Abstract
Epigenetic aberrations are recognized as having pivotal roles in cancer etiology and progression. Histone deacetylases are among the most studied epigenetic modulators in various cancer types. The expression levels of class I histone deacetylase isoforms 1, 2, and 3 in patient-derived primary osteosarcoma cells (6 cases) was investigated, comparing them to normal bone graft-derived osteoblasts (6 cases) using the immunoblotting technique. Expression profiles of histone deacetylases in high-grade osteosarcoma tissue of 89 patients were examined and their association with clinicopathologic parameters and the patient survival was evaluated. Histone deacetylases were immunohistochemically stained on formalin-fixed paraffin-embedded biopsied tissue. Primary osteosarcoma cells expressed higher levels of histone deacetylase 1 and histone deacetylase 2, but lower levels of histone deacetylase 3 compared to benign osteoblasts. Overall, 82, 99, and 93% of 89 osteosarcomas showed nuclear expression of the histone deacetylase isoforms 1, 2, and 3, respectively. Low levels of histone deacetylase 1 were significantly associated with a high Enneking stage (P=0.014) and the presence of initial metastasis (P=0.040), while low levels of histone deacetylase 3 were significantly correlated with age >15 years (P=0.026). Univariate survival analysis found significantly shorter survival in the patients with a high Enneking stage (P<0.001), axial location (P=0.009), presence of initial metastasis (P<0.001), low-histone deacetylase 1 expression (P=0.038), and low-all-histone deacetylases expression (P=0.016). Multivariate survival analysis showed that only axial location (P=0.011) and low-all-histone deacetylases expression (P=0.039) were independent prognostic factors. In subgroup analysis of stage IIB patients (n=45), only axