Abstract
Asian rust affects the physiology of soybean plants and causes losses in yield. Repeatability coefficients may help breeders to know how many measurements are needed to obtain a suitable reliability for a target trait. Therefore, the objectives of this study were to determine the repeatability coefficients of 14 traits in soybean plants inoculated with Phakopsora pachyrhizi and to establish the minimum number of measurements needed to predict the breeding value with high accuracy. Experiments were performed in a 3x2 factorial arrangement with three treatments and two inoculations in a random block design. Repeatability coefficients, coefficients of determination and number of measurements needed to obtain a certain reliability were estimated using ANOVA, principal component analysis based on the covariance matrix and the correlation matrix, structural analysis and mixed model. It was observed that the principal component analysis based on the covariance matrix out-performed other methods for almost all traits. Significant differences were observed for all traits except internal CO2 concentration for the treatment effects. For the measurement effects, all traits were significantly different. In addition, significant differences were found for all Treatment x Measurement interaction traits except coumestrol, chitinase and chlorophyll content. Six measurements were suitable to obtain a coefficient of determination higher than 0.7 for all traits based on principal component analysis. The information obtained from this research will help breeders and physiologists determine exactly how many measurements are needed to evaluate each trait in soybean plants infected by P. pachyrhizi with a desirable reliability.
Introduction
Soybean (Glycine Max (L.) Merrill) is one of the most important cereal crops worldwide. Originally from East Asia, it is currently cultivated around the world for a variety of uses in animal and human food and for industrial applications such as oil and biofuel production [1]. Moreover, soybean is one of the most important commodities for exporter countries, and it is one of the most important food staples of importer countries. World production reached 312 million tons in the 2015/2016 season harvested from 119 million hectares [2]. Brazil is the second worldwide soybean producer behind the United States, with a production in the 2015/2016 season reaching 95 million tons harvested from 33 million hectares [3].
Although the worldwide soybean production is high, the yield is not suitable (2.62 tons ha-1). One of the most important reasons for the lack of yield is disease. Currently, more than 100 pathogens infect soybean plants. A disease that stands out, mainly in South America, is Asian soybean rust (SBR). This disease is caused by the fungal pathogen Phakopsora pachyrhizi. In Brazil, this disease has caused more than R$40 billion in crop losses since it was first detected in 2001 [4, 5]. Finding natural products that work in disease control has a number of economic and environmental benefits. The global threat to crop productivity represented by gap between agricultural production and food demand is steadily increasing year by year [6, 7]. Three strategies have been used to control SBR: applying chemical fungicides, employing specific cultivation practices, and most recently breeding or engineering of SBR-resistant soybean cultivars [8–10].
Several compounds may influence P. pachyrhizi infection. Isoflavonoids such as daidzein, genistein, and glyceollin efficiently reduce P. pachyrhizi uredospore germination in vitro [11]. Chung and Singh [12] described that SBR resistance in a Glycine tomentella accession was correlated with the presence of a flavonoid that also inhibited P. pachyrhizi spore germination. The high potential of phytoalexins in defeating SBR is further supported by medicarpin accumulating in P. pachyrhizi infected Medicago truncatula, a non-host of P. pachyrhizi [13]. SBR also reduces soybean photosynthetic function in susceptible soybean plants. Since resistant lines have reduced fungal growth, the resistance genes may protect these plants against injury to leaf photosynthesis [14]. Therefore, research evaluating phenolic compounds, proteins and compounds involved in photosynthesis is needed to understand the mechanism of P. pachyrhizi infection in soybean, and will consequently help breeders select resistant genotypes.
An important analysis is to identify how many measurements should be done to get a suitable accuracy to obtain useful results. This analysis is performed by using a repeatability coefficient [15]. The repeatability coefficient measures the ability of organisms to repeat character expression over a long period. This allows the verification of genotype behavior during the breeding cycle for different traits. High values of repeatability coefficients predict that the breeding value can be based on a few measurements [16].
The repeatability coefficient can be influenced by the genetic architecture of the trait and the environmental conditions under which the experiment is performed [15]; these factors make the study of repeatability coefficients important to estimate how many measurements should be done for each trait to guarantee a high accuracy. Several authors have investigated the changes related to enzymatic activities and photosynthetic compounds in soybean plants infected by Phakopsora pachyrhizi [17–20]. Although the biochemical and physiological changes caused by P. pachyrhizi in soybean plants are well documented, these studies have measured traits such as enzymatic activity, phenolic compound content and gas exchange at different hours after inoculation, that is, they use different measurements for these traits. To date, there are no known referenced studies that investigate how many measurements are needed to accurately assess enzymatic activities and photosynthetic compounds in soybean. Therefore, the objectives of this study were: (i) to determine the repeatability coefficients of the 14 traits evaluated in soybean plants inoculated with P. pachyrhizi; and (ii) to establish, based on the coefficient of repeatability and determination, the minimum number of measurements needed to predict the breeding value for each trait.
Materials and methods
Experimental design
The experiments were performed in the greenhouse of EMBRAPA Soybean, Londrina, State of Parana, Brazil. Climate conditions during the experiment are displayed in S1 Fig. The genotype BRS 361 was used in this study as a susceptible control to Asian Rust Soybean (SBR). Seeds were obtained from the germplasm bank in the EMBRAPA Soybean. Plants were maintained in the greenhouse through the V5 stage when different chemical products were applied using a CO2 pressurized sprayer.
Experiments were designed in a 3x2 factorial arrangement, with three treatments (water, Tween 20 and methyl jasmonate), and two inoculations (plants inoculated or non-inoculated). Each plant was evaluated four times (48, 96, 144, and 192 hours after inoculation—HAI). Experiments were performed in a randomized block design with six replications, and each plot was composed of one pot with five plants.
Chemical products used
The chemical products used were: water as control treatment; a 0.02% surfactant (Tween 20—polyoxyethylene (20) sorbitan monolaurate; and 1.25 mM methyl jasmonate plus Tween 20 (Methyl jasmonate + tween 20). Methyl jasmonate is known as a regulator volatile [21], therefore plants treated with methyl jasmonate were maintained in a separate greenhouse for 24 hours using the same temperature and humidity to avoid affecting other treatments.
