Abstract
Deposition of secondary cell wall in the xylem elements is controlled by a subgroup of NAC (NAM, ATAF, CUC) family, known as vascular-related NAC transcription factors (VNDs). In the present study, we analyzed the 5’ upstream regulatory region of two banana NAC transcription factors (MusaVND6 and MusaVND7) for tissue specific expression and presence of 19-bp secondary-wall NAC binding element (SNBE)-like motifs. Transgenic banana plants of Musa cultivar Rasthali harboring either PMusaVND7::GUS or PMusaVND6::GUS showed specific GUS (β-D-Glucuronidase) activity in cells of the xylem tissue. Approximately 1.2kb promoter region of either MusaVND6 or MusaVND7 showed presence of at least two SNBE-like motifs. This 1.2kb promoter region was retarded in a gel shift assay by three banana VND protein (VND1,VND2 and VND3). The banana VND1-VND3 could also retard the mobility of isolated SNBE-like motifs of MusaVND6 or MusaVND7 in a gel shift assay. Transcript levels of MusaVND6 and MusaVND7 were elevated in transgenic banana overexpressing either banana VND1, VND2 or VND3. Present study suggested a probable regulation of banana VND6 and VND7 expression through direct interaction of banana VND1- VND3 with SNBE-like motifs. Our study also indicated two promoter elements for possible utilization in cell wall modifications in plants especially banana, which is being recently considered as a potential biofuel crop.
Introduction
Evolution of secondary cell wall deposition was a prominent feature which not only provided the mechanical strength required for the vertical growth of plants but also helped in the long distance transport of water and minerals. This secondary cell wall is laid in cells of xylem tissue and is composed of cellulose, hemicelluloses, lignin and xylan. Water transport in plants from roots to other organs is through the channels of xylem tissue composed of tracheids and vessel elements. These tracheids develop an array of secondary wall depositions in the form of reticulate, pitted, helical and annular thickenings. Secondary cell wall deposition is regulated and highly coordinated for systematic deposition of multiple components by many genes among which a subgroup of NAC transcription factors, VNDs (vascular related NAC transcription factors) are most important. Secondary cell wall deposition in tracheids is accompanied with programmed cell death, hence regulation of the activity of secondary cell wall associated genes in specialized cells is of prime importance for proper functioning and homeostasis of plants.
Our earlier work identified seven VND (VND1-VND7) genes from banana genomic database and analyzed their expression pattern during the time course of invitro tracheary element differentiation from banana embryogenic cells in a brassinolide supplemented medium [1]. During tracheary element formation, transcript of banana VND1-VND3 and VND6-VND7 were detected as early as third day after induction with brassinolide [1]. Overexpression of banana VND1-VND3 (MusaVND1, MusaVND2 and MusaVND3) genes in transgenic banana indicated ectopic secondary wall deposition and tracheids development suggesting the important roles of these genes in tracheary element formation [2,3].
In Arabidopsis, roles of vascular related NAC domain transcription factors (VND1-VND7) in secondary wall deposition have been documented in detail [4–5].A pioneering work carried out by Kubo et al. (2005) showed that ArabidopsisVND6 and VND7 are master regulators of secondary wall deposition as their overexpression resulted in transdifferentiation of cells into tracheids [6]. However recent work on VND1-VND5 from Arabidopsis has established that these genes also carry the potential to independently trigger the secondary wall deposition and tracheid differentiation [4]. Apart from VND1-VND7, other members of NAC group involved in secondary wall deposition have also been characterized.SECONDARY WALL ASSOCIATED NAC DOMAIN PROTEIN1 (SND1) and NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1 (NST1)are independent master regulators of secondary wall deposition and functions redundantly in differentiation of xylem fibers in Arabidopsis[7–8]. Secondary wall thickening in anther endothecium isregulated by NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1 (NST1)and NST2and T-DNA insertion lines for NST1 and NST2 are defective in anther dehiscence [9].
A common motif in the promoters of secondary wall associated genes for their regulation has been identified recently which has been named as secondary-wall NAC binding element (SNBE)- motif [10,11]. Further, some secondary wall associated transcription factors, VND6, VND7 and SND1 have been shown to regulate the expression of their downstream target genes by SNBE-like motif [10,11]. The sequence of SNBE motif has been worked out as an imperfect palindrome of 19bp with consensus sequence (T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T) (A/T) and has been identified in the 5’upstream regulatory regions of multiple downstream target genes [10].ArabidopsisSND1, VND6, VND7, NST1 and NST2 have been shown to regulate the expression of downstream genes like MYB46, MYB83, MYB103, SND3 and KNAT7 through such SNBE-like motifs in their promoter region [10]. Furthermore, ArabidopsisSND1, VND6 and VND7 also regulate the expression of programmed cell death related gene like XCP1(tracheary elements related cysteine proteases) through direct binding to SNBE motif [10]. These findings suggest that SNBE-like motifs are common regulatory cis-element for direct regulation of secondary wall associated genes. Recently a potential regulation of ArabidopsisVND7 expression was shown by binding of ArabidopsisVNDs to VND7 promoter and itsSNBE motif [12].
Lignocellulosic biomass which is primarily composed of secondary cell wall is an important source of renewable energy, paper and pulp production [13]. Hence, genetic factors with potential to modulate the biochemical properties of lignocellulosic biomass can be of great importance in the field of renewable energy and pulp production. However, most of the studies conducted on regulation of lignocellulosic biomass by secondary wall associated NAC transcription factors has resulted in unwanted side effect of growth reduction, possibly due to transdifferentiation of cells into tracheary elements [7,8]. Hence, a tighter control over the expression of VNDs and preferably limiting their overexpression in vascular tissue of transgenic plants will provide a suitable way for desired modification of plant lignocellulosic biomass. Hence, characterization of promoter region with specific activation in xylem vessel elementis indispensable. This study has characterized two such regulatory regions, PMusaVND7 and PMusaVND6and demonstrated their xylem specific activation.
In this study we report the characterization of 5’upstream regulatory region of two banana VNDs (MusaVND6 and MusaVND7). The tissue specific expression of these was identified by cloning their promoter region upstream of GUS and analyzing the transgenic banana plants by GUS staining. We analyzed sequence of MusaVND6 and MusaVND7promoter and identified SNBE-like sites which indicated their potential regulation by this important cis-element. Our study demonstrated that banana VND1, VND2 and VND3 could bind to 5’upstream regulatory region (approximately 1.2 kb) of banana VND6 and VND7 in a gel shift assay and also to SNBE-like motifs (present in PMusaVND7 and PMusaVND6). Moreover, elevated transcript level of MusaVND6 and MusaVND7 were detected in the corm tissue of banana overexpressing either banana VND1, VND2 or VND3. Banana VND1-VND3 could trigger the expression of GUS from PMusaVND7 and PMusaVND6 indicating that these transcription factors could activate the expression of MusaVND6 and MusaVND7. The present work indicated a potential regulation of banana VND6 and VND7 transcription factors through SNBE-like motif and also identified two xylem specific regulatory regions which can serve aspotential DNA elements for xylem specific cell wall modification. This work will also enhance our present knowledge about the regulation of secondary wall thickening in plants especially in monocots.
Material and methods
PCR amplification of 5’ up-stream region and cloning upstream of GUS
Genomic DNA of banana cultivar Rasthali was isolated from leaf tissue usingGenElute Plant Genomic-DNA Miniprep-Kit (Sigma,USA) following the instruction provided with the kit. DNA specific primers were designed and used for PCR amplification using the commercially available PCR master mix (Thermofisher scientific) and purified genomic DNA as template. PCR was carried out as follows: 98°C(10min), 35-cycles of [98°C(1min), 58°C(1min), 72°C(1.5min)] followed by final extension of 72°C(20min). PMusaVND7 was amplified with the forward primer (AACTGCAGCCTCGAAAGGAAAAGGTCAT PstI underlined) and reverse primer (AACCATGGTGTGGATTCACCTGGAGAC NcoI underlined) and further PCR purified. Forward primer (AATCTAGAAAAGCATTCATTCCACGTACT XbaI underlined) and reverse primer AACCATGGTCATGCGAAGTGATTTAAAGTG NcoI underlined) were used for amplification of PMusaVND6. Binary vector pCAMBIA 1301 was digested with PstI or XbaI and NcoI (to release the CaMV35S promoter) and then used for ligation with PstI/NcoI digested PMusaVND7 and XbaI/NcoI digested PMusaVND6 to generate pCAMBIA1301- PMusaVND7:: GUS and pCAMBIA1301- PMusaVND6:: GUS respectively. The ligation reaction was confirmed by digestion and sequencing of recombinant binary vector. The sequences generated have been deposited in the NCBI database as MF429830 (for PMusaVND7) and MF429831 (for PMusaVND6).
