Abstract
Recent advancements in 3D bioprinting have led to the fabrication of more complex, more precise, and larger printed tissue constructs. As the field continues to advance, it is critical to develop quantitative benchmarks to compare different bio-inks for key cell-biomaterial interactions, including (1) cell sedimentation within the ink cartridge, (2) cell viability during extrusion, and (3) cell viability after ink curing. Here we develop three simple protocols for quantitative analysis of bio-ink performance. These methods are used to benchmark the performance of two commonly used bio-inks, poly(ethylene glycol) diacrylate (PEGDA) and gelatin methacrylate (GelMA), against three formulations of a novel bio-ink, Recombinant-protein Alginate Platform for Injectable Dual-crosslinked ink (RAPID ink). RAPID inks undergo peptide-self-assembly to form weak, shear-thinning gels in the ink cartridge and undergo electrostatic crosslinking with divalent cations during curing. In the one-hour cell sedimentation assay, GelMA, the RAPID inks, and PEGDA with xanthan gum prevented appreciable cell sedimentation, while PEGDA alone or PEGDA with alginate experienced significant cell settling. To quantify cell viability during printing, 3T3 fibroblasts were printed at a constant flowrate of 75 μl/min and immediately tested for cell membrane integrity. Less than 10% of cells were damaged using the PEGDA and GelMA bio-inks, while less than 4% of cells were damaged using the RAPID inks. Finally, to evaluate cell viability after curing, cells were exposed to ink-specific curing conditions for five minutes and tested for membrane integrity. After exposure to light with photo-initiator at ambient conditions, over 50% of cells near the edges of printed PEGDA and GelMA droplets were damaged. In contrast, fewer than 20% of cells found near the edges of RAPID inks were damaged after a 5-minute exposure to curing in a 10 mM CaCl2 solution. As new bio-inks continue to be developed, these protocols offer a convenient means to quantitatively benchmark their performance against existing inks.
Introduction
As the field of 3D bioprinting continues to expand, so too has the development of new bio-inks for cell-laden additive manufacturing [1, 2]. To make cell-laden tissue constructs, a suitable bio-ink must be printable, cell compatible during printing, and cell compatible post-printing. Recent development of new bio-inks has focused primarily on the printability of the material and the cell compatibility post-printing, often overlooking the viability of the cells during printing. These studies have enabled proof-of-concept demonstrations for many different applications in tissue engineering and regenerative medicine[3–8], tissue modeling [6, 7, 9, 10], and stem cell biology [11]. As the field expands beyond proof-of-concept studies, it will be increasingly important to also consider the bio-ink compatibility with the cells during the fabrication process to make 3D bioprinting scalable and cost efficient. Towards this goal, here three simple assays are developed that enable quantitative assessment of a bio-ink’s cell compatibility during the printing process. These assays are used to benchmark a new family of bio-inks against an array of commonly used bio-inks.
A wide range of hydrogels have been developed for injectable drug- and cell-delivery applications either through the use of in situ crosslinking [12–14] or through the use of thixotropic and self-healing rheological properties [15–17]. To date, much of the development of bio-inks has focused on translating these strategies for clinically injectable hydrogels for use as extrudable, printable materials [1]. However, as the bioprinting community begins to develop complex tissue constructs with high cell densities that more closely mimic the structure as well as the function of native tissue, the viability of cells during printing will become increasingly important. This is due in part to the costly, time intensive nature of cell expansion for many key cell types [18]. Additionally, functional tissue mimics often require a high cell density, as cell density influences cell phenotype for several cell types [19–22]. Furthermore, the delivery of viable cells can be important in maintaining the health and function of the printed construct, as dead cells or cell fragments from printing could release byproducts that may influence neighboring cells [23].
As we move towards printing full-scale tissues and organs, the print times required may reach hours to days [7]. Because of this, the cells used may need to remain suspended in the bio-ink within the cartridge for long time periods. Therefore, utilizing a biomaterial that maintains a homogeneous solution of encapsulated cells with minimal cell sedimentation is desirable. In addition to more precise control of cell density, cell sedimentation can also be detrimental to bio-ink printability due to printhead clogging. Here we developed a protocol to quantify cell sedimentation and used it to evaluate two different strategies to prevent sedimentation: the use of thickening agents for solution (sol) phase inks such as poly(ethylene glycol) diacrylate (PEGDA) and the use of gel phase inks such as gelatin methacrylate (GelMA) (Fig. 1A). These two bio-inks are common in the bioprinting field because they demonstrate excellent cell compatibility as “traditional” 3D biomaterials, which resemble the final printed construct [5, 7, 24–28].
Figure 1.

Schematic of cell/bio-ink interactions that occur during pressure-driven bioprinting and can be quantified through simple assays. A. Cell sedimentation, which can lead to inhomogeneous cell distribution and needle clogging, is more likely to occur in sol phase bio-inks compared to gel phase bio-inks. B. When traveling through an extrusion needle, cells experience different types of flow profiles in different types of bioinks, which can lead to cell membrane damage and death. C. Different bio-inks often require different curing conditions, which can compromise cell viability through various mechanisms, for example from dehydration or exposure to crosslinking reagents.
Another key aspect of bioprinting we chose to address is the viability of cells during extrusion. Cell viability is often measured at 1–7 days post printing, which provides important metrics for long-term cell compatibility and potential bio-ink effects on cell proliferation. This measurement, however, does not provide information on the cell compatibility during printing, such as acute cell damage that may occur during the fabrication process. This is a fundamentally different measurement, as non-adherent, dead cells can condense and fragment after death [29], allowing for diffusion out of the construct. Thus, cell viability measurements at later time points do not adequately quantify cell death that occurs during printing and curing. Cell death during extrusion printing and curing is important because printing dead or dying cells can affect the health and function of surrounding cells [30]. During extrusion in sol phase bio-inks, cells may undergo Pouseille flow, with a force profile that can lead to cell membrane damage and death [31]. Some gel phase bio-inks, in comparison, allow cells to experience plug flow, with a more cell-protective force profile (Fig. 1B). Here we present a simple method to explicitly quantify cell membrane damage during extrusion printing in different bio-inks.
