Abstract
Increasing evidence shows that tumor clonal architectures are often the consequence of a complex branching process, yet little is known about the expected dynamics and extent to which these divergent subclonal expansions occur. Here we develop and implement more than 88,000 instances of a stochastic evolutionary model simulating genetic drift and neoplastic progression. Under different combinations of population genetic parameter values, including those estimated for colorectal cancer and glioblastoma multiforme, the distribution of sizes of subclones carrying driver mutations had a heavy right tail at the time of tumor detection, with only 1–4 dominant clones present at ≥10% frequency. In contrast, the vast majority of subclones were present at <10% frequency, many of which had higher fitness than currently dominant clones. The number of dominant clones (≥10% frequency) in a tumor correlated strongly with the number of subclones (<10% of the tumor). Overall, these subclones were frequently below current standard detection thresholds, frequently harbored treatment-resistant mutations and were more common in slow-growing tumors.
Keywords: cancer evolution, stochastic modeling, subclonal composition, micro and macro heterogeneity, drug resistance
Introduction
Cancer is a subclonal evolutionary process and is governed by the dynamic interplay of mutation, stochastic drift, and selection [4–6]. Although most mutations that steadily accumulate in our cells are probably neutral or weakly deleterious, a fraction of these mutations, especially in genes and regulatory elements, can confer a selective advantage to the cell by increasing its fitness [7–9]. In cancers, these mutations can result in increased survival of a clone [7,10]. In the field of cancer biology, the term “driver mutation” is often used to refer to mutations that increase a cell’s fitness (and thus are increased in frequency due to positive selection) [7,11]. The term “passenger mutation” is used for mutations that are neutral or deleterious [7,11,12], and increase in frequency due to hitchhiking alongside driver mutations, bottleneck events or genetic drift. A common model for the evolutionary process of tumor growth envisions driver mutations causing clonal expansions that sweep through the cancer cell population and reach fixation (100% frequency) [1,13]. If such a driver mutation did reach fixation, it would appear as a “trunk” mutation, present in all the tumor cells. However, the experimental evidence points to tumor clonal architectures that are the consequence of a complex branching processes, with divergent subclones evolving simultaneously [14–19].
While a few clones may dominate the composition of a tumor, minor subclones, often below current detection thresholds, can determine the clinical course of disease progression and recurrence [20–24]. For example, in patients with chronic lymphocytic leukemia (CLL) who received chemotherapy, the presence of detectable subclones harboring one or more cancer-driver genes in the primary leukemia adversely impacted clinical outcome [20]. More recently, similar findings have shown that there is a clear association between a greater number of detectable subclonal populations and poorer clinical outcome in lower grade glioma, and cancers of the prostate, kidney, head and neck, breast, and lung [25–28]. The generation of genetic variation and subclonal diversity may be an indicator of the potential of a tumor to adapt under different selective pressures and has important implications on disease progression and drug resistance [29]. When studying cancers in patients who have relapsed, several studies have revealed that tumor cells in the relapsed-associated clone were often present as an undetected subclone in the primary tumor before the initiation of therapy, which suggests that mutations contributing to recurrence are selected for during treatment [30–33].
Current standard sequencing methods have low sensitivity and high false positive rates when detecting mutations below 10% frequency in the DNA extracted from the tumor sample [34]. Many subclones carrying driver mutations can remain rare and undetectable because their abundance falls below the detection limit of standard genome or exome sequencing techniques [16,21,34–36]. Both ultra-deep sequencing [16] and high density sampling [37] have shown that large numbers of rare subclones at <10% frequency are often present in a neoplasm and even normal tissue. However, quantitative assessment of this subclonal diversity across cancers, and theoretical expectations are needed to understand the dynamics of neoplastic progression and therapeutic resistance.
Here, we developed and implemented a computational model to gain insight into the dynamics of the subclonal evolution of cancer, and to assess the extent to which heterogeneous subclonal expansions occur. We simulated tumor growth via a birth-and-death branching process, where we keep track of all subclones that arise, die out, are maintained, and grow during the evolutionary process. We include both driver mutations that increase the fitness of a clone, and passenger mutations that confer therapeutic resistance.
Materials and Methods
Subclonal evolutionary model of cancer cell populations
Previous dynamical models developed to study tumor evolution [1,38–40] assume that each driver mutation affects the fitness of a tumor cell lineage equally (with the exception of refs. [39,41]). They also assume that the fitness effect of a driver mutation is independent of the other driver mutations carried by the cell. However, epistatic interactions are a central aspect of the dynamics of adaption of asexual populations [42], and should be relevant to asexual tumor populations as well [43]. Moreover, as it is a computationally prohibitive task, cancer evolution models have not studied the extent of heterogeneous subclonal expansions that can occur simultaneously during the neoplastic process, nor variation across multiple convergent tumors with a distribution of starting parameter values.
By employing an optimized algorithm on a Hadoop cluster (see Materials and Methods for details), we are able to keep track of all subclones (branches) that arise and die out, or are maintained and grow during the evolutionary process. Given the limited quantitative knowledge of parameter values across cancers, we test a range of values for μd and s̄. The ranges we explored are centered on values obtained from the literature, which have been estimated from experimental data. For example, the mean selection coefficient for glioblastoma multiforme (GBM) has been estimated by fitting a mathematical model to GBM sequencing data from 14 tumor samples [1]. The values selected for the driver mutation rate, μd, are: 1 × 10−8, 1 × 10−7, 1 × 10−6, and 1 × 10−5 mutations per cell division [1,8,40,44]. And the values chosen for the mean, s̄, of the exponential distribution of fitness effects are: 0.1, 0.01, and 0.005 [1,40,45]. Because of our optimized algorithm, we are able to simulate more than 100 tumors for each combination of parameter values, allowing us to consider variation across tumors of each combination of parameter values.
