Abstract
The transporter associated with antigen processing 2 (TAP2) is involved in the development of multidrug resistance and the etiology of immunological diseases. In this study, we investigated whether the expression of TAP2 can be perturbed by single nucleotide polymorphisms (SNPs) located in 3′-untranslated region (3′-UTR) of the gene via interactions with microRNAs. Using a series of in silico assays, we selected the candidate microRNAs (miRNAs) with the potential to interact with functional SNPs of TAP2. The SNP rs241456—located in the 3′-UTR of TAP2—resides in a potential binding site for hsa-miR-1270 and hsa-miR-620. HEK 293 cells, from a human kidney cell line, were used to characterize the extent of binding of miRNAs to each polymorphic allele of the SNP by a luciferase reporter gene assay. RNA electrophoretic mobility shift assays were used to evaluate the interaction between the miRNAs and each allele sequence of the SNP. We found that hsa-miR-1270 inhibited luciferase activity by binding to the T allele of the SNP in an allele-specific manner. A negative correlation was also found between the expression of hsa-miR-1270 and the T allele of the SNP in kidney tissues. Our findings support the hypothesis that hsa-miR-1270 suppresses the production of TAP2 by binding to this SNP in the 3′-UTR of this gene.
Keywords: hsa-miR-1270, TAP2, genetics, epigenetics, transporter, microRNA
INTRODUCTION
The transporter associated with antigen processing 2 (TAP2), or the ATP-binding cassette subfamily B member 3 (ABCB3), is located in the major histocompatibility complex II (MHC-II) locus of chromosome 6 and is 8–12 kb in size (Trowsdale et al., 1991). Drug resistance, usually leading to unsuccessful treatment, is a major challenge for the treatment of cancer patients. Multidrug resistance for anticancer drugs has been associated with the overexpression of TAP2 (Lage et al., 2001). For instance, the overexpression of TAP2-mediated drug resistance in breast cancer and ovarian cancer has been reported (Park et al., 2006; Ricciardelli et al., 2013). On the other hand, as a transporter, TAP2 also plays a vital role in the delivery of some antigenic peptides to the surface of cytotoxic T lymphocytes. Loss of function of the gene results in a loss of expression of MHC class I molecules on the cell surface due to the absence of appropriate antigenic peptides (Cerundolo et al., 1990; Hosken and Bevan, 1990). Some abnormal cells may escape immune surveillance due to loss of the expression of MHC class I molecules (Zeidler et al., 1997; Lankat-Buttgereit and Tampe, 2002). Despite these intriguing observations, the regulation of TAP2 gene expression has not been fully investigated.
Epigenetic modifications can produce changes in the expression of genes, without changes in the DNA sequences. There are three major mechanisms for epigenetic regulation: DNA methylation, histone acetylation, and noncoding RNA modulation. For example, DNA methylation can inhibit the expression of TAP1 in cervical intrae-pithelial neoplasia (CIN) tissue. Decreased expression of TAP2 at the protein level has also been observed in CIN, although abnormal DNA methylation in the promoter region of the gene was not detected (Hasim et al., 2012). Histone deacetylases (HDACs) can catalyze histone deacetylation, which typically decreases the expression of genes (Buchwald et al., 2009). Histone deacetylase inhibitors (HDACi) are in clinical use as a new class of anti-cancer drugs. Trichostatin A (TSA), a type of HDACi used to treat carcinoma patients, can increase the expression of TAP2 (Setiadi et al., 2008) through epigenetic modulation. Genetic variants of TAP2 have also been examined for their associations with a number of diseases (Ishihara et al., 1997; Kumagai et al., 1997; Kuzushita et al., 1999). Only a few studies have examined the relationship between microRNAs (miRNAs) and TAP2 (Albanese et al., 2016).
The functional consequences of SNPs located in the 3′-untranslated regions (3′-UTRs) of genes have received attention. Genetic variation in the 3′-UTR can affect gene expression by interfering with miRNA binding (Popp et al., 2016). The miRNAs, a group of noncoding RNAs with a length of 20–24 nucleotides, can bind to the target sequences of genes to silence the gene expression (Saito et al., 2014). MiRNAs usually suppress the gene expression by targeting partially complementary sequences located in the 3′-UTR or coding region of mRNA transcripts. SNPs residing in putative miRNA binding sites located in the 3′-UTR of the TAP2 gene are predicted to have a functional effect when examined using publically available prediction models (Wei et al., 2012). However, functionality of those sites requires empirical confirmation.
Our previous studies demonstrated that miRNAs can modulate the expression of genes encoding drug metabolizing enzymes and transporters (Yu et al., 2010, 2015a, 2015b, 2015c; Jin et al., 2016; Chen et al., 2017; Wang et al., 2017; Zeng et al., 2017). In this study, we hypothesized that a SNP in the 3′-UTR of TAP2 interferes with miRNA binding, thus inhibiting the expression of the gene. We applied in silico assays to select a putative functional SNP and the corresponding miRNAs involved in the interaction. The SNP- rs241456 (C>T), located at 3′-UTR of TAP2, was shown to be a potential binding site for hsa-miR-620 or hsa-miR-1270. Furthermore, TAP2 mRNA levels are correlated negatively with the expression of these miRNAs in brain tissues (Wei et al., 2012). Thus, we selected the rs241456, hsa-miR-620 and hsa-miR-1270 for this study. Because altered TAP2 expression is involved in renal disorders, such as renal cell carcinoma, and rejection of renal transplantation (Chevrier et al., 1995; Hodson et al., 2003; George and Mittal, 2011), we conducted our analyses in the kidney tissues. We also found that hsa-miR-1270 can bind to the T allele of rs241456 SNP in an allele-specific manner. In addition, the T allele was negatively correlated with the hsa-miR-1270 expression in kidney tissues. Data from in vitro experiments suggested that hsa-miR-1270 decreases the expression of TAP2 through interacting with the T allele of rs241456 in the 3′-UTR of TAP2.
METHODS AND MATERIALS
Normal Kidney Samples
Normal kidney samples were obtained from the University of Arkansas for Medical Sciences Tissue Bank under Internal Review Board Exemption 4.
In Silico Analysis
The PolymiRTS database 3.0 (http://compbio.uthsc.edu/miRSNP) was screened for common SNPs (minor allele frequency, MAF>0.1) within potential miRNA target sites in the 3′-UTR of the TAP2 transcript (Refseq: NM_000544). Pairwise linkage disequilibrium (LD) analysis was performed based on the 1000 Genomes data for the CEU population using the SNP Annotation and Proxy (SNAP) tool (http://www.broadinstitute.org/mpg/snap). The RNA hybrid program (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) was employed to calculate the minimum free energy (MFE) of hybridization between miRNAs and their potential target sequences. The SNP (rs241456) was selected for further analysis.
In Silico Analysis of the Allele-Specific Correlation Between TAP2 Expression and Hsa-miR-1270
TAP2 gene expression, hsa-miR-1270 expression and B allele frequency (BAF) values of rs241456 were downloaded from the dataset of GSE49280 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49280). We defined rs241456 with BAF > 0.65 as the genotype CC (n = 21), rs241456 with BAF < 0.35 as the genotype TT (n = 7) and rs241456 with 0.35 ≤ BAF ≤ 0.65 as the genotype CT (n = 5). The correlation between TAP2 expression and hsa-miR-1270 expression in both CC and CT/TT genotypes was analyzed using Graph-Pad Prism (https://www.graphpad.com/scientific-software/prism/).