DNA gel-shift assay (EMSA)
The SNBE-like sites identified from the PMusaVND7 and PMusaVND6were synthesized as complementary oligonucleotides and annealed together at 37°C after heating at 90°C in a thermal cycler machine. The oligonucleotides containing SNBE-like site from PMusaVND7 are 5’-AATTCGATAGCCTTTGGCGTGCACTCAAAGAAATTGGT-3’; and 5’-ACCAATTTCTTTGAGTGCACGCCAAAGGCTATCGAATT-3’(19bp SNBE-like site spanning from -186 to -167 is italicized and consensus resides are underlined). Complementary oligonucleotides with SNBE-like site from PMusaVND6 are 5’-AATTCGATAGCCTTAGTTGTAGGGCTCAAGATTTTGGT-3’ and 5’-ACCAAAATCTTGAGCCCTACAACTAAGGCTATCGAATT-3’ (19bp SNBE-like site spanning from -220 to -201 is italicized and consensus resides are underlined).The annealed oligonucleotides or the PCR amplified promoter fragments was incubated with either banana VND1, VND2 or VND3 protein in a EMSA buffer (10mM Tris-buffer, pH8, 5mM MgCl2 and 10mM KCl) for 20 minutes. The reaction was later resolved on a 1.5% agarose-gel to visualize the DNA-protein interaction. Mutated SNBE-like(mSNBE) oligonucleotides were annealed as mentioned above and utilized in EMSA reaction as discussed before. The sequences of oligonucleotides for mSNBE were 5’-CCTTCGGAAAGCACTCATTTAGGTTGG -3’ and 5’- CCAACCTAAATGAGTGCTTTCCGAAGG -3’(for -186 to -167 position SNBE-like site of PMusaVND7; consensus residues of SNBE site were mutated and are underlined). Sequence of oligonucleotides used for mutated SNBE –like site of PMusaVND6 were 5’-CCTTCGTAAAAGGGCTCTTTAGGTTGG-3’and 5’- CCAACCTAAAGAGCCCTTTTACGAAGG-3’ (consensus residues of SNBE site were mutated and are underlined).A 250bp DNA fragment (-287 to -37 upstream of transcription start site) spanning the SNBE-like site (-186 to -167) was PCR amplified from PMusaVND7 using oligonucleotides 5’-GAGCATGTGGAAACCATAGC-3’ and 5’-AAGAACATCCCATACTTGGAA-3’ and was utilized in gel shift assay with the VND1-VND3 proteins. Similarly a 224bp DNA fragment (-388 to -164 upstream of transcription start site) encompassing the SNBE-like sites in PMusaVND6 was PCR amplified (using oligonucleotides 5’-CATCACCTTCTGTGGTATTCTTC -3’ and 5’- AGAGGAGCAACAAGTGATTGA -3’)for gel shift assay with VND1-VND3 proteins. EMSA reaction with these DNA was carried out as discussed above.
Quantitative RT-PCR analysis
Transgenic banana cultivar overexpressing MusaVND1(KT356872),MusaVND2 (KP335047) or MusaVND3 (KP666170) have been described earlier by us[2,3]. Tissue of three independent plants was mixed in equal amount and then utilized for RNA isolation. Total RNA was isolated from the leaves and corm of the banana plant using Concert plant RNA reagent (Invitrogen, USA) as described in the manufacturer’s protocol. Total RNA was then cleaned using RNA binding column of RNeasy plant mini kit (Qiagen, Germany) and genomic DNA contamination was removed using on-column DNAase-digestion (Qiagen, Cat. No.79254) set as per the protocol supplied in the kit. Total RNA was analyzed on 1% agarose gel and then utilized for first strand cDNA synthesis using thermoscript AMV-RT (Invitrogen: Cat. No.12236-014).Sequence of the primers utilized are: FP 5’-AGGGATGGGTTGTGTGTAGG-3’ and RP 5’-TGCAGGTGGAGAAGATCTGG-3’ for MusaVND6 (GSMUA_Achr8T11590_001); FP 5’-AGAGCAAAGCGAGTGGTACT-3’ and RP 5’-ACCCATCCTTCTTCCTGAGC-3’ for MusaVND7 (GSMUA_Achr3T22360_001); FP 5’-CCGATTGTGCTGTCCTCATT-3’ and RP 5’-TTGGCACGAAAGGAATCTTCT-3’ for MusaEF1α (GSMUA_Achr6T02020_001). Transcript analysis was carried out on a rotor gene Q platform (Qiagen, Germany) using 2x SYBR Green Jump Start Taq Ready Mix (Sigma, USA) with 1:50 diluted cDNA. Reaction carried out under following conditions: 95°C(4min), 32-cycles of 95°C(15sec), 60°C(20sec), 72°C(20sec) ending with a melting curve analysis step. The Ct-values were normalized with the expression of housekeeping gene MusaEF1α and fold change in expression of MusaVND6 and MusaVND7 was calculated by comparative Ct method (2−ΔΔCt) as described earlier [14].
Transformation and generation of transgenic banana plants harboring either PMusaVND7:: GUS or PMusaVND6:: GUS
Binary vectors (pCAMBIA1301- PMusaVND7:: GUS and pCAMBIA1301- PMusaVND6:: GUS) were used for transformation of Agrobacterium strain EHA105 [15] by electroporation. The recombinant colonies of EHA105 were selected on LB-agar plate supplemented with kanamycin (50μg/ml) at 27°C in an incubator for 3 days. The recombinant EHA105 was confirmed by colony PCR for presence of the binary vector. Overnight grown Agrobacterium was cultivated to OD of 0.6 and then induced by acetosyringone for 3 hours. Induced Agrobacterium was utilized for transformation of embryogenic cell suspension of banana cultivar Rasthali as described previously [16]. Banana embryogenic cells (0.5mlpacked cell volume for each co-cultivation tube) were co-cultivated with induced Agrobacterium for 30 minutes in presence of acetosyringone. Complete co-cultivation medium was aspirated under pressure upon glass fiber filtersusing a filter assembly. The glass fiber filters with banana embryogenic cells were then cultured in dark for three days on M2-medium supplemented with acetosyringone. Further, the cells were scrapped from the filters and the transformed cells were then selected for hygromycin resistanceafter culturing on banana embryo development medium (BEM) supplemented with cefotaxime (400mg/l) and hygromycin (5mg/l) as described earlier [16]. Developing embryos were cultured on fresh BEM with cefotaxime (400mg/l) and hygromycin (5mg/l) every 3 weeks until well developed embryos were visible (usually after 3 months of culture). Well developed embryos with potential to transform into shootswere converted into banana shootby culturing on embryo germination medium supplemented with 6-benzylaminopurine (0.5mg/l BAP) [16]. Putatively transformed banana shoots were then multiplied on banana multiplication medium (MS-medium with 2mg/l BAP and 30mg/l adenine sulphate). Rooting of the banana shoots were carried on MS-medium supplemented with NAA (1mg/l). Well rooted banana plants were hardened in sterile soil in a contained green house facility. Genomic DNA of transgenic banana lines was isolated from leaf tissue with the help of GenElute Plant Genomic-DNA Miniprep-Kit (Sigma,USA) following the instructions provided with the kit. The genomic DNA was utilized for PCR analysis of T-DNA integration in these banana plants. PCR was carried out for amplification of promoter-GUS fusion (~3.2kb) and promoter alone (~1.2kb for PMusaVND7and ~1.1 kb for PMusaVND6). For promoter-GUS fusion amplification, forward primer of either PMusaVND6and PMusaVND7and a reverse primer (CACGTGGTGGTGGTGGTGGTGGCTA) in GUS coding region was utilized while for amplification of either PMusaVND6and PMusaVND7, primers utilized for their cloning were used. PCR running condition was 98°C(10min), 35-cycles of [98°C(1min), 58°C(1min), 72°C(3min)] followed by final extension of 72°C(20min).
GUS staining of the histological sections and GUS activity analysis
The activation of PMusaVND7 and PMusaVND6in different organ and tissue was carried out by staining for GUS (β-D-Glucuronidase) activity. GUS staining buffer was prepared as follows: 1mM X-Gluc (5-Bromo-4-chloro-3-indolyl Glucuronide), 0.1% triton X-100, 0.5mM potassium ferricyanide and 0.5mM potassium ferrocyanide prepared in 100mM sodium phosphate buffer with pH 7.0. GUS staining was carried out at 37°C for maximum of 12 hours. Before recording the observations, chlorophyll of green tissue was removed by repeated incubation in 70% ethanol at 37°C till satisfactory resultwas obtained. Image of GUS stained sections of different organ was observed and recorded in Eclipse 80i, Nikon microscope.Estimation of GUS activity was carried out as picomoles 4-MU per min per μg soluble protein according to protocoldescribed earlier using 4-methylumbelliferyl-b-D-glucuronide (MUG) and soluble extract of plant [17].
Transcriptional activation assay of PMusaVND7and PMusaVND6
The activation of PMusaVND7and PMusaVND6by VND1-VND3 was assayed in terms of GUS activity in banana embryogenic cells. Construct pCAMBIA1301- PMusaVND7:: GUS and pCAMBIA1301- PMusaVND6:: GUS generated above were utilized as reporter construct. MusaVND1, MusaVND2 and MusaVND3were cloned in pCAMBIA1302 in place of GFPutilizing the restriction sites NcoI and PmlI. Primers utilized are: FP 5’-AAATCATGAAATCGTGTGTTCCTCCT -3’ and RP 5’- AACACGTGTCATTGCTCAAATATACAGATGC-3’ for MusaVND1; FP 5’-AATCATGAAATCGTGTGTTCCTC -3’ and RP 5’-AACACGTGTCACTGCTCGAATATACAGATG -3’ for MusaVND2; FP 5’-AATCATGAATGCATTCCCTCATGT -3’ and RP 5’-AACACGTGTCATTTCCACAGTTCGACTT -3’ for MusaVND3(BspHI and PmlI sites underlined). PCaMV35S:MusaVND1, PCaMV35S:MusaVND2 andPCaMV35S:MusaVND3 served as effector constructs. Agrobacterium strain EHA105 harboring different reporter and effector constructswere used for transient co-transformation of embryogenic cells of banana cultivar Rasthali as described earlier [3]. The cells aspirated onto glass fiber filters were analyzed for the GUS activity after fifth day of transformation. Banana cells transformed with either of the reporter construct served as control. For each replication 0.5ml packed cell volume was used for transformation and complete cells on one filter was utilized for GUS estimation. The experiment was repeated atleast three times.
