SUMMARY
Circadian behavior in mammals is orchestrated by neurons within the suprachiasmatic nucleus (SCN), yet the neuronal population necessary for the generation of timekeeping remains unknown. We show that a subset of SCN neurons expressing the neuropeptide neuromedin S (NMS) plays an essential role in generation of daily rhythms in behavior. We demonstrate that lengthening period within Nms neurons is sufficient to lengthen period of the SCN and behavioral circadian rhythms. Conversely, mice without a functional molecular clock within Nms neurons lack synchronous molecular oscillations and coherent behavioral daily rhythms. Interestingly, we found that mice lacking Nms and its closely-related paralog, Nmu, do not lose in vivo circadian rhythms. However, blocking vesicular transmission from Nms neurons with intact cell-autonomous clocks disrupts the timing mechanisms of the SCN, revealing that Nms neurons define a subpopulation of pacemakers that control SCN network synchrony and in vivo circadian rhythms through intercellular synaptic transmission.
INTRODUCTION
The SCN in the hypothalamus controls near 24 hr (circadian) rhythms in biochemical, physiological, and behavioral processes (Saper, 2013; Welsh et al., 2010). At the molecular level, genetic approaches have successfully uncovered autoregulatory feedback loops that form the basis of circadian rhythms in the SCN and in extra-SCN oscillators (Lowrey and Takahashi, 2011). The transcription factors, CLOCK and BMAL1, heterodimerize to drive the expression of Period (Per1, Per2) and Cryptochrome (Cry1, Cry2), which in turn feedback inhibit their own transcription by CLOCK and BMAL1. This inhibition is slowly relieved as PER and CRY are degraded by the proteasome through association with ubiquitin ligase complexes. At the cellular level, the SCN, like other nuclei within the hypothalamus, contain a heterogenous population of ~20,000 neurons which secrete more than 100 identified neurotransmitters, neuropeptides, cytokines, and growth factors (Abrahamson and Moore, 2001; Lee et al., 2010). Some of these signaling molecules have been implicated to play important roles in cell-cell coupling, a process that synchronizes period length and phase relationships among SCN neurons (Evans et al., 2013; Herzog, 2007; Mohawk and Takahashi, 2011). However, the roles of specific SCN cell types and the mode of intercellular signaling mechanisms engaged in the generation of behavioral circadian rhythms are poorly understood, in part due to the paucity of genetic tools available to restrictively interrogate subpopulations of SCN neurons.
Neuromedin S (NMS), named for its restricted expression within the SCN, is a 36-amino acid neuropeptide that was identified from rat brain extracts as an endogenous ligand for neuromedin U (NMU) receptors type 1 (NMUR1) and 2 (NMUR2) (Mori et al., 2005). Due to its SCN-restricted expression, Nms represents an attractive genetic target for exploiting and understanding the organization of the central circadian clock. Here, we describe the generation of a BAC transgenic mouse line containing a bicistronically expressed Cre recombinase (iCre) inserted into the 3′-UTR of Nms. Using several complementary genetic approaches, we found that a subset of SCN neurons marked by Nms is capable of dictating circadian period length and required for the generation of behavioral circadian rhythms. We further show that daily behavioral rhythms of mutant mice with non-rhythmic Nms neurons can be transiently restored by light entrainment. This short-lived restoration, however, is dependent on signaling mediators from Nms neurons because blocking synaptic transmission from Nms neurons leads to the loss of network synchrony and the disruption of in vivo circadian rhythms.
RESULTS
Neuromedin S (Nms) is Restrictively Expressed in a Subset of Neurons in the Mouse SCN
Individual SCN neurons, which can act as independent cell-autonomous oscillators when isolated in vitro (Webb et al., 2009; Welsh et al., 1995), normally function in a unified coupled manner in vivo. How heterogenous neurons in the SCN coordinate with one another to generate circadian rhythms in behavior is not well understood. To explore whether a subpopulation of neurons in the SCN can control the entire neuronal network, we searched for candidate cell markers that would allow us to target neuronal subpopulations within the SCN in a restricted manner and consequently picked Nms as a likely candidate (Figure 1). To determine the expression of Nms in the mouse brain, we performed in situ hybridization on C57BL/6J mouse brain sections using anti-sense Nms riboprobe and found that Nms is highly expressed within the mouse SCN with little to no detectable signals in other areas of the brain (Figure S1). This result is consistent with a report showing robust levels of Nms expression within the rat SCN with only minimal expression detected by quantitative RT-PCR in other regions of the rat brain (Mori et al., 2005).
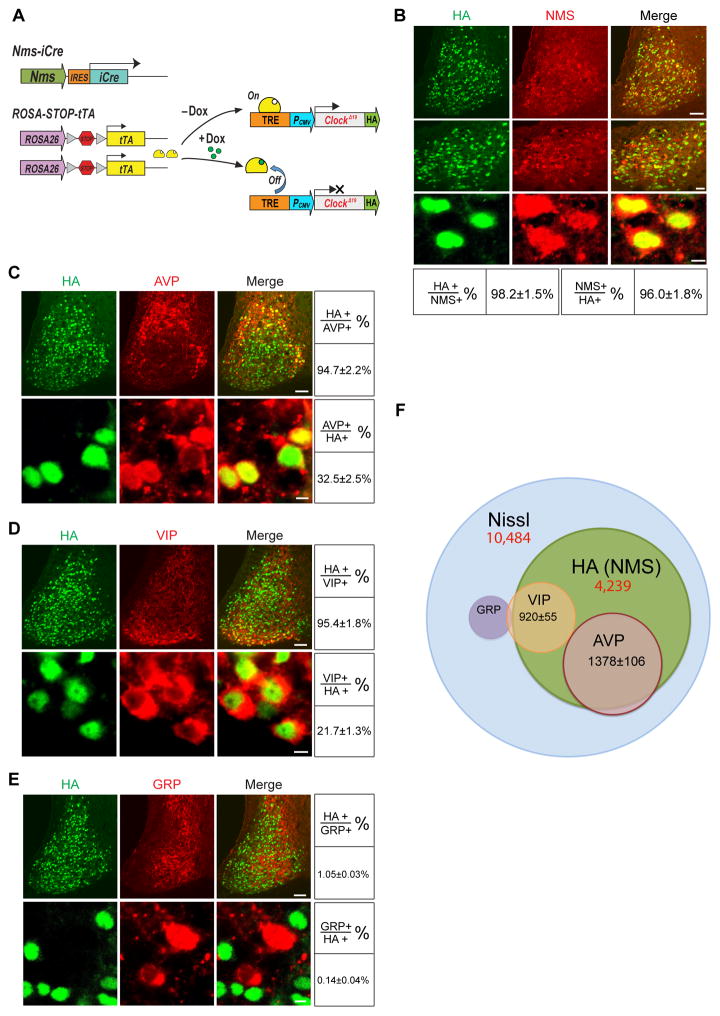
Figure 1. Distribution of HA (NMS)-Expressing Neurons in the Nms-ClockΔ19 Mouse SCN.
(A) Diagram showing the Tet-Off system and constructs used to generate the Nms-ClockΔ19 mice. The Nms promoter drives the bicistronic expression of iCre, which excises the loxP flanked stop codon of ROSA-STOP-tTA, allowing tTA to be expressed. tTA binds to the Tetracycline Response Element (TRE) in the absence of Dox but not in its presence, resulting in the transcriptional activation or repression of the HA-tagged tetO-ClockΔ19 transgene, respectively.
(B) Double staining of HA (green) and NMS (red) on SCN sections of Nms-ClockΔ19 mice. Scale bars are 50 μm (top), 20 μm (middle), and 5 μm (bottom). Quantitative data are shown at the bottom.
(C) Double staining of HA (green) and AVP (red) on SCN sections of Nms-ClockΔ19 mice. Scale bars are 50 μm (top) and 5 μm (bottom). Quantitative data are shown at the right.
(D) Double staining of HA (green) and VIP (red) on SCN sections of Nms-ClockΔ19 mice. Scale bars same as (B).
(E) Double staining of HA (green) and GRP (red) on SCN sections of Nms-ClockΔ19 mice. Scale bars same as (B).
(F) Venn diagram displaying the overlap between HA (NMS), VIP, AVP, and GRP. HA (NMS) and Nissl cell numbers are from stereological counts listed in (F). The number of cells that overlap with NMS was extrapolated from the relative % of co-localization in Figure 1C to 1E based on the number of HA (NMS) neurons from stereological counts.
Having observed that Nms is restrictively expressed within the mouse SCN, we generated a BAC transgenic mouse line that bicistronically expresses codon-improved iCre under the promoter of Nms (hereafter, Nms-iCre). All offspring of this mouse line appear healthy with daily locomotor rhythms not different from wild-type C57BL/6J mice (data not shown). We then crossed Nms-iCre to ROSA-STOP-tTA mice (Wang et al., 2008) and HA epitope-tagged tetO-ClockΔ19 transgenic mice (Hong et al., 2007) to generate mice that reversibly overexpress ClockΔ19 in Nms-producing neurons (hereafter, Nms-ClockΔ19) (Figure 1A, Figure S2, and Table S1). In these mice, iCre induces the expression of the tetO promoter transactivator (tTA; Tet-Off) in Nms neurons by removing a stop cassette from the tTA transgene knocked into the ROSA26 locus (ROSA-STOP-tTA). The tetO-ClockΔ19 transgene is therefore controlled transcriptionally by the ROSA26 promoter-regulated tTA in Nms neurons of the SCN. This was done for two reasons: (i) to assess the phenotypic effects of overexpressing the ClockΔ19 transgene in Nms neurons (to be discussed below) and (ii) to use the HA epitope tag as a convenient marker to assess the distribution of Nms neurons within the mouse SCN.
First, in order to confirm that the HA-tagged ClockΔ19 transgene is expressed in the endogenous Nms neurons in the SCN of Nms-ClockΔ19 mice, we performed double immunostaining using anti-HA and anti-NMS antibodies and found that ~98% of NMS-positive neurons express HA while ~96% of HA-immunoreactive cells overlap with NMS (Figure 1B). Accounting for the detection limitations of the staining procedure, we conclude that in the Nms-ClockΔ19 mouse line, the ClockΔ19 transgene is faithfully expressed only in SCN neurons marked by NMS. Outside of the SCN, sparse HA immunoreactivity was observed in other hypothalamic regions, the ventromedial thalamic nucleus, the median preoptic nucleus, and the Edinger-Westphal nucleus (Figure S3).