Luciferase Reporter Gene Assay
We modified the pGL3-Control vector (Promega, Madison, WI) by adding the Universal USER Cassette (New England Biolabs, Ipswich, MA) according to our previous research (Yu et al., 2015a). The cloning primers TAP2-C-F: 5′AATCATTATC CCCAACCCTATGAGGT-3′, and TAP2-C-R: 5′GTTGGGGATAATGATTAAAAACGAT-3′ were used to amplify the core region of TAP2 3′-UTR spanning the putative binding sites of hsa-miR-1270 and hsa-miR-620. The USER enzyme (New England Biolabs) was used to digest the PCR products. The digested products were then subcloned into the modified pGL3-Control vector. The prepared plasmid was designated as TAG2-C-CU. Using the site-directed mutagenesis, we constructed TAG -T-CU that contains the rs241456 T allele sequence by using TAP2-T-F: 5′-AATCATTATC TCCAACCCTA TGAGGT-3′, and TAP2-T-R: 5′-GTTGGAGATAATGAT TAAAAACGAT-3′ primers. The authenticity of constructed plasmid was confirmed by DNA sequencing.
HEK 293 cells (ATCC, Manassas, VA), a human kidney cell line, were used for the luciferase reporter gene assay. The cells were cultured in Dulbecco’s Modified Eagle medium with 10% fetal bovine serum (FBS) at 378C in a humidified 5% CO2 atmosphere and 4 × 104 cells were seeded per well in 96-multiwell plates. After the cells reached 70%–80% confluence, we used Lipofectamine 2000 reagent (Life Technologies, Carlsbad, CA) to transfect the prepared TAP2-C-CU orTAP2-T-CU (100 ng), 50 nmol/L hsa-miR-1270 mimics, hsa-miR-620 mimics, or an miRNA negative control (Thermo Scientific, Tewksbury, MA) into the cells. Three independent transfection experiments were performed, and each transfection was carried out in triplicate.
RNA Electrophoretic Mobility Shift Assays (RNA EMSAs)
RNA EMSAs were carried out by the Light Shift Chemiluminescent RNA EMSA Kit (Thermo Scientific) as described previously (Popp et al., 2016). The infrared dye-D800 labeled miRNA oligonucleotide probes used for hsa-miR-1270 and hsa-miR-620 were D800-MIR1270 (rCrUrGrGrArGrArUrArUrGrGrArArGrArGrCrUrGrUrGrU), and D800-MIR620 (rArUrGrGrArGrArUrArGrArUrArUrAr GrArArArU), respectively. The sequences of infrared dye-D700 labeled probes containing the C allele or the T allele of rs241456 are D700-TAP2-T (mAmAmUmCmAm UmUmAmUmCmUmCmCmAmAmCmCmCmUmAmUm GmAmGmGmU) and D700-TAP2-C (mAmAmUmCm AmUmUmAmUmCmCmCmCmAmAmCmCmCmUmAm UmGmAmGmmU), respectively. All the probes were purchased from Integrated DNA Technologies.
Briefly, after heating the oligonucleotides for 5 min at 808C, they were placed on ice for 5 min, and then incubated at 258C for 20 min. The oligonucleotides were mixed with 1× REMSA binding buffer, 5% glycerol, 200 mMKCl, 100 mM MgCl2, and 200 nmol synthetic miRNA and/or cognate mRNA oligonucleotides to a 20 µL basic reaction mixture. Bound complexes and/or unbound probes were separated on an 8% polyacrylamide gel electrophoresis (PAGE) that was carried out at 4°C for 2.5 h. The resulted images for binding assays were then viewed by an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
RNA Extraction and Quantitative Reverse-Transcription PCR (qRT-PCR)
The qRT-PCR was performed as described previously (Yu et al., 2015b). Briefly, the miRNeasy Mini Kit (Qiagen, Valencia, CA) was used to extract total RNA from 63 normal kidney tissues (43 males, 20 females, with a mean age of 60.4 ± 11.3years). The cDNA was prepared using a QuantiTect Reverse Transcription Kit (Qiagen) and an NCode™ miRNA First-Strand cDNA Synthesis Kit (Life Technologies). An ABI Prism7900 Sequence Detection System (Applied Biosystems) was used to detect the expression of TAP2 and hsa-miR-1270 with the SYBR Green method by QuantiFast SYBR1 Green RT-PCR Kit (Qiagen). The primers for the TAP2 gene are TAP2-F: 5′-ACGGCTGAGCTCGGATACCAC-3′ and TAP2-R: 5′-CC TCGGCCCCAAAACTGC-3′. The primers for the GAPDH gene are GAPDH-F: 5′-GAA ATCCCATCACCATCTTC-CAGG-3′ and GAPDH-R: 5′-GAGCCCCAGCCT TCTC CATG-3′. The primers for hsa-miR-1270 and U6 were miR214-F: 5′-CTGGAGATATGGAAGAGCTGTGT-3′ and U6-F: 5′-CTCGCTTCGGCAGCACA-3′, and U6-R: 5′-AACGCTTCACGA ATT TGCGT-3′, respectively. The fold changes of TAP2 or hsa-miR-1270 expression were calculated relative to expression of GAPDH or U6 small nuclear RNA respectively, using the comparative CT method (2−ΔΔCT).
Statistical Analyses
The correlations between the mRNA levels of each allele of the SNP for TAP2 and the levels of the hsa-miR-1270 were analyzed by Spearman correlation in human kidney tissues. Student’s t tests were used to compare results from luciferase reporter gene assays between subgroups. The differences were considered significant at a P value <0.05. Statistical analyses were performed using SPSS software (SPSS 17.0; SPSS, Inc., USA).
