Abstract
The transforming growth factor–β (TGF-β) and bone morphogenetic protein (BMP) family of cytokines critically regulates vascular morphogenesis and homeostasis. Impairment of TGF-β or BMP signaling leads to heritable vascular disorders, including hereditary hemorrhagic telangiectasia (HHT). Drosha, a key enzyme for microRNA (miRNA) biogenesis, also regulates the TGF-β and BMP pathway through interaction with Smads and their joint control of gene expression through miRNAs. We report that mice lacking Drosha in the vascular endothelium developed a vascular phenotype resembling HHT that included dilated and disorganized vasculature, arteriovenous fistulae, and hemorrhages. Exome sequencing of HHT patients who lacked known pathogenic mutations revealed an over-representation of rare nonsynonymous variants of DROSHA. Two of these DROSHA variants (P100L and R279L) did not interact with Smads and were partially catalytically active. In zebrafish, expression of these mutants or morpholino- directed knockdown of Drosha resulted in angiogenesis defects and abnormal vascular permeability. Together, our studies point to an essential role of Drosha in vascular development and the maintenance of vascular integrity, and reveal a previously unappreciated link between Drosha dysfunction and HHT.
INTRODUCTION
Vascular morphogenesis and homeostasis are controlled by various signaling molecules, such as vascular endothelial growth factor, ephrin, fibroblast growth factor, Notch and Delta-like 4, and angiopoietin (1). It has been increasingly evident that signaling mediated by the transforming growth factor–β (TGF-β) family of cytokines [TGF-βs and bone morphogenetic proteins (BMPs)] is essential for regulating vascular development and function (1). TGF-β and BMPs bind to cell surface heterotetrameric type I and type II receptor complexes, which are trans-membrane dual-specificity kinases. Upon ligand binding, the type I receptor kinase is activated and phosphorylates Smad proteins, which then bind with co-Smad Smad4 and translocate to the nucleus and bind DNA and modulate gene transcription (2). Alternatively, Smad proteins are incorporated into the Drosha microprocessor complex through association with p68 (also known as DDX5) and DGCR8 and promote the production of a subset of microRNAs (miRNAs), which, in turn, target various mRNAs (3). Drosha, a ribonuclease (RNase) III enzyme, cleaves the primary transcripts of miRNAs (pri-miRNAs) to generate precursor miRNAs (pre-miRNAs) in the nucleus. Pre-miRNAs subsequently give rise to mature miRNAs of ~22 nucleotides (nt), which are noncoding RNAs that regulate gene expression by directing target mRNAs toward degradation or translational repression (4). Drosha is essential for the biogenesis of nearly all miRNAs, with the exception of a few intronic miRNAs (5–10). In addition, Drosha has functions other than gene regulation. For example, Drosha directly associates with a hairpin structure within target mRNAs, leading to their cleavage and degradation (11, 12). Drosha also associates with the promoter- proximal regions of genes and regulates their transcription (13).
Deregulation of the TGF-β or BMP signaling pathway causes hereditary hemorrhagic telangiectasia (HHT; MIM187300), a vascular disease also known as Rendu-Osler-Weber syndrome (MIM600376) (1). HHT is an autosomal dominant disease with aberrant vascular development that occurs in ~1 in 5000 to 8000 individuals (14). The characteristic lesions found in patients with HHT include epistaxis (nosebleeds) and dilated and often tortuous blood vessels that lack a normal capillary bed between artery and vein (14). Clinical manifestations of HHT include recurrent epistaxis; telangiectasias of the lips, hands, and mucosa of the nose and gastrointestinal (GI) tract; and arteriovenous malformations (AVMs) in the lung, liver, and central nervous system (15). In general, the development of vascular lesions in HHT is associated with abnormalities in the development and function of endothelial cells (16). Therefore, understanding the link between the TGF-β signaling pathway and endothelial cell homeostasis is critical to uncover the molecular etiology of HHT.
Causal mutations for HHT have been reported to occur within genes encoding components of the TGF-β signaling pathway, such as the ligand (BMP9, also known as GDF2), receptors (endoglin and activin A type II–like 1, also known as ALK1), and signal transducer (Smad4) of the TGF-β and BMP pathways (17). Most HHT mutations in these genes are thought to be inactivating mutations (18). Endoglin and Alk1 are predominantly found in endothelial cells. In particular, Alk1 binds preferentially to BMP9 and BMP10 and plays a critical role in endothelial cell homeostasis and function (18, 19). Only BMP10, however, is present during embryonic development in mouse (20). Collectively, the genetic evidence suggests that HHT is caused by the impairment of the TGF-β or BMP pathways in endothelial cells.
In the present study, biochemical analyses revealed that the missense mutations Pro100→Leu (P100L) and Arg279→Leu (R279L) in the N terminus of DROSHA had partial loss of function that resulted in a markedly reduced interaction with Smads upon BMP4 treatment. Furthermore, Drosha-deficient zebrafish showed abnormalities in vascular development, and mice with an endothelial-specific knockout of Drosha exhibited disorganized, dilated vasculature and hemorrhage, which resemble the clinical presentations of HHT patients. Finally, we found an overrepresentation of missense variants of DROSHA in HHT patients compared to the control population. These observations suggest an essential role of Drosha in the development and homeostasis of the endothelium and suggest that a hypomorphic DROSHA allele might be linked to the pathogenesis of HHT.
RESULTS
Drosha morphants show vascular abnormalities
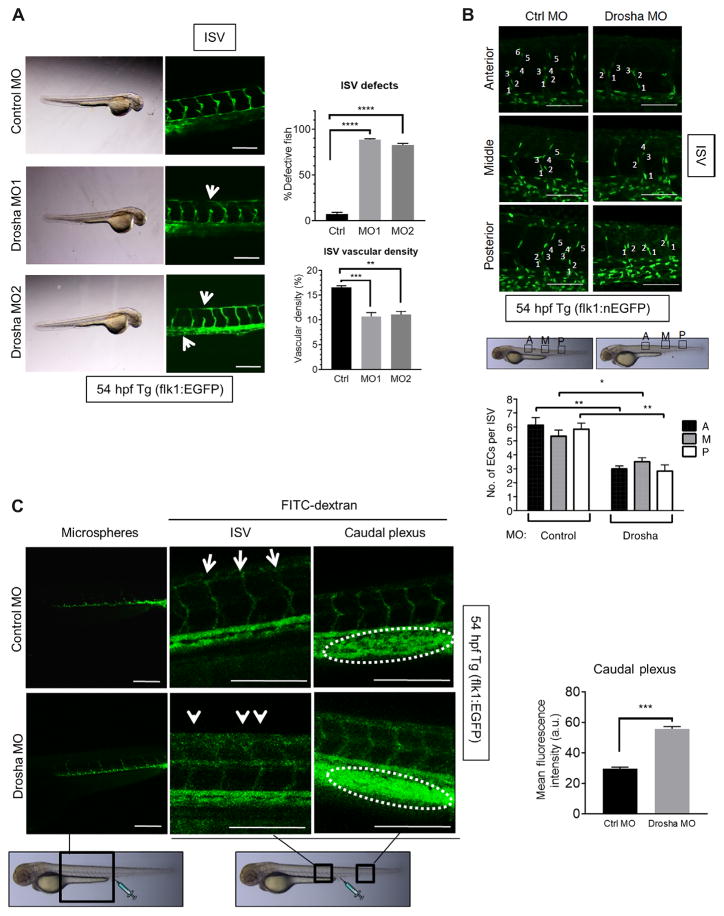
To study the function of Drosha during vascular development, two different morpholino antisense oligonucleotides (MO1 and MO2) against Drosha were injected into transgenic [Tg(flk1:egfp)s843; Tg(gata1: dsRed)sd2] zebrafish (Danio rerio) embryos at the one- to two-cell stage to knock down Drosha expression (hereafter called “Drosha morphants”). In this transgenic fish, enhanced green fluorescence protein (EGFP) labels endothelial cells and dsRed labels hematopoietic cells (21). MO1 targets the translation initiation (ATG) and MO2 targets a splicing site. At 1 ng of MOs, the gross morphology and blood circulation of Drosha morphants were normal, except for the development of a mild pericardial edema (Fig. 1A, bright-field images). No abnormalities in vasculogenesis were detected in Drosha morphants; however, ~40% of the intersegmental vessels (ISVs) of the trunk, which are among the first vessels to form, were prematurely truncated in ~90% Drosha morphants (Fig. 1A), and vascular density was decreased (Fig. 1A). Coinjection of in vitro–transcribed human DROSHA mRNA, but not control RNA, rescued the vascular phenotypes in Drosha morphants (fig. S1, A and B), further supporting the idea that the phenotype of Drosha morphants was due to depletion of Drosha. Furthermore, all Drosha morphants displayed underdeveloped cranial vessels (fig. S1C). To quantitate the ISV phenotype, Drosha or control MO was injected into the Tg(fli1:nEGFP)y7 zebrafish, which expresses EGFP in the nuclei of all endothelial cells (21) to allow counting of cell numbers (Fig. 1B). The anterior, middle, and posterior region of ISVs contained 45% less endothelial cells on average in Drosha morphants compared to control MO–injected zebrafish (Fig. 1B), indicating early angiogenesis defects in Drosha morphants.
Fig. 1. Drosha morphants exhibit vascular abnormalities with increased vascular permeability.