To determine the regional distribution of Nms in the SCN, we used anti-vasoactive intestinal peptide (VIP), anti-arginine vasopressin (AVP), and anti-gastrin-releasing peptide (GRP) antibodies individually with anti-HA to double immunostain the SCN of Nms-ClockΔ19 mice. VIP and AVP, which exhibit no co-localization with one another, delineate the core and shell regions of the SCN, respectively, while GRP is expressed in the core and central regions of the SCN (Abrahamson and Moore, 2001; Morin et al., 2006). We found that HA overlaps with ~95% of AVP and VIP-immunoreactive cells, while AVP and VIP overlap with ~33% and ~22% of HA-immunoreactive cells, respectively (Figure 1C and 1D). In contrast, GRP expression exhibits minimal co-localization with HA with only ~0.1–1% of cells overlapping (Figure 1E). Although VIP and GRP co-localization has been demonstrated in the rat SCN (Romijn et al., 1998), to our knowledge, the amount of VIP and GRP neurons that co-localize in the mouse SCN is unknown. We used anti-VIP and anti-GRP to double-stain the SCN of C57BL/6J mice and found that ~7% of VIP neurons express GRP, while ~22% of GRP cells express VIP (Figure S4). GRP and AVP have been reported to exhibit no co-localization in C57BL/6J mice (Karatsoreos et al., 2006). Taken together, our histological results demonstrate that within the mouse SCN, Nms is expressed in the majority of AVP+ and VIP+ neurons but not in GRP+VIP− or GRP+VIP+ neurons.
We next utilized unbiased stereological counting (West et al., 1991) to estimate the total number of Nms-expressing neurons in the SCN. Using NeuroTrace Nissl staining, we estimated a total number of 10,484 ±109 neurons per unilateral SCN, which corresponds well with previously reported figures (Abrahamson and Moore, 2001; Atkins et al., 2010). Using anti-HA staining on the SCN of Nms-ClockΔ19 mice, the total number of Nms neurons were estimated to be 4239±112 per unilateral SCN (Figure 1F). Therefore, Nms neurons represent ~40% of all SCN neurons and encompass most but not all neurons expressing AVP and VIP (Figure 1F and 1G).
Inducible and Reversible Overexpression of the ClockΔ19 Transgene in Nms Neurons Causes Period Lengthening
Having characterized the restricted expression of Nms within a subset of cells in the SCN, we next asked whether Nms neurons are capable of acting as pacemaking cells that control behavioral circadian rhythms. To examine this hypothesis, we assessed whether lengthening the molecular oscillation of Nms neurons by overexpression of the ClockΔ19 transgene can lengthen the in vivo circadian period. The ClockΔ19 mutation was initially identified as a dominant negative mutation resulting from a single nucleotide transversion (King et al., 1997; Vitaterna et al., 1994). Heterozygous ClockΔ19 mutant and transgenic mice that express tetO-ClockΔ19 transgene under the regulation of the secretogranin II (Scg2) promoter, which is expressed in essentially all of SCN neurons, exhibit a lengthened circadian period of ~24.4–24.5 h (Hong et al., 2007).
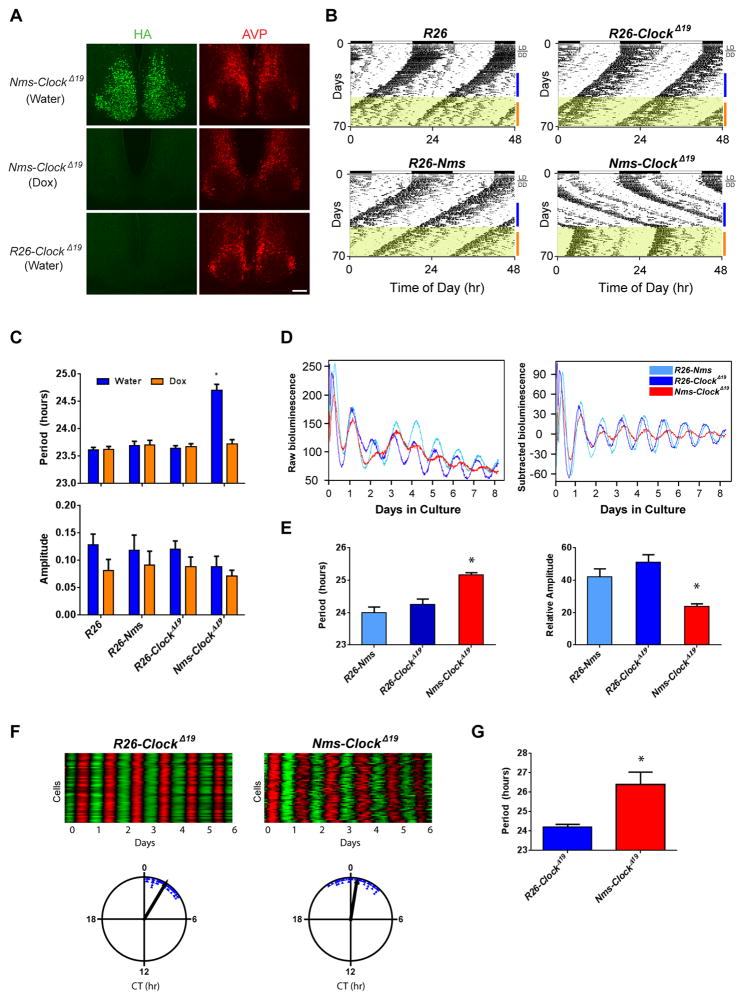
In Nms-ClockΔ19 mice, the ROSA26 promoter-driven tTA overexpresses the tetO-ClockΔ19 transgene in Nms neurons in a manner repressible by doxycycline (Dox). Control mice contain the homozygous ROSA-STOP-tTA loci but lack either the Nms-iCre transgene (referred to as R26-ClockΔ19), the tetO-ClockΔ19 transgene (referred to as R26-Nms), or both transgenes (referred to as R26). To determine whether the ClockΔ19 transgene can be properly turned off, Dox water was given for 7 days prior to immunostaining using anti-HA antibody. By day 7, the CLOCKΔ19-HA protein was nearly undetectable (Figure 2A). As expected, Nms-ClockΔ19 mice administered with water exhibited high levels of transgene expression while the control R26-ClockΔ19 mice had no detectable expression in the SCN. Therefore, the ClockΔ19 transgene can be conditionally regulated by Dox in the SCN of Nms-ClockΔ19 mice.
Figure 2. Reversible Overexpression of ClockΔ19 in Nms Neurons Lengthens Circadian Period.
(A) Immunostaining of HA (green) on SCN sections of Nms-ClockΔ19 mice administered with water (top), 1 week of Dox (middle), and R26-ClockΔ19 mice given water (bottom). Immunostaining of AVP peptide (red) was performed as a staining control. The scale bar represents 100 μm.
(B) Representative actograms of Nms-ClockΔ19 transgenic (bottom-right) and genetic controls (R26: top-left; R26-ClockΔ19: top-right; R26-Nms: bottom-left). All mice were initially placed in 12:12 LD for at least 7 days then transferred into DD, as indicated on the plot. Dox (10 μg/ml) water was administered during the intervals highlighted in yellow. Colored bars to the right of the actograms represent the days of analyses presented in (C).
(C) Quantification of circadian activity in Nms-ClockΔ19 transgenic and genetic controls. A significantly longer freerunning period was observed in water-treated Nms-ClockΔ19 mice compared to all other groups (two-way ANOVA, *p< 0.05 by Tukey’s post-hoc test). Mean freerunning amplitude was not different among water- or Dox-treated mice across all genotypes (two-way ANOVA, p>0.05 by Tukey’s post-hoc test). Values are means ± SEM (R26, n=6; R26-Nms, n=6; R26-ClockΔ19, n=12; Nms-ClockΔ19, n=7).
(D) Representative PER2::LUC bioluminescence records from the SCN of Nms-ClockΔ19;Luc mice and controls. Raw and baseline-subtracted plots are both displayed (R26-Nms: light blue trace; R26-ClockΔ19: dark-blue trace; Nms-ClockΔ19: red trace).
(E) Quantification of average SCN period and amplitude in Nms-ClockΔ19 and control mice. Nms-ClockΔ19 SCN exhibit longer mean circadian period and lower circadian amplitude compared to R26-Nms and R26-ClockΔ19 SCN (one-way ANOVA, *p< 0.05 by Tukey’s post-hoc test). Values are means ± SEM (R26-Nms, n=9; R26-ClockΔ19, n=7; Nms-ClockΔ19, n=8).
(F) Representative heatmap and Rayleigh plots of PER2::LUC oscillation in 50 neurons from the SCN of adult R26-ClockΔ19 and Nms-ClockΔ19 mice. In the heatmap, the red corresponds to peak bioluminescence and the green to trough. The length of the arrow in the Rayleigh plot represents the strength of synchronization.
(G) Quantification of average circadian period of individual SCN neurons from R26-ClockΔ19 and Nms-ClockΔ19 mice (Student’s t test, *p<0.0001). Values are as means ± SD (R26-ClockΔ19, n=50; Nms-ClockΔ19, n=50).
We next assessed the wheel-running activity of Nms-ClockΔ19 mice along with controls to examine their behavioral circadian rhythms. As expected, all mice entrained to light/dark (LD) 12:12 cycle, exhibiting primarily nocturnal activity. Upon release into constant darkness (DD), however, Nms-ClockΔ19 mice notably displayed a lengthened circadian period approximately one hour longer than the three control groups of mice without any effect on the robustness of the rhythm (24.70 ± 0.11 hr vs. 23.61 ± 0.05, 23.69 ± 0.08, and 23.65 ± 0.05 hr respectively; Figures 2B and C). To determine whether this lengthened period is dependent on the expression of the ClockΔ19 transgene, Dox was given to turn off transgene expression. During Dox administration, control mice continued to run with an unaltered circadian period, while Nms-ClockΔ19 mice displayed a rapid reversal of the lengthened period to a mean circadian period of 23.72 ± 0.08 hours, comparable with the control mice (23.62 ± 0.06, 23.71 ± 0.08, 23.68 ± 0.05; Figure 2B and 2C). Therefore, the conditional expression of the ClockΔ19 transgene exclusively in SCN neurons marked by Nms is sufficient to lengthen the daily rhythms in locomotor activity, suggesting that Nms neurons are capable of dictating behavioral circadian period.
