Abstract
Boar taint is an offensive odour and/or taste from a proportion of non-castrated male pigs caused by skatole and androstenone accumulation during sexual maturity. Castration is widely used to avoid boar taint but is currently under debate because of animal welfare concerns. This study aimed to identify expression quantitative trait loci (eQTLs) with potential effects on boar taint compounds to improve breeding possibilities for reduced boar taint. Danish Landrace male boars with low, medium and high genetic merit for skatole and human nose score (HNS) were slaughtered at ~100 kg. Gene expression profiles were obtained by RNA-Seq, and genotype data were obtained by an Illumina 60K Porcine SNP chip. Following quality control and filtering, 10,545 and 12,731 genes from liver and testis were included in the eQTL analysis, together with 20,827 SNP variants. A total of 205 and 109 single-tissue eQTLs associated with 102 and 58 unique genes were identified in liver and testis, respectively. By employing a multivariate Bayesian hierarchical model, 26 eQTLs were identified as significant multi-tissue eQTLs. The highest densities of eQTLs were found on pig chromosomes SSC12, SSC1, SSC13, SSC9 and SSC14. Functional characterisation of eQTLs revealed functions within regulation of androgen and the intracellular steroid hormone receptor signalling pathway and of xenobiotic metabolism by cytochrome P450 system and cellular response to oestradiol. A QTL enrichment test revealed 89 QTL traits curated by the Animal Genome PigQTL database to be significantly overlapped by the genomic coordinates of cis-acting eQTLs. Finally, a subset of 35 cis-acting eQTLs overlapped with known boar taint QTL traits. These eQTLs could be useful in the development of a DNA test for boar taint but careful monitoring of other overlapping QTL traits should be performed to avoid any negative consequences of selection.
Introduction
Boar taint is an offensive odour and/or taste observed in cooked meat from a proportion of non-castrated male pigs undergoing sexual maturity and is primarily related to two compounds: skatole (3-methylindole) from the hindgut and androstenone (5α-androst-16-ene-3-one) from the testis [1]. The compounds accumulate in adipose tissue and are correlated with sexual maturity at the time of slaughter [2, 3] and/or the amount of liver degradation [4, 5]. In the liver, a high concentration of androstenone prevents breakdown of skatole by inhibiting enzymes responsible for skatole metabolism [6]. Furthermore, androstenone itself causes boar taint, and previous research has shown that the compound is primary in the hierarchy of boar taint development [7]. To avoid boar taint, castration is usually performed at an early age, but this strategy is under debate because of animal welfare concerns [8]. As an alternative to castration, gene-based selection [9] through large-scale breeding programmes using predictive biomarkers for reduced boar taint in non-castrated male pigs has been proposed [10–15]. Skatole and androstenone are moderate to highly heritable traits [16–18] depending on the breed [4], and previous work has established low and mostly favourable genetic correlations between boar taint compounds and production traits such as meat quality [19]. Furthermore, selection could be performed from birth in both sexes, not only after sexual maturity [20], which could accelerate genetic gain [21]. Hence, gene-based selection should be possible without negatively affecting important male fertility traits [19] or sexual maturation [6].
To accelerate genetic control of boar taint and other economically important traits, vast numbers of quantitative trait loci (QTLs) have been identified [4]. Currently, the Pig Quantitative Trait Locus (QTL) Database (PigQTLdb) [22] has information on 25,473 QTLs from 593 publications that represent 646 different traits (as of December 2017). Importantly, QTLs may provide information on genes that could affect phenotype (positional candidate gene approach) [23]. Most of the published data derive from studies involving experimental crosses using pig breeds exhibiting extremes for the phenotypes of interest [24]. Previous studies on QTLs for levels of skatole, androstenone and indole have shown that these traits are highly polygenic [10, 11, 21, 25], and efforts to increase precision and accuracy have led to complementation of QTL studies by genome-wide association studies (GWAS) through high-density single-nucleotide polymorphism (SNP) panels. These studies are usually performed with Illumina Porcine SNP60 Genotyping BeadChips, which allow for the identification of significant association with the phenotype of interest. In contrast to traditional QTL mapping, GWAS provides unlinked individual genes or SNPs, which are easy to separate for analysis, but the caveat may be a large number of false positives [26]. Previous research involving one thousand Danish Landrace boars with divergent levels of skatole and androstenone identified a number of associated SNPs for skatole on pig chromosome (SSC) 14 and for androstenone on SSC5 [15]. A number of other breeds have been subjected to GWAS for SNPs associated with androstenone and/or skatole levels, including Pietrain [27], Duroc and Norwegian Landrace [12].
In recent years, extensive efforts have been made to incorporate next-generation sequencing into the elucidation of the genetic determinants of economically important traits. These new technologies, such as RNA sequencing (RNA-Seq), provide an unprecedented level of accuracy and precision for measurements of gene expression profiles (transcriptome) [28]. To accommodate new technologies into the framework of genomics and breeding development, the concept of “systems genomics” was introduced by Kadarmideen, von Rohr [29]. The concept enables the identification of potential biologically associated genes and variants for traits or diseases by studying genetic variation in the context of gene expression profiles and/or other high-throughput molecular measurements (methylome, proteome and metabolome) [30].
In the current study, systems genomics was applied by identifying expression quantitative trait loci (eQTLs) [31, 32] in liver and testis. The tissues were selected based on their biological roles within the boar taint condition: the liver is responsible for the breakdown of both skatole and androstenone, and the testis is the primary synthesis site for androstenone and other odour-causing androgens [4]. The analysis of eQTLs enables identification of cis-acting and trans-acting elements, e.g., SNPs located within gene promoters and/or distant transcription factors [33]. From the identified eQTLs, manageable lists of positional candidate SNPs and genes can be created for further investigation as potential biomarkers [34]. The strategy has been applied for traits such as hypertension [35] and obesity [33, 36] in humans and meat quality traits in pigs [37, 38]. A disadvantage of eQTL analysis is the computation time, which can amount to weeks or months because of the enormous data calculations required. Recently, an ultra-fast software package that employs large matrix operations was described for rapid eQTL identification [39]. Furthermore, the authors published a multi-tissue eQTL identification package that utilises a Bayesian hierarchical model to find eQTLs that are shared between tissues [40] and thus are highly applicable as candidate biomarkers for a given trait.
The aims of this study were to i) identify and analyse single- and multi-tissue eQTLs significantly associated with estimated breeding values (EBVs) of skatole and human nose scores (indicators of boar taint) in Danish Landrace pigs, ii) functionally characterise the SNPs and genes of eQTLs and iii) find candidate eQTLs associated with low-EBV genotypes and evaluate any possible selection effects by comparison of genomic loci with those of important production traits.
Materials and methods
A complete illustration of the study design is available in S1 File..
Animals and data
The animals in this study were obtained from a previously published study by our group [41]. Briefly, commercial Danish Landrace male pigs (n = 114) were produced from sires with known genetic merit of boar taint, assessed by estimated breeding values (EBVs) of skatole concentrations and human nose scores (HNS). The EBVs of the sires were obtained from the Danish pig breeding database (Pig Research Centre, SEGES, Copenhagen, Denmark). The sire EBVs were corrected for age and weight by a previously published methodology [19] to account for variances in sexual maturation of the boars and to obtain full steroidogenic potential. The pigs were produced and housed at the testing station “Bøgildgård” in Denmark (Mallingvej 1, 8620 Kjellerup, Denmark, CVR license 12739664) operated by the Pig Research Center, SEGES (Copenhagen, Denmark) with ad libitum feed and water supply. The authors of this study were not responsible for animal husbandry, diet and care as the testing station is a commercial facility. Furthermore, Animal Care and Use Committee accreditation was not obtained for this study because the animals were commercial slaughter pigs and tissue samples were obtained from the commercial slaughter facility “Danish Crown Pork A/S” (Danmarksgade 22, 7400 Herning, Denmark, CVR license 26121264).
Before slaughter, the pigs were grouped into one of three groups: high, medium and low genetic merit of boar taint. The grouping was performed according to the summarised EBVs of skatole and HNS obtained from the sire of each pig. The low and high groups comprised the lowest and highest sire summarised EBVs, whereas the medium group comprised pigs with sire summarised EBVs closest to the overall mean. Finally, a total of 48 pigs were selected for analysis, with 16 pigs in each of the three groups. From the selected pigs, the mean summarised EBVs were 0.71 (± 0.19), -0.01 (± 0.09) and -0.38 (± 0.17) for high, medium and low genetic merit of boar taint groups, respectively. Full information on the animals and their corresponding summarised EBVs of the sires are accessible as supplementary information in the previously published study by our group [41].
Tissue collection and preparation
Pigs were slaughtered at a weight of ~100 kg at a commercial slaughterhouse (Danish Crown, Herning, Denmark, CVR 26121264). Slaughter was performed by submersion into CO2 until unconsciousness ensued followed by exsanguination. Following slaughter, liver and ham muscle was extracted from the carcass and 150 mg of tissue were retrieved by punch biopsy and immediately immersed into 1.5 ml RNAlater (QIAGEN, Hilden, Germany) in 2 ml Eppendorf tubes (Eppendorf, Hamburg, Germany). The carcasses were kept in a cold room at 4°C for approximately 1.5 h before 150 mg testis tissue were retrieved by punch biopsy and immersed into 1.5 ml RNAlater (QIAGEN). All samples (n = 96) were stored at -20°C for 14 days until RNA extraction and sequencing at a commercial facility (AROS A/S, Aarhus, Denmark).
RNA extraction and sequencing
RNA extraction, sequencing and gene counting was performed as described in a previous study from our group [41]. Briefly, total RNA was extracted from the 96 samples by RNeasy Mini Kit (QIAGEN) following instructions of the manufacturer. Concentration of RNA was measured by Nanodrop 2000 (ThermoScientific, Massachusetts, USA) and quality was evaluated with a Bioanalyzer (Agilent, California, USA). Sequencing libraries were prepared from 400 ng RNA using either the TruSeq stranded mRNA (Illumina, San Diego, USA) kit following manufacturer’s instructions or the TruSeq total stranded RNA (Illumina) kit following manufacturer’s instructions according to RNA quality. Libraries were subjected to quality control with respect to size profile by test on an Agilent DNA 1000 (Agilent, California, USA) and library concentration by KAPA quantitative PCR (qPCR) kit and three independent 106-fold dilutions of libraries following instructions of the manufacturer (Kapa Biosystems, Massachusetts, USA). The samples were sequenced with an Illumina HiSeq 2500 (Illumina, San Diego, USA) which amounted to a theoretical 40 million reads per sample and demultiplexed into FastQ-files by CASAVA-software (Illumina, San Diego, USA).
Quality control and gene counting
Quality control (QC) of RNA-Seq reads was conducted with FastQC (v. 0.11.3) [42]. Reads were trimmed for known Illumina TruSeq adapter sequences using the software CutAdapt (v. 1.8.1) [43]. Poor reads were trimmed by the software Trimmomatic (v. 0.33) [44] using default parameters. The trimmed reads were then mapped to the Sus scrofa reference genome (10.2, version 79) by the STAR aligner (v. 020201) [45] using default parameters. Post-mapping QC was performed with Qualimap [46]. The mapped reads were counted to each gene by HTSeq count (v. 0.6.0) [47] using default parameters. All subsequent statistical analysis were performed in R (v. 3.1.0) [48]. Only genes with a mean count of more than five were included in the gene count matrices. Normalisation of gene counts was performed by voom variance-stabilization function with sample quality weights [49] implemented in the R package limma (v. 3.30.3) [50].
Genotyping and filtering
Genomic DNA was extracted from ham muscle by DNA Minikit (QIAGEN) following manufactures instructions and genotyped using the Illumina PorcineSNP60 Beadchip (San Diego, CA, USA) [51]. Variants were called using Illumina GenomeStudio (v. 2011.1) using the Genotyping module (v. 1.9.4). To export the data into a PLINK-readable format, the PLINK Input Report plugin (v. 2.1.3) by Illumina, Inc. was used. For filtering of genotype data, PLINK [52] (v. 1.90b3.44) was used. The variants were included in the genotype data when they achieved a call rate above 0.95, a minor allele frequency (MAF) of above 0.05 and were in Hardy-Weinberg equilibrium (P > 0.0001). To remove variants which were in linkage disequilibrium (LD), LD-based variant pruning was performed with a window size and step size of 5 Kb and an r2 threshold of 0.8. Finally, the variants were converted to their genomic coordinates and Reference SNP cluster ID (rs) accession numbers which was obtained from SNPchiMp v. 3 database [53].
Analysis of single- and multi-tissue eQTLs
Identification of single- and multi-tissue eQTLs
Identification of cis- and trans-acting single-tissue eQTLs was performed by the software Matrix eQTL (v. 2.1.1) [39]. The maximum distance at which a gene-SNP pair was considered a local cis-acting eQTL was defined as 1 Mb which was consistent with threshold used in previous research on eQTLs for obesity in pigs [33]. P-value thresholds were 1 × 10−3 for cis-acting eQTLs and 1 × 10−6 for trans-acting eQTLs. In accordance with guidelines for Matrix eQTL, different thresholds for each eQTL type was applied [39]. This was due to a limitation in the Matrix eQTL software as it was not possible to apply FDR thresholds. Hence, diverging P value thresholds were set to avoid excessive large output files as only a small amount of the identified eQTLs passed the FDR threshold (FDR < 0.05). The grouping of the pigs as low, medium or high boar taint by their summarised EBVs of skatole and HNS was loaded into the software as covariates in the model.