Software required
We used the following open-source platforms and programming languages for tumor simulation, monitoring and analysis: Apache Hadoop (HortonWorks 2.6.0); Apache Hive (1.2.1 spark HiveMetastoreConnection version 1.2.1, interactive hive-cli-0.14) – external data warehousing stacked on Hadoop, provides simulation monitoring, data summarization, query and analysis; Apache Scala (2.10.5) – functional programming language that utilizes the JVM (Java Virtual Machine) for platform independency, controls tumor simulation logic; Apache Spark (1.6.0 with a min of 1.4.0) – distributed computing framework originally developed at UC Berkeley AMPLab (https://amplab.cs.berkeley.edu), tracks tumor array memory across multiple machines; Bash (Sun AMD64 Linux 2.6.32–504.el6.x86_64) – for monitoring, analysis and data export to spreadsheets or other visualizations; Tableau (public 9.1 to 9.3) – for visualization of subclonal composition of simulated tumors; YARN (2.2.4.2–2) – Yet Another Resource Manager, manages Hadoop data and hardware resources. We used a hierarchical data structure to store common attributes for all cells within the same subclonal population.
Run environment
The computation and data intensive piece includes a 44 node HDP 2.3 cluster on Dell PowerEdge 720xd servers. Each of the 40 worker nodes has 128GB ram, 2x Intel E5-2640 6 core processors and 22TB of disk. The cluster backbone network consists of 10Ge HA top of rack switching combined with Intel x520 10Ge NICs in each server. Although the tumor simulator can run parallel jobs utilizing multiple resources, the demands upon the hadoop NameNode (worker, memory, disk resource managment) are quite exhaustive; therefore, it is suggested to run sequential jobs on a single node for as many images as needed to emulate parallelization.
Statistical analysis
We created scripts on RStudio (Version 0.99.891) to analyze the data sets, perform statistical analysis, and generate most of the figures (with the exception of the figures displaying subclonal composition of simulated tumors).
Code accessibility
The computer code for simulations, tumorsim.scala is available at: https://github.com/WilsonSayresLab/TumorHeterogeneity. For details on the steps necessary to run tumorsim application see Supplementary Material and the readme section on GitHub. All the R scripts for analysis are also available at https://github.com/WilsonSayresLab/TumorHeterogeneity.
Results
Drift dominates early neoplastic dynamics
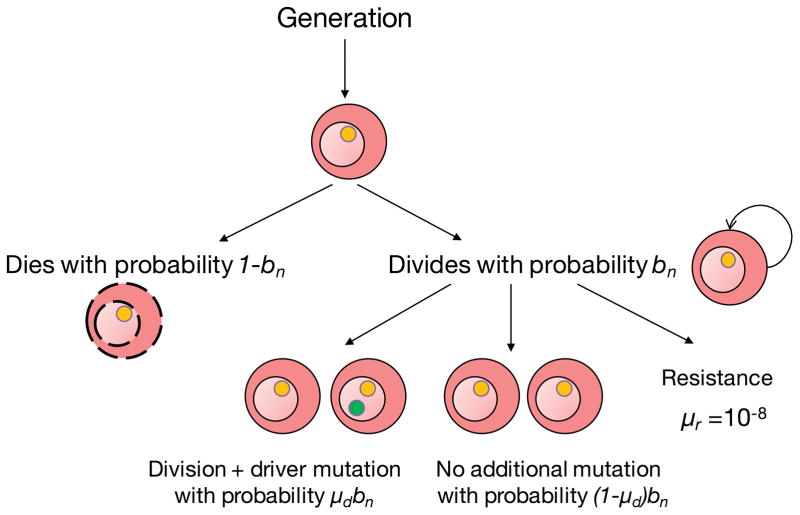
A necessary step in neoplastic initiation is that the first mutated cell lineage survives stochastic drift to result in a clone growing at the expense of its normal neighbors. The growth of the first clone is important in increasing the number of cells in which a second driver mutation could occur, and subsequently, another clone could emerge from the cell with the second driver, and so on, until a clinically detectable tumor is formed (Fig. 1). To quantify the effect of stochastic drift in neoplastic initiation we ran our simulations until we generated at least 100 clinically detectable simulated tumors (defined as a tumor cell population reaching ≈ 109 cells) for each combination of the chosen parameter values, for a total of 88,265 simulations of the process (Table 1). Overall, we observed that out of the total number of realizations executed, only 1,432 became clinically detectable tumors, despite each simulation being initiated with a driver mutation. Thus, on average, ≈ 98% of all the initiating mutated cell lineages carrying a driver mutation spontaneously regress (Table 1), which is in line with theoretical expectations [46]. This result highlights the importance of genetic drift affecting neoplastic initiation, even after a first driver mutation has occurred.
Figure 1.
Branching evolutionary process of cancer. Schematic representation of the process developed to simulate the subclonal evolution of cancer is presented below. For details of the process and assumptions, see main Quick Guide to Equations and Assumptions.
Table 1.
