, Huerta-Espino J, Lillemo M, Viccars L, Milne R, Periyannan S, Kong X, Spielmeyer W, Talbot M, Bariana H, Patrick JW, Dodds P, Singh R, Lagudah W. A recently evolved hexose transporter variant confers resistnace to multiple pathogens in wheat. Nature Genetics. 2015;47:1494–1498. doi: 10.1038/ng.3439. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501–1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, Pevzner SJ, Donovan SE, Ghamsari L, Santhanam B, Romero V, Poulin MM, Gebreab F, Gutierrez BJ, Tam S, Monachello D, Boxem M, Harbort CJ, McDonald N, Gai L, Chen H, He Y, Vandenhaute J, Roth FP, Hill DE, Ecker JR, Vidal M, Beynon J, Braun P, Dangl JL European Union Effectoromics Consortium. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Kubo Y, Hatakeyama K, Imamura J, Ezura H, Nanasato Y, Tabei Y, Takano Y, Shirasu K, Narusaka Y. Interfamily transfer of dual NB-LRR genes confers resistance to multiple pathogens. PLoS ONE. 2013;8:e55954. doi: 10.1371/journal.pone.0055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela G, Williamson VM, Muñiz M. The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant Microbe Interact. 2003;16:645–649. doi: 10.1094/MPMI.2003.16.7.645. [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell. 1997;9:521–532. doi: 10.1105/tpc.9.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D, de Guillen K, Cesari S, Chalvon V, Gracy J, Padilla A, Kroj T. Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell. 2017;29:156–168. doi: 10.1105/tpc.16.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul JW, III, Qi Y. CRISPR/Cas9 for plant genome editing: accomplishments, problems and prospects. Plant Cell Rep. 2016;35:1417–1427. doi: 10.1007/s00299-016-1985-z. [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJ, Luu DD, Chen H, Bahar O, Daudi A, De Vleesschauwer D, Caddell D, Zhang W, Zhao X, Li X, Heazlewood JL, Ruan D, Majumder D, Chern M, Kalbacher H, Midha S, Patil PB, Sonti RV, Petzold CJ, Liu CC, Brodbelt JS, Felix G, Ronald PC. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv. 2015;1:e1500245. doi: 10.1126/sciadv.1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Dubiella U, Kim SH, Sloss DI, Dowen RH, Dixon JE, Innes RW. Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on S-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol. 2014;164:340–351. doi: 10.1104/pp.113.227686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Leonelli L, Dahlbeck D, Zhao B, Staskawicz BJ. Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc Natl Acad Sci U S A. 2008;105:1091–1096. doi: 10.1073/pnas.0711215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S, Moscou MJ, Ward ER, Horvath DM. Engineering plant disease resistance based on TAL effectors. Annu Rev Phytopathol. 2013;51:383–406. doi: 10.1146/annurev-phyto-082712-102255. [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Bahar O, Thomas N, Holton N, Nekrasov V, Ruan D, Canlas PE, Daudi A, Petzold CJ, Singan VR, Kuo R, Chovatia M, Daum C, Heazlewood JL, Zipfel C, Ronald PC. Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog. 2015;11:e1004809. doi: 10.1371/journal.ppat.1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Smith PG. Embryo culture of a tomato species hybrid. Proc Amer Soc Hort Sci. 1944;44:413–416. [Google Scholar]
- Sohn KH, Lei R, Nemri A, Jones JD. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell. 2007;19:4077–4090. doi: 10.1105/tpc.107.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonah H, Deshmukh RK, Bélanger RR. Computational prediction of effector proteins in fungi: opportunities and challenges. Front Plant Sci. 2016;7:126. doi: 10.3389/fpls.2016.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- Steuernagel B, Periyannan SK, Hernández-Pinzón I, Witek K, Rouse MN, Yu G, Hatta A, Ayliffe M, Bariana H, Jones JD, Lagudah ES, Wulff BB. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol. 2016;34:652–655. doi: 10.1038/nbt.3543. [DOI] [PubMed] [Google Scholar]
- Stork W, Kim JG, Mudgett MB. Functional analysis of plant defense suppression and activation by the Xanthomonas core type III effector XopX. Mol Plant Microbe Interact. 2015;28:180–194. doi: 10.1094/MPMI-09-14-0263-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013;200:808–819. doi: 10.1111/nph.12411. [DOI] [PubMed] [Google Scholar]