Identification of multiple tissue (multi-tissue) eQTLs which comprised eQTLs identified in both liver and testis was performed using the Matrix eQTL extension named Empirical Bayesian Hierarchical Model [40]. Subsequently, all eQTLs were ordered according to their Benjamini-Hochberg adjusted P-values (FDR). An association analysis was performed on both single- and multi-tissue eQTLs to test for association with: i) estimated breeding values (EBVs) for skatole and human nose score (HNS) of the animals and ii) the expression profile of the gene associated with the eQTL. This was performed by comparing the summarised EBVs and expression profiles from the animals that were naturally grouped by one of the three available genotypes (AA / AB / BB) in each individual eQTL by a linear regression model. To account for any batch or slaughter line-derived biological effects, tissue yield (mg), quality control data from the genotyping machine and the RNA integrity number (RIN) of each sample were included as interacting covariates in the model. Subsequently, test statistics and P-values were computed with ANOVA tests by the function anova() in the R package stats (v. 3.3.1) [48]. Finally, the P values were subjected to stringent multiple testing corrections by the FDR procedure. The eQTLs with significant (FDR < 0.05) association to summarised EBVs and gene expression were defined as the “filtered eQTLs”. The filtered cis-acting eQTLs from both single- and multi-tissue were used for functional characterisation and evaluation as candidate eQTLs.
Chromosomal statistics of eQTLs
To provide descriptive statistics on the eQTLs, chromosomal locations together with lengths of the Sus scrofa chromosomes, were obtained by the R package biomaRt (v. 2.30.0) [54, 55] from the Sus scrofa genome build (10.2, version 79). The chromosomal locations of the SNPs associated with the eQTLs were obtained from SNPchiMp (v. 3) database [53]. The chromosomal densities of eQTLs were calculated by the number of either unfiltered or filtered eQTLs per chromosome divided by length in bases of the respective chromosome.
Functional characterisation and pathway analysis
Functional characterisation was performed by the Cytoscape (v. 3.2.1) [56] plug-in ClueGO (v. 2.2.5) [57]. By providing the genes associated with the filtered cis-acting eQTLs from liver and testis, ClueGO performed an overrepresentation test which provided enriched functions and pathways in the gene subset. Furthermore, the tool visualised significantly (FDR < 0.05) enriched gene ontology (GO) terms of each gene. Finally, ClueGO visualised the expression profiles of “low” and “high” boar taint groups by a scale of red to green. Red colour represented upregulation and green colour represented downregulation, as compared to the mean expression values of all three groups. Results from enrichment test were subjected to multiple testing corrections of the P values by Benjamini-Hochberg (FDR) correction.
Correlation analysis between multi-tissue eQTLs
Correlation analysis was performed to reveal co-expression patterns of genes associated with multi-tissue eQTLs. The analysis was performed in ClueGO. Correlations were calculated Spearman rank correlation between expression levels of all multi-tissue eQTLs from both liver and testis. The correlations were visualised as a network plot in Cytoscape. Each node represented a gene associated with multi-tissue eQTLs and edges represented the results from the correlation analyses. Edges from correlations calculated from both liver and testis expression data were included and coloured identically. However, positive correlations were coloured red whereas negative correlations were coloured green, consistent with the colours used by other network co-expression analyses tools, e.g. Weighted Gene Co-expression Network Analysis (WGCNA) [58]. All positive and negative correlations with R2 ≥ 0.5 were visualised.
Genomic location analysis and QTL trait enrichment test
To find and evaluate possible selection effects, genomic coordinates from known traits represented by their quantitative trait loci (QTLs) were obtained from the Animal Genome PigQTLdb database where all previous research on QTLs are curated. The known QTL traits from the database were tested for enrichment by the number of genomic overlaps of the filtered cis-acting eQTLs from both liver and testis. This was done using the R package binom.test.QTL [59]. Briefly, the package tests each trait in a global QTL dataframe obtained from the PigQTLdb [22] against SNPs associated with the eQTLs. In this study, a total of 646 unique porcine traits were obtained from the PigQTLdb and used in the enrichment test, which were represented by a total of 25,473 QTLs and their corresponding genomic positions (updated December 2017). The enrichment test was performed by obtaining the number of expected overlaps between the filtered multi-tissue eQTLs and the known traits and comparing the expected overlaps by a binominal test with the full length of the porcine genome as the background set. The expected overlaps were calculated as the number of intersections between the genomic locations of the SNPs associated with the filtered multi-tissue eQTLs and the summarised length of the QTLs which were associated with each database-derived trait. The background set was defined as the full length (2.7 Gb) of the porcine genome. Finally, the P-values were subjected to multiple testing corrections by the Benjamini-Hochberg (FDR) procedure. In order to obtain overlaps from the most relevant breeds and filter out esoteric and local breeds, only QTLs represented by breeds with a minimum of a thousand QTLs were included in the analysis. After filtering, a total of 577 traits represented by 19,417 QTLs were included in the final analysis. The most represented breeds were Duroc × Erhulian, Duroc × Pietrain and Duroc × Yorkshire/Landrace. The package also provided the category of each of the enriched trait. To adjust for any bias due to higher research interests in particular production traits, the sum of enriched traits from each category were divided with the sum of all traits from the corresponding category in the current Animal Genome PigQTLdb (release 32).
Selection of candidate eQTLs
In order to select the most interesting subset of eQTLs with potential usefulness in the development of candidate biomarkers for gene-based selection schemes towards lowered boar taint, a list of candidate eQTLs was generated from the filtered cis-acting eQTLs that fulfilled the two criteria: i) associated with genotype(s) with a median of summarised EBVs equal to or lower than the maximum summarised EBVs of the low boar taint pig group (low group EBVmax = -0.070) and ii) significantly (FDR < 0.05) overlapping a known boar taint relevant QTL trait by its genomic coordinates. Finally, the candidate eQTLs were visualised plotted with expression levels from liver and testis and its candidate genotype associated with low summarised EBVs for skatole and HNS.
Results
Quality control and filtering
RNA-Seq data
All RNA-Seq data was obtained from a previous study on differentially expressed genes (DEG) associated with the summarised estimated breeding values (EBVs) of skatole and human nose scores (HNS) [41]. Briefly, post-mapping quality control revealed a mean of 33.25 million reads successfully mapped to the Sus scrofa Ensembl reference genome (10.2, version 79). Furthermore, reads aligned to genes (± SD) were 7.76 (± 1.61) million and 7.04 million (± 2.44) in liver and testis, respectively. To remove genes with none or very low expression levels, only genes with mean expression levels of more than five counts were included in the analysis. This filtering approach was consistent with previous eQTL analysis in the porcine genome [33] and consistent with previous warrants to obtain the highest statistical power [60]. The final numbers of genes were 10,545 for liver and 12,731 for testis, respectively.
Genotype data
Genotype data comprised a total of 59,319 SNPs with a total genotyping rate of 0.997751. Due to missing genotype data, 567 variants removed. Furthermore, 68 SNPs were removed due to Hardy-Weinberg exact test filtering and 19,843 SNPs were removed due to minor allele threshold. Linkage disequilibrium (LD) based variant pruning removed 13,186 variants due to high LD (r2 ≥ 0.8) and a final subset of 20,827 SNPs was used for the analysis.
Analysis of single-tissue eQTLs
Identification of single-tissue eQTLs in liver
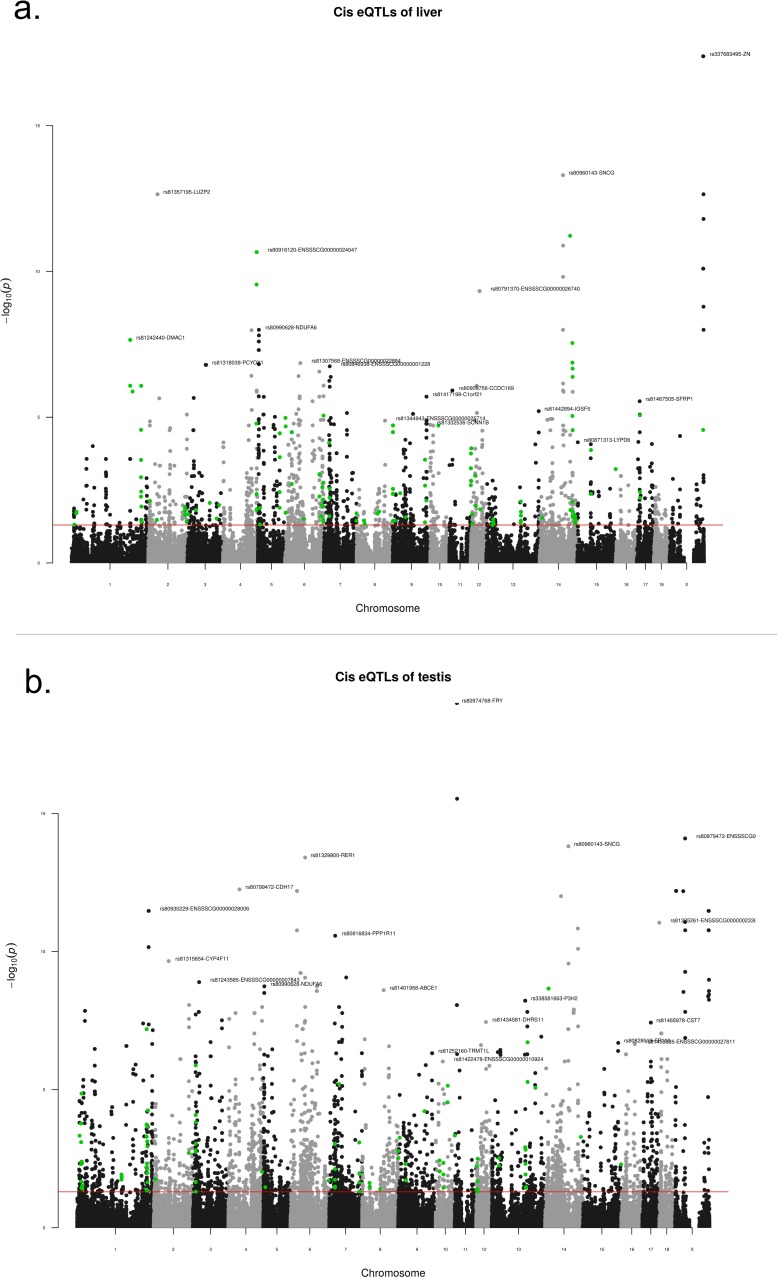
A total of 1,949 cis-acting and 851 trans-acting expression quantitative trait loci (eQTLs) with significant (FDR < 0.05) gene-SNP relationship were identified in liver (Fig 1A). Association analysis revealed 142 cis- and 63 trans-acting eQTLs with significant association (FDR < 0.05) to the summarised EBVs of skatole and HNS. These eQTLs were defined as the “filtered eQTLs” of liver and selected for functional analysis and QTL trait enrichment analysis for candidate eQTLs. The cis- and trans-acting filtered eQTLs were associated with 85 and 17 genes, respectively. The top 30 cis-acting eQTLs were associated with the genes ARMC7, UROS, ZNF662, BAIAP2, FAM198A, ENSSSCG00000024047, TEN1, ENSSSCG00000020732, ENSSSCG00000005474, NEB, HEXDC, ATP13A3, LSG1, BUB3, ACADSB and GSTA4 (Table 1). Full results table are available in S2 File.
Fig 1.
Identification of significant cis-acting eQTLs in porcine (a) liver and (b) testis. Y-axis indicates the negative logarithm of False Discovery Rate for each eQTL, X-axis indicates chromosome number. Only the X chromosome is shown, as no eQTLs were discovered on Y chromosome. Red line indicates a FDR threshold of 0.05 which defined the eQTLs with a significant gene-SNP association. For each chromosome, the eQTLs with lowest FDR values are illustrated with SNP rs number and gene symbol. Green circles indicate “filtered eQTLs” which had a significant (FDR < 0.05) association with sire-obtained summarised estimated breeding values for skatole and human nose scores and were used in the subsequent analyses.
Table 1. Top 30 cis-acting filtered eQTLs in porcine liver.
| SNP | Gene | FDR-eQTL | FDR- EBV | SNP- Chr | Distance (bp) | GO term |
|---|---|---|---|---|---|---|
| rs81265837 | ARMC7 | 0.045 | 0.000 | 12 | -871 | molecular function |
| rs81435284 | ARMC7 | 0.045 | 0.000 | 12 | -92982 | molecular function |
| rs80892090 | UROS | 0.038 | 0.002 | 14 | 702737 | biological process |
| rs80822699 | ZNF662 | 0.044 | 0.002 | 13 | -48202 | organelle |
| rs81444147 | ZNF662 | 0.044 | 0.002 | 13 | -205961 | organelle |
| rs81347708 | ZNF662 | 0.044 | 0.002 | 13 | -411021 | organelle |
| rs81444056 | ZNF662 | 0.047 | 0.002 | 13 | 160293 | organelle |
| rs81305046 | BAIAP2 | 0.000 | 0.002 | 12 | -111914 | cell morphogenesis |
| rs81438833 | BAIAP2 | 0.000 | 0.002 | 12 | -404940 | cell morphogenesis |
| rs81439307 | BAIAP2 | 0.000 | 0.002 | 12 | -552982 | cell morphogenesis |
| rs81440749 | BAIAP2 | 0.000 | 0.002 | 12 | -1009083 | cell morphogenesis |
| rs81268762 | FAM198A | 0.032 | 0.003 | 13 | 979078 | organelle |
| rs80871264 | ENSSSCG00000024047 | 0.004 | 0.006 | 4 | 348015 | |
| rs81433762 | TEN1 | 0.021 | 0.006 | 12 | 238431 | biosynthetic process |
| rs81434107 | TEN1 | 0.021 | 0.006 | 12 | 183314 | biosynthetic process |
| rs80878429 | ENSSSCG00000020732 | 0.004 | 0.007 | 5 | 634366 | |
| rs81444193 | ZNF662 | 0.042 | 0.008 | 13 | -800329 | organelle |
| rs81351832 | ENSSSCG00000005474 | 0.000 | 0.009 | 1 | 214191 | |
| rs80905121 | ZNF662 | 0.040 | 0.011 | 13 | -6078 | organelle |
| rs81456636 | NEB | 0.001 | 0.013 | 15 | 27586 | extracellular region |
| rs81311681 | HEXDC | 0.001 | 0.013 | 12 | -10300 | molecular function |
| rs81434341 | HEXDC | 0.001 | 0.013 | 12 | -315178 | molecular function |
| rs81271924 | HEXDC | 0.001 | 0.013 | 12 | -394460 | molecular function |
| rs81286188 | ATP13A3 | 0.024 | 0.014 | 13 | -753004 | biological process |
| rs81286188 | LSG1 | 0.036 | 0.014 | 13 | -469274 | biological process |
| rs80871231 | BUB3 | 0.032 | 0.015 | 14 | -853556 | molecular function |
| rs80871231 | ACADSB | 0.043 | 0.015 | 14 | -967074 | molecular function |
| rs80799070 | ENSSSCG00000005474 | 0.004 | 0.016 | 1 | 255068 | |
| rs81257157 | ENSSSCG00000020732 | 0.013 | 0.016 | 5 | 768093 | |
| rs80957762 | GSTA4 | 0.020 | 0.016 | 7 | 1429 | molecular function |
SNP = Single nucleotide polymorphism, FDR-eQTL = False Discovery Rate from association test between gene and SNP in the eQTL calculated by Matrix eQTL software, FDR-EBV = False Discovery Rate from association test between eQTL and summarised estimated breeding values (EBVs) of skatole and human nose scores, SNP-Chr = Chromosome of the SNP associated with the eQTL, Distance = Genomic distance between gene-SNP pair in bases. Negative number indicates upstream location of SNP respective to gene, GO Term = Gene Ontology term associated with the eQTL.