Number of simulations performed for each combination of parameter values (s̄, μd). The mean time from initiation to clinical detection of a simulated tumor is shown. The generation time assigned in the simulations is 4 days.
| s̄ | μd | Number of realizations | Number of simulations that reached 109 cells | Percentage of simulations that reached 109 cells | Mean number of generations to detection | Mean time to detection (years) |
|---|---|---|---|---|---|---|
| 0.1 | 1 × 10−8 | 10155 | 162 | 1.6% | 1596.66 | 17.50 |
| 0.1 | 1 × 10−7 | 1948 | 112 | 5.7% | 475.08 | 5.21 |
| 0.1 | 1 × 10−6 | 748 | 134 | 17.9% | 158.54 | 1.74 |
| 0.1 | 1 × 10−5 | 748 | 111 | 14.8% | 147.50 | 1.62 |
| 0.01 | 1 × 10−8 | 6867 | 125 | 1.8% | 1807.63 | 19.80 |
| 0.01 | 1 × 10−7 | 6866 | 113 | 1.6% | 1406.75 | 15.41 |
| 0.01 | 1 × 10−6 | 6866 | 120 | 1.7% | 1263.80 | 13.85 |
| 0.01 | 1 × 10−5 | 6865 | 115 | 1.7% | 1018.40 | 11.16 |
| 0.005 | 1 × 10−8 | 11951 | 102 | 0.9% | 2552.70 | 27.97 |
| 0.005 | 1 × 10−7 | 11751 | 112 | 1.0% | 2546.85 | 27.91 |
| 0.005 | 1 × 10−6 | 11750 | 126 | 1.1% | 2046.78 | 22.43 |
| 0.005 | 1 × 10−5 | 11750 | 100 | 0.9% | 1668.07 | 18.28 |
| 88265 | 1432 | 1.6% | ||||
To test how this model works with parameter values from a known cancer type, we use estimates from colorectal cancer. It has been experimentally estimated that colorectal cancer cells divide every 4 days [1,47]. Assuming this cell division time in the simulations, we find that the average expected time from onset to clinical detection of the simulated tumors ranges from 1.64 years to 27.97 years, depending on the values for s̄ and μd (Table 1). Additionally, using the parameter values s̄ = 0.005 and μd = 1 × 10−5 per cell division, which have been estimated for colorectal cancer [1], our model predicts that it would take an average of 18.28 years for a colorectal tumor to grow to a detectable size after the first driver mutation appears (Table 1). This estimate is concordant with previous estimates of tumor development in colorectal cancer [1,47].
Relationship between selection and mutation on tumor growth
By having generated a total of 1,432 clinically detectable simulated tumors under a wide range of parameter values, we can determine the general contribution of each of the parameters to initiation and neoplastic progression. We find, consistent with a previous report [40], that selection has a larger effect on neoplastic initiation than the driver mutation rate (Table 1). Moreover, the average expected time from initiation to detection of a tumor increases with decreasing the average fitness effect of driver mutations and with decreasing the driver mutation rate (Table 1). However, we also find that there is a non-linear relationship in the effect of selection and mutation. When the mean selective coefficient is low, mutation rate does not have a large effect on the mean time to detection, but as the mean selective coefficient increases, mutation rate has an increasingly large effect on the mean time to detection (Table 1).
Extent of intratumor subclonal variation at detection
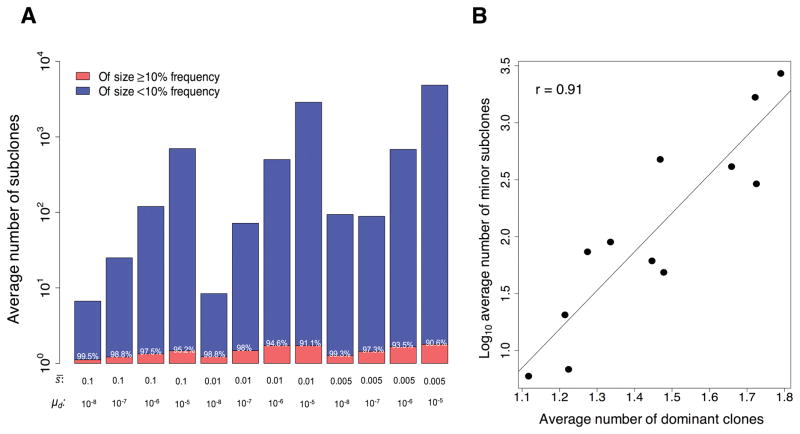
To gain insight into the extent of subclonal populations within a tumor at the time of detection and to determine how the different evolutionary parameters impact the subclonal composition, we analyzed all the 1,432 detectable tumors generated by our model. In most tumors, we find that the number of dominant clones, defined here as a subclone present at ≥10% frequency in a tumor, ranges from 1 to 4 (Table S1); however, the average number of dominant clones in a tumor tends to increase with decreasing the average fitness effect of driver mutations and with increasing the driver mutation rate (Fig. 2A and Table S1). Across all the 1,432 detectable simulated tumors, the average number of dominant clonal populations is 1.46 (Table S1). Importantly, we find that even though only a few dominant clones compose the majority of the cancer cell population in a simulated tumor (range 90.6% – 99.5%; Fig. 2A), the number of minor subclones present at <10% frequency is substantial (Fig. 2A and Table S1); this number increases with decreasing the average fitness effect of drivers and increases with the driver mutation rate (range 0 – 6,734; Fig. 2A and Table S1).
Figure 2.