Results
Isolation and sequence analysis of PMusaVND7and PMusaVND6
Expression of both MusaVND6 and MusaVND7was elevated along with other genes during an invitro xylogenesis of banana embryogenic cell suspension in brassinolide supplemented medium indicating that they are important NAC transcription factors for tracheary element differentiation in banana [1]. 5-upstream region of the MusaVND6 (GSMUA_Achr8T11590_001) and MusaVND7 (GSMUA_Achr3T22360_001) were identified from the banana genome database (http://banana-genome-hub.southgreen.fr/) and primers were designed for PCR amplification of at least 1kb DNA region utilizing genomic DNA of banana cultivar Rasthali as template. These sequences were designated as PMusaVND7(1237 bp) and PMusaVND6(1143 bp) and were deposited in the NCBI database. Putative TATA box and transcription start site (+1 TSS) was determined by insilico analysis at TSSplant server (http://www.softberry.com/berry.phtml?topic=tssplant&group=programs&subgroup=promoter)Sequence analysis of either PMusaVND7 or PMusaVND6 revealed presence of at least two 19bp perfect SNBE-like sequences which were identified by the consensus sequence of SNBE site[(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T) (A/T)] reported earlier [10]. In PMusaVND7 first SNBE-like site (TGGCGTGCACTCAAAGAAA on plus strand; consensus residues are underlined) was positioned at -186 to -167 while the second SNBE-like site (TATCGTACGTAGTACGCTT on complementary strand; consensus residues are underlined) was positioned at -926 to -907 (S1 Fig). In case of PMusaVND6 first SNBE-like site was detected at -220 to -201 (AGTTGTAGGGCTCAAGATT on complementary strand; consensus residues are underlined) and second SNBE-like site was positioned at -372 to -353(TCCCTTTACACAGAAGAAT on complementary strand; consensus residues are underlined) relative to transcription start site (S2 Fig).
Banana VND1-VND3 proteins bind to PMusaVND7 and PMusaVND6
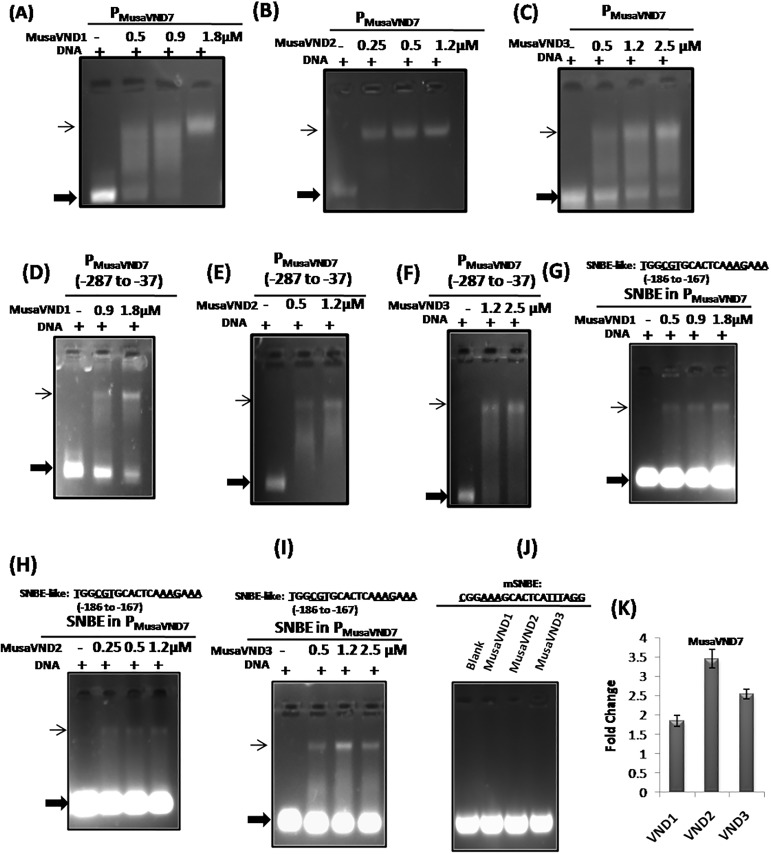
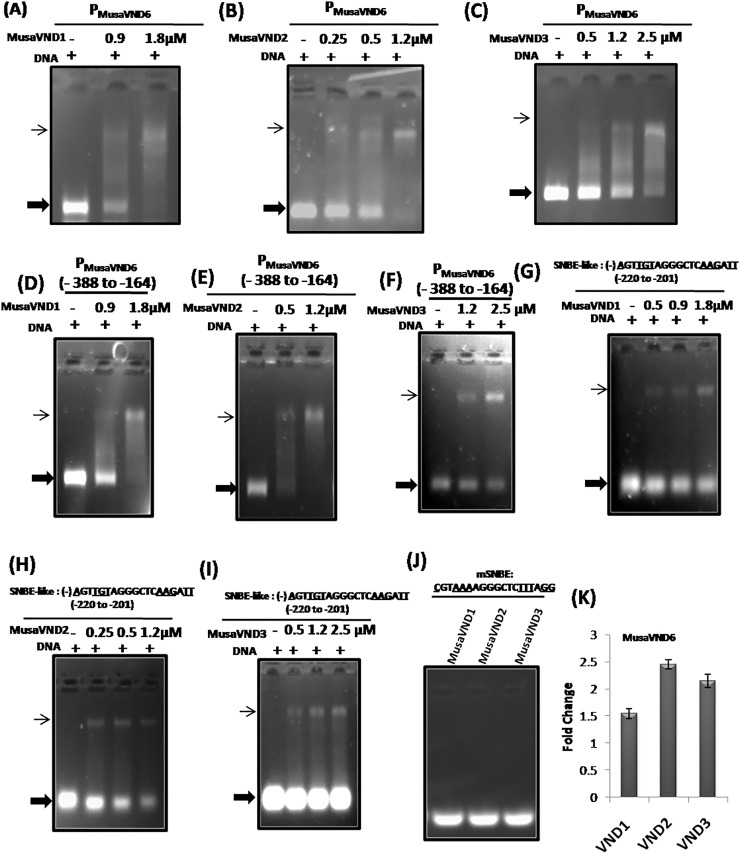
Vascular related NAC domain containing proteins(VNDs) are reported to regulate the expression of genes by binding to the SNBE-motifs in the promoter region[10]. Moreover, Arabidopsis VND1-VND7 proteinsare shown to bind SNBE site in isolation or as a part of ArabidopsisVND7 promoter for its potential regulation [12]. We also analyzed whether banana VND proteins have similar potential to recognize the SNBE site of PMusaVND7 and PMusaVND6in a gel shift assay. For this a 1.2 kb 5’upstream region of banana VND7 was incubated with different concentration of banana VND1-VND3 proteins in appropriate reaction condition and the interaction was analyzed after resolving on a gel. Mobility of PMusaVND7 in gel shift assay was retarded by all the three banana VNDs (VND1, VND2 and VND3) proteinsuggesting that PMusaVND7 is a potential target gene for regulation by banana VND1- VND3 (Fig 1A, 1B and 1C). Similarlyfor analyzing whether, banana VND1- VND3 could recognize and bind the 5’upstream region of banana VND6, a 1.1kb region of PMusaVND6was incubated with different concentration of either banana VND1, VND2 or VND3 protein in EMSA buffer and the interaction was visualized after resolving the reaction on a gel. Our results indicated that banana VND1- VND3 could retard the mobility of a 1.1kb region of PMusaVND6indicating that PMusaVND6is also a potential gene regulated by banana VND1- VND3 (Fig 2A, 2B and 2C). We also checked whether banana VND1-VND3 could retard a small fragment of PMusaVND7 and PMusaVND6withSNBE-like site.Results indicated that VND1-VND3 were able to retard a 250bp PMusaVND7 fragment (-287 to -37 relative to transcription start site) containing the SNBE–like site present at -186 to -167 (Fig 1D, 1E and 1F). Similarly the mobilityof a 224bp PMusaVND6 fragment (-388 to -164 relative to transcription start site) encompassing the SNBE–like sites was retarded by banana VND1- VND3 protein in gel shift assay (Fig 2D, 2E and 2F). These results suggested that PMusaVND7 and PMusaVND6 are potential target genes for regulation by VND1-VND3.
Fig 1. Regulation of PMusaVND7by banana VND1-VND3.
(A-C) Retardation of PMusaVND7(approximately 1.2 kb region) by banana VND1- VND3 protein. The amount of protein utilized is indicated on the top of the figure. + sign indicates the presence of equal amount of DNA in each reaction. (D-F) Retardation of a 250 bp PMusaVND7 fragment (-287 to -37 relative to transcription start site) containing the SNBE–like site by banana VND1- VND3 protein. The amount of protein utilized is indicated on the top of the figure. + sign indicatesthe presence of equal amount of DNA in each reaction.(G-I) SNBE–like site in the PMusaVND7 (-186 to -167) was used for gel shift analysis with banana VND1- VND3 protein. The conserved residues in the SNBE site (19bp) are underlined. The SNBE-like site was retarded in the gel by VND1- VND3 protein. Thin arrow indicates the retarded DNA as protein-DNA complex while thick arrow indicates the unbound DNA. (J) Banana VND1-VND3 failed to bind the mutated SNBE-like site (mSNBE). The mutated residues in SNBE –like site are underlined. (K) Transcript level of MusaVND7 in the corm tissue of the transgenic banana overexpressing either banana VND1, VND2 or VND3. Fold change in control was kept at one and the data was normalized by the expression of banana EF1α.