Plant inoculation with P. pachyrhizi
Twenty-four hours after the application of the chemical products, soybean plants were inoculated. P. pachyrhizi inoculum was primarily maintained in soybean cultivar BRS 316 in the greenhouse of EMBRAPA soybean. Spores were collected with light beats on leaves with urediniospores onto white paper and then packed in microtubes. A suspension was then made with distilled water and 0.01% Tween 20 and adjusted to a final concentration of 1.4x105 urediniospores/mL. After the inoculation, plants were maintained on nebulization to stimulate the infection, with the temperature ranging from 23°C to 25°C, humidity greater than 95% in the dark for 12 hours. Non-inoculated plants were treated with water + Tween 20 and maintained in the same conditions as inoculated plants.
Evaluation of the SBR severity
The SBR severity was evaluated 16 days after inoculation in soybean plants in the V2 stage using the scale proposed by Godoy, Koga [22]. This scale is based on the percentage of infected leaf area, and ranged from 0.6% to 78.5% (S2 Fig).
Enzymatic activity evaluation
Leaves from plants inoculated and non-inoculated with P. Pachyrhizi in the V4 and V5 stages were collected to determine enzymatic activities. Samples were composed of two leaves, and samples were collected at four times: 48, 96, 144, and 192 hours after inoculation (HAI). Samples were packed individually in aluminum foil packages, immediately frozen in liquid nitrogen and then stored in the ultrafreezer (-80°C). Chitinase (CHI) and β-1,3-glucanase (GLU) activities were determined. The total protein concentration for each sample was calculated following the Bradford method Bradford [23].
Foliar tissue was crushed in a pillowcase using liquid nitrogen plus 2% polyvinylpolypyrrolidone. The leaf extract was obtained by maceration of 0.3 g foliar tissue. The leaf powder was homogenized using 2 mL of 50 mM sodium phosphate buffer, pH = 6.5 and 1 mM phenylmethylsulfonyl fluoride. This mixture was centrifuged for 25 min at 20,000 X g at a temperature of 4°C. The supernatant was used to determine the enzymatic activity.
Chitinase activity was determined according to the method proposed by Roberts and Selitrennikoff [24] and modified by Harman, Hayes [25], in which p-nitrophenyl-β-D-N,N'-diacetylchitobiose (PNP) was used as a substrate. The incubation system consisted of the addition of a 20 μL aliquot of the supernatant obtained as described above, with 470 μL of 50 mM sodium acetate buffer, pH 5.0, and 10 μL of 2 mg mL-1 PNP. This mixture was stored at 37°C for 2 hours. The reaction was stopped by adding 0.5 mL of 0.2 M sodium carbonate and the absorbances were determined in the wave-length of 410 nm. A molar extinction coefficient of 7 x 104 mM-1.cm-1 was used to calculate the chitinase activity expressed in mM of the p-nitrophenyl produced per min-1 mg-1 protein.
β-1,3-glucanase activity was determined according to the method described by Lever [26], with a slight modification: 3,5-dinitrosalicylic acid substituted P-hydroxybenzoic acid [27]. Reaction was composed of 230 μL of 100 mM sodium acetate buffer, pH 5.0, 250 μL of 4 mg mL-1 laminarin substrate solution and 20 μL of foliar extract. This mixture was incubated for 30 min at 45°C, followed by adding 1 mL of 3,5-dinitrosalicylic acid, and heated to 100°C for 5 min. In the next step, the mixture was cooled to 30°C and the absorbance was determined by a wavelength of 540 nm. The results were expressed in units of absorbance.min-1.mg-1 per protein.
Phenolic compound analysis
Samples for phenolic compound analysis were composed of V5 soybean leaves collected 48, 96, 144 and 192 HAI. Inoculated and non-inoculated plants were collected in the same period. Samples were stored individually in the aluminum foil packages in liquid nitrogen in the freezer (-20°C) through the extraction and analysis. Phenolic compounds were analyzed according to a standard methodology proposed by the laboratory of chemistry ecology of EMBRAPA Soybean. Foliar tissue was crushed in a pillowcase using liquid nitrogen and stored in a falcon tube. Five hundred mg of leaves were then weighed and mixed with 5 mL of 90% methanol and conditioned in an ultrasonic bath for 20 min. Samples were centrifuged for 12 min at 9,880 G at a temperature of 4°C, dried under vacuum, solubilized using 1.5 mL of 80% methanol and homogenized manually. After that, leaf extract was filtered using a Millipore® 0.45 μm membrane in a high performance liquid chromatography (HPLC), Shimadzu, Prominence model.
The methanol extracts from the samples were analyzed using a C18 column (250 mm long and 4.6 mm internal diameter, 5 μM particles). Aliquots of 10 μL were injected automatically; the instrument was equipped with a CBM-20ª controller, SPD-20A detector, DGU 20A5 degasser, LC-20AT pump, SIL-20A automatic sampler and CTO 20A furnace. The mobile phase was composed of two solvents: (A) 2% acetic acid (HOAc) and (B) A mixture of methanol, acetic acid and MilliQ® water (MeOH:HOAc:H2O; 18:1:1). The linear gradient system used in the analysis began with 75% of solvent A and 25% of solvent B, and by 40 min, the solvents were inverted using 25% of solvent A and 75% of solvent B for 5 min. After 45 min, it returned to the initial condition where it remained for 5 min before the next injection. The solvent flow was 1 mL min-1 and ultraviolet (UV) analysis was obtained at wavelengths of 260 and 280 nm.
Concentration of isoflavone aglycones (Genistein–GEI in μg.g-1) in the form 7-O-glycosides (daidzein–DZI in μg.g-1, and genistin–GI in μg.g-1), glycosidic malonyl (malonyl daidzin–MD in μg.g-1, and malonyl genistin–MG in μg.g-1), flavonoid (quercetin-3-O-rutinoside–R in μg.g-1) and phytoalexin coumestrol (C in μg.g-1) were identified based on spectra comparison between standards spectra, sample spectra, and retention time.
For each compound, the mean peak area was measured and multiplied by the correction factor based on each standard compound, which was the initial tissue weight and volume of solubilization. Different standard concentrations of isoflavones and phenolic acids (6,25; 12,5; 25; 50; 100 μg.mL-1) were tested in the HPLC to obtain the correction factor for each compound, which led to a dispersion graphic that obtained a linear regression equation and the correction factor.