Intratumor subclonal variation. (A) Bar plots of the log10 average number of dominant clones present at ≥10% frequency in the simulated tumors (red) and the log10 average number of minor subclones that are present at <10% frequency (blue). The value shown over each red bar represents the average percentage tumor cell population composed by the dominant clones. Note that the extent of intratumor subclonal variation is greatly affected by both the mean selective coefficient, s̄, of the exponential distribution of fitness effects associated with the driver mutations and the driver mutation rate, μd. (B) Correlation between the average number of dominant clones and the log10 average number of minor subclones in the simulated tumors for each combination of parameter values used as in (A).
Given the parameter values s̄ = 0.005 and μd = 1 × 10−5, which have been estimated for glioblastoma multiforme and colorectal cancer [1], the model predicts that, on average, 1.8 (range 1 –4) dominant clones and 2,705 (range 1,190 – 6,734) minor subclonal populations carrying driver mutations compose a tumor (Fig. 2A and Table S1). We also find that there is a strong, statistically significant correlation between the average number of dominant clones and the average number of minor subclones across all simulated tumors (Pearson r = 0.91, P < 0.001; Fig. 2B). This result suggests that the number of detectable clonal populations by standard sequencing techniques may serve as a crude proxy for the extent of undetectable minor subclones in a tumor.
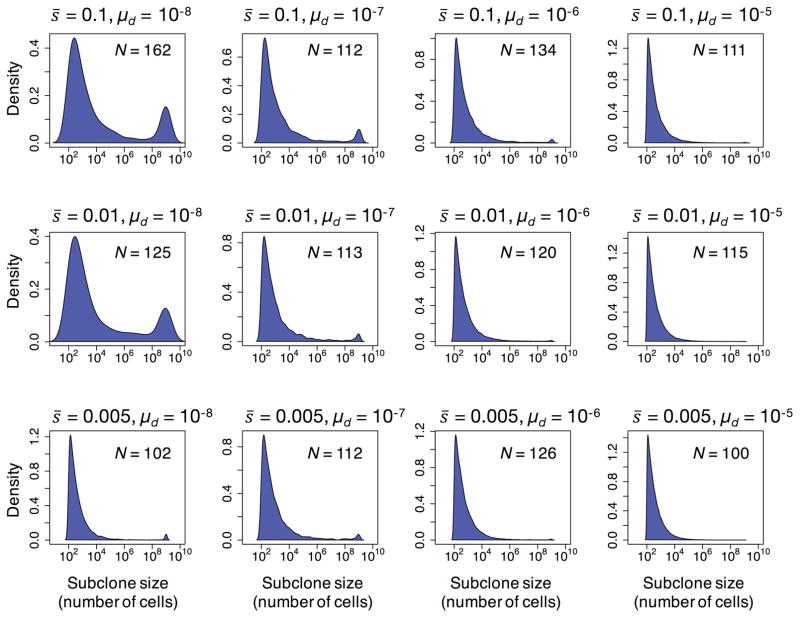
For each combination of parameter values, we computed the probability density function for the subclone sizes present in the simulated tumors (Fig. 3). Overall, we find that the distribution of subclone sizes harboring driver mutations has a heavy right tail, with only a few clones present at ≥10% frequency in the tumor, and with most subclones present at frequencies as low as 10−9 (Fig. 3).
Figure 3.
Probability density function for the log10 sizes of subclones carrying driver mutations present in the clinically detectable simulated tumors for each combination of parameter values; N is the number of clinically detectable tumors generated by the model. The distribution has a heavy right tail with only a few dominant clones present at ≥10% frequency, composing most of the tumor cell population (Fig. 2A), and with the majority of subclones present at frequencies below the detection limit of standard sequencing techniques.
Differential fitness between dominant and minor subclones
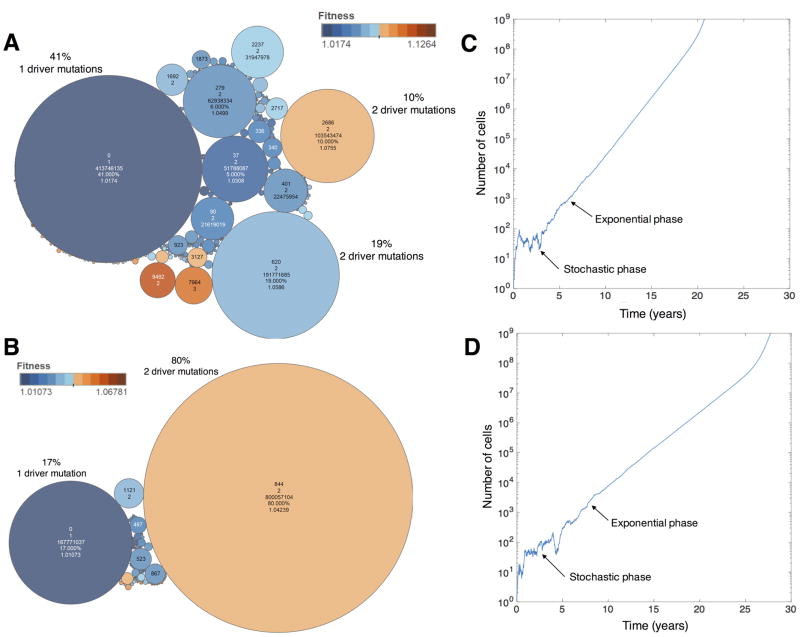
A key aspect of our computational model is that it simulates and tracks subclonal heterogeneity. For illustrative purposes, we show the subclonal composition and their corresponding fitness values in two clinically detectable simulated tumors using the parameter values s̄ = 0.01 and μd = 1 × 10−5 (Fig. 4A); and s̄ = 0.005 and μd = 1 × 10−5 (Fig. 4B). The corresponding population dynamics of both simulated tumors are shown as well (Fig 4 C and D, respectively). The simulated tumor presented in Fig. 4A has 3 dominant clones present at 41%, 19%, and 10% frequency in the tumor, carrying 1–2 driver mutations, when it reached a clinically detectable size. On the other hand, the simulated tumor shown in Fig. 4B has only two dominant clones preset at 80% and 17% frequency, harboring 2 and 1 driver mutations, respectively. Additionally, there is substantial intra- and inter- subclonal variation in both cases (Fig. 4 A and B). On average, the relative fitness of some minor subclones is greater than the fitness of the dominant clonal populations (Fig. 4 A and B and Table S2). These results show that, even though some subclones have acquired additional driver mutations that provide a fitness advantage over the dominant clones, they have not yet swept through the tumor population when the tumor is detected. Consequently, a substantial number of minor – and often fitter – subclones harboring driver mutations are present in the tumor at very low frequency.