- Studholme DJ, Glover RH, Boonham N. Application of high-throughput DNA sequencing in phytopathology. Annu Rev Phytopathol. 2011;49:87–105. doi: 10.1146/annurev-phyto-072910-095408. [DOI] [PubMed] [Google Scholar]
- Sugimoto T, Kato M, Yoshida S, Matsumoto I, Kobayashi T, Kaga A, Hajika M, Yamamoto R, Watanabe K, Aino M, Matoh T, Walker DR, Biggs AR, Ishimoto M. Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breeding Sci. 2012;61:511–522. doi: 10.1270/jsbbs.61.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Goverse A. How to build a pathogen detector: structural basis of NB-LRR function. Curr Opin Plant Biol. 2012;15:375–384. doi: 10.1016/j.pbi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Thakur S, Weir BS, Guttman DS. Phytopathogen genome announcement: draft genome sequences of 62 Pseudomonas syringae type and pathotype strains. Mol Plant Microbe Interact. 2016;29:243–246. doi: 10.1094/MPMI-01-16-0013-TA. [DOI] [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH. Of PAMPs and Effectors: The blurred PTI-ETI dichotomy. Plant Cell. 2011;23:4–15. doi: 10.1105/tpc.110.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S. Agricultural sustainability and intensive production practices. Nature. 2002;418:671–677. doi: 10.1038/nature01014. [DOI] [PubMed] [Google Scholar]
- Tiwari JK, Siddappa S, Singh BP, Kaushik SK, Chakrabarti SK, Bhardwaj V, Chandel P. Molecular markers for late blight resistance breeding of potato: an update. Plant Breeding. 2013;132:237–245. [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G. Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol. 2016;54:419–441. doi: 10.1146/annurev-phyto-080615-100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CM, Dorrance AE, Dou D, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee MK, McDonald WH, Medina M, Meijer HJ, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JK, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BW, Terry A, Torto-Alalibo TA, Win J, Xu Z, Zhang H, Grigoriev IV, Rokhsar DS, Boore JL. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- van Schie CC, Takken FL. Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol. 2014;52:551–581. doi: 10.1146/annurev-phyto-102313-045854. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, Oliver RP. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol Plant Microbe Interact. 2014;27:196–206. doi: 10.1094/MPMI-10-13-0313-IA. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, Raffaele S, Vossen JH, Champouret N, Oliva R, Segretin ME, Rietman H, Cano LM, Lokossou A, Kessel G, Pel MA, Kamoun S. Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol. 2011;49:507–531. doi: 10.1146/annurev-phyto-072910-095326. [DOI] [PubMed] [Google Scholar]
- Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, Fierens-Onstenk J, de Both M, Peleman J, Liharska T, Hontelez J, Zabeau M. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol. 1998;16:1365–1369. doi: 10.1038/4350. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yau YY, Perkins-Balding D, Thomson JG. Recombinase technology: applications and possibilities. Plant Cell Rep. 2011;30:267–285. doi: 10.1007/s00299-010-0938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PRJ. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- Witek K, Jupe F, Witek AI, Baker D, Clark MD, Jones JD. Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat Biotechnol. 2016;34:656–660. doi: 10.1038/nbt.3540. [DOI] [PubMed] [Google Scholar]
- Yasuda N, Mitsunaga T, Hayashi K, Koizumi S, Fujita Y. Effects of pyramiding quantitative resistance genes pi21, Pi34, and Pi35 on rice leaf blast disease. Plant Dis. 2015;99:904–909. doi: 10.1094/PDIS-02-14-0214-RE. [DOI] [PubMed] [Google Scholar]
- Zhao B, Lin X, Poland J, Trick H, Leach J, Hulbert S. A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci U S A. 2005;102:15383–15388. doi: 10.1073/pnas.0503023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BY, Ardales E, Brasset E, Claflin LE, Leach JE, Hulbert SH. The Rxo1/Rba1 locus of maize controls resistance reactions to pathogenic and non-host bacteria. Theor Appl Genet. 2004;109:71–79. doi: 10.1007/s00122-004-1623-y. [DOI] [PubMed] [Google Scholar]
- Zhu S, Li Y, Vossen JH, Visser RG, Jacobsen E. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 2012;21:89–99. doi: 10.1007/s11248-011-9510-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–60. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]