In addition to cell death during extrusion printing, bio-ink curing can also be detrimental to cells (Fig. 1C). Previously, bioprinting studies have modified traditional 3D printing of thermoplastics onto dry substrates to make the fabrication process more cell compatible. Unlike additive manufacturing of plastics, one cannot use large temperature changes to melt and cure the ink. Instead, light-induced curing is very common due to its long history of use in traditional 3D tissue engineering applications and good long-term cell viability. Additionally, light-based curing is fairly rapid, which improves print resolution of the inks. However, during the bioprinting fabrication process, cells experience much longer exposure to photo-initiators, which can be cytotoxic [32]. Furthermore, these light-based curing methods are often performed in air to maintain print fidelity and to allow high transmission of light. This long exposure to air can lead to dehydration and cell death. In response, a variety of different bio-ink systems with alternative curing processes recently have been reported [32–34]. Thus, another important evaluation criterion for different bio-inks is cell viability during the ink-specific curing process.
Our group recently designed a proof-of-concept, dual cross-linked hydrogel that can be printed into an aqueous environment as an alternative to in-air printing methods [35]. This gel-phase ink, called Recombinant-protein Alginate Platform for Injectable Dual-crosslinked ink, or RAPID ink, consists of two components that undergo an initial cross-linking mechanism that exploits reversible, hetero-assembly of complementary peptide-binding domains (Fig. 2). The first component is a recombinant protein (C7) with seven repeats of a complementary peptide (C) that hetero-assembles with a proline-rich peptide (P) tethered to the second component, an alginate biopolymer [35, 36]. The 1:1 stoichiometric binding between C and P peptides results in a reversible, shear-thinning network with a relatively low modulus in the absence of calcium ions [35]. The secondary cross-linking mechanism exploits electrostatic alginate-Ca2+ interactions. Upon printing the gel-phase ink into a calcium ion-rich bath, these secondary cross-links stabilize and reinforce the scaffold. In this study, we present a family of RAPID inks with varying alginate molecular weight and P peptide avidity. Specifically, we benchmark these novel bio-inks against commercially available bio-inks (PEGDA and GelMA) using our three simple, quantitative assays for cell compatibility during bioprinting. While GelMA [5, 7, 37], PEGDA [28, 38], alginate [31, 39, 40], and recombinant protein hydrogels both with [35] and without alginate [36, 41] are known to have excellent, longterm cell compatibility when used as 3D biomaterials with encapsulated cells, their ability to maintain short-term cell viability and homogeneity during the printing and curing processes is relatively unknown. We find that our family of materials performs as well or better than the comparison bio-inks in these three criteria.
Figure 2.
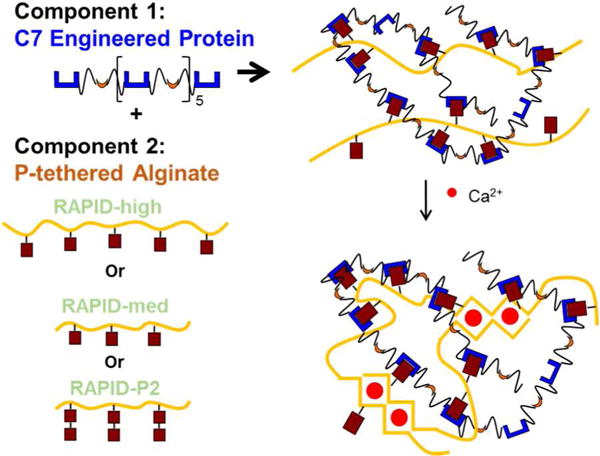
Schematic of RAPID inks. Component 1, termed C7, is a recombinant engineered protein with seven repeats of peptide C (shown in blue). Component 2 is high or low molecular weight alginate covalently modified with one or two repeats of peptide P (shown in crimson). Upon mixing the two components together, the C and P peptides hetero-assemble to form a weak network. Upon exposure to calcium cations, the alginate undergoes additional electrostatic crosslinking.
Materials and Methods
Material Synthesis
RAPID ink
Both P1 peptide (EYPPYPPPPYPSGC, 1536 g/mol) and P2 peptide (EYPPYPPPPYPSGGGGGEYPPYPPPPYPSGC, 3234 g/mol) were purchased from Genscript Corp (Piscataway, NJ, USA), and alginate was purchased from Novamatrix (Sandvika, Norway) in the LVG (MW 75–200 kDa) and MVG (MW > 200 kDa) formulations. P1 and P2 tethered alginate was synthesized using sulfo-NHS bioconjugation chemistry as previously reported [29]. The number of tethered peptides was quantified using a bicinchoninic acid (BCA) assay as previously reported [35] (see Table S1). The C7 protein was synthesized recombinantly as previously reported; full amino acid sequence is shown in Fig. S1 [30]. Alginate-P1 or alginate-P2 (2 wt/v% in Milli-Q water) was mixed in a 1:1 ratio with C7 protein (10 wt/v% in PBS) to create a gel-phase bioink with a final concentration of 1 wt/v% Alginate-P and 5 wt/v% C7. For bioprinting, cells were suspended in the C7 component before mixing with the Alginate-P component. Secondary crosslinking occurred in a 10 mM calcium chloride solution.
PEGDA
The PEGDA bio-inks were prepared as described by Hockaday et al.[28]. Briefly, 700 Da average molecular weight PEGDA (Sigma-Aldrich, Milwaukee, WI) was dissolved at room temperature into phosphate-buffered saline (PBS) at 20 wt/v% with rigorous stirring for 1 hr. The solution was heated to 50°C, and alginate (LVG version, Novamatrix) was dissolved into the solution at 1 wt/v% to generate the PEGDA + Alg bio-ink. Alternatively, 0.5 wt/v% xanthan gum (Sigma-Aldrich) was mixed into the PEGDA solution at 37°C to generate the PEGDA + XG bio-ink. The photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP, Biobots, Inc, Philadelphia, PA) was dissolved into both bio-inks at 0.1 wt%. LAP was chosen due to its improved cell compatibility over Irgacure 2959 [24, 42]. The storage vials were covered with foil after adding photoinitiator.
GelMA
The GelMA bio-ink was prepared as described by Bertassoni et al. and Kolesky et al. [5, 7] Briefly, gelatin (type A porcine skin, Sigma-Aldrich) was dissolved into Dulbecco’s phosphate buffered saline (DPBS, Thermo Fisher Scientific, Waltham, MA) at 10 wt/v% at 60°C. The temperature was then lowered to 50°C and methacrylic anhydride (Sigma-Aldrich) was added at 8 v/v% at 1 mL/min. The reaction was quenched after 3 hours via 5× dilution in DPBS and dialyzed with 15-kDa cutoff dialysis tubing in Milli-Q purified water for 5 days at 37°C. The solution was then lyophilized for 1 week. The GelMA, which was a white foam, was dissolved at 15 wt/v% in PBS at 60°C to create the bio-ink. LAP photoinitiator was dissolved at 0.1 wt%, and the storage vials were covered with foil.