(A) Two different Drosha morpholino oligonucleotides (MO) (MO1 or MO2) or control MO (Ctrl) were injected into Tg(flk1:egfp)s843; Tg(gata1:dsRed)sd2 zebrafish at the one- to two-cell stage. Representative images of intersegmental vessel (ISV) are shown. n = 33 zebrafish for Ctrl MO, 34 zebrafish for MO1, and 35 zebrafish MO2. The fraction (%) of fish with defective ISVs and vascular density are shown as means ± SEM. White arrow, defective ISVs. **P < 0.01, ***P < 0.001, ****P < 0.0001, significant by one-way analysis of variance (ANOVA) with post hoc Tukey’s test. (B and C) Ctrl MO (2 ng) or Drosha MO (1 ng of MO1 + 1 ng of MO2) was injected into Tg(fli1:nEGFP)y7 (B) or Tg(flk1:egfp)s843 (C) zebrafish at the one- to two-cell stage. (B) Representative images of the anterior (A), middle (M), or posterior (P) regions of the ISVs. The number of endothelial cells (ECs) per ISV was shown as means ± SEM. n = 33 zebrafish for Ctrl MO and 28 zebrafish for Drosha MO. (C) Representative angiography images of ISVs and the caudal plexus are shown. Mean fluorescence density of the caudal plexus is presented as means ± SEM. n = 18 zebrafish for Ctrl MO and 20 for Drosha MO. a.u., arbitrary units. *P < 0.05, **P < 0.01, ***P < 0.001, significant by two-tailed unpaired Student’s t test. Scale bars (A to C), 200 μm.
To further examine the phenotype in the absence of Drosha, clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 genome editing was applied to transgenic [Tg(flk1:egfp)s843; Tg(gata1: dsRed)sd2] zebrafish (D. rerio) to generate genetic Drosha null zebrafish, and several Drosha mutant lines were established (fig. S2A). One of the mutant lines in which 4 nt in exon 2 of the Drosha gene was deleted, resulting in a frameshift mutation at asparagine (N) 40 of Drosha and premature termination (hereafter referred to as N40fs), was used for subsequent analysis (fig. S2, A and B). Upon mating N40fs heterozygous mutant (N40fs/+) fish (F3), which developed normally and had normal life spans, N40fs homozygous mutant (N40fs/N40fs) fish were produced in Mendelian ratios and exhibited no developmental abnormalities or angiogenesis defects up to 3 days postfertilization (dpf) (fig. S2C). At 10 dpf, however, N40fs/N40fs fish exhibited a developmental delay (fig. S2D), and all N40fs/N40fs fish died between 14 and 16 dpf, similar to Dicer1 knockout fish (22). At 3 dpf, the amount of Drosha mRNA in N40fs/N40fs fish was indistinguishable from N40fs/+ and control (+/+) fish (fig. S2E). This was due to the persistent expression of maternal Drosha after the maternal-to-zygotic transition. Furthermore, miRNA abundance in N40fs/N40fs fish at 3 dpf was unchanged from control (+/+) fish (fig. S2E). However, at 10 dpf, Drosha mRNA abundance in N40fs/N40fs fish was 50% of the littermate controls (fig. S2E). Consistent with the reduction of Drosha, all miRNAs examined were significantly reduced in N40fs/N40fs fish compared to the littermate controls at 10 dpf (fig. S2E). In Drosha morphants injected with MO1 and MO2, however, the depletion of miRNAs was evident as early as 3 dpf (fig. S2F) because MOs target both maternal and zygotic Drosha mRNA. Although the phenotype of Drosha morphants should be confirmed by generating the maternal-zygotic mutant of Drosha in which maternally derived Drosha is ablated by the germline replacement technique to exclude the possibility of off-target effects of MOs (23), Dicer maternal-zygotic mutants exhibit vascular defects similar to Drosha morphants (24). Thus, we examined Drosha morphants to assess the effect of the depletion of Drosha and miRNAs on developmental angiogenesis.
To examine vascular permeability in Drosha morphants, fluorescein isothiocyanate (FITC)–conjugated dextrans (diameter, ~10 nm) were injected into the caudal plexus at 54 hours postfertilization (hpf). Twenty to thirty minutes after the injection, the surface of both anterior and posterior ISVs was marked by FITC-dextrans in control Tg(flk1:egfp)s843 zebrafish, indicating a normal vasculature (Fig. 1C, control MO; FITC- dextran). In Drosha morphants, however, the surface of ISVs and caudal veins appeared rough, indicating leakage of FITC from vasculature (Fig. 1C, white arrows). The intensity of FITC-dextran signal in the caudal plexus area of Drosha morphants was 1.9-fold higher than that in control MO–injected fish, indicating a higher permeability of the vasculature in Drosha morphants resulting in rapid accumulation of FITC- dextran in the caudal plexus area (Fig. 1C, right). At 45 min after injection, FITC-dextran signal was nearly undetectable in the vasculature of Drosha morphants. In contrast, FITC-dextran signal was detectable in the control zebrafish (fig. S3). However, when polymer microspheres (diameter, ~25 nm), which are larger than dextrans and cannot easily leak from the vasculature, were injected into Drosha morphants, the microspheres remained in the vasculature for up to 2 hours, suggesting a lack of gross defects in the vascular wall in Drosha morphants (Fig. 1C, left). Thus, we conclude that the depletion of Drosha in zebrafish produces an increased vascular permeability.
Endothelial cell–specific deletion of Drosha in mice results in vascular defects similar to HHT
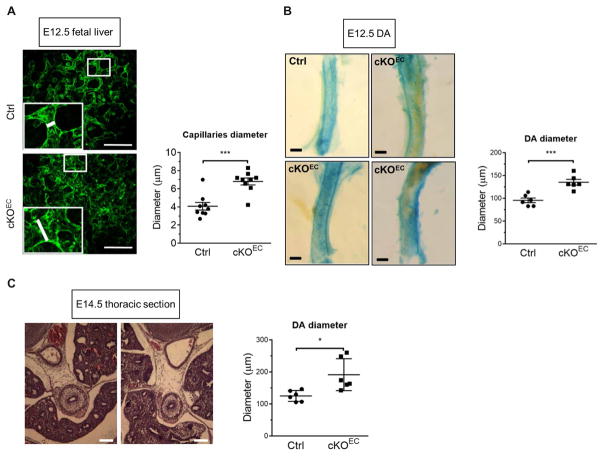
To examine the role of Drosha in the development and homeostasis of endothelium in mammals, an endothelial cell–specific Drosha knockout mouse (Drosha cKOEC) was generated by crossing a Drosha-floxed (Droshafl/fl) mouse (25) with a Cdh5-Cre transgenic mouse line (26). Cre recombinase in the Cdh5-Cre mouse is expressed specifically in CD31+ (a pan-endothelial marker) endothelial cells in the yolk sac since as early as embryonic day (E) 7.5 (26). To determine whether recombination was successful and resulted in depletion of Drosha in endothelial cells, total RNA was prepared from CD31+CD45− cells isolated from E11.5 embryos by flow cytometry and subjected to quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis. Drosha mRNA in endothelial cells from cKOEC embryos was reduced to 33% of that in control littermates (Droshafl/fl, Droshafl/+, or Droshafl/+; Cdh5-Cre+) (fig. S4A), demonstrating efficient deletion of Drosha in endothelial cells. Vasculogenesis and angiogenesis appeared superficially normal in Drosha cKOEC embryos; however, all cKOEC embryos died around E14.5 to E15.5 due to a hematopoiesis defect (27). Immunofluorescence staining of the endothelial marker CD31 showed that branching of blood vessels was reduced by 20% in the yolk sacs of cKOEC embryos at E13.5 compared to those of control embryos (fig. S4B), suggesting that early endothelial patterning was mildly affected by Drosha deletion. Whole-mount CD31 staining revealed twisted, disorganized, and dilated vasculature in the liver of cKOEC embryos at E12.5 (Fig. 2A). Consistently, CD31 staining of E13.5 liver sections revealed tortuous, disorganized blood vessels only in cKOEC embryos (fig. S4C). Hematoxylin and eosin (H&E) staining of E14.5 liver sections in cKOEC embryos revealed the presence of erythrocytes in the extravascular area, which is evidence of vascular leakage (fig. S4D, circles). Furthermore, immunofluorescence staining for zonula occludens–1 (ZO-1), an essential component of tight junctions, was weak or undetectable in the liver vasculature in cKOEC embryos (fig. S4E, circles). This result indicates a disruption of the endothelial barrier and increased permeability of blood vessels in the fetal liver and is consistent with vascular leakage observed in the liver sections from cKOEC embryos (fig. S4D, circles) (28). Dorsal aortas (DAs) in cKOEC embryos were also dilated: 41.6% larger than littermate controls at E12.5 (Fig. 2B) and 52.9% larger by E14.5 (Fig. 2C), indicating a progression of DA dilation during cKOEC embryonic development. 5-Ethynyl- 2′-deoxyuridine (EdU) staining did not reveal differences in the proliferation rate of endothelial cells from cKOEC or control mice (fig. S5A), suggesting that the DA dilation in cKOEC mice was not due to increased endothelial cell numbers. Similar vascular abnormalities were found in the extraembryonic vasculature of cKOEC embryos, such as tortuous vasculature (fig. S5B), misalignment of artery and vein in the yolk sac (fig. S5C), and dilated umbilical arteries (fig. S5D). These results further support an essential role of Drosha- dependent miRNAs in the control of endothelial homeostasis and alignment of arteries and veins.
Fig. 2. Depletion of Drosha in the endothelium in mice results in embryonic developmental vascular abnormalities similar to HHT.