Fig 2. EMSA analysis of PMusaVND6 with banana VND1-VND3.
(A-C) The PMusaVND6(~1.2 kb) was amplified from genomic DNA of banana cultivar Rasthali and incubated with the indicated amount of the protein (either banana VND1, VND2 or VND3). The amount of the retarded DNA (thin arrow) increased with the increasing amount of the protein. Presence of equal amount of DNA in each reaction is indicated by +signs. (D-F) Retardation of a 224 bp PMusaVND6 fragment (-388 to -164 relative to transcription start site) containing the SNBE–like sites by banana VND1- VND3 protein. The amount of protein utilized is indicated on the top of the figure. + sign indicates the presence of equal amount of DNA in each reaction. (G-I) SNBE–like site (-220 to -201) detected in the PMusaVND6was retarded in a gel shift assay by VND1- VND3 protein. The minus sign indicates the presence of SNBE site on the complementary strand. Thin arrow indicates the retarded DNA as protein-DNA complex while thick arrow indicates the unbound DNA. (J) Banana VND1-VND3 failed to bind the mutated SNBE-like site (mSNBE). The mutated residues in SNBE –like site are underlined. (K) Quantitative RT-PCR analysis of MusaVND6 in the corm tissue of transgenic banana overexpressing either of banana VND1, VND2 or VND3. Fold change in control was kept at one and the data was normalized by the expression of banana EF1α.
Banana VND1-VND3 proteins binds to SNBE-like sequences of PMusaVND7 and PMusaVND6
As SNBE-like sitewas detected in both PMusaVND7 and PMusaVND6and mobility of PMusaVND7 and PMusaVND6was retarded by VND1- VND3, we analyzed whether VND1- VND3 could bind to these SNBE-like sites. A 38bp double stranded DNA containing SNBE-like site of PMusaVND7 (TGGCGTGCACTCAAAGAAA positioned at -186 to -167) was incubated with either banana VND1- VND3 protein and the interaction was analyzed on a gel. All the three VND1- VND3 strongly retard the mobility of SNBE-like site of PMusaVND7 indicating that banana VND1- VND3 recognize the SNBE-site in PMusaVND7(Fig 1G, 1H and 1I). Similarly the SNBE-like site in PMusaVND6was also bound by banana VND1- VND3 in a gel shift assay indicating that banana VND1- VND3 bindSNBE-like site in PMusaVND6(Fig 2G, 2H and 2I). However, banana VND1-VND3 failed to bind a mutated version of the SNBE-likesite (mSNBE where the consensus residues of SNBE site were mutated) in PMusaVND7 and PMusaVND6 indicating that VND1-VND3 specifically interact with PMusaVND7 and PMusaVND6 and the interaction is mediated by SNBE-like sites (Figs 1J and 2J).
Overexpression of banana VND1-VND3 induce MusaVND6 and MusaVND7
As the banana VND1- VND3 could bind the ~1.2kb of PMusaVND7 and PMusaVND6 as well as SNBE-like site within them, suggesting expression of these genes may be altered after overexpression of either VND1, VND2 or VND3. We analyzed the expression of MusaVND6 and MusaVND7 in leaves and corm tissue of transgenic plants overexpressing either banana VND1-VND3 by quantitative RT-PCR [2,3]. Results indicated that expression of both MusaVND7 and MusaVND6 was elevated in corm tissue of transgenic banana plants overexpressing either banana VND1, VND2 or VND3 (Figs 1K and 2K). These results suggested that banana VND1-VND3 regulate the expression of banana VND6-VND7 through SNBE-like motifs.
Banana VND1-VND3 activate expression from PMusaVND7 and PMusaVND6
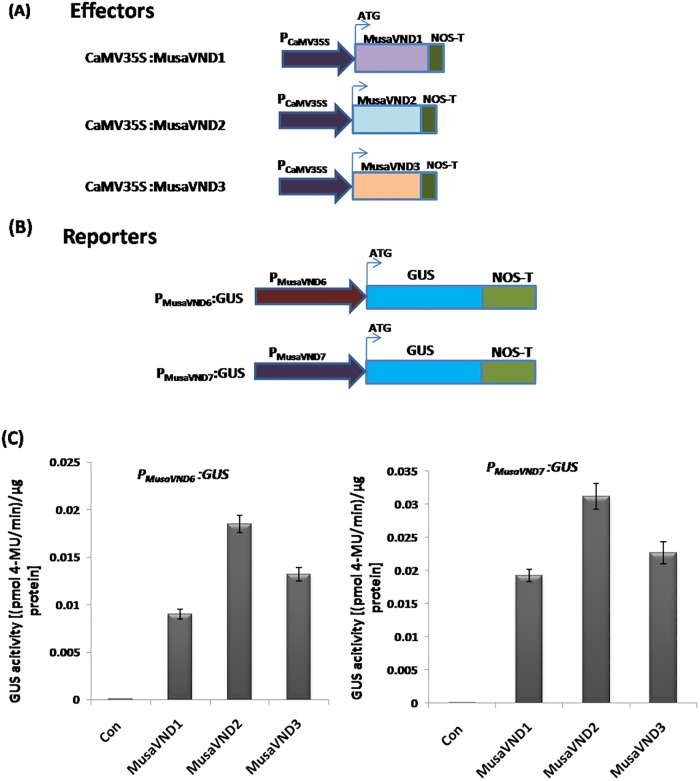
To test whether the elevated transcript levels of MusaVND6 and MusaVND7 in transgenic banana overexpressing VND1-VND3 is due to transcriptional activation of PMusaVND7 and PMusaVND6 by VND1-VND3, we performed a transient activation assay using GUS as reporter gene. Effector constructs contain CaMV35S promoter driving the expression of VND1-VND3 (Fig 3A). Reporter constructs harbor PMusaVND7 and PMusaVND6 fused upstream of GUS coding sequence (Fig 3B).Different combination of effector and reporter constructs were utilized for analyzing the transcriptional activation of PMusaVND7 and PMusaVND6 by VND1-VND3 and the GUS activity was estimated after fifth day of the transient co-transformation. Data obtained indicated that transformation of banana embryogenic cell suspension with reporter and effector constructs (containing VND1-VND3) remarkably elevated the GUS activity (Fig 3C). Cells transformed with reporter alone showed negligible GUS activity as PMusaVND7 and PMusaVND6are inactive in embryogenic cells of banana.
Fig 3. Transcriptional activation assay.
VND1-VND3 induce the VND6 and VND7 promoter. (A) Diagrammatic representation of the effector constructs used for the assay. The VND1-VND3 coding sequence were driven by the CaMV35S promoter. (B) Different reporter constructs employed for the assay. PMusaVND7 and PMusaVND6 fused upstream of GUS in pCAMBIA1301 served as reporter constructs. (C) GUS activity obtained after fifth day of transient activation assay. Data was represented as mean±SD of three replication and represented as (pmol 4-MU/min)/μg protein.
Generationand molecular analysis of banana plants harboring either PMusaVND7::GUS, PMusaVND6::GUS
Vascular related NAC domain genes are regulator of secondary wall deposition and xylem element development and VND1-VND3 regulate the expression of banana VND6-VND7, we analyzed the tissue specific activation of PMusaVND7 and PMusaVND6. Both PMusaVND7 and PMusaVND6 were cloned upstream of GUS in pCAMBIA1301 and transgenic banana plants harboring either PMusaVND7::GUS, PMusaVND6::GUS were generated. Agrobacterium harboring binary vectorpCAMBIA1301- PMusaVND7:: GUS and pCAMBIA1301- PMusaVND6:: GUSwas used for transformation of banana embryogenic cells of cultivar Rasthali and transformed cells were selected in culture medium supplemented with 5mg/l hygromycin (S3A and S4A Figs). Growth of transformed cells on embryo development medium resulted in emergence of white and opaque somatic embryos (S3B and S4B Figs). Developing somatic embryos showed different stageof development like globular to torpedo shaped stage and further resulted in emergence of secondary embryos indicating appropriate development pathway toward generation of banana plants (S3C and S4C Figs). Transgenic banana plants were cultured on banana multiplication medium for clonal propagation (S3D and S4D Figs) and individual shoots were separated and rooted on rooting medium (S3E and S4E Figs). Appropriately rooted banana plants harboring either PMusaVND7::GUS, PMusaVND6::GUSwas hardened in a contained green house for further analysis (S3F and S4F Figs).The T-DNA insertion in these transgenic banana plants was analyzed by PCR amplification of either PMusaVND7-GUS fusion or PMusaVND6-GUS fusion and PMusaVND7 or PMusaVND6from the leaf genomic DNA. PCR analysis showed positive amplification of promoter-GUS fusion product and promoter alone indicating successful integration of PMusaVND7::GUSandPMusaVND6::GUS in genome of these banana plants (S5A and S5B Fig).These plants were then analyzed for tissue specific activity of GUS controlled by activation of PMusaVND7 or PMusaVND6.