To investigate the molecular effects of ClockΔ19 transgene expression in Nms neurons of the SCN, we crossed Nms-ClockΔ19 to mice carrying the PER2::LUC reporter (Yoo et al., 2004) (referred to as Nms-ClockΔ19;Luc). In concordance with the in vitro rhythms of heterozygous ClockΔ19 mutant SCN (Vitaterna et al., 2006), SCN explants from Nms-ClockΔ19;Luc mice expressed a longer period and lower amplitude in PER2::LUC oscillation compared to SCN cultures from R26-Nms and R26-ClockΔ19 control mice carrying the PER2::LUC reporter (Figures 2D and 2E). Similarly, on a single-cell level, we observed that Nms-ClockΔ19;Luc SCN neurons exhibit a longer circadian period compared to control SCN neurons (Figures 2F and 2G). Since Nms is expressed in only ~40% of the entire SCN, these findings suggest that Nms neurons are capable of reorganizing and lengthening the rhythmic oscillation of the entire SCN network to elicit a longer in vivo circadian period.
Overexpression of the ClockΔ19 Transgene in Vip Neurons Does Not Lengthen Behavioral Circadian Period
While properties of the SCN neuronal circuitry are still being elucidated, VIP signaling has been shown to play critical roles in the SCN circuitry (Aton et al., 2005; Colwell et al., 2003; Harmar et al., 2002; Maywood et al., 2011). Since the majority of Nms neurons express VIP (Figure 1D), we next asked whether lengthening the molecular clock of Vip-expressing neurons is sufficient to lengthen in vivo circadian period. Utilizing an analogous strategy used to generate Nms-ClockΔ19 mice, mice with Cre recombinase knocked into the Vip locus (Vip-Cre) (Taniguchi et al., 2011) were crossed to ROSA-STOP-tTA and tetO-ClockΔ19 mice to generate transgenic mice that conditionally overexpress the ClockΔ19 transgene in Vip cells (Vip-ClockΔ19). Double immunostaining using anti-HA and anti-VIP antibodies found that ~97% of Vip neurons express HA (Figure S5A and S5B), indicating that the ClockΔ19 transgene is faithfully overexpressed in endogenous Vip neurons in the SCN. As expected, no HA-immunoreactive cells co-localized with AVP-expressing cells (Figure S5C). Next, we assessed the locomotor activity of Vip-ClockΔ19 mice along with control mice that lack the ClockΔ19 transgene (referred to as R26-Vip). Interestingly, no differences in freerunning period, amplitude, or average activity counts were observed between the two groups under DD (Figure S5D and S5E). These results reveal that altering the intracellular clock of Vip neurons is insufficient to modulate behavioral circadian rhythms and that the long period phenotype of Nms-ClockΔ19 is not simply due to the overexpression of the ClockΔ19 transgene in Vip neurons.
The Loss of Bmal1 in Nms Neurons Abolishes Behavioral Circadian Rhythms
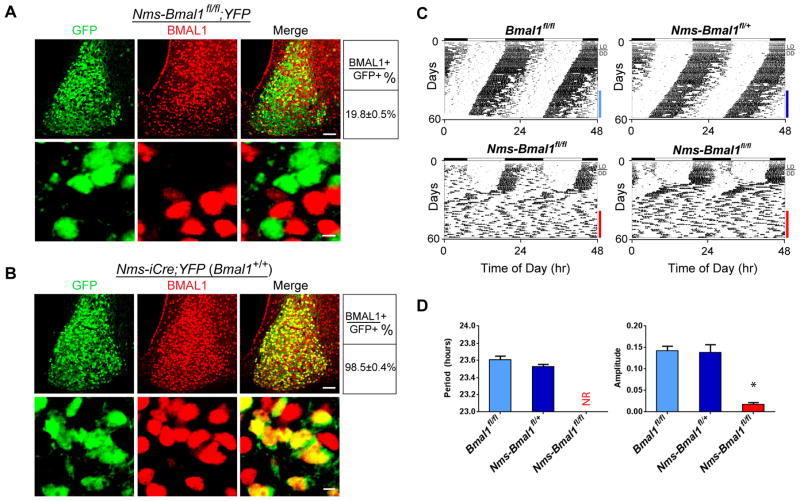
Having found that Nms neurons are capable of dictating in vivo circadian period, we next asked whether intracellular clock oscillations in Nms neurons are necessary for the generation of behavioral circadian rhythms. To address this, we generated mice lacking a functional molecular clock in Nms neurons by breeding mice with floxed Bmal1 alleles (Bma1fl/fl) (Storch et al., 2007) to Nms-iCre transgenic mice (Nms-Bmal1fl/fl). Conventional Bmal1−/− mice exhibit a loss of behavioral circadian rhythmicity along with a variety of phenotypes such as decreased viability/survivability, presumably due to the loss of BMAL1 in peripheral tissues (Bunger et al., 2000; Kondratov et al., 2006; McDearmon et al., 2006; Sun et al., 2006). We find that Nms-Bmal1fl/fl mice appear normal in appearance and were produced with expected Mendelian ratios, a finding in agreement with the restricted expression pattern of Nms. To confirm that the BMAL1 protein is deleted in Nms neurons of Nms-Bmal1fl/fl mice, we crossed Nms-Bmal1fl/fl mice to ROSA-STOP-YFP reporter mice (Srinivas et al., 2001) to generate mice that express YFP and two floxed alleles of Bmal1 in Nms neurons (Nms-Bmal1fl/fl;YFP) (Figure 3). Prior to this, we confirmed that YFP-expressing neurons recapitulate NMS-expressing neurons by crossing Nms-iCre to ROSA-STOP-YFP mice (Nms-iCre;YFP). Double immunostaining with anti-NMS and anti-GFP (which recognizes YFP) reveals ~95–99% overlap between YFP- and NMS-expressing neurons (Figure S6). Next, using anti-BMAL1 and anti-GFP antibodies, we double-stained Nms-Bmal1fl/fl;YFP SCN and found that BMAL1 immunoreactivity only remains in ~20% of YFP-positive neurons (Figure 3A). This verifies that BMAL1 is specifically deleted in ~80% of Nms neurons in Nms-Bmal1fl/fl SCN. As a control, we also double-stained Nms-iCre;YFP SCN, which contains both wild-type Bmal1 alleles, with anti-BMAL1 and anti-GFP and found that nearly all of YFP-positive neurons express BMAL1 (Figure 3B). To examine the behavioral effect of the loss of Bmal1 in Nms neurons, we monitored the wheel-running activity of Nms-Bmal1fl/fl along with controls including Bmal1fl/fl, which lack the Nms-iCre transgene, and Nms-Bmal1fl/+ mice, which have one intact Bmal1 allele. Under 12:12 LD, all mice exhibit normal bouts of restricted activity during the dark phase. Under DD, control mice exhibit normal freerunning period and amplitude while Nms-Bmal1fl/fl mice (14 of 14) displayed a dramatic loss of daily behavioral rhythms with a significant reduction in relative power in the circadian range (Figure 3C and 3D). Interestingly, unlike conventional Bmal1−/− mice, Nms-Bmal1fl/fl mice did not lose behavioral rhythms immediately but exhibited sustained rhythmicity for an average of 12 days upon release from LD to DD, suggesting the presence of mechanisms capable of transiently compensating for the loss of molecular clocks in 80% of Nms neurons. Nevertheless, these data reveal that a functional molecular clock in Nms neurons is necessary for the sustained generation of behavioral circadian rhythms.
Figure 3. Loss of Bmal1 in Nms Neurons Abolishes Behavioral Circadian Rhythms.
(A) Double staining of YFP (green) and BMAL1 (red) on SCN sections of Nms-Bmal1fl/fl;YFP mice using anti-GFP and anti-BMAL1 antibodies, respectively. The scale bars are 50 μm (top) and 5 μm (bottom). Quantitative data are shown at the right.
(B) Double staining of YFP (green) and BMAL1 protein (red) on SCN sections of Nms-iCre;YFP mice using anti-GFP and anti-BMAL1 antibodies, respectively.
(C) Representative actograms of Nms-Bmal1fl/fl (bottom) along with Bmal1fl/fl (top-left) and Nms-Bmal1fl/+ mice (top-right). Colored bars represent the days of analyses presented in (D).
(D) Quantification of circadian activity in Nms-Bmal1fl/fl, Bma11fl/fl, Nms-Bmal1fl/+ mice. Nms-Bmal1fl/fl exhibited no coherent rhythms (NR) and a low average amplitude in the circadian range compared to Bmal1fl/fl and Nms-Bmal1fl/+ control mice (one-way ANOVA, *P<0.05 by Tukey’s post-hoc test). Values are mean ± SEM (Bmal1fl/fl, n=7; Nms-Bmal1fl/+, n=6; Nms-Bmal1fl/fl, n=14).
