SUMMARY
Hedgehog (Hh) pathway inhibitors such as vismodegib are highly effective for treating basal cell carcinoma (BCC); however, residual tumor cells frequently persist and regenerate the primary tumor upon drug discontinuation. Here, we show that BCCs are organized into 2 molecularly and functionally distinct compartments. Whereas interior Hh+/Notch+ suprabasal cells undergo apoptosis in response to vismodegib, peripheral Hh+++/Notch− basal cells survive throughout treatment. Inhibiting Notch specifically promotes tumor persistence without causing drug resistance, while activating Notch is sufficient to regress already established lesions. Altogether, these findings suggest that the 3-dimensional architecture of BCCs establishes a natural hierarchy of drug response in the tumor, and that this hierarchy can be overcome, for better or worse, by modulating Notch.
INTRODUCTION
Basal cell carcinoma (BCC) is the most common cancer in North America, with >1 million new cases diagnosed each year. Although these tumors rarely metastasize, the ubiquity of this disease imposes a significant economic burden on our healthcare system. At an individual level, BCCs often cause local tissue damage, which is especially problematic for Gorlin syndrome patients who are genetically predisposed to BCC. These individuals can develop hundreds of lesions throughout the skin—potentially spending a lifetime having their tumors excised, only for new ones to invariably appear.
The singular defining feature of all BCCs is dysregulation of Hedgehog (Hh) signaling, a key developmental pathway that is also critical for adult tissue homeostasis and regeneration (Briscoe and Therond, 2013; Epstein, 2008; Lum and Beachy, 2004). In the absence of ligand, Hh signaling is normally silenced by Patched1 (PTCH1), which inhibits Smoothened (SMO), the major upstream activator of the pathway. Upon binding Hh ligands, PTCH1 is itself inhibited, enabling SMO to transduce downstream signals via GLI transcription factors. In BCCs, it is this delicate balance of PTCH1-mediated silencing, and SMO-mediated activation, that is perturbed: Roughly 80–90% of tumors are caused by loss-of-function mutations in PTCH1, while the remainder are largely driven by gain-of-function mutations in SMO (Gailani et al., 1996; Hahn et al., 1996; Johnson et al., 1996; Reifenberger et al., 2005; Xie et al., 1998).
As SMO is the central upstream activator of canonical Hh signaling, small molecule inhibitors targeting this protein represent a promising new class of therapeutics for combating Hh-dependent tumors such as BCC and medulloblastoma (Metcalfe and Sauvage, 2011). Vismodegib (vismo), an FDA-approved SMO inhibitor for treating advanced BCC, has been reported to induce therapeutic responses in roughly half of sporadic BCCs and in nearly all Gorlin BCCs (Axelson et al., 2013; Hoff et al., 2009; Sekulic et al., 2012; Tang et al., 2012). These successes, however, have been tempered by the fact that a significant fraction of sporadic tumors either do not respond to vismo, or respond initially but subsequently recur during treatment (Brinkhuizen et al., 2014; Chang and Oro, 2012). Importantly, most resistant BCCs maintain high level Hh signaling by expressing mutant SMO variants that cannot be inactivated by vismo (Atwood et al., 2015; Brinkhuizen et al., 2014; Pricl et al., 2015; Sharpe et al., 2015).
A problem less insidious than drug resistance, but far more pervasive, is the observation that Hh pathway inhibitors rarely eliminate all tumor cells. Even among responding tumors, residual tumor cells can be detected in 17–100% of skin biopsies, and nearly all Gorlin BCCs will reappear at their original sites following drug discontinuation (Skvara et al., 2011; Tang et al., 2016; Tang et al., 2012). This issue is especially pertinent given that patients on vismo frequently interrupt treatment due to likely on-target side effects including muscle cramps, loss of taste and fatigue (Sekulic et al., 2012; Tang et al., 2012). Thus, even clinically resolved BCCs can be regarded as a form of persistent disease in which tumor regrowth must be suppressed by continuous treatment. Whereas drug resistance is clearly caused by a failure to quell high level Hh pathway activation, the genetic basis for why some tumor cells persist even upon near-complete Hh suppression currently remains unclear.
Clues for answering this question may come from recent exome sequencing studies, which have revealed that BCC is the most highly mutated human cancer, with 50–75 mutations/Mb in sporadic tumors and 21–33 mutations/Mb in Gorlin BCCs (Atwood et al., 2015; Jayaraman et al., 2014; Sharpe et al., 2015). Although not regarded as oncogenic drivers for this disease, TP53 and NOTCH1/2 are among the most commonly mutated genes, each occurring in ~50% of BCCs (Bonilla et al., 2016; Jayaraman et al., 2014). Notch signaling, in particular, is a central regulator of epidermal differentiation and typically becomes activated as normal keratinocytes lose contact with the basement membrane (Watt et al., 2008). Notch1-deficient mice are susceptible to forming SCCs and, to a lesser extent, BCCs (Nicolas et al., 2003), but whether Notch modulates BCCs driven by Hh pathway mutations has not been tested.
We and others have previously demonstrated that loss of Ptch1 in hair follicle stem cells leads to formation of microscopic Gorlin BCC-like tumors (Kasper et al., 2011; Peterson et al., 2015; Wang et al., 2011). Using Ptch1 conditional mice, here we model BCC formation and vismo-induced regression to identify mechanisms that likely enable drug-treated tumors to persist, enter dormancy, and subsequently recur upon stopping treatment.
RESULTS
A mouse model of vismo-induced tumor regression and reactivation
Our previous studies utilized mice expressing a tamoxifen-inducible, Gli1 promoter-driven Cre recombinase (Gli1-CreERT2) to target deletion of Ptch1 to stem cells residing within hair follicle and mechanosensory touch dome epithelia (Gli1;Ptch1 mice) (Peterson et al., 2015; Uhmann et al., 2007). Within 5 weeks of induction, Gli1;Ptch1 mice develop abundant BCC-like tumors manifesting a classic human BCC feature termed “palisading,” where peripheral tumor cells in direct contact with the basement membrane (here, termed “basal cells”) take on a columnar appearance (Figure 1A). In contrast, cells within the tumor interior (here, termed “suprabasal cells”) display a more loosely-packed morphology.
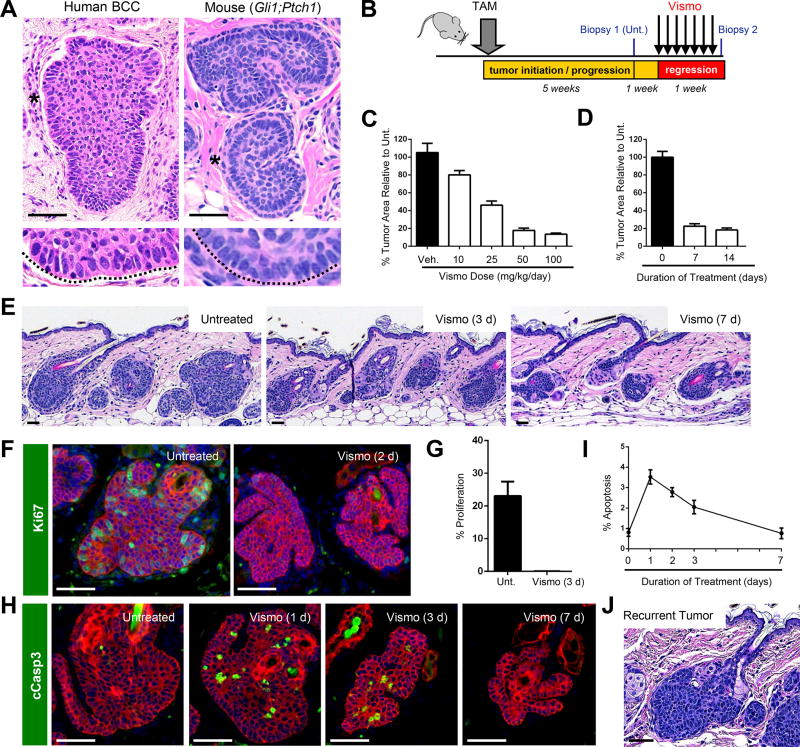
Figure 1. Vismo regresses tumors in a mouse model of BCC.
A. Nodular human BCC (left) and mouse Gli1;Ptch1 tumor (right). Magnified views of palisaded basal cells (asterisks) are depicted in the lower panels. B. Schematic for vismo treatment. Unt., untreated. TAM, tamoxifen. C. Tumor regression in response to varying doses of daily vismo for 1 week. Veh., vehicle control. D. Tumor regression in response to 1 or 2 weeks of daily vismo at a dose of 50 mg/kg animal weight. E. Histology of untreated or treated Gli1;Ptch1 tumors. F. Vismo completely inhibits proliferation (green). G. Quantitation for F. H. Vismo causes tumor apoptosis, as assessed by cleaved Caspase 3 (cCasp3) staining (green). I. Quantitation for H, where apoptosis is expressed as a percentage of total K5+ tumor cells. J. Recurrent Gli1;Ptch1 tumor, treated for 1 week with vismo, then left untreated for an additional 1 week. Red, co-staining for K5. Error bars, SE. Scale bars, 100 µm. See also Figure S1.