Identification of single-tissue eQTLs in testis
A total of 4,442 cis-acting and 1,437 trans-acting eQTLs with significant (FDR < 0.05) gene-SNP relationship were identified in testis (Fig 1B). Association analysis revealed 77 cis- and 32 trans-acting eQTLs with significant association (FDR < 0.05) to the summarised EBVs of skatole and HNS. These eQTLs were defined as the “filtered eQTLs” of testis and selected for functional analysis and QTL trait enrichment analysis for candidate eQTLs. The cis- and trans-acting filtered eQTLs were associated with 47 and 11 genes, respectively. The top 30 cis-acting eQTLs were associated with the genes GTF2I, ENSSSCG00000025739, NPTX1, TEPSIN, CCDC170, SERPINA3, NWD2, SNX30, ESR1, ENSSSCG00000028881 and FBP2 (Table 2). Full results table are available in S2 File.
Table 2. Top 30 cis-acting filtered eQTLs in porcine testis.
| SNP | Gene | FDR-eQTL | FDR- EBV | SNP- Chr | Distance (bp) | GO term |
|---|---|---|---|---|---|---|
| rs81265837 | ARMC7 | 0.003 | 0.000 | 12 | -871 | molecular function |
| rs81435284 | ARMC7 | 0.003 | 0.000 | 12 | -92982 | molecular function |
| rs81286957 | GTF2I | 0.002 | 0.003 | 3 | 432935 | cytoplasm |
| rs81371954 | GTF2I | 0.002 | 0.006 | 3 | 343502 | cytoplasm |
| rs81223031 | GTF2I | 0.050 | 0.006 | 3 | -299269 | cytoplasm |
| rs80892090 | UROS | 0.001 | 0.007 | 14 | 702737 | biological process |
| rs81444403 | ENSSSCG00000025739 | 0.003 | 0.008 | 13 | -387191 | |
| rs81305046 | NPTX1 | 0.006 | 0.008 | 12 | 285025 | cell-cell signalling |
| rs81438833 | NPTX1 | 0.006 | 0.008 | 12 | -8001 | cell-cell signalling |
| rs81439307 | NPTX1 | 0.006 | 0.008 | 12 | -156043 | cell-cell signalling |
| rs81440749 | NPTX1 | 0.006 | 0.008 | 12 | -612144 | cell-cell signalling |
| rs81435983 | ARMC7 | 0.021 | 0.008 | 12 | -348155 | molecular function |
| rs81321757 | ARMC7 | 0.021 | 0.008 | 12 | -491525 | molecular function |
| rs81305046 | TEPSIN | 0.042 | 0.008 | 12 | -222102 | molecular function |
| rs81438833 | TEPSIN | 0.042 | 0.008 | 12 | -515128 | molecular function |
| rs81439307 | TEPSIN | 0.042 | 0.008 | 12 | -663170 | molecular function |
| rs80814798 | CCDC170 | 0.005 | 0.009 | 1 | 896311 | molecular function |
| rs81348764 | CCDC170 | 0.005 | 0.009 | 1 | -25868 | molecular function |
| rs80871264 | ENSSSCG00000024047 | 0.009 | 0.009 | 4 | 348015 | |
| rs81348677 | CCDC170 | 0.021 | 0.009 | 1 | 499103 | molecular function |
| rs81328558 | CCDC170 | 0.021 | 0.009 | 1 | 349781 | molecular function |
| rs80982470 | SERPINA3 | 0.001 | 0.020 | 7 | -228084 | lysosome |
| rs81398873 | NWD2 | 0.024 | 0.020 | 8 | -114091 | |
| rs80993397 | CCDC170 | 0.036 | 0.020 | 1 | -263557 | molecular function |
| rs81351832 | SNX30 | 0.000 | 0.022 | 1 | 61251 | cell |
| rs80836463 | ESR1 | 0.000 | 0.022 | 1 | 686066 | signal transducer activity |
| rs81348579 | ESR1 | 0.000 | 0.022 | 1 | 566976 | signal transducer activity |
| rs81348614 | ESR1 | 0.001 | 0.022 | 1 | 261590 | signal transducer activity |
| rs81420611 | ENSSSCG00000028881 | 0.004 | 0.022 | 10 | -88026 | |
| rs81288871 | FBP2 | 0.005 | 0.022 | 10 | 1026726 | ion binding |
SNP = Single nucleotide polymorphism, FDR-eQTL = False Discovery Rate from association test between gene and SNP in the eQTL calculated by Matrix eQTL software, FDR-EBV = False Discovery Rate from association test between eQTL and summarised estimated breeding values (EBVs) of skatole and human nose scores, SNP-Chr = Chromosome of the SNP associated with the eQTL, Distance = Genomic distance between gene-SNP pair in bases. Negative number indicates upstream location of SNP respective to gene, GO Term = Gene Ontology term associated with the eQTL.
Chromosomal statistics of single-tissue eQTLs
Pig chromosomes have the following densities of all eQTLs, ordered from highest to lowest: SSC6 (8.06), SSC14 (7.89), SSC12 (4.97), SSC4 (4.87), SSC7 (4.08), SSC5 (3.87), SSC3 (3.35), SSC17 (3.34), SSC2 (2.87), SSC9 (2.79), SSC18 (2.52), SSC1 (2.29), SSC13 (2.12), SSC8 (2.05), SSC10 (1.83), SSC16 (1.83), SSC11 (1.37), SSCX (1.24), SSC15 (1.17) and SSCY (0). Pig chromosomes have the following densities of filtered cis- and trans-acting eQTLs, ordered from highest to lowest: SSC12 (0.519), SSC1 (0.415), SSC13 (0.338), SSC9 (0.299), SSC14 (0.279), SSC10 (0.265), SSC2 (0.215), SSC3 (0.173), SSC7 (0.171), SSC6 (0.165), SSC4 (0.146), SSC5 (0.135), SSC17 (0.0717), SSC15 (0.0571), SSC8 (0.0539), SSCX (0.0139), SSC11 (0.0114), SSC18 (0), SSC16 (0) and SSCY (0).
Functional characterisation and pathway analysis of the single-tissue eQTLs
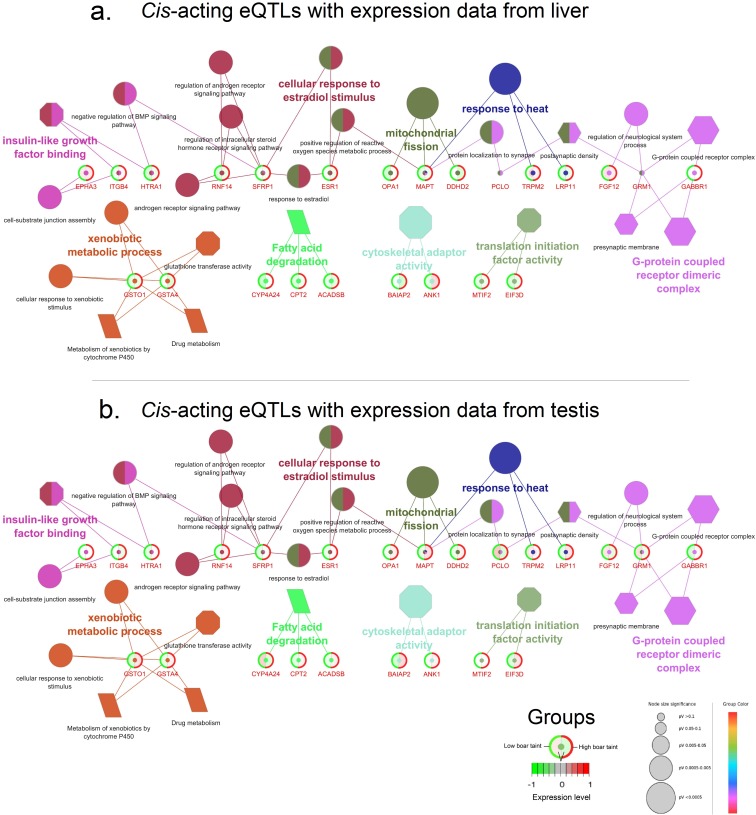
Functional characterisation revealed 31 and three GO terms and KEGG pathways to be significantly (FDR < 0.05) enriched from the filtered single-tissue cis-acting eQTLs, respectively (Fig 2). Of these, biological relevant GO terms sorted according to FDR included “Xenobiotic metabolic process”, “Cellular response to xenobiotic stimulus”, “Cellular response to estradiol stimulus”, “Regulation of androgen receptor signalling pathway”, “Glutathione transferase activity”, “Response to estradiol” “Androgen receptor signalling pathway” and “Regulation of intracellular steroid hormone receptor signaling pathway” (Fig 2). Pathway analysis revealed three KEGG pathways to be significantly (FDR < 0.05) enriched: “Metabolism of xenobiotics by cytochrome P450”, “Drug metabolism” and “Fatty acid degradation”. Full results table are available in S3 File.
Fig 2. Functional characterisation and pathway analysis of the filtered cis-acting eQTLs.
Filtered cis-acting eQTLs with expression data (a) liver and (b) testis were subjected to functional characterisation and pathway analysis. Functional characterisation is represented by gene ontology (GO) terms and pathways are represented by their Kyoto Encyclopedia Genes and Genomes (KEGG) pathways. Each function/pathway is visualised by a uniquely coloured symbol. Each gene is represented by a circle where the colour in the centre indicates the function of the gene from the corresponding GO term. The diameter of the circle is negatively correlated with the adjusted P value (FDR) of the GO term from the functional enrichment test made by ClueGO software. Each node represents a gene where the expression levels are indicated in the circle where the green group (left side) represents the expression level in low boar taint pigs and the red group (right side) represents the expression level in high boar taint pigs. Ellipse symbol: Biological process; hexagon symbol: cellular component; octagon symbol: molecular function; parallelogram: KEGG pathway.
Multi-tissue eQTL analysis
Identification of multi-tissue eQTLs from liver and testis
By a Bayesian Hierarchical Model approach implemented in Matrix eQTL software, eQTLs shared between liver and testis was identified. These were defined as “multi-tissue eQTLs”. A total of cis-acting multi-tissue eQTLs were identified as significant (FDR < 0.05) by their gene-SNP relationship. Association analysis revealed a total of 26 eQTLs as significantly associated (FDR < 0.05) with summarised EBVs for skatole and HNS. These were defined as the “filtered multi-tissue eQTLs” and were associated with the genes ARMC7, UROS, ENSSSCG00000024047, NEB, ENSSSCG00000029075, TMEM41B, SYTL3, ZNF185, DMAC1, ENSSSCG00000025800, KLHDC4 and CES1 (Table 3). Full results table are available in S4 File.