Inducible Overexpression of the Per2 Transgene in Nms Neurons Reversibly Disrupts Behavioral Circadian Rhythms
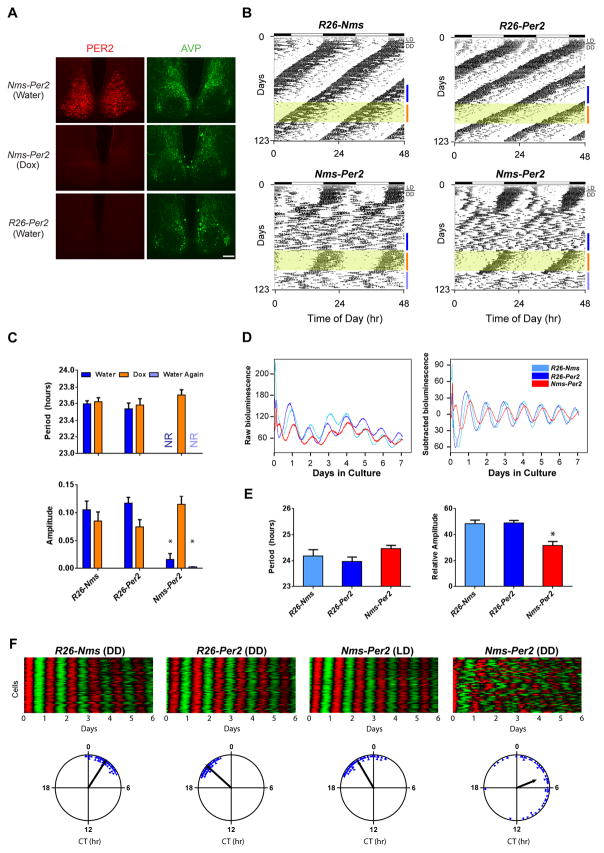
We hypothesized that light entrainment plays a key role in driving the transient behavioral rhythmicity of Nms-Bmal1fl/fl since this phenotype occurred after release from LD into DD. To investigate this, we took another genetic approach in order to reversibly disrupt the molecular clock of Nms neurons in DD. Nms-iCre was crossed to ROSA-STOP-tTA and tetO-Per2 transgenic mice (Chen et al., 2009a) to generate mice that overexpress the Per2 transgene in Nms neurons (Nms-Per2). The constitutive overexpression of Per2 abolishes circadian rhythms at a cellular and organismal level by clamping the molecular oscillations of the core clock (Chen et al., 2009a). First, to validate that the Per2 transgene can be overexpressed in a reversible manner, the SCN of water and Dox-administered Nms-Per2 mice along with control mice lacking the Nms-iCre transgene (R26-Per2) were immunostained using anti-PER2 antibody. The SCN of these mice were collected in the morning when the endogenous PER2 is normally at its nadir (Field et al., 2000). As expected, water-maintained Nms-Per2 mice had high levels of PER2 expression while Dox-administered mice exhibit minimal PER2 immunoreactivity that is comparable to the R26-Per2 control (Figure 4A).
Figure 4. Reversible Overexpression of Per2 in Nms Neurons Causes the Loss of Behavioral Circadian Rhythms and Reduced Network Synchrony.
(A) Immunostaining of the PER2 (red) on SCN sections of Nms-Per2 mice administered with water (top), 1 week of Dox (middle), and R26-Per2 mice given water (bottom). Immunostaining of AVP (green) is shown for staining control. The scale bar represents 100 μm.
(B) Representative actograms of Nms-Per2 (bottom) and genetic controls (R26-Nms: top-left; R26-Per2: top-right). Dox (10 μg/ml) was administered during the intervals highlighted in yellow. Colored bars represent the days of analyses presented in (C).
(C) Quantification of circadian activity in Nms-Per2 transgenic and genetic controls. Nms-Per2 mice administered with water displayed no coherent rhythms (NR) and a low average amplitude in the circadian spectrum (two-way ANOVA, *p< 0.05 by Tukey’s post-hoc test). No significant differences in mean circadian period or amplitude were found between Dox-administered Nms-Per2 mice and control mice (two-way ANOVA, p> 0.05 by Tukey’s post-hoc test). Values are mean ± SEM (R26-Nms, n=13; R26-Per2, n=9; Nms-Per2, n=13).
(D) Representative PER2::LUC bioluminescence records from the SCN of Nms-Per2 and control mice maintained in DD. Raw and baseline-subtracted plots are both displayed (R26-Nms: light-blue trace; R26-Per2: dark blue trace; Nms-Per2: red trace).
(E) Quantification of average SCN period and amplitude in Nms-Per2 and controls. Mean circadian period was not significantly different between genotypes (one-way ANOVA, p>0.05 by Tukey’s post-hoc test). The mean circadian amplitude of Nms-Per2 SCN is significantly lower compared to R26-Nms and R26-Per2 SCN (one-way ANOVA, *p< 0.05 by Tukey’s post-hoc test). Values are mean ± SEM (R26-Nms, n=6; R26-Per2, n=11; Nms-Per2, n=6).
(F) Representative raster and Rayleigh plots of PER2::LUC oscillation in 50 individual neurons from the SCN of adult R26-Nms, R26-Per2, and Nms-Per2 mice. R26-Nms and R26-Per2 SCN were cultured from mice taken out of DD while Nms-Per2 SCN collected from mice maintained in LD and in DD are both displayed here. In the raster plot, the red corresponds to peak bioluminescence and the green to trough. The length of the arrow in the Rayleigh plot represents the strength of synchronization.
Next, we subjected Nms-Per2 and control mice that lack either tetO-Per2 or Nms-iCre (R26-Nms and R26-Per2, respectively) to wheel-running. In DD, as expected, the controls exhibited normal freerunning period and amplitude both in water and Dox-treated conditions. In contrast, much like the Nms-Bmal1fl/fl mice, Nms-Per2 mice (13 of 13) displayed a complete loss of normal circadian rhythms after an average of 18 days of transient rhythmicity post-release into DD from LD (Figure 4B and 4C). During Dox administration, Nms-Per2 mice displayed a rapid recovery of circadian rhythms to a mean period comparable to that of the genetic controls (Figures 4B and 4C). To abolish the molecular rhythms of Nms neurons while in DD, we then re-administered regular water to turn on the Per2 transgene again. Interestingly, Nms-Per2 mice rapidly lost coherent daily rhythms without the transient rhythmic activity observed when they were initially released into DD from LD (Figure 4B). These results suggest that LD entrainment can transiently sustain rhythmicity in mice lacking a functional molecular clock in Nms neurons.
Disrupting the Molecular Clock of Nms Neurons Causes Reduced Cellular Synchrony
Given that SCN cells are normally coupled together to synchronously express coherent rhythmic outputs in behavior, we speculated that the loss of behavioral circadian rhythms in Nms-Per2 (and Nms-Bmal1fl/fl) mice may be due to a disrupted SCN cellular network that cannot maintain synchrony without a molecularly rhythmic Nms neuronal population. To test this hypothesis, we crossed Nms-Per2 mice to mice carrying the PER2::LUC reporter (Nms-Per2;Luc) (Yoo et al., 2004) in order to examine the molecular clock on the level of the SCN. We observed that SCN cultures from Nms-Per2;Luc mice that have lost coherent behavioral rhythms in DD displayed notably lower amplitudes in PER2::LUC oscillation compared to controls that lack either the Nms-iCre or the tetO-Per2 transgene (R26-Nms;Luc and R26-Per2;Luc, respectively) (Figure 4D and 4E). The presence of less robust rhythms in the Nms-Per2 SCN suggests that the disruption of the molecular clock in Nms neurons does not completely abolish synchronous oscillations among the remaining unaltered neurons but compromises cellular synchrony enough such that daily rhythms on a behavioral level are lost. To explore this possibility, we imaged real-time PER2::LUC bioluminescence of Nms-Per2;Luc SCN along with the controls on a single cell level. As expected, neurons of R26-Nms;Luc and R26-Per2;Luc SCN exhibited synchronized oscillations as represented by the raster and Rayleigh plots (Figure 4F). Interestingly, similar to the controls, SCN neurons from Nms-Per2;Luc mice maintained in LD prior to sacrifice also displayed synchronous oscillations (Figure 4F), which suggests that LD entrainment preserves synchrony in Nms-Per2 mice and may be responsible for the transient behavioral rhythmicity of Nms-Per2 mice after release from LD into DD (Figure 4B). In stark contrast, Nms-Per2;Luc SCN collected in DD after the loss of daily behavioral rhythms exhibited desynchronized and less robust single-cell rhythms (Figure 4F). Likewise, imaging of SCN neurons from Nms-Bmal1fl/fl mice crossed to PER2::LUC also displayed desynchronized and unstable single-cell rhythms compared to Bmal1fl/fl mice crossed to PER2::LUC (Figure S6). These results suggest that network synchrony of the SCN is compromised when the molecular clock in Nms neurons is disrupted and reveal that cell-autonomous oscillations in Nms neurons are required for proper synchrony within the SCN cellular network.
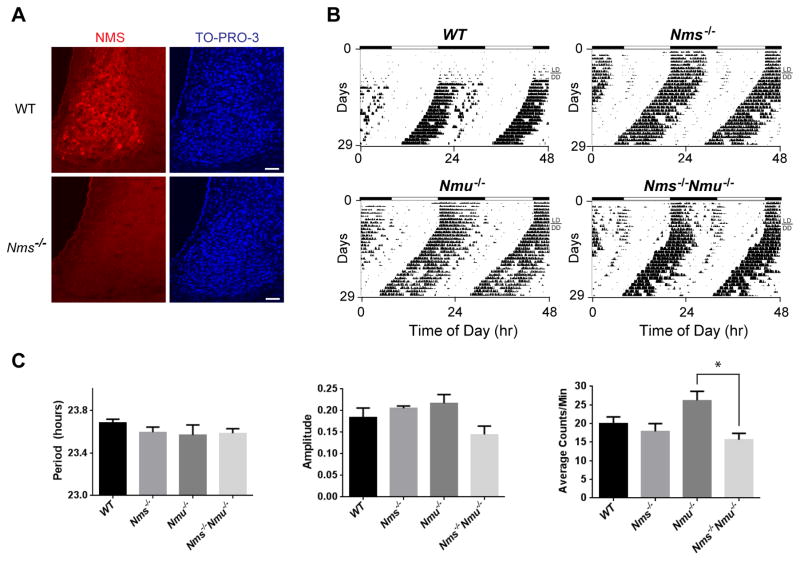
Nms/Nmu signaling is not essential for the generation of behavioral circadian rhythms
Intercellular signaling molecules in the SCN involve neuropeptides, neurotransmitters, gap junctions, cytokines, and growth factors (Lee et al., 2010). To elucidate the signaling mechanisms by which Nms neurons communicate with other neurons to coordinate synchrony among the SCN network, we generated knockout mice lacking the Nms gene (Nms−/−). After verifying that no NMS peptide was detected in the SCN of Nms−/− mice (Figure 5A), we produced mutant mice that lack both Nms and its closely related paralog, neuromedin U (Nmu) by breeding Nms−/− to Nmu−/− mice (Hanada et al., 2004). To determine the effects of the loss of Nms/Nmu signaling on behavioral circadian rhythms, we monitored wheel-running activity in LD and in DD. Nms−/−, Nmu−/−, and Nms−/−Nmu−/− mice all exhibited normal freerunning periods and amplitudes similar to wild-type (WT) littermates (Figure 5B and 5C), suggesting that Nms/Nmu signaling is not required for the generation of behavioral circadian rhythms.
