Abstract
Introduction:
Plasma concentrations of lipids (i.e., total cholesterol, high-density cholesterol, low-density cholesterol, and triglycerides) are amenable to therapeutic intervention and remain important factors for assessing risk of cardiovascular diseases. Some of the observed variability in serum lipid concentrations has been associated with genetic and epigenetic variants among cohorts with European ancestry (EA). Serum lipid levels have also been associated with genetic variants in multiethnic populations.
Methods:
The purpose of this study was to determine whether single-nucleotide polymorphisms (SNPs) and DNA methylation (DNAm) differences contribute to lipid variation among African Americans ([AAs], N = 739) in the Genetic Epidemiology Network of Arteriopathy (GENOA) study.
Results:
Previous meta-analyses identified 161 SNPs that are associated with lipid traits in populations of EA. We evaluated these SNPs and 66 DNAm sites within the genes containing the SNPs in the GENOA cohort using linear mixed-effects modeling. We did not identify any significant associations of SNPs or DNAm with serum lipid levels. These results suggest that the SNPs identified as being significant for lipid levels through the EA genome-wide association studies may not be significant across AA populations.
Conclusions:
Reductions in morbidity and mortality due to variation in lipids among AAs may be achieved through a better understanding of the genetic and epigenetic factors associated with serum lipid levels for early and appropriate screening. Further large-scale studies specifically within AA and other non-EA populations are warranted.
Keywords: African Americans, cholesterol, DNA methylation, genetic research, GENOA, lipids
In the United States, African Americans (AAs), or those of African ancestry, face significant disparities for multiple chronic health conditions. When compared to those of European ancestry (EA), AAs fare worse over many indices associated with cardiovascular disease including (1) highest incidence and prevalence of hypertension, obesity, and diabetes; (2) highest death rates from heart disease and stroke; and (3) shorter life expectancy (Meyer, Yoon, & Kaufmann, 2013). Individuals’ health outcomes and life expectancy are strongly influenced by the characteristics of their environment (Yoon, Bastian, Anderson, Collins, & Jaffe, 2014). Accordingly, health disparities among AAs have been associated with genomic underpinnings (Taylor, Sun, Hunt, & Kardia, 2010), social inequalities (Taylor et al., 2012), disproportionate burdens of pollution (Taylor, Wright, & Housman, 2016), and unequal access to quality health care (Hynes & Lopez, 2012).
Serum lipid levels (i.e., total cholesterol [cholesterol], low-density lipoprotein [LDL] cholesterol, high-density lipoprotein [HDL] cholesterol, and triglycerides) differ between AAs and their counterparts of EA (Wright, Housman, & Taylor, 2016). AA individuals tend to have lower levels of triglycerides and cholesterol and higher levels of HDL than EA individuals (de Ferranti et al., 2016; Mozaffarian et al., 2015). Variations in lipid levels between these groups may be attributed to genetic or epigenetic differences. Although a number of genome-wide association studies (GWAS) have identified independent effects of risk alleles for hypertension and other chronic diseases among AA populations, very few studies have used multiple omic methods together, such as single-nucleotide polymorphisms (SNPs) and DNA methylation (DNAm), to explore the contribution of both genetic and environmentally mediated (via DNAm) influences on phenotypic expression of disease (Taylor, Wright, Crusto, & Sun, 2016).
In large-scale GWASs evaluating genetic variations that contribute to the heritability of serum lipid levels in populations of EA (N = 100,184; Teslovich et al., 2010) and mixed ancestry (N = 188,577; Willer et al., 2013), researchers identified 161 SNPs that are significantly associated with serum lipid levels. In their replication GWAS study, Willer et al. also identified significant associations between some of these 161 SNPs and various clinical outcomes including body mass index (BMI), blood pressure, and Type 2 diabetes. Teslovich et al., however, in one of the original studies that assessed replicability among other populations, found that a lower proportion of these SNPs were associated with abnormal lipids in groups of AA compared to groups of South and East Asian ancestry groups. To date, no follow-up on the modest SNP replication from the most recent GWAS studies of serum lipid levels in AA cohorts has been published. These data are alarming as diseases associated with altered lipids (e.g., stroke, heart disease, obesity) disproportionately affect AAs, and lipids in AAs are less likely to be responsive to therapy than they are in their counterparts of EA (Goff, 2006). Given that plasma concentrations of lipids are amenable to therapeutic intervention and remain among the most important factors for assessing risk of cardiovascular diseases, it is important to elucidate molecular mechanisms that may contribute to variation among AA cohorts (Teslovich et al., 2010; Willer et al., 2013).