To confirm the requirement for Hh in these tumors, we administered vismo by oral gavage to Gli1;Ptch1 mice, and measured tumor area prior to and after treatment (Figure 1B). We first generated a dose response profile for vismo, and determined that 50 mg/kg of drug, administered daily for 1 week, reduced tumor area by ~80%, with higher doses providing only marginal additional benefit (Figure 1C). We next determined that at this drug concentration, tumor shrinkage largely occurred during the first week of treatment, since longer dosing regimens did not further reduce tumor size (Figure 1D–E). We also observed that vismo was equally effective at regressing tumors that originated from either Gli1+ or Lrig1+ hair follicle stem cells, suggesting that tumor-drug response likely does not depend upon cellular origin or genetic background (Figure S1A). It is important to note that Gli1;Ptch1 mice exhibit transformation throughout the skin that is likely more extensive than what is typically seen in humans, with possible confounding effects on the adjacent stromal microenvironment and on the hair cycle. Keeping these limitations in mind, for the remainder of this study we treat Gli1;Ptch1 mice with 50 mg/kg of vismo daily and focus on cellular events that occur during the first week.
Previous studies have reported that Hh suppression can inhibit proliferation and induce apoptosis in BCC (Hutchin et al., 2005; Williams et al., 2003). We therefore performed time course studies on vismo-treated Gli1;Ptch1 mice, and determined that proliferation is fully halted within 2–3 days after drug administration (Figure 1F–G). Furthermore, while apoptotic cells were rarely observed in untreated Gli1;Ptch1 tumors, similar to human BCCs, vismo induced apoptotic cell death, which peaked within 1 day after treatment and declined subsequently (Figure 1H–I). Upon discontinuing vismo, residual tumor cells re-entered mitosis within 3 days (Figure 1J and S1B). Thus, our mouse model mimics vismo-induced regression of Gorlin BCCs, where residual tumor cells persist even upon higher drug doses and longer treatment regimens, and dormant cells become reactivated soon after drug stoppage.
Basal and suprabasal tumor cells respond differently to vismo
To better understand how residual tumor cells persist in the face of anti-Hh therapy, we began by testing the requirement for p53 in mediating regression. Although TP53 is frequently mutated in BCC, its role in modulating drug response in these tumors remains unclear (Ponten et al., 1997; Sharpe et al., 2015). We first examined the localization of p53 in Gli1;Ptch1 mice and observed protein accumulation predominantly in basal tumor cells (Figure 2A). However, upon deleting both copies of this gene in our Gli1;Ptch1 model, neither tumor progression nor drug response was affected (Figure 2B).
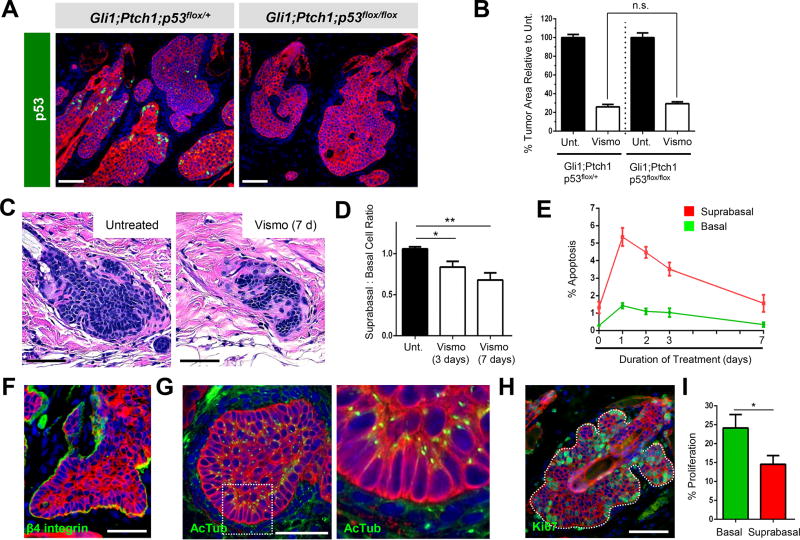
Figure 2. Gli1;Ptch1 tumors are comprised of 2 compartments.
A. p53 (green) is detected in Gli1;Ptch1;p53flox/+ tumors (left), but not in tumors that deleted the allele (right). B. Loss of p53 does not affect tumor response to vismo. Unt., untreated. C. Histology of untreated (left) or regressed tumor (right) after 1 week of vismo. D. Ratio of suprabasal:basal cells in untreated and treated tumors. E. Quantitation of tumor apoptosis, as in Figure 1I, divided by tumor compartment. F. Basal tumor cells express β4 integrin (green). G. Polarized basal cells display apical primary cilia, as marked by acetylated α Tubulin (AcTub, green), whereas suprabasal cells exhibit random ciliary orientation. The right panel is magnified from the boxed area. H. Basal tumor cells exhibit a modest increase in proliferation (green). I. Quantitation for H. Red, co-staining for K5. Error bars, SE. *, p < 0.05; **, p < 0.01; n.s., not significant. Scale bars, 100 µm.
In the process of characterizing tumor regression, we noticed that residual tumor nests often appeared histologically distinct from untreated tumors. Whereas untreated tumors were comprised of both basal and suprabasal compartments, regressed tumors often appeared as elongated strands with relatively fewer suprabasal cells (Figure 2C–D). Given that vismo induces rapid tumor cell death (Figure 1H–I), we compared apoptosis rates between the basal and suprabasal compartments. Although vismo induced apoptosis in both populations, suprabasal cells were roughly 3 times more likely to undergo cell death (Figure 2E). Consistent with these findings, when either Ptch1-deficient primary keratinocytes or ASZ BCC cells were grown as adherent monolayers in culture (Aszterbaum et al., 1999), these cells did not undergo apoptosis even when Hh signaling was fully suppressed by 2 days of vismo treatment (Figure S1C–D). This suggests that vismo’s ability to induce cell death is manifest by the 3-dimensional architecture—specifically, in suprabasal cells—of tumors that develop in vivo.
This functional difference between basal and suprabasal cells prompted us to examine these 2 compartments more closely. We confirmed that basal cells, but not suprabasal cells, expressed the adhesion receptor β4 integrin (Figure 2F). Basal tumor cells were also highly polarized, with primary cilia extending towards the inside of the tumor, marking the apical surface of these cells (Figure 2G). In contrast, suprabasal cells manifest cilia that were not oriented in any specific direction, suggesting that these interior cells were less polarized. Finally, we noted that basal tumor cells exhibited modestly increased proliferation relative to suprabasal cells (Figure 2H–I). Altogether, these observations suggest that Gli1;Ptch1 tumors are comprised of 2 histologically and functionally distinct compartments. Whereas Hh pathway inhibitors efficiently eliminate interior suprabasal cells, peripheral basal cells in contact with the basement membrane persist in dormancy and likely regenerate the primary tumor upon drug discontinuation.
Basal tumor cells display high level Hh signaling
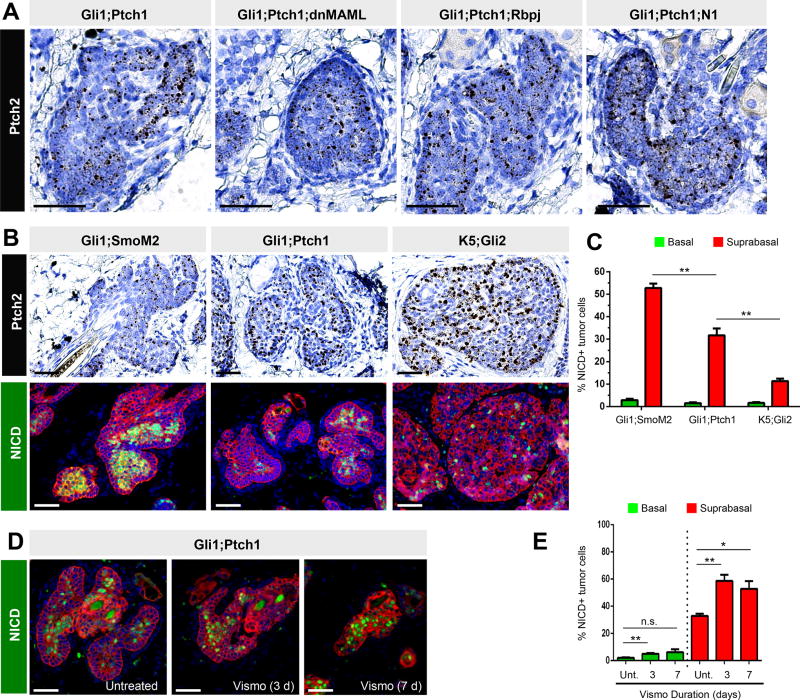
Given the divergent responses of basal and suprabasal tumor cells to vismo, we next queried whether these 2 populations differed molecularly. To isolate cells for gene expression comparisons, we initially generated Gli1-CreERT2 mice that possessed a conditional oncogenic allele of Smo fused to EYFP (Gli1;SmoM2 mice) (Mao et al., 2006). These mice formed tumors over 25 weeks (Figure 3A), and we sorted EYFP+ tumor cells from EYFP− bulk keratinocytes. We further subdivided these into α6high (basal) and α6low (suprabasal) populations, confirming proper cell enrichment by qPCR (Figure 3B). Interestingly, we observed that basal tumor cells displayed elevated expression of canonical Hh target genes, relative to suprabasal tumor cells (Figure 3C). These canonical targets, including Gli1, Ptch1, Ptch2 and Hhip1, were also increased in both tumor compartments, compared to non-tumor basal cells (Figure 3C).