Table 3. Filtered multi-tissue eQTLs from liver and testis.
| SNP | Gene | FDR-expression | FDR- EBV |
SNP Chr | Distance (bp) | GO term |
|---|---|---|---|---|---|---|
| rs81265837 | ARMC7 | 0.005 | 0.000 | 12 | -871 | molecular function |
| rs81435284 | ARMC7 | 0.005 | 0.000 | 12 | -92982 | molecular function |
| rs80892090 | UROS | 0.038 | 0.001 | 14 | 702737 | biological process |
| rs80871264 | ENSSSCG00000024047 | 0.000 | 0.003 | 4 | 348015 | |
| rs81456636 | NEB | 0.000 | 0.008 | 15 | 27586 | extracellular region |
| rs80947338 | ENSSSCG00000029075 | 0.000 | 0.021 | 14 | -673630 | |
| rs81411161 | TMEM41B | 0.000 | 0.021 | 9 | -299322 | biological process |
| rs81313669 | TMEM41B | 0.000 | 0.021 | 9 | -435492 | biological process |
| rs339471111 | TMEM41B | 0.000 | 0.021 | 9 | -681330 | biological process |
| rs81270995 | TMEM41B | 0.000 | 0.021 | 9 | 231435 | biological process |
| rs81411123 | TMEM41B | 0.000 | 0.021 | 9 | -51275 | biological process |
| rs81477176 | TMEM41B | 0.000 | 0.021 | 9 | -721167 | biological process |
| rs80932898 | SYTL3 | 0.035 | 0.021 | 1 | 512807 | molecular function |
| rs81479582 | ZNF185 | 0.000 | 0.022 | X | 965559 | molecular function |
| rs80916120 | ENSSSCG00000024047 | 0.000 | 0.022 | 4 | 382514 | |
| rs80919384 | ENSSSCG00000024047 | 0.000 | 0.022 | 4 | 128566 | |
| rs337275357 | ENSSSCG00000024047 | 0.000 | 0.022 | 4 | -140288 | |
| rs81242440 | DMAC1 | 0.000 | 0.022 | 1 | 503055 | cytoplasm |
| rs80922101 | DMAC1 | 0.000 | 0.022 | 1 | 311763 | cytoplasm |
| rs80830287 | ENSSSCG00000025800 | 0.001 | 0.022 | 14 | -440700 | |
| rs80948915 | ENSSSCG00000024047 | 0.000 | 0.022 | 4 | 550515 | |
| rs80906401 | ENSSSCG00000025800 | 0.000 | 0.023 | 14 | 201338 | |
| rs81323702 | KLHDC4 | 0.000 | 0.026 | 6 | 89447 | molecular function |
| rs81380844 | ENSSSCG00000024047 | 0.000 | 0.041 | 4 | 982688 | |
| rs81320759 | KLHDC4 | 0.000 | 0.041 | 6 | -812563 | molecular function |
| rs81395038 | CES1 | 0.000 | 0.041 | 6 | -150074 | intracellular |
SNP = Single nucleotide polymorphism, FDR-expression = False Discovery Rate from association test between eQTL and expression profile of the associated gene, FDR-EBV = False Discovery Rate from association test between eQTL and summarised estimated breeding values (EBVs) of skatole and human nose scores, SNP Chr = Chromosome of the SNP associated with the eQTL, Distance = Genomic distance between gene and SNP pair in the eQTL in bases. Negative number indicates upstream location of SNP respective to gene, GO Term = Gene Ontology of the gene associated with the eQTL.
Correlation analysis between expression levels of multi-tissue eQTLs
To identify multi-tissue eQTLs that were co-expressed with other multi-tissue eQTLs, a correlation analysis by Spearman rank correlation analysis was performed on all multi-tissue eQTLs identified with significant gene-SNP relationship which included the filtered multi-tissue eQTLs. The filtered multi-tissue eQTLs were indicated with red letters in the illustration. Correlation analysis revealed multi-tissue eQTLs associated with the genes PPP1R11, NEU1, SMARCC2, UROS and CIAPIN1 to contain most correlations to and from other multi-tissue eQTLs (threshold: R2 ≥ 0.5) (Fig 3).
Fig 3. Correlations analysis between expression levels of multi-tissue eQTLs.
To identify co-expression patterns between multi-tissue eQTLs, correlations of expression levels of genes associated with multi-tissue eQTLs was analysed by Spearman rank correlation analysis. Correlations were calculated from expression data from both liver and testis. Edges indicating correlation are shown for both liver and testis expression data. Green circles indicate filtered multi-tissue eQTLs with significant association to summarised EBVs for skatole and HNS. The two types of correlations are discerned by both colour and shape. Positive correlations between expression levels are indicated with dashed red line. Negative correlations between expression levels are indicated by a dashed green line. Boldness of lines indicates the degree of correlation. Only edges representing Spearman rank correlations above R2 > 0.5 or below R2 < -0.5 are shown.
Genomic location analysis and QTL trait enrichment test
Enrichment test for selection effects revealed 89 QTL traits from Animal Genome PigQTLdb to be significantly (FDR < 0.05) enriched by genomic overlaps from a total of 206 filtered cis-acting eQTLs from liver and testis. The top five enriched traits were “Palmitic acid to myristic acid ratio”, “Vertebra number”, “Loin muscle area”, “Ear area” and “Ear weight” (Table 4).
Table 4. Top 30 enriched QTL traits from the filtered cis-acting eQTLs of liver and testis.
| Known QTL trait | Genes from eQTLs with significant overlap | FDR |
|---|---|---|
| Palmitic acid to myristic acid ratio | ENSSSCG00000020784, LATS1 | 5.816E-16 |
| Vertebra number | ENSSSCG00000029150, CCDC170, SNX30, ESR1, ZWILCH, TIPIN, ENSSSCG00000004117, GRM1, ENSSSCG00000005474, TNC, ENSSSCG00000020784, SYTL3, LATS1, ASTN2, LRP11, GOLM1, ENSSSCG00000028881, FBP2, CTSL, ARMC7, BAIAP2, TEN1, HEXDC, INTS2, AOC2, ATP13A3, ENSSSCG00000029252, ENSSSCG00000011817, KALRN, OPA1, ACADSB, ENSSSCG00000029075, ENSSSCG00000025800, ENSSSCG00000010634, ENSSSCG00000009663, UROS, EXTL3 | 1.009E-15 |
| Loin muscle area | ENSSSCG00000005474, ENSSSCG00000022796, TMEM242, CCDC170, SNX30, ESR1, ZWILCH, TIPIN, ENSSSCG00000004117, GRM1, TNC, ENSSSCG00000020784, CTSL, TEN1, HEXDC, MAPT, BAIAP2, ARMC7, NPTX1, TEPSIN, ITGB4, ZNF662, ATP13A3, LSG1, PLCD1, EPHA3, KALRN, UROS, BUB3, EXTL3, NEB, DDHD2, MAP3K20, ANK1, BBOX1, NAALADL1, FAM49A, FNDC4, MTIF2, AP5Z1, CARHSP1, ENSSSCG00000024047 | 5.074E-09 |
| Ear area | FBP2, NAALADL1, CARHSP1, ENSSSCG00000022090 | 1.359E-08 |
| Ear weight | ENSSSCG00000029252, ENSSSCG00000011817, KALRN, OPA1, NAALADL1, ENSSSCG00000022090 | 1.946E-06 |
| PH for Biceps femoris | CCDC170, SNX30, ESR1, ZWILCH, TIPIN, ENSSSCG00000004117, GRM1, ENSSSCG00000005474, TNC, ENSSSCG00000020784 | 2.762E-06 |
| pH for Semispinalis Dorsi | CCDC170, SNX30, ESR1, ZWILCH, TIPIN, ENSSSCG00000004117, GRM1, ENSSSCG00000005474, TNC, ENSSSCG00000020784 | 2.762E-06 |
| Average backfat thickness | ENSSSCG00000005474, NUP43, TNC, ENSSSCG00000026492, DMAC1, ENSSSCG00000022796, TMEM242, ENSSSCG00000029150, CCDC170, SNX30, ESR1, ZWILCH, TIPIN, ENSSSCG00000004117, GRM1, ENSSSCG00000020784, FBP2, CTSL, ARMC7, MAPT, BAIAP2, TEPSIN, ITGB4, ZNF662, ATP13A3, LSG1, PLCD1, ENSSSCG00000029252, ENSSSCG00000011817, ENSSSCG00000025800, ENSSSCG00000029075, ENSSSCG00000010634, HTRA1, GSTO1, EXTL3, NEB, BBOX1, NAALADL1, FNDC4, ENSSSCG00000024047, ENSSSCG00000022090 | 5.032E-06 |
| Thoracic vertebra number | CCDC170, SNX30, ESR1, ZWILCH, TIPIN, ENSSSCG00000004117, GRM1, ENSSSCG00000005474, TNC, ENSSSCG00000020784, LATS1, ASTN2, LRP11, GOLM1, ENSSSCG00000028881, FBP2, CTSL, EXTL3, NEB, MAP3K20, ENSSSCG00000022090 | 7.896E-06 |
| External fat on ham | ENSSSCG00000022796, TMEM242, ARMC7, ZNF662, ATP13A3, LSG1, ENSSSCG00000029252, ENSSSCG00000011817, NEB, ENSSSCG00000024047 | 8.209E-06 |
| Shoulder external fat weight | ENSSSCG00000022796, TMEM242, ENSSSCG00000029150, CCDC170, PLCD1, ENSSSCG00000029252, ENSSSCG00000011817, NEB, NAALADL1, ENSSSCG00000024047 | 8.209E-06 |
| Myristic acid content | LATS1, ASTN2, SNX30, LRP11, ENSSSCG00000020784, TNC, GOLM1, ENSSSCG00000028881, FBP2 | 1.773E-05 |
| Feed intake | ENSSSCG00000022796, TMEM242, ARMC7, ZNF662, ATP13A3, LSG1, PLCD1, ENSSSCG00000010634, GSTO1, ENSSSCG00000024047 | 5.929E-05 |
| cis-11-Eicosenoic acid content | LATS1, ASTN2, SNX30, LRP11, NUDT22 | 6.655E-05 |
| Plateletcrit | ENSSSCG00000005474, CARHSP1, GTF2I, RCC1L | 6.655E-05 |
| Neck weight | ENSSSCG00000005474, NUP43, TNC, ENSSSCG00000026492, DMAC1, ENSSSCG00000022796, ENSSSCG00000024047 | 8.269E-05 |
| Conductivity 24 hours postmortem (loin) | ENSSSCG00000022796, TMEM242, SNX30, CTSL, TEPSIN, ITGB4, EXTL3, NEB | 1.006E-04 |
| Liver weight | ENSSSCG00000022796, TMEM242, NEB, BBOX1, NAALADL1, FNDC4, CLDN4, RCC1L, ENSSSCG00000024047 | 1.225E-04 |
| PRRSV antibody titer | ENSSSCG00000022796, TMEM242 | 1.272E-04 |
| Loin muscle depth | ENSSSCG00000022796, TMEM242, CTSL, TEN1, HEXDC, NPTX1, ARMC7, TEPSIN, ATP13A3, PLCD1, NEB, BBOX1, NAALADL1 | 1.803E-04 |
| Muscle moisture percentage | FBP2, CTSL, BAIAP2, ATP13A3, NEB, DDHD2, MAP3K20, ANK1, SFRP1, BBOX1, NAALADL1, ENSSSCG00000024047 | 2.647E-04 |
| CD8-negative leukocyte percentage | SYTL3, TNC, ENSSSCG00000020784, EPHA3, KALRN | 2.791E-04 |
| CD8-positive leukocyte percentage | SYTL3, TNC, ENSSSCG00000020784, EPHA3, KALRN | 2.791E-04 |
| Off-Flavor Score | ATP13A3, PLCD1, EPHA3, KALRN, ENSSSCG00000029252, ENSSSCG00000011817, ENSSSCG00000024047 | 5.498E-04 |
| CD2-positive leukocyte number | ENSSSCG00000022796, TMEM242 | 6.638E-04 |
| Inside ham weight | FBP2, ENSSSCG00000029429, ENSSSCG00000011035, ARMC7, ENSSSCG00000025800, ENSSSCG00000029075, ENSSSCG00000010634, HTRA1, GSTO1 | 7.146E-04 |
| CD4-positive leukocyte number | ENSSSCG00000022796, TMEM242, CARHSP1 | 8.192E-04 |
| Abdominal fat weight | ENSSSCG00000022796, TMEM242, MAPT, BAIAP2, ZNF662, ATP13A3, LSG1, PLCD1, P3H2, ENSSSCG00000029252, ENSSSCG00000011817, KALRN, OPA1, NEB, BBOX1, NAALADL1, FAM49A, FNDC4, MTIF2, AP5Z1, CARHSP1, ENSSSCG00000024047, ENSSSCG00000022090 | 9.920E-04 |
| Days to 105 kg | GTF2I, RCC1L, CLDN4 | 9.920E-04 |
| Linolenic acid content | LRP11, ENSSSCG00000020784, TNC, GOLM1, ENSSSCG00000028881, FBP2, ENSSSCG00000025800, ENSSSCG00000029075, ENSSSCG00000010634, HTRA1, GSTO1 | 1.164E-03 |
Table of known QTL traits enriched from the genomic coordinates of the filtered cis-acting eQTLs. Bold letters indicate traits with boar taint relevance. Known QTL trait column indicates the name of the trait from the Animal Genome PigQTLdb database. Gene column indicates genes associated with eQTLs with significant genomic overlap with known traits. FDR column indicates Benjamini-Hochberg adjusted P-values obtained from QTL enrichment test. Briefly, a binominal test was performed on the intersections between the genomic locations of each eQTL compared with the genomic locations of all known QTLs on a background defined as the full length (2.7 Gb) of the porcine genome.
Grouped by their corresponding categories of QTLs, the enriched traits contained 54 meat and carcass traits, 22 health traits, 6 exterior traits, 5 production traits and 2 reproduction traits. To adjust for any bias due to uneven distribution of the total number of QTL categories in the Animal Genome PigQTLdb (release 32), the frequencies of the QTL categories was calculated as the number of enriched traits in the summarised category divided by total number of the corresponding summarised category from database. The trait category with highest adjusted frequency was meat and carcass with 0.004 and the lowest was reproduction with 0.001 (Table 5). All categories were constructed and named by the Animal Genome PigQTLdb database [22] and not by the authors of this study.
Table 5. Summary of categories from the enriched QTL traits.
| Trait category | Number of enriched traits | Total number of QTLs in category (December 2017) |
Adjusted number of enriched traits |
|---|---|---|---|
| Meat and carcass | 54 | 13,392 | 0.004 |
| Health | 22 | 6,011 | 0.004 |
| Production | 5 | 1,875 | 0.003 |
| Exterior | 6 | 2,366 | 0.003 |
| Reproduction | 2 | 1,966 | 0.001 |
Categories of enriched traits significantly overlapped by filtered cis-acting eQTLs. Total number of traits in categories was obtained from Animal Genome PigQTLdb (release 32) in December 2017. Adjusted number of enriched traits was calculated as the number of enriched traits divided by total number of the traits in the category.