(A) Representative images of a whole-mount liver from control (Ctrl) or Drosha cKOEC embryos at embryonic day (E) 12.5 stained with CD31 are shown. Quantitative analysis of the diameter of fetal liver capillaries is shown as means ± SEM. Scale bar, 100 μm. n = 4 Ctrl embryos and 3 cKOEC embryos. (B) β-Galactosidase staining was performed to visualize the dorsal aorta (DA) isolated from Ctrl or cKOEC embryo at E12.5. Quantitative analysis of the DA diameter is shown as means ± SEM. ***P < 0.001, significant by two-tailed unpaired Student’s t test. Scale bar, 50 μm. n = 4 Ctrl embryos and 4 cKOEC embryos. (C) Representative hematoxylin and eosin (H&E) stain images of a 7-μm transverse thoracic section of Ctrl or cKOEC embryos at E14.5. Scale bar, 100 μm. The diameter of DA was quantified by ImageJ and presented as means ± SEM. n = 3 Ctrl embryos and 3 cKOEC embryos. *P < 0.05, significant by two-tailed unpaired Student’s t test. Hemorrhagic telangiectasia, HHT.
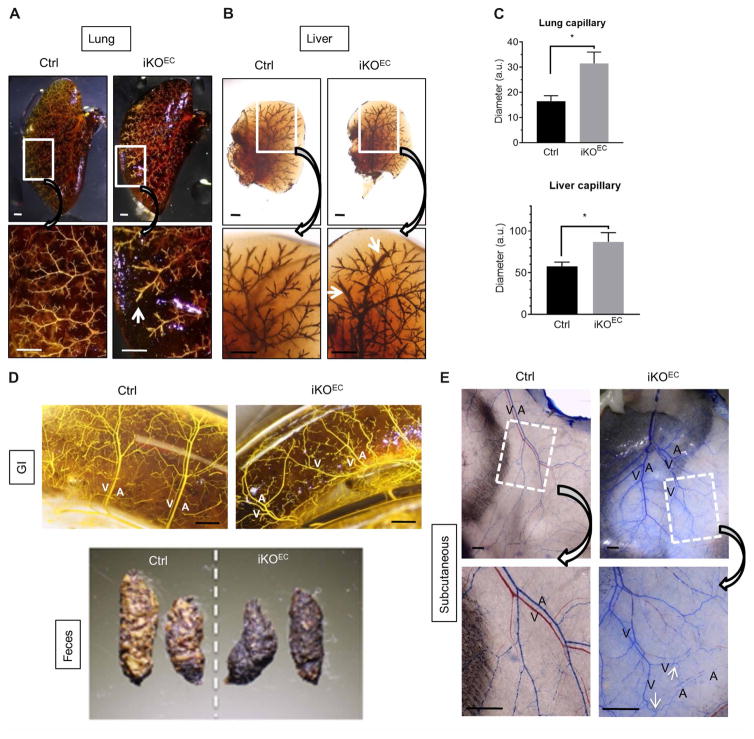
To examine Drosha function during postnatal vascular development, a tamoxifen- inducible endothelial-specific Drosha knockout (Drosha iKOEC) mouse (Droshaflox/flox; Cdh5-cre/ERT2+) was generated by crossing the Cdh5-cre/ERT2 transgenic mouse line (29) with Droshafl/fl mice. Tamoxifen was injected at postnatal day (P) 2 and P4 to induce the excision of the Drosha gene in endothelial cells. Drosha iKOEC mice were viable, had a normal life span, and had no substantial differences in gross morphology compared to tamoxifen-injected littermate controls (fig. S6A). Immunofluorescence staining indicated that Drosha protein had been successfully depleted in endothelial cells in iKOEC mice (fig. S6B). qRT-PCR analysis confirmed reductions in Drosha mRNA and miRNAs in endothelial cells sorted from the livers of iKOEC mice (fig. S6C). Casting analysis showed dilated capillaries in the lungs and livers of iKOEC mice (Fig. 3, A to C). The distal capillaries of lung and liver exhibited bulges and roughness (30), suggesting increased vascular permeability in iKOEC mice compared to Ctrl mice (Fig. 3, A and B). No AVMs or arteriovenous fistulas were detected in the lungs (Fig. 3A) or livers (Fig. 3B) of iKOEC or Ctrl mice. Similar to the yolk sac vasculature of cKOEC mice (fig. S5C), the arteries and veins in the GI tract of iKOEC mice were misaligned, unlike those in control mice, which ran in parallel (Fig. 3D, top). In addition, the feces from iKOEC mice appeared darker than those from control mice (Fig. 3D, bottom). A fecal blood test validated the presence of blood in the feces of iKOEC but not of control mice (fig. S6D). Together with the disruption of tight junctions in the intestinal vasculature (fig. S7B, ZO-1 staining, white arrows), these results suggest that the presence of blood in the feces of iKOEC mice is due to intestinal bleeding, a phenotype also exhibited by endothelial cell–specific Alk1 knockout (Alk1fl/fl; L1-cre) mice (30). Casting analysis of the subcutaneous vasculature in iKOEC mice was not possible when the blue casting agent was injected from the left ventricle, because it did not flow through the veins. Therefore, the veins remained red, whereas the arteries turned blue in control mice (Fig. 3E, Ctrl). However, the casting agent could flow from artery to vein in ~50% of the iKOEC mice, which resulted in both arteries and veins turning blue because of the formation of arteriovenous fistulae (Fig. 3E, iKOEC, arrows). In addition, the casted subcutaneous tissue in iKOEC mice acquired a blue tint, indicating leakage of the casting agent from the vasculature (Fig. 3E). To validate this result, a Microfil of different gravity was used to cast iKOEC and control mice. The casted subcutaneous tissue in iKOEC mice turned yellow because of the leakage of Microfil from the vasculature (fig. S7A), consistent with the effect of the blue casting agent (Fig. 3E). Immunofluorescence staining of the skin showed that the abundance of the tight junction protein ZO-1 in endothelial cells was reduced in iKOEC mice compared to control mice (fig. S6E), indicating the disruption of endothelial cell barriers in iKOEC mice, which leads to vascular leakage. Together, these results demonstrate that endothelial cell–specific deletion of Drosha in mouse leads to multiple vascular defects, including tortuous and disorganized vasculature, vascular dilation, and AVMs, which mimic the phenotypes of the endothelial cell–specific Alk1 or Eng gene knockout mice and the symptoms of human HHT patients.
Fig. 3. Postnatal depletion of Drosha in the endothelium in mice results in HHT-like vascular abnormalities.
(A) Representative images of lungs from 10-week-old Drosha iKOEC or Ctrl mice. White arrow, dilated capillary. n = 4 mice for each genotype. Scale bar, 1 mm. (B) Representative images of livers from 8-month-old Drosha iKOEC or Ctrl mice. White arrow, dilated capillary. Scale bar, 1 mm. n = 3 mice for each genotype. (C) The diameter of lung and liver capillaries was quantified by ImageJ and presented as means ± SEM. *P < 0.05, significant by two-tailed unpaired Student’s t test. (D) Representative images of intestines from 8-month-old Drosha iKOEC or Ctrl mice and feces from 13-month-old Drosha iKOEC or Ctrl mice. A, artery; V, vein. n = 4 Ctrl and 3 iKOEC mice. Scale bar, 1 mm. (E) Representative images of the subcutaneous vasculature in the back of 8-month-old Drosha iKOEC and Ctrl mouse n = 4 mice for each genotype. Scale bar, 500 μm.
Rare DROSHA variants are identified among HHT patients
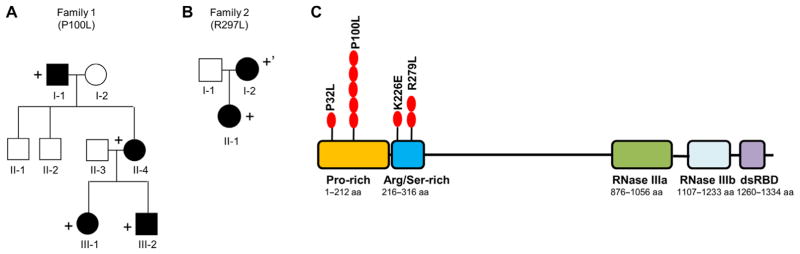
Because of the similarities of the vascular lesions between Drosha cKOEC and iKOEC mice and human HHT patients, we hypothesized that patients with HHT without known causal mutations may carry an inactivating or hypomorphic mutant allele in the DROSHA gene. Thus, we performed exome sequencing in 23 affected individuals from 9 families and 75 probands suspected to have HHT who lacked mutations in known HHT genes (ENG, ALK1, SMAD4, or BMP9). The majority of individuals evaluated (74 of 98 individuals, 75.5%) had three or more clinical diagnostic criteria for HHT, and the remaining 24 individuals had two HHT clinical diagnostic criteria, typically epistaxis and telangiectasia. Consistent with our hypothesis, the exome data revealed an overrepresentation of rare missense variants in the DROSHA gene among individuals with HHT compared to the control population (Table 1). Three rare heterozygous, nonsynonymous substitutions (P32L, P100L, and K226E) were identified in the DROSHA gene in 7.14% (7 out of 98) of HHT patients, whereas they were present in only 0.04% of individuals in the regular population, a significantly lower percentage (Table 1). The P100L variant was found in one pro-band (Table 1, P4) and in four affected individuals of family 1 (F1-I-1, F1-II-4, F1-III-1, and F1-III-2) spanning three generations (Fig. 4A). The other two variants (P32L and K226E) were found in two additional probands with HHT (P1 and P5 in Table 1). Clinical findings for each are listed in Table 1. All patients but one (P1) have epistaxis, most have telangiectasias, and several have solid organ AVMs. In addition, all these three rare variants were predicted to be “damaging” by at least one of the computational algorithms, such as SIFT (sorting intolerant from tolerant), PolyPhen-2, or Mutation Taster, which predict the effect of an amino acid substitution based on evolutionary conservation and structural impact. In particular, the Pro100-to-Leu substitution was predicted to be damaging by all three computational algorithms (Table 1), and Pro100 is conserved from human to zebrafish. Thus, these three variants are likely to substantially change Drosha’s structure and possibly its function. Because of the overrepresentation of missense variants in the DROSHA gene among individuals with HHT compared to the control population, it is plausible to speculate that these rare DROSHA variants might be linked to the development of vascular phenotypes of HHT.