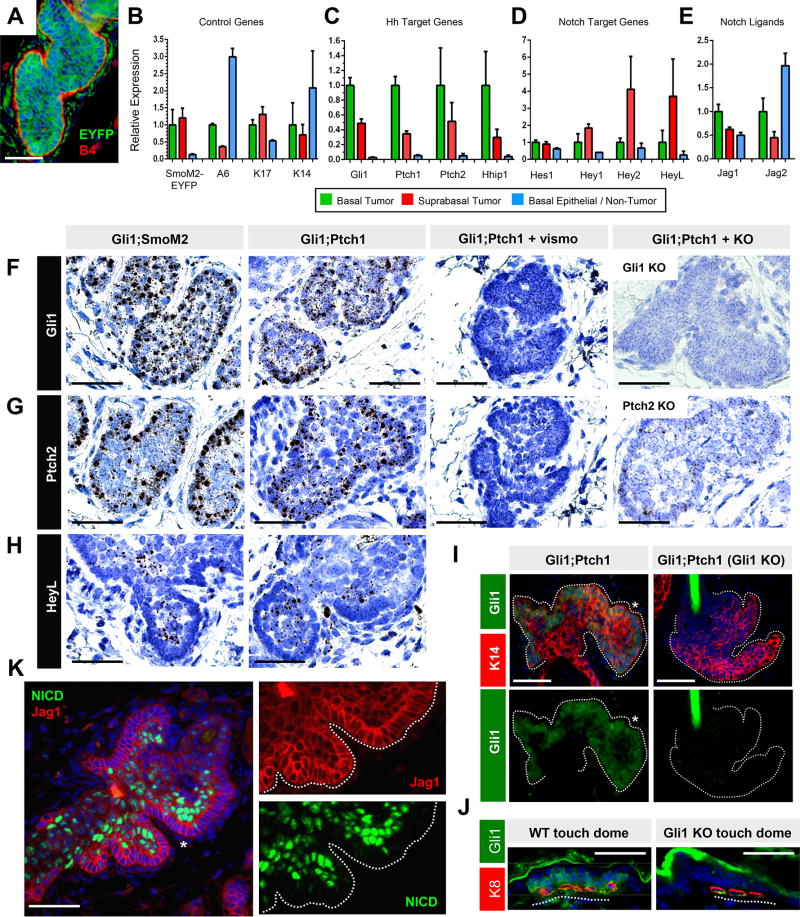
Figure 3. Tumor basal and suprabasal cells are molecularly distinct.
A. Gli1;SmoM2 tumor, 25 weeks after induction, with fluorescent tumor cells (green) and β4 integrin at the periphery (red). B. Validation of flow sorting of Gli1;SmoM2 tumor and non-tumor populations by qPCR for the indicated control genes. A6, α6 integrin. C. Basal Gli1;SmoM2 tumor cells display increased Hh target gene expression. D. Suprabasal Gli1;SmoM2 tumor cells display elevated Hey2 and HeyL. E. Basal Gli1;SmoM2 tumors exhibit increased Jag1 and Jag2. F. Single molecule in situ staining (black/brown dots) for Gli1 in different tumor models, as indicated. Gli1 is modestly enriched in basal tumor cells, lost upon vismo treatment, and absent in tumors that formed in a Gli1 null background (right). G. Single molecule in situ staining for Ptch2, which is also increased in basal tumor cells, lost upon vismo treatment, and largely absent in tumors that developed in a Ptch2 null background (right). Residual staining is likely due to abortive transcripts from the Ptch2-disrupted locus. H. In situ staining for HeyL, which is enriched in suprabasal cells. I. IHC for Gli1 (green) in Gli1;Ptch1 tumors that had developed in an otherwise wild-type or Gli1 null background. Staining is enriched at the tumor periphery (asterisk). Lower panels are single-channel views of the above images. J. Validation of Gli1 IHC (green) in touch dome epithelia from wild-type or Gli1 null mice. Merkel cells are identified by K8 (red). K. IHC for NICD (green) and Jag1 (red) in Gli1;Ptch1 tumors. Right panels are single-channel views of the area indicated by the asterisk. Error bars, SE. Scale bars, 100 µm. See also Figure S2–3.
We next performed single molecule RNA in situ analysis and confirmed that the Hh target gene Ptch2 and, to a lesser extent, Gli1 were both enriched in basal cells from both Gli1;SmoM2 and Gli1;Ptch1 tumors, as evidenced by increased staining at the tumor periphery (Figure 3F–G, quantitated in Figure S1E). In contrast, expression of a ubiquitous gene, Polr2a, was not enriched in either compartment (Figure S1F). Probe specificity was further validated by the absence of signal in Gli1;Ptch1 tumors that had formed in either a Gli1 null or Ptch2 null background, while both Hh targets were downregulated by vismo, as expected (Figure 3F–G). Although we observed a clear tumor basal:suprabasal bias for Hh target gene expression, both compartments displayed markedly elevated Hh signaling compared to surrounding non-tumor cells (Figure S1G). We also generated an allele-specific probe against exons 8–9 of Ptch1, which are specifically deleted upon Cre-mediated recombination (Uhmann et al., 2007). Although this probe detected Ptch1 in growing hair follicles from Ptch1flox/flox mice lacking Cre expression, Gli1;Ptch1 tumor cells fully deleted these exons (Figure S1H). This eliminated the possibility that non-recombined (Ptch1+) keratinocytes with low Hh pathway activity had entered the tumor and posed as oncogenic suprabasal cells.
Consistent with our in situ results, GLI1 protein was also elevated in Gli1;Ptch1 basal tumor cells, as assessed by immunohistochemistry (IHC) (Figure 3I). As before, we confirmed antibody specificity using Gli1 null skin (Figure 3I–J). Altogether, these findings suggest that tumor basal and suprabasal cells differ molecularly in terms of Hh pathway activation. Since palisaded basal cells exhibit elevated Hh activity, this might explain, at least partially, why this compartment is less likely to apoptose during drug treatment (Figure S2).
Suprabasal tumor cells display increased Notch activation
We next assessed whether other signaling pathways could modulate tumor persistence. Since Notch is activated in differentiating suprabasal cells of the skin (Watt et al., 2008), we examined Notch target gene expression in sorted α6high basal and α6low suprabasal cells from Gli1;SmoM2 tumors, similar to above. Indeed, we detected increased expression of some canonical Notch targets (HeyL and Hey2), but not others (Hes1 and Hey1), in suprabasal tumor cells (Figure 3D). As canonical Notch signaling is induced by receptor-ligand engagement between adjacent cells, we also determined that expression of genes encoding the Notch ligands Jagged1 (Jag1) and Jagged2 (Jag2) was elevated in the basal compartment of Gli1;SmoM2 tumors, compared to suprabasal cells (Figure 3E).
Turning back to Gli1;Ptch1 tumors, we next assessed the Notch status of these lesions by performing IHC against the cleaved activated Notch1 intracellular domain (NICD), a canonical marker of upstream pathway activation. Indeed, we observed strong NICD specifically in the tumor suprabasal compartment (Figure 3K). Furthermore, these NICD+ suprabasal cells were frequently juxtaposed to NICD− basal cells that expressed Jagged1 (Figure 3K). To examine downstream Notch, we again performed single molecule RNA in situ analysis, this time against HeyL, and found enriched staining in the suprabasal compartments of both Gli1;Ptch1 and Gli1;SmoM2 tumors (Figure 3H, quantitated in Figure S1I). Jagged1 can also be expressed by regulatory T cells (Tregs) that modulate hair cycling (Ali et al., 2017); however, we did not observe major differences in the abundance or localization of Tregs during tumor growth and vismo-induced regression (Figure S3). Our results therefore suggest that Jagged1/2-expressing basal cells likely signal to adjacent Notch receptor-expressing suprabasal cells to induce pathway activation.
Inhibiting downstream Notch promotes tumor persistence
Since Gli1;Ptch1 suprabasal tumor cells display elevated Notch as well as increased apoptosis in response to vismo, we next asked whether Notch functionally modulates drug response. To do this, we utilized 2 genetic approaches to inhibit downstream signaling. During canonical Notch pathway activation, NICD enters the nucleus and complexes with co-factors such as Mastermind and RBP-J to induce target gene expression. Therefore, we generated Gli1;Ptch1 tumors which additionally expressed a GFP-tagged, dominant-negative form of Mastermind-like 1 (Gli1;Ptch1;dnMAML). Alternatively, we also generated tumors which conditionally deleted Rbp-j (Gli1;Ptch1;Rbpj) to block canonical downstream activity.