A total of four boar taint relevant QTL traits were enriched: “Fat androstenone level", "indole, laboratory", "Off-Flavor Score" and "Overall impression, sensory panel". The traits were significantly overlapped by a total of 35 filtered cis-acting eQTLs associated with the genes ARMC7, BAIAP2, TEN1, HEXDC, INTS2, ATP13A3, ENSSSCG00000004117, ENSSSCG00000024047, AOC2, MAPT, EPHA3, PLCD1, BBOX1, KALRN, NAALADL1, ENSSSCG00000029252 and ENSSSCG00000011817. Full results table from QTL trait enrichment test is available in S5 File.
Candidate eQTLs
Identification of candidate eQTLs for gene-based selection was performed by analysing the filtered cis-acting single- and multi-tissue eQTLs for genotypes with lowest EBVs for boar taint. The criteria for candidacy was defined on two parameters: i) a filtered cis-acting eQTL containing a genotype with a median of summarised EBV, equal to or lower than the maximum summarised EBV of the low boar taint pigs (EBVmax = -0.0699208) and ii) significantly overlapping a known boar taint relevant QTL trait by its genomic coordinates. A total of 35 filtered cis-acting eQTLs associated with 17 unique genes with low EBVs for skatole and human nose score were selected as candidate eQTLs for gene-based selection (Table 6). Full results table of the candidate eQTLs are available in S6 File.
Table 6. Candidate cis-acting eQTLs for gene-based selection.
| SNP | Gene | Candidate genotype | Mean EBVs genotype | FDR-EBV | Tissue | Sign. overlapping known QTL trait |
|---|---|---|---|---|---|---|
| rs81342651 | BAIAP2 | CC | -0.732 | 0.029 | liver | Fat androstenone level |
| rs81265837 | ARMC7 | AA | -0.696 | 0.000 | both | Fat androstenone level |
| rs81435284 | ARMC7 | AA | -0.696 | 0.000 | both | Fat androstenone level |
| rs81441228 | EPHA3 | GG | -0.412 | 0.040 | testis | Fat androstenone level |
| rs80817135 | KALRN | CC | -0.805 | 0.045 | testis | Fat androstenone level |
| rs81448091 | ENSSSCG00000029252 | AA | -0.805 | 0.048 | testis | Fat androstenone level |
| rs81357053 | BBOX1 | AA | -0.412 | 0.044 | liver | Fat androstenone level |
| rs81305789 | BBOX1 | AA | -0.412 | 0.044 | liver | Fat androstenone level |
| rs81340851 | NAALADL1 | TC | -0.412 | 0.045 | liver | Fat androstenone level |
| rs81212188 | NAALADL1 | AG | -0.412 | 0.045 | liver | Fat androstenone level |
| rs81359616 | NAALADL1 | TC | -0.412 | 0.045 | liver | Fat androstenone level |
| rs81309447 | ATP13A3 | AG | -0.248 | 0.020 | liver | Off-Flavor Score |
| rs81478384 | ATP13A3 | TT | -0.805 | 0.020 | liver | Off-Flavor Score |
| rs81303987 | ATP13A3 | AC | -0.248 | 0.020 | liver | Off-Flavor Score |
| rs80803537 | PLCD1 | AG | -0.499 | 0.042 | liver | Off-Flavor Score |
| rs81448091 | ENSSSCG00000011817 | AA | -0.805 | 0.048 | testis | Off-Flavor Score |
| rs80919384 | ENSSSCG00000024047 | TC | -0.412 | 0.028 | liver | Off-Flavor Score |
| rs337275357 | ENSSSCG00000024047 | AG | -0.412 | 0.028 | liver | Off-Flavor Score |
| rs81433762 | TEN1 | AA | -0.805 | 0.006 | liver | Overall impression, sensory panel |
| rs81434107 | TEN1 | AA | -0.805 | 0.006 | liver | Overall impression, sensory panel |
| rs81311681 | HEXDC | TT | -0.768 | 0.013 | liver | Overall impression, sensory panel |
| rs81293055 | ENSSSCG00000004117 | AG | -0.294 | 0.025 | testis | indole, laboratory |
| rs80806865 | ENSSSCG00000004117 | AG | -0.294 | 0.025 | testis | indole, laboratory |
| rs81305046 | BAIAP2 | AA | -0.412 | 0.002 | liver | indole, laboratory |
| rs81438833 | BAIAP2 | CC | -0.412 | 0.002 | liver | indole, laboratory |
| rs81439307 | BAIAP2 | CC | -0.412 | 0.002 | liver | indole, laboratory |
| rs81440749 | BAIAP2 | GG | -0.412 | 0.002 | liver | indole, laboratory |
| rs81434341 | HEXDC | AA | -0.768 | 0.013 | liver | indole, laboratory |
| rs81271924 | HEXDC | TT | -0.768 | 0.013 | liver | indole, laboratory |
| rs81283210 | HEXDC | AA | -0.768 | 0.017 | liver | indole, laboratory |
| rs81233143 | INTS2 | TT | -0.412 | 0.018 | liver | indole, laboratory |
| rs327709564 | AOC2 | AG | -0.294 | 0.028 | liver | indole, laboratory |
| rs80864895 | HEXDC | GG | -0.732 | 0.029 | liver | indole, laboratory |
| rs81238764 | MAPT | AG | -0.412 | 0.029 | liver | indole, laboratory |
| rs80864895 | BAIAP2 | GG | -0.732 | 0.029 | liver | indole, laboratory |
SNP = Single nucleotide polymorphism, Candidate genotype = The best genotype for gene-based selection associated with animals with lowest summarised EBVs for skatole and human nose scores, Mean EBVs genotype. = Mean summarised EBVs in animals with the candidate genotype, FDR-EBV = False Discovery Rate of association test with summarised estimated breeding values (EBVs) of skatole and human nose scores for the eQTL, Tissue = Tissue of eQTL identification, Sign. overlapping known QTL trait = The QTL trait which was significantly overlapped by the candidate eQTL from its genomic coordinates.
The candidate eQTLs contained a total of 56 candidate genotypes which were distributed as follows, indicated by the gene associated with eQTLs and the number of candidate genotypes associated with the gene in parentheses: HEXDC (9), ATP13A3 (6), BAIAP2 (6), NAALADL1 (6), ARMC7 (4), ENSSSCG00000004117 (4), ENSSSCG00000024047 (4), TEN1 (4), AOC2 (2), BBOX1 (2), ENSSSCG00000011817 (2), ENSSSCG00000029252 (2), EPHA3 (1), INTS2 (1), KALRN (1), MAPT (1) and PLCD1 (1) (Fig 4).
Fig 4. Candidate eQTLs for selection.
The left Y-axis indicates summarised estimated breeding values (EBVs) for skatole and human nose score. The right Y-axis indicates the mean normalised expression levels where liver is symbolised with a square and testis is symbolised by a triangle. Due to filtering of low gene counts, some eQTLs only have expression levels from one tissue presented. The X-axis indicates the three possible genotypes of each eQTL. The title indicates the rs code of SNP and gene name associated with the eQTL. Red box indicates upregulation and blue box indicates downregulation of the eQTL at each genotype. A green barplot indicates candidate genotypes for gene-based selection, defined as genotypes with a median of summarised EBV equal or lower than the maximum summarised EBV of the low boar taint pigs.
Discussion
Advantages and limitations
This study applied a systems genomics approach to investigate genetic variants affecting genome-wide gene expression levels in multiple porcine tissues from pigs with known genetic merits of boar taint. We identified and functionally characterised single- and multi-tissue cis- and trans-acting expression quantitative trait loci (eQTLs) from liver and testis of Danish Landrace pigs. Subsequently, we filtered for significant association with sire-obtained estimated breeding values (EBVs) of skatole and human nose scores (HNS) and gene expression profiles of liver and testis obtained from each animal. Hence, we obtained only the most relevant eQTLs (filtered eQTLs) containing genotypes with actual association with levels of summarised EBVs. These eQTLs were subjected to functional characterisation of enriched functions and pathways and tested for enrichment of known QTL traits curated by the global Pig Quantitative Trait Locus (QTL) Database (PigQTLdb) [22]. The enrichment test revealed filtered cis-acting eQTLs with significant genomic overlap of boar taint QTL traits. These eQTLs were extracted and designated as candidate eQTLs. Furthermore, the enrichment test revealed potential QTL traits that could be affected by gene-based selection performed with genetic markers residing on or near the loci of the eQTLs. To the best of our knowledge, this is the first study of its kind to report candidate eQTLs associated with EBVs for skatole and HNS. The results indicate actual SNP genotypes for the identification of animals with low EBVs with respect to two important boar taint parameters with the potential for development of an inexpensive and rapid DNA test.
A limitation of the study is related to the definition of a cis- and trans-acting eQTL. The threshold of the cis-distance was obtained from a previous study on obesity that was conducted in the porcine genome [33]. In the aforementioned study, the authors defined a cis-acting eQTL as having a threshold distance of 1 Mb between gene and SNP. The threshold was calculated based on genome comparisons among species [33]. Hence, we used the same physical distance of 1 Mb as a threshold to define the cis -acting eQTLs. Ideally, the definition of a local (cis-acting) eQTL should also consider the linkage disequilibrium (LD) patterns and the persistence of LD phase in the animal model used in a study. LD structure has been estimated for a variety of pig breeds, e.g., US and Finnish breeds [61, 62]. Knowledge regarding LD structure is important in performing genetic mapping on economically important traits because gene-based selection relies on the LD between causative variants and genetic markers [63]. Furthermore, examining the persistence of LD phase in breeds is also important. The persistence of phase is a measure of consistency of LD phase for pairs of SNPs between two populations [61]. Persistence of phase explains why linkage between a marker and a QTL may differ between populations [64]. In a recent study on three Danish pig breeds (Duroc, Landrace and Yorkshire), the authors found local patterns of LD to vary between breeds because of “old inbreeding” of Duroc, but the three breeds showed similar patterns of LD on the chromosomal level [65]. In this study, we filtered for LD structure as part of genotype filtering, but ideally, the examination of LD should be included in the definition of a local cis-acting eQTL. However, we provided several layers of stringent eQTL filtering downstream of the eQTL identification, which should counter any breed-specific differences in LD patterns.
General findings
Tissue differences
Before filtering, the number of eQTLs in testis was found to be twice the number of eQTLs in liver (n = 5,879 vs. n = 2,800). However, by filtering of the eQTLs for association with EBVs for skatole and HNS, the number of eQTLs was reduced ten-fold, and the differences between tissues were relatively small. Interestingly, liver then had a larger number of eQTLs (n = 205) than testis (n = 109). The reason for the differences between tissues is unclear. The number of mapped reads was nearly the same across the two tissues (7.76 vs 7.04 million reads in liver and testis, respectively). The numbers of mapped reads were mostly similar to those reported in previous research using RNA-Seq on porcine tissue, ranging from 8.5 to 9.1 million reads [66, 67]. In another study on candidate genes of boar taint, tissue differences between differentially expressed genes (DEG) were also found but with more DEG in testis than in liver (n = 46 vs. n = 25) [68]; however, identification of DEG cannot be compared with association of gene expression with genotypes. The disparity between the tissues in our study is likely related to technical differences in sample preparation, as both tissues were prepared using punch biopsies at the slaughter line. Finally, we did not use the raw eQTLs for analysis, for which the numeric disparity was nearly double, but only the eQTLs with significant association with the EBVs where the disparity was close to negligible.
Chromosomal densities of eQTLs
The densities of unfiltered eQTLs were highest on pig chromosomes SSC6, SSC14, SSC12, SSC4 and SSC7, in descending order. In comparing the densities of filtered eQTLs, SSC12, SSC1, SSC13, SSC9 and SSC14 showed the highest densities. The chromosomes with the highest densities of unfiltered eQTLs should be interpreted as chromosomes with a high prevalence of regulating loci affecting many traits and not only boar taint. The results are indeed interesting within candidate biomarker research. It appears that SSC6 and SSC14 are heavily inhabited by eQTLs, which could be possible hubs of regulatory control of a multitude of genes. In the context of boar taint, SSC1 and SSC14 had the highest densities of filtered eQTLs. Interestingly, previous literature employing genome-wide association study (GWAS) on Danish Landrace boars identified SNPs with significant association with skatole levels on mainly SSC14 and SSC6 [15]. In a similar GWAS study on commercial Duroc-based boars, SSC1 and SSC6 were found to contain major genetic factors that affected androstenone levels [13]. Identification of QTLs associated with skatole and indole in crossbred pigs (Meishan and Landrace) revealed the highest frequency of QTLs to be located on SSC14. Furthermore, the authors found QTLs affecting androstenone levels on SSC2, SS4, SSC6, SSC7 and SSC9, with SSC6 being the location of the QTL that causes the unpleasant “boar taste” of meat [11]. Another study located the QTLs for androstenone levels in fat tissue on SSC3, SSC4, SSC7, SSC14 and the ends of short arms of SSC6 and SSC9 [10]. A third study found SSC6 as the location for skatole levels but found no QTLs for androstenone levels, likely because of the lack of sexual maturity in the sampled pigs [25]. In conclusion, previous studies found loci affecting skatole and androstenone levels located mainly on SSC14 and SSC6. Thus, a high degree of similarity was evident between locations of known loci associated with boar taint and our findings on chromosomal densities of filtered eQTLs. Although we did not use EBVs for androstenone, previous research has documented a high correlation between HNS phenotype and androstenone levels [69], which might have contributed to the similarity. We cannot conclude that EBVs of skatole and HNS are mainly regulated by SSC14, as this chromosome also showed the highest densities of unfiltered eQTLs, which introduce a bias into the results. However, it appears reasonable to confirm that SSC14 (as well as SSC1) is associated with important loci controlling the development of boar taint.