Epigenetic differences may also be important risk factors for lipid variations that contribute to multiple disease processes. Cell types have unique epigenetic signatures that add further programming information with strong consequences for cellular activity with downstream effects such as which proteins will be produced (Jenuwein & Allis, 2001). Researchers conducting epigenome-wide association studies in AA cohorts have identified DNAm differences associated with age (Smith et al., 2014), sex (Sun et al., 2010), BMI (Demerath et al., 2015), cigarette smoking (Joehanes et al., 2016; Klebaner et al., 2016; Sun, Smith, et al., 2013; Taylor, Schwander, et al., 2016), and inflammatory markers (Bomotti et al., 2013; Ligthart et al., 2016; Sun, Lazarus, et al., 2013). However, few studies have evaluated the relationships between DNAm and serum lipid levels among AAs. Of the studies that have been published, none were designed to evaluate DNAm related to lipids specifically within an AA cohort.
We hypothesized that interactions between genetic and epigenetic factors contribute to serum lipid variation among AAs. In this study, we (1) examined the influence of 161 genetic loci previously identified as being associated with lipids among those of EA in an AA cohort from the Genetic Epidemiology Network of Arteriopathy (GENOA) study, (2) examined the influence of DNAm at the genes associated with those 161 loci on lipids, and (3) integrated findings from (1) and (2) into a multivariable model of the joint effects of lipid SNP variants, DNAm, and clinical outcomes associated with variation in serum lipid levels including evaluation of interactions.
Material and Method
Sample
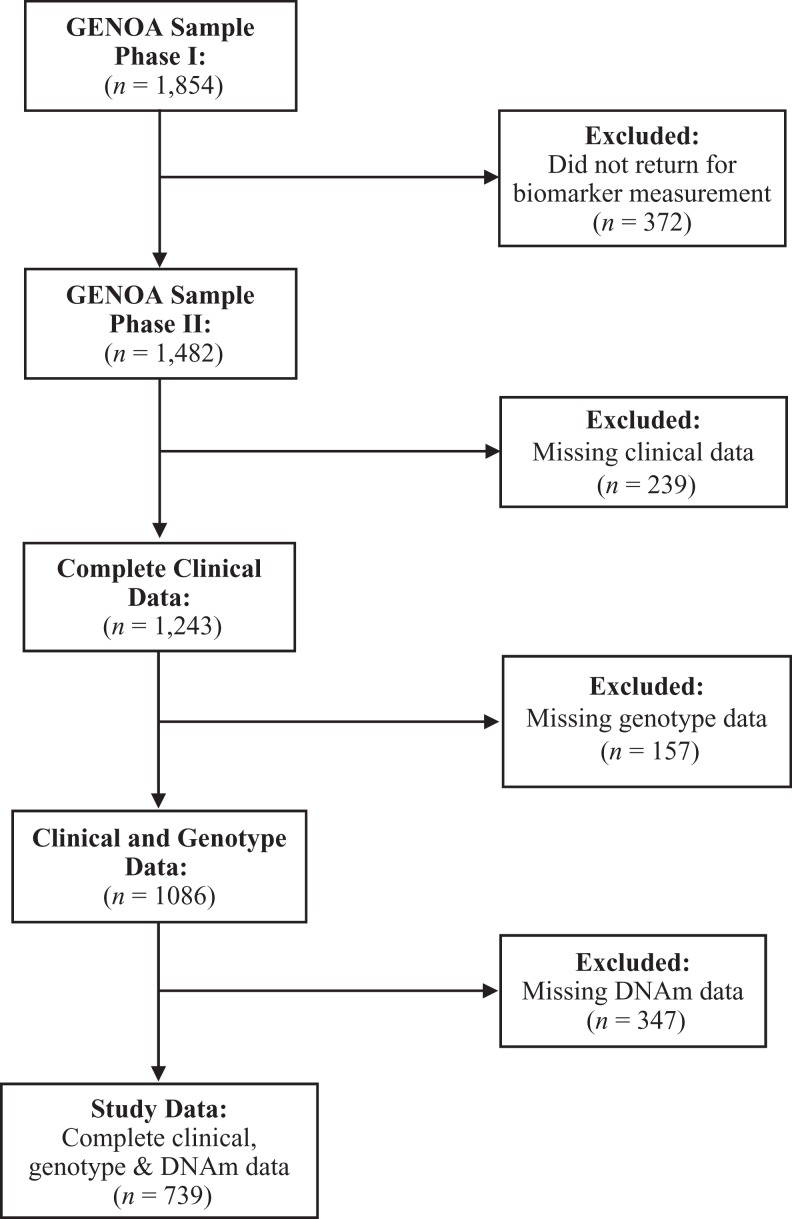
We completed this secondary data analysis on a subset of participants from the GENOA study. The GENOA study had previously obtained institutional review board approval via the University of Mississippi Medical Center and University of Michigan. Data collection methods were completed in accordance with the approved guidelines. GENOA is a community-based prospective study that recruited participants from sibships with two or more siblings who were diagnosed with primary hypertension prior to age 60 and self-identified as AA. All members of the sibship were invited to participate, regardless of hypertension status. In Phase I (1995–2000), 1,854 AA participants from 683 sibships from the Jackson, Mississippi, area were recruited. In Phase II (2000–2005), 1,482 of the initial subjects returned. Study visits were made in the morning after an overnight fast. Each participant was interviewed by trained study personnel to collect demographic and medical history data as previously described (Daniels et al., 2004). A peripheral blood sample was also collected at Phase II, which was used to measure serum lipid and DNA methylation levels.