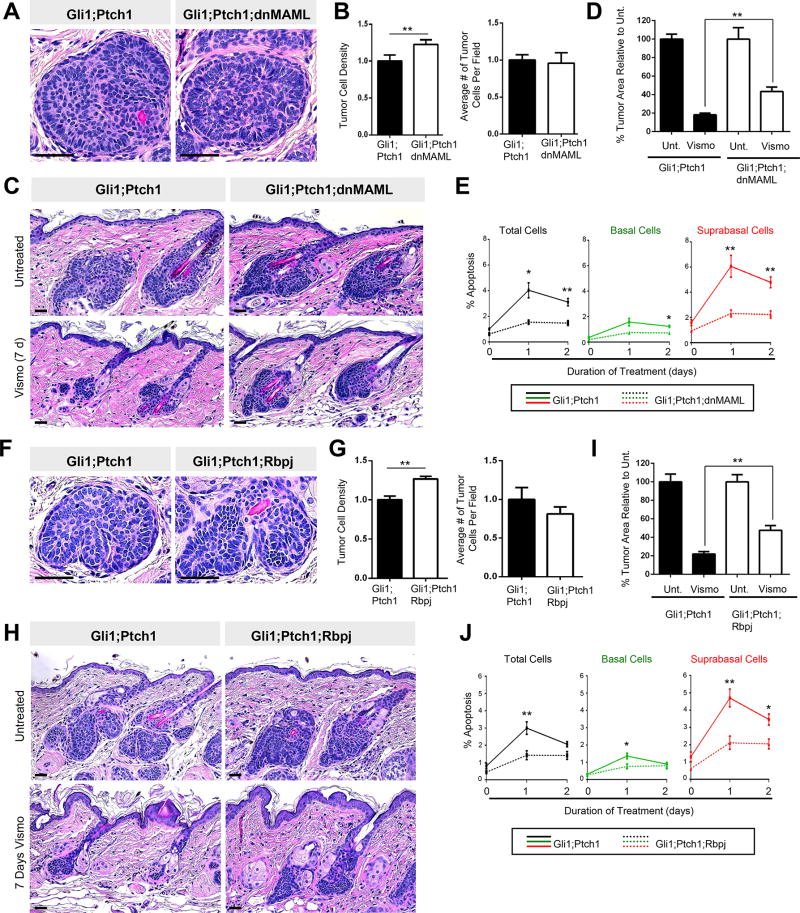
Histologically, Gli1;Ptch1;dnMAML tumors appeared slightly denser than control Gli1;Ptch1 tumors; however, dnMAML expression did not enhance tumor formation, since overall cell numbers were unaffected (Figure 4A–B). When we treated these tumors with vismo, we observed that dnMAML conferred partial protection against Hh inhibition, roughly doubling the number of residual tumor cells relative to control tumors (Figure 4C–D). This effect was not due to dnMAML-induced changes in proliferation, since vismo suppressed Ki67 in all tumors (Figure S4A). dnMAML expression also did not prevent vismo from inhibiting Hh signaling (Figure S4B). Rather, Gli1;Ptch1;dnMAML tumors possessed significantly fewer apoptotic cells, particularly in the suprabasal compartment, after vismo (Figure 4E). Finally, since dnMAML-expressing tumor cells are marked by GFP, we traced the fate of these cells relative to their non-fluorescent peers, which arose due to incomplete Cre-mediated recombination. Indeed, the proportion of dnMAML-expressing GFP+ tumor cells increased between 3–7 days after vismo, reinforcing the notion that blocking downstream Notch provides a survival advantage for these cells during treatment (Figure S4C–D).
Figure 4. Suppressing downstream Notch promotes tumor persistence.
A. Histology of Gli1;Ptch1 (left) and Gli1;Ptch1;dnMAML (right) tumors. B. Quantitation of tumor cell density (left) and number per field (right). C. Histology showing that dnMAML expression (right panels) partially protects tumors against vismo. D. Quantitation for C. Unt., untreated. E. Tumors expressing dnMAML (dotted lines) exhibit less apoptosis, both overall (black lines), as well as in the basal (green) and suprabasal (red) compartments, relative to control tumors (solid lines). F. Histology of Gli1;Ptch1 (left) and Gli1;Ptch1;Rbpj (right) tumors. G. Quantitation of tumor cell density (left) and number per field (right). H. Histology showing that deletion of Rbpj (right panels) partially protects tumors against vismo. I. Quantitation for H. J. Tumors which delete Rbpj (dotted lines) exhibit less apoptosis, both overall (black lines), as well as in the basal (green) and suprabasal (red) compartments, relative to control tumors (solid lines). Error bars, SE. *, p < 0.05; **, p < 0.01. Scale bars, 100 µm.
We made similar observations in Gli1;Ptch1;Rbpj tumors. As was seen with dnMAML expression, loss of Rbpj increased tumor density, although overall cell numbers were largely unchanged (Figure 4F–G). After 7 days of vismo treatment, roughly twice the number of residual tumors cells were observed in Gli1;Ptch1;Rbpj tumors, relative to control tumors (Figure 4H–I). As before, vismo effectively inhibited cell proliferation and Hh target gene expression regardless of Rbpj status; however, Gli1;Ptch1;Rbpj tumors displayed fewer apoptotic cells during treatment (Figure 4J and S4A–B). Therefore, in both Gli1;Ptch1;Rbpj and Gli1;Ptch1;dnMAML tumors, interfering with downstream Notch signaling neither affects tumor progression nor vismo’s ability to suppress Hh. Rather, these observations suggest that Notch specifically modulates BCC persistence upon challenge with Hh pathway antagonists.
Inhibiting upstream Notch promotes tumor persistence
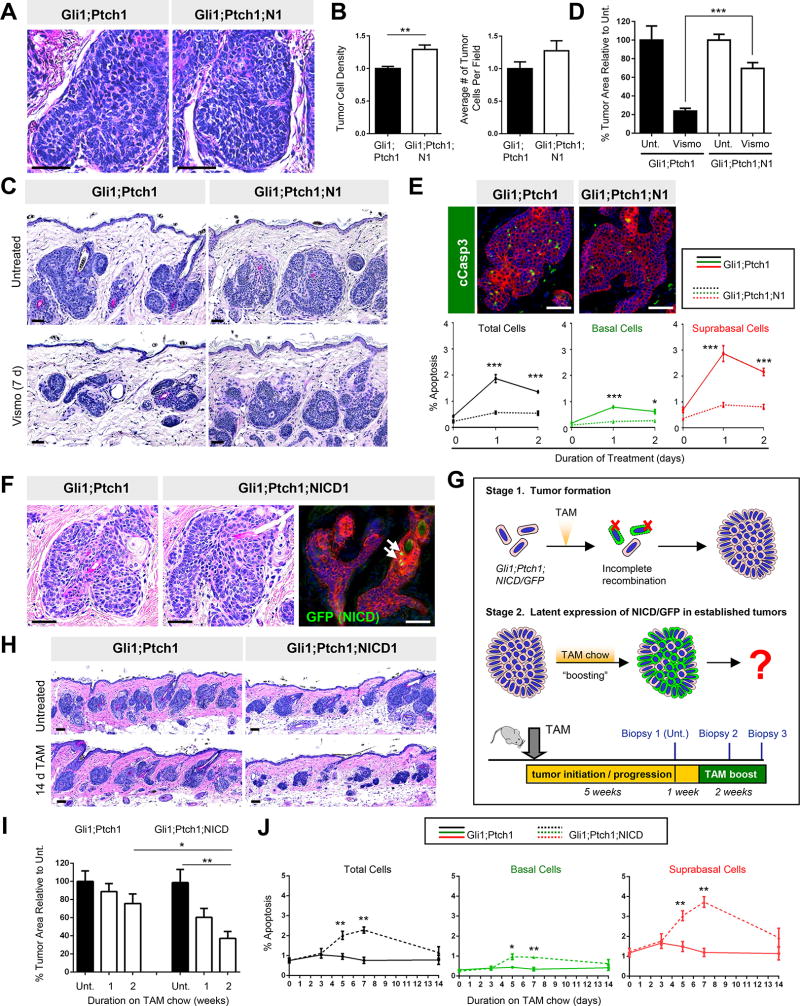
Although our findings suggest that manipulating downstream Notch can affect tumor-drug response, human BCCs more commonly possess mutations in genes encoding upstream components of the pathway, particularly NOTCH1 or NOTCH2 receptors (Bonilla et al., 2016; Jayaraman et al., 2014). Therefore, to better recapitulate the genetic background underlying human disease, we generated Gli1;Ptch1 tumors that harbored conditional loss-of-function alleles for Notch1 (Gli1;Ptch1;N1).
Similar to inhibiting downstream Notch signaling, disrupting upstream Notch1 did not affect tumor progression (Figure 5A–B). As before, Gli1;Ptch1;N1 tumors stopped proliferating and downregulated Hh target genes in response to vismo (Figure S4A–B). However, after 7 days of vismo treatment, Gli1;Ptch1;N1 tumors hardly regressed and were ~3-fold larger than control Gli1;Ptch1 tumors (Figure 5C–D). Consistent with results seen in dnMAML and Rbpjflox tumors, this effect was associated with a strong reduction in vismo-induced apoptosis in Gli1;Ptch1;N1 tumors, relative to control tumors (Figure 5E). These findings suggest that Notch1 loss-of-function mutations commonly seen in human BCCs can promote tumor persistence, independent of Hh-driven drug resistance.
Figure 5. Deleting Notch1 promotes tumor persistence, while NICD overexpression regresses established tumors.