Analysis of eQTLs
Liver
In liver, we identified CYP2R1 to be associated with the filtered eQTLs. CYP2R1 encodes a member of cytochrome P450 superfamily (CYP) enzymes. These enzymes are known to catalyse many reactions involved in the synthesis of cholesterols, steroids and lipids and many have direct implications for the development of boar taint (as reviewed by Rasmussen and Zamaratskaia [70]). Specifically, CYP2R1 encodes a vitamin D 25-hydroxylase participating in the synthesis of active vitamin D [71] and has been listed as a candidate gene for boar taint [16]. Furthermore, previous research from our group on the same animals identified CYP2R1 as a differentially expressed gene in testis [41]. Our findings suggest that eQTLs regulating CYP2R1 could play important roles in the development of boar taint, and the gene should be the subject of further research. Other CYP Family 4 members were also found to be associated with the filtered eQTLs: CYP4A22 in cis-acting eQTLs from liver and CYP4F11 in trans-acting eQTLs from testis. Both CYPs of this family encode monooxygenases involved in drug metabolism and synthesis of cholesterol, steroids and other lipids [72, 73]. In addition, CYP4V2 was found to be a differentially expressed genes in testis [41]. In conclusion, CYPs are a particularly interesting group of enzymes, but none were found as candidate eQTLs with significant overlap with boar taint QTL traits. Additional testing is needed to confirm a relationship between the low-EBV genotypes of the CYP eQTLs and relevant boar taint traits.
GSTO1 and GSTA4 encode a member of glutathione S-transferases (GSTs) and were found to be associated with the filtered eQTLs. GSTs are known for their role in the catalysis of conjugation reactions in endogenous substances, haeme, fatty acids, xenobiotics and products of oxidative processes [74]. Previous work by our group on the same animals found GSTO1 as a candidate biomarker in liver [41]. A close relative, GSTO2, has been speculated to be involved in excretion of skatole from the porcine body [75]. Both GSTO1 and GSTA4 showed downregulation in the high-boar-taint group in liver. This pattern could be explained by decreased excretion of boar taint compounds such as skatole, which would be consistent with previous findings pertaining to GSTO1 in liver [76]. Interestingly, we found that GSTO1 and GSTA4 were highly upregulated in testis, which was consistent with previous findings on GSTO1 in Duroc pigs [77]. In this study, the authors hypothesised that the upregulation of GSTO1 and GSTs in general in the testis was caused by the biological role of GTSs as transporters of intracellular steroids to their site of action [74]. The authors also found breed-specific expression patterns of GSTs. Finally, we found eQTLs associated with GSTO1 to enrich a number of QTL traits related to growth and health parameters, including backfat thickness, feed intake, ham weight and linolenic acid content. Hence, selection using eQTLs associated with the gene should be monitored carefully.
Testis
In testis, RCC1L was represented within the filtered cis-acting eQTLs. RCC1L encodes a protein associated with a regulator of chromosome condensation 1-like repeats and may function as a guanine nucleotide exchange factor [78]. Recently, the protein was found to be an essential component in oxidative phosphorylation, in which it forms a 16S rRNA regulatory module [79]. Previous studies have found an association between oxidative phosphorylation and boar taint in the testis of pigs [80, 81]. In the current study, we found eQTLs associated with RCC1L to enrich a number of QTL traits, including positive enrichments for “Days to 105 kg”, “Conductivity 45 minutes post-mortem” and “Carcass temperature (24 hr post-mortem)”. Because of the lack of previous knowledge on the subject and significant genomic overlaps with known boar taint QTL traits, it is not clear whether the eQTLs associated with RCC1L are actually associated with boar taint. However, the eQTLs are located in genomic loci where important production traits also reside, which represent interesting results and motivation for further research.
UROS encodes an enzyme that catalyses a step in porphyrin biosynthesis in the haeme biosynthetic pathway. Porphyrins are cofactors for a multitude of enzymes with a diverse range of roles, such as methionine synthesis (vitamin B12) or oxygen transport (haeme). Deficiency in enzyme activity because of autosomal recessive inheritance can lead to congenital erythropoietic porphyria (CEP) [82]. It is not clear why UROS was associated with filtered eQTLs in liver and testis. The eQTLs associated with UROS enriched known QTL traits such as “Base excess”, “Creatinine level”, “Loin muscle area” and “Vertebra number”. Interestingly, expression of UROS was found to be co-expressed by correlation (R2 > 0.5) with the expression profiles of four other genes. However, the occurrence of UROS appears strange, and to our knowledge, no biological relationship with boar taint is currently evident.
Functional characterisation
Functional characterisation of the filtered cis-acting eQTLs identified from liver revealed association with androgen and oestradiol receptor signalling and regulation. The eQTLs associated with these functions contained the genes SFRP1, ESR1, MAPT and RNF14. SFRP1 encodes an inhibitor of the Wnt/β-catenin pathway. Previous research concluded that Wnt/β-catenin signalling is necessary for the development and masculinisation of male genitalia [83]. Furthermore, Wnt/β-catenin has been found to be associated with proliferation and self-renewal of mouse and human spermatogonia [84]. Wnt/β-catenin signalling is regulated by a decrease in expression of Wnt inhibitors, such as DKK2 and SFRP1 [83]. In this study, SFRP1 was found to be downregulated in the high-boar-taint group in liver. This finding was expected as androgen signalling leads to decreased expression of Wnt/β-catenin inhibitors. Oddly, the gene was associated with a filtered eQTL in liver but not in testis. We speculate that increased androstenone is associated with an increased level of androgen signalling and, in turn, a decrease in the general tissue expression of SFRP1. However, this relationship does not explain the lack of finding an eQTL associated with the gene in testis, where spermatogonia are developed. Furthermore, the Wnt inhibitor DKK2 was recently found to be the highest upregulated gene in testis of high androstenone pigs [68], in contrast with the expression pattern of SFRP1 found in this study. Finally, eQTLs associated with SFRP1 were found to enrich known QTL traits of two important muscle fibre characteristics: “Muscle moisture percentage” and “Semimembranosus angle”. Because of the ambiguous results, more research is needed on these important Wnt inhibitors and their relationship with boar taint development and meat quality. ESR1 encodes an oestrogen receptor that binds steroid hormones and was found to be associated with cis-acting eQTLs from testis. Oestrogen is secreted in large amounts from the testis [85], and concentrations have been found to be correlated with androstenone in adipose tissue [86, 87]. Interestingly, eQTLs associated with ESR1 enriched a number of QTL traits for muscle, carcass and the fertility trait of teat numbers but no overlapping boar taint QTL traits. Finally, RNF14 encodes a RING zinc finger that interacts with the androgen receptor and may function as a coactivator and transcriptional regulator [88]. RNF14 was found to play an important role in the regulation of mitochondrial and immune-related genes in skeletal muscle [89]. The eQTLs associated with the gene enriched five important production and muscling QTL traits, including “Cortisol level variations” and “Potassium level”. To our knowledge, no research has provided a link between RNF14 and boar taint, but an association is highly likely because of the gene’s regulation of androgen receptors.
Enrichment test for possible selection effects
An enrichment test to reveal enriched known QTL traits was performed to identify possible selection effects using the filtered cis-acting eQTLs and to identify candidate eQTLs. The test revealed 89 enriched traits, which were mainly related to meat and carcass features and production. The results provide an approximation of which traits may be affected if gene-based selection is performed on the genomic coordinates of the cis-acting eQTLs. Two enriched traits were in the reproduction category: “Left teat number” and “Litter size”. The traits were enriched by eQTLs associated with a total of 30 unique genes, including six genes intersecting with the candidate eQTLs: INTS2, AOC2, HEXDC, BBOX1, KALRN and EPHA3. A common concern regarding selection for lowered levels of boar taint compounds is negative consequences on male fertility [90]. Androstenone is synthesised from pregnenolone during sexual maturity together with other testicular steroids, such as testosterone and oestrogens [91, 92]. These have been established to be important sex steroids affecting male fertility [93, 94]. An unfavourable genetic correlation between androstenone levels and sex steroids was reported [18]. Nevertheless, the possibility of selection for reduced boar taint was found to be a viable solution in a large-scale genetic study involving direct measures of fertility and concentrations of boar taint compounds [90]. We speculate that our finding of two enriched traits for reproduction would confirm the viability of selection with little chance of a negative effect on fertility if one applies careful monitoring. However, we did find several eQTLs in liver and testis associated with SFRP1, which is related to male reproduction and fertility. Thus, future studies should focus on the roles of these genes and whether any causal relationship between them and male fertility exists. To the authors’ knowledge, we are the first to report eQTLs associated with EBVs of skatole and HNS in Danish Landrace pigs and known QTL traits that could possibly be affected by selection on the filtered eQTLs or loci/genes in their vicinity. However, it should be clearly noted that the findings of the QTL enrichment test are highly speculative, and no experimental testing, other than a statistical association test, has been performed to confirm the associations. Furthermore, the QTL traits are obtained from a large number of publications that differ in their methodologies. Finally, the QTL regions curated by the Animal Genome PigQTL db database are very large, likely because of the difficulty of translating cM into genomic coordinates in bp, which is confirmed in the disclaimer on the Animal Genome website. Validation and verifications must be performed when these data are used as a basis for further research.
Candidate eQTLs for selection of low EBV animals
A total of 35 candidate eQTLs were identified from the filtered cis-acting eQTLs, which were selected based on the criteria of significant association with EBVs and genomic overlap with boar taint QTL traits. The candidate eQTLs were sorted according to the number of candidate genotypes for low EBVs (displayed in parentheses): HEXDC (9), ATP13A3 (6), BAIAP2 (6), NAALADL1 (6), ARMC7 (4), ENSSSCG00000004117 (4), ENSSSCG00000024047 (4), TEN1 (4), AOC2 (2), BBOX1 (2), ENSSSCG00000011817 (2), ENSSSCG00000029252 (2), EPHA3 (1), INTS2 (1), KALRN (1), MAPT (1) and PLCD1 (1). The candidate eQTLs associated with HEXDC comprised the largest number of candidate genotypes. HEXDC encodes a hexosaminidase involved in metabolism and glycan degradation. These enzymes have been shown to play key roles in cellular physiology and health and are also considered essential for the development of mammals [95]. More importantly, hexoaminidases are present in very high concentrations in the epididymis of the male pig reproductive organs [96] where hexosaminidases have been described as key players in post-testicular sperm maturation [97]. In this study, we found eQTLs associated with HEXDC to enrich two both boar-taint-related QTL traits (“Overall impression, sensory panel” and “indole, laboratory”) but also the fertility trait “Left teat number”. Hence, it appears plausible to assume that the gene is associated, to a degree, with fertility; interestingly, however, the eQTLs associated with the genes were identified in the liver. The gene was upregulated with increasing EBVs, which could indicate a correlation between hexoaminidases and steroid hormone production. However, further validation in large trials should be performed to assess the role of the gene with respect to the boar taint condition and fertility. ATP13A3 encodes a P-type ATPase family that transports cations across membranes [98]. The gene has been analysed for its potential as a biomarker for early detection of pancreatic cancer [99]. In the context of boar taint, other ATPases involved in ATP synthesis have been reported as candidate genes in boar taint development [75, 100], but no data are available for ATP13A3. However, the eQTLs associated with ATP13A3 enrich a total of 24 QTL traits on many different parameters including boar-taint-relevant QTLs. It could be speculated that the gene is somehow affected by increased skatole metabolism, as the level of gene expression is clearly correlated with high- and low-EBV genotypes. ARMC7 encodes an essential nuclear protein and has been found to be amplified in several cancer tissues and cell lines [101]. Within cancer research, ARMC7 has been the subject of research as a prognostic marker for breast cancer [102], but to our knowledge, no studies have shown a relationship between the gene and boar taint. Interestingly, the gene was markedly upregulated in high-EBV genotypes and vice versa in low-EBV genotypes, implying association with boar taint. Furthermore, the eQTLs associated with ARMC7 enriched known QTL traits for boar taint, such as “Fat androstenone level” and “indole, laboratory”. However, the eQTLs also enriched a number of important meat quality, production and growth traits, such as “Juiciness score”, “Conductivity 45 minutes post-mortem” and “Inside ham weight”. Hence, it is plausible to assume that the eQTLs are located within QTL-rich genomic areas of SSC12. Careful monitoring of the enriched traits should be performed if selection is performed on this subset of eQTLs. Finally, AOC2 encodes a copper amine oxidase that catalyses oxidative conversion of amines to aldehydes and ammonia. The eQTLs associated with AOC2 were found to enrich a QTL trait for indole concentrations but also enrich the reproductive trait of “Left teat number”. Interestingly, the enzyme relative of aldehyde oxidase (AO) was considered negatively correlated with levels of skatole in fat because of its role in skatole metabolism in liver [103]. The authors proposed that a high hepatic enzyme activity of AO was associated with low levels of skatole, but other reports found AO to be uncorrelated with skatole in plasma and fat [104]. Interestingly, our findings show a contrary pattern of gene expression pattern proposed for AO as AOC2 is upregulated with high EBVs for skatole and HNS. This finding could be explained by an activating effect of skatole as a xenobiotic compound, but further research should be performed to investigate the role of AOC2 in hepatic clearance of skatole.