Xylem specific activation of 5’-upstream regulatory region of MusaVND7
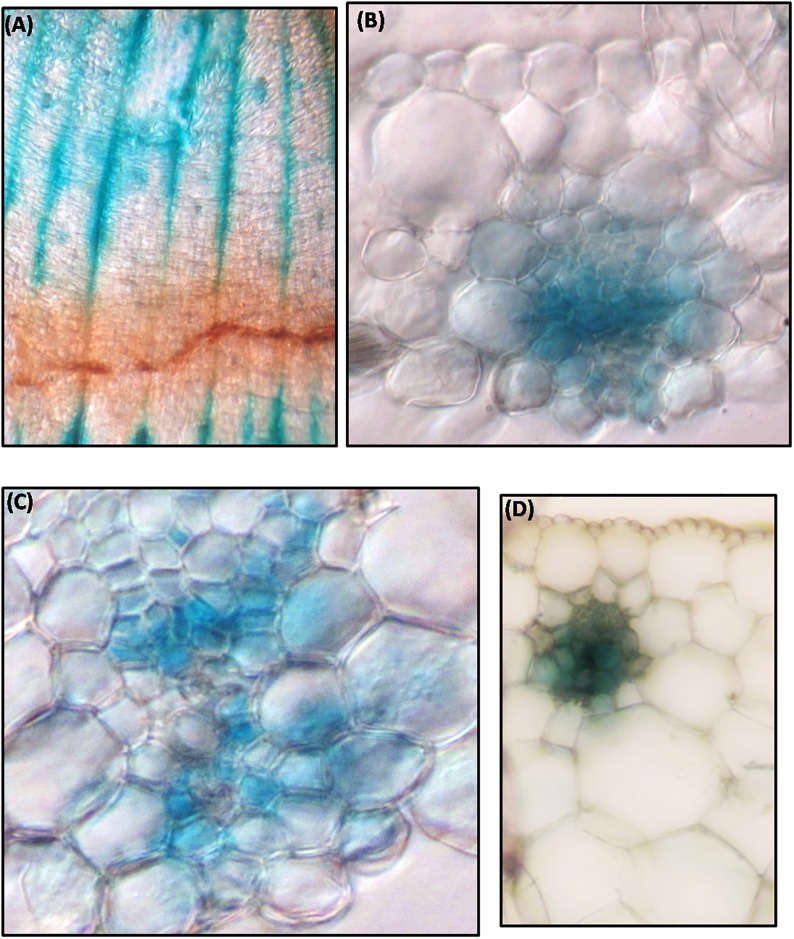
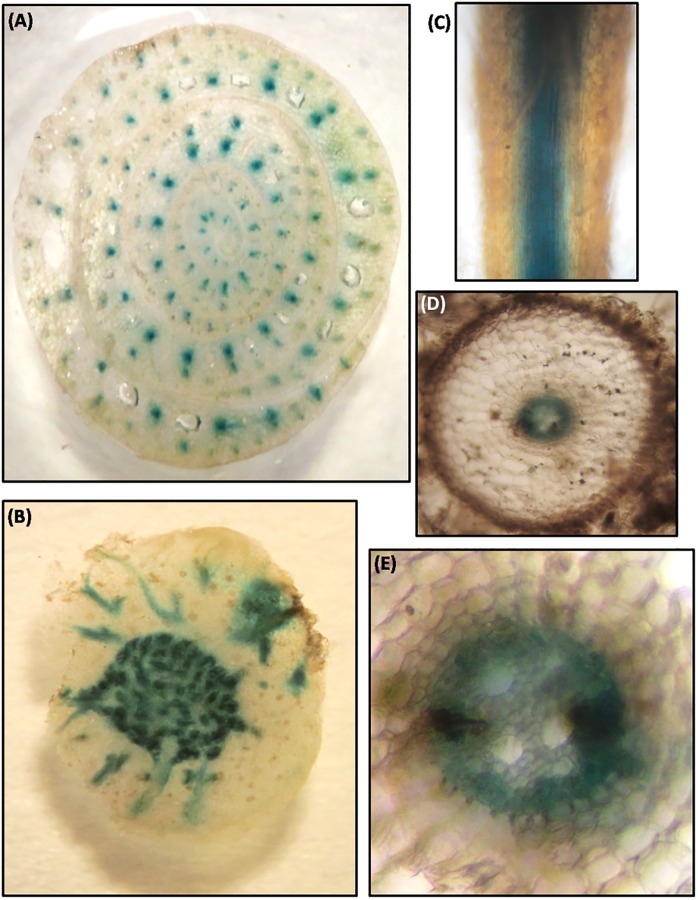
Tissue specific activation of PMusaVND7 was ascertained by analyzing the expression of GUSin banana plants harboring the PMusaVND7::GUS. At least three independent transgenic lines were analyzed and all of them showed similar GUS expression pattern and representative resultwas shown. GUS staining of leaf tissue showed GUS expression in longitudinal veins as well as in commissural veins (Fig 4A). Analysis of transverse section of the leaf suggested prominent GUS expression in vascular bundles while other tissues do not show any GUS staining indicating very specific activation of PMusaVND7 in xylem tissue (Fig 4B). Close-up of one of the leaf vascular bundle showed evidently strong GUS expression in xylem vessels and surrounding tracheary elements (Fig 4C). Similarly transverse section of the petiole was analyzed which also indicated vascular specific activation of PMusaVND7 as GUS expression was precisely observed in vessels and tracheids (Fig 4D). In pseudostem, GUS expression was observed in all the vascular bundle of different leaf sheath and moreover, the GUS expression appeared to increase from inner to outer sheath indicating thatPMusaVND7 activation increased with age suggesting important role of MusaVND7 in vascular tissue development (Fig 5A). In corm, specific and intense GUS activity was observed in vascular strands of the central stele region as well as in the vascular strands traversing from central vascular cylinder to the outwards growing roots (Fig 5B). GUS staining of the whole roots displayed GUS expression in the central cylinder (Fig 5C and 5D) and in cross section, the GUS expression appeared to predominate in the protoxylem and metaxylem vessels as well as nearby tracheids (Fig 5E). These results indicated that MusaVND7 showed xylem specific expression and is a probable important regulator of xylem development.
Fig 4. Expression analysis of GUS under the control of PMusaVND7 in leaves and petiole.
(A) GUS staining observed in banana leaf due to activity of PMusaVND7. (B) Transverse section of GUS stained leaf showing prominent and specific GUS staining in cells of the vascular bundle. Note the absence of staining in other cells. (C) Close-up of one of the leaf vascular bundle showing GUS staining in vessels and tracheids of the xylem. (D) Transverse section of the petiole indicating the vascular specific activity of the MusaVND7 promoter. Note the GUS activity in the central vessel cells and surrounding tracheary elements.
Fig 5. Activity of MusaVND7 promoter in pseudostem, corm and roots of banana.
(A) Cross section of the pseudostem demonstrating the vascular bundle specific activation of the PMusaVND7. Note the absence of GUS staining from tissues other than vascular bundles of the pseudostem. (B) Activity of MusaVND7 promoter in corm of the banana. PMusaVND7got activated only in vascular tissues. Note the specific and intense GUS activity in vascular strands of the central cylinder. Also note the GUS activity in the vascular strands traversing from central vascular cylinder to the outwards growing roots. (C) GUS staining observed in the central vascular cylinder in the roots of banana harboring PMusaVND7:: GUS. (D) Cross section of the GUS stained root. (E) Close-up view of the central vascular cylinder showing activation of PMusaVND7in protoxylem as well as metaxylem vessels and surrounding tracheids.