Table 1. Rare DROSHA variants identified in HHT patients.
Sequencing was performed by Sanger sequencing. All nucleotide substitutions are heterozygous. F, family; P, proband; E, epistaxis; T, telangiectasia; GI-T, gastrointestinal telangiectasia; C, cerebral arteriovenous malformation (AVM); H, hepatic AVM; and P, pulmonary AVM. The presence of P32L (rs202053700; frequency, 0.0001), P100L (rs199846087; frequency, 0.0002), and K226E (rs762758438; frequency, 0.00009) was significantly lower in the Exome Aggregation Consortium (ExAC) database (which spans 60,706 unrelated individuals) compared to HHT patients using Fisher’s exact test at P < 0.01. NA, not available.
| Individual | Clinical description | Nucleotide change | Protein change | Presence in population* | Family segregation | Predicted effect | Other mutation |
|---|---|---|---|---|---|---|---|
| P1 | P, mother with H, sister and grandmother have E | c.95C>T | p.P32L | 0.0001 | NA | Damaging† | None |
| F1-I-1 | Severe E, (cauterized age 10) | c.299C>T | p.P100L | 0.0002 | Yes | Damaging‡ | None |
| F1-II-4 | Severe E, T | c.299C>T | p.P100L | 0.0002 | Yes | Damaging‡ | None |
| F1-III-1 | E, T | c.299C>T | p.P100L | 0.0002 | Yes | Damaging‡ | None |
| F1-III-2 | E, C (ruptured) | c.299C>T | p.P100L | 0.0002 | Yes | Damaging‡ | None |
| P4 | E, T | c.299C>T | p.P100L | 0.0002 | NA | Damaging‡ | None |
| P5 | E, T, P, | c.676A>G | p.K226E | 0.00009 | NA | Damaging§ | None |
| F2-I-2 | E, T, GI-T, P, H, liver shunts | c.836G>T | p.R279L | Absent | Yes | Damaging¶ | ENG|| |
| F2-II-1 | E, T, GI-T, multiple P | c.836G>T | p.R279L | Absent | Yes | Damaging¶ | ENG |
Presence in population refers to the presence of the variant in the ExAC database.
Mutation is predicted to be damaging by SIFT.
Mutation is predicted to be damaging by SIFT, PolyPhen-2, and Mutation Taster.
Mutation is predicted to be damaging by Mutation Taster.
Mutation is predicted to be damaging by PolyPhen-2 and Mutation Taster.
Affected individuals in family 2 that carries a mosaic ENG c.1311+1G>A splice site mutation.
Fig. 4. HHT pedigrees with DROSHA nonsynonymous substitution alleles and a map of DROSHA variants.
(A) The pedigree of family 1 carrying the DROSHA P100L variant. Circles and squares indicate females and males, respectively. Black symbols indicate individuals affected with HHT. The + sign indicates carriers of the rare DROSHA variant. (B) The pedigree of family 2 who carry the DROSHA R279L variant. Circles and squares indicate females and males, respectively. The + sign indicates carrier of the DROSHA variant and an ENG c.1311+1G>A mutation. The +’ sign indicates carrier of the DROSHA variant and mosaic carrier of an ENG c.1311+1G>A mutation. (C) Schematic representation of DROSHA protein and variants associated with HHT. Red circles indicate the position of variants and the number of HHT patients. dsRBD, double-stranded RNA–binding domain. Red ovals indicate the number of HHT patients carrying the respective variant. RNase, ribonuclease.
In a separate HHT family (family 2) who carries a pathogenic splice site mutation in the ENG gene (c.1311+1G>A) (Table 1 and Fig. 4B), we found an additional DROSHA missense mutation in the Arg/Ser-rich domain, R279L, which was predicted to be damaging by two computational algorithms (PolyPhen-2 and Mutation Taster) (Table 1). The affected parent (F2-I-2) was mosaic for the ENG gene alleles because the mutation was detected in the hair bulb but not in the peripheral blood cells (Fig. 4B). Both affected individuals (F2-I-2 and F2-II-1) exhibited epistaxis of unusual frequency and duration for their age and multiple pulmonary AVMs that required treatment (Table 1), which is atypical for their ages. Furthermore, the parent (F2-I-2) exhibited hepatic involvement (Table 1). It is plausible to speculate that the R279L variant modifies the severity of the clinical manifestations mediated by the ENG gene alleles.
The N-terminal missense DROSHA mutants are partially inactive
All four variants identified in HHT patients were located in the N terminus of the DROSHA protein, which contains two evolutionarily conserved domains of unknown function, the Pro-rich domain (amino acids 1 to 212) and the Arg/Ser-rich domain (amino acids 219 to 316) (Fig. 4C). This region of the polypeptide has not been thoroughly studied, because the C terminus contains all the domains essential for Drosha’s processing activity, such as two RNase III domains and the double-stranded RNA–binding domain (dsRBD) (Fig. 4C) (31). An N-terminal truncation mutant of Drosha, which lacks amino acids 1 to 390, shows significantly reduced processing activity compared to full-length Drosha when expressed at equivalent levels, indicating that the N-terminal region (amino acids 1 to 390) of Drosha is required for the full activity of Drosha (32). In addition, the conservation of the Pro-rich and Arg/Ser-rich domains in Drosha among all vertebrates also supports the belief that the N terminus is functionally important.
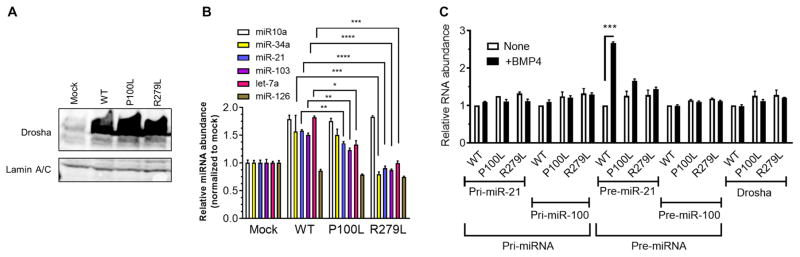
To understand the function of the Pro-rich and Arg/Ser-rich N-terminal domains, mouse embryonic fibroblasts (MEFs) stably expressing human DROSHA wild type (WT) or variants (P100L and R279L) were subjected to miRNA expression analysis. The amount of all three DROSHA proteins (WT, P100L, or R279L) was similar (Fig. 5A). Exogenous expression of the WT DROSHA increased the abundance of miR-10a, miR-34a, miR-21, miR-103, and let-7a by 1.7-fold on average (Fig. 5B, mock compared to WT). In contrast, exogenous expression of P100L and R279L variants increased the abundance of these miRNAs only 1.4-fold and 1.1-fold, respectively. miR-126 abundance was unchanged because its processing does not require Drosha (Fig. 5B) (33). Next, an in vivo pri-miRNA processing assay was performed to assess DROSHA activity in nuclear extracts from MEFs stimulated with BMP4. The abundance of three DROSHA variants (WT, P100L, and R279L), pri-miR-21, and pri-miR-100 was similar (Fig. 5C). The amount of pre-miR-21 was induced 2.67-fold upon BMP4 treatment in WT cells; however, the abundance of pre-miR-21 in P100L and R279L cells was 1.66-fold and 1.44-fold, respectively (Fig. 5C). Consistent with this result, the DROSHA mutants produced less pre-miR-21 than WT DROSHA according to an in vitro processing assay (fig. S8A). Unlike miR-21, the amount of pre-miR-100, whose processing is independent of the BMP4-Smad pathway, was similar regardless of whether WT or the mutant DROSHA proteins were present (Fig. 5C). These results suggest that both variants are less active than WT Drosha and indicate an important function of the N-terminal Pro-rich and Arg/Ser-rich domain in the processing of pri-miRNAs.
Fig. 5. The N-terminal missense mutants of DROSHA are partially inactive.
(A) Drosha protein abundance in the nuclear extracts of mouse embryonic fibroblasts (MEFs) expressing DROSHA WT, P100L, R279L, or mock was analyzed by immunoblotting. Lamin A/C was used as a loading control. n = 3 independent experiments. (B) The abundance of different microRNAs (miRNAs) (miR-10a, miR-34a, miR-21, miR-103, let-7a, and miR-126) relative to U6 small nuclear RNA was quantitated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis in MEFs expressing DROSHA WT, P100L, R279L, or mock (empty vector) and shown as means ± SEM. n = 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, significant by one-way ANOVA with post hoc Tukey’s test. (C) In vivo miRNA processing assay. The nuclear extracts from MEFs expressing DROSHA WT, P100L, R279L, or mock were treated with or without BMP4 for 2 hours. Total RNA was extracted and subjected to qRT-PCR analysis of primary transcripts of miRNAs (pri-miRNA) and precursor miRNAs (pre-miRNA) of miR-21 and miR-100, and DROSHA mRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase. RNA abundance relative to nontreated WT MEFs was plotted as means ± SD. n = 3 independent experiments. ***P < 0.001, significant by two-tailed unpaired Student’s t test.