Measures
Demographic, anthropometric, and clinical measures
Complete clinical data for variables of interest in participants who had fasted for greater than 10 hr prior to peripheral blood sample collection were available for 1,243 participants from Phase II. Clinical variables included in this study were age, sex, prescription lipid/cholesterol medication use (yes/no), current cigarette smoker (yes/no), height (by wall stadiometer), weight (by electronic balance), serum cholesterol (mg/dl), serum HDL (mg/dl), serum LDL (mg/dl), and serum triglycerides (mg/dl). Serum cholesterol, HDL, and triglycerides were measured by standard enzymatic methods on a Hitachi 911 Chemistry Analyzer (Roche Diagnostics, Indianapolis, IN), and LDL cholesterol levels were calculated using the Friedewald formula (Friedewald, Levy, & Fredrickson, 1972).
Genotype measures
Of the 1,854 AA participants enrolled in Phase I, genotyping data were available for 1,599 participants. Participants were genotyped on the Affymetrix Genome-Wide Human SNP Array 6.0 or the Illumina Human 1M-Duo BeadChip. Samples were removed if they had a missing call rate ≥0.05 or a value ≥6 standard deviations from the mean of the first 10 genome-wide principal components (PCs) from the genotype data. SNPs with a missing call rate ≥0.05 were removed. Imputation for the Affymetrix-genotyped and Illumina-genotyped samples was performed separately. For each, samples were prephased using the Segmented HAPlotype Estimation and Imputation Tool (SHAPEIT), Version v2.r, using HapMap Phase II b37. Imputation was performed using IMPUTE, Version 2. The imputation reference panels are from the 1,000 Genomes Project’s Phase I integrated variant set release (v3) in NCBI build 37 (hg19) coordinates (released in March 2012). Since the two genotyping platforms contain only a small number of overlapping SNPs (∼200,000), association analyses were performed using imputed data only. We used the aforementioned overlapping SNPs to calculate the genetic PCs instead of a limited number of ancestry informative markers. The top four genetic PCs were estimated and used in analyses to control for population stratification.
Methylation measures
Peripheral blood leukocytes were isolated from stored blood samples of 1,008 Phase II AA participants and used to measure DNAm levels. The EZ DNA Methylation Gold Kit (Zymo Research, Orange, CA) was used for bisulfite conversion. The methylation assay was performed at the Mayo Clinic Advanced Genomics Technology Center using Illumina® Infinium HumanMethylation27 BeadChips and Illumina BeadXpress reader. As a quality control, seven samples were excluded from analysis due to poor bisulfite conversion efficiency (intensity <4,000). An additional 28 samples were removed because of poor background signals, leaving a total of 973 samples. The lumi package (Du, Kibbe, & Lin, 2008) in R software (lumi package) was used for background adjustment, color balance adjustment, and quantile normalization. Thirty samples were removed because <95% of probes had a detection p value <.01, and two were excluded because the predicted gender based on DNA methylation did not match with the reported gender, leaving a total of 941 samples. Analyses for this study were conducted on participants who had completed clinical, genotyping, and DNAm data (Figure 1, N = 739).
Figure 1.
Data inclusion process. The analyses for this study were conducted on individuals who had complete clinical, single-nucleotide polymorphism (SNP), and DNA methylation (DNAm) data. GENOA = Genetic Epidemiology Network of Arteriopathy study.
Gene Selection
We selected a total of 161 index SNPs that were significantly associated (p < 5 × 10−8) with at least one lipid trait (total cholesterol, HDL, LDL, or triglycerides) from two large GWAS meta-analyses among EA cohorts (Teslovich et al., 2010; Willer et al., 2013). We then selected all DNAm sites that were within genes that contained an index SNP (i.e., SNP was within the gene or within 5 kB of the start or stop position of the gene), for a total of 66 DNAm sites in 36 genes.