Gas exchange and chlorophyll pigment evaluation
Gas exchange and chlorophyll pigment evaluations were performed in the central V4 stage leaflets, previously at 39, 87, 135, and 183 HAI, between 9 am and 10 am. The leaf gas exchange parameters net CO2 assimilation rate (A), stomatal conductance to water vapor (GS), internal CO2 concentration (Ci) and transpiration rate (E) were measured by using a portable open-flow gas exchange system (LCpro-SD; ADA from Laboratory of Ecophysiology Vegetal in the EMBRAPA Soybean). At those periods, A was at its maximum under artificial photosynthetically active radiation (i.e., 1044 μmol photons m-2.s-1 at the leaf level).
Chlorophyll pigment contents were estimated indirectly by the SPAD index, calculated using a chlorophyll meter SPAD-502 and the following equation. The equation was generated based on calibration and validation tests performed by EMBRAPA soybean.
In which: CLO is the chlorophyll contents expressed in mg.cm-2.
Statistical analysis
Analysis of variance was performed for each trait according to the following statistical model:
In which: Yij is the phenotypic value in the jtth replication, evaluated in the ith treatment; μ is the overall mean; Bj is the fixed effect of the jth replication; Ti is the random effect of the ith treatment; Mk is the random effect of the kth measurement; TxMik is the interaction between the ith treatment with the kth measurement; and eij is the random error associated with the phenotypic value Yij.
After that, five methods were used to estimate the repeatability coefficient: analysis of variance, principal component analysis based on the correlation matrix, principal component analysis based on the covariance matrix, structural analysis based on the correlation matrix, and mixed model.
Analysis of variance was calculated based on the following model:
In which: yij is the phenotypic value for the ith treatment in the jth environment; μ é the general mean; Ti is the random effect of the ith treatment influenced by permanent environment (i = 1, 2,…,p); Aj is the random effect of the temporal environment in the jth measurement; TAij is the interaction between the ith treatment influenced by permanent environment and the temporal environment in the jth measurement; and εij is the residual vector.
Heritability (h2) for each trait in each measurement was estimated by:
In which: is the genetic variance and is the residual variance.
The repeatability coefficient (r) by analysis of variance was estimated by:
The repeatability coefficient based on principal component analysis associated with correlation matrix was estimated by:
In which: λ1 is the highest eigenvalue of the correlation matrix (R) associated with eigenvector in which all elements have the same direction and similar magnitude, and η is the number of measurements.
λ1 is estimated by:
in which: ρ is the correlation between two measurements based on the correlation matrix (R).
In which: the R dimension is η x η.
The repeatability coefficient based on principal component analysis associated with covariance matrix is estimated by:
In which: is the phenotypic variance.
In this case, λ1 is the highest eigenvalue of the covariance matrix (Γ) associated with eigenvector which all elements have the same direction and similar magnitude.
and λ1 is estimated by:
The repeatability coefficient based on structural analysis associated with the correlation matrix is estimated by:
In which rjj′ is the phenotypic correlation between treatments during the measurements.
Mixed model equations were used according to Resende [28] using the software Selegen [29]
Estimation of the number of measurements needed to obtain a certain coefficient of determination, the following equation was used:
Analyses were performed using software R [30] and GENES [31].
Results
Analysis of variance and estimation of genetic and environmental parameters
The analysis of variance was performed to verify significant differences among treatments for 14 traits evaluated in soybean plants inoculated or non-inoculated with Phakopsora pachyrhizi during four consecutive measurements (48, 96, 144, 192 hours after inoculation—HAI). Significant differences were observed for all traits except Ci for the treatment effect (Table 1). The measurement effects of all traits were significantly different. In addition, significant differences were found for all traits for the interaction Treatment x Measurement, except C, CHI and CLO.
Table 1. Analysis of variance for the 14 traits evaluated in the soybean plants inoculated or non-inoculated with Phakopsora pachyrhizi during four consecutive measurements (48, 96, 144, 192 hours after inoculation).
| Trait | MSTrat | MSMea | MSTratxMea | MSRes |
|---|---|---|---|---|
| DZI | 6175.67** | 849.71** | 2762.43** | 110.78 |
| GI | 2312.22** | 2590.58** | 826.39** | 224.19 |
| MD | 110494.54** | 16210.89** | 41313.28** | 607.88 |
| R | 2077.38** | 742.43** | 284.12* | 137.87 |
| MG | 49942.03** | 27327.93** | 20220.51** | 551.27 |
| GEI | 1.39** | 3.56** | 1.34** | 0.12 |
| C | 18.98** | 3.03** | 3.30ns | 0.50 |
| GLU | 33997.61** | 5503.93** | 776.32* | 380.52 |
| CHI | 754.57** | 349.31** | 6.66ns | 6.44 |
| CLO | 4x10-5* | 2.1x10-4** | 2x10-5ns | 2x10-5 |
| Ci | 6574.69ns | 94792.24** | 6555.60* | 3620.35 |
| E | 1.26** | 3.19** | 0.46** | 0.11 |
| GS | 0.46** | 0.10* | 0.13** | 0.03 |
| A | 67.07** | 247.01** | 3.79* | 3.49 |
MSTrat–Mean Squares for treatment; MSMea–Mean Squares for measurement; MSTratxMea–Mean squares for the interaction TreatmentxMeasurement; DZI—Daidzein; GI—Gensitin; MD–Malonyl Daidzin; R—Quercetin-3-O-Rutinoside; MG–Malonyl Genistin; GEI–Genistein; C–Coumestrol; GLU– β-1,3-glucanase; CHI—Chitinase; CLO–Chlorophyll; Ci—internal CO2 concentration; E—transpiration rate; GS—stomatal conductance to water vapor; and A—CO2 assimilation rate
ns: not significance
*: significant at 5% probability
**: significant at 1% probability.
It was observed that the mean decreased with the plant growth for GI, GLU, and CHI depending on the different measurements (Table 2). In addition, the mean for DZI, MD, MG, C, and Ci decreased as well, except for the second measurement when the mean was lower than the third measurement. The mean for all traits decreased when we compared the first and the fourth measurements, except for CLO which was constant during the measurements.