Figure 5. Nms−/−/Nmu−/− Mice Exhibit Normal Freerunning Circadian Rhythms.
(A) Immunostaining of NMS (red) on SCN sections of C57BL/6J wild-type (WT) mice and Nms−/− mice. NMS immunoreactivity was not detected in Nms−/− mice. The scale bar is 50 μm.
(B) Representative actograms of WT (top-left panel), Nms−/− (top-right), Nmu−/− (bottom-left), and Nms−/−/Nmu−/− mice (bottom-right).
(C) Quantification of circadian activity in WT, Nms−/−, Nmu−/−, and Nms−/−/Nmu−/− mice. Mean freerunning periods and amplitudes were not significantly different between genotypes (one-way ANOVA, p>0.05 by Tukey’s post-hoc test). Total activity counts of Nmu−/− mice were significantly greater than that of Nms−/−/Nmu−/− mice (one-way ANOVA, *p<0.05 by Tukey’s post-hoc test). Values are mean ± SEM (WT, n=5; Nms−/−, n=8; Nmu−/−, n=5; Nms−/−Nmu−/−, n=13).
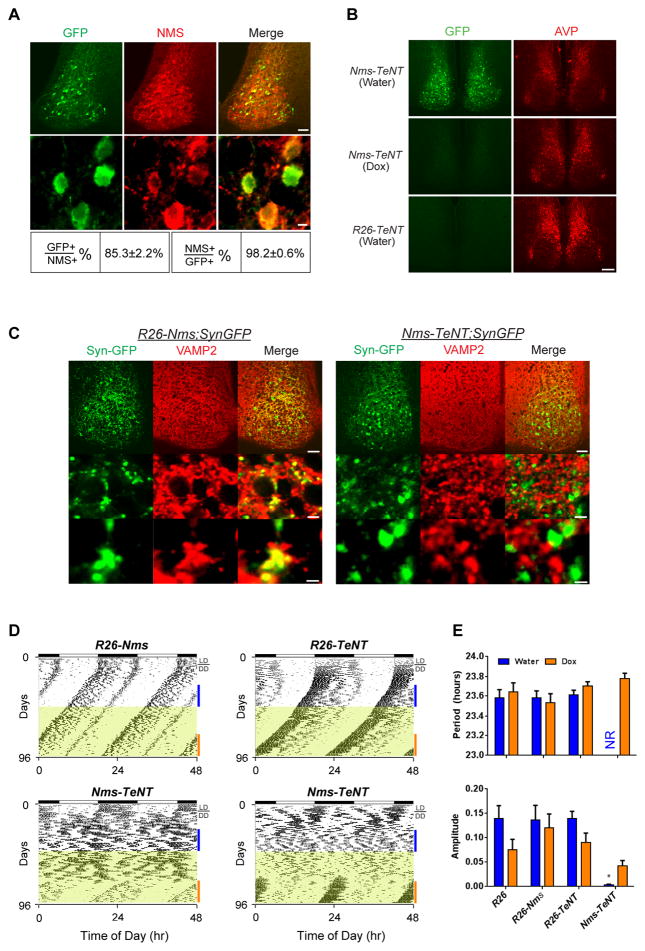
Reversible Inhibition of Synaptic Transmission from Nms Neurons Abolishes Behavioral Circadian Rhythms
Recent evidence suggests that neuropeptides and neurotransmitters (i.e. GABA) packaged in synaptic vesicles play prominent but likely redundant roles in the synchronization of SCN neurons. In vitro, for example, AVP and GRP can restore synchronous rhythms in SCN neurons lacking VIP (Brown et al., 2005; Maywood et al., 2011). In vivo, only ~25–60% of mice lacking Vip or Vipr2 lose circadian rhythmicity (Aton et al., 2005; Colwell et al., 2003; Hughes et al., 2004), suggesting that additional synchronizing agents can compensate for the loss of Vip. To evaluate the consequences of blocking the release of additional signaling mediators from synaptic vesicles of Nms neurons, we crossed Nms-iCre to ROSA-STOP-tTA and tetO-tetanus toxin (TeNT) mice (Yamamoto et al., 2003) that express a fusion gene of TeNT light chain and EGFP under the tetO promoter to generate mice deficient in vesicular synaptic transmission from Nms neurons (referred to as Nms-TeNT) (Figure 6). TeNT is an endopeptidase that specifically cleaves VAMP2, which is required for activity-dependent synaptic release of neurotransmitters and neuropeptides (Schoch et al., 2001). In accordance with anti-GFP immunostaining showing minimal TeNT expression outside of the SCN (Figure S7), the resulting Nms-TeNT mice are viable and fertile with no readily apparent health detriments. Double staining of GFP and NMS shows that TeNT is specifically expressed in Nms neurons with ~85% of NMS-positive cells expressing GFP and ~98% of GFP-expressing cells overlapping with NMS (Figure 6A). To assess the reversibility of the tetO-TeNT transgene, we used anti-GFP immunostaining to verify that TeNT expression is minimal in the SCN of Dox-administered Nms-TeNT mice compared to the SCN of water-treated Nms-TeNT and mice lacking the Nms-iCre transgene (R26-TeNT) (Figure 6B). Next, to confirm that VAMP2 is cleaved in the synapses of Nms neurons of Nms-TeNT mice, we crossed Nms-TeNT to mice that express synaptophysin-GFP (SynGFP) under the tetO promoter (tetO-SynGFP) (Li et al., 2010) to generate mice that express SynGFP and TeNT in the synapses of Nms neurons (Nms-TeNT;SynGFP) along with controls that express SynGFP but not TeNT in Nms synapses (R26-Nms;SynGFP). Double staining using anti-VAMP2 and anti-GFP shows that while VAMP2 is intact in both non-GFP and the GFP-expressing synapses in the R26-Nms;SynGFP control, VAMP2 is only expressed in non-GFP synapses of Nms-TeNT;SynGFP SCN (Figure 6C), indicating that VAMP2 is cleaved specifically in Nms neurons. Altogether, these results indicate that TeNT can reversibly block vesicular synaptic signaling (Nakashiba et al., 2008) from Nms neurons in the Nms-TeNT SCN.
Figure 6. Reversible Overexpression of TeNT in Nms Neurons Abolishes Behavioral Circadian Rhythms.
(A) Double staining of GFP (green) and NMS (red) on SCN sections of Nms-TeNT mice. Scale bars are 50 μm (top panels) and 5 μm (bottom panels).
(B) Immunostaining of GFP (green) on SCN sections of Nms-TeNT mice administered with water (top), 1 week of Dox (middle), and R26-TeNT mice given water (bottom). Immunostaining of AVP (red) is shown as a staining control. The scale bar represents 100 μm.
(C) Double staining of GFP (green) and VAMP2 (red). VAMP2 is intact in Nms synapses marked by synaptophysin-GFP (Syn-GFP) in R26-Nms;SynGFP mice (left panels) but undetectable in GFP-positive synapses of Nms-TeNT;SynGFP SCN (right panels). GFP can be observed in the cell body (site of synthesis) and in the synapses. Scales bars are 50 μm (top), 5 μm (middle), and 2 μm (bottom).
(D) Representative actograms of Nms-TeNT transgenic (bottom panels) and genetic controls (R26-Nms: top-left; R26-TeNT: top-right; R26: not displayed). Dox (20 μg/ml) was administered during the intervals highlighted in yellow. Colored bars represent the days of analyses presented in (E).
(E) Quantification of circadian activity in Nms-TeNT and genetic controls. Water-treated Nms-TeNT mice displayed no coherent rhythms (NR) in the circadian range and a low mean amplitude compared to other genotypes (two-way ANOVA, *p< 0.05 by Tukey’s post-hoc test). No significant differences in average circadian period or amplitude were found between Dox-administered Nms-TeNT mice and Dox-administered controls (two-way ANOVA, *p> 0.05 by Tukey’s post-hoc test). Values are mean ± SEM (R26, n=7; R26-Nms, n=5; R26-TeNT, n=10; Nms-TeNT, n=9).
To examine the in vivo effects of inhibiting synaptic transmission from Nms neurons, we assessed the locomotor activity rhythms of Nms-TeNT mice along with genetic controls that lack either the tetO-TeNT transgene (R26-Nms) or the Nms-iCre transgene (R26-TeNT). In DD, interestingly, Nms-TeNT mice exhibited a loss of coherent daily rhythms (9 of 9) while control mice displayed normal freerunning periods and amplitudes (Figure 6D and 6E). To examine whether circadian rhythmicity can be recovered when synaptic transmission is restored, Dox was given to turn off the TeNT expression. During Dox administration, Nms-TeNT mice displayed a complete recovery of circadian rhythmicity after an average waiting time of 28 days. Once recovered, the mean period and amplitude of the recovered rhythmicity was comparable to that of the genetic controls on Dox (Figures 6D and 6E). Thus, synaptic transmission from Nms neurons is necessary for coherent behavioral circadian rhythms.