Statistical Methods
For all outcome and independent variables, we examined the distributions of continuous measures. The serum HDL and triglyceride levels were not normally distributed and were natural log transformed prior to analysis to reduce the skewness and kurtosis. GENOA SNP allele frequencies were compared to the previous allele frequencies reported in EA GWAS studies looking at lipids (Teslovich et al., 2010; Willer et al., 2013) and to the 1,000 Genomes Project’s build 37 SNP allele frequencies for Americans of African descent (African Ancestry in Southwest US (ASW), http://grch37.ensembl.org/Homo_sapiens/Info/Index, Supplementary Table 1).
Then, we examined the 161 SNPs from previous lipid GWAS studies completed in individuals with EA to determine whether the findings were replicated in this AA cohort. We completed a series of linear mixed-effects models for the four outcome variables: total cholesterol, serum LDL, serum HDL, and triglycerides. We analyzed minimal- (Model 1) and full-adjustment (Model 2) models. Model 1 included age, sex, and the top four genetic PCs to control for population stratification and also included sibship modeled as a random effect. Model 2 also included BMI and lipid medication use. SNP genotypes were dummy coded to represent the additive and dominance deviation of each variation (i.e., for genotypes BB, Bb, and bb, we created two dummy variables X 1 and X 2, where X 1 = 1, 0, −1 and X 2 = 0, −1, 0; Falconer & Mackay, 1996) and were tested for association with each lipid measure separately.
We also examined the association between each of the 66 DNAm sites and lipids (Supplementary Table 2). M values, calculated as the log2 ratio of the intensities of methylated probe versus unmethylated probe, were calculated for each DNAm site. Positive M values mean that more molecules are methylated than unmethylated, while negative M values mean the opposite (Du et al., 2010). We used the M value of DNA methylation levels because the β value has a bounded range from 0 to 1 that violates the Gaussian distribution assumption (Du et al., 2010). We first adjusted each DNAm site for peripheral blood cell heterogeneity using the Houseman correction method and technical covariates (i.e., DNAm chip and position; Houseman et al., 2012; Houseman, Molitor, & Marsit, 2014). We then used linear mixed effect models to analyze each adjusted DNAm site, using Models 1 and 2 as outlined above but also controlling for smoking status in both models.
To assess interaction effects, we carried forward SNP and DNAm sites with p values <.1. We chose, a priori, to carryforward sites with a less stringent p value because if there is a statistical interaction, a marginally significant main effect would be expected. Second, we suspected we would not have a high number of sites reaching significance with a false discovery rate (FDR) <0.05 because our sample size is small relative to previous GWAS studies. For genes that had both an SNP and a DNAm value of p < .1, we evaluated interaction effects using Model 2. All analyses were completed using R statistical computing environment (https://www.r-project.org/). To control for multiple comparisons, we determined significance using an FDR of <0.05 (Benjamini & Hochberg, 1995).
Results
The sample was comprised of self-identified AA men and women. Demographic data are listed in Table 1. Participants were predominately female, older, obese, and had lipid levels consistent with those previously described among AAs (Meyer et al., 2013). Although there were statistically significant differences in total cholesterol and HDL levels by sex, the differences were not of clinical significance (i.e., trait levels were not pathologic in one group vs. nonpathologic in the other), nor were there differences in SNP or DNAm findings (data not shown) when analyses were run separately by sex. Results presented here include both males and females within the same cohort, controlling for sex statistically as a covariate, per models described above. We did not identify any significant associations between the 161 SNPs and any of the four serum lipid levels, nor between DNAm and serum lipid levels after controlling for multiple comparisons (Table 2). Three SNP with corresponding DNAm sites had nominal p values of <.1 within the same gene (two with serum HDL and one with serum LDL) and were carried forward for interaction analyses. We detected no significant interaction effects among these SNPs and DNAm sites (Table 3).
Table 1.
Participant Characteristics.
| Variable | M (SD) |
|---|---|
| Age, years | 66.95 (7.34) |
| BMI, kg/m2 | 31.44 (6.46) |
| Total cholesterol, mg/dl | 204.90 (41.44) |
| LDL, mg/dl | 122.85 (38.63) |
| HDL, mg/dl | 58.66 (18.22) |
| Triglycerides, mg/dl | 116.95 (55.80) |
| n (%) | |
| Sex, female | 536 (72.5) |
| Smoker, yes | 77 (10.4) |
| Lipid medication, yes | 155 (21.0) |
| Clinically significant lipid levels | |
| Total cholesterol > 240 mg/dl | 138 (18.7) |
| LDL > 160 mg/dl | 121 (16.4) |
| HDL < 40 mg/dl | 98 (13.3) |
| Triglycerides >150 mg/dl | 154 (20.8) |
Note. N = 739. BMI = body mass index; HDL = serum high-density lipoprotein; LDL = serum low-density lipoprotein.
Table 2.