Figure 4.
Variability in subclonal composition, differential fitness among subclones, and population dynamics of progression of two simulated tumors. (A and C) Parameter values used are: s̄ = 0.01 and μd =1 × 10−5. (B and D) Parameter values used are: s̄ = 0.005 and μd = 1 × 10−5. Each circle represents a subclone carrying a certain number of driver mutations. The size of a circle is proportionate to the number of cells composing the subclone. The color of a circle corresponds to the fitness of the subclone. Note that the relative fitness of some minor subclones is greater than the fitness of the dominant clones. And, as would be expected, there is substantial intra- and inter- subclonal variation between the two simulated tumors. The numbers within each circle indicate the identifier of the subclone, the number of driver mutations, the percentage of cells relative to the total number of cells in the tumor, and its fitness value. The plots depicting the population dynamics of cancer progression use a log10 scale for the y-axis. In both cases, the generation time used is T = 4 days.
Resistant subclones carrying driver mutations are present at low frequency when the tumor is detected
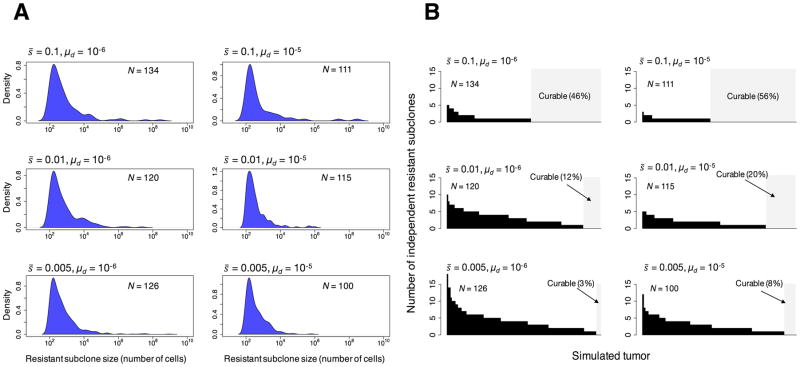
Populations can adapt to novel environments in two different ways – selection can act on pre-existing genetic variants or on de novo mutations [48]. Adaptation from standing genetic variation is faster than adaptation from novel mutations, not only because beneficial mutations are immediately available in the new environment, but also because they may start at higher frequencies [48]. We tested whether subclones carrying driver mutations could also carry at least one resistance mutation in the clinically detectable simulated tumors. To test this, we assume that multiple different mutations can independently cause resistance [49], and assume a resistance mutation, rate, μr, of 1 × 10−8 [49] during cell division (Fig. 1). Additionally, we assume that the resistance mutation does not affect fitness in the absence of therapy. Under these assumptions and parameter values, we find that the majority of resistant subclones are often present at very low frequency, below the detection limit of standard DNA sequencing techniques (Fig. 5A). We then calculated the number of independent resistant subclones within each clinically detectable simulated tumor (Fig. 5B). We find that the number of independent resistant subclones ranges from 0 to 18 at the time the tumor is detected (Fig. 5B). Overall, we find that the number of independent resistant subclones in a simulated tumor increases with decreasing the average fitness effect of the drivers (Fig. 5B). In addition, Fig. 5B shows that the probability that a tumor is curable, i.e., that there are no resistant clones present when the tumor is detected, is higher for fast-growing tumors (larger fitness effects of driver mutations) and for high driver gene mutation rates. We finally “treated” all clinically detectable simulated tumors with a hypothetical targeted drug, killing all non-resistant cells, and calculated the average time from the moment when the drug is applied to the time at disease recurrence (cancer cell population rebounds to ≈ 109 cells). Our analysis suggests that there is on average an eight-fold decrease in the time from the start of treatment to the time at recurrence relative to the average time from initiation to clinical detection of the simulated tumors (Table 1 and S3).
Figure 5.
Resistant subclones in clinically detectable simulated tumors. (A) Probability density functions for the log10 sizes of therapeutically resistant subclones within in the primary simulated tumors. (B) Number of independent resistant subclones in each simulated primary tumor. Each bar represents a tumor. The area in light gray represents those simulated tumors with no resistant subclones, which should be curable. For each set of parameter values used, N is the number of tumors generated by the model. It is assumed that a resistant mutation occurs at a rate of 1 × 10−8 per cell division. Resistant mutations are assumed not to affect fitness.