A. Histology of Gli1;Ptch1 (left) and Gli1;Ptch1;N1 (right) tumors. B. Quantitation of tumor cell density (left) and number (right). C. Histology showing that Notch1 deletion (right panels) protects tumors against vismo. D. Quantitation for C. Unt., untreated. E. Top, tumors that delete Notch1 are less apoptotic (green) after 1 day of vismo. Bottom, quantitation of overall apoptosis levels (black), or subdivided by basal (green) and suprabasal (red) compartments. F. Histology showing that tumors harboring a Notch1 activated allele (Gli1;Ptch1;NICD) (middle) resemble control tumors (left). Right, only rare cells expressed GFP (green, arrows) in Gli1;Ptch1;NICD tumors, indicating incomplete recombination. G. Strategy for tamoxifen (TAM) “boosting” to induce latent expression of NICD/GFP (green) in established tumors. H. Histology of Gli1;Ptch1;NICD tumors (right) and control tumors (left) after 2 weeks of tamoxifen boosting. I. Quantitation of tumor area after boosting. J. TAM boosting induces apoptosis in Gli1;Ptch1;NICD tumors (dotted lines), with more subtle effects in control tumors (solid lines). Overall tumor apoptosis rates are shown (black), and subdivided between basal (green) and suprabasal (red) compartments. Red, co-staining for K5. Error bars, SE. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Scale bars, 100 µm. See also Figure S4.
Latent Notch activation regresses established tumors
If disrupting Notch protects BCCs against anti-Hh therapy, can activating Notch eliminate tumors? Since pharmacological in vivo Notch agonists have not been described, we generated Gli1;Ptch1 mice that also expressed a conditional activated allele of Notch1 (NICD) harboring an IRES-driven GFP reporter (Gli1;Ptch1;NICD). These tumors developed similarly to Gli1;Ptch1 tumors, but upon staining for GFP, we determined that fluorescent cells were rarely visible in Gli1;Ptch1;NICD tumors (Figure 5F). This suggests that activated Notch may be incompatible with tumor initiation, and that the tumors which did form failed to undergo full recombination to express NICD/GFP.
To circumvent this problem, we took advantage of the fact that non-fluorescent Gli1;Ptch1;NICD tumors nonetheless continued to harbor the conditional NICD/GFP allele (Figure 5G). Furthermore, these tumors also likely continued expressing Gli1-CreERT2, since Gli1 is a canonical Hh target gene. We therefore predicted that if we allowed Gli1;Ptch1;NICD tumors to form over 5 weeks, subsequent “boosting” with tamoxifen would cause latent recombination at the NICD/GFP locus (Figure 5G). Indeed, we determined that tamoxifen boosting induced GFP expression in established Gli1;Ptch1;NICD tumors (Figure S4E). This was accompanied by increased apoptosis, which caused tamoxifen-boosted Gli1;Ptch1;NICD tumors to regress by ~60% over 2 weeks of treatment (Figure 5H–J and S4F). Tamoxifen boosting also slightly regressed control Gli1;Ptch1 tumors, but to a lesser extent than tumors which expressed NICD/GFP (Figure 5I). In total, these findings indicate that Notch signaling is highly correlated with tumor-drug response: Low Notch levels protect tumors against therapy, whereas high level Notch causes cell death.
Hh signaling modulates Notch activity
Up to this point, we have shown that Gli1;Ptch1 tumors possess a bi-compartmental organization that can influence whether cells undergo apoptosis or persist in response to therapy. Given the complementary molecular features that define these compartments—Hh+++/Notch− tumor basal cells, and Hh+/Notch+ suprabasal cells—we next asked whether these 2 pathways might act in opposition to each other.
To assess whether Notch can impinge upon Hh, we began by examining Hh target gene expression in our 3 Gli1;Ptch1 tumor models where Notch was inhibited. In spite of highly efficient deletion of Rbpj or Notch1, or heterogeneous overexpression of dnMAML (Figure S5A–B, S4C), these untreated tumors retained a clear basal:suprabasal bias in Hh target gene expression, similar to control Gli1;Ptch1 tumors (Figure 6A). Notch inhibition also did not alter tumor cell polarity, and we again confirmed that these tumors were comprised of cells that had fully deleted Ptch1 (Figure S5C–D). Together, this suggests that manipulating Notch does not affect Hh in these tumors.
Figure 6. Hh signaling opposes Notch activation.
A. Hh pathway activity, assessed by Ptch2, is unaffected in Gli1;Ptch1 tumors when Notch is suppressed, as indicated. B. Gli1;SmoM2 tumors display low level Hh pathway activation, as assessed by Ptch2 (top left). Gli1;Ptch1 tumors display intermediate Hh (top middle), while tumors expressing Keratin 5 promoter-driven Gli2 (K5;Gli2) display high Hh activity (top right). Lower panels, the abundance of NICD+ cells (green) is inversely associated with Hh activity in these tumors. C. Quantitation of NICD+ cells in different models, subdivided by basal (green) and suprabasal (red) compartments. D. Gli1;Ptch1 tumors treated with vismo display increased NICD+ cells. E. Quantitation of NICD+ cells in the tumor basal (green) and suprabasal (red) compartments, in untreated (Unt.) and vismo-treated tumors. Red, co-staining for K5. Error bars, SE. *, p < 0.05; **, p < 0.01; n.s., not significant. Scale bars, 100 µm. See also Figure S5.
To test the alternative possibility that Hh inhibits Notch, we assessed NICD levels in 3 independent mouse models of BCC that vary in Hh signaling amplitude. Previous studies have demonstrated that tumors induced by mutant GLI2 overexpression display high level Hh pathway activity, whereas SmoM2-induced tumors display far weaker downstream signaling (Grachtchouk et al., 2011). We confirmed these differences, while further noting that Gli1;Ptch1 tumors displayed an intermediate level of Hh activation (Figure 6B). Using this panel of tumors, we observed that Notch activation was inversely associated with Hh activity: Gli1;SmoM2 tumors with low level Hh were abundantly populated with NICD+ cells; Gli1;Ptch1 tumors with intermediate Hh also displayed intermediate numbers of NICD+ cells; and mutant GLI2-induced tumors with high level Hh possessed few NICD+ cells (Figure 6B–C). In all cases, these differences in NICD occurred specifically in the tumor suprabasal compartments, whereas the tumor basal compartments exhibited low levels of NICD, regardless of Hh signaling amplitude.
Extending these findings further, we wondered whether reducing Hh might elevate Notch. We therefore compared Gli1;Ptch1 tumors before and during vismo, and indeed observed an increased percentage of NICD+ cells in the suprabasal compartment of Hh-inhibited tumors (Figure 6D–E). In contrast, basal tumor cells exhibited subtler differences in Notch upon vismo treatment. Altogether, these experiments provide evidence that Hh signaling can suppress Notch signaling in the tumor suprabasal compartment, whereas basal tumor cells possess low level Notch regardless of Hh status.
Human BCCs exhibit compartment-specific marker expression
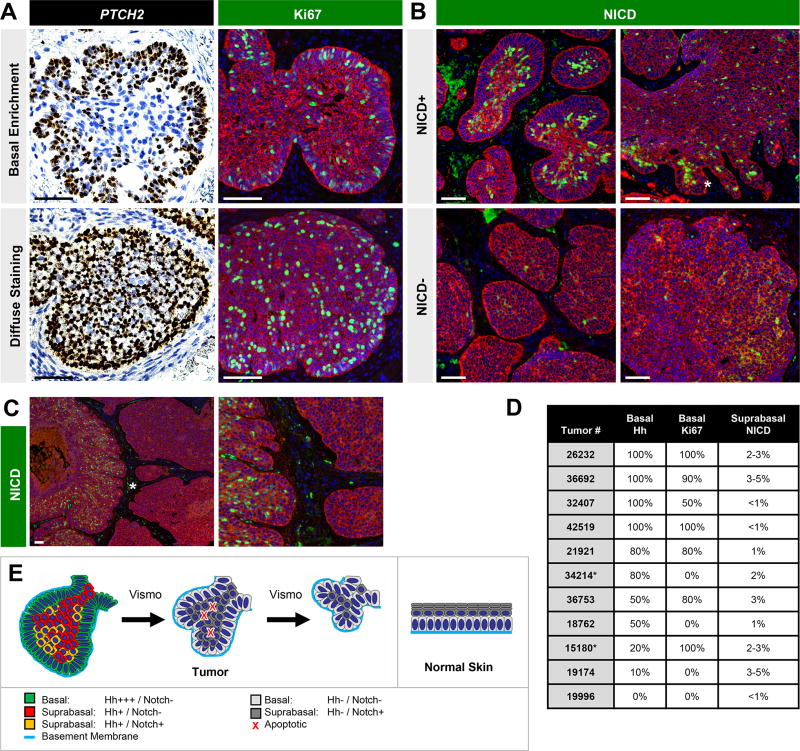
Finally, in order to place our findings in the context of human tumors, we characterized Hh, Notch and cell proliferation in a cohort of 11 untreated nodular BCCs. Consistent with our observations in Gli1;Ptch1 tumors, 6/11 BCCs displayed enriched Hh target gene expression at the periphery (Figure 7A). In contrast, diffuse Hh activation was seen in 3/11 cases, while 2 tumors displayed a mixed pattern. In 6/11 BCCs, we also noticed enhanced cell proliferation at the tumor periphery (Figure 7A). 4/11 tumors exhibited proliferation throughout the tumor, and 1 tumor was comprised of a roughly equal mix of basally-enriched and non-enriched Ki67. A similar distribution of tumors displaying biased or unbiased Hh pathway activation and proliferation was also seen in a second, independent cohort of human BCCs that were either untreated or treated with vismo (Figure S6). Finally, we confirmed that the ubiquitous gene POLR2A did not exhibit biased expression in any tumor compartment (Figure S6B).