Table 2. Mean, heritability (h2) and coefficient of variation (CV, %) for the 14 traits evaluated in soybean plants inoculated or non-inoculated with Phakopsora pachyrhizi during four consecutive measurements (48, 96, 144, 192 hours after inoculation).
| Trait | Mean | h2 | CV (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 48 | 96 | 144 | 192 | 48 | 96 | 144 | 192 | 48 | 96 | 144 | 192 | |
| DZI | 22.04 | 11.35 | 18.76 | 13.58 | 0.98 | 0.94 | 0.95 | 0.94 | 47.26 | 103.57 | 67.47 | 61.93 |
| GI | 70.06 | 59.05 | 58.44 | 49.34 | 0.93 | 0.82 | 0.71 | 0.63 | 15.56 | 24.38 | 22.13 | 39.19 |
| MD | 97.36 | 49.59 | 86.41 | 66.33 | 0.99 | 0.88 | 0.99 | 0.98 | 30.06 | 47.95 | 28.01 | 37.72 |
| R | 34.68 | 23.78 | 27.36 | 28.18 | 0.88 | 0.53 | 0.95 | 0.58 | 31.34 | 56.27 | 26.46 | 45.05 |
| MG | 125.92 | 77.22 | 84.66 | 61.27 | 0.98 | 0.96 | 0.96 | 0.95 | 22.83 | 25.54 | 28.93 | 35.73 |
| GEI | 0.60 | 1.27 | 0.97 | .049 | 0.86 | 0.73 | 0.96 | 0.80 | 45.80 | 36.59 | 36.61 | 70.11 |
| C | 1.22 | 1.28 | 0.96 | 0.64 | 0.96 | 0.95 | 0.57 | 0.82 | 60.01 | 53.56 | 81.76 | 95.63 |
| GLU | 132.57 | 123.07 | 122.65 | 103.12 | 0.95 | 0.96 | 0.95 | 0.96 | 17.09 | 15.89 | 11.98 | 17.12 |
| CHI | 20.27 | 18.95 | 18.35 | 13.17 | 0.97 | 0.97 | 0.96 | 0.96 | 12.17 | 12.83 | 14.13 | 17.43 |
| CLO | 0.01 | 0.01 | 0.01 | 0.01 | 0.24 | 0.11 | 0.20 | .053 | 30.94 | 10.00 | 35.77 | 32.85 |
| Ci | 419.16 | 296.05 | 376.61 | 374.30 | 0.63 | 0.79 | 0.41 | 0.81 | 3.67 | 6.48 | 31.75 | 3.23 |
| E | 2.21 | 2.25 | 1.69 | 1.74 | 0.49 | 0.97 | 0.47 | 0.99 | 6.17 | 6.88 | 33.75 | 5.15 |
| GS | 0.58 | 0.52 | 0.62 | 0.50 | 0.75 | 0.86 | 0.83 | 0.96 | 19.15 | 27.65 | 44.77 | 15.94 |
| A | 13.77 | 14.90 | 8.97 | 11.53 | 0.11 | 0.84 | 0.80 | 0.93 | 15.67 | 13.05 | 21.65 | 11.56 |
DZI—Daidzein; GI—Gensitin; MD–Malonyl Daidzin; R—Quercetin-3-O-Rutinoside; MG–Malonyl Genistin; GEI–Genistein; C–Coumestrol; GLU– β-1,3-glucanase; CHI—Chitinase; CLO–Chlorophyll; Ci—internal CO2 concentration; E—transpiration rate; GS—stomatal conductance to water vapor; and A—CO2 assimilation rate.
Heritability was high (greater than 0.70) for all traits except CLO in all measurements: R in the second measurement, C in the third measurement, Ci and E in the first and third measurements, and A in the first measurement (Table 2). The highest heritability was observed for GLU and CHI, which were greater than 0.95 for all measurements.
The coefficients of variation were low (CV less than 20%) for GLU and CHI for all measurements, and Ci, E and A for the first, second and fourth measurements (Table 2). In addition, the coefficients of variation were medium (ranged from 21% from 40%) in all measurements for the GI, MG, CLO. However, at least one measurement had a coefficient of variation greater than 41% for other traits.
Estimate of repeatability coefficients and coefficients of determination
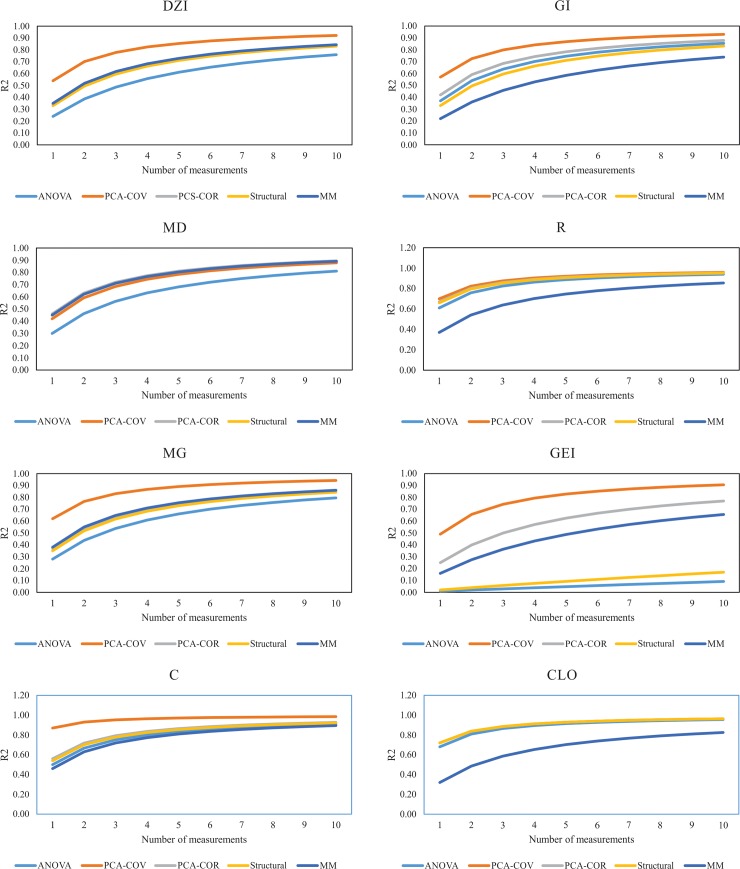
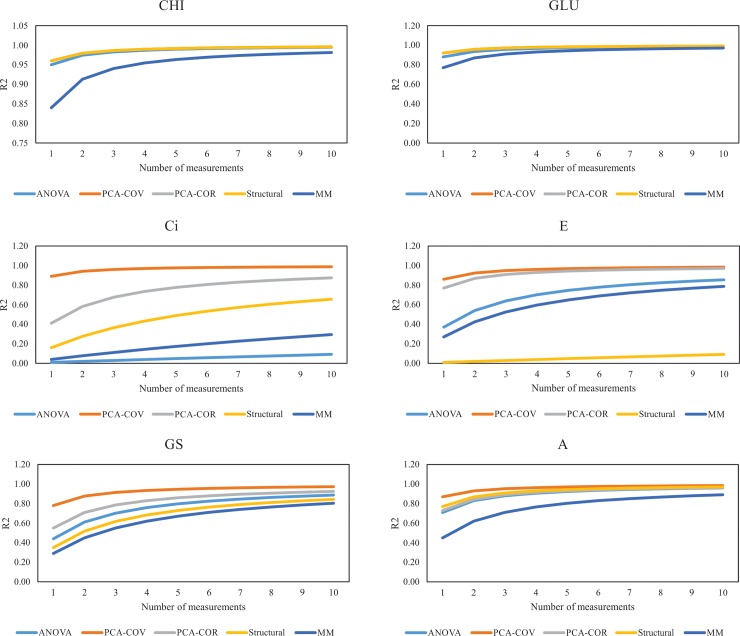
Five methods were used to estimate the repeatability coefficients and coefficients of determination for 14 traits evaluated in soybean plants inoculated or non-inoculated with P. pachyrhizi during four consecutive measurements (48, 96, 144, 192 hours after inoculation). The principal component analysis based on the covariance matrix out-performed other methods for all traits, except MD in which the principal component analysis based on the correlation matrix showed the highest repeatability coefficient (Table 3).