Rheological Characterization
An 8-mm diameter, parallel plate on a stress-controlled rheometer (AR-G2, TA Instrument, New Castle, DE) was used for rheological measurements. For primary cross-linking of RAPID, components were mixed immediately before loading on rheometer. For secondary cross-linking of RAPID, materials were allowed to cure for 10 minutes in a 10 mM calcium chloride solution before loading on the rheometer. PEGDA + Alg, PEGDA + XG, and GelMA bio-inks were exposed to light (λ = 400–410 nm) for 5 minutes for curing before loading on the rheometer. Strain and frequency sweeps were measured for all materials. For viscosity measurements, a 20-mm diameter, cone geometry on a stress-controlled rheometer was used, measuring the viscosity over a strain sweep from a strain of 0.001 to 100. Viscosity was determined in the linear region of the strain sweep.
Cell Culture
NIH 3T3 cells (Sigma-Aldrich) were cultured in high-glucose DMEM medium with L-glutamine, 10% FBS (fetal bovine serum), and 1% penicillin/streptomycin. Cultures were maintained following the supplier’s protocols. A cell concentration of 10,000 cells/μL was used for all bio-ink assays.
Cell Sedimentation Assay
3T3 cells were labeled with Cell Vybrant DiO tracker (Thermo Fisher Scientific) according to manufacturer’s protocol prior to encapsulation in bio-inks. Bio-ink with cells (total volume of 100 μL) were then added to a 70-μL cuvette (BrandTech manufacturer number 759220), which is then closed with a styrofoam cap and placed in the incubator for 1 hour. After the incubation period, the samples were rotated 90 degrees and immediately imaged using confocal microscopy (Leica SPE with a 10× objective) along the entire height of the cuvette chamber (3.5 mm). Images were then stitched together using Leica Application Suite software and divided into four quadrants each of 0.89 mm. The cell number in each quadrant was counted using ImageJ (NIH freeware). The sedimentation coefficients δ was defined as:
Where n is the number of zones and ci is the cell density in each zone. Three independent replicates were performed, and statistical significance was determined using ANOVA with Tukey post-test.
Cell Viability during Printing Assay
Extrusion and photocuring was done using a BioBots printer (BioBots, Philadelphia, PA). Encapsulated cells were loaded into a 10-mL syringe with a 32-gauge, blunt-tip nozzle (Jensen Global, Santa Barbara, CA). Extrusion pressure was optimized to print at a rate of 60 μl/min or 75 μl/min for each material to standardize flow rate between different materials (see Table S2 for extrusion pressures for each bio-ink). Each bio-ink was printed directly into PBS to maintain cell hydration. Cell viability was then immediately measured (without exposing the bio-inks and cells to any curing steps) using a LIVE/DEAD viability/cytotoxicity kit according to the manufacturer’s protocol (Molecular Probes, Eugene, OR). Labeled cells were imaged using a confocal microscope (Leica SPE with 10× objective), and at least three fields of view were imaged for each sample. Three independent replicates were performed, and statistical significance was determined using ANOVA with Tukey post-test.
Cell Viability during Curing Assay
For curing experiments, RAPID ink formulations were extruded directly into a 10 mM calcium chloride solution, while PEGDA and GelMA formulations were extruded directly onto a microscope slide in air before exposure to light (λ = 400-410 nm) for 5 minutes. A total of 5 μL of each material was extruded for each sample, and all bio-inks were extruded slowly by hand using a 10-μL pipette to avoid cell membrane damage associated with extrusion printing at high flow rates. To mimic long print-times, a subset of samples were incubated for a further 60 minutes, with RAPID inks exposed to 10 mM calcium chloride solution and PEGDA and GelMA bio-inks exposed to air and ambient light. Cell viability was then measured using a LIVE/DEAD viability/cytotoxicity kit (Molecular Probes, Eugene, OR). Labeled cells were imaged using a confocal microscope (Leica SPE with 10× objective), and at least three fields of view were imaged near the edge (within 25 μm) and within the middle of each sample. Three independent replicates were performed, and statistical significance was determined using ANOVA with Tukey post-test.
Cell Viability during Irgacure Exposure
3T3 cells were mixed with PEGDA + XG bio-ink in the absence or presence (0.025 wt/v% or 0.05 wt/v%) of the photoinitiator Irgacure 2959 (Sigma-Aldrich). After one hour, cell viability was measured (without exposing the bio-inks and cells to any curing steps) using a LIVE/DEAD viability/cytotoxicity kit according to the manufacturer’s protocol (Molecular Probes, Eugene, OR). Labeled cells were imaged using a confocal microscope (Leica SPE with 10× objective), and at least three fields of view were imaged for each sample. Three independent replicates were performed, and statistical significance was determined using ANOVA with Tukey post-test.
Results and Discussion
Rheological Characterization of Bio-inks
In this study, we compared the effects of several different sol phase and gel phase bio-inks on three cell compatibility metrics for bioprinting. We first evaluated the mechanical properties of the bio-inks pre- and post-curing by measuring shear moduli (Fig. 3) and viscosity (Table S3). PEGDA + Alg is a viscous fluid pre-curing, whereas PEGDA + XG behaves as a weak gel due to polymer entanglements. Both materials have a storage modulus, G′, around 5 kPa after curing with LAP and light for 5 minutes. GelMA, a thermoresponsive bio-ink, shows weak, unstable gel behavior at 37°C and forms a stiffer gel at room temperature with a modulus around 500 Pa. After curing using LAP and light for 5 minutes, GelMA reaches a modulus over 10 kPa. All of these values are consistent with previously published reports for these bio-inks [5, 28].
Figure 3.

Rheological frequency sweep of storage (G′) and loss (G″) moduli for various bio-inks. All bio-inks were tested at room temperature except for GelMA, which was measured at room temperature and 37°C. PEGDA and GelMA bio-inks were cured by exposure to light (λ = 400–410 nm) for 5 minutes. RAPID inks were cured by exposure to a 10 mM calcium chloride solution for 10 minutes.
For our novel family of RAPID inks, three different formulations were compared. All three RAPID inks contain 5 wt/v% of the same C7 component and 1 wt/v% of P-tethered alginate. RAPID-high contains a high molecular weight alginate (> 200 kDa), while RAPID-med contains a medium molecular weight alginate (75-200 kDa), both tethered with peptides bearing a single repeat of the P domain (Fig. 2). RAPID-P2 contains medium molecular weight alginate tethered with peptides bearing two repeats of the P domain. All three variants showed similar rheological properties pre-curing, with storage moduli consistently greater than loss moduli (G′>G″), indicating a stable, weak gel network has formed through the hetero-assembly of C and P peptides present on the two components. Previous work with gels consisting of C7 and P-tethered PEG molecules had found that increasing the avidity of the P peptide (i.e. increasing from P1 to P2) resulted in moderately stiffer hetero-assembled gels [43]. However, here we discovered that increasing the peptide length from P1 to P2 decreased the conjugation efficiency of the peptide to alginate (Table S1), which likely compensated for the effect of avidity, resulting in gels of similar stiffness.