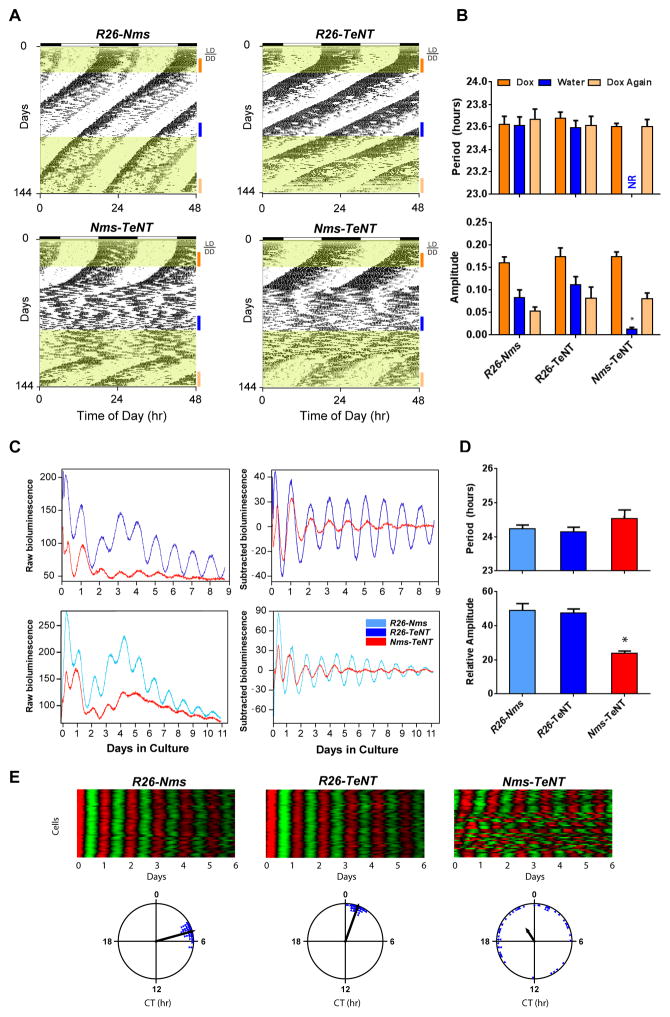
Post-developmental Inhibition of Synaptic Transmission from Nms Neurons Resets the Freerunning Phase of Behavioral Circadian Rhythms
To further exclude the possibility that the circadian phenotype seen in Nms-TeNT mice is due to a developmental defect, we raised Nms-TeNT mice along with R26-Nms and R26-TeNT controls on Dox from the start of conception. In 12:12 LD, all mice of adult age entrained properly. In DD, Dox-maintained Nms-TeNT mice along with genetic controls exhibited normal freerunning rhythms (Figure 7A and 7B). After >2 weeks of DD, Dox was replaced with water for the first time to turn on the TeNT transgene in Nms-TeNT mice. All Nms-TeNT mice lost coherent behavioral circadian rhythms as expected, after an average of 26 days of sustained rhythmicity. Subsequently, the TeNT transgene was turned off again by Dox re-administration. After an average of 28 days on Dox, normal rhythms re-emerged, interestingly, at a phase different from the prior phase (Figure 7A). These results suggest that synaptic transmission from Nms neurons is required for the intrinsic timekeeping machinery of the adult mouse SCN.
Figure 7. Reversible Overexpression of TeNT in Nms Neurons Causes Resetting of the Clock and Reduced Network Synchrony.
(A) Representative actograms of Nms-TeNT (bottom) and genetic controls (R26-Nms: top-left; R26-TeNT: top-right). Dox was administered from conception until the switch to water during wheel running recording. Dox administration is highlighted in yellow. Colored bars represent the days of analyses presented in (B).
(B) Quantification of circadian activity in Nms-TeNT and genetic controls collected from mice maintained in DD. Water-treated Nms-TeNT mice displayed no coherent rhythms (NR) in the circadian range and a low average amplitude in the circadian spectrum (two-way ANOVA, *p<0.05 by Tukey’s post-hoc test). Re-administration of Dox (20 μg/ml) (Dox Again) rescues the circadian rhythms of Nms-TeNT mice. No significant differences in mean circadian period or amplitude were found between Nms-TeNT and controls under the same Dox treatment (two-way ANOVA, *p> 0.05 by Tukey’s post-hoc test). Values are mean ± SEM (R26-Nms, n=6; R26-TeNT, n=8; Nms-TeNT, n=13).
(C) Representative bioluminescence records from the SCN of Nms-TeNT mice and controls maintained in DD. Raw and baseline-subtracted plots are both displayed (R26-Nms: light-blue trace; R26-TeNT: dark-blue trace; Nms-TeNT: red trace).
(D) Quantification of average SCN period and amplitude in Nms-TeNT and controls. No significant differences in mean circadian period were found between genotypes. Nms-TeNT SCN exhibited lower amplitudes of PER2::LUC rhythms compared to controls (one-way ANOVA, *p< 0.05 by Tukey’s post-hoc test). Values are mean ± SEM (R26-Nms, n=12; R26-TeNT, n=9; Nms-TeNT, n=12).
(E) Representative raster and Rayleigh plots of PER2::LUC oscillation in 50 individual neurons from the SCN of adult R26-Nms, R26-TeNT, and Nms-TeNT mice. In the raster plot, the red corresponds to peak bioluminescence and the green to trough. The length of the arrow in the Rayleigh plot represents the strength of synchronization.
Synaptic Transmission from Nms Neurons is Required for Synchronization of the SCN Network
We hypothesized that the disruption of SCN network synchrony underlies the phenotype observed in Nms-TeNT mice. To examine the molecular effects of disrupting synaptic transmission in Nms neurons, we crossed Nms-TeNT mice to PER2::LUC mice (Nms-TeNT;Luc). Similar to the Nms-Per2;Luc mice, PER2::LUC oscillation in SCN cultures from DD-maintained Nms-TeNT;Luc mice exhibited lower amplitudes compared to tissues from mice lacking either the Nms-iCre or the tetO-TeNT transgene (R26-Nms;Luc and R26-TeNT;Luc, respectively) (Figure 7C and 7D). The period of Nms-TeNT;Luc SCN was not significantly different from that of genetic controls (Figure 7C). To examine the SCN network synchrony, we next imaged the PER2::LUC oscillation of individual neurons from Nms-TeNT;Luc SCN along with controls maintained and sacrificed under DD. We observed that, like the neurons of Nms-Per2;Luc SCN, a subset of neurons (~half) in the Nms-TeNT;Luc SCN were desynchronized; however, the remaining SCN neurons were rhythmic and coherent in phase (Figure 7E). This partial disruption of the single-cell population is consistent with the ensemble average rhythms seen in the entire SCN (Figure 7C and 7D). These results suggest that synaptic transmission from Nms neurons generates the proper network synchrony required for the generation of behavioral circadian rhythms.
DISCUSSION
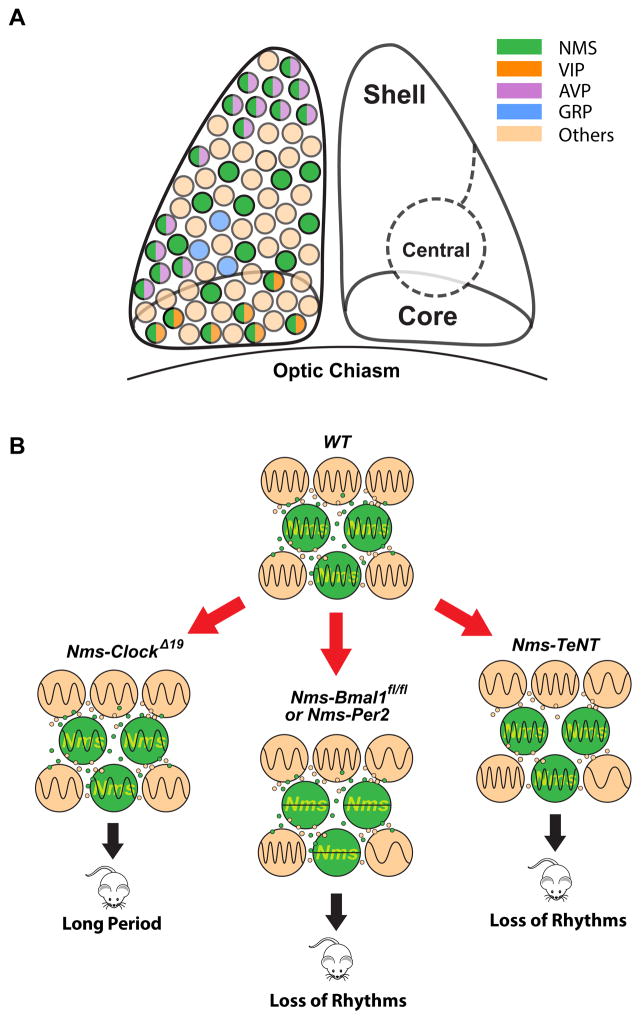
Circadian timekeeping in the SCN represents an exceptional system to dissect how a complex, evolved behavior is organized by a well-defined neuronal circuit with cell-autonomous and emergent network-level properties. A remarkable amount of heterogeneity can be found within the SCN cellular network, and a long-standing question of neurobiological interest has been whether a particular SCN cell type can dictate the intricate internal representation of time. Here, we have shown that lengthening the molecular clock of Nms neurons lengthens the SCN network along with the behavioral circadian period and disrupting the intracellular clock of Nms neurons abolishes coherent SCN and behavioral daily rhythms. We further demonstrated that blocking the synaptic transmission from Nms neurons also disrupts SCN network synchrony and leads to the loss of normal behavioral circadian rhythms, which when rescued, runs at a de novo phase. Our findings (summarized in Figure 8) reveal that Nms neurons define a population of pacemaking cells that are (i) capable of dictating in vivo circadian periodicity, (ii) required for the generation of behavioral circadian rhythms, and (iii) necessary for the synchronized timekeeping machinery of the SCN.
Figure 8. Schematic Summary of Findings.
(A) NMS-expressing neurons are localized in the core, central, and shell regions of the SCN and encompass the majority of VIP- and AVP-expressing neurons but do not overlap with GRP-producing neurons. A small number of VIP- or AVP-expressing neurons (~5% each) that do not express NMS are not depicted here.
(B) Green cells represent Nms neurons. Lengthening the intracellular circadian period of Nms neurons lengthens behavioral circadian period. Abolishing molecular rhythmicity of Nms neurons or blocking synaptic transmission from Nms neurons leads to the loss of coherent circadian rhythms.
Neuromedin S Defines a Set of Specialized Pacemaking Neurons in SCN
Here, we report that NMS is expressed in the majority of the VIP- and AVP-expressing neurons of the mouse SCN. In rats, it has been reported that NMS is only expressed in the core region of the SCN (Mori et al., 2005), although double immunostaining using core markers such as VIP was not shown. This discrepancy in localization is likely due to species differences in the expression of NMS. Although we found that the Nms−/− mouse exhibits normal behavioral circadian rhythms, intracerebroventricular (ICV) administration of NMS has been reported to elicit circadian phase shifts in rats (Mori et al., 2005), suggesting that NMS may be among several synchronizing agents that act in partially redundant manners in cellular communication.