Table 3. Repeatability coefficients (r) and coefficients of determination (R2) obtained by different statistical methods for 14 traits evaluated in soybean plants inoculated or non-inoculated with Phakopsora pachyrhizi during four consecutive measurements (48, 96, 144, 192 hours after inoculation).
| Trait | ANOVA | PCA-COV | PCA-COR | Structural | MM | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| r | R2 | r | R2 | r | R2 | r | R2 | r | R2 | |
| DZI | 0.24 | 56.85 | 0.54 | 82.67 | 0.34 | 68.07 | 0.33 | 66.70 | 0.35 | 0.68 |
| GI | 0.37 | 70.86 | 0.57 | 84.13 | 0.42 | 74.37 | 0.33 | 67.08 | 0.22 | 0.52 |
| MD | 0.30 | 63.66 | 0.42 | 74.57 | 0.46 | 77.73 | 0.45 | 77.17 | 0.45 | 0.76 |
| R | 0.61 | 86.69 | 0.70 | 90.53 | 0.67 | 89.06 | 0.66 | 88.76 | 0.37 | 0.70 |
| MG | 0.28 | 60.87 | 0.62 | 87.05 | 0.37 | 70.76 | 0.35 | 68.81 | 0.38 | 0.71 |
| GEI | 0.01 | 2.75 | 0.49 | 79.54 | 0.25 | 58.18 | 0.02 | 10.90 | 0.16 | 0.43 |
| C | 0.50 | 80.54 | 0.87 | 96.53 | 0.56 | 84.05 | 0.54 | 82.60 | 0.46 | 0.77 |
| GLU | 0.88 | 96.91 | 0.92 | 98.05 | 0.92 | 98.12 | 0.92 | 98.10 | 0.77 | 0.93 |
| CHI | 0.95 | 98.91 | 0.96 | 99.05 | 0.96 | 99.07 | 0.96 | 99.07 | 0.84 | 0.95 |
| CLO | 0.68 | 89.72 | 0.72 | 91.26 | 0.72 | 91.30 | 0.72 | 91.26 | 0.32 | 0.65 |
| Ci | 0.01 | 0.69 | 0.89 | 97.25 | 0.41 | 74.24 | 0.16 | 70.90 | 0.04 | 0.14 |
| E | 0.37 | 70.28 | 0.86 | 96.11 | 0.77 | 93.24 | 0.01 | 4.58 | 0.27 | 0.60 |
| GS | 0.44 | 76.02 | 0.78 | 93.65 | 0.55 | 83.22 | 0.35 | 68.78 | 0.29 | 0.62 |
| A | 0.71 | 91.12 | 0.87 | 96.43 | 0.73 | 91.85 | 0.77 | 91.22 | 0.45 | 0.77 |
DZI—Daidzein; GI—Gensitin; MD–Malonyl Daidzin; R—Quercetin-3-O-Rutinoside; MG–Malonyl Genistin; GEI–Genistein; C–Coumestrol; GLU– β-1,3-glucanase; CHI—Chitinase; CLO–Chlorophyll; Ci—internal CO2 concentration; E—transpiration rate; GS—stomatal conductance to water vapor; and A—CO2 assimilation rate.
On the other hand, the repeatability coefficient estimated by mixed model and ANOVA were the lowest compared to principal component analysis and structural analysis. The same shape was observed for the coefficient of determination, which principal component analysis based on the covariance matrix estimated the highest values for almost all traits, followed by principal component analysis based on the correlation matrix and structural analysis (Table 3).
Calculating the number of measurements needed to obtain a reasonable coefficient of determination
An estimate of the optimal number of measurements needed to obtain an acceptable coefficient of determination was calculated from one to ten measurements for each trait and plotted.
The coefficients of determination were different among methods for the same number of measurements (Figs 1 and 2). Four measurements were suitable to obtain a coefficient of determination higher than 0.7 for almost all traits (except MD) based on principal component analysis via covariance matrix. Moreover, six measurements were enough to obtain a coefficient of determination greater than 0.7 based on principal component analysis via correlation matrix for all traits (Figs 1 and 2).
Fig 1. Graphic analysis of the minimum number of measurements required to reach a determined degree of certainty for phenolic compounds and chlorophyll according to different methods: ANOVA–analysis of variance; PCA-COV—principal component analysis associated with covariance matrix; PCA-COR—principal component analysis associated with correlation matrix; structural analysis; MM–mixed model.
DZI—Daidzein; GI—Gensitin; MD–Malonyl Daidzin; R—Quercetin-3-O-Rutinoside; MG–Malonyl Genistin; GEI–Genistein; C–Coumestrol; and CLO–Chlorophyll.
Fig 2. Graphic analysis of the minimum number of measurements required to reach a determined degree of certainty for enzymatic activities and photosynthetic performance according to different methods: ANOVA–analysis of variance; PCA-COV—principal component analysis associated with covariance matrix; PCA-COR—principal component analysis associated with correlation matrix; structural analysis; MM–mixed model.