After curing with calcium ions, all three RAPID inks experienced significant increases in gel stiffness due to electrostatic crosslinking of the alginate. As expected, the RAPID-high had a higher G′ compared to RAPID-med (4 kPa and 1 kPa, respectively). Also as expected, RAPID-med and RAPID-P2, which have similar alginate molecular weights, had similar gel properties post-curing. After secondary crosslinking, the mechanical properties are largely dominated by the alginate component with little effect from changes in P1 avidity after exposure to calcium. Together, these data suggest that we could further alter the post-curing mechanical properties of RAPID inks by tuning alginate molecular weight.
Cell Sedimentation
For all of our assays, we have selected to use the commercially available 3T3 mouse fibroblast cell line so that results can be easily replicated and compared across different laboratories. In clinically relevant applications of 3D bioprinting, it may be necessary to load billions of cells into the bio-ink cartridge and to keep these cells homogeneously suspended during print times of multiple hours or longer [7]. Thus, cell sedimentation is an important criterion that must be considered during the development of bio-inks. Cell sedimentation is unfavorable both because printing and processing is more challenging if there is printer clogging, but additionally because the cell density of the printed structure is uncontrolled. Here we develop and report a sedimentation coefficient as a quantitative metric to compare different bio-inks. The bio-inks were first mixed with cells at 10,000 cells/μL, loaded into a 70-μL cuvette and left vertical at 37°C for 1 hour. Confocal microscope images were then taken along the vertical axis (3.5 mm long), which was arbitrarily divided into 4 zones, and the cell density in each zone was quantified with ImageJ software (Fig. 4A). The sedimentation coefficient, δ, was defined as:
where n is the number of zones and ci is the cell density in each zone. Using a larger number of zones did not impact the sedimentation coefficient (data not shown). This coefficient is the second-moment average divided by the first-moment average and is analogous to the dispersity index of polymers [44]. Thus, δ = 1 for a system with no cell sedimentation and equal cell density across all zones.
Figure 4. Cell sedimentation assay.
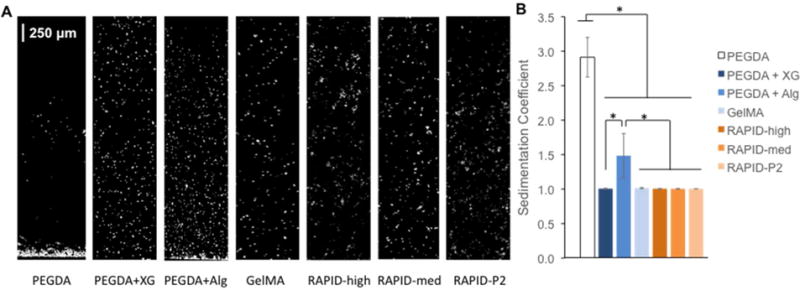
A. Confocal images of 3T3 cells encapsulated in various bio-inks for 1 hour. B. Quantified sedimentation coefficient for each bio-ink demonstrates that xanthan gum (XG) is an effective thickening agent for PEGDA bio-ink and that gel-phase inks (GelMA and RAPID) prevent cell sedimentation.
PEGDA (20 wt/v%) has a low viscosity of 0.01 Pa*s (Table S3), thus, as expected cells can quickly sediment within a solution of PEGDA (δ = 2.912 ± 0.287, Fig. 4A,B). Due to this, it is common to add various thickening agents to PEGDA, and here we evaluated both xanthan gum (0.5 wt/v%) and alginate (1 wt/v%) for their ability to prevent cell sedimentation [26, 28, 45, 46]. Interestingly, while both thickening agents significantly inhibited cell sedimentation relative to PEGDA alone, PEGDA + XG resulted in significantly more homogeneous cell distributions compared to PEGDA + Alg (δ = 1.007 ± 0.006 vs. 1.484 ± 0.321). These data are consistent with our rheological analysis that demonstrated PEGDA + XG is a weak gel of entangled polymers precuring, while PEGDA + Alg is a viscous solution (Fig. 3). For all of the gel phase bio-inks tested (GelMA and the three RAPID ink formulations), negligible cell sedimentation was observed. Taken together, these data demonstrate the potential advantage of using a bio-ink with weak, gel phase rheology in the ink cartridge to maintain a uniform cell distribution. While the sedimentation time used in these experiments was 1 hour, for future applications that require longer print times, it will be necessary to evaluate cell sedimentation over longer time scales.
Cell Viability During Printing
In addition to cell sedimentation, cells experience forces during the printing process that may impact cell viability. Our previous work on injectable hydrogels for cell transplantation demonstrated that syringe needle extrusion forces can cause cell membrane damage leading to significant cell death [31, 36, 47]. As cells in pressure-based bioprinting undergo a similar process (i.e. transport through a syringe barrel and a small gauge needle), we hypothesized that they too can experience detrimental mechanical forces during the extrusion process [29]. When developing a protocol to quantify this “print viability”, one can standardize across various bio-inks based either on applied pressure or flow rate. Our previous work has shown that cell viability does not directly correlate with the applied pressure for typical syringe needle flow rates [31]; therefore, we instead chose to standardize the flow rate for our protocol. Typical flow rates for GelMA at 12 and 10 PSI were measured to be 75 and 60 μL/min, respectively, in our printer setup. We then individually optimized the pressure for each of the other bio-inks to achieve similar flow rates (see Table S2 for a listing of all pressures). Cell viability was immediately quantified without exposing any of the bio-inks or cells to curing conditions.
The higher flow rate, 75 μL/min, showed the greatest significant differences between groups, with the commercially available inks showing between 5–9% cell death in all groups (Fig. 5). In comparison, the three RAPID inks experienced between 2–4% cell death at the same flow rate. Previously, our group demonstrated that weak, physical gels can protect cells during syringe needle flow, presumably by shear-thinning at the edges, which results in bulk plug flow through the needle [31, 35, 47]. Similar trends were observed at a flow rate of 60 μL/min, though with less statistical significance. This suggests that hetero-assembled, peptide-specific physical gels can significantly protect cells from the damaging mechanical forces experienced during 3D bioprinting.
Figure 5. Cell print viability assay.