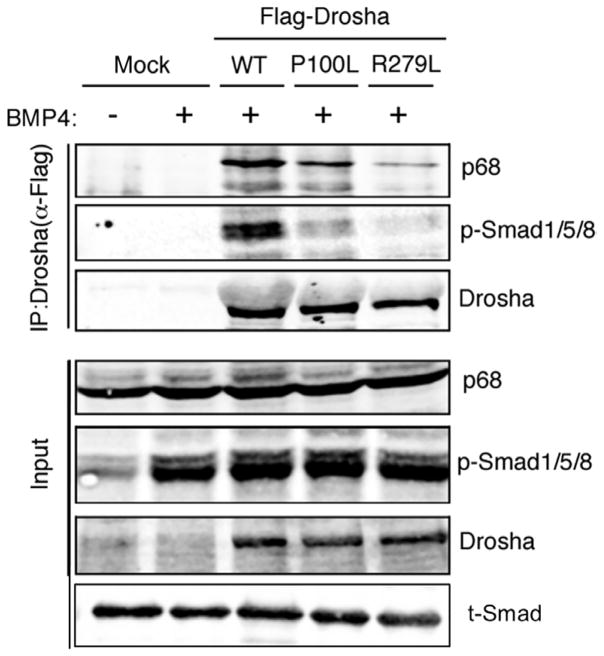
Drosha activity is controlled by different cofactors, including the BMP signal transducers Smad1, Smad5, and Smad8 (Smad1/5/8). Upon BMP4 treatment, Smad1/5/8 are phosphorylated by the type I BMP receptor, translocate to the nucleus, and interact with Drosha in complex with RNA helicase p68 (also known as DDX5), which promotes the processing of a subset of miRNAs (3). After BMP4 stimulation, the amount of phosphorylated Smad1/5/8 was comparable between WT and P100L- or R279L-expressing MEFs (Fig. 6, input, p-Smad1/5/8). The amount of phosphorylated Smad1/5/8 was similar in the endothelial cells isolated from control or E13.5 Drosha cKOEC embryos (fig. S8B). Similarly, BMP9 treatment induced similar amounts of phosphorylated SMAD1/5/8 in human microvascular endothelial cells (hMVECs) (fig. S8C), demonstrating that the DROSHA mutants did not affect the BMP-SMAD pathway. However, in MEFs, the amount of p68 and phosphorylated Smad1/5/8 that precipitated with the P100L and R279L mutants was decreased compared to WT DROSHA, despite similar immunoprecipitation efficiencies for all DROSHA proteins (Fig. 6). These results suggest that the Pro-rich and Arg/Ser-rich domains in the N terminus of Drosha are required for the stable interaction with p68 and phosphorylated Smad1/5/8. We conclude that compared to WT DROSHA, P100L and R279L are less active enzymes in the processing of the BMP-Smad–dependent miRNAs.
Fig. 6. The N-terminal missense mutants of DROSHA show reduced binding activity with cofactors.
The amount of Drosha cofactors (p68 and Smad1/5/8 associated with Drosha) was examined using nuclear extracts from MEFs stably expressing Flag-tagged WT, P100L, or R279L. Exogenously expressed DROSHA was immunoprecipitated by anti-Flag antibody. Immunoprecipitates were immuno-blotted for p68, p-Smad1/5/8, or Drosha. Representative images are shown. n = 3 independent experiments.
Expression of the DROSHA mutants in zebrafish mimics the vascular phenotypes found in the Drosha morphants
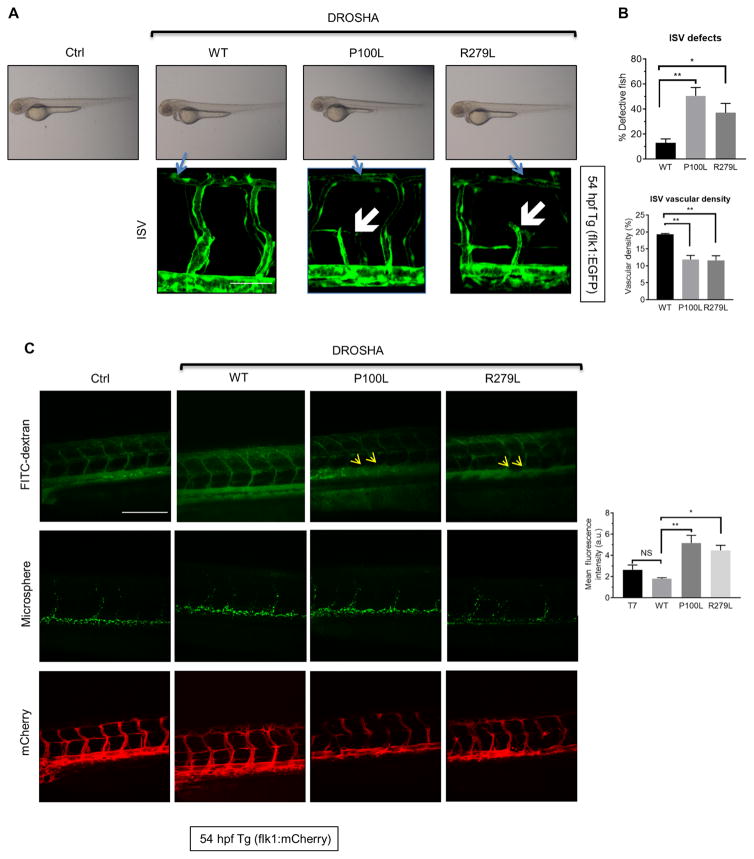
To examine their in vivo activity, mRNAs encoding DROSHA mutants (P100L and R279L), WT or control (T7) RNA transcribed in vitro were injected into Tg(flk1:egfp)s843 embryos at the one- to two-cell stage. The morphology of the vasculature in P100L- and R279L-injected embryos was normal at 54 hpf (Fig. 7A, top). However, 45% of P100L and 37% of R279L mRNA-injected fish showed ISVs that failed to connect to the DLAV [Fig. 7, A (white arrows) and B], similar to the Drosha morphants (Fig. 1A, white arrows). These results suggest that partially inactive DROSHA mutants interfere with endogenous Drosha in zebrafish. Unlike WT, the expression of P100L or R279L in Drosha morphants did not rescue the ISV phenotype (fig. S9, top right) or the vascular density of Drosha morphants (fig. S9, bottom right), demonstrating the compromised activity of the mutants. To examine the effect on vascular permeability, DROSHA (WT, P100L, or R279L) or control (T7) mRNA were injected into one- to two-cell stage Tg(flk1:mcherry) zebrafish in which endothelial cells are labeled by mCherry, and FITC-dextran was injected into the caudal plexus at 54 hpf. The ISVs and DAs in the DROSHA WT mRNA– or T7 RNA–injected zebrafish were demarcated by FITC-dextran and did not exhibit signs of vascular leakage (Fig. 7C). In contrast, in the zebrafish injected with Drosha mutants, the surface of the DA appeared rough (Fig. 7C, yellow arrows), a sign of FITC-dextran leakage, similar to the result of Drosha morphants (Fig. 1C, white arrows). These results demonstrate that exogenous expression of DROSHA missense mutants phenocopies angiogenesis defects mediated by the depletion of Drosha. Expression of these mutants impairs endothelial cell development and barrier function in vivo, underscoring an essential role of Drosha activity in vascular development and homeostasis in vertebrates. Although further genetic studies are necessary, our study suggests a potential link between DROSHA missense mutations and heritable vascular diseases such as HHT.
Fig. 7. Exogenous expression of DROSHA missense mutants in zebrafish exhibits vascular phenotypes similar to Drosha morphants.
(A) Control (T7) RNA and DROSHA WT, P100L, or R279L mRNA were injected into Tg(flk1:egfp)s843; Tg(gata1:dsRed)sd2 transgenic zebrafish at the one- to two-cell stage. Representative images of the whole zebrafish (top) and ISVs (bottom) at 54 hours postfertilization (hpf) are shown. n = 36 zebrafish for Ctrl, 30 zebrafish for WT, 28 zebrafish for P100L, and 33 zebrafish for R279L. Scale bar, 50 μm. (B) The fraction (%) of fish with ISV defects (top) and vascular density (bottom) are shown as means ± SEM. *P < 0.05, **P < 0.01, significant by one-way ANOVA with post hoc Tukey’s test. (C) Control (T7) RNA and DROSHA WT, P100L, or R279L mRNA were injected into Tg(flk:mcherry) zebrafish. At 54 hpf, angiography was performed, and representative images are shown (left). Mean fluorescence density was quantified by ImageJ and presented as means ± SEM (right). n = 20 zebrafish for Ctrl, 17 zebrafish for WT, 20 zebrafish for P100L, and 22 zebrafish for R279L. Scale bar, 200 μm. NS, not significant. *P < 0.05, **P < 0.01, significant by one-way ANOVA with post hoc Tukey’s test.
DISCUSSION
Our study demonstrated that Drosha was essential for normal endothelial development and promoted vascular development and homeostasis in vertebrates. It also sheds light on a potential involvement of DROSHA dysfunction in human vascular disorders. On the basis of the overrepresentation of DROSHA variants in HHT patients that are rare among control populations, combined with analysis of these variants in vitro and in vivo, we speculate that these missense DROSHA mutants exhibit a substantially reduced activity that contribute to the development of vascular abnormalities similar to HHT. Somatic mutations in the C-terminal RNase III domains of DROSHA have been identified in Wilms tumors (34, 35). Rare DROSHA missense mutations enriched in patients with HHT were found in the N-terminal noncatalytic domains, demonstrating that the N-terminal region is indispensable for the full activity of DROSHA.