Discussion
In this study, we developed and implemented a stochastic evolutionary model to study the subclonal dynamics of cancer progression. First, we show that despite the selective advantage of the driver mutation, drift substantially affects neoplastic initiation (Table 1). This result is expected from population genetics theory, as selection is less efficient in small populations [46]. That said, we find that the mean selective coefficient has a much larger effect on neoplastic initiation than the driver mutation rate (Table 1). Based on this, we hypothesize that the fitness effects conferred by driver genetic alterations in certain sporadic childhood cancers, which arise within a few years, should be greater than those associated with drivers in sporadic adult cancers, and those rapidly growing tumors should be more likely to be curable than adult cancers.
A statistical strength of this study is that we simulated 88,265 tumor initiation events for a range of biologically realistic parameters, resulting in more than 1,400 clinically detectable tumors, from which we could analyze and gain important biological insights.
Across all the combinations of the chosen evolutionary parameter values, our analysis show that the distribution of sizes of subclones carrying driver mutations has a heavy right tail at the time of tumor detection. We find that the vast majority of subclones are predicted to be present at <10% frequency in the tumor, and only 1–4 dominant clones are present at ≥10% frequency, composing the majority of the tumor cell population (Fig. 2A). Additionally, we find that there is a strong, statistically significant correlation between the average number of numerically dominant clones and the average number of minor subclones across all simulated tumors (Fig. 2B).
The distribution of subclones sizes inferred by our model is qualitatively similar to that found in a recent empirical study where the authors used ultra-deep sequencing technology to detect mutations in specific known cancer genes in biopsies of sun-exposed eyelids from different individuals [37]. The authors found that aged sun-exposed skin has already thousands of subclones with driver mutations subjected to selection, with some clones as large as several square millimeters in surface area [37]. Moreover, the predicted range by the model on the number of dominant clonal populations is largely concordant with a recent pan-cancer analysis of intratumor heterogeneity [26]. In this study, the authors analyzed whole exome sequencing mutational data from over 3,300 tumors from The Cancer Genome Atlas (TCGA) to infer that 92% of all the tumors had between 1 and 4 clonal populations at detectable levels by standard sequencing methods [26].
The assumptions that each tumor simulation begins with a single cell, that at each time step a cell can die, divide and not incur a mutation, or divide and incur a mutation, and that we sample selection coefficients from an exponential distribution are consistent with current expectations about cell division, tumorigenesis, and selective coefficients from population genetics. Our assumption that each driver mutation increases fitness means that we are only modeling positive selection, and not negative selection. Tumors, while affected by negative selection, have been shown to be much more affected by positive selection [50], and so we expect this assumption is valid for our initial model. One of the major extensions we provide in this computational modeling framework is that we track subclonal heterogeneity, where each subclone evolves as a cellular lineage (a branch) with its own fitness, which is dependent on the fitness effects conferred by the driver mutations present in the cell and their epistatic interactions - we think this is very likely to match tumorigenesis. We also assume that the fitness advantages confered by driver mutations in our model scale by how near the current fitness background is to a hypothesized upper fitness boundary. This specific form of fitness advantage generates some qualitative features commonly observed in adaptive evolution ([3,51,52].
While some tumors may have a substantial number of subclones (Figure 2), these should not necessarily be interpreted as these subclones having unique mutations, but rather unique combinations of mutations. In our simulated tumors each of these subclones carry only a handful of drivers. The number of passengers in a given cell lineage should be much larger, which should theoretically be proportional to the number of cell divisions. In some combinations of parameter values, a simulated tumor can have many subclones. Of note, as mentioned above, each subclone has only a handful of driver mutations. And each of these subclones should be viewed as a unique combination of mutations, not that there are thousands of affected genes. We do not keep track of unique mutations, and in fact in this model, even unique mutations would not make sense, because we have a model of diminishing returns epistasis, where the order of mutations is important.
Understanding the expected levels of subclonal variation is important for treating primary cancers and predicting recurrence. By employing ultra-deep sequencing technology to study intratumor subclonal diversity in patients with CLL, it has been shown the presence of rare subclones at frequencies down to the limit of detection for depth of this specific sequencing method, 10−4 [16]. Our model predicts that minor subclones carrying driver mutations can be present in a tumor at lower frequencies, 10−7 (Fig. 3). Importantly, we find that these commonly undetectable minor subclones can harbor therapeutically resistant mutations (Fig. 5A). These results are in line with multiple previous reports showing that tumor cells corresponding to the relapsed clone were often present as a rare subclone in the diagnostic tumor before the initiation of treatment [30,31,33]. Additionally, we find that the number of independent treatment-resistant subclones ranges from 0 to 18 at the time the tumor is detected (Fig. 5B).
Given that cancer is the result of a complex evolutionary process, our approach has some limitations. First, we have not taken spatial structure into consideration in our computational model, which may restrict the expansion of certain subclonal populations during the neoplastic process. Thus, our modeling framework may be more suitable for cancers that develop in the absence of spatial constraints. Second, we assumed that resistant mutations are neutral. While there is often a fitness cost of resistance [53], in the absence of drug, it is well known in population genetics that deleterious mutations can grow to substantial frequencies in expanding populations [12]. Third, the host immune system has not been taken into account, which has been demonstrated to have an important role for both cancer suppression and promotion [54]. Fourth, future studies should extend this work and consider non-cell autonomous interactions, as well as “public good” factors, which are likely to be a strong influence on subclonal dynamics [55]. Despite these caveats, our model captures some essential features of the dynamics of subclonal evolution of cancer progression. The subclonal dynamics predicted by our model are consistent with the “Big Bang” model [56], where clonal driver mutations and most detectable subclonal drivers occurred relatively early during tumor growth. This result is in contrast with the traditional clonal selection model, where sequential stepwise accumulation of drivers leads to fitter clones that sweep through the population [1,13].
In conclusion, our modeling provides a theoretical framework for tumor growth and spontaneous regression, predicting that a substantial number of subclonal populations carrying driver mutations will be rare and undetectable within a tumor because their abundance has not yet grown above the detection limit of standard genome sequencing methods. Additionally, these minor and often undetectable subclones can harbor treatment-resistant mutations, which present a major challenge for personalized medicine and clinical management. These results suggest that driver mutations that have been identified in individual tumors through standard genome sequencing [57] are likely to constitute only the “tip of the iceberg”, with many mutations never rising above very low frequencies, but that can expand post-treatment and are critical for the evolutionary dynamics of cancer progression and relapse. The strong correlation between macrodiversity (the diversity of clones present at ≥10% frequency) [34] and microdiversity (the diversity of clones present at <10% frequency), supports efforts to predict the probability that a resistant minor subclone is present based on the measures of macrodiversity. Altogether, our findings help explain why tumors with greater numbers of detectable clonal populations are associated with poorer prognosis across multiple cancer types [20,25,26].
Supplementary Material
Major Findings.
Our modeling analysis suggests that most tumors harbor significant heterogenetiy at levels lower than the sensitivity of current assays. Moreover we predict that slow-growing tumors harbor numerous resistant subclones at sub-detectable levels, while fast-growing tumors are expected to be more representative of the clonality typically modeled for tumors.
Quick Guide to Equations and Assumptions.
Our computational model is a non-spatial stochastic model based on a branching evolutionary process. The major assumptions of our modeling framework are stated as follows:
Assumption 1: Similar to the framework presented in [1], each simulation begins with a single cell carrying a single initiating driver mutation (the potential founder of a primary tumor).
Assumption 2: At each time step, a cell may either die or divide. If it divides, it can acquire an additional driver mutation in one of the daughter cells at rate μd.
Assumption 3: For each driver mutation, we randomly sample a selection coefficient from an exponential distribution of mean s̄ [2], and update fitness f.
-
Assumption 4: Each subsequent driver mutation increases the probability of cell division, defined as:
The exponential distribution of mean s̄ is truncated for only si < 1. The probability of cell death depends on the fitness of the cell, and is defined as 1–b.
Assumption 5: The parameter wwt represents the fitness background of the wild-type cell in which the first driver occurs, and without loss of generality, it is assumed to be 1.
Assumption 6: Driver mutations have intrinsic effects. However, rather than assuming fitness differences are background independent (like the additive model) or that differences scale by background fitness (like the multiplicative model), fitness advantages in our model scale by how near the current fitness background is to a hypothesized upper fitness boundary. Without loss of generality, we assume that the maximum possible fitness gain through adaptation is d = 1 [3]. The fitness change in a cell produced by a driver is thus dependent of the other driver mutations and the temporal order at which they occur in the cell.
Assumption 7: The parameters s1, s2,…, sn characterize the fitness effects associated with each of n driver mutations that a cell lineage carries.
Assumption 8: The modeling process is terminated when a clinically detectable simulated tumor is generated (defined as a tumor cell population reaching ≈ 109 cells).
Assumption 9: A subclone is defined as a subpopulation of cells that descended from another clone but then diverged by accumulating another driver mutation.
Acknowledgments
We thank Jay Taylor, Sergey Kryazhimskiy, and Andriy Marusyk for their helpful comments and insightful discussions. We also thank ASU Research Computing for providing computational resources, and Lisa Faiss who provided technical expertise that assisted the research. This research was partially supported through startup funds from the ASU School of Life Sciences and the Biodesign Institute to M.A. Wilson Sayres. Grant-based support includes the Flinn Foundation to M.A. Wilson Sayres and C.C. Maley. This work was also supported in part by the Breast Cancer Research Foundation to K.S. Anderson, NIH grants R21 CA196460 and 5R21 CA196460 to K.S. Anderson, NIH grants P01 CA91955, R01 CA149566, R01 CA170595, R01 CA185138 and R01 CA140657 as well as CDMRP Breast Cancer Research Program Award BC132057 C.C. Maley. The findings, opinions and recommendations expressed here are those of the authors and not necessarily those of the universities where the research was performed or the National Institutes of Health.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A. 2010;107:18545–50. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orr HA. The distribution of fitness effects among beneficial mutations. Genetics. 2003;163:1519–1526. doi: 10.1016/j.jtbi.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagel AC, Joyce P, Wichman HA, Miller CR. Stickbreaking: A novel fitness landscape model that harbors epistasis and is consistent with commonly observed patterns of adaptive evolution. Genetics. 2012;190:655–667. doi: 10.1534/genetics.111.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 6.Frank SA. Dynamics of cancer: incidence, inheritance, and evolution. Princeton University Press; 2007. [PubMed] [Google Scholar]
- 7.Martinocorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 8.Lynch M. Mutation and Human Exceptionalism: Our Future Genetic Load. Genetics. 2016;202:869–875. doi: 10.1534/genetics.115.180471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Cell. Vol. 144. Elsevier Inc; 2011. Hallmarks of cancer: The next generation; pp. 646–674. [DOI] [PubMed] [Google Scholar]