RESULTS
In Silico Prediction and Selection of Potentially Functional SNPs
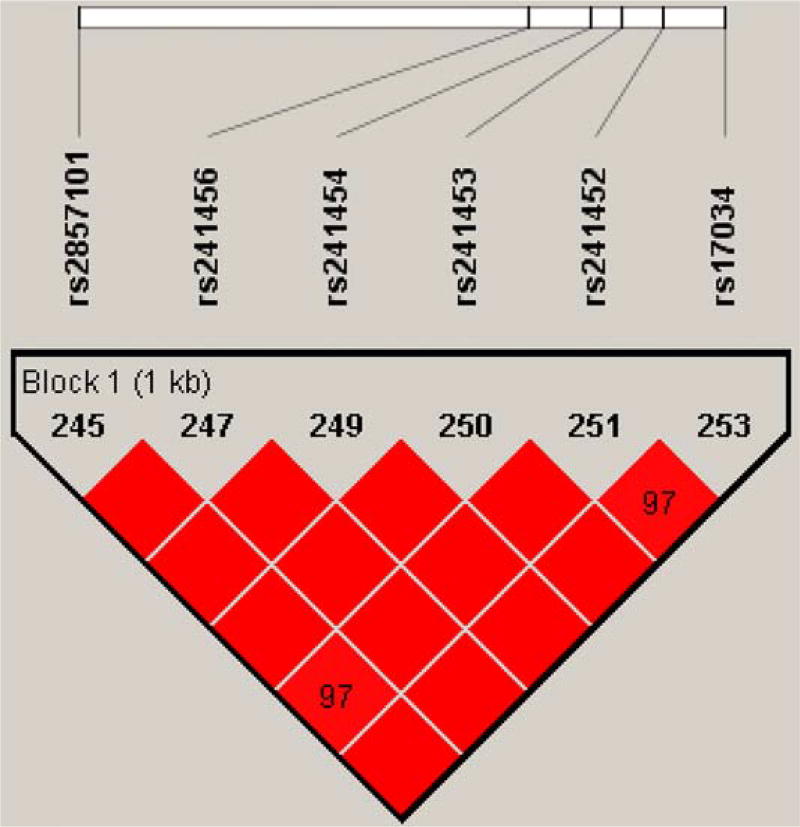
The PolymiRTS database was screened for common SNPs (MAF > 0.1) within potential miRNA target sites in the TAP2 3′-UTR. Six common SNPs were identified as miRNA target site SNPs (Table I). All the SNPs are in strong linkage disequilibrium (LD) (r2> 0.8). Particularly, rs241454 T>C and rs241456 C>T were in a complete LD (r2 = 1.0) with the other five SNPs (Fig. 1). As shown in Table I, a total of 33 miRNAs were predicted to interact with the six SNPs. To determine if a specific SNP can affect miRNA binding, the minimum free energy (MFE) was calculated for the common and variant alleles of each of these SNPs. The difference of MFE (ΔMFE) indicated the change of binding affinity between the miRNA and the mRNA transcript. Remarkable changes in MFE were observed for common and variant transcripts. The variant “T” allele of rs241456 significantly increased the MFE of binding for hsa-miR-1270 and hsa-miR-620, as compared to the common “C” allele. Therefore, hsa-miR-1270 and hsa-miR-620 were selected for examination of their ability to interact with rs241456 C > T.
TABLE I.
MiRNA Target Site SNPs in the 3′-UTR of the TAP2 Gene
| MFE (kcal/mol)
|
|||||
|---|---|---|---|---|---|
| SNP | MAF | Putative miRNAs | Major Allele | Minor Allele | ΔMFE (kcal/mol) |
| rs241452 A>G | 0.302 | hsa-miR-1206 | −18.4 | −18.0 | 0.4 |
| hsa-miR-6516-3p | −14.4 | −14.0 | 0.4 | ||
| rs241454 T>C | 0.301 | hsa-miR-4533 | −20.2 | −21.8 | −1.6 |
| hsa-miR-6797-5p | −26.1 | −26.7 | −0.6 | ||
| rs241456 C>T | 0.301 | hsa-miR-2110 | −18.8 | −20.5 | −1.7 |
| hsa-miR-3150a-3p | −26.6 | −26.8 | −0.2 | ||
| hsa-miR-4450 | −25.8 | −23.4 | 2.4 | ||
| hsa-miR-450a-2–3p | −16.4 | −16.9 | −0.5 | ||
| hsa-miR-6763-5p | −22.6 | −25.1 | −2.5 | ||
| hsa-miR-6810-5p | −21.4 | −21.4 | 0.0 | ||
| hsa-miR-6857-5p | −19.5 | −19.8 | −0.3 | ||
| hsa-miR-92a-1–5p | −19.9 | −19.9 | 0.0 | ||
| hsa-miR-1270 | −21.7 | −16.8 | 4.9 | ||
| hsa-miR-4531 | −14.5 | −12.5 | 2.0 | ||
| hsa-miR-4683 | −18.1 | −14.4 | 3.7 | ||
| hsa-miR-620 | −19.2 | −14.3 | 4.9 | ||
| rs17034 G>A | 0.298 | hsa-miR-1271-3p | −23.2 | −28.0 | −4.8 |
| hsa-miR-4763-5p | −20.2 | −24.3 | −4.1 | ||
| hsa-miR-4772-3p | −18.7 | −23.6 | −4.9 | ||
| hsa-miR-550a-3–5p | −17.6 | −24.1 | −6.5 | ||
| hsa-miR-550a-5p | −20.7 | −27.2 | −6.5 | ||
| hsa-miR-6894-3p | −15.3 | −21.6 | −6.3 | ||
| hsa-miR-4327 | −25.5 | −25.2 | 0.3 | ||
| hsa-miR-636 | −25.3 | −25.0 | 0.3 | ||
| rs241453 C>T | 0.287 | hsa-miR-1302 | −17.6 | −14.7 | 2.9 |
| hsa-miR-4298 | −31.0 | −28.1 | 2.9 | ||
| hsa-miR-3134 | −18.6 | −18.6 | 0.0 | ||
| hsa-miR-4534 | −18.8 | −22.0 | −3.2 | ||
| hsa-miR-8082 | −18.7 | −22.8 | −4.1 | ||
| rs2857101 A>G | 0.287 | hsa-miR-126-5p | −11.6 | −11.6 | 0.0 |
| hsa-miR-4795-3p | −12.2 | −12.2 | 0.0 | ||
| hsa-miR-944 | −12.7 | −12.7 | 0.0 | ||
| hsa-miR-6731-3p | −10.9 | −12.8 | −1.9 | ||
Abbreviations: MAF, minor allele frequency; MFE, minimum free energy; ΔMFE, difference in MFE between wild-types and variants.
Fig. 1.
Linkage disequilibrium (LD) map of the TAP2 3′-UTR showing the degree of linkage disequilibrium between the six genotyped SNPs. The LD plot is displayed using an r2 black and white color scheme. Black: r2 = 1; white: r2 = 0; shades of grey: 0 < r2 <1.