Number of Significant SNPs and DNAm Sites in Sample by Outcome.a
| Outcome | p < .1 | p < .05 | p < .01 |
|---|---|---|---|
| Number of SNPs (%) | |||
| Total cholesterol | |||
| Model 1b | 21 (13) | 13 (8) | 2 (1) |
| Model 2c | 23 (14) | 14 (9) | 3 (2) |
| HDL | |||
| Model 1 | 14 (9) | 7 (4) | 2 (1) |
| Model 2 | 14 (9) | 6 (4) | 3 (2) |
| LDL | |||
| Model 1 | 17 (11) | 11 (7) | 1 (1) |
| Model 2 | 17 (11) | 10 (6) | 1 (1) |
| Triglycerides | |||
| Model 1 | 18 (11) | 7 (4) | 0 (0) |
| Model 2 | 16 (10) | 8 (5) | 0 (0) |
| Number of DNAm sites (%)d | |||
| Total cholesterol | |||
| Model 1 | 7 (11) | 4 (6) | 0 (0) |
| Model 2 | 8 (12) | 4 (6) | 1 (2) |
| HDL | |||
| Model 1 | 11 (17) | 8 (12) | 1 (2) |
| Model 2 | 11 (17) | 6 (9) | 2 (3) |
| LDL | |||
| Model 1 | 9 (14) | 5 (8) | 0 (0) |
| Model 2 | 10 (15) | 6 (9) | 0 (0) |
| Triglycerides | |||
| Model 1 | 3 (5) | 1 (2) | 0 (0) |
| Model 2 | 3 (5) | 1 (2) | 0 (0) |
Note. DNAm = DNA methylation; HDL = high-density lipoprotein; LDL = low-density lipoprotein; SNP = single-nucleotide polymorphism.
aNone of the associations were significant when controlling for multiple testing (false discovery rate [FDR] < 0.05). bModel 1 controls for age, sex, sibship, and genetic principal components. cModel 2 controls for Model 1 covariates and body mass index, lipid medication, and smoking. dDNAm sites were adjusted for peripheral blood cell heterogeneity using the Houseman correction method and technical covariates (DNAm chip and position).
Table 3.
Results From SNP-DNAm Interaction Models for SNPs and DNAm Sites From the Same Gene With p Value <.1 in Univariate Association Analyses.
| Trait, Gene, and Variants | SNP-Only Modela |
DNAm-Only Modelb |
SNP × DNAm Interaction Model |
|||
|---|---|---|---|---|---|---|
| β | p Value | β | p value | β | p Value | |
| HDL | ||||||
| ERGIC3 | ||||||
| rs2277862 | 0.03 | .06 | ||||
| cg00340102 | 0.02 | .05 | ||||
| rs2277862 × cg00340102 | 0.01 | .56 | ||||
| ST3GAL4 | ||||||
| rs11220462 | 0.08 | .04 | ||||
| cg08203715 | −0.02 | .05 | ||||
| rs11220462 × cg08203715 | 0.02 | .49 | ||||
| LDL | ||||||
| RAB3GAP1 | ||||||
| rs7570971 | 5.59 | .09 | ||||
| cg12813922 | −3.54 | .03 | ||||
| rs7570971 × cg12813922 | 2.85 | .38 | ||||
Note. DNAm = DNA methylation; HDL = high-density lipoprotein; LDL = low-density lipoprotein; SNP = single-nucleotide polymorphism.
aAdjusted models control for age, body mass index, ancestry, sibship, sex, lipid medications, and smoking. bDNAm sites were adjusted for peripheral blood cell heterogeneity using the Houseman correction method and technical covariates (DNAm chip and position).
Discussion
It is well known that ancestry contributes to genetic risk of disease. However, most studies evaluating genetic and epigenetic risk of disease are primarily completed using cohorts with EA. Similar to a study conducted by Deo and colleagues (2009), who did not find any strong contribution to lipid levels among AAs of genetic variants found to be associated with lipid levels in EA cohorts, we did not identify any significant associations between the 161 SNPs significant to lipid levels among the EA cohorts in our AA cohort. Deo and colleagues suggest that additional fine mapping is necessary, specifically within AA cohorts, because both global and local ancestries may alter how strongly SNPs contribute to phenotypic variation. For example, they found that ancestry did have a significant association with serum levels of triglycerides and LDL, but they could not identify any genes with strong associations to explain the variation. They postulate that gene–gene epistatic effects could explain some of the difficulty associating specific genetic variants with variation in lipid levels between ancestry groups. Conversely, Dumitrescu and colleagues (2011) determined that approximately half of the SNPs identified in earlier studies among EA individuals could be generalized to an AA population. However, similar to Deo et al.’s findings, Dumitrescu et al. conceded that the SNPs identified and indexed in EA studies may only represent tagging SNPs among individuals with EA and may not be functional variants for lipids. If the SNPs identified in previous EA studies are not of functional relevance, it could also explain some of the limited replicability of these findings observed in genetic studies among AA cohorts. The differing patterns of linkage disequilibrium that exist between ancestry backgrounds add credence to the argument that additional genetic studies must be completed specifically within non-EA cohorts.