- 10.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose Sa, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer Genome Landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFarland CD, Korolev KS, Kryukov GV, Sunyaev SR, Mirny LA. Impact of deleterious passenger mutations on cancer progression. Proc Natl Acad Sci U S A. 2013;110:2910–5. doi: 10.1073/pnas.1213968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fearon EF, Vogelstein B. A Genetic Model for Colorectal Tumorigenesis. 1989;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 14.Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–14. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1406617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burrell Ra, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 19.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Cell. Vol. 152. Elsevier Inc; 2013. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia; pp. 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt MW, Loeb LA, Salk JJ. Nat Rev Clin Oncol. Nature Publishing Group; 2015. The influence of subclonal resistance mutations on targeted cancer therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–73. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 23.Chowell D, Boddy AM, Mallo D, Tollis M, Maley CC. When (distant) relatives stay too long: implications for cancer medicine. Genome Biol Genome Biology. 2016;17:1–4. doi: 10.1186/s13059-016-0906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bozic I, Nowak MA. Timing and heterogeneity of mutations associated with drug resistance in metastatic cancers. Proc Natl Acad Sci U S A. 2014;111:15964–8. doi: 10.1073/pnas.1412075111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–9. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris LGT, Riaz N, Desrichard A, Şenbabaoğlu Y. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. 2016:7. doi: 10.18632/oncotarget.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2015;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, Sedlak JC, Srinivas R, Creixell P, Pritchard JR, Tidor B, et al. Exploiting Temporal Collateral Sensitivity in Tumor Clonal Evolution. Cell. 2016;165:234–246. doi: 10.1016/j.cell.2016.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrissy AS, Garzia L, Shih DJH, Zuyderduyn S, Huang X, Skowron P, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 2016 doi: 10.1038/nature16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diaz La, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. Nature. Nature Publishing Group; 2012. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers; pp. 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roche-Lestienne C, Lai JL, Darre S, Facon T, Preudhomme C. A mutation conferring resistance to imatinib at the time of diagnosis of chronic myelogenous leukemia. NEnglJ Med. 2003;348:2265–2266. doi: 10.1056/NEJMc035089. [DOI] [PubMed] [Google Scholar]
- 33.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Nature. Vol. 481. Nature Publishing Group; 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing; pp. 506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barber LJ, Davies MN, Gerlinger M. Curr Opin Genet Dev. Vol. 30. Elsevier Ltd; 2015. Dissecting cancer evolution at the macro-heterogeneity and micro-heterogeneity scale; pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tirosh I, Izar B, Prakadan SM, Ii MHW, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerstung M, Beisel C, Rechsteiner M, Wild P, Schraml P, Moch H, et al. Nat Commun. Vol. 3. Nature Publishing Group; 2012. Reliable detection of subclonal single-nucleotide variants in tumour cell populations; p. 811. [DOI] [PubMed] [Google Scholar]
- 37.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waclaw B, Bozic I, Pittman ME, Hruban RH, Vogelstein B, Nowak Ma. Spatial model predicts dispersal and cell turnover cause reduced intra-tumor heterogeneity. Nature. 2015;525:261–267. doi: 10.1101/016824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durrett R, Foo J, Leder K, Mayberry J, Michor F. Intratumor heterogeneity in evolutionary models of tumor progression. Genetics. 2011;188:461–477. doi: 10.1534/genetics.110.125724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, Velculescu VE, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:2239–2246. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostadinov R, Maley CC, Kuhner MK. Bulk Genotyping of Biopsies Can Create Spurious Evidence for Hetereogeneity in Mutation Content. PLoS Comput Biol. 2016;12:e1004413. doi: 10.1371/journal.pcbi.1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, et al. Nature. Vol. 500. Nature Publishing Group; 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations; pp. 571–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprouffske K, Merlo LMF, Gerrish PJ, Maley CC, Sniegowski PD. Curr Biol. Vol. 22. Elsevier Ltd; 2012. Cancer in light of experimental evolution; pp. R762–R771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shendure J, Akey JM. The origins, determinants, and consequences of human mutations. Orig Determinants Consequences Hum Mutat. 2015;349:1478–1483. doi: 10.1126/science.aaa9119. [DOI] [PubMed] [Google Scholar]
- 45.Vermeulen L, Morrissey E, Heijden M, van der Nicholson AM, Sottoriva A, Buczacki S, et al. Defining Stem Cell Dynamics in Models of Intestinal Tumor Initiation. Science. 2013;342:995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- 46.Charlesworth B. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 47.Jones S, Chen W-D, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–8. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Komarova NL, Burger JA, Wodarz D. Evolution of ibrutinib resistance in chronic lymphocytic leukemia (CLL) Proc Natl Acad Sci U S A. 2014;111:13906–11. doi: 10.1073/pnas.1409362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostrow SL, Barshir R, DeGregori J, Yeger-Lotem E, Hershberg R. Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes. PLOS Genet. 2014;10:e1004239. doi: 10.1371/journal.pgen.1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kryazhimskiy S, Rice DP, Jerison E, Desai MM. Global Epistasis Makes Adaptation Predictable Despite Sequence-Level Stochasticity. 2014:344. doi: 10.1101/001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiser MJ, Ribeck N, Lenski RE, Littell JS, Muller CJ, Dunne Ka, et al. Long-Term Dynamics of Adaptation in Asexual Populations. 2013;342:1364–1367. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]
- 53.Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69:4894–4903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schreiber RD, Old LJ, Smyth MJ. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 55.Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Nature. Vol. 514. Nature Publishing Group; 2015. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity; pp. 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, et al. Nat Genet. Vol. 47. Nature Publishing Group; 2015. A Big Bang model of human colorectal tumor growth; pp. 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.