5’-upstream sequence of MusaVND6 is active in vascular tissues
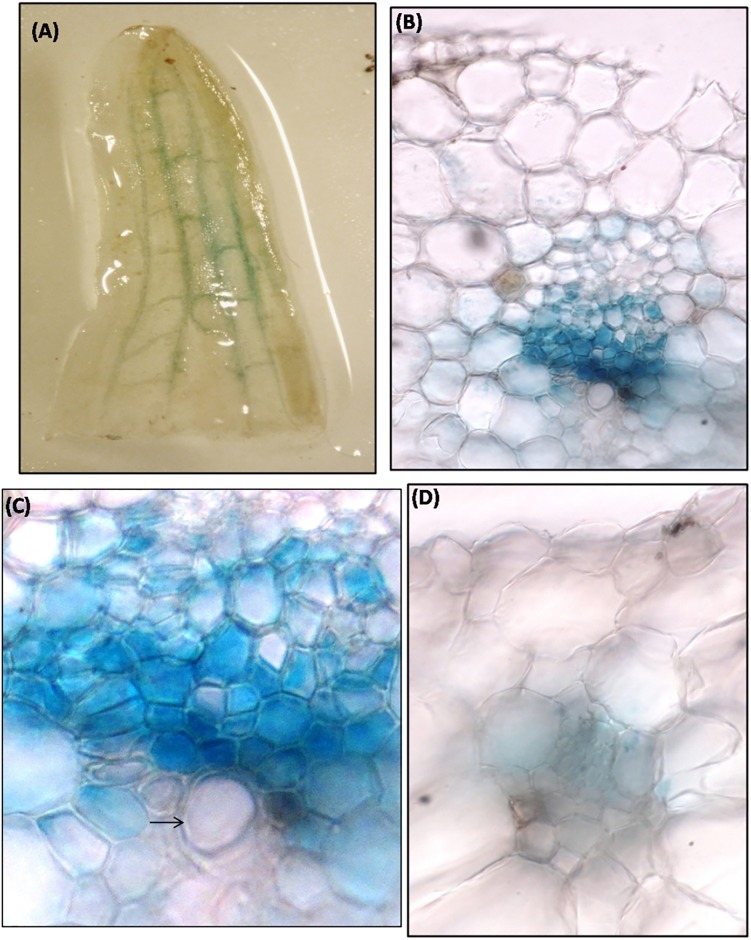
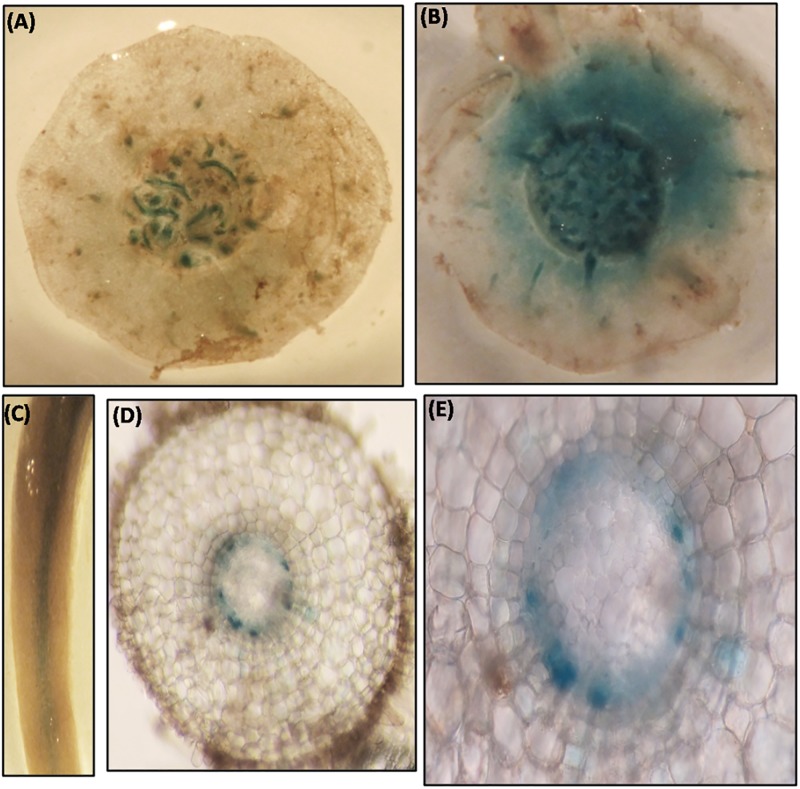
Transgenic banana harboring PMusaVND6::GUS were generated to analyze the tissue specific expression pattern of MusaVND6. Three independent transgenic lines showed similar GUS expression pattern and representative images were recorded. GUS expression in leaves was specifically observed in longitudinal veins (Fig 6A) which was confirmed by GUS staining of transverse section which showed explicit and strong GUS expression in vascular bundles with no GUS expression in epidermal or cortical cells (Fig 6B). In leaves vascular bundle, the GUS expression was prominent in tracheary elements surrounding the xylem vessel (Fig 6C). GUS staining of the petiole showed activation of PMusaVND6 in cells of vascular bundle while it was undetectable in other tissue typeindicating important role of MusaVND6 in xylem development (Fig 6D). In pseudostem, activation of MusaVND6 promoter was observed in central cylinder, and scattered vascular bundles of leaf sheaths (Fig 7A). In corm the activation of PMusaVND6 was observed in vascular bundles of central stele however, GUS staining was also observed in cells of the cortical region suggesting a probable differential activation of MusaVND6 in different organs (Fig 7B). In roots the GUS expression was observed in central vascular region (Fig 7C and 7D) however, the GUS expression was prominent in peripheral protoxylem elements while it is absent from the central metaxylem region (Fig 7E). These results suggested that MusaVND6 have important role in xylem development.
Fig 6. Analysis of activation of PMusaVND6in leaf and petiole of transgenic banana harboring PMusaVND6:: GUS.
(A) GUS staining observed in veins of banana leaf due to activation of PMusaVND6. (B) Transverse section of the leaf showing specific activation of the PMusaVND6 in cells of the vascular tissue. Note the absence of the staining from the other tissue of the leaf. (C) Close-up of the leaf vascular bundle showing the GUS activity in the tracheids surrounding the vessel element (indicate by black arrow). (D) GUS staining in the petiole of banana. Note the GUS staining in vascular bundle.
Fig 7. GUS activity in the pseudostem, corm, and roots of the banana harboring PMusaVND6:: GUS.
(A) Transverse section of the pseudostem displaying the GUS activity in the vascular strands. (B) GUS staining observed in the corm of the banana. (C) Root of transgenic banana harboring PMusaVND6:: GUS showing GUS activity in the central vascular region. (D) Transverse section of the root showing specific GUS staining in the central vascular region. (E) Close-up of central vascular cylinder. Note the GUS staining in the peripheral protoxylem elements while it is absent from the central metaxylem region.
GUS activity under the control of PMusaVND6andPMusaVND7
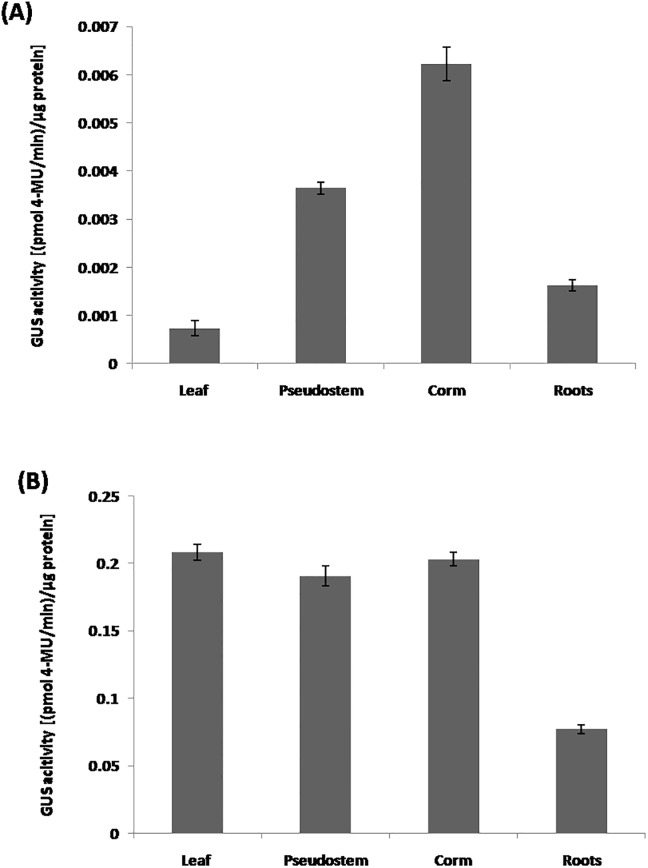
We estimated the GUS activity in various organs of banana harboring either PMusaVND6:: GUS or PMusaVND7:: GUS to quantitate the tissue specific activation of PMusaVND6and PMusaVND7. PMusaVND6and PMusaVND7 could drive the GUS expression in all the organs of the banana albeit at different level suggesting qualitative difference in the activity of these promoter in banana organs. PMusaVND6drives highest GUS expression in banana corm and pseudostem followed by roots and leaves (Fig 8A). While the extent of GUS activation by PMusaVND7 was almost similar in leaves, pseudostem and corm of transgenic banana and lower activityof GUS was observed in root tissue compared to other organs(Fig 8B).
Fig 8. GUS activity in banana due to PMusaVND6:: GUS or PMusaVND7:: GUS.
(A) GUS estimation in different organ of transgenic banana transformed with PMusaVND6:: GUS. (B) GUS activity estimated by fluorometric analysis in different organ of banana harboring PMusaVND7:: GUS. Data was represented as mean±SD of three replication and represented as (pmol 4-MU/min)/μg protein.