Conclusions
This study applied an integrative systems genomics approach to identify and characterise single-tissue eQTLs associated with EBVs for skatole and HNS and with a number of known genes, such as CYP2R1, GSTO1, GSTA4, RCC1L, UROS, SFRP1 and RNF14. Furthermore, the study provided descriptive statistics of densities of eQTLs on chromosomes of Danish Landrace pigs, among which SSC1, SSC12 and SSC14 were confirmed as the main chromosomes of boar taint regulation. Using a QTL enrichment test, we identified 89 known QTL traits to be enriched from the genomic coordinates of the filtered eQTLs. The enriched traits included important meat and carcass, production, exterior and reproduction traits. Any selection process must closely monitor both positive and negative effects on these traits, which included muscle, fat and weight gain. Furthermore, reproduction-associated eQTLs were found, which warrants further consideration of careful monitoring of any consequences for fertility in future breeding schemes. From four enriched boar taint QTL traits, a total of 35 filtered cis-acting eQTLs with genotypes for low EBVs of skatole and HNS were extracted and designated as candidate eQTLs. The candidate eQTLs are an interesting subset of readily available genes and SNPs with strong association with lowered EBVs and genomic locations in confirmed boar taint regions. Future work should focus on the validation of the candidate eQTLs in large populations as well as in breeds other than Danish Landrace pigs to develop a sensitive and specific DNA test for optimised gene-based animal breeding under industrial settings.
Supporting information
Liver, testis and ham muscle was obtained from non-castrated Danish Landrace male pigs. The liver and testis was subjected to RNA extraction and RNA-sequencing (RNA-Seq) to obtain gene expression profiles. DNA was extracted from ham muscle and subjected to genotyping by Illumina Porcine 60K SNP-chip. The software Matrix eQTL identified single- and multi-tissue eQTLs which were subsequently filtered by a statistical model comparing EBVs and expression profiles from animals grouped by the three genotypes available of each eQTL. The filtered single- and multi-tissue eQTLs were subjected to functional characterisation and a QTL trait enrichment test to find potential known QTL traits that could be affected by selection. Finally, eQTLs enriching boar taint QTL traits from PigQTLdb by significant (FDR < 0.05) genomic overlaps were identified and evaluated as candidate eQTLs for future biomarker development.
(TIF)
Result table obtained from analysis of eQTLs with significant (FDR < 0.05) association with the summarised estimated breeding values of skatole and human nose scores.
(XLS)
Result table obtained from functional characterisation of liver and testis eQTLs with significant (FDR < 0.05) association with the summarised estimated breeding values of skatole and human nose scores. Gene symbols are included for each GO term and KEGG pathway.
(XLS)
Result table obtained from analysis of multi-tissue eQTLs with significant (FDR < 0.05) association with the summarised estimated breeding values of skatole and human nose scores.
(XLS)
Result table obtained from enrichment test with using filtered cis-acting eQTLs on known QTL traits from Animal Genome PigQTLdb. The test revealed QTL traits which were significantly (FDR < 0.05) overlapped by the eQTLs and the genes associated with the eQTLs that overlapped the QTL traits.
(XLS)
The candidate eQTLs were defined as filtered cis-acting eQTLs which enriched known boar taint QTL traits from the PigQTLdb. The candidate eQTLs could be useful in future development of biomarkers for gene-based selection schemes of pigs with lowered boar taint. The spreadsheet includes rs codes for SNPs, gene names and the genotypes associated with lowest values of summarised estimated breeding values for skatole and human nose scores.
(XLS)
Acknowledgments
We thank the Danish Pig Research Centre (VSP-SEGES) for animals and breeding data used in this study and the staff of Danish Crown Herning for assistance during sample collection at the slaughter house. Furthermore, we acknowledge the Green Development and Demonstration Programme (GUDP, grant number VETENH/ANHJ NR. 14029) and Veterinary Department of the Ministry of Food, Agriculture and Fisheries, Denmark and University of Copenhagen, Faculty of Health and Medical Sciences, Denmark for funding.
Data Availability
The datasets generated during and/or analysed during the current study are available in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE93734 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93734).
Funding Statement
This “AGES” project was funded by the Green Development and Demonstration Programme (GUDP, grant number VETENH/ANHJ NR. 14029) and Veterinary Department of the Ministry of Food, Agriculture and Fisheries, Denmark and University of Copenhagen, Faculty of Health and Medical Sciences, Denmark.
References
- 1.Babol J, Squires EJ, Lundström K. Relationship between metabolism of androstenone and skatole in intact male pigs. J Anim Sci. 1999;77:84–92. [DOI] [PubMed] [Google Scholar]
- 2.Zamaratskaia G, Babol J, Andersson H, Lundström K. Plasma skatole and androstenone levels in entire male pigs and relationship between boar taint compounds, sex steroids and thyroxine at various ages. Livest Prod Sci. 2004;87:91–8. [Google Scholar]
- 3.Babol J, Zamaratskaia G, Juneja R, Lundström K. The effect of age on distribution of skatole and indole levels in entire male pigs in four breeds: Yorkshire, Landrace, Hampshire and Duroc. Meat Sci. 2004;67:351–8. doi: 10.1016/j.meatsci.2003.11.008 [DOI] [PubMed] [Google Scholar]
- 4.Robic A, Larzul C, Bonneau M. Genetic and metabolic aspects of androstenone and skatole deposition in pig adipose tissue: A review. Genet Sel Evol. 2008;40:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wesoly R, Weiler U. Nutritional influences on skatole formation and skatole metabolism in the pig. Animals. 2012;2:221–42. doi: 10.3390/ani2020221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tajet H, Andresen Ø. Estimation of genetic parameters of boar taint; skatole and androstenone and their correlations with sexual maturation. Acta Vet Scand. 2006;48:1. [Google Scholar]
- 7.Squires EJ, Lundström K. Relationship between cytochrome P450IIE1 in liver and levels of skatole and its metabolites in intact male pigs. J Anim Sci. 1997;75:2506–11. [DOI] [PubMed] [Google Scholar]
- 8.European Declaration on alternatives to surgical castration of pigs 2010. Available from: http://ec.europa.eu/food/animals/welfare/practice/farm/pigs/castration_alternatives_en.
- 9.Hayes B, Goddard M. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintanilla R, Demeure O, Bidanel J, Milan D, Iannuccelli N, Amigues Y, et al. Detection of quantitative trait loci for fat androstenone levels in pigs. J Anim Sci. 2003;81:385–94. doi: 10.2527/2003.812385x [DOI] [PubMed] [Google Scholar]
- 11.Lee G, Archibald A, Law A, Lloyd S, Wood J, Haley C. Detection of quantitative trait loci for androstenone, skatole and boar taint in a cross between Large White and Meishan pigs. Anim Genet. 2005;36:14–22. doi: 10.1111/j.1365-2052.2004.01214.x [DOI] [PubMed] [Google Scholar]
- 12.Moe M, Lien S, Aasmundstad T, Meuwissen TH, Hansen MH, Bendixen C, et al. Association between SNPs within candidate genes and compounds related to boar taint and reproduction. BMC Genet. 2009;10(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duijvesteijn N, Knol EF, Merks JW, Crooijmans RP, Groenen MA, Bovenhuis H, et al. A genome-wide association study on androstenone levels in pigs reveals a cluster of candidate genes on chromosome 6. BMC Genet. 2010;11:42 doi: 10.1186/1471-2156-11-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Squires E, Schenkel F, editors. Managing boar taint: focus on genetic markers. Proceedings of the 10th London Swine Conference–Focus on the Future; 2010.
- 15.Rowe SJ, Karacaören B, De Koning D-J, Lukic B, Hastings-Clark N, Velander I, et al. Analysis of the genetics of boar taint reveals both single SNPs and regional effects. BMC Genomics. 2014;15:424 doi: 10.1186/1471-2164-15-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grindflek E, Lien S, Hamland H, Hansen MH, Kent M, van Son M, et al. Large scale genome-wide association and LDLA mapping study identifies QTLs for boar taint and related sex steroids. BMC Genomics. 2011;12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sellier P, Le Roy P, Fouilloux M, Gruand J, Bonneau M. Responses to restricted index selection and genetic parameters for fat androstenone level and sexual maturity status of young boars. Livest Prod Sci. 2000;63:265–74. [Google Scholar]
- 18.Grindflek E, Aasmundstad T, Hamland H, Hansen M, Nome T, Kent M, et al. Revealing genetic relationships between compounds affecting boar taint and reproduction in pigs. J Anim Sci. 2011;89:680–92. doi: 10.2527/jas.2010-3290 [DOI] [PubMed] [Google Scholar]
- 19.Strathe AB, Velander I, Mark T, Kadarmideen H. Genetic parameters for androstenone and skatole as indicators of boar taint and their relationship to production and litter size traits in Danish Landrace. J Anim Sci. 2013;91:2587–95. doi: 10.2527/jas.2012-6107 [DOI] [PubMed] [Google Scholar]
- 20.Lukić B, Pong‐Wong R, Rowe S, Koning D, Velander I, Haley C, et al. Efficiency of genomic prediction for boar taint reduction in Danish Landrace pigs. Anim Genet. 2015;46:607–16. doi: 10.1111/age.12369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zadinová K, Stupka AS, Čítek J, Vehovský DU. Boar taint–the effects of selected candidate genes associated with androstenone and skatole levels–a review. Anim Sci Pap Rep. 2016;34:107–28. [Google Scholar]
- 22.Hu Z-L, Park CA, Wu X-L, Reecy JM. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013;41:D871–D9. doi: 10.1093/nar/gks1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamaratskaia G, Squires E. Biochemical, nutritional and genetic effects on boar taint in entire male pigs. Animal. 2009;3:1508–21. doi: 10.1017/S1751731108003674 [DOI] [PubMed] [Google Scholar]
- 24.Ernst CW, Steibel JP. Molecular advances in QTL discovery and application in pig breeding. Trends Genet. 2013;29:215–24. doi: 10.1016/j.tig.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 25.Varona L, Vidal O, Quintanilla R, Gil M, Sanchez A, Folch J, et al. Bayesian analysis of quantitative trait loci for boar taint in a Landrace outbred population. J Anim Sci. 2005;83:301–7. doi: 10.2527/2005.832301x [DOI] [PubMed] [Google Scholar]
- 26.Miles C, Wayne M. Quantitative trait locus (QTL) analysis. Nature Education. 2008;1:208. [Google Scholar]
- 27.Große-Brinkhaus C, Storck LC, Frieden L, Neuhoff C, Schellander K, Looft C, et al. Genome-wide association analyses for boar taint components and testicular traits revealed regions having pleiotropic effects. BMC Genet. 2015;16:36 doi: 10.1186/s12863-015-0194-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadarmideen HN, von Rohr P, Janss LL. From genetical genomics to systems genetics: potential applications in quantitative genomics and animal breeding. Mamm Genome. 2006;17:548–64. doi: 10.1007/s00335-005-0169-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadarmideen HN. Genomics to systems biology in animal and veterinary sciences: progress, lessons and opportunities. Livest Sci. 2014;166:232–48. [Google Scholar]
- 31.Breitling R, Li Y, Tesson BM, Fu J, Wu C, Wiltshire T, et al. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 2008;4:e1000232 doi: 10.1371/journal.pgen.1000232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westra H-J, Franke L. From genome to function by studying eQTLs. Biochim Biophys Acta. 2014;1842:1896–902. doi: 10.1016/j.bbadis.2014.04.024 [DOI] [PubMed] [Google Scholar]
- 33.Kogelman LJ, Zhernakova DV, Westra H-J, Cirera S, Fredholm M, Franke L, et al. An integrative systems genetics approach reveals potential causal genes and pathways related to obesity. Genome Med. 2015;7:105 doi: 10.1186/s13073-015-0229-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wayne ML, McIntyre LM. Combining mapping and arraying: an approach to candidate gene identification. Proc Natl Acad Sci U S A. 2002;99:14903–6. doi: 10.1073/pnas.222549199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–53. doi: 10.1038/ng1522 [DOI] [PubMed] [Google Scholar]
- 36.Kogelman LJ, Pant SD, Fredholm M, Kadarmideen HN. Systems genetics of obesity in an F2 pig model by genome-wide association, genetic network, and pathway analyses. Front Genet. 2014;5:214 doi: 10.3389/fgene.2014.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.González-Prendes R, Quintanilla R, Cánovas A, Manunza A, Cardoso TF, Jordana J, et al. Joint QTL mapping and gene expression analysis identify positional candidate genes influencing pork quality traits. Sci Rep. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ponsuksili S, Murani E, Trakooljul N, Schwerin M, Wimmers K. Discovery of candidate genes for muscle traits based on GWAS supported by eQTL-analysis. Int J Biol Sci. 2014;10:327–37. doi: 10.7150/ijbs.8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–8. doi: 10.1093/bioinformatics/bts163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G, Shabalin AA, Rusyn I, Wright FA, Nobel AB. An empirical Bayes approach for multiple tissue eQTL analysis. arXiv. 2013:arXiv:1311.2948. [DOI] [PMC free article] [PubMed]
- 41.Drag M, Skinkyte-Juskiene R, Do D, Kogelman L, Kadarmideen H. Differential expression and co-expression gene networks reveal candidate biomarkers of boar taint in non-castrated pigs. Sci Rep. 2017;7:12205 doi: 10.1038/s41598-017-11928-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrews S. FastQC: a quality control tool for high throughput sequence data 2010. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 43.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17 doi: 10.14806/ej.17.1.200 pp. 10–12. [Google Scholar]
- 44.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014:btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–4. doi: 10.1093/bioinformatics/btv566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014:btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Team RC. R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing; 2014. Available from: http://www.R-project.org/. [Google Scholar]
- 49.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015:gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramos AM, Crooijmans RP, Affara NA, Amaral AJ, Archibald AL, Beever JE, et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One. 2009;4:e6524 doi: 10.1371/journal.pone.0006524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolazzi EL, Caprera A, Nazzicari N, Cozzi P, Strozzi F, Lawley C, et al. SNPchiMp v. 3: integrating and standardizing single nucleotide polymorphism data for livestock species. BMC Genomics. 2015;16:283 doi: 10.1186/s12864-015-1497-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–40. doi: 10.1093/bioinformatics/bti525 [DOI] [PubMed] [Google Scholar]
- 55.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–91. doi: 10.1038/nprot.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559 doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansen MB. binom.test.QTL: Binomial test of SNP QTL 2017. Available from: https://github.com/MathiasBryggerHansen/binom.test.QTL.