Similar to our SNP findings, we did not detect any significant association between DNAm and lipid levels within our cohort. In a recent study using an EA cohort, Hedman et al. (2017) observed associations of 33 DNAm sites with serum lipid levels. In their initial analysis of DNAm data produced using the Illumina 450 K array, the authors discovered an association with lipid levels at 193 DNAm sites. The number of DNAm sites that remained significant after controlling for BMI decreased to 80, nine of which were unique to the BMI-adjusted analysis. Hedman and colleagues repeated the analyses in three separate EA cohorts and determined that 33 DNAm sites replicated across all cohorts. Similar to the studies analyzing SNPs related to lipids, these researchers also found that 15% of the DNAm sites were associated with expression changes in genes adjacent to where the differential methylation occurred. They also noted that these results suggest that there are underlying linkages between the genes and methylation networks that contribute to lipid levels as opposed to a single variant altering expression alone. The large sample size of the study allowed for the team to capture some variants within SNP and DNAm linked networks (i.e., cis-meQTL SNPs); however, the study was not designed to evaluate these findings among diverse populations. The results from our study suggest that DNAm that may be associated with lipids among AA cohorts may differ from that identified in studies of populations with EA.
We sought to identify potential explanations for the lack of reproducibility between the EA GWAS studies and our AA cohort. First, our cohort of 739 individuals may not have been large enough to detect significant contributions of SNP or DNAm variation to serum lipid variability. However, we did not take a genome-wide approach to either the SNP or DNAm analysis, greatly reducing the number of independent tests. Nonetheless, even using a targeted approach, we did not observe significant contributions to serum lipid levels by the selected set of SNPs or corresponding DNAm sites. Second, our minor allele frequencies differed from those of the original studies, as would be expected. Of note, 20% (33/161 SNPs) of the alleles significantly associated with lipid levels among EA cohorts are not the same alleles among AAs in the 1,000 Genomes Project or our GENOA cohort; meaning that the minor allele in the EA population is the major allele in the AA population or vice versa. Additionally, 12% (20/161 SNPs) of the alleles associated with lipid levels in the EA population are present in less than 5% of AAs in our GENOA cohort, which would make it difficult to detect an association in a small cohort of 739 individuals (see Supplementary Table 1 for detailed information of allele frequencies among EA and AA cohorts). Lastly, our data set contains DNAm array data from the 27 K array, which interrogates a fraction of the sites found on the more recent Illumina 450 K and EPIC arrays. The sites associated with lipid levels in the more recent study may not have been included on the 27 K array or associated with the SNP sites we evaluated.
Conclusions
Although the results in this study were not statistically significant among the loci examined, they do provide a platform for future work in this important area of inquiry among AAs disproportionately affected by health disparities associated with lipids. The approach we applied in this study may be used to evaluate omic variation that contributes to cardiovascular disease via different mechanisms, such as angiotensinogen expression. In fact, scientists conducting work in health disparities have noted the emergent need for increased testing and screening among AAs for serum lipid levels (Wright et al., 2016). Inclusivity approaches with diverse populations and future multi-omic work is critical for understanding the physiological and environmental factors influencing health disparities among AAs and other non-EA groups. Once the key omic and environmental factors are identified, health providers will be better equipped to develop and implement interventions that are based on individuals’ unique needs. Nurses and nurse scientists are well trained to examine health disparities using omic approaches to improve health (Starkweather et al., 2017; Taylor, Wright, Hickey, & Housman, 2017). Their unique skills in clinical translational science would be well utilized in omics-based care for reductions in health disparities. Future multi-omic studies that take into account environmental and omic data will help to identify variants in additional pathways that could contribute to differences in disease risk among different racial and ethnic groups.
Supplementary Material
Footnotes
Author Contribution: Michelle L. Wright contributed to conception, design, analysis, and interpretation; drafted the manuscript; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Erin B. Ware contributed to conception, design, analysis, and interpretation; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Jennifer A. Smith contributed to conception, design, acquisition, analysis, and interpretation; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Sharon L. R. Kardia contributed to conception, design, acquisition, analysis, and interpretation; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Jacquelyn Y. Taylor contributed to conception, design, acquisition, analysis, and interpretation; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Support for the Genetic Epidemiology Network of Arteriopathy (GENOA) was provided by the National Heart, Lung and Blood Institute (HL054457, HL100185, HL 087660, HL119443, and HL133221); additional support for this project was provided by the National Institute of Nursing Research (NINR), R01NR013520.