Discussion
Modifications of secondary cell wall including reduction of lignin especially in plants important for biofuel and pulp production holds great importance. The efficient conversion of lignocellulosic biomass (chiefly made up of secondary cell wall) into ethanol through fermentation process is deterred by high lignin content [18]. Hence reduction of lignin and increased deposition of hydrolysable cellulose through elevated deposition of secondary cell wall can be achieved byNAC transcription factors and other potential genetic factors. Moreover, many important crop plants like banana are prone to lodging during fruiting stage (due to heavy bunch weight), which can be improved through increased mechanical strength of pseudostem by elevatingsecondary cell wall deposition [19,20]. Information on regulation of secondary wall thickening especially in context of VNDs in plants like Arabidopsis, poplar and Brachypodium among otherhave been generated suggesting that these transcription factors regulate a number of downstream genes for coordinated regulation of secondary cell wall deposition [21–24]. In Arabidopsis seven members have been included in VND subgroup of NAC family and VND6 and VND7are suggested to be master regulators for secondary wall synthesis [5,6]. Attempts to overexpress these genes for secondary cell wall modification have often resulted in unwanted effect of biomass reduction probably through transdifferentiation of essential tissue into tracheids as these genes were overexpressed under constitutive promoter [5,7]. Hence, identification and characterization of potential xylem specific regulatory DNA element for regulated expression of VNDs is indispensable [18]. In this study we have characterized two 5’upstream regulatory region (PMusaVND7 and PMusaVND6) with specific activation in xylem tissue element which can be utilized in future for biomass engineering. Some examples of utilization of xylem specific promoter element have been reported in the past. In Arabidopsis, the expression of C4H (cinnamate 4-hydroxylase), an important gene for lignin synthesis was restricted under the activation of VND6 promoter (a vessel specific promoter) which reduced the lignin content (without plant growth reduction), thus increasing the hydrolysis of cellulose for sugar release [18]. Another study has reduced lignin content through expression of bacterial 3-dehydroshikimate dehydratase (QsuB) under the PC4Hand emphasized the need of fiber cell specific promoter for higher stringency in lignin reduction [25]. Moreover, studies on genetic factors regulating secondary wall deposition in banana are important due to the fact that banana has recently being researched as a potential second generation biofuel crop due to high amount of lignocellulosic biomass being left out after the fruit harvest [26–28]. Quantitative RT-PCR analysis to determine the transcript levels of VND6 and VND7 in native banana indicated differential expression of MusaVND6 and MusaVND7 in different organs. Transcript levels of both VND6 and VND7was high in corm and pseudostem compared to other organs (S6 Fig). Though the transcript level of VND7was higher than VND6 in different organ, but it does not correlate with the relative GUS activity observed in transgenic banana harboringPMusaVND7::GUS and PMusaVND6::GUS. Lower GUS activity in transgenic banana harboring PMusaVND6::GUS than transgenic banana harboring PMusaVND7::GUS might be due to reasons such as integration of T-DNA in less active chromatin regions or requirement of additional upstream DNA element for achievement of full potential inexpression of VND6. In the present study we have successfully analyzed the xylem specific activity of PMusaVND6and PMusaVND7.
Expression pattern of the seven VNDgenes during tracheary element differentiation was different suggesting that these genes may function in different aspects of tracheid development [1,6]. Multiple secondary wall associated transcription factors like VNDs and SND1 regulate downstream genes by binding to a 19bp imperfect palindrome known as SNBE-like motif and its consensus sequence was worked out to be (T/A)NN(C/T) (T/C/G)TNNNNNNNA(A/C)GN(A/C/T) (A/T) suggesting a common mode of regulation of a set of genes involved in tracheids differentiation [10]. ArabidopsisSND1 and VND7 have been shown to regulate the expression of multiple class of genes like MYB transcription factors (MYB46, MYB83, MYB58/72, MYB48/59, MYB52 and MYB85), cell wall associated genes like IRX genes (irregular xylem) and programmed cell death related genes like XCP1/2 (xylemcysteine peptidase) through SNBE-like motifs [29–31]. Cross regulation of VND genes is relatively unknown, however, a recent work on analysis of SNBE-like motif in promoter of ArabidopsisVND7 and its regulation by VND1-VND7 proteins suggested potential regulation of VND7throughSNBE-like cis-elements besides other known and unknown mechanisms [12]. Some studies on regulation of VND7 have been carried out suggesting multiple level of regulation of its expression.VND-INTERACTING2 (VNI2), is a transcriptional repressor which binds with VND proteins including VND7 protein and repress the expression of the secondary wall associated genes directly controlled by VND7 [32]. Amount of Arabidopsis VND7 in cells is regulated by proteasome mediated degradation as accumulation of VND7 protein was observed upon treatment of cells with MG-132,which is a potent proteasome inhibiting chemical [33]. Two ASYMMETRIC LEAVES2 (AS2)/LATERALORGANBOUNDARIESDOMAIN (LBD) gene family members ASL19/LBD30 and ASL20/LBD18were reported to regulate the expression of VND6 and VND7 via a positive feedback loop as the expression of ASL19/LBD30 and ASL20/LBD18 is dependent on VND6 and VND7 and plants expressing PASL20:ASL20-SRDX showed reduced expression of VND6 and VND7 [34]. Recently other genetic factors like GATA5, GATA12, ANAC075 LBD15/ASL11are shown to induce the VND7 promoter suggesting additional regulations over VND7 expression [12].
Our work showed that banana VND6 and VND7 expression is positively regulated by banana VND1- VND3 protein through direct binding withSNBE-like motif in the PMusaVND7 and PMusaVND6.Our report is in line with study on ArabidopsisVND7 wherein its promoter was identified and induced by VND1-VND7 proteins [12] suggesting cross regulation of VND genes among themselves. In our previous work, we had successfully purified banana VND1-VND3 proteins and demonstrated their regulation over multiple downstream target genes including MYBtranscription factors and IRX genes by binding to SNBE-like sites in their regulatory region [35]. Our work also identified presence of SNBE-like sites in the regulatory regions of banana VND6 and VND7 prompting us to analyze whether these genes can also be regulated by other banana VNDs. Here we show that banana VND1- VND3 could also bind to PMusaVND7 and PMusaVND6 as well as SNBE-like motif in them using a agarose based gel shift assay [36]. The binding of the VND1-VND3 get abolished by mutating the SNBE consensus residues indicating that the interaction of these three transcription factors with the PMusaVND7 and PMusaVND6 is specific and is governed by the presence of SNBE-like motifs. Elevated transcript levels of VND6 or VND7was detected in the corm but not in leaves tissue of transgenic banana overexpressing either VND1, VND2 or VND3. Moreover results obtained from transcriptional activation assay indicated that banana VND1-VND3 could activate transcription from the PMusaVND6 and PMusaVND7 confirming that these transcription factors not only bind but also could activate the expression of both MusaVND6 and MusaVND7.
Present work showed the specific activation of PMusaVND6 and PMusaVND7 in sclerenchymatous cells (xylem tissue) suggesting that these promoters are inactive in non-sclerenchymatous cells. Overexpression of VND1-VND3 in banana transdifferentiate many non sclerenchymatous cells into sclerenchymatous type and our previous work has indicated that compared to other organs, corm tissue showed remarkably higher transdifferentiation [2,3]. Moreover, banana corm contain abundant xylem tissue compared to other organs suggesting that elevation of VND6 and VND7 in VND1-VND3 overexpression plants probably occurred specifically in native sclerenchymatous as well as transdifferentiated sclerenchymatous cells of transgenic corm. Besides, there is a possibility of tissue specific regulation of VND6 and VND7 by VND1-VND3 as suggested by differential up regulation in transgenic banana overexpressing VND1-VND3. Previous work on regulation of ArabidopsisVND7 by VND1-VND7 through SNBE motifs has suggested possible presence of some unknown inhibitory factors regulating the expression of VND7 and possibly its transcript stability in non-sclerenchymatous cells of plants overexpressing VND1-VND7 and thus restricting its expression [12]. Further studies on identification of these possible inhibitors and the probable differential regulation of VND6-VND7 transcript are warranted.Besides studies on regulation of transcript levels of VND6 and VND7 due to expression of VNDs in non-xylem tissue need to be carried out in detail. Nonetheless this study identified two potential xylem specific promoter regionswhich will be useful for crop improvement and presented an interesting mechanism for regulation of VND6 and VND7expression in banana.
Supporting information
Sequence of 5’ upstream regulatory region of MusaVND7. The PMusaVND7was amplified from genomic DNA of Musa cultivar Rasthali and analyzed for the presence of SNBE-like sites. SNBE–like sites in the PMusaVND7are boxed in green. Putative TATA box is shown in red and the transcription start site (+1 TSS) is underlined.
(PDF)
Sequence of 5’ upstream regulatory region of MusaVND6 from Musa cultivar Rasthali. TATA box is boxed in red and transcription start site (+1 TSS) is indicated by an underline. SNBE–like sites detected in the sequence are boxed in green.
(PDF)
(A) Embryogenic cells of banana cultivar Rasthali transformed with pCAMBIA1301- PMusaVND7:: GUS and growing on banana embryo development medium supplemented with hygromycin (5mg/l). (B) Emergence of putatively transformed embryos on hygromycin supplemented medium after 2 months of growth. (C) Close-up of embryos showing growth in various stages of development as well as emergence of secondary embryos. (D) Multiplication of transgenic shoots on banana shoot multiplication medium. (E) Rooting of transgenic banana plants on banana rooting medium containing NAA(1mg/l). (F) Rooted banana plants were hardened in a green house for GUS analysis.
(PDF)
(A) Embryogenic cells of banana cultivar Rasthali growing on banana embryo development medium supplemented with hygromycin (5mg/l) after transformation with pCAMBIA1301- PMusaVND6:: GUS. (B) White and opaque embryos developed from continuous culturing of the transformed embryogenic cells on embryo development medium. (C) Developing embryos appeared globular to torpedo shaped in close-up. (D) Transformed banana shoots were multiplied on shoot multiplication medium to generate multiple lines. (E) Rooting of transgenic banana plants on banana rooting medium containing NAA(1mg/l). (F) Rooted banana plants were hardened in a green house for GUS analysis.
(PDF)
(A) PCR analysis of banana plants harboring PMusaVND7::GUS. (B) PCR analysis of banana plants harboring PMusaVND6::GUS. Two different primer pairs were utilized in PCR analysis. 1: FP in either PMusaVND7 or PMusaVND6 and RP in GUS; 2–4:FP and RP in either PMusaVND7 or PMusaVND6 2–4: analysis of three independent transgenic banana lines. M: 1 KB DNA ladder. 1kb and 3 kb band size are indicated. (FP: forward primer; RP: reverse primer).