The anatomical localization and functional connectivity of the Nms neuronal population appears to confer its ability to dictate behavioral circadian period and its requirement for the generation of in vivo circadian rhythms. Localized within core, central, and shell regions of the SCN, Nms cells are well situated to release VAMP2-dependent slow-acting neuropeptides and fast-acting neurotransmitters to neighboring neurons in order to coordinate the rhythms of other populations of oscillators in the SCN (Mohawk and Takahashi, 2011). It is apparent that not all sparsely distributed SCN neuronal populations are specialized for the generation of behavioral rhythms since the loss of Bmal1 from ~30% of SCN neurons using a Nestin-Cre promoter did not lead to the loss of behavioral circadian rhythms (Mieda and Sakurai, 2011). Another study that highlighted the difficulty in achieving circadian arrhythmicity showed that the loss of 65% of BMAL1 from a null/floxed Bmal1 (Bmal1fl/−) mice using a heterozygous pan-neuronal Synaptotamin-Cre (Syt10Cre/+) driver did not lead to arrhythmicity while the loss of 83% of BMAL1 using a homozygous Syt10Cre/Cre driver caused the loss of coherent rhythms (Husse et al., 2011). In contrast, in our study, the deletion of ~80% of Bmal1 from Nms neurons (Figure 3A), which represents a loss of Bmal1 in ~32% of all SCN neurons, leads to the loss of normal circadian rhythms (Figure 3C). This suggests that the disruption of daily rhythms observed in mice lacking a functional clock in Nms neurons cannot simply be attributed to the loss of a certain percentage (~30–40%) of cells within the SCN. In fact, partial lesions of the SCN had revealed that the survival of only a small percentage of SCN cells is adequate for behavioral circadian rhythms (Harrington et al., 1993; Meijer and Rietveld, 1989; Van den Pol and Powley, 1979). Transplantation studies have also shown that a small amount of SCN tissue is sufficient to restore rhythms in animals with complete SCN lesions (Ralph et al., 1990; Silver et al., 1990, 1996). These findings suggest that random ablation of any ~30–40% of SCN cells is unlikely to cause the disrupted daily rhythms seen in Nms-Bmal1fl/fl, Nms-Per2, and Nms-TeNT mice. Interestingly, it has been reported that lesioning an anatomically small region of the hamster SCN containing calbindin-D28 (CalB) leads to the loss of daily behavioral rhythms (Kriegsfeld et al., 2004). Although CalB is not expressed in adult mice and lesion studies are not cell-type specific, this report supports the presence of specific neuronal populations that are critical for the generation of circadian rhythms. It is worth noting that using mouse aggregation chimeras composed of wild-type and ClockΔ19 mutant cells, we have previously shown that with a few exceptions, the proportion of either cell type in mice determines the circadian behavior (Low-Zeddies and Takahashi, 2001). In contrast, we have shown here that ClockΔ19 transgene expression in a minority of SCN neurons marked by Nms dictate the circadian period. One difference between the two studies is that unlike the chimeric ClockΔ19 mice which contain either wild-type or mutant copies of Clock, Nms-ClockΔ19 mice are composed of the overexpressed ClockΔ19 transgene on a background of two wild-type Clock alleles.
Although nearly all Vip neurons in the SCN co-express Nms, the loss of coherent daily rhythms in Nms-TeNT mice cannot be merely attributed to the loss of VIP signaling. Nms-TeNT mice have a phenotype distinct from mouse models deficient in Vip signaling. Behaviorally, Vip−/− and Vipr2−/− mice exhibit impairments in light entrainment, advanced phase angles of activity onset, and losses of circadian rhythmicity in ~25–60% of mice in DD (Aton et al., 2005; Colwell et al., 2003; Hughes et al., 2004). Nms-TeNT mice, on the other hand, show normal LD entrainment and loss of coherent daily rhythms in all mice in DD. VIP signaling alone also cannot explain the capability of Nms neurons to dictate periodicity in vivo, since we found that lengthening the molecular rhythms of Vip neurons by ClockΔ19 transgene expression was insufficient to affect in vivo circadian period (Figure S4). Interestingly, a recent report found that activation of Gq signaling by expressing DREADD (designer receptor exclusively activated by designer drug) in Vip neurons increased the period of PER2::LUC oscillation across the SCN (Brancaccio et al., 2013). However, unlike the overexpression of the ClockΔ19 transgene which cell-autonomously interferes with the intracellular molecular feedback loop, Gq receptor activation leads to supraphysiological neuronal excitability and neurotransmitter release due to the upregulation of intracellular Ca2+ (Rogan and Roth, 2011). It will be interesting to see whether DREADD expression in Vip neurons can modulate circadian rhythms in vivo. Regardless, our results reveal for the first time in vivo that Nms neurons encompass the essential signaling factors necessary for the generation of coherent circadian rhythms.
Photic Signals Transiently Sustain Network and Behavioral Rhythms in Mice with a Disrupted Nms Cellular Pacemaker
Our data reveal that light entrainment can temporarily synchronize and rescue a SCN cellular network that lacks a functional Nms cellular pacemaker. Ex vivo, without light entrainment, the SCN network synchrony is lost when the intracellular clocks of Nms neurons are disrupted. When exposed to LD entrainment, however, network synchrony is preserved for at least 6 days in culture (Figure 4F). Consistent with this, behaviorally, transient rhythmicity was observed after the release of Nms-Bmal1fl/fl and Nms-Per2 from LD into DD. This phenotype is similar to what has been reported for some Clock Δ19 mutants (Vitaterna et al., 1994) and Per mutant lines in a 129/sv or mixed background (Bae et al., 2001; Zheng et al., 1999) [C57BL/6J Per1 or Per2 mutants do not become arrhythmic (Pendergast et al., 2009; Xu et al., 2007)]. Notably, however, transitory rhythmicity was not observed in Nms-Per2 mice when the disruption of the molecular clock occurred during DD. Since the effects of light on the circadian clock is thought to be mediated via Per1 and Per2 induction (Shigeyoshi et al., 1997; Yan and Okamura, 2002), it appears unlikely that LD exposure can directly rescue the rhythmicity of Nms neurons that are deficient in the core molecular clock. Rather, it appears more likely that the direct recipients of photic entraining signals are non-Nms neurons which transiently sustain the disrupted clocks of Nms neurons via coupling. Indeed, intercellular coupling among SCN neurons can rescue the rhythmicity of defective oscillators (Liu et al., 2007). Ultimately, however, intercellular signaling outputs from Nms neurons are required for the expression of LD-sustained rhythmicity since water-raised Nms-TeNT mice exhibit minimal delay in the loss of coherent rhythms after release into DD from LD (Figure 5D). Although further experimentation will be necessary, we speculate that light input to the retinorecipient Grp neuronal population (Aida et al., 2002; Karatsoreos et al., 2004), which are largely devoid of Nms expression (Figure 1E), could elicit this transient entrainable signaling to Nms neurons.
Synaptic Transmission from Nms Neurons is Required for SCN Timekeeping
We have shown that synaptic transmission from Nms neurons encompasses the intercellular signaling mediators critical for network synchrony and the in vivo timekeeping mechanisms of the SCN. Ex vivo, network synchrony is lost when synaptic vesicle release from Nms neurons are inhibited (Figure 6E). In vivo, Dox suppression of the TeNT transgene rescues behavioral circadian rhythms with a re-emerged phase different from the phase of prior rhythms (Figure 7A), further suggesting that the inhibition of synaptic transmission from Nms neurons cause desynchronization of the SCN. One key difference between our model and the studies using TTX (Schwartz et al., 1987; Yamaguchi et al., 2003) is that the recovery of synaptic transmission in our model is a gradual process and therefore neurons likely have sufficient time to drift apart in phase before eventually coalescing in a new phase. In the Nms-TeNT mice, it takes a number of weeks for the conditional loss and the restoration of daily rhythms to occur. This appears to be due to the kinetics of molecular events that must occur for synaptic transmission to be modulated rather than the kinetics of coupling/decoupling since the disruption and recovery of daily rhythms occur rapidly in DD-maintained Nms-Per2 mice (Figure 4B). The delay in the disruption of the rhythms in Dox-raised Nms-TeNT is likely a function of the kinetics of TeNT upregulation after Dox withdrawal since water-raised Nms-TeNT mice promptly lost coherent daily rhythms in DD (Figure 5D). The delay in the recovery of rhythmicity, on the other hand, can be attributed to the slow recovery of exocytosis after TeNT exposure, which can take longer than 3 weeks in vitro (Habig et al., 1986). Nevertheless, the specificity, reversibility, and non-invasiveness that the Nms-TeNT mouse model offers significantly outweigh this time-consuming idiosyncrasy. Furthermore, Nms-TeNT provides a model system that allows one to reversibly abolish coherent daily rhythms without the deletion of transcription factors/genes and without significant impact on normal development of mice. This feature may be of great future utility given that the studies linking circadian disruption to medical disorders have largely been conducted using mouse models deficient in transcription factors that are involved in physiology beyond circadian rhythms.
In summary, our results demonstrate that Nms neurons define a subset of bona fide SCN pacemakers that dictate and generate SCN network synchrony and in vivo circadian rhythms through intercellular synaptic transmission. Furthermore, our SCN-restricted genetic approach uncovered a potential mechanistic insight to how light entrainment information and network synchrony are organized in the master circadian clock in the SCN. Nms neurons may serve as a population of cells worthy of targeting for treatment of diseases related to circadian dysfunction.
EXPERIMENTAL PROCEDURES
Animals
All procedures were in accordance with UT Southwestern Medical Center guidelines for animal care and use. The generation of mice carrying the tetO-ClockΔ19, tetO-Per2, tetO-TeNT, ROSA-STOP-tTA, Vip-Cre, Bmal1fl/fl, PER2::LUC, ROSA-STOP-YFP, tetO-SynGFP, Nmu−/− alleles has been described previously (Chen et al., 2009b; Hanada et al., 2004; Hong et al., 2007; Li et al., 2010; Srinivas et al., 2001; Storch et al., 2007; Taniguchi et al., 2011; Wang et al., 2008; Yamamoto et al., 2003; Yoo et al., 2004). See Supplemental Experimental Procedures for a detailed description of the generation of Nms-iCre and Nms−/− mice along with other compound transgenic mice used in this paper.