Allele-Specific Expression of TAP2 and Hsa-miR-1270 Using Silico Analysis
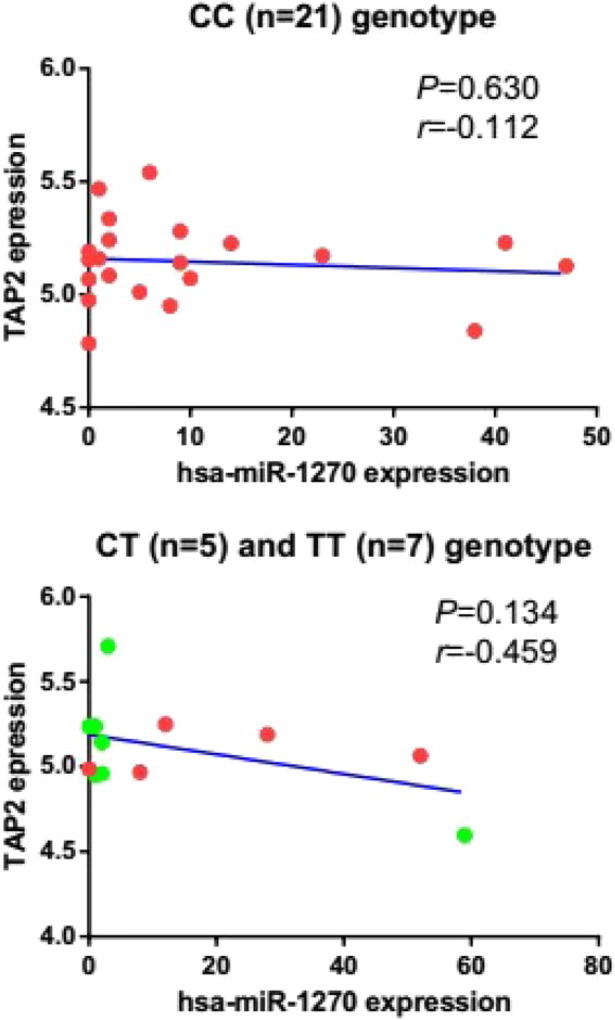
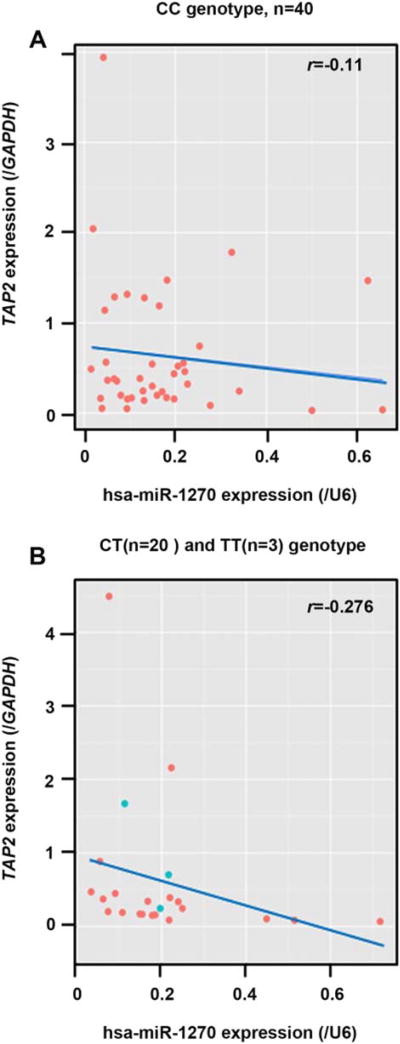
Using the GSE49280 dateset, we did not find an allele specific correlation between CC (Pearson’s correlation, r = −0.112 and P = 0.630) or CT/TT genotypes (Pearson’s correlation, r = −0.459 and P = 0.134) of TAP2 transcripts with the expression of hsa-miR-1270 (Fig. 2). However, the trend of “allele specific” correlation suggested the need of correlation analysis using human kidney tissues (see results below).
Fig. 2.
Correlation between the expression of CC and CT/TT genotypes of TAP2 and hsa-miR-1270 expression. The expression levels of (A) CC (n = 21) genotype (r = −0.112, P = 0.630) and (B) CT (n = 5)/TT (n = 7) genotype (r = −0.459, P = 0.134) of TAP2 were not statistically significantly correlated with hsa-miR-1270 expression levels in the dataset of GSE49280 using Pearson’s correlation analysis.
Allele-Specific Suppression of TAP2 by Hsa-miR-1270
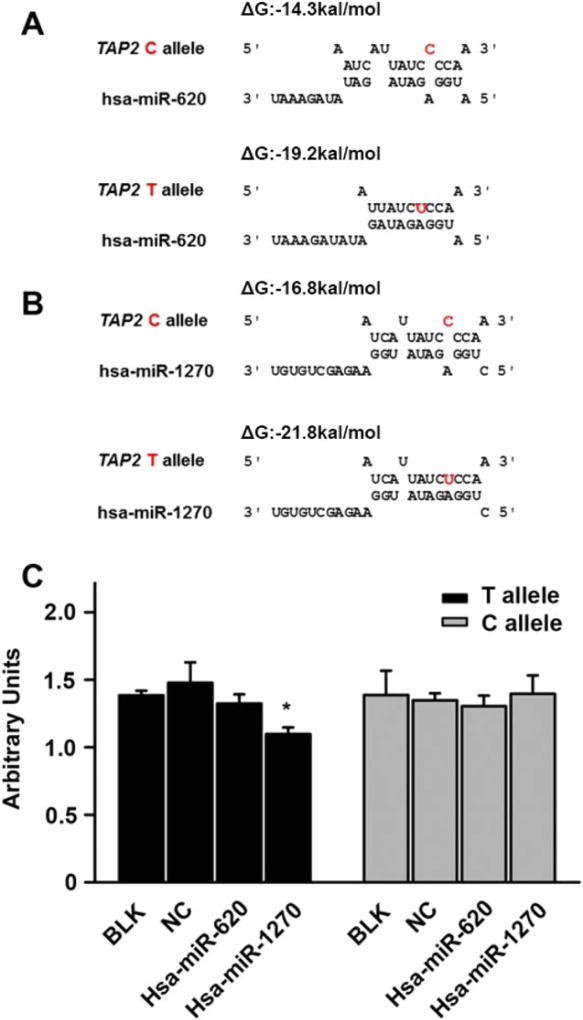
Using the RNA hybrid program to predicted free energy between the sequence of C-allele or T-allele and hsa-miR-1270 or hsa-miR-620, we found that the T-allele sequence exhibited a lower free energy for binding than the C allele sequence by both hsa-miR-1270 (−26.7 kcal/ mol vs −16.8 kcal/mol) and hsa-miR-620 (−19.2 kcal/ mol vs −14.3 kcal/mol) (Fig. 3A,B). The results suggested that hsa-miR-1270 or hsa-miR-620 binds more readily to the T allele sequence than that of C allele. Next, the rs241456 T or C allele sequence and the hsa-miR-1270, hsa-miR-620 mimics, or the miRNA negative control, were transfected to HEK 293 cells, respectively. The reporter gene assays showed that the expression of the T-allele harboring luciferase was efficiently suppressed by hsa-miR-1270 compared with the miRNA negative control (25.7%, 1.1 vs 1.48). However, the C allele harboring luciferase was not suppressed by hsa-miR-1270 and neither allele was suppressed by hsa-miR-620 (Fig. 3C).
Fig. 3.
The hsa-miR-1270 suppressed reporter gene expression in an allele special manner. (A) Targeting prediction of hsa-miR- 620 to the 3′-UTR of TAP2 based on rs241456 allele. The free energy for binding of the miRNA to the T allele is stronger than the C allele. (B) Targeting prediction of hsa-miR-1270 to the 3′ -UTR of TAP2 based on rs241456 allele. The free energy for binding of the miRNA to the T allele is stronger than the C allele. (C) In HEK 293 cells, using luciferase reporter assay to detect the effect of each allele of rs241456. After HEK 293 cells were transiently transfected with a plasmid containing either the T or C allele for rs241456 together with hsa-miR-1270 mimics, hsa-miR-620 mimics, or an miRNA-negative control (NC). Renilla luciferase was measured as an internal control for the transfection efficiency, and firefly luciferase was measured to compare the gene expression levels. All experiments were carried out in triplicate. *P <0.05.