- 60.Łabaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics. 2011;27(13):i383–i91. doi: 10.1093/bioinformatics/btr247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Badke YM, Bates RO, Ernst CW, Schwab C, Steibel JP. Estimation of linkage disequilibrium in four US pig breeds. BMC Genomics. 2012;13:24 doi: 10.1186/1471-2164-13-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uimari P, Tapio M. Extent of linkage disequilibrium and effective population size in Finnish Landrace and Finnish Yorkshire pig breeds. J Anim Sci. 2011;89:609–14. doi: 10.2527/jas.2010-3249 [DOI] [PubMed] [Google Scholar]
- 63.Hayes BJ, Bowman PJ, Chamberlain A, Goddard M. Invited review: Genomic selection in dairy cattle: Progress and challenges. J Dairy Sci. 2009;92:433–43. doi: 10.3168/jds.2008-1646 [DOI] [PubMed] [Google Scholar]
- 64.De Roos A, Hayes BJ, Spelman R, Goddard ME. Linkage disequilibrium and persistence of phase in Holstein–Friesian, Jersey and Angus cattle. Genetics. 2008;179:1503–12. doi: 10.1534/genetics.107.084301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, Sørensen P, Janss L, Ostersen T, Edwards D. Genome-wide and local pattern of linkage disequilibrium and persistence of phase for 3 Danish pig breeds. BMC Genet. 2013;14:115 doi: 10.1186/1471-2156-14-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen C, Ai H, Ren J, Li W, Li P, Qiao R, et al. A global view of porcine transcriptome in three tissues from a full-sib pair with extreme phenotypes in growth and fat deposition by paired-end RNA sequencing. BMC Genomics. 2011;12:448 doi: 10.1186/1471-2164-12-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Esteve-Codina A, Kofler R, Palmieri N, Bussotti G, Notredame C, Pérez-Enciso M. Exploring the gonad transcriptome of two extreme male pigs with RNA-seq. BMC Genomics. 2011;12:552 doi: 10.1186/1471-2164-12-552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gunawan A, Sahadevan S, Neuhoff C, Große-Brinkhaus C, Gad A, Frieden L, et al. RNA deep sequencing reveals novel candidate genes and polymorphisms in boar testis and liver tissues with divergent androstenone levels. PLoS One. 2013b;8:e63259 doi: 10.1371/journal.pone.0063259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mathur P, Ten Napel J, Bloemhof S, Heres L, Knol E, Mulder H. A human nose scoring system for boar taint and its relationship with androstenone and skatole. Meat Sci. 2012;91:414–22. doi: 10.1016/j.meatsci.2012.02.025 [DOI] [PubMed] [Google Scholar]
- 70.Rasmussen MK, Zamaratskaia G. Regulation of porcine hepatic cytochrome p450—implication for boar taint. Comput Struct Biotechnol J. 2014;11:106–12. doi: 10.1016/j.csbj.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci U S A. 2004;101:7711–5. doi: 10.1073/pnas.0402490101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hiratsuka M, Nozawa H, Katsumoto Y, Moteki T, Sasaki T, Konno Y, et al. Genetic polymorphisms and haplotype structures of the CYP4A22 gene in a Japanese population. Mutat Res. 2006;599:98–104. doi: 10.1016/j.mrfmmm.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 73.Edson KZ, Prasad B, Unadkat JD, Suhara Y, Okano T, Guengerich FP, et al. Cytochrome P450-dependent catabolism of vitamin K: ω-hydroxylation catalyzed by human CYP4F2 and CYP4F11. Biochemistry. 2013;52(46):8276–85. doi: 10.1021/bi401208m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Listowsky I, Abramovitz M, Homma H, Niitsu Y. Intracellular binding and transport of hormones and xenobiotics by glutathiones-transferases. Drug Metab Rev. 1988;19:305–18. doi: 10.3109/03602538808994138 [DOI] [PubMed] [Google Scholar]
- 75.Gunawan A, Sahadevan S, Cinar MU, Neuhoff C, Große-Brinkhaus C, Frieden L, et al. Identification of the novel candidate genes and variants in boar liver tissues with divergent skatole levels using RNA deep sequencing. PLoS One. 2013a;8:e72298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moe M, Lien S, Bendixen C, Hedegaard J, Hornshøj H, Berget I, et al. Gene expression profiles in liver of pigs with extreme high and low levels of androstenone. BMC Vet Res. 2008;4:1 doi: 10.1186/1746-6148-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moe M, Meuwissen T, Lien S, Bendixen C, Wang X, Conley LN, et al. Gene expression profiles in testis of pigs with extreme high and low levels of androstenone. BMC Genomics. 2007;8:1 doi: 10.1186/1471-2164-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Merla G, Ucla C, Guipponi M, Reymond A. Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum Genet. 2002;110:429–38. doi: 10.1007/s00439-002-0710-x [DOI] [PubMed] [Google Scholar]
- 79.Arroyo JD, Jourdain AA, Calvo SE, Ballarano CA, Doench JG, Root DE, et al. A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab. 2016;24:875–85. doi: 10.1016/j.cmet.2016.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sahadevan S, Gunawan A, Tholen E, Große-Brinkhaus C, Tesfaye D, Schellander K, et al. Pathway based analysis of genes and interactions influencing porcine testis samples from boars with divergent androstenone content in back fat. PLoS One. 2014;9:e91077 doi: 10.1371/journal.pone.0091077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lervik S, Kristoffersen AB, Conley L, Oskam I, Hedegaard J, Ropstad E, et al. Gene expression during testis development in Duroc boars. Animal. 2015;9:1832–42. doi: 10.1017/S1751731115000907 [DOI] [PubMed] [Google Scholar]
- 82.Clavero S, Bishop DF, Giger U, Haskins ME, Desnick RJ. Feline congenital erythropoietic porphyria: two homozygous UROS missense mutations cause the enzyme deficiency and porphyrin accumulation. Mol Med. 2010;16:381 doi: 10.2119/molmed.2010.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miyagawa S, Satoh Y, Haraguchi R, Suzuki K, Iguchi T, Taketo MM, et al. Genetic interactions of the androgen and Wnt/β-catenin pathways for the masculinization of external genitalia. Mol Endocrinol. 2009;23:871–80. doi: 10.1210/me.2008-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Üren A, Dym M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction. 2009;138:151–62. doi: 10.1530/REP-08-0510 [DOI] [PubMed] [Google Scholar]
- 85.Claus R, Hoffmann B. Oestrogens, compared to other steroids of testicular origin, in bloodplasma of boars. Acta Endocrinol. 1980;94:404–11. [DOI] [PubMed] [Google Scholar]
- 86.Zamaratskaia G, Rydhmer L, Chen G, Madej A, Andersson H, Lundström K. Boar taint is related to endocrine and anatomical changes at puberty but not to aggressive behaviour in entire male pigs. Reprod Domest Anim. 2005;40:500–6. doi: 10.1111/j.1439-0531.2005.00613.x [DOI] [PubMed] [Google Scholar]
- 87.Chen G, Bourneuf E, Marklund S, Zamaratskaia G, Madej A, Lundström K. Gene expression of 3β-hydroxysteroid dehydrogenase and 17β-hydroxysteroid dehydrogenase in relation to androstenone, testosterone, and estrone sulphate in gonadally intact male and castrated pigs. J Anim Sci. 2007;85:2457–63. doi: 10.2527/jas.2007-0087 [DOI] [PubMed] [Google Scholar]
- 88.Xu K, Shimelis H, Linn DE, Jiang R, Yang X, Sun F, et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer cell. 2009;15(4):270–82. doi: 10.1016/j.ccr.2009.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ingham AB, Osborne SA, Menzies M, Briscoe S, Chen W, Kongsuwan K, et al. RNF14 is a regulator of mitochondrial and immune function in muscle. BMC Syst Biol. 2014;8:10 doi: 10.1186/1752-0509-8-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Strathe AB, Velander I, Mark T, Ostersen T, Hansen C, Kadarmideen H. Genetic parameters for male fertility and its relationship to skatole and androstenone in Danish Landrace boars. J Anim Sci. 2013;91:4659–68. doi: 10.2527/jas.2013-6454 [DOI] [PubMed] [Google Scholar]
- 91.Bonneau M. Compounds responsible for boar taint, with special emphasis on androstenone: a review. Livest Sci. 1982;9:687–705. [Google Scholar]
- 92.Andresen Ø. Boar taint related compounds: Androstenone/skatole/other substances. Acta Vet Scand. 2006;48:S5. [Google Scholar]
- 93.Hess RA. Estrogen in the adult male reproductive tract: a review. Reprod Biol Endocrinol. 2003;1:52 doi: 10.1186/1477-7827-1-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walker WH. Non-classical actions of testosterone and spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1557–69. doi: 10.1098/rstb.2009.0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alteen MG, Oehler V, Nemčovičová I, Wilson IB, Vocadlo DJ, Gloster TM. Mechanism of Human Nucleocytoplasmic Hexosaminidase D. Biochemistry. 2016;55(19):2735–47. doi: 10.1021/acs.biochem.5b01285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gatti J-L, Castella S, Dacheux F, Ecroyd H, Metayer S, Thimon V, et al. Post-testicular sperm environment and fertility. Anim Reprod Sci. 2004;82:321–39. doi: 10.1016/j.anireprosci.2004.05.011 [DOI] [PubMed] [Google Scholar]
- 97.Dacheux J-L, Castella S, Gatti JL, Dacheux F. Epididymal cell secretory activities and the role of proteins in boar sperm maturation. Theriogenology. 2005;63:319–41. doi: 10.1016/j.theriogenology.2004.09.015 [DOI] [PubMed] [Google Scholar]
- 98.Habtemichael N, Kovacs G. Cloning the AFURS1 gene which is up-regulated in senescent human parenchymal kidney cells. Gene. 2002;283(1):271–5. [DOI] [PubMed] [Google Scholar]
- 99.Madan M, Patel A, Skruber K, Geerts D, Altomare DA, Phanstiel O IV. ATP13A3 and caveolin-1 as potential biomarkers for difluoromethylornithine-based therapies in pancreatic cancers. Am J Cancer Res. 2016;6:1231 [PMC free article] [PubMed] [Google Scholar]
- 100.Leung MC, Bowley K-L, Squires EJ. Examination of testicular gene expression patterns in Yorkshire pigs with high and low levels of boar taint. Anim Biotechnol. 2010;21:77–87. doi: 10.1080/10495390903500607 [DOI] [PubMed] [Google Scholar]
- 101.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, Angers S, et al. Systematic discovery and classification of human cell line essential genes. bioRxivorg. 2015:015412. [Google Scholar]
- 102.Tuna M, Smid M, Martens JW, Foekens JA. Prognostic value of acquired uniparental disomy (aUPD) in primary breast cancer. Breast Cancer Res Treat. 2012;132:189–96. doi: 10.1007/s10549-011-1579-y [DOI] [PubMed] [Google Scholar]
- 103.Diaz GJ, Squires EJ. Role of aldehyde oxidase in the hepatic in vitro metabolism of 3-methylindole in pigs. J Agric Food Chem. 2000;48:833–7. [DOI] [PubMed] [Google Scholar]
- 104.Lanthier F, Lou Y, Squires E. Skatole metabolism in the intact pre-pubescent male pig: The relationship between hepatic enzyme activity and skatole concentrations in plasma and fat. Livest Sci. 2007;106:145–53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Liver, testis and ham muscle was obtained from non-castrated Danish Landrace male pigs. The liver and testis was subjected to RNA extraction and RNA-sequencing (RNA-Seq) to obtain gene expression profiles. DNA was extracted from ham muscle and subjected to genotyping by Illumina Porcine 60K SNP-chip. The software Matrix eQTL identified single- and multi-tissue eQTLs which were subsequently filtered by a statistical model comparing EBVs and expression profiles from animals grouped by the three genotypes available of each eQTL. The filtered single- and multi-tissue eQTLs were subjected to functional characterisation and a QTL trait enrichment test to find potential known QTL traits that could be affected by selection. Finally, eQTLs enriching boar taint QTL traits from PigQTLdb by significant (FDR < 0.05) genomic overlaps were identified and evaluated as candidate eQTLs for future biomarker development.
(TIF)
Result table obtained from analysis of eQTLs with significant (FDR < 0.05) association with the summarised estimated breeding values of skatole and human nose scores.
(XLS)
Result table obtained from functional characterisation of liver and testis eQTLs with significant (FDR < 0.05) association with the summarised estimated breeding values of skatole and human nose scores. Gene symbols are included for each GO term and KEGG pathway.
(XLS)
Result table obtained from analysis of multi-tissue eQTLs with significant (FDR < 0.05) association with the summarised estimated breeding values of skatole and human nose scores.
(XLS)
Result table obtained from enrichment test with using filtered cis-acting eQTLs on known QTL traits from Animal Genome PigQTLdb. The test revealed QTL traits which were significantly (FDR < 0.05) overlapped by the eQTLs and the genes associated with the eQTLs that overlapped the QTL traits.
(XLS)
The candidate eQTLs were defined as filtered cis-acting eQTLs which enriched known boar taint QTL traits from the PigQTLdb. The candidate eQTLs could be useful in future development of biomarkers for gene-based selection schemes of pigs with lowered boar taint. The spreadsheet includes rs codes for SNPs, gene names and the genotypes associated with lowest values of summarised estimated breeding values for skatole and human nose scores.
(XLS)
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE93734 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93734).