Endothelial-specific inactivation of Eng or Alk1 in mice generates the most robust HHT animal models. Alk1 endothelial-specific inactivation using the L1-Cre transgenic mouse (Alk1fl/fl; L1-cre) shows tortuous, disorganized, enlarged vasculature; loss of microvessels; and AV shunts (30, 36). We pursued a similar approach in investigating the role of DROSHA in HHT etiology. Similar to the endothelial cell–specific Alk1 knockout mouse, Drosha cKOEC mice displayed an enlarged DA and disorganized and dilated hepatic and extraembryonic vasculature. Drosha iKOEC mice showed loss of microvessels, enlarged capillaries, and AV shunts in the skin vasculature. Thus, endothelial-specific inactivation of Drosha has a damaging impact on endothelial cell development and homeostasis that partially resembles the effect of Alk1 inactivation. However, Alk1 endothelial cell–specific inactivation produces robust AVMs in the E16.5 yolk sac and in the GI and brain vasculature by P3 (30, 36). These features were not observed in Drosha cKOEC or Drosha iKOEC mice, suggesting that Drosha inactivation in the vascular endothelium causes HHT-like vascular defects that are partially distinct from those observed in Alk1fl/fl; L1-cre mice. Cdh5-cre is expressed earlier in a broader range of tissues compared with L1-cre, which is driven by the Alk1 promoter (30, 36), thus suggesting that the difference in cre-drivers might contribute to the differential vascular phenotype. Unlike Alk1 (30, 36), endothelial-specific deletion of Drosha did not affect the BMP-Smad signaling pathway (fig. S8B). Therefore, it is likely that DROSHA mutations in endothelial cells have a different effect from that of ALK1 or ENG mutations and that they alter the clinical spectrum or severity of HHT and possibly other vascular disorders. The co-occurrence of DROSHA and ENG mutant alleles in HHT family 2 (Fig. 3A) suggests that DROSHA mutant allele might lead to a more severe manifestation of HHT, possibly due to their combined effects on Smad-dependent transcriptional regulation (through Eng) and miRNA-dependent posttranscriptional regulation (through Drosha), resulting in broader alterations in the gene expression profile in vascular endothelium during developmental and post-natal angiogenesis.
It is intriguing that inactivation or missense mutations of ubiquitously expressed DROSHA mediated primarily vascular abnormalities in humans. Endothelial cells may be more sensitive to changes in the abundance of miRNAs than other tissues. For example, inactivation of endothelial cell–specific miR-126 is sufficient to cause ISV defects similar to those observed in Drosha morphants in zebrafish (33). However, miR-126 biogenesis is independent of Drosha (33), and the abundance of miR-126 is not affected in Drosha morphants or Drosha cKOEC mice (27). Thus, future studies are necessary to identify endothelial cell–specific miRNAs that are essential for the vascular development and homeostasis. Upon identification of these miRNAs, it is possible that treatment with miRNA mimics or antagonists could ameliorate vascular lesions in patients with HHT or other vascular abnormalities.
MATERIALS AND METHODS
Ethical statement concerning the use of human subjects
The HHT exome cohort consisted of 23 affected individuals from 9 families and 75 additional probands for a total of 84 probands (Institutional Review Board #00020480 at the University of Utah). The majority of the cohort was of Caucasian descent. All individuals were suspected to have HHT but harbored no mutations among the known genes associated with HHT. An additional HHT family 2, who carried a novel variant of DROSHA and a mosaic mutation in the ENG gene, was also included, because these patients have more severe clinical symptoms than is normal. Informed consent was obtained from all individuals.
Mouse husbandry and handling
All animal experiments were conducted in compliance with University of California at San Francisco (UCSF) Laboratory Animal Research Committee guidelines and performed under the approved protocols (AN108100-03). Cdh5-Cre (26), Droshatm1Littfloxed lines (25), Gt(ROSA) 26Sortm4(ACTB-tdTomato,-EGFP)Luo lines (37), and Cdh5-Cre/ERT2 (29) have been previously described. Embryos were dated by the presence of a vaginal plug in the female mouse at E0.5. Genomic DNA was generated from day 12 pups’ tail tips or conceptus yolk sacs and genotyped with regular PCR. Primers were as follows: Drosha, 5′-GCAGAAAGTCTC-CCACTCCTAACCTTC-3′ (forward) and 5′-CCAGGGGAAATTA-AACGAGACTCC-3′ (reverse); Cre, 5′-TGCCACGACCAAGTGACAG-CA-3′ (forward) and 5′-AGAGACGGAAATCCATCGCTC-3′ (reverse).
Zebrafish husbandry and handling
Zebrafish (D. rerio) were maintained according to standard protocol (http://zfin.org) in compliance with local animal welfare regulations (AN151574). The following transgenic (Tg) zebrafish lines were used: Tg(flk1:mCherry), Tg(fli1:nEGFP)y7, Tg(flk1:egfp)s843, and Tg(flk1: GFP)s843;Tg(gata1:dsRed)sd2 transgenic zebrafish (21, 38, 39). The culture was conducted in conformity with UCSF Institutional Animal Care and Use Committee and Association for Assessment and Accreditation of Laboratory Animal Care International guidelines.
Next-generation sequencing
Genomic DNA was extracted from peripheral blood using a Gentra Pure gene Blood Kit (Qiagen), and 3 μg was sheared to 180-bp fragments. Illumina adapters were added using the Bravo automated instrument and SureSelect XT kit reagents (Agilent Technologies). Adapter-ligated DNA underwent hybridization with biotinylated RNA baits designed to target the whole exome for 24 hours at 65°C. Hybridized DNA targets of interest were captured using streptavidin-coated magnetic beads and were eluted and barcoded/indexed after a series of washes to remove the nontargeted, unbound genome. DNA quality and quantity were assessed using the Bioanalyzer (Agilent Technologies), and samples were pooled and sequenced on a HiSeq2500 instrument (Illumina) using 2 × 100 paired-end sequencing. Sequences were aligned to the human genome reference (hg19) sequence using the Burrows-Wheeler Alignment tool (BWA 0.5.9) with default parameters (40). PCR duplicates were removed using the SAMtools package (41), and base quality score recalibration, local realignment, and variant calling were performed using the Genome Analysis Toolkit (GaTK v1.3) (42). Variants with a less than 0.02% frequency in the general population were analyzed further. Ingenuity pathway analysis was used to identify biologically relevant gene variants (43). The prevalence of rare or previously unknown biologically relevant gene variation was compared to the prevalence in the control cohort to exclude genes that were not specific to the vascular HHT phenotype. Variants of interest were confirmed using Sanger sequencing. Primer sequences are available upon request.
Quantitation of miRNAs and in vitro pri-miRNA processing assay
Quantitative RT-PCR analysis of miRNAs and in vitro pri-miRNA processing assay were performed as previously reported (3, 44). MEFs were stimulated by BMP4 for 2 hours. Nuclear extracts were then mixed with in vitro–transcribed pri-miR-21 (~100 nt) labeled with biotinylated uridine 5′-triphosphate, followed by the processing reaction and visualization of pri-miR-21 and the processing product pre-miR-21 (72 nt) by streptavidin staining.
Casting assay
The casting assay was performed as previously described (45, 46). Mice were anesthetized with 2% isoflurane and oxygen at 2 ml/min. The thoracic cavity was open, and the left ventricle was cannulated and perfused with 1× phosphate-buffered saline (PBS) at 80 mmHg. The right atrium was cut to allow outflow of PBS until all blood was washed out. Microfil casting agent (Flowtech Microfil MV-122 yellow or Flowtech Microfil MV-120 blue) was then perfused until vessels in the gut, stomach, brain, and liver were full. For lung casting, blood was cleared from the lungs by transecting the carotid artery. PBS (3 ml) was infused into the inferior vena cava. The Microfil casting agent was infused into the pulmonary artery by hand injection. After casting, mice were left for 90 min at room temperature or overnight at 4°C to cure. Organs were then dissected and dehydrated through a series of graded ethanol treatments and cleared in methyl salicylate for 12 to 24 hours.
Tamoxifen injection
Tamoxifen (Sigma-Aldrich T5648) was dissolved in corn oil at a concentration of 10 mg/ml. For adult Drosha iKOEC mice analysis, 100 μg of tamoxifen was intraperitoneally injected into P2 and P4 pups. For miRNA and Drosha qPCR analysis, 2 mg of tamoxifen was injected into pregnant female mice on E11.5 and E13.5, and the livers of P1 pups were subjected to flow cytometry sorting for subsequent RNA preparation and qPCR analysis.
Whole-mount immunofluorescence staining
Yolk sacs were dissected at E13.5 and fixed in 4% paraformaldehyde solution overnight, dehydrated in a series of methanol/PBS washes, and stored at −20°C. The aorta-gonad-mesonephros (AGM) was re-hydrated through a series of graded methanol/PBS washes, blocked at 3% milk with 0.1% Tween-20 in PBS for 2 hours at room temperature, and then incubated with primary antibody for 48 hours at 4°C. AGMs were then washed with 3% milk with 0.1% Tween 20 in PBS five times, 1 hour each time, and the last wash was overnight at 4°C. Secondary antibody was incubated for 48 hours at 4°C. AGMs were then dehydrated through methanol/PBS washes, cleared, and mounted with benzyl alcohol/benzyl benzoate. Images were taken with a Leica SPE confocal microscope and compiled with ImageJ software. The following antibodies directed against mouse antigens were used: CD31 (BD Pharmaceuticals, 550274) and anti-rat 594 (Thermo Fisher Scientific, A-21209).
Cryostat and immunofluorescence staining
Livers from E13.5 mice or guts from adult mice were dissected and fixed in 4% paraformaldehyde solution overnight, dehydrated with 30% sucrose in PBS until tissues sunk to the bottom of the tube. Tissues were then embedded in Tissue-Tek OCT compound (Sakura Finetek, 4583). Cryosections (40 μm) were done with Leica CM1850V. Slides were dried for 2 hours at room temperature and stored at −80°C before immunofluorescence staining. Cryosections were warmed to room temperature, washed in PBS, permeabilized in 0.5% Triton X-100 in PBS for 10 min and blocked in 3% bovine serum albumin (BSA) and 0.05% Triton X-100 in PBS for 1 hour at room temperature. Slides were incubated with primary antibodies at 4°C overnight, followed by PBS wash and secondary antibody incubation for 1 to 2 hours at room temperature. Images were taken with Leica SPE confocal microscope and compiled with ImageJ software.