Allele-Specific Interaction between TAP2 mRNA and Hsa-miR-1270
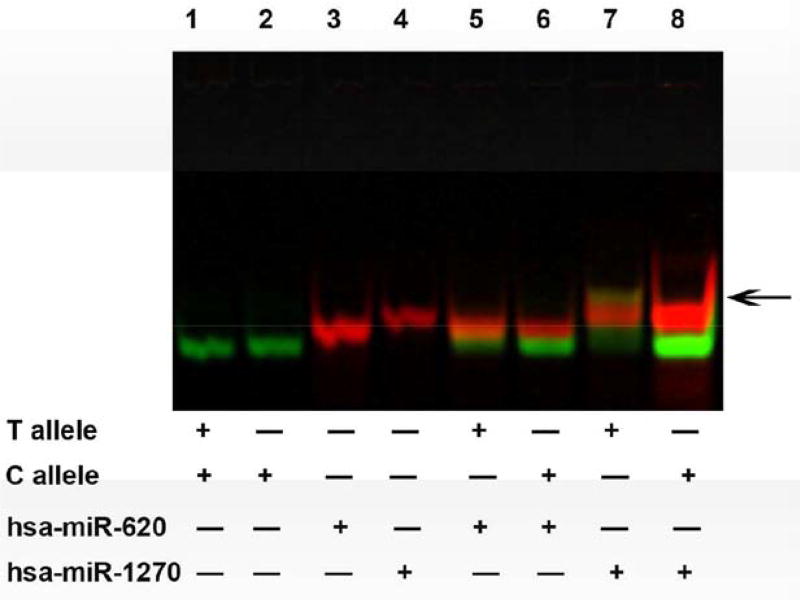
To determine if the miRNA molecule indeed interacts with its targeted mRNA sequence, and if it recognizes the cognate mRNA in an allele specific manner, we conducted mRNA:miRNA EMSA experiments. A complex was formed by adding the hsa-miR-1270 and mRNA oligonucleotides containing rs241456 T allele sequence (Fig. 4, Lane 7) in vitro using RNA EMSA, but no such a complex was observed from the interaction of hsa-miR-1270 and rs241456 C allele sequence (Lane 8). In addition, we did not detect any complexes formed between the C-allele or T-allele oligonucleotides and hsa-miR-620 (Lanes 5 and, 6). The result indicated an interaction between miR-1270 and the T allele (rs241456) at 3′-UTR of the TAP2 transcript.
Fig. 4.
Interaction of hsa-miR-1270 with different allele sequence of rs142456. Lanes 1–4 show the mobility of the labeled nucleotides. Lanes 5–6 show the mobility of hsa-miR-620 with each allele of rs241456 in TAP2. Lanes 7–8 show the mobility of hsa-miR- 1270 with each allele of rs241456 in TAP2. Arrow indicated the complex of hsa-miR1270 and T allele of the SNP241456 in yellow.
Correlation between the Expression of Hsa-miR-1270 and TAllele or C Allele of TAP2 Transcripts in Human Kidney Tissues
Tissue expression correlation analysis was conducted to confirm the biological relevance of miR-1270 in the regulation of TAP2 in human tissues. As shown in Figure 5B, significant negative correlations between hsa-miR-1270 levels and the T allele of TAP2 mRNA expression were found in kidney tissues (r = −0.276; P = 0.018). However, the C allele of TAP2 mRNA was not correlated (r = −0.111; P = 0.468) with the hsa-miR-1270 level in the same tissues (Fig. 5A). Although the inverse correlation between hsa-miR-1270 levels and the expression of T allele in TAP2 mRNA transcripts is modest owing to the heterogeneity of human samples achieved from an environmentally diversified population, together with the results of RNA-EMSA and luciferase assays, we showed that hsa-miR-1270 can regulate the expression of TAP2 by targeting the cognitive sequence containing the T allele of rs241456.
Fig. 5.
Correlation between the expression of hsa-miR-1270 and each allele of rs241456 for TAP2 transcripts. The levels of hsa-miR-1270 was negatively correlated with the mRNA levels for T allele of rs241456 in TAP2 (r = −0.111, 63 normal kidney samples) but not for C allele of rs241456 (r = −0.276) using Spearman correlation analysis.
DISCUSSION
The interindividual variability in the expression of genes encoding drug-metabolizing enzymes and transporters was documented (Ning et al., 2008; Yang et al., 2013), and it may influence drug absorption, metabolism, and pharmacokinetics, and play an important role in drug efficacy, safety, and adverse drug reactions (Evans and Relling, 1999; Spear et al., 2001; Wilkinson, 2005). Genetic changes of transporter genes associated with some diseases have also been demonstrated (Dietrich and Geier, 2014) and can be clinically significant (Brambila-Tapia, 2013; De Iuliis et al., 2015). An SNP is a common genetic variation in the germline and they have been extensively studied due to an important role in the pathology of diseases (Bodmer and Bonilla, 2008). Recently, the functional effects of genetic variations in 3′-UTR of the genes have received attention (Ning et al., 2014; Relling and Evans, 2015). Epigenetic regulation is an important mechanism for modifying gene expression (Wang et al., 2015, 2016). As an important part of epigenetics, miRNAs play a vital role in regulation of transporter gene expression through binding the 3′-UTR of genes (Koturbash et al., 2015; Wan et al., 2015; Engstrom et al., 2016; Kap et al., 2016).
In this study, we selected hsa-miR-1270 and hsa-miR-620 as candidate miRNAs to elucidate the interactions between the miRNA and a functional SNP in the 3′-UTR of TAP2 using in silico analysis. Employing luciferase reporter gene assays, we found that hsa-miR-1270 can reduce luciferase gene activity in an allele-specific manner in vitro. RNA EMSA also showed complex formation of hsa-miR-1270 and the T allele of rs241456 but not with the C allele. The expression of hsa-miR-1270 and different alleles of rs241456 of TAP2 in kidney tissues were detected. The expression levels of hsa-miR-1270 were negatively correlated with the C allele of rs241456 of TAP2 mRNA levels using the Spearman correlation analysis (r = −0.276), but not the T allele. These results show that hsa-miR-1270 negatively regulates TAP2 expression in an allele-specific manner in kidney tissues. Due to large interindividual variability in the expression of both TAP2 and miR-1270, and the small human samples available for use in this study, the correlation between the expression of the T allele at TAP2 (rs241456) and miR-1270 is not strong, but does indicate a trend of negativity. It is a common phenomenon that the correlations between miRNAs and their cognate mRNAs in human samples are possibly confounded due to the complexity of gene regulation and the diversity of genetic and environmental background among human populations(Yu et al., 2010, 2015a, 2015b, 2015c; Jin et al., 2016; Chen et al., 2017; Wang et al., 2017; Zeng et al., 2017).