Figure 7. Human BCCs exhibit compartment-specific marker expression.
A. Single molecule in situ staining for PTCH2 (brown, left panels) or IHC for Ki67 (green, right panels) in untreated human nodular BCCs. Top row shows tumors with basally enriched staining, whereas bottom row shows tumors with diffuse staining. Red, IHC for K5. B. Top row, some tumors contain concentrated areas of NICD+ suprabasal cells (green), whereas other regions are sparse for NICD+ cells (bottom row). Top right, abundant NICD+ cells (asterisk) in regions extending from a tumor mass. C. Tumor containing NICD+ nodule directly adjacent to NICD− nodule. Right box is a magnified view of the area denoted by the asterisk. D. Table summarizing findings. As all BCCs were comprised of multiple discrete tumor nodules, an estimate of the percent of total tumor periphery displaying basally enriched Hh or Ki67, or the overall percent of tumor cells displaying suprabasal NICD, is shown. As an example for Hh or Ki67, an estimate of “80%” indicates that 80% of the total tumor periphery displayed basal enrichment for a given marker, whereas the remaining 20% displayed diffuse staining. Asterisk, nodular tumors admixed with infiltrative regions. E. Resemblance of BCC tumor architecture (left) with architecture of normal skin (right). In both cases, basement membrane contact likely confers basal cell polarity and modulates Notch signaling. Suprabasal cells that do not contact the basement membrane typically activate Notch. In BCC, vismo treatment suppresses Hh signaling and increases the proportion of Notch-activated suprabasal cells, some of which undergo apoptosis to cause tumor shrinkage. Scale bars, 100 µm. See also Figure S6.
Regarding Notch, we observed that 6/11 untreated tumors from the first cohort possessed NICD+ cells specifically in the suprabasal compartment, whereas NICD was largely absent from 3/11 BCCs (Figure 7B). It should be noted that even for tumors scored as positive for Notch, NICD+ cells comprised a minority population (~2–5% overall), but were often unevenly distributed and highly concentrated (>25% of cells locally) in smaller tumor islands or outgrowths extending out from larger masses (Figure 7B). In some cases, tumor nodules containing abundant NICD+ suprabasal cells were situated directly adjacent to nodules which completely lacked NICD, possibly reflecting the extensive genetic heterogeneity of these tumors (Sharpe et al., 2015) (Figure 7C). As summarized in Figure 7D–E, these findings indicate that a sizeable fraction of human BCCs manifest compartment-specific expression of Hh, Notch and cell proliferation markers, consistent with our observations in Gli1;Ptch1 tumors.
DISCUSSION
Although pharmacological Hh pathway inhibitors are effective at treating BCC, these therapies by themselves are rarely curative. Drug resistance is observed in a significant fraction of tumors, and is invariably associated with high level Hh pathway activity even in the face of treatment (Basset-Seguin et al., 2015). Distinct from drug resistance, tumor persistence, the focus of our studies, represents an even more common problem. Persistent tumors exist in a largely dormant state characterized by suppressed Hh signaling and the potential to reactivate growth upon drug withdrawal. Since even clinically resolved tumors can reappear at their original sites after treatment is stopped (Tang et al., 2016; Tang et al., 2012), drugs such as vismo must be administered continuously to reliably prevent recurrence.
Consistent with findings in other BCC mouse models (Hutchin et al., 2005; Williams et al., 2003), we have shown here that Hh suppression causes rapid cell death in Gli1;Ptch1 tumors. Following an initial phase of tumor shrinkage, however, apoptosis is rarely detected in residual tumors. One reason why tumors persist may be due to incomplete Hh pathway suppression, as has been observed in some human BCCs that nonetheless exhibited a clinical response to vismo (Atwood et al., 2015) (also Figure S6C). In Gli1;Ptch1 mice, escalating drug dosages yielded a more thorough downregulation of Hh pathway activity, but even at the highest doses, residual target gene expression could still be detected (Figure S2). Whether complete Hh suppression can eliminate persistent tumor cells remains unclear; however, we also observed that increasing drug dosages were associated, after a certain point, with diminishing returns in the ability to abolish residual tumors (Figure 1C). These findings are concordant with previous studies showing that suppression of Hh signaling in a doxycycline-inactivating model of BCC yielded residual tumor cells that persisted for at least 5 months (Hutchin et al., 2005). Although further studies are needed, we speculate that vismo or other Hh pathway inhibitors may never fully eradicate all tumor cells, even if Hh is completely blocked. Given that Hh suppression also does not directly induce apoptosis in vitro (Figure S1C–D), together, these observations suggest that BCCs may not be “oncogenically addicted” to Hh in the conventional sense. Rather, the requirement for this pathway in promoting cell survival is likely a function of the 3-dimensional architecture of tumors grown in vivo.
For BCC, the typical architectural features of this tumor include a bi-compartmental structure where palisaded basal cells encapsulate less polarized suprabasal cells at the interior. Tumor basal cells in contact with the basement membrane display high level Hh pathway activity, and since Hh can function upstream of Wnt in BCC, this may explain why nuclear β-catenin is also enriched in this compartment (Yang et al., 2008; Youssef et al., 2012). Other markers, including those associated with the hair follicle (e.g. Edar, Runx1, Lrig1 and Sox9) and cell proliferation (e.g. Ki67), have similarly been reported to be elevated at the tumor edge (Quist et al., 2016; Yang et al., 2008; Youssef et al., 2012). Overall, these findings are highly reminiscent of normal skin structure (Figure 7E), where basal cell contact with the basement membrane is all-important for establishing normal skin architecture, conferring stem cell identity, and modulating Notch signaling (Joost et al., 2016; Watt et al., 2008).
Given the extensive mutational burden reported for BCC (Atwood et al., 2015; Bonilla et al., 2016; Jayaraman et al., 2014; Sharpe et al., 2015), genetic factors likely also modulate drug response. In this regard, deep sequencing studies have recently found that aberrations in NOTCH1/2 are among the most commonly detected mutations in BCC and normal skin, even though these mutations are not thought to be drivers for this disease (Bonilla et al., 2016; Jayaraman et al., 2014; Martincorena et al., 2015). Consistent with this, we have previously observed that Notch-inhibited keratinocytes spread rapidly throughout the skin without forming tumors in mice (Vagnozzi et al., 2015). Since loss of Notch does not prevent vismo from inhibiting Hh signaling in Gli1;Ptch1 tumors, this suggests that Notch signaling specifically modulates tumor persistence, but not drug resistance.
In human BCCs, we found that some tumors displayed compartment-specific features resembling Gli1;Ptch1 tumors. These features included enhanced Hh signaling and cell proliferation in tumor basal cells, and Notch activation in a subset of suprabasal cells. While it remains unclear why some tumors manifest these characteristics, whereas others do not, previous studies have demonstrated that the magnitude of Hh pathway activation in mice can influence tumor phenotype, including whether cell proliferation is diffuse or confined to the periphery (Grachtchouk et al., 2011; Grachtchouk et al., 2003). At the same time, we observed that elevating Hh inhibits Notch specifically in suprabasal tumor cells, suggesting multiple mechanisms for tumors to suppress this pathway (Shi et al., 2017). How tumors achieve varying levels of Hh may depend on the nature of the instigating mutation in PTCH1 or SMO, as well as whether alterations in downstream components such as GLI2, SUFU and aPKC-ι/λ are present (Atwood et al., 2013; Reifenberger et al., 2005; Sharpe et al., 2015). Synergism with other pathways such as EGFR may also play a role (Eberl et al., 2012), and together all these components may act in concert with polarity proteins to shape the magnitude of Hh signaling as well as the consequent tumor phenotype.
Incorporating these different elements, our findings suggest a mechanistic basis for understanding how BCCs respond to treatment (Figure 7E). Peripheral palisaded basal cells are highly polarized, which we hypothesize may account for elevated Hh and decreased Notch. As these basal tumor cells detach from the basement membrane and fill the expanding tumor interior, these now-suprabasal cells exhibit reduced polarity and only modest Hh pathway activity, possibly causing elevated Notch. Upon further suppression of Hh by vismo, we speculate that this combination of factors—low level Hh, high level Notch, and absence of basement membrane contacts—ultimately dooms tumor suprabasal cells. In contrast, neighboring basal cells remain anchored to the basement membrane, do not highly activate Notch in spite of vismo-mediated Hh pathway suppression, and persist in a largely dormant state. Persistence may also be conferred by incomplete Hh suppression and/or loss-of-function mutations in Notch, thereby effectively blunting or delaying the overall drug response.
In light of all this, the next question becomes: What can be done to eradicate persistent tumor cells? Based on our findings, alternate first- or second-line approaches may one day entail formulating novel activators of Notch for topical or intralesional delivery to the skin; developing strategies for converting highly polarized basal cells into non-polarized suprabasal cells; or designing therapies that more thorough suppress Hh signaling, or that target the very survival pathways that tumors rely upon to subsist in dormancy. Until these approaches become realized, however, the issue of persistent tumor cells will likely remain a lingering problem for BCC therapy.