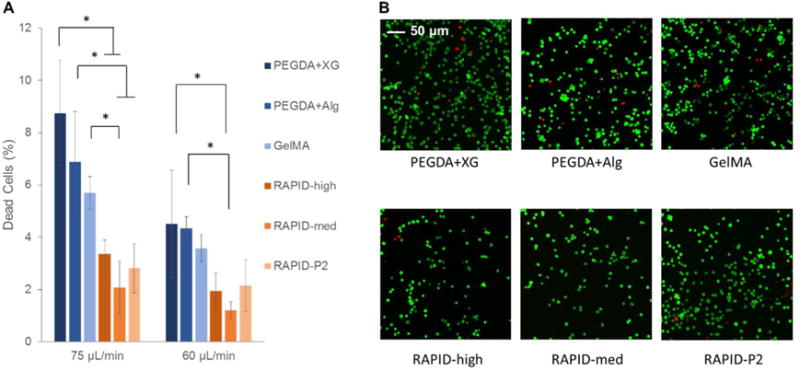
A. Quantification of LIVE/DEAD staining of 3T3 cells printed through a 32-gauge nozzle at average speeds of 75 and 60 μL/min. All bio-inks show less than 10% cell death during printing, while RAPID inks show less than 4% cell death. B. Representative LIVE/DEAD confocal images of 3T3 cells printed at a flow rate of 75 μL/min immediately after print extrusion and before any curing steps.
Cell Viability during Curing
After the cell-laden ink is printed, a curing period is typically required to stabilize the mechanical properties of the printed construct. Both GelMA and PEGDA use photocrosslinking to maintain the structure after printing. Photocrosslinking can be ideal for printing due to the fast curing time [48–50], and photocrosslinking of a variety of polymers has been used with great success for tissue engineering applications [51, 52]. However, photocrosslinking reactions require exposing cells to photoinitiators, which are known to be harmful to cell viability, particularly over long times. We show that exposure to as little as 0.05% Irgacure 2959, a common photoinitiator for tissue engineering studies [53], for one hour can kill over 20% of the encapsulated 3T3 fibroblast cells (Fig. S1). These data are consistent with previous studies exposing human adipose-derived mesenchymal stem cells to Irgacure [54]. Thus, for our further studies we chose to use the LAP photoinitiatior, which is known to be less cytotoxic than Irgacure [55].
In addition to cell death from photocrosslinking reagents, cells can also undergo dehydration during the curing process, as this often requires exposing the printed construct to air [56]. To assess the effects of the curing process on cell viability, we directly pipetted 5 μL of a cell-laden bio-ink onto a glass microscope slide to avoid any cell death due to extrusion. After 5 minutes of curing using LAP and light, cells encapsulated in all of the photo-cured bio-inks (PEGDA + XG, PEGDA + Alg, and GelMA) experienced about 50% cell death at the edge of the construct (Fig. 6, left). Interestingly, at the center of the construct, the cell death ranged from about 25–40%, suggesting that dehydration is a significant cause of cell death and may be altered by the relative hygroscopy of the bio-ink. In contrast, RAPID inks are cured by a 5-minute exposure to a 10 mM calcium chloride solution to induce electrostatic crosslinking. Calcium is the ideal divalent cation to use for bioprinting due to its fast gelation kinetics relative to magnesium [57] and lower cell toxicity compared to barium [58]. Due to its role in various cell functions, high extracellular calcium concentration can have a negative effect on cell viability or proliferation [59], however, here we limit the calcium concentration to 10 mM to limit the potential cellular effects of calcium crosslinking. This curing process was found to result in 15–20% cell death at the edges of the construct and 5–12% in the center of the construct, which was significantly less than all of the commercially available bio-inks.
Figure 6. Cell curing viability assay.
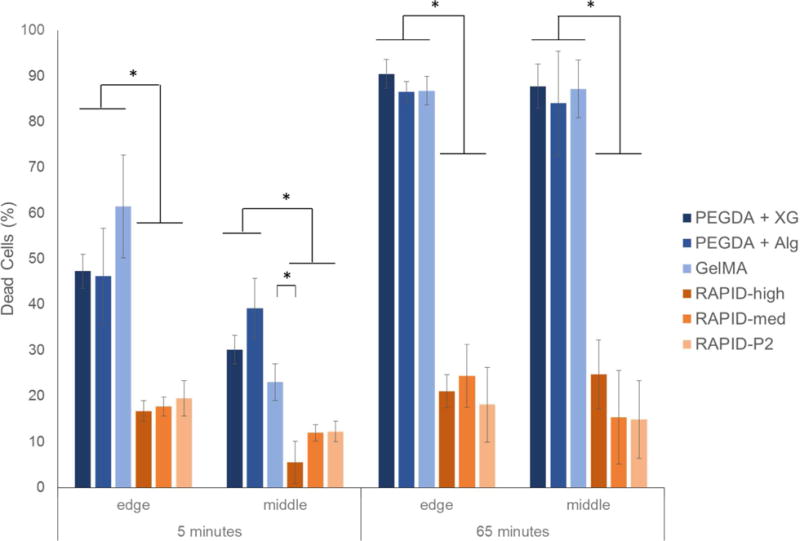
3T3 cells encapsulated in various bio-inks were pipetted onto a microscope slide and allowed to cure for 5 minutes. PEGDA and GelMA bio-inks were cured with exposure to light in air, while RAPID inks were cured with exposure to 10 mM calcium chloride solution. Samples were either immediately assayed using LIVE/DEAD reagents (left) or, to mimic longer print times, were further exposed to air (PEGD and GelMA inks) or calcium chloride (RAPID inks) for 60 minutes before viability quantification (right). Bio-inks that use photocrosslinking showed significantly more cell death than the RAPID inks.
Next, to mimic what would occur during longer print times of larger structures, we kept the photocured bio-inks in air for an additional 60 minutes with ambient light exposure (i.e. a total of 65 minutes passed between printing and viability quantification). This resulted in a dramatic increase in cell death for all of the photocured bio-inks, with over 80% cell death throughout the constructs, presumably due to dehydration. For comparison, the RAPID inks were exposed to 10 mM calcium chloride for an additional 60 minutes to mimic long print times. Interestingly, the cell viability after 5 minutes or 65 minutes of calcium chloride exposure along the edges of the construct was not significantly different. Unlike at 5 minutes, when cell viability at the center of the RAPID ink constructs was higher than at the edges, at 65 minutes the cell viability was similar throughout the entire construct, presumably due to the equilibration of calcium and chloride ions throughout the construct. These data suggest 3T3 cells survive better during long-term exposure to calcium chloride solution compared to dehydration during air exposure. Together, our data demonstrate that as the bioprinting community begins to scale-up this technology to print larger constructs, we will either need to dramatically reduce the overall print time or develop printing techniques that prevent cell dehydration. Alternative techniques to prevent construct dehydration have been explored by groups printing into either colloidal slurries [32, 34] or into a shear-thinning gel bath [33]. Additionally, one could design a humidified print chamber or hygroscopic bio-inks to help offset the effects of dehydration during curing.