Supplemental Material: Supplementary material is available for this article online.
References
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B, 57, 289–300. [Google Scholar]
- Bomotti S. M., Smith J. A., Zagel A. L., Taylor J. Y., Turner S. T., Kardia S. L. (2013). Epigenetic markers of renal function in African Americans. Nursing Research and Practice, 2013, 687519 doi:10.1155/2013/687519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels P. R., Kardia S. L., Hanis C. L., Brown C. A., Hutchinson R., Boerwinkle E., Turner S. T. (2004). Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) study. American Journal of Medicine, 116, 676–681. doi:10.1016/j.amjmed.2003.12.032 [DOI] [PubMed] [Google Scholar]
- de Ferranti S. D., Rodday A. M., Mendelson M. M., Wong J. B., Leslie L. K., Sheldrick R. C. (2016). Prevalence of familial hypercholesterolemia in the 1999 to 2012 United States National Health and Nutrition Examination Surveys (NHANES). Circulation, 133, 1067–1072. doi:10.1161/CIRCULATIONAHA.115.018791 [DOI] [PubMed] [Google Scholar]
- Demerath E. W., Guan W., Grove M. L., Aslibekyan S., Mendelson M., Zhou Y.-H.…Boerwinkle E. (2015). Epigenome-wide asociation study (EWAS) of BMI, BMI change, and waist circumference in African American adults identifies multiple replicated loci. Human Molecular Genetics, 24, 4464–4479. doi:10.1093/hmg/ddv161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo R. C., Reich D., Tandon A., Akylbekova E., Patterson N., Waliszewska A.…Wilson J. G. (2009). Genetic differences between the determinants of lipid profile phenotypes in African and European Americans: The Jackson Heart Study. PLOS Genetics, 5, e1000342 doi:10.1371/journal.pgen.1000342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Kibbe W. A., Lin S. M. (2008). Lumi: A pipeline for processing Illumina microarray. Bioinformatics, 24, 1547–1548. doi:10.1093/bioinformatics/btn224 [DOI] [PubMed] [Google Scholar]
- Du P., Zhang X., Huang C.-C., Jafari N., Kibbe W. A., Hou L., Lin S. M. (2010). Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics, 11, 587–587. doi:10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu L., Carty C. L., Taylor K., Schumacher F. R., Hindorff L. A., Ambite J. L.…Crawford D. C. (2011). Genetic determinants of lipid traits in diverse populations from the Population Architecture Using Genomics and Epidemiology (PAGE) Study. PLOS Genetics, 7, e1002138 doi:10.1371/journal.pgen.1002138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C. (1996). Introduction to quantitative genetics (4th ed). Harlow, England: Longman. [Google Scholar]
- Friedewald W. T., Levy R. I., Fredrickson D. S. (1972). Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clinical Chemistry, 18, 499–502. [PubMed] [Google Scholar]
- Goff D. C. (2006). Dyslipidemia prevalence, treatment, and control in the Multi-Ethnic Study of Atherosclerosis (MESA): Gender, ethnicity, and coronary artery calcium. Circulation, 113, 647–656. doi:10.1161/CIRCULATIONAHA.105.552737 [DOI] [PubMed] [Google Scholar]
- Hedman A. K., Mendelson M. M., Marioni R. E., Gustafsson S., Joehanes R., Irvin M. R.…Ingelsson E. (2017). Epigenetic patterns in blood associated with lipid traits predict incident coronary heart disease events and are enriched for results from genome-wide association studies. Circulation: Cardiovascular Genetics, 10, e001487 doi:10.1161/CIRCGENETICS.116.001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E. A., Accomando W. P., Koestler D. C., Christensen B. C., Marsit C. J., Nelson H. H.…Kelsey K. T. (2012). DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 2–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E. A., Molitor J., Marsit C. J. (2014). Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics (Oxford, England), 30, 1431–1439. doi:10.1093/bioinformatics/btu029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes H. P., Lopez R. (2012). Cumulative risk and a call for action in environmental justice communities. Journal of Health Disparities Research and Practice, 1, 29–57. [Google Scholar]
- Jenuwein T., Allis C. D. (2001). Translating the histone code. Science, 293, 1074–1080. doi:10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Joehanes R., Just A. C., Marioni R. E., Pilling L. C., Reynolds L. M., Mandaviya P. R.…London S. J. (2016). Epigenetic signatures of cigarette smoking. Circulation: Cardiovascular Genetics, 9, 436–447. doi:10.1161/CIRCGENETICS.116.001506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebaner D., Huang Y., Hui Q., Taylor J. Y., Goldberg J., Vaccarino V., Sun Y. V. (2016). X chromosome-wide analysis identifies DNA methylation sites influenced by cigarette smoking. Clinical Epigenetics, 8, 20–20. doi:10.1186/s13148-016-0189-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligthart S., Marzi C., Aslibekyan S., Mendelson M. M., Conneely K. N., Tanaka T.…Dehghan A. (2016). DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biology, 17, 255 doi:10.1186/s13059-016-1119-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P. A., Yoon P. W., Kaufmann R. B. (2013). CDC health disparities and inequalities report—United States, 2013. Morbidity and Mortality Weekly Report, 62, 1–189. [PubMed] [Google Scholar]