(PDF)
Transcript level of MusaVND6 and MusaVND7 in tissue of different organs of wild type banana was analyzed by quantitative RT-PCR. Expression of VND6 and VND7 in different tissue types is shown after normalization of the data by the expression of banana EF1α. Data was represented as mean±SD of three replications.
(PDF)
Acknowledgments
Authors thank, Head, Nuclear Agriculture and Biotechnology Division, BARC for his constant encouragement
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Department of Atomic Energy, Govt. of India funded the study. The funder had no role in study design, data collection and analysis, decisionto publish, or preparation of the manuscript.
References
- 1.Negi S, Tak H, Ganapathi TR (2015) In vitro xylem vessel elements formation from banana embryogenic cells and expression analysis of vessel development-related genes. Plant Biotechnol Rep 9:47–54 [Google Scholar]
- 2.Negi S, Tak H, Ganapathi TR (2015) Cloning and functional characterization of MusaVND1 using transgenic banana plants. Transgenic Res. 24: 571–85 doi: 10.1007/s11248-014-9860-6 [DOI] [PubMed] [Google Scholar]
- 3.Negi S, Tak H, Ganapathi TR (2015) Functional characterization of secondary wall deposition regulating transcription factors MusaVND2 and MusaVND3 in transgenic banana plants. Protoplasma doi: 10.1007/s00709-015-0822-5 [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, Zhong R, Ye ZH (2014) Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9:e105726 doi: 10.1371/journal.pone.0105726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi M, Goué N, Igarashi H, Ohtani M, Nakano Y, Mortimer JC et al. (2010) VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153:906–914 doi: 10.1104/pp.110.154013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J et al. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 16: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 doi: 10.1105/tpc.106.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong R, Richardson EA, Ye ZH (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225:1603–1611 doi: 10.1007/s00425-007-0498-y [DOI] [PubMed] [Google Scholar]
- 9.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulates secondary wall thickening and are required for anther dehiscence. Plant Cell 17: 2993–3006 doi: 10.1105/tpc.105.036004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong R, Lee C, Ye ZH (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant. 3:1087–103 doi: 10.1093/mp/ssq062 [DOI] [PubMed] [Google Scholar]
- 11.Ohashi-Ito K, Oda Y, Fukuda H (2010) Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22:3461–73 doi: 10.1105/tpc.110.075036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endo H, Yamaguchi M, Tamura T, Nakano Y, Nishikubo N, Yoneda A et al. (2015) Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56:242–54 doi: 10.1093/pcp/pcu134 [DOI] [PubMed] [Google Scholar]
- 13.Ona T, Sonoda T, Ito K, Shibata M, Tamai Y, Kojima Y et al. (2001) Investigation of relationships between cell and pulp properties in Eucalyptus by examination of within-tree property variations. Wood Sci Technol 35:229–243 [Google Scholar]
- 14.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–8 doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 15.Hood EE, Gelvin SB, Melchers LS, Hoekama A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2:208–218 [Google Scholar]
- 16.Ganapathi TR, Higgs NS, Balint Kurti PJ, Arntzen CJ, May GD et al. (2001) Agrobacterium mediated transformation of embryogenic cell suspensions of the banana cultivar Rasthali (AAB). Plant Cell Rep. 20:157–162 [DOI] [PubMed] [Google Scholar]
- 17.Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405 [Google Scholar]
- 18.Yang F, Mitra P, Zhang L, Prak L, Verhertbruggen Y, Kim JS et al. (2013) Engineering secondary cell wall deposition in plants. Plant Biotechnol J. 11:325–35 doi: 10.1111/pbi.12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ookawa T, Yasuda K, Kato H, Sakai M, Seto M, Sunaga K et al. (2010) Biomass Production and Lodging Resistance in ‘Leaf Star’, a New Long-Culm Rice Forage Cultivar. Plant Prod Sci 13: 58–66 [Google Scholar]
- 20.Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q et al. (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol. 69:685–697 doi: 10.1007/s11103-008-9448-8 [DOI] [PubMed] [Google Scholar]
- 21.Valdivia ER, Herrera MT, Gianzo C, Fidalgo J, Revilla G, Zarra I et al. (2013) Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodiumdistachyon. J Exp Bot. 64:1333–43 doi: 10.1093/jxb/ers394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi M, Demura T (2010) Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnology 27: 237–242 [Google Scholar]
- 23.Zhong R and Ye Z H (2007) Regulation of cell wall biosynthesis. Current Opinion in Plant Biology 10: 564–572 doi: 10.1016/j.pbi.2007.09.001 [DOI] [PubMed] [Google Scholar]
- 24.Zhong R, Lee C, Ye ZH (2010) Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 152:1044–55 doi: 10.1104/pp.109.148270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eudes A, Sathitsuksanoh N, Baidoo EE, George A, Liang Y, Yang F et al. (2015) Expression of a bacterial 3-dehydroshikimate dehydratase reduces lignin content and improves biomass saccharification efficiency. Plant Biotechnol J. doi: 10.1111/pbi.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke WP, Radnidge P, Lai TE, Jensen PD, Hardin MT (2008) Digestion of waste bananas to generate energy in Australia. Waste Manag 28:527–533 doi: 10.1016/j.wasman.2007.01.012 [DOI] [PubMed] [Google Scholar]
- 27.Ingale Snehal, Joshi Sanket J., Gupte Akshaya (2014) Production of bioethanol using agricultural waste: Banana pseudo stem. Braz J Microbiol. 45: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma P, Mishra AA (2015) Biofuel production from banana peel by using micro wave. Int. J. Sci. Eng. Technol 3: 1015–1018 [Google Scholar]
- 29.Zhong R, Richardson EA, Ye ZH (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–92 doi: 10.1105/tpc.107.053678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy RL, Zhong R, Ye ZH (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50:1950–64 doi: 10.1093/pcp/pcp139 [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21:248–66 doi: 10.1105/tpc.108.063321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H et al. (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell. 22:1249–63 doi: 10.1105/tpc.108.064048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55:652–64 doi: 10.1111/j.1365-313X.2008.03533.x [DOI] [PubMed] [Google Scholar]
- 34.Soyano T, Thitamadee S, Machida Y, Chua NH (2008) ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell. 20:3359–73 doi: 10.1105/tpc.108.061796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Negi S, Tak H, Ganapathi TR (2017) Native vascular related NAC transcription factors are efficient regulator of multiple classes of secondary wall associated genes in banana. Plant Sci. 265:70–86 doi: 10.1016/j.plantsci.2017.09.018 [DOI] [PubMed] [Google Scholar]
- 36.Rodgers JT, Patel P, Hennes JL, Bolognia SL, Mascotti DP (2000) Use of biotin-labeled nucleic acids for protein purification and agarose-based chemiluminescent electromobility shift assays. Anal Biochem. 277:254–9 doi: 10.1006/abio.1999.4394 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of 5’ upstream regulatory region of MusaVND7. The PMusaVND7was amplified from genomic DNA of Musa cultivar Rasthali and analyzed for the presence of SNBE-like sites. SNBE–like sites in the PMusaVND7are boxed in green. Putative TATA box is shown in red and the transcription start site (+1 TSS) is underlined.
(PDF)
Sequence of 5’ upstream regulatory region of MusaVND6 from Musa cultivar Rasthali. TATA box is boxed in red and transcription start site (+1 TSS) is indicated by an underline. SNBE–like sites detected in the sequence are boxed in green.
(PDF)
(A) Embryogenic cells of banana cultivar Rasthali transformed with pCAMBIA1301- PMusaVND7:: GUS and growing on banana embryo development medium supplemented with hygromycin (5mg/l). (B) Emergence of putatively transformed embryos on hygromycin supplemented medium after 2 months of growth. (C) Close-up of embryos showing growth in various stages of development as well as emergence of secondary embryos. (D) Multiplication of transgenic shoots on banana shoot multiplication medium. (E) Rooting of transgenic banana plants on banana rooting medium containing NAA(1mg/l). (F) Rooted banana plants were hardened in a green house for GUS analysis.
(PDF)
(A) Embryogenic cells of banana cultivar Rasthali growing on banana embryo development medium supplemented with hygromycin (5mg/l) after transformation with pCAMBIA1301- PMusaVND6:: GUS. (B) White and opaque embryos developed from continuous culturing of the transformed embryogenic cells on embryo development medium. (C) Developing embryos appeared globular to torpedo shaped in close-up. (D) Transformed banana shoots were multiplied on shoot multiplication medium to generate multiple lines. (E) Rooting of transgenic banana plants on banana rooting medium containing NAA(1mg/l). (F) Rooted banana plants were hardened in a green house for GUS analysis.
(PDF)
(A) PCR analysis of banana plants harboring PMusaVND7::GUS. (B) PCR analysis of banana plants harboring PMusaVND6::GUS. Two different primer pairs were utilized in PCR analysis. 1: FP in either PMusaVND7 or PMusaVND6 and RP in GUS; 2–4:FP and RP in either PMusaVND7 or PMusaVND6 2–4: analysis of three independent transgenic banana lines. M: 1 KB DNA ladder. 1kb and 3 kb band size are indicated. (FP: forward primer; RP: reverse primer).
(PDF)
Transcript level of MusaVND6 and MusaVND7 in tissue of different organs of wild type banana was analyzed by quantitative RT-PCR. Expression of VND6 and VND7 in different tissue types is shown after normalization of the data by the expression of banana EF1α. Data was represented as mean±SD of three replications.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.