GLU– β-1,3-glucanase; CHI—Chitinase; Ci—internal CO2 concentration; E—transpiration rate; GS—stomatal conductance to water vapor; and A—CO2 assimilation rate.
Based on ANOVA and mixed models, more than 10 measurements were needed to obtain a coefficient of determination greater than 0.70 for DZI, Gi, GEI, CLO, Ci, E and GS (Figs 1 and 2). GLU and CHI were the traits that needed fewer measurements (just one) to obtain a coefficient of determination greater than 0.70, followed by C and A with four and five measurements respectively.
Discussion
Analysis of variance and estimation of genetic and environmental parameters
There were significant treatment effects for all traits, except Ci (Table 1). This indicates that the inoculation of P. pachyrhizi made changes to the enzymatic activities, phenolic compounds, and gas exchange in soybean plants. Therefore, it is important to understand the biochemical and physiological mechanisms and their relation to the Soybeanx ASR interaction because it will help breeders to select resistant genotypes or to find methodologies that will help the farmers to manage the pathogen more efficiently.
Findings in this study agree with previous research which proved that methyl jasmonate can efficiently reduce disease severity in many crops such as sweet cherry fruit [32], Alternaria brassicicola, Botrytis cinerea and Plectosphaerella cucumerina in Arabidopsis thaliana [33], Monilinia fructicola in peach [34], Erysiphe necator in grapes [35] and Colletotrichum acutatum in plum [36]. Moreover, methyl jasmonate can affect disease severity in two ways: directly via fungal growth suppression as well as spore germination suppression, which were reported for Alternaria alternata [37], and indirectly via fungal growth suppression as observed in A. brassicicola on Arabidopsis thaliana [38].
The significant effects of the measurement factors for all traits indicated that the concentration of those substances changed over time. These results were expected because the fungal latency period likely increased several physiological changes due to the high spore production that consequently caused changes in the values of the traits (Table 2). These results reinforce the need to study how many measurements are suitable to select superior genotypes in ASR-tolerant soybean, high yield, and other important agronomic traits. In addition, the evaluation of enzymatic activities, phenolic compounds and gas exchange is necessary to better understand how ASR can affect yield and possibly use these traits to indirectly select ASR-resistant genotypes.
The estimation of heritability allows breeders to know the fraction of phenotypic variation among treatments is explained by genetic factors [15]. The estimation of this parameter presented high or moderate magnitude for all traits, except CLO (Table 2). These results indicated that the CLO evaluation can be significantly influenced by environmental conditions.
The measurement 48 hours after inoculation performed the smallest CV estimate (Table 2). However, evaluating biochemical and physiological traits in advanced infected stages can contribute most to the discrimination of resistant genotypes for P. pachyrhizi, although the experimental precision decreased during the measurements. Repeatability analysis can be used to fix the problem of experimental precision since repeatability coefficients and coefficients of determination estimated by several methodologies can indicate how many measurements are needed to obtain a suitable reliability for each trait [16].
Estimate of repeatability coefficients and coefficients of determination
Principal component analysis based on the covariance matrix estimated the highest repeatability coefficient for all traits, except MD; this resulted in fewer measurements needed to identify superior soybean genotypes for traits related with ASR infection. These results are similar to the results observed by Matsuo, Sediyama [39] that evaluated the repeatability of oidium (Erysiphe diffusa) severity in several soybean genotypes. In addition, several studies revealed the efficiency of the principal component analysis based on the covariance matrix to estimate the repeatability coefficients for agronomic traits in perennial species [40–43] and annual species [44–46]. This type of analysis can help breeders, physiologists and phytopathologists to find experimental strategies to evaluate traits with accurate results.
The significance of the measurement effect indicates that many changes occurred in the physiological and biochemical mechanisms of the soybean plants during the evaluation [15]. This effect varied among treatments, and ANOVA did not allow the separate analysis of this effect; therefore, it was consequently mixed with the residual. This fact can lead to the underestimation of the repeatability and consequently the overestimation of the number of measurements needed. Therefore, principal component analysis that considers cyclic trait behavior is most recommended for the estimation of the repeatability coefficient. This is because the eigenvector estimated by principal component analysis expresses the progeny trend and their capacity to maintain their relative position during the evaluation period [47].
This type of analysis might be very important to understand how the physiological changes occur in soybean plants affected by ASR over a period of time. For instance, according to Cruz, Rodrigues [48] chitinase and β‑1,3‑glucanase are important to decrease the P. pachyrhizi colonization in soybean leaf tissue. These enzymes are involved in plant defense against the pathogen by the chitin and β-1,3-glucan hydrolytic action present on the fungal cell wall, and consequently the oligosaccharide liberation that may act as elicitors of the plant defense mechanism against the pathogen [49]. The overexpression of genes affecting chitinase and β‑1,3‑glucanase in plants have increased the resistance to several plant pathogens [50].
Number of measurements needed to obtain a reasonable coefficient of determination
Sustainable food production to feed a growing world population is a major challenge for plant scientists, especially given the unpredictable and dynamic nature of global climatic conditions. These changes affect the dynamics of diseases in crops such as soybeans. Therefore, the development of effective strategies to assess the severity of these diseases will be essential for their control in a sustainable way [6, 7].
A coefficient of determination greater than 0.70 is desirable in plant breeding because it improves the selection accuracy, i.e., breeders can select superior genotypes more accurately. Based on this parameter, the method of principal component analysis based on the covariance matrix indicated that six measurements are needed for all traits. These results are similar to results reported by Matsuo, Sediyama [39], which concluded that four measurements are needed to obtain a suitable prediction, i.e., a prediction with a confidence greater than 70% for the severity in the foliates infected by oidium.
Based on the results revealed in this study, breeders, physiologists and phytopathologists will be able to evaluate phenolic compounds, enzymatic activities and gas exchange more accurately to select superior soybean genotypes for resistance to P. pachyrhizi. Additional research should be done evaluating these traits in several genotypes and the selection of resistant genotypes based on these traits.
Conclusion
The number of measurements affect the reliability in traits related to enzymatic activity, phenolic compound content and gas exchange in soybean plants infected by Asian rust soybean. Six measurements are suitable to obtain a coefficient of determination higher than 0.7 for all traits based on principal component analysis.
Supporting information
The days in which gas exchange was evaluated were highlighted.