Histology
E14.5 embryos were harvested by cesarean section, rinsed in PBS (pH 7.4), fixed in 4% paraformaldehyde overnight at 4°C, embedded in paraffin, sectioned at a thickness of 7 μm, and stained with H&E.
β-Galactosidase staining
E12.5 embryos were dissected and fixed in 4% fixature (2% formaldehyde, 0.02% NP-40, 0.2% glutaraldehyde, 5 mM EDTA, and 2 mM magnesium chloride in PBS) for 1 hour at room temperature, washed in PBST (0.1% Triton X-100 and 2 mM magnesium chloride in PBS), and then stained in staining solution (2 mM magnesium chloride, 0.01% sodium deoxycholate, 0.02% NP-40, 5 mM potassium ferricyanide crystalline, 5 mM potassium ferricyanide trihydrate, and 1 mg/ml X-Gal in PBS) at 37°C for 3 to 5 hours. The DA was subsequently dissected for imaging.
Flow analysis of EdU incorporation
The rate of cell proliferation in vivo was measured by EdU incorporation into DNA followed by the Click-iT EdU Alexa Flour 488 Flow Cytometry Assay Kit (Invitrogen/Molecular Probes C10425). E10.5 pregnant female mice were injected with EdU (50 mg/kg) 1 hour before euthanization, and embryos were harvested in fluorescence-activated cell sorting (FACS) buffer. AGMs were pipetted into single-cell suspensions, washed with PBS, and stained with fixable viability dye eFluor 450 (eBioscience, 65-0863-14). Cells were then fixed, permeabilized, and stained with Click-iT EdU reaction cocktail, followed by wash and cell surface marker staining and flow analysis.
Stable expression of DROSHA WT or mutant in MEFs
Flag-tagged DROSHA WT, P100L, or R279L was subcloned into the pBABE-puro plasmid. To produce virus, Bosc23 cells were co-transfected with the pBABE-DROSHA WT, P100L, or R279L expression constructs and the ecotropic packaging vector pCL-L at 3:1 ratio using Lipofectamine 2000 (11668027, Invitrogen) according to the manufacturer’s instructions. Medium was collected 48 hours later, filtered with a 0.45-μm filter and buffered with 10 mM Hepes (pH 7.5). MEFs were infected with a 1:5 dilution of virally conditioned medium at 50% confluence with polybrene (2 μg/ml). After 2 days of culture, MEFs were subsequently split and selected with puromycin (2.5 μg/ml).
Analysis of primary endothelial cells
E13.5 livers were harvested in FACS buffer, pipetted into single-cell suspension, and stained with CD31 and CD45 for 1.5 hours at 4°C. Endothelial cells (CD31+CD45−) were then sorted by a BD FACSAria III Cell Sorter. Cells from E13.5 livers of four embryos of the same genotype were pooled and lysed in lysis buffer (10 mM tris-HCl, 1% SDS, and 0.2 mM phenylmethylsulfonyl fluoride) supplemented with phosphatase inhibitor (Roche, PhosSTOP, 04906837001) and protease inhibitor (Roche, complete mini EDTA free, 04693159001). Samples were then boiled at 95°C for 10 min and centrifuged at 10,000g for 30 min. The supernatants were subjected to Western blot. For miRNA and Drosha qPCR analysis, 2 mg of tamoxifen was injected into pregnant female mice on E11.5 and E13.5. P1 liver was harvested and sorted as described above. Endothelial cells were sorted into Trizol LS (Invitrogen, 10296010), and total RNA was prepared and subjected to miRNA qRT-PCR analysis.
hMVEC culture, adenovirus infection, and BMP9 treatment
hMVECs were cultured in EGM-2 MV BulletKit medium (Lonza, CC-4147) in 5% CO2 at 37°C. Adenoviruses encoding DROSHA WT and mutants were produced with AdEasy Adenoviral Vector System (Agilent Technologies). Complementary DNA (cDNA) of human DROSHA WT, P100L, or R279L was cloned into pShuttle-CMV vector. Adenovirus was then produced following the manufacturer’s manual and amplified with ade-293 cell lines. Infection of MVECs at ~80% confluence was carried out at 37°C and 5% CO2 at a multiplicity of infection of 50 for 6 hours. Infected MVECs were cultured overnight in fresh medium. Infection of MVECs was carried out again on the next day for 6 hours before cells were treated with BMP9 and then lysed for nuclear extract. For BMP9 treatment, BMP9 or vehicle (0.1% BSA in 4 mM HCl) was added into basal EGM-2 medium at a final concentration of 1 nM for 2 hours at 37°C and 5% CO2.
MO and mRNA injection
For Drosha morphant analysis, MO1 (5′-TCCACGGCCAGCATG-GAAAGACATC-3′) or MO2 (5′-AGAGGTGAAGCACATTACCT-GCTGC-3′) was synthesized by Gene Tools, LLC. One nanogram of DroshaMO1 (which targeted translation initiation) or DroshaMO2 (which targeted mRNA splicing) was injected into the fertilized egg of Tg(flk1:GFP)s843;Tg(gata1:dsRed)sd2 transgenic zebrafish within 45 min after fertilization. The endothelial cells are labeled with EGFP, and hematopoietic cells are labeled with dsRed in this zebrafish line. For angiography analysis, 1 ng of MO1 and 1 ng of MO2 were combined and injected into Tg(flk1:mCherry) transgenic zebrafish within 45 min after fertilization. The endothelial cells are labeled with mCherry in this zebrafish line. For mRNA expression, human DROSHA WT, P100L, and R279L mutant cDNAs were cloned into pcDNA3.1 vector and in vitro–transcribed with the mMACHINE T7 Transcription Kit (Ambion, AM1344). Human DROSHA mRNA is resistant to MO1 and MO2. One hundred picograms of control T7 or DROSHA WT, P100L, or R279L mRNA was injected into Tg(flk1:GFP)s843;Tg(gata1: dsRed)sd2 transgenic zebrafish at the one- to two-cell stage. For MO rescue, 1 ng of MO1 + 1 ng of MO2 + 100 pg of control T7 mRNA or DROSHA WT mRNA were mixed and injected into Tg(flk1:GFP)s843; Tg(gata1:dsRed)sd2 transgenic zebrafish at the one- to two-cell stage. At 54 hpf, the vasculature of zebrafish was analyzed. Images were taken with a Leica SPE confocal microscope and compiled with ImageJ software. Images were taken with a Leica SPE confocal microscope and compiled with ImageJ software.
Angiography
Polymer microspheres (Thermo Fisher Scientific, 3020A) or FITC- conjugated dextran (2,000,000 molecular weight, ~10-nm diameter; Molecular Probes, D7137) was diluted in PBS (1:10) and injected into the caudal plexus of 54-hpf flk:mCherry transgenic zebrafish. Zebrafish injected with dextran were imaged 20 min after the injection, and zebrafish injected with microspheres were imaged 2 hours after the injection with a Leica SPE confocal microscope and images were compiled with ImageJ software.
Stool guaiac test
Hemoccult SENSA and Hemoccult II SENSA (Beckman Coulter Inc.) were used to detect the presence of occult blood in the feces of Drosha iKO mice or littermate controls. Some modifications were made to the manual to test fecal samples from mice: 15 μl of PBS was injected into feces to soften it. Five pieces of softened feces from each mouse were then pooled, smeared, and applied into the sample box. Five minutes later, two drops of Hemoccult SENSA Developer was applied to guaiac paper directly over the smear.
Image quantitation
Vascular density was calculated as the percentage of endothelial cell area in total area. For ISV vascular density, the area between four continuous ISV was calculated as the total area, and the area of vessels within the total area (four ISVs) was calculated as the endothelial cell area. For craniofacial vascular density, the area between the middle cerebral vein, the primordial hindbrain channel, and the posterior cerebral vein was calculated as the total area, and the area of all the vessels within the total area was calculated as the endothelial cell area. All the area calculations were carried out by ImageJ software. Mean fluorescence density was quantitated by ImageJ. Quantification of ISV defects was calculated as the percentage of fish with ISV defects in the total fish.
Statistical analysis
A Fisher’s exact test was used to determine the statistical significance of the prevalence of rare DROSHA variants in the HHT compared to the Exome Aggregation Consortium database. Statistical analysis was performed using the Prism 5.01 GraphPad package. Statistical tests and significance are denoted in the figure legends.
Supplementary Material
Fig. S1. Drosha morphants develop angiogenesis defects.
Fig. S2. Generating Drosha mutant fish by CRISPR/Cas9.
Fig. S3. Drosha morphants showed vascular leakiness.
Fig. S4. Drosha cKOEC embryos showed mild angiogenesis defects and a sign of hemorrhage.
Fig. S5. Drosha cKOEC embryos showed disorganized and dilated vasculature.
Fig. S6. Drosha iKOEC mice show vascular leakage.
Fig. S7. The distribution of the tight junction protein ZO-1 is disrupted in the endothelial cells of Drosha iKOEC mice.
Fig. S8. In vitro processing assay of pri-miR-21 and the BMP-Smad signaling pathway in cells expressing DROSHA mutants or depleted of Drosha.
Fig. S9. ISV defects mediated by the knockdown of Drosha by morpholino oligos are rescued by coinjection of wild-type (WT) Drosha mRNA, but not by mutant mRNA.