STAR METHODS TEXT
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sunny Wong (sunnyw@umich.edu).
dnMAML and Rpb-jflox mice were obtained under an MTA and can be distributed only after formal approval is granted from their institute of origin.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal Models
Gli1;Ptch1 mice were induced with 5 mg tamoxifen per 40 g body weight at 4.5 weeks of age. Five weeks later, dorsal skin biopsies were collected to serve as a baseline before treatment. One week subsequently, vismodegib was administered orally, typically at a dose of 50 mg/kg/day, diluted in 100 µL PEG 400/5% dextrose in water (75:25 v/v). Tamoxifen “boosting” was performed by switching mice onto chow containing 400 mg/kg tamoxifen citrate for 2 weeks. For information regarding animal strains, please refer to the Key Resources Table. All studies were performed on mice of both genders in a mixed genetic background, using littermate animals for comparisons when possible, and in accordance with regulations established by the University of Michigan Unit for Laboratory Animal Medicine.
Human Studies
Human BCCs were obtained with informed consent under IRB HUM00042233 (untreated nodular BCCs) and HUM00075822 (vismodegib-BCC patient study), in accordance with procedures approved by the Institutional Review Board at the UM Medical School. Biopsies were processed for histology and de-identified, and thus not regulated as per IRB guidelines (exempt IRB protocol HUM00051875).
Cell culture
Primary keratinocytes were isolated from newborn pups expressing Keratin 5 promoter-driven Cre recombinase and Ptch1flox/flox alleles, using standard techniques and as previously described (Peterson et al., 2015). Cells were grown in complete Cnt-57 keratinocyte media for 3–4 days at 37° C/5% CO2, then either maintained in complete media or switched to basal starve media (CnT-BM1.500) for an additional 2 days to induce Hh activation. Subsequently, cells were treated with either 2 µg/mL vismo for 2 days or 1 µM staurosporine (Santa Cruz) for 4.5 hours, both diluted into starve media. ASZ cells (Aszterbaum et al., 1999) were cultured in 154-CF media with 4% fetal bovine serum (6.7 parts chelexed:1 part unchelexed). These cells were starved in 154-CF media lacking serum, and treated with vismo or staurosporine, similar to primary keratinocytes. Sex of cells was not determined, as this is not a known determining factor for Hh signaling in vitro. ASZ cells were authenticated based on epithelial morphology and robust Hh pathway activity upon serum starvation, as previously reported (Aszterbaum et al., 1999).
METHOD DETAILS
Immunofluorescence
Skin biopsies were fixed in 3.7% formalin overnight for paraffin embedding. For frozen sections, samples were fixed in 3.7% paraformaldehyde at 4° C for 1 hour, rinsed in PBS, sunk in 30% sucrose overnight and embedded into OCT. Frozen sections were utilized for β4-integrin, GFP/YFP, Gli1 and K8/14/17 staining; all other markers were detected using paraffin sections. Antibodies against the following markers were diluted between 1:100–1:200 for staining: cCasp3, CD4, Foxp3, Ki67 and Rbpj. Antibodies against the following markers were diluted between 1:300–1:500 for staining: β4-integrin, Gli1, K8, K14, NICD and Notch1. Antibodies against the following markers were diluted between 1:1,000–1:1,500 for staining: Acetylated tubulin, GFP/YFP, Jag1 and K17. Antibodies against the following markers were diluted between 1:4,000–1:5,000 for staining: K5 and P53. For p53, NICD and Notch1 staining, signal amplification was performed for 2 minutes, 3 minutes or 4 minutes, respectively, using the TSA Fluorescein Plus kit following manufacturer’s instructions.
Western Blot
Cells were extracted in Laemmli sample buffer. SDS electrophoresis was performed using Mini-Protean TGX precast gels (BioRad #456-1096S), and Western blot was performed using standard protocols. Antibodies against cCasp3, cPARP and Gli1 were diluted 1:1,000 prior to use. Antibodies against Actin were diluted 1:5,000.
Flow cytometry and RNA analysis
Twenty-five weeks after tamoxifen induction, bulk keratinocytes were isolated from Gli1;SmoM2 mice by digesting dorsal skin in 0.25% trypsin overnight at 4° C. Keratinocytes were scraped from the dermis, and cells were resuspended in 0.1% BSA/PBS solution. This suspension was filtered through 100 and 40 µm strainers, and cell viability was determined by trypan blue staining (Invitrogen). Cells were resuspended at a concentration of 106 cells/100 µl and incubated with 0.15 µL antibodies against α6-integrin (CD49f conjugated to Alexa Fluor 647) per 100 µl cell suspension for 1 hour on ice with gentle shaking, and DAPI was added just prior to analysis. Cells from YFP-negative control mice were used to set gates for sorting YFP+ tumor cells. For each population, 30,000 cells were directly sorted into RLT buffer containing β-mercaptoethanol. RNA was isolated using the RNeasy Micro Kit (Qiagen), and half of the total yield was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Invitrogen). Quantitative RT-PCR was performed on 1:5 diluted cDNAs, using Power SYBR Green PCR Master Mix (Applied Biosystems). Please refer to the Supplemental Table for primer sequences. All experiments were performed on 3 independent biological replicates, individually normalized to Hprt loading control, and then averaged.
QUANTIFICATION AND STATISTICAL ANALYSIS
Tumor Measurements
For tumor area measurements, H&E images were taken spanning the entire biopsy (~1 cm) and quantitated using ImageJ software. The average tumor area per field was then calculated, and expressed as a single value for each animal/biopsy. Tumor regression was calculated by dividing ‘tumor area after treatment’ by ‘tumor area before treatment’ and expressed as a single value for each individual animal. Tumor cells in the basal or suprabasal compartment were scored visually, with basal cells defined as being located at the tumor periphery and in contact with the basement membrane, and suprabasal cells constituting all other tumor cells interior to basal cells. Tumor density was calculated by dividing K5+ tumor cell number by K5+ tumor area on stained sections. For each mouse, 4 random fields were assessed, and a single average tumor density value was calculated for each animal. Similarly, quantitation of apoptosis, proliferation, NICD, or dnMAML/YFP abundance was performed on 4 random fields per sample, using identical imaging conditions, and expressed as a percentage normalized to the overall number of K5+ tumor cells. For quantitating in situ staining, the Color Threshold function in ImageJ was first used to filter out background from the counterstain. Afterwards the tumor basal and suprabasal regions were denoted, and the total signal area from each region was measured using the Analyze Particles function in ImageJ. The total signal area was then divided by the total number of cells in each region, which were counted manually, to obtain the average signal area per cell in each region. Finally, these values were expressed relative to the tumor basal cell signal, which was set to ‘1’.
Statistics
All data are shown as means from independent biological replicates, and all in vivo tumor drug studies utilized, at a minimum, 4 independent experimental and control samples. More typically, most studies utilized 5–7 independent animals per group. Unpaired t-tests were performed using GraphPad Prism 6, and results from these tests are indicated in the Figures and Figure Legends.
Supplementary Material
SIGNIFICANCE.
Although therapeutics targeting the Hedgehog signaling pathway are highly effective at treating basal cell carcinoma (BCC), cancer cells frequently persist and regenerate the primary tumor once treatment is stopped. Our findings suggest that persistent cancer cells likely originate from the tumor periphery and display low Notch pathway activity. In contrast, cancer cells located at the interior of the tumor mass activate Notch and are efficiently eliminated by drug treatment. These findings suggest that the cellular architecture of BCC, likely determined by contact with the surrounding basement membrane, may influence whether tumor cells persist or are destroyed in response to therapy.
HIGHLIGHTS.
Hedgehog pathway inhibitors are effective against BCC, but tumor cells often persist.
Tumor basal and suprabasal cells differ in gene expression and drug response.
Inhibiting Notch promotes tumor persistence, but not drug resistance, upon treatment.
Latently activating Notch is sufficient to regress already-established tumors.
Acknowledgments
We are grateful to Drs. L. Samuelson, B. Allen, X. Fan and I. Maillard for sharing mice. S.Y.W. acknowledges the support of the NIH (R21CA209166, R00AR059796, R01AR065409); the UM Department of Dermatology; the Biological Sciences Scholars Program; the Center for Organogenesis; and the John S. and Suzanne C. Munn Cancer Fund. A.A.D. was funded by the NIH (R01CA87837 and P30CA046592). M.L.F. was funded by a Dermatology Foundation Clinical Career Development Award in Dermatologic Surgery.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization and Methodology, M.E. and S.Y.W.; Investigation, M.E., D.M., J.B.S., M.E.V., M.L.F., and S.Y.W.; Formal Analysis, P.W.H.; Writing – Original Draft, Review & Editing: M.E. and S.Y.W.; Funding Acquisition, M.L.F., A.A.D., and S.Y.W.; Resources, M.E.V. and P.W.H.; Supervision, A.A.D. and S.Y.W.
DECLARATION OF INTERESTS
The authors have nothing to declare.