When considering all three assays together, these quantitative benchmarks provide a comprehensive evaluation of cell-bio-ink interactions during the construct fabrication process. We have developed these assays to be simple and affordable, so that they can be readily adopted by other labs to enable direct comparison between different bio-ink formulations. We also developed the assays to decouple the various effects of different cell-bio-ink interactions; however, during an actual bioprinting experiment, all three types of cell-bio-ink interactions would be occurring simultaneously. Thus, the “acute” cell viability immediately following construct fabrication (i.e. after extrusion printing and bio-ink curing) would likely be much lower than the values reported here for a single assay. After several days, membrane compromised cells can fragment and diffuse out of the construct, while surviving cells may proliferate to repopulate the construct. For this reason, quantification of cell viability in printed constructs should include both “acute” and “long-term” measurements.
While the assays developed here have focused on cell viability during fabrication, the long-term effects of these different printing processes on cell function are currently unknown. For instance, forces experienced during the extrusion printing process or cellular stress responses to dehydration could induce long-term changes to cell behavior that would be unobserved using only the metrics outlined here. Furthermore, these long-term cellular responses are likely to be different for different cell types; thus, future assays for cellular function will likely need to be developed for specific 3D bioprinting applications. In addition, while this paper only develops benchmarks for single-cell-type printing, precise control over cell density will become even more important when printing multiple cell types, as the ratio of cell types in co-cultures is essential in achieving the desired cell interactions for the tissue type of interest [60–62]. Thus, a further study of the effects of sedimentation on multi-cell-type printing will be important in the future. The development of simple, reproducible protocols to quantitatively benchmark various bio-ink formulations against one another should prove helpful to the field in evaluating the appropriate bio-ink for specific printing applications.
Conclusion
In this paper, we have developed three simple assays that provide quantitative benchmarks to compare various bio-inks developed for 3D bioprinting applications, with a focus on cell compatibility during the fabrication process. These quantitative assays allow biomaterials scientists to directly compare different formulation strategies when developing bio-inks for a specific bioprinting application. These assays were used to benchmark three novel RAPID ink formulations against commonly used bio-inks, PEGDA and GelMA. First, an assay was developed to determine a cell sedimentation coefficient for each bio-ink. Two different strategies (1. the use of polymer thickeners for sol phase bio-inks or 2. the use of gel phase bio-inks) were found to efficiently prevent cell sedimentation and maintain cell homogeneity for up to one hour. Second, an assay was developed to quantify the amount of cell membrane damage that occurs during typical extrusion printing protocols. While cell death was 9% or less for all tested bio-inks, the peptide-assembled RAPID inks were found to provide significant protection to the encapsulated cells, resulting in 4% or less of damaged cells. Finally, an assay was developed to quantify cell death that occurs during the bio-ink curing process. Even when using a photocuring protocol known to be relatively cell compatible (LAP with light λ = 400–410 nm), cell death after 5 minutes of curing was found to be about 50% near the edges of the constructs. In contrast, curing of RAPID inks with a 10 mM calcium chloride solution was found to result in significantly less cell death, likely due to inhibition of cell dehydration. By defining benchmarks to quantify important bio-ink interactions with cells during the fabrication process, we allow future researchers to assess the suitability of potential bio-inks for specific cell-laden 3D printing applications.
Supplementary Material
Acknowledgments
The authors acknowledge funding support from the National Science Foundation (DMR 1508006), the National Institutes of Health (U19 AI116484), Stanford Bio-X Bowes Fellowship (KD), the Stanford Summer Research Program and the Amgen Scholars Program (AT). The authors would like to thank Daniel Hunt for materials synthesis and helpful discussions; Chris Lindsay, BioBots, Inc., and the BioBots beta-testers community for bioprinter troubleshooting assistance; and Dorothy Tovar, Samar Fahmy, and Dr. Terrance Mayes for encouragement in pursuing this project.
References
- 1.Malda J, Visser J, Melchels FPW, Jungst T, Hennink WE, Dhert WJA, Groll J, Hutmacher DW. Engineering Hydrgels for Biofabrication. Adv Mater. 2013;25:5011–28. doi: 10.1002/adma.201302042. [DOI] [PubMed] [Google Scholar]
- 2.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32:773–85. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 3.Morris VB, Nimbalkar S, Younesi M, McClellan P, Akkus O. Mechanical Properties, Cytocompatibility and Manufacturability of Chitosan: PEGDA Hybrid-Gel Scaffolds by Stereolithography. Ann Biomed Eng. 2016;45 doi: 10.1007/s10439-016-1643-1. [DOI] [PubMed] [Google Scholar]