Circadian behavioral recording and analyses
Running wheel activity was recorded by ClockLab (Actimetrics) as described previously (Siepka and Takahashi, 2005). All mice used were adult ≥8 weeks of age. Mice were entrained to a light/dark (LD) 12:12 cycle for a minimum of 7 days prior to release into constant darkness. Freerunning period was calculated by χ2 periodogram using the 14 days or 21 days before a change in condition (water vs. Dox; light vs. dark) or before the end of experimentation. Fast Fourier transform (FFT) was used to calculate the relative amplitude of the circadian component from 18–30 hours. Losses of circadian rhythms were assessed based on visual inspection of the activity record in conjunction with FFT and χ2 periodogram analysis (Low-Zeddies and Takahashi, 2001). Animals with a loss of circadian rhythms were excluded from period calculations. All values are displayed as the mean ± the standard error of the mean (SEM).
Supplementary Material
Acknowledgments
We thank Shuzhang Yang, Jennifer Mohawk, Seung-Hee Yoo, Shin Yamazaki, Hidetoshi Kumagai, Makito Sato, Yuka Lee, Teresa Ricado for discussions and technical assistance; Randal Floyd and Marcus Thornton for help with mouse husbandry; Shigetada Nakanishi for tetO-TeNT mice; Z. Josh Huang for Vip-Cre mice; Masayasu Kojima for Nmu−/− mice. M.Y. and J.S.T. are Investigators in the Howard Hughes Medical Institute.
Footnotes
Supplemental Information includes eight figures, one table and Supplemental Experimental Procedures and can be found online at X.
AUTHOR CONTRIBUTIONS
ITL, ASC, YI, JST, and MY conceived and designed the experiments. ITL performed the majority of the experiments. ASC produced the founder Nms-iCre transgenic mouse line and Nms−/− mice. MM contributed significantly to behavioral, anatomical and bioluminescence experiments. TM, SD, YS, JF, MI and JES contributed to technical procedures and analysis tools. ITL, MM, JST, and MY analyzed the data. ITL, JST, and MY wrote the manuscript with contributions from ASC, MM, and YS.
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S. Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of Period gene in mice. Mol Pharmacol. 2002;61:26–34. doi: 10.1124/mol.61.1.26. [DOI] [PubMed] [Google Scholar]
- Atkins N, Mitchell JW, Romanova EV, Morgan DJ, Cominski TP, Ecker JL, Pintar JE, Sweedler JV, Gillette MU. Circadian integration of glutamatergic signals by little SAAS in novel suprachiasmatic circuits. PLoS One. 2010;5:e12612. doi: 10.1371/journal.pone.0012612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon ASI, Hastings MH. A Gq-Ca(2+) axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 2013;78:714–28. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Hughes AT, Piggins HD. Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J Neurosci. 2005;25:11155–11164. doi: 10.1523/JNEUROSCI.3821-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R939–49. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 2013;80:973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MD, Maywood ES, O’Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Habig WH, Bigalke H, Bergey GK, Neale EA, Hardegree MC, Nelson PG. Tetanus toxin in dissociated spinal cord cultures: long-term characterization of form and action. J Neurochem. 1986;47:930–7. doi: 10.1111/j.1471-4159.1986.tb00700.x. [DOI] [PubMed] [Google Scholar]
- Hanada R, Teranishi H, Pearson JT, Kurokawa M, Hosoda H, Fukushima N, Fukue Y, Serino R, Fujihara H, Ueta Y, et al. Neuromedin U has a novel anorexigenic effect independent of the leptin signaling pathway. Nat Med. 2004;10:1067–1073. doi: 10.1038/nm1106. [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Harrington ME, Rahmani T, Lee CA. Effects of damage to SCN neurons and efferent pathways on circadian activity rhythms of hamsters. Brain Res Bull. 1993;30:655–669. doi: 10.1016/0361-9230(93)90097-u. [DOI] [PubMed] [Google Scholar]
- Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- Hong HK, Chong JL, Song W, Song EJ, Jyawook AA, Schook AC, Ko CH, Takahashi JS. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS Genet. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AT, Fahey B, Cutler DJ, Coogan AN, Piggins HD. Aberrant gating of photic input to the suprachiasmatic circadian pacemaker of mice lacking the VPAC2 receptor. J Neurosci. 2004;24:3522–3526. doi: 10.1523/JNEUROSCI.5345-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husse J, Zhou X, Shostak A, Oster H, Eichele G. Synaptotagmin10-Cre, a driver to disrupt clock genes in the SCN. J Biol Rhythms. 2011;26:379–389. doi: 10.1177/0748730411415363. [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN, Yan L, LeSauter J, Silver R. Phenotype matters: identification of light-responsive cells in the mouse suprachiasmatic nucleus. J Neurosci. 2004;24:68–75. doi: 10.1523/JNEUROSCI.1666-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatsoreos IN, Romeo RD, McEwen BS, Silver R. Diurnal regulation of the gastrin-releasing peptide receptor in the mouse circadian clock. Eur J Neurosci. 2006;23:1047–1053. doi: 10.1111/j.1460-9568.2006.04633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegsfeld LJ, LeSauter J, Silver R. Targeted microlesions reveal novel organization of the hamster suprachiasmatic nucleus. J Neurosci. 2004;24:2449–2457. doi: 10.1523/JNEUROSCI.5323-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Atkins N, Hatcher NG, Zamdborg L, Gillette MU, Sweedler JV, Kelleher NL. Endogenous peptide discovery of the rat circadian clock: a focused study of the suprachiasmatic nucleus by ultrahigh performance tandem mass spectrometry. Mol Cell Proteomics. 2010;9:285–297. doi: 10.1074/mcp.M900362-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tasic B, Micheva KD, Ivanov VM, Spletter ML, Smith SJ, Luo L. Visualizing the distribution of synapses from individual neurons in the mouse brain. PLoS One. 2010;5:e11503. doi: 10.1371/journal.pone.0011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, Brien JAO, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci U S A. 2011;108:14306–11. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer JH, Rietveld WJ. Neurophysiology of the suprachiasmatic circadian pacemaker in rodents. Physiol Rev. 1989:671–707. doi: 10.1152/physrev.1989.69.3.671. [DOI] [PubMed] [Google Scholar]
- Mieda M, Sakurai T. Bmal1 in the nervous system is essential for normal adaptation of circadian locomotor activity and food intake to periodic feeding. J Neurosci. 2011;31:15391–15396. doi: 10.1523/JNEUROSCI.2801-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Takahashi JS. Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci. 2011;34:349–358. doi: 10.1016/j.tins.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Miyazato M, Ida T, Murakami N, Serino R, Ueta Y, Kojima M, Kangawa K. Identification of neuromedin S and its possible role in the mammalian circadian oscillator system. EMBO J. 2005;24:325–335. doi: 10.1038/sj.emboj.7600526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin LP, Shivers KY, Blanchard JH, Muscat L. Complex organization of mouse and rat suprachiasmatic nucleus. Neuroscience. 2006;137:1285–1297. doi: 10.1016/j.neuroscience.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Nakashiba T, Young JZ, McHugh TJ, Buhl DL, Tonegawa S. Transgenic inhibition of synaptic transmission reveals role of CA3 output in hippocampal learning. Science. 2008;319:1260–1264. doi: 10.1126/science.1151120. [DOI] [PubMed] [Google Scholar]
- Pendergast JS, Friday RC, Yamazaki S. Endogenous rhythms in Period1 mutant suprachiasmatic nuclei in vitro do not represent circadian behavior. J Neurosci. 2009;29:14681–14686. doi: 10.1523/JNEUROSCI.3261-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Rogan SC, Roth BL. Remote control of neuronal signaling. Pharmacol Rev. 2011;63:291–315. doi: 10.1124/pr.110.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romijn HJ, Sluiter AA, Wortel J, Van Uum JF, Buijs RM. Immunocytochemical evidence for a diurnal rhythm of neurons showing colocalization of VIP with GRP in the rat. J Comp Neurol. 1998;405:397–405. [PubMed] [Google Scholar]
- Saper CB. The central circadian timing system. Curr Opin Neurobiol. 2013;23:747–51. doi: 10.1016/j.conb.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, Deák F, Königstorfer A, Mozhayeva M, Sara Y, Südhof TC, Kavalali ET. SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science. 2001;294:1117–1122. doi: 10.1126/science.1064335. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Gross RA, Morton MT. The suprachiasmatic nuclei contain a tetrodotoxin-resistant circadian pacemaker. Proc Natl Acad Sci U S A. 1987;84:1694–1698. doi: 10.1073/pnas.84.6.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Takahashi JS. Forward genetic screens to identify circadian rhythm mutants in mice. Methods Enzymol. 2005;393:219–229. doi: 10.1016/S0076-6879(05)93007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R, Lehman MN, Gibson M, Gladstone WR, Bittman EL. Dispersed cell suspensions of fetal SCN restore circadian rhythmicity in SCN-lesioned adult hamsters. Brain Res. 1990;525:45–58. doi: 10.1016/0006-8993(90)91319-c. [DOI] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–3. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, Weitz CJ. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yang Z, Niu Z, Wang W, Peng J, Li Q, Ma MY, Zhao Y. The mortality of MOP3 deficient mice with a systemic functional failure. J Biomed Sci. 2006;13:845–851. doi: 10.1007/s11373-006-9108-4. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Kvitsani D, Fu Y, Lu J, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Pol A, Powley T. A fine-grained anatomical analysis of the role of the rat suprachiasmatic nucleus in circadian rhythms of feeding and drinking. Brain Res. 1979;160:307–326. doi: 10.1016/0006-8993(79)90427-x. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, Schook A, Antoch MP, Turek FW, Takahashi JS. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci U S A. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–25. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sharma K, Deng HX, Siddique T, Grisotti G, Liu E, Roos RP. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiol Dis. 2008;29:400–408. doi: 10.1016/j.nbd.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Webb AB, Angelo N, Huettner JE, Herzog ED. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci U S A. 2009;106:16493–16498. doi: 10.1073/pnas.0902768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, Slomianka L, Gundersen HJ. Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat Rec. 1991;231:482–497. doi: 10.1002/ar.1092310411. [DOI] [PubMed] [Google Scholar]
- Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptácek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Wada N, Kitabatake Y, Watanabe D, Anzai M, Yokoyama M, Teranishi Y, Nakanishi S. Reversible suppression of glutamatergic neurotransmission of cerebellar granule cells in vivo by genetically manipulated expression of tetanus neurotoxin light chain. J Neurosci. 2003;23:6759–6767. doi: 10.1523/JNEUROSCI.23-17-06759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.