(TIF)
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by CAPES (Coordenacao de Aperfeicoamento de Pessoal do Ensino Superior), Cnpq (National Counsel of Technological and Scientific Development), FAPEMIG (Fundacao de Amparo a Pesquisa de Minas Gerais), Funarbe (Fundacao Arthur Bernardes) to TB, and Federal University of Vicosa for financial support. We also thank the Biometric Lab (Federal University of Vicosa, Brazil) where all analyses were performed by remote access. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Milfont MdO, Rocha EEM, Lima AON, Freitas BM. Higher soybean production using honeybee and wild pollinators, a sustainable alternative to pesticides and autopollination. Environmental chemistry letters. 2013;11(4):335–41. [Google Scholar]
- 2.USDA. World Agricultural Supply and Demand Estimates. United States Department of Agriculture, 2017. [Google Scholar]
- 3.CONAB. Companhia Nacional de Abasteciento—safra 2016/2017 2017 [cited 2017 Junho 06th]. Available from: http://www.conab.gov.br/.
- 4.Silva ACd, Souza PEd, Amaral DC, Zeviani WM, Pinto JEBP. Essential oils from Hyptis marrubioides, Aloysia gratissima and Cordia verbenacea reduce the progress of Asian soybean rust. Acta Scientiarum Agronomy. 2014;36(2):159–66. [Google Scholar]
- 5.Yorinori J, Paiva W, Frederick R, Costamilan L, Bertagnolli P, Hartman G, et al. Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Disease. 2005;89(6):675–7. [DOI] [PubMed] [Google Scholar]
- 6.Abatepaulo ARR, Caetano AR, Mendes CT Jr, Carvalho WA, Ferreira BR, de Miranda Santos IKF. Detection of SNPs in bovine immune‐response genes that may mediate resistance to the cattle tick Rhipicephalus (Boophilus) microplus. Animal genetics. 2008;39(3):328–9. doi: 10.1111/j.1365-2052.2008.01713.x [DOI] [PubMed] [Google Scholar]
- 7.Abdelrahman M, Jogaiah S, Burritt DJ, Tran LSP. Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant, cell & environment. 2018. [DOI] [PubMed] [Google Scholar]
- 8.Hartman GL, Miles MR, Frederick RD. Breeding for resistance to soybean rust. Plant disease. 2005;89(6):664–6. [DOI] [PubMed] [Google Scholar]
- 9.Kendrick MD, Harris DK, Ha B-K, Hyten DL, Cregan PB, Frederick RD, et al. Identification of a second Asian soybean rust resistance gene in Hyuuga soybean. Phytopathology. 2011;101(5):535–43. doi: 10.1094/PHYTO-09-10-0257 [DOI] [PubMed] [Google Scholar]
- 10.Godoy CV, Bueno AdF, Gazziero DLP. Brazilian Soybean Pest Management and Threats to its Sustainability. Outlooks on Pest Management. 2015;26(3):113–7. [Google Scholar]
- 11.Lygin AV, Li S, Vittal R, Widholm JM, Hartman GL, Lozovaya VV. The importance of phenolic metabolism to limit the growth of Phakopsora pachyrhizi. Phytopathology. 2009;99(12):1412–20. doi: 10.1094/PHYTO-99-12-1412 [DOI] [PubMed] [Google Scholar]
- 12.Chung G, Singh RJ. Broadening the genetic base of soybean: a multidisciplinary approach. Critical Reviews in Plant Sciences. 2008;27(5):295–341. [Google Scholar]
- 13.Ishiga Y, Upplapapti SR, Mysore KS. Expression analysis reveals a role for hydrophobic or epicuticular wax signals in pre-penetration structure formation of Phakopsora pachyrhizi. Plant signaling & behavior. 2013;8(11):e26959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumudini S, Godoy C, Kennedy B, Prior E, Omielan J, Boerma H, et al. Role of host-plant resistance and disease development stage on leaf photosynthetic competence of soybean rust infected leaves. Crop science. 2010;50(6):2533–42. [Google Scholar]
- 15.Cruz CD, Regazzi AJ, Carneiro PCS. Modelos biométricos aplicados ao melhoramento genético. 5, editor. Viçosa: UFV; 2012. 480 p. [Google Scholar]
- 16.Laviola BG, e Oliveira AMC, Bhering LL, Alves AA, Rocha RB, Gomes BEL, et al. Estimates of repeatability coefficients and selection gains in Jatropha indicate that higher cumulative genetic gains can be obtained by relaxing the degree of certainty in predicting the best families. Industrial Crops and Products. 2013;51:70–6. [Google Scholar]
- 17.Pereira MR, Gouvêa BC, Marcelino-Guimarães FC, Ramos HJdO, Moreira MA, Barros EGd. Proteomic analysis of soybean leaves in a compatible and an incompatible interaction with Phakopsora pachyrhizi. Organelles Proteomics. 2014;1(1):16–27. [Google Scholar]
- 18.Choi JJ, Alkharouf NW, Schneider KT, Matthews BF, Frederick RD. Expression patterns in soybean resistant to Phakopsora pachyrhizi reveal the importance of peroxidases and lipoxygenases. Functional & integrative genomics. 2008;8(4):341–59. [DOI] [PubMed] [Google Scholar]
- 19.Tremblay A, Hosseini P, Alkharouf NW, Li S, Matthews BF. Transcriptome analysis of a compatible response by Glycine max to Phakopsora pachyrhizi infection. Plant Science. 2010;179(3):183–93. [Google Scholar]
- 20.Srivastava P, George S, Marois JJ, Wright DL, Walker DR. Saccharin-induced systemic acquired resistance against rust (Phakopsora pachyrhizi) infection in soybean: Effects on growth and development. Crop protection. 2011;30(6):726–32. [Google Scholar]
- 21.Seo HS, Song JT, Cheong J-J, Lee Y-H, Lee Y-W, Hwang I, et al. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proceedings of the National Academy of Sciences. 2001;98(8):4788–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godoy CV, Koga LJ, Canteri MG. Diagrammatic scale for assessment of soybean rust severity. Fitopatologia Brasileira. 2006;31(1):63–8. [Google Scholar]
- 23.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72(1–2):248–54. [DOI] [PubMed] [Google Scholar]
- 24.Roberts WK, Selitrennikoff CP. Plant and bacterial chitinases differ in antifungal activity. Microbiology. 1988;134(1):169–76. [Google Scholar]