- 4.Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Pi Q, Byambaa B, Dokmeci M, Shin SR, Khadhemhosseini A. Direct 3D bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials. 2016;106:58–68. doi: 10.1016/j.biomaterials.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertassoni LE, Cardosa JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, Zorlutuna P, Vrana NE, Ghaemmaghami AM, Dokmeci MR. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels. Biofabrication. 2014;6:024105. doi: 10.1088/1758-5082/6/2/024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W, Xin Q, Zhu J, Ma X, Patel S, Liu J, Wang P, Sun C, Lai E, Gou M, Xu Y, Zhang K, Chen S. Direct 3D bioprinting of prevascularized tissue constructs with complex microarchitecture. Biomaterials. 2017;124:106–15. doi: 10.1016/j.biomaterials.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. 3D Bioprinting of Vascularized, Heterogeneous Cell-Laden Tissue. Adv Mater. 2014;26:3124–30. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 8.Gu BK, Choi DJ, Park SJ, Kim MS, Kang CM, Kim C-H. 3-dimensional bioprinting for tissue engineering applications. Biomater Res. 2016;20:12. doi: 10.1186/s40824-016-0058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Nowicki M, Cui H, Zhu W, Fang X, Miao S, Lee S-J, Keidar M, Zhang LG. 3D bioprinted graphene oxide-incorporated matrix for promoting chondrogenic differentiation of human bone marrow mesenchymal stems cells. Carbon. 2017;116:615–24. [Google Scholar]
- 10.Tabet A, Gardner M, Swanson S, Crump S, McMeekin A, Gong D, Tabet R, Hacker B, Nestrasil I. Low-cost, rapidly-developed, 3D printed in vitro corpus callosum model for mucopolysaccharidosis type I. F1000Res. 2017;5:2811. doi: 10.12688/f1000research.9861.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao G, Zhang X-F, Hubbel K, Cui X. NR2F2 regulates chondrogenesis of human mesenchymal stem cells in bioprinted cartilage. Biotechnol Bioeng. 2016;114:208–16. doi: 10.1002/bit.26042. [DOI] [PubMed] [Google Scholar]
- 12.Kretlow JD, Klouda L, Mikos AG. Injectable matrices and scaffolds for drug delivery in tissue engineering. Advanced drug delivery reviews. 2007;59:263–73. doi: 10.1016/j.addr.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Yang JA, Yeom J, Hwang BW, Hoffman AS, Hahn SK. In situ-forming injectable hydrogels for regenerative medicine. Prog Polym Sci. 2014;39:1973–86. [Google Scholar]
- 14.Du YK, Shinde UP, Yeon B, Jeong B. Recent progress of in situ formed gels for biomedical applications. Prog Polym Sci. 2013;38:672–701. [Google Scholar]
- 15.Marquardt LM, Heilshorn SC. Design of Injectable Materials to Improve Stem Cell Transplantation. Curr Stem Cell Rep. 2016;2:207–20. doi: 10.1007/s40778-016-0058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster AA, Marquardt LM, Heilshorn SC. The diverse roles of hydrogel mechanics in injectable stem cell transplantation. Curr Op Chem Eng. 2017;15:15–23. doi: 10.1016/j.coche.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guvendiren M, Lu HD, Burdick JA. Shear-thinning hydrogels for biomedical applications. Soft Matter. 2012;8:260–72. [Google Scholar]
- 18.Ikada Y. Challenges in tissue engineering. J R Soc Interface. 2006;3 doi: 10.1098/rsif.2006.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bitar M, Brown RA, Salih V, Kidane AG, Knowles JC, Nazhat SN. Effect of cell density on osteoblastic differentiation and matrix degradation of biomimetic dense collagen scaffolds. Biomacromolecules. 2008;9:129–35. doi: 10.1021/bm701112w. [DOI] [PubMed] [Google Scholar]
- 20.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;2:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 21.Watt F. Epidermal stem cells in culture. J Cell Science. 1988;10:85–94. doi: 10.1242/jcs.1988.supplement_10.7. [DOI] [PubMed] [Google Scholar]
- 22.Glowacki J, Trepman E, Folkman J. Cell Shape and Phenotypic Expression in Chondrocytes. Proc Soc Exp Biol Med. 1983;172:93–8. doi: 10.3181/00379727-172-41533. [DOI] [PubMed] [Google Scholar]
- 23.Kawamoto Y, Nakajima Y-I, Kuranaga E. Apoptosis in Cellular Society: Communication between Apoptotic Cells and Their Neighbors. Int J Mol Sci. 2016;17:2144. doi: 10.3390/ijms17122144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Abdulla R, Parker B, Samanipour R, Ghosh S, Kim K. A simple and high-resolution stereolithography-based 3D bioprinting system using visible light crosslinkable bioinks. Biofabrication. 2015;7:045009. doi: 10.1088/1758-5090/7/4/045009. [DOI] [PubMed] [Google Scholar]
- 25.Shanjani Y, Pan C C, Elomaa L, Yang Y. A novel bioprinting method and system for forming hybrid tissue engineering constructs. Biofabrication. 2015;7:045008. doi: 10.1088/1758-5090/7/4/045008. [DOI] [PubMed] [Google Scholar]
- 26.Kang LH, Armstrong PA, Lee LJ, Duan B, Kang KH, Butcher JT. Optimizing Photo-Encapsulation Viability of Heart Valve Cell Types in 3D Printable Composite Hydrogels. Ann of Biomed Eng. 2017;45:360–77. doi: 10.1007/s10439-016-1619-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Constantini M, Idaszek J, Szoke K, Jaroszewicz J, Dentini M, Barbetta A, Birinchmann J, Swieszkowski W. 3D bioprinting of BM-MSCs-loaded ECM biomimetic hydrogels for in vitro neocartilage formation. Biofabrication. 2016;8:035002. doi: 10.1088/1758-5090/8/3/035002. [DOI] [PubMed] [Google Scholar]
- 28.Hockaday LA, Kang KH, Colangelo NW, Cheung PYC, Malone E, Wu J, Girardi LN, Bonassar LJ, Lipson H, Chu CC, Butcher JT. Rapid 3D printing of anatomically accurate and mechanicalls heterogeeous aortic valve hydrogel scaffolds. Biofabrication. 2012;4:035005. doi: 10.1088/1758-5082/4/3/035005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzanne M, Stellar H. Shaping organisms with apoptosis. Cell Death Differ. 2013;20:669–75. doi: 10.1038/cdd.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.BA A, W M, J S, KJ L, SC H. Improving Viability of Stem Cells During Syringe Needle Flow Through the Design of Hydrogel Cell Carriers. Tissue Eng Part A. 2012;18:806–15. doi: 10.1089/ten.tea.2011.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, Ramadan MH, Hudson AR, Feinberg AW. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci Adv. 2015;1:e1500758. doi: 10.1126/sciadv.1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Highley CB, Rodell CB, Burdick JA. Direct 3D printing of shear-thinning hydrogels into self-healing hydrogels. Adv Mater. 2015;27:5075–9. doi: 10.1002/adma.201501234. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharjee T, Gil CJ, Marshall SL, Uruena JM, O’Brayn CS, Carstens M, Keselowsky B, Palmer GD, Ghivizzani S, Gibbs CP, Sawyer WG, Angelini TE. Liquid-like solids support cells in 3D ACS. Biomater Sci Eng. 2016;2:1787–95. doi: 10.1021/acsbiomaterials.6b00218. [DOI] [PubMed] [Google Scholar]