- Mozaffarian D., Benjamin E. J., Go A. S., Arnett D. K., Blaha M. J., Cushman M.…Turner M. B. (2015). Heart disease and stroke statistics—2016 Update: A report From the American Heart Association. Circulation, 133, e38–e360. doi:10.1161/CIR.0000000000000350 [DOI] [PubMed] [Google Scholar]
- Smith J. A., Zagel A. L., Sun Y. V., Dolinoy D. C., Bielak L. F., Peyser P. A.…Kardia S. L. R. (2014). Epigenomic indicators of age in African Americans. Hereditary Genetics: Current Research, 3, 137 doi:10.4172/2161-1041.1000137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkweather A., Coleman B., Barcelona de Mendoza V., Fu M., Taylor J., Henderson W.…Anderson C. (2017). Policy brief: Improve coverage of newborn genetic screening to include the recommended uniform screening panel and newborn screening registry. Nursing Outlook, 65, 480–484. doi:10.1016/j.outlook.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. V., Lazarus A., Smith J. A., Chuang Y.-H., Zhao W., Turner S. T., Kardia S. L. R. (2013). Gene-specific DNA methylation association with serum levels of C-reactive protein in African Americans. PLoS One, 8, e73480–e73480. doi:10.1371/journal.pone.0073480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. V., Smith A. K., Conneely K. N., Chang Q., Li W., Lazarus A.…Kardia S. L. R. (2013). Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Human Genetics, 132, 1027–1037. doi:10.1007/s00439-013-1311-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. V., Turner S. T., Smith J. A., Hammond P. I., Lazarus A., Van De Rostyne J. L.…Kardia S. L. R. (2010). Comparison of the DNA methylation profiles of human peripheral blood cells and transformed B-lymphocytes. Human Genetics, 127, 651–658. doi:10.1007/s00439-010-0810-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. Y., Schwander K., Kardia S. L. R., Arnett D., Liang J., Hunt S. C.…Sun Y. V. (2016). A genome-wide study of blood pressure in African Americans accounting for gene-smoking interaction. Scientific Reports, 6, 18812–18812. doi:10.1038/srep18812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. Y., Sun Y. V., Hunt S. C., Kardia S. L. R. (2010). Gene-environment interaction for hypertension among African American women across generations. Biological Research For Nursing, 12, 149–155. doi:10.1177/1099800410371225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. Y., Wright M. L., Crusto C. A., Sun Y. V. (2016). The intergenerational impact of genetic and psychological factors on blood pressure (InterGEN) study: Design and methods for complex DNA analysis. Biological Research For Nursing, 18, 521–530. doi:10.1177/1099800416645399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. Y., Wright M. L., Hickey K. T., Housman D. E. (2017). Genome sequencing technologies and nursing: What is the role of nurses and nurse scientists? Nursing Research, 66, 198–205. doi:10.1097/NNR.0000000000000211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. Y., Wright M. L., Housman D. (2016). Lead toxicity and genetics in Flint, MI. NPJ Genomic Medicine, 1, 16018–16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. Y., Wu C. Y., Darling D., Sun Y. V., Kardia S. L. R., Jackson J. S. (2012). Gene-environment effects of SLC4A5 and skin color on blood pressure among African American women. Ethnicity & Disease, 22, 155–161. [PMC free article] [PubMed] [Google Scholar]
- Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M.…Kathiresan S. (2010). Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. doi:10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S.…Abecasis G. R. (2013). Discovery and refinement of loci associated with lipid levels. Nature Genetics, 45, 1274–1283. doi:10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M. L., Housman D., Taylor J. Y. (2016). A perspective for sequencing familial hypercholesterolaemia in African Americans. NPJ Genomic Medicine, 1, 16012–16012. doi:10.1038/npjgenmed.2016.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon P. W., Bastian B., Anderson R. N., Collins J. L., Jaffe H. W. (2014). Potentially preventable deaths from the five leading causes of death—United States, 2008–2010. Morbidity and Mortality Weekly Report, 63, 369–374. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.