- 25.Harman GE, Hayes CK, Lorito M, Broadway RM, Di Pietro A, Peterbauer C, et al. Chitinolytic enzymes of Trichoderma harzianum: purification of chitobiosidase and endochitinase. Phytopathology. 1993;83(3):313–8. [Google Scholar]
- 26.Lever M. A new reaction for colorimetric determination of carbohydrates. Analytical biochemistry. 1972;47(1):273–9. [DOI] [PubMed] [Google Scholar]
- 27.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical chemistry. 1959;31(3):426–8. [Google Scholar]
- 28.Resende MDV. Genética biométrica e estatística no melhoramento de plantas perenes Brasília; 2002. [Google Scholar]
- 29.Resende MDVd. Software Selegen-REML/BLUP: a useful tool for plant breeding. Crop Breeding and Applied Biotechnology. 2016;16(4):330–9. [Google Scholar]
- 30.Team RC. R: A language and environment for statistical computing R Foundation for Statistical Computing; Vienna: Austria; 2014. [Google Scholar]
- 31.Cruz CD. GENES—a software package for analysis in experimental statistics and quantitative genetics. Acta Scientiarum Agronomy. 2013;35(3):271–6. doi: 10.4025/actasciagron.v35i3.21251 [Google Scholar]
- 32.Yao H, Tian S. Effects of pre-and post-harvest application of salicylic acid or methyl jasmonate on inducing disease resistance of sweet cherry fruit in storage. Postharvest Biology and Technology. 2005;35(3):253–62. [Google Scholar]
- 33.Thomma BP, Eggermont K, Broekaert WF, Cammue BP. Disease development of several fungi on Arabidopsis can be reduced by treatment with methyl jasmonate. Plant Physiology and Biochemistry. 2000;38(5):421–7. [Google Scholar]
- 34.Yao H, Tian S. Effects of a biocontrol agent and methyl jasmonate on postharvest diseases of peach fruit and the possible mechanisms involved. Journal of Applied Microbiology. 2005;98(4):941–50. doi: 10.1111/j.1365-2672.2004.02531.x [DOI] [PubMed] [Google Scholar]
- 35.Belhadj A, Saigne C, Telef N, Cluzet S, Bouscaut J, Corio-Costet M-F, et al. Methyl jasmonate induces defense responses in grapevine and triggers protection against Erysiphe necator. Journal of agricultural and food chemistry. 2006;54(24):9119–25. doi: 10.1021/jf0618022 [DOI] [PubMed] [Google Scholar]
- 36.Cao S, Zheng Y, Yang Z, Tang S, Jin P, Wang K, et al. Effect of methyl jasmonate on the inhibition of Colletotrichum acutatum infection in loquat fruit and the possible mechanisms. Postharvest Biology and Technology. 2008;49(2):301–7. [Google Scholar]
- 37.Chen J, Zou X, Liu Q, Wang F, Feng W, Wan N. Combination effect of chitosan and methyl jasmonate on controlling Alternaria alternata and enhancing activity of cherry tomato fruit defense mechanisms. Crop Protection. 2014;56:31–6. [Google Scholar]
- 38.Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, et al. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences. 1998;95(25):15107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo É, Sediyama T, Barros HB, Cruz CD, de Cássia Teixeira R, Boldt AF, et al. Análise de repetibilidade da ocorrência de oídio em genótipos de soja. Bioscience Journal. 2009;25(2). [Google Scholar]
- 40.da Silva J CA, Eduardo To F, Borges do C, Lempp B, Eduardo Te P, dos Santos A. Minimum Number of Measurements for Accurate Evaluation of Qualitative Traits in Urochloa brizantha. Journal of Agronomy. 2015;14(3):180–4. doi: 10.3923/ja.2015.180.184 [Google Scholar]
- 41.da Silva Negreiros JR, Neto RdCA, Miqueloni DP, Lessa LS. Estimativa de repetibilidade para caracteres de qualidade de frutos de laranjeira‑doce. Pesquisa Agropecuária Brasileira. 2014;49(1):40–8. [Google Scholar]
- 42.Danner MA, Raseira MdCB, Sasso SAZ, Citadin I, Scariot S. Repetibilidade de caracteres de fruto em araçazeiro e pitangueira. Ciência Rural. 2010;40(10):2086–91. [Google Scholar]
- 43.Teodoro PE, Costa RD, Rocha RB, Laviola BG. Número mínimo de medições para a avaliação acurada de características agronômicas de pinhão-manso. Pesquisa Agropecuária Brasileira. 2016;51(2):112–9. doi: 10.1590/s0100-204x2016000200003 [Google Scholar]
- 44.Carvalho LP, Farias FJ, Morello CL, Rodrigues JI, Teodoro PE. Number of repetitions for evaluating technological traits in cotton genotypes. Genetics and molecular research: GMR. 2016;15(3). doi: 10.4238/gmr.15038557 . [DOI] [PubMed] [Google Scholar]
- 45.Torres FE, Sagrilo E, Teodoro PE, Ribeiro LP, Cargnelutti Filho A. Número de repetições para avaliação de caracteres em genótipos de feijão-caupi. Bragantia. 2015;74(2):161–8. [Google Scholar]
- 46.Ribeiro L, Teodoro P, Correa C, da Luz Junior R, Torres F. Number of replications required to differentiate maize hybrids. Revista Brasileira de Milho e Sorgo. 2015;14(3):392–9. [Google Scholar]
- 47.Abeywardena V. An application of principal component analysis in genetics. Journal of Genetics. 1972;61(1):27–51. doi: 10.1007/bf02984099 [Google Scholar]
- 48.Cruz MFAd, Rodrigues FÁ, Polanco LR, Curvêlo CRdS, Nascimento KJT, Moreira MA, et al. Inducers of resistance and silicon on the activity of defense enzymes in the soybean-Phakopsora pachyrhizi interaction. Bragantia. 2013;72(2):162–72. [Google Scholar]
- 49.Mohammadi M, Roohparvar R, Torabi M. Induced chitinase activity in resistant wheat leaves inoculated with an incompatible race of Puccinia striiformis f. sp. tritici, the causal agent of yellow rust disease. Mycopathologia. 2002;154(3):119–26. [DOI] [PubMed] [Google Scholar]
- 50.Van Loon L. Induced resistance in plants and the role of pathogenesis-related proteins. European Journal of Plant Pathology. 1997;103(9):753–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The days in which gas exchange was evaluated were highlighted.
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.