Genetic variation in the coding region of TAP2 has been associated with tuberculosis (TB) infection in some population-based studies (Rajalingam et al., 1997; Gomez et al., 2006; Roh et al., 2015; Thu et al., 2016). However, there are some differences in the results reported for those studies. The reasons for discordant results may be the heterogeneity between different cohorts or weak genetic effects. We speculated that other unknown factors could play vital roles in regulating gene expression. For example, the expression of TAP2 can be reduced by Epstein– Barr virus (EBV) miRNAs through targeting its transcript (Albanese et al., 2016). We examined the hypothesis that the rs241456 SNP had functional consequences on miR-NAs targeting. We identified two miRNAs that could potentially bind to the SNP-harboring site using in silico analysis. Calculations demonstrated that the free energy of the two miRNAs binding the T allele is less than that of the C allele of the rs241456 SNP. The results indicate that the T allele binds more readily to the two miRNAs due to a more favorable free energy profile (less than 220 kcal/mol) (Yu et al., 2015c). Using luciferase assays, we further confirmed the inhibition of T allele expression in the TPA2 gene, based on the promise that any change greater than 15%–35% should be detected at the enzyme activity or protein production levels (Chen et al., 2017; Wang et al., 2017; Zeng et al., 2017).
Alterations of the expression of TAP2 can result in important impacts on human health. The ATP-binding cassette transporter, a heterodimer of TAP1 and TAP2, mainly translocates peptides from the cytosol to MHC class I and then presents the trimeric MHC complex to the cell-surface of immune cells including T lymphocytes and natural killer cells. Decreased TAP accumulation results in surface HLA accumulation, thus impairing immune functions (Karttunen et al., 2001). A study suggested that alterations in TAP2 stoichiometry could change MHC processing properties in decidual stromal cells, thus interfering with maternal–fetal interactions during pregnancy (Mika and Lynch, 2016). On the other hand, ATP-binding cassette transporters are involved in xenobiotic metabolism. In concordance to the observation that upregulation of multiple drug resistance gene expression is associated with breast cancer patients’ resistance to neoadjuvant chemotherapy, the deletion of TAP2 expression is favorable for patients to respond to neoadjuvant chemotherapy (Litviakov et al., 2016). In addition, with a bacterial model, Lerebours et al. (2016) demonstrated that the alteration of TAP2 expression could change the metal accumulation level and survival ratio in E. coli cells, indicating the significance of TAP2 in metal detoxification.
It should be helpful to understand what degree of TAP2 inhibition would lead to a biologically significant outcome. Are there logical positive and negative controls that can help set the boundaries? Answering this question is extremely challenging, because interindividual variability in the expression of genes encoding human drug metabolizing enzymes and transporters (DMETs) is dependent on multifactorial components, including genetic, epigenetic, environmental factors, and individual’s health status (Koturbash et al., 2015). As to what degree a miRNA can modulate DMET expression, our previous studies (for more than 10 DMETs and corresponding miRNAs) and literature results suggested a level of ~15%–35% inhibition could be achieved by miRNAs in the expression of DMETs. The small amount of inhibition in DMETs could be biologically significant in drug efficacy and safety if the magnitude of miRNA inhibition is intensified by drug–drug interaction, gene–gene, and gene-environmental interaction (Koturbash et al., 2015; Chen et al., 2017; Wang et al., 2017; Zeng et al., 2017).
RNA-ESMA is an effective way to observe the interaction between a miRNA and its target sequences in vitro (Yu et al., 2015c). In this work, the binding complex can be detected between the T allele of rs241456 SNP and hsa-miR-1270, but not for the C allele and the miRNA. The allele-specific manner of binding has been extensively studied by several researchers. For instance, hsa-miR-548s and hsa-miR-4480 suppressed the expression of IL-17A in an allele-specific manner in age-related macular degeneration (AMD) (Popp et al., 2016). This is consistent with our results that miRNA can inhibit target gene expression through binding the SNP in an allele-specific manner.
Mutations and/or polymorphism within TAP2 could alter the efficacy of the immune response and contribute to the etiology of renal cell carcinoma (Hodson et al., 2003). We detected the expression of each allele of rs241456 for TAP2 and hsa-miR-1270. Spearman correlation analysis showed hsa-miR-1270 was negatively correlated with mRNA levels of the T allele in TAP2, but not the C allele. These results indicate that miRNA plays an important role in regulating gene expression. Importantly, the overexpression of TAP2 has been demonstrated to associate with drug resistance in some cancers. In MCF7/ AdVp300 cells, a drug resistance breast cancer cell line, the expression of TAP2 was elevated 2.1-fold, compared to the parental MCF7 (Liu et al., 2005). It was also reported that the expression of TAP2 was significantly increased in recurrent malignant ovarian tissues compared to benign tissues (Auner et al., 2010). In this study, hsa-miR-1270 can inhibit the expression of TAP2 in an allele-dependent manner, specifically with a higher inhibitory efficiency towards the T allele of the SNP rs241456, which may help to explain the individual variability of drug metabolisms and drug resistances for some drugs that are transported by TAP2.
In conclusion, based on the current results, rs241456 is a functional SNP in TAP2 and the different alleles of the SNP can alter the ability of hsa-miR-1270 to bind and degrade TAP2 mRNA in vitro. However, as the transcripts of TAP2 can be targeted by many miRNAs, our study does not exclude the possibility that other miRNAs binding to the gene decreases expression of TAP2. There are several limitations with this study. For example, the allele frequency at the population level is not known. Nevertheless, our study provides new insight into the role of miRNAs in the modulation of TAP2, which may impact the design of personalized medicine strategies for patients based upon genotype.
Acknowledgments
Grant sponsor: University of Arkansas for Medical Sciences; Grant number: 76788.
Grant sponsor: National Center for Toxicological Research of the U.S. Food and Drug Administration; Grant numbers: E0752601 and E0731301.
Footnotes
Disclaimer: The information in these materials is not a formal dissemination of information by the U.S Food and Drug Administration and does not represent agency position or policy.
AUTHOR CONTRIBUTIONS
Participated in Study design: Ning, Knox, and Kadlubar.
Conducted experiments: Knox, Rogers, Wang, Yu, and Chen.
Performed data analysis: Xuan, Knox, Wang, Yu, Guan, Shi, Ning, and Kadlubar.
Wrote or contributed to the writing of the manuscript: Wang, Knox, Kadlubar, Xuan, Rogers, and Ning.
CONFLICT OF INTEREST
The authors have no conflict of interest.