References
- Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong HA, Lai K, Ahn R, Corbin K, Lowe MM, Scharschmidt TC, et al. Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell. 2017;169:1119–1129. doi: 10.1016/j.cell.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszterbaum M, Epstein J, Oro A, Douglas V, LeBoit PE, Scott MP, Epstein EH., Jr Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat. Med. 1999;5:1285–1291. doi: 10.1038/15242. [DOI] [PubMed] [Google Scholar]
- Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase Cl/gamma regulates the growth of basal cell carcinomas. Science. 2013;494:484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Sarin KY, Whitson RJ, Li JR, Kim G, Rezaee M, Ally MS, Kim J, Yao C, Chang AL, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27:342–353. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelson M, Liu K, Jiang X, He K, Wang J, Zhao H, Kufrin D, Palmby T, Dong Z, Russell AM, et al. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin. Cancer Res. 2013;19:2289–2293. doi: 10.1158/1078-0432.CCR-12-1956. [DOI] [PubMed] [Google Scholar]
- Basset-Seguin N, Sharpe HJ, de Sauvage FJ. Efficacy of Hedgehog pathway inhibitors in basal cell carcinoma. Mol. Cancer Ther. 2015;14:633–641. doi: 10.1158/1535-7163.MCT-14-0703. [DOI] [PubMed] [Google Scholar]
- Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, Seplyarskiy VB, Sharpe HJ, McKee T, Letourneau A, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- Brinkhuizen T, Reinders MG, van Geel M, Hendriksen AJ, Paulussen AD, Winnepenninckx VJ, Keymeulen KB, Soetekouw PM, van Steensel MA, Mosterd K. Acquired resistance to the Hedgehog pathway inhibitor vismodegib due to smoothened mutations in treatment of locally advanced basal cell carcinoma. J. Am. Acad. Dermatol. 2014;71:1005–1008. doi: 10.1016/j.jaad.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Chang AL, Oro AE. Initial assessment of tumor regrowth after vismodegib in advanced Basal cell carcinoma. Arch. Dermatol. 2012;148:1324–1325. doi: 10.1001/archdermatol.2012.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl M, Klingler S, Mangelberger D, Loipetzberger A, Damhofer H, Zoidl K, Schnidar H, Hache H, Bauer HC, Solca F, et al. Hedgehog-EGFR cooperation response genes determine the oncogenic phenotype of basal cell carcinoma and tumor-initiating pancreatic cancer cells. EMBO Mol. Med. 2012;4:218–233. doi: 10.1002/emmm.201100201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat. Rev. Cancer. 2008;8:743–754. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailani MR, Ståhle-Bäckdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, Undén AB, Dean M, Brash DE, Bale AE, et al. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat. Genet. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- Grachtchouk M, Pero J, Yang SH, Ermilov AN, Michael LE, Wang A, Wilbert D, Patel RM, Ferris J, Diener J, et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J. Clin. Invest. 2011;121:1768–1781. doi: 10.1172/JCI46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachtchouk V, Grachtchouk M, Lowe L, Johnson T, Wei L, Wang A, de Sauvage F, Dlugosz AA. The magnitude of hedgehog signaling activity defines skin tumor phenotype. EMBO J. 2003;22:2741–2751. doi: 10.1093/emboj/cdg271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- Holtz AM, Peterson KA, Nishi Y, Morin S, Song JY, Charron F, McMahon AP, Allen BL. Essential role for ligand-dependent feedback antagonism of vertebrate hedgehog signaling by PTCH1, PTCH2 and HHIP1 during neural patterning. Development. 2013;140:3423–3434. doi: 10.1242/dev.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchin ME, Kariapper MS, Grachtchouk M, Wang A, Wei L, Cummings D, Liu J, Michael LE, Glick A, Dlugosz AA. Sustained Hedgehog signaling is required for basal cell carcinoma proliferation and survival: conditional skin tumorigenesis recapitulates the hair growth cycle. Genes Dev. 2005;19:214–223. doi: 10.1101/gad.1258705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman SS, Rayhan DJ, Hazany S, Kolodney MS. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J. Invest. Dermatol. 2014;134:213–220. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH, Jr, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lönnerberg P, Linnarsson S, Kasper M. Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 2016;3:221–237. doi: 10.1016/j.cels.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper M, Jaks V, Are A, Bergström Å, Schwäger A, Barker N, Toftgård R. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc. Natl. Acad. Sci. USA. 2011;108:4099–4104. doi: 10.1073/pnas.1014489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, McMahon AP. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Loo PV, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 2011;71:5057–5061. doi: 10.1158/0008-5472.CAN-11-0923. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Peterson SC, Eberl M, Vagnozzi AN, Belkadi A, Veniaminova NA, Verhaegen ME, Bichakjian CK, Ward NL, Dlugosz AA, Wong SY. Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches. Cell Stem Cell. 2015;16:400–412. doi: 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten F, Berg C, Ahmadian A, Ren ZP, Nister M, Lundeberg J, Uhlen M, Ponten J. Molecular pathology in basal cell cancer with p53 as a genetic marker. Oncogene. 1997;15:1059–1067. doi: 10.1038/sj.onc.1201435. [DOI] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pricl S, Cortelazzi B, Col VD, Marson D, Laurini E, Fermeglia M, Licitra L, Pilotti S, Bossi P, Perrone F. Smoothened (SMO) receptor mutations dictate resistance to vismodegib in basal cell carcinoma. Mol. Oncol. 2015;9:389–397. doi: 10.1016/j.molonc.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quist SR, Eckardt M, Kriesche A, Gollnick HP. Expression of epidermal stem cell markers in skin and adnexal malignancies. Br. J. Dermatol. 2016;175:520–530. doi: 10.1111/bjd.14494. [DOI] [PubMed] [Google Scholar]
- Reifenberger J, Wolter M, Knobbe CB, Köhler B, Schönicke A, Scharwächter C, Kumar K, Blaschke B, Ruzicka T, Reifenberger G. Somatic mutations in the Ptch, Smoh, Sufuh and Tp53 genes in sporadic basal cell carcinomas. Br. J. Dermatol. 2005;152:43–51. doi: 10.1111/j.1365-2133.2005.06353.x. [DOI] [PubMed] [Google Scholar]
- Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med. 2012;366:2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Sequin N, Modrusan Z, Januario T, Tsui V, Durham AB, Dlugosz AA, Haverty PM, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27:327–341. doi: 10.1016/j.ccell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi FT, Yu M, Zloty D, Bell RH, Wang E, Akhoundsadegh N, Leung G, Haegert A, Carr N, Shapiro J, et al. Notch signaling is significantly suppressed in basal cell carcinomas and activation induces basal cell carcinoma cell apoptosis. Mol. Med. Rep. 2017 doi: 10.3892/mmr.2017.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvara H, Kalthoff F, Meingassner JG, Wolff-Winiski B, Aschauer H, Kelleher JF, Wu X, Pan S, Mickel L, Schuster C, et al. Topical treatment of basal cell carcinomas in nevoid basal cell carcinoma syndrome with a smoothened inhibitor. J. Invest. Dermatol. 2011;131:1735–1744. doi: 10.1038/jid.2011.48. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JY, Ally MS, Chanana AM, Mackay-Wiggan JM, Aszterbaum M, Lindgren JA, Ulerio G, Rezaee MR, Gildengorin G, Marji J, et al. Inhibition of the hedgehog pathway in patients with basal-cell nevus syndrome: final results from the multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2016;17:1720–1731. doi: 10.1016/S1470-2045(16)30566-6. [DOI] [PubMed] [Google Scholar]
- Tang JY, Mackay-Wiggan JM, Aszterbaum M, Yauch RL, Lindgren J, Chang K, Coppola C, Chanana AM, Marji J, Bickers DR, et al. Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N. Engl. J. Med. 2012;366:2180–2188. doi: 10.1056/NEJMoa1113538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, Pear WS. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhmann A, Dittmann K, Nitzki F, Dressel R, Koleva M, Frommhold A, Zibat A, Binder C, Adham I, Nitsche M, et al. The Hedgehog receptor Patched controls lymphoid lineage commitment. Blood. 2007;110:1814–1823. doi: 10.1182/blood-2007-02-075648. [DOI] [PubMed] [Google Scholar]
- Vagnozzi AN, Reiter JF, Wong SY. Hair follicle and interfollicular epidermal stem cells make varying contributions to wound regeneration. Cell Cycle. 2015;14:3408–3417. doi: 10.1080/15384101.2015.1090062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hoff DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- Wang GY, Wang J, Mancianti ML, Epstein EH., Jr Basal cell carcinomas arise from hair follicle stem cells in Ptch1+/− mice. Cancer Cell. 2011;19:1–11. doi: 10.1016/j.ccr.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: differentiation, cancer and adhesion. Curr. Opin. Cell Biol. 2008;20:171–179. doi: 10.1016/j.ceb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Guicherit OM, Zaharian BI, Xu Y, Chai L, Wichterle H, Kon C, Gatchalian C, Porter JA, Rubin LL, et al. Identification of a small molecule inhibitor of the hedgehog signaling pathway: effects on basal cell carcinoma-like lesions. Proc. Natl. Acad. Sci. USA. 2003;100:4616–4621. doi: 10.1073/pnas.0732813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Tanigaki K, Han H, Hiai H, Honjo T. Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr. Biol. 2003;13:333–338. doi: 10.1016/s0960-9822(03)00081-2. [DOI] [PubMed] [Google Scholar]
- Yang SH, Andl T, Grachtchouk V, Wang A, Liu J, Syu LJ, Ferris J, Wang TS, Glick AB, Millar SE, et al. Pathological responses to oncogenic Hedgehog signaling in skin are dependent on canonical Wnt/beta-catenin signaling. Nat. Genet. 2008;40:1130–1135. doi: 10.1038/ng.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Youssef KK, Lapouge G, Bouvree K, Rorive S, Brohee S, Appelstein O, Larsimont JC, Sukumaran V, de Sande BV, Pucci D, et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat. Cell Biol. 2012;14:1282–1294. doi: 10.1038/ncb2628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.