- 35.Dubbin K, Hori Y, Lewis KK, Heilshorn SC. Dual-Stage Crosslinking of a Gel-Phase Bioink Improves Cell Viability and Homogeneity for 3D Bioprinting. Adv Healthcare Mater. 2016;5:2488–92. doi: 10.1002/adhm.201600636. [DOI] [PubMed] [Google Scholar]
- 36.Wong Po Foo C, Lee JS, Mulyasasmita W, Parisi-Amon A, Heilshorn SC. Two-component protein-engineered physical hydrogels for cell encapsulation. PNAS. 2009;106:22067–72. doi: 10.1073/pnas.0904851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visser J, Melchels FPW, Jeong JE, van Bussel EM, Kimpton LS, Byrne HM, Dhert WJ, Dalton PD, Hutmacher DW, Malda J. Reinforcement of hydrogels using three-dimensionally printed microfibres. Nature Communications. 2015;6:6933. doi: 10.1038/ncomms7933. [DOI] [PubMed] [Google Scholar]
- 38.Chan V, Zorlutuna P, Jeong JH, Bashir R. Three-dimensional photopatterning of hydrogels using stereolithography for long-term cell encapsulation. Lab on a Chip. 2010;10:2062–70. doi: 10.1039/c004285d. [DOI] [PubMed] [Google Scholar]
- 39.Shi P, Laude A, Yeong WY. Investigation of cell viability and morphology in 3D bioprinted alginate constructs with tunable stiffness. Journal of Biomedical Materials Research Part A. 2017;105:1009–18. doi: 10.1002/jbm.a.35971. [DOI] [PubMed] [Google Scholar]
- 40.Tabriz AG, Hermida MA, Leslie NR, Shu W. Three-dimensional bioprinting of complex cell laden alginate hydrogel structures. Biofabrication. 2015;7:045012. doi: 10.1088/1758-5090/7/4/045012. [DOI] [PubMed] [Google Scholar]
- 41.Mulyasasmita W, Lee JS, Heilshorn SC. Molecular-level engineering of protein physical hydrogels for predictive sol-gel phase behavior. Biomacromolecules. 2011;12:3406–11. doi: 10.1021/bm200959e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin H, Zhang D, Alexander PG, Yang G, Tan J, Cheng AWM, RS T. Application of visible light-based projection stereolithography for live cell-scaffold fabrication with designed architecture. Biomaterials. 2013;34:331–9. doi: 10.1016/j.biomaterials.2012.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulyasasmita W, Cai L, Dewi RE, Jha A, Ullmann SD, Luong RG, Huang NF, Heilshorn SC. Avidity-controlled hydrogels for injectable co-delivery of induced pluripotent stem cell- derived endothelial cells and growth factors. J Controlled Release. 2014;191:71–81. doi: 10.1016/j.jconrel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heimenz PC, Lodge TP. Polymer Chemistry 2007 [Google Scholar]
- 45.Hong S, Sycks D, Chan HF, Lin S, Lopez GP, Farshid G, Leong KW, Zhao X. 3D Printing: 3D Printing of highly stretchable and tough hydrogels into complex, cellularized structures. Adv Mater. 2015;27:4034. doi: 10.1002/adma.201570182. [DOI] [PubMed] [Google Scholar]
- 46.Freyer JP, Wilder ME, Schor PL, Coulter J, Raju MR. A Simple Electronic Volume Cell Sorter for Clonogenicity Assays. Cytometry. 1989;10:273–81. doi: 10.1002/cyto.990100306. [DOI] [PubMed] [Google Scholar]
- 47.Cai L, Dewi RE, Heilshorn SC. Injectable hydrogels with in situ double network formation enhance retention of transplanted stem cells. Adv Functional Mater. 2015;25:1344–51. doi: 10.1002/adfm.201403631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen KT, West JL. Photopolymerizable hydrogel for tissue engineering application. Biomaterials. 2002;23:4307–14. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 49.Skardal A, Atala A. Biomaterials for Integration with 3-D Bioprinting. Ann Biomed Eng. 2015;43:730–46. doi: 10.1007/s10439-014-1207-1. [DOI] [PubMed] [Google Scholar]
- 50.Pereira RF, Bartolo PJ. 3D bioprinting of photocrosslinkable hydrogel constructs. App polym sci. 2015;132:42458. [Google Scholar]
- 51.El-Sherbiny I, Yacoub MH. Hydrogel scaffolds for tissue engineering: progress and challenges. Glob Cardiol Sci Pract. 3:316–42. doi: 10.5339/gcsp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drury JL, Mooney DJ. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials. 2003;24:4337–51. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 53.Fedorovich NE, Oudshoorn MH, van Geemen D, Hennink WE, Alblas J, Dhert WJ. The effect of photopolymerization on stem cells embedded in hydrogels. Biomaterials. 2009;30:344–53. doi: 10.1016/j.biomaterials.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 54.Sabnis A, Rahimi M, Chapman C, Nguyen KT. Cytocompatibility Studies of an in situ photopolymerized thermoresponsive hydrogel nanoparticle system using human aortic smooth muscle cells. J Biomed Mater Res A. 2009;91:52–9. doi: 10.1002/jbm.a.32194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials. 2009;30:6702–7. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahn BY, Duoss EB, Motala MJ, Guo X, Park S-I, Xiong Y, Yoon J, Nuzzo RG, Rogers JA Lewis JA. Omnidirectional printing of flexible, stretchable, and spanning silver microelectrodes. Science. 2009;323:5921. doi: 10.1126/science.1168375. [DOI] [PubMed] [Google Scholar]
- 57.Topuz F, Henke A, Richtering W, Groll J. Magnesium ions and alginate do form hydrogels: a rheological study. Soft Matter. 2012;8:4877–81. [Google Scholar]
- 58.Zimmermann U, Mimietz S, Zimmermann H, Hillgartner M, Schneider H, Ludwig J, Hasse C, Haase A, Rhothmund M, Fuhr G. Hydrogel-based non-autologous cell and tissue therapy. Biotechniques. 2000;29:564–72. doi: 10.2144/00293rv01. [DOI] [PubMed] [Google Scholar]
- 59.Cao N, Chen XB, Schreyer DJ. Influence of calcium ions on cell survival and proliferation in the context of an alginate hydrogel. ISRN Chemical Engineering. 2012;2012:516461. [Google Scholar]
- 60.Ma J, van den Beucken JJJP, Yang F, Both SK, Cui F-Z, Pan J, Jansen JA. Coculture of Osteablasts and Endothelial Cells: Optimization of Culture Medium and Cell Ratio. Tissue Eng Part C. 2011;17:2011. doi: 10.1089/ten.TEC.2010.0215. [DOI] [PubMed] [Google Scholar]
- 61.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Microfabrication of Hepatocyte/Fibroblast Co-cultures: role of homotypic cell interactions. Biotechnol Progress. 1998;14:378–87. doi: 10.1021/bp980036j. [DOI] [PubMed] [Google Scholar]
- 62.Bhatia SN, Yarmush ML, Toner M. Controlling cell interactions by micropatterning in co-cultures: hepatacytes and 3T3 fibroblasts. J Biomed Mater Res. 1997;34:189–99. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