References
- Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J, Hastreiter M, Hayes M, Sugden B, Martin L, Moosmann A, Hammerschmidt W. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc Natl Acad Sci USA. 2016;113:E6467–E6475. doi: 10.1073/pnas.1605884113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auner V, Sehouli J, Oskay-Oezcelik G, Horvat R, Speiser P, Zeillinger R. ABC transporter gene expression in benign and malignant ovarian tissue. Gynecol Oncol. 2010;117:198–201. doi: 10.1016/j.ygyno.2009.10.077. [DOI] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambila-Tapia AJ. MDR1 (ABCB1) polymorphisms: Functional effects and clinical implications. Rev Invest Clin. 2013;65:445–454. [PubMed] [Google Scholar]
- Buchwald M, Kramer OH, Heinzel T. HDACi-targets beyond chromatin. Cancer Lett. 2009;280:160–167. doi: 10.1016/j.canlet.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Cerundolo V, Alexander J, Anderson K, Lamb C, Cresswell P, McMichael A, Gotch F, Townsend A. Presentation of viral antigen controlled by a gene in the major histocompatibility complex. Nature. 1990;345:449–452. doi: 10.1038/345449a0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zeng L, Wang Y, Tolleson WH, Knox B, Chen S, Ren Z, Guo L, Mei N, Qian F, Huang K, Liu D, Tong W, Yu D, Ning B. The expression, induction and pharmacological activity of CYP1A2 are post-transcriptionally regulated by microRNA hsa-miR-132-5p. Biochem Pharmacol. 2017 doi: 10.1016/j.bcp.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier D, Giral M, Braud V, Bourbigot B, Muller JY, Bignon JD, Soulillou JP. Effects of MHC-encoded TAP1 and TAP2 gene polymorphism and matching on kidney graft rejection. Transplantation. 1995;60:292–296. doi: 10.1097/00007890-199508000-00015. [DOI] [PubMed] [Google Scholar]
- De Iuliis F, Salerno G, Taglieri L, Scarpa S. Are pharmacogenomic biomarkers an effective tool to predict taxane toxicity and outcome in breast cancer patients? Literature review. Cancer Chemother Pharmacol. 2015;76:679–690. doi: 10.1007/s00280-015-2818-4. [DOI] [PubMed] [Google Scholar]
- Dietrich CG, Geier A. Effect of drug transporter pharmacogenetics on cholestasis. Expert Opin Drug Metab Toxicol. 2014;10:1533–1551. doi: 10.1517/17425255.2014.963553. [DOI] [PubMed] [Google Scholar]
- Engstrom K, Love TM, Watson GE, Zareba G, Yeates A, Wahlberg K, Alhamdow A, Thurston SW, Mulhern M, McSorley EM, Strain JJ, Davidson PW, Shamlaye CF, Myers GJ, Rand MD, van Wijngaarden E, Broberg K. Polymorphisms in ATP-binding cassette transporters associated with maternal methylmercury disposition and infant neurodevelopment in mother-infant pairs in the Seychelles Child Development Study. Environ Int. 2016;94:224–229. doi: 10.1016/j.envint.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, Relling MV. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- George GP, Mittal RD. Genetic polymorphisms in MHC-encoded antigen processing gene TAP2: A case-control study in end-stage renal disease patients of North India. Transpl Immunol. 2011;24:220–223. doi: 10.1016/j.trim.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Gomez LM, Camargo JF, Castiblanco J, Ruiz-Narvaez EA, Cadena J, Anaya JM. Analysis of IL1B, TAP1, TAP2 and IKBL polymorphisms on susceptibility to tuberculosis. Tissue Antigens. 2006;67:290–296. doi: 10.1111/j.1399-0039.2006.00566.x. [DOI] [PubMed] [Google Scholar]
- Hasim A, Abudula M, Aimiduo R, Ma JQ, Jiao Z, Akula G, Wang T, Abudula A. Post-transcriptional and epigenetic regulation of antigen processing machinery (APM) components and HLA-I in cervical cancers from Uighur women. PLoS One. 2012;7:e44952. doi: 10.1371/journal.pone.0044952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson I, Bock M, Ritz U, Brenner W, Huber C, Seliger B. Analysis of the structural integrity of the TAP2 gene in renal cell carcinoma. Int J Oncol. 2003;23:991–999. [PubMed] [Google Scholar]
- Hosken NA, Bevan MJ. Defective presentation of endogenous antigen by a cell line expressing class I molecules. Science. 1990;248:367–370. doi: 10.1126/science.2326647. [DOI] [PubMed] [Google Scholar]
- Ishihara M, Ohno S, Ishida T, Naruse T, Kagiya M, Mizuki N, Maruya E, Saji H, Inoko H. Analysis of allelic variation of the TAP2 gene in sarcoidosis. Tissue Antigens. 1997;49:107–110. doi: 10.1111/j.1399-0039.1997.tb02722.x. [DOI] [PubMed] [Google Scholar]
- Jin Y, Yu D, Tolleson WH, Knox B, Wang Y, Chen S, Ren Z, Deng H, Guo Y, Ning B. MicroRNA hsa-miR-25-3p suppresses the expression and drug induction of CYP2B6 in human hepatocytes. Biochem Pharmacol. 2016;113:88–96. doi: 10.1016/j.bcp.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kap EJ, Seibold P, Scherer D, Habermann N, Balavarca Y, Jansen L, Zucknick M, Becker N, Hoffmeister M, Ulrich A, Benner A, Ulrich CM, Burwinkel B, Brenner H, Chang-Claude J. SNPs in transporter and metabolizing genes as predictive markers for oxaliplatin treatment in colorectal cancer patients. Int J Cancer. 2016;138:2993–3001. doi: 10.1002/ijc.30026. [DOI] [PubMed] [Google Scholar]
- Karttunen JT, Lehner PJ, Gupta SS, Hewitt EW, Cresswell P. Distinct functions and cooperative interaction of the subunits of the transporter associated with antigen processing (TAP) Proc Natl Acad Sci USA. 2001;98:7431–7436. doi: 10.1073/pnas.121180198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Tolleson WH, Guo L, Yu D, Chen S, Hong H, Mattes W, Ning B. microRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark Med. 2015;9:1153–1176. doi: 10.2217/bmm.15.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai S, Kanagawa S, Morinobu A, Takada M, Nakamura K, Sugai S, Maruya E, Saji H. Association of a new allele of the TAP2 gene, TAP2*Bky2 (Val577), with susceptibility to Sjogren’s syndrome. Arthritis Rheum. 1997;40:1685–1692. doi: 10.1002/art.1780400919. [DOI] [PubMed] [Google Scholar]
- Kuzushita N, Hayashi N, Kanto T, Takehara T, Tatsumi T, Katayama K, Ohkawa K, Ito A, Kasahara A, Moribe T, Sasaki Y, Hori M. Involvement of transporter associated with antigen processing 2 (TAP2) gene polymorphisms in hepatitis C virus infection. Gastroenterology. 1999;116:1149–1154. doi: 10.1016/s0016-5085(99)70018-1. [DOI] [PubMed] [Google Scholar]
- Lage H, Perlitz C, Abele R, Tampe R, Dietel M, Schadendorf D, Sinha P. Enhanced expression of human ABC-transporter tap is associated with cellular resistance to mitoxantrone. FEBS Lett. 2001;503:179–184. doi: 10.1016/s0014-5793(01)02722-3. [DOI] [PubMed] [Google Scholar]
- Lankat-Buttgereit B, Tampe R. The transporter associated with antigen processing: Function and implications in human diseases. Physiol Rev. 2002;82:187–204. doi: 10.1152/physrev.00025.2001. [DOI] [PubMed] [Google Scholar]
- Lerebours A, To VV, Bourdineaud JP. Danio rerio ABC transporter genes abcb3 and abcb7 play a protecting role against metal contamination. J Appl Toxicol. 2016;36:1551–1557. doi: 10.1002/jat.3313. [DOI] [PubMed] [Google Scholar]