Acknowledgments
We thank R. Wang and C. Nielsen (UCSF) for assisting in vasculature casting. We also thank R. Adams (Max Planck Institute) for the Cdh5-CreERT2 mouse, N. Speck (University of Pennsylvania) for the Cdh5-Cre mouse, and S. Sumanas (University of Cincinnati College of Medicine), D. Stanier (Max Planck Institute), and B. Weinstein (Eunice Kennedy Shriver National Institute of Child Health and Human Development) for the Tg(flk1:mCherry), Tg(flk1:egfp)s843, and Tg(gata1:dsRed)sd2 transgenic zebrafish line, respectively. Finally, we thank all members of the Hata laboratory, in particular Y. Peng for technical support.
Funding: This work was supported by grants from the NIH (HL116191 and HL132058) and the LeDucq foundation to A.H. and the ARUP Institute for Clinical and Experimental Pathology to P.B.-T.
Footnotes
Author contributions: X.J., W.L.W.-D., J.M., P.B.-T., G.L., and A.H. designed, performed, and analyzed most experiments, conceived the project, and drafted the manuscript. P.G. assisted with the genotyping of mice. M.B., M.S., and D.H. designed, performed, and analyzed zebrafish MO injections. H.C. and S.C. designed, performed, and analyzed angiography in zebrafish.
Data and materials availability: Drosha knockout mouse and zebrafish lines will be available upon completion of a Material Transfer Agreement with UCSF.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Goumans M-J, Zwijsen A, ten Dijke P, Bailly S. Bone morphogenetic proteins in vascular homeostasis and disease. Cold Spring Harb Perspect Biol. 2017:a031989. doi: 10.1101/cshperspect.a031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hata A, Chen Y-G. TGF-β signaling from receptors to Smads. Cold Spring Harb Perspect Biol. 2016;8:a022061. doi: 10.1101/cshperspect.a022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellano L, Stebbing J. Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Res. 2013;41:3339–3351. doi: 10.1093/nar/gks1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Šestan N, Steitz JA. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolando C, Erni A, Grison A, Beattie R, Engler A, Gokhale PJ, Milo M, Wegleiter T, Jessberger S, Taylor V. Multipotency of adult Hippocampal NSCs in vivo is restricted by Drosha/NFIB. Cell Stem Cell. 2016;19:653–662. doi: 10.1016/j.stem.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Gromak N, Dienstbier M, Macias S, Plass M, Eyras E, Cáceres JF, Proudfoot NJ. Drosha regulates gene expression independently of RNA cleavage function. Cell Rep. 2013;5:1499–1510. doi: 10.1016/j.celrep.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards-Yutz J, Grant K, Chao EC, Walther SE, Ganguly A. Update on molecular diagnosis of hereditary hemorrhagic telangiectasia. Hum Genet. 2010;128:61–77. doi: 10.1007/s00439-010-0825-4. [DOI] [PubMed] [Google Scholar]
- 15.McDonald J, Bayrak-Toydemir P, Pyeritz RE. Hereditary hemorrhagic telangiectasia: An overview of diagnosis, management, and pathogenesis. Genet Med. 2011;13:607–616. doi: 10.1097/GIM.0b013e3182136d32. [DOI] [PubMed] [Google Scholar]
- 16.Mahmoud M, Upton PD, Arthur HM. Angiogenesis regulation by TGFβ signalling: Clues from an inherited vascular disease. Biochem Soc Trans. 2011;39:1659–1666. doi: 10.1042/BST20110664. [DOI] [PubMed] [Google Scholar]
- 17.McDonald J, Wooderchak-Donahue W, VanSant Webb C, Whitehead K, Stevenson DA, Bayrak-Toydemir P. Hereditary hemorrhagic telangiectasia: Genetics and molecular diagnostics in a new era. Front Genet. 2015;6:1. doi: 10.3389/fgene.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdalla SA, Letarte M. Hereditary haemorrhagic telangiectasia: Current views on genetics and mechanisms of disease. J Med Genet. 2006;43:97–110. doi: 10.1136/jmg.2005.030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuis-Girod S, Bailly S, Plauchu H. Hereditary hemorrhagic telangiectasia: From molecular biology to patient care. J Thromb Haemost. 2010;8:1447–1456. doi: 10.1111/j.1538-7836.2010.03860.x. [DOI] [PubMed] [Google Scholar]
- 20.Neuhaus H, Rosen V, Thies RS. Heart specific expression of mouse BMP-10 a novel member of the TGF-β superfamily. Mech Dev. 1999;80:181–184. doi: 10.1016/s0925-4773(98)00221-4. [DOI] [PubMed] [Google Scholar]
- 21.Roman BL, Pham VN, Lawson ND, Kulik M, Childs S, Lekven AC, Garrity DM, Moon RT, Fishman MC, Lechleider RJ, Weinstein BM. Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development. 2002;129:3009–3019. doi: 10.1242/dev.129.12.3009. [DOI] [PubMed] [Google Scholar]
- 22.Wienholds E, Koudijs MJ, van Eeden FJM, Cuppen E, Plasterk RHA. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 23.Stainier DYR, Raz E, Lawson ND, Ekker SC, Burdine RD, Eisen JS, Ingham PW, Schulte-Merker S, Yelon D, Weinstein BM, Mullins MC, Wilson SW, Ramakrishnan L, Amacher SL, Neuhauss SCF, Meng A, Mochizuki N, Panula P, Moens CB. Guidelines for morpholino use in zebrafish. PLOS Genet. 2017;13:e1007000. doi: 10.1371/journal.pgen.1007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 25.Chong MMW, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Hawkins JS, Lee J, Lizama CO, Bos FL, Zape JP, Ghatpande P, Peng Y, Louie J, Lagna G, Zovein AC, Hata A. Let-7 microRNA-dependent control of leukotriene signaling regulates the transition of hematopoietic niche in mice. Nat Commun. 2017;8:128. doi: 10.1038/s41467-017-00137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tornavaca O, Chia M, Dufton N, Almagro LO, Conway DE, Randi AM, Schwartz MA, Matter K, Balda MS. ZO-1 controls endothelial adherens junctions, cell–cell tension, angiogenesis, and barrier formation. J Cell Biol. 2015;208:821–838. doi: 10.1083/jcb.201404140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sörensen I, Adams RH, Gossler A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 2009;113:5680–5688. doi: 10.1182/blood-2008-08-174508. [DOI] [PubMed] [Google Scholar]
- 30.Park SO, Wankhede M, Lee YJ, Choi EJ, Fliess N, Choe SW, Oh SH, Walter G, Raizada MK, Sorg BS, Oh SP. Real-time imaging of de novo arteriovenous malformation in a mouse model of hereditary hemorrhagic telangiectasia. J Clin Invest. 2009;119:3487–3496. doi: 10.1172/JCI39482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DYR, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rakheja D, Chen KS, Liu Y, Shukla AA, Schmid V, Chang TC, Khokhar S, Wickiser JE, Karandikar NJ, Malter JS, Mendell JT, Amatruda JF. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun. 2014;2:4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrezan GT, Ferreira EN, Nakahata AM, Barros BDF, Castro MTM, Correa BR, Krepischi ACV, Olivieri EHR, Cunha IW, Tabori U, Grundy PE, Costa CML, de Camargo B, Galante PAF, Carraro DM. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun. 2014;5:4039. doi: 10.1038/ncomms5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SO, Lee YJ, Seki T, Hong KH, Fliess N, Jiang Z, Park A, Wu X, Kaartinen V, Roman BL, Oh SP. ALK5- and TGFBR2-independent role of ALK1 in the pathogenesis of hereditary hemorrhagic telangiectasia type 2. Blood. 2008;111:633–642. doi: 10.1182/blood-2007-08-107359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 38.Proulx K, Lu A, Sumanas S. Cranial vasculature in zebrafish forms by angioblast cluster-derived angiogenesis. Dev Biol. 2010;348:34–46. doi: 10.1016/j.ydbio.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 39.Jin SW, Herzog W, Santoro MM, Mitchell TS, Frantsve J, Jungblut B, Beis D, Scott IC, D’Amico LA, Ober EA, Verkade H, Field HA, Chi NC, Wehman AM, Baier H, Stainier DYR. A transgene-assisted genetic screen identifies essential regulators of vascular development in vertebrate embryos. Dev Biol. 2007;307:29–42. doi: 10.1016/j.ydbio.2007.03.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The sequence alignment/ map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuervo H, Nielsen CM, Simonetto DA, Ferrell L, Shah VH, Wang RA. Endothelial notch signaling is essential to prevent hepatic vascular malformations in mice. Hepatology. 2016;64:1302–1316. doi: 10.1002/hep.28713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miniati D, Jelin EB, Ng J, Wu J, Carlson TR, Wu X, Looney MR, Wang RA. Constitutively active endothelial Notch4 causes lung arteriovenous shunts in mice. Am J Physiol Lung Cell Mol Physiol. 2010;298:L169–L177. doi: 10.1152/ajplung.00188.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Drosha morphants develop angiogenesis defects.
Fig. S2. Generating Drosha mutant fish by CRISPR/Cas9.
Fig. S3. Drosha morphants showed vascular leakiness.
Fig. S4. Drosha cKOEC embryos showed mild angiogenesis defects and a sign of hemorrhage.
Fig. S5. Drosha cKOEC embryos showed disorganized and dilated vasculature.
Fig. S6. Drosha iKOEC mice show vascular leakage.
Fig. S7. The distribution of the tight junction protein ZO-1 is disrupted in the endothelial cells of Drosha iKOEC mice.
Fig. S8. In vitro processing assay of pri-miR-21 and the BMP-Smad signaling pathway in cells expressing DROSHA mutants or depleted of Drosha.
Fig. S9. ISV defects mediated by the knockdown of Drosha by morpholino oligos are rescued by coinjection of wild-type (WT) Drosha mRNA, but not by mutant mRNA.