- Litviakov NV, Cherdyntseva NV, Tsyganov MM, Slonimskaya EM, Ibragimova MK, Kazantseva PV, Kzhyshkowska J, Choinzonov EL. Deletions of multidrug resistance gene loci in breast cancer leads to the down-regulation of its expression and predict tumor response to neoadjuvant chemotherapy. Oncotarget. 2016;7:7829–7841. doi: 10.18632/oncotarget.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Peng H, Zhang JT. Expression profiling of ABC transporters in a drug-resistant breast cancer cell line using AmpArray. Mol Pharmacol. 2005;68:430–438. doi: 10.1124/mol.105.011015. [DOI] [PubMed] [Google Scholar]
- Mika KM, Lynch VJ. An ancient fecundability-associated polymorphism switches a repressor into an enhancer of endometrial TAP2 expression. Am J Hum Genet. 2016;99:1059–1071. doi: 10.1016/j.ajhg.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning B, Dial S, Sun Y, Wang J, Yang J, Guo L. Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J Biomol Screen. 2008;13:194–201. doi: 10.1177/1087057108315513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning B, Su Z, Mei N, Hong H, Deng H, Shi L, Fuscoe JC, Tolleson WH. Toxicogenomics and cancer susceptibility: Advances with next-generation sequencing. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2014;32:121–158. doi: 10.1080/10590501.2014.907460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Shimizu C, Shimoyama T, Takeda M, Ando M, Kohno T, Katsumata N, Kang YK, Nishio K, Fujiwara Y. Gene expression profiling of ATP-binding cassette (ABC) transporters as a predictor of the pathologic response to neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res Treat. 2006;99:9–17. doi: 10.1007/s10549-006-9175-2. [DOI] [PubMed] [Google Scholar]
- Popp NA, Yu D, Green B, Chew EY, Ning B, Chan CC, Tuo J. Functional single nucleotide polymorphism in IL-17A 3’ untranslated region is targeted by miR-4480 in vitro and may be associated with age-related macular degeneration. Environ Mol Mutagen. 2016;57:58–64. doi: 10.1002/em.21982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam R, Singal DP, Mehra NK. Transporter associated with antigen-processing (TAP) genes and susceptibility to tuberculoid leprosy and pulmonary tuberculosis. Tissue Antigens. 1997;49:168–172. doi: 10.1111/j.1399-0039.1997.tb02731.x. [DOI] [PubMed] [Google Scholar]
- Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526:343–350. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardelli C, Ween MP, Lokman NA, Tan IA, Pyragius CE, Oehler MK. Chemotherapy-induced hyaluronan production: A novel chemoresistance mechanism in ovarian cancer. BMC Cancer. 2013;13:476. doi: 10.1186/1471-2407-13-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh EY, Yoon JH, Shin S, Song EY, Park MH. Association of TAP1 and TAP2 genes with susceptibility to pulmonary tuberculosis in Koreans. APMIS. 2015;123:457–464. doi: 10.1111/apm.12373. [DOI] [PubMed] [Google Scholar]
- Saito Y, Saito H, Liang G, Friedman JM. Epigenetic alterations and microRNA misexpression in cancer and autoimmune diseases: A critical review. Clin Rev Allergy Immunol. 2014;47:128–135. doi: 10.1007/s12016-013-8401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiadi AF, Omilusik K, David MD, Seipp RP, Hartikainen J, Gopaul R, Choi KB, Jefferies WA. Epigenetic enhancement of antigen processing and presentation promotes immune recognition of tumors. Cancer Res. 2008;68:9601–9607. doi: 10.1158/0008-5472.CAN-07-5270. [DOI] [PubMed] [Google Scholar]
- Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–204. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- Thu KS, Sato N, Ikeda S, Naka-Mieno M, Arai T, Mori S, Sawabe M, Muramatsu M, Tanaka M. Association of polymorphisms of the transporter associated with antigen processing (TAP2) gene with pulmonary tuberculosis in an elderly Japanese population. APMIS. 2016;124:675–680. doi: 10.1111/apm.12562. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Ragoussis J, Campbell RD. Map of the human MHC. Immunol Today. 1991;12:443–446. doi: 10.1016/0167-5699(91)90017-n. [DOI] [PubMed] [Google Scholar]
- Wan W, Xu X, Zhao DB, Pang YF, Wang YX. Polymorphisms of uric transporter proteins in the pathogenesis of gout in a Chinese Han population. Genet Mol Res. 2015;14:2546–2550. doi: 10.4238/2015.March.30.13. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li F, Zhang G, Kang L, Guan H. Ultraviolet-B induces ERCC6 repression in lens epithelium cells of age-related nuclear cataract through coordinated DNA hypermethylation and histone deacetylation. Clin Epigenetics. 2016;8:62. doi: 10.1186/s13148-016-0229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li F, Zhang G, Kang L, Qin B, Guan H. Altered DNA methylation and expression profiles of 8-oxoguanine DNA glycosylase 1 in lens tissue from age-related cataract patients. Curr Eye Res. 2015;40:815–821. doi: 10.3109/02713683.2014.957778. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yu D, Tolleson WH, Yu LR, Green B, Zeng L, Chen Y, Chen S, Ren Z, Guo L, Tong W, Guan H, Ning B. A systematic evaluation of microRNAs in regulating human hepatic CYP2E1. Biochem Pharmacol. 2017;138:174–184. doi: 10.1016/j.bcp.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R, Yang F, Urban TJ, Li L, Chalasani N, Flockhart DA, Liu W. Impact of the interaction between 3’-UTR SNPs and microRNA on the expression of human xenobiotic metabolism enzyme and transporter genes. Front Genet. 2012;3:248. doi: 10.3389/fgene.2012.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson GR. Drug metabolism and variability among patients in drug response. N Engl J Med. 2005;352:2211–2221. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- Yang L, Price ET, Chang CW, Li Y, Huang Y, Guo LW, Guo Y, Kaput J, Shi L, Ning B. Gene expression variability in human hepatic drug metabolizing enzymes and transporters. PLoS One. 2013;8:e60368. doi: 10.1371/journal.pone.0060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, Fuscoe J, Pogribny I, Ning B. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015a;5:8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Green B, Tolleson WH, Jin Y, Mei N, Guo Y, Deng H, Pogribny I, Ning B. MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. Biochem Pharmacol. 2015b;98:215–223. doi: 10.1016/j.bcp.2015.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Tolleson WH, Knox B, Jin Y, Guo L, Guo Y, Kadlubar SA, Ning B. Modulation of ALDH5A1 and SLC22A7 by micro-RNA hsa-miR-29a-3p in human liver cells. Biochem Pharmacol. 2015c;98:671–680. doi: 10.1016/j.bcp.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Dhakal IB, Beggs M, Edavana VK, Williams S, Zhang X, Mercer K, Ning B, Lang NP, Kadlubar FF. Functional genetic variants in the 3’-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010;118:391–403. doi: 10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler R, Eissner G, Meissner P, Uebel S, Tampe R, Lazis S, Hammerschmidt W. Downregulation of TAP1 in B lymphocytes by cellular and Epstein-Barr virus-encoded interleukin-10. Blood. 1997;90:2390–2397. [PubMed] [Google Scholar]
- Zeng L, Chen Y, Wang Y, Yu LR, Knox B, Chen J, Shi T, Chen S, Ren Z, Guo L, Wu Y, Liu D, Huang K, Tong W, Yu D, Ning B. MicroRNA hsa-miR-370-3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol. 2017;140:139–149. doi: 10.1016/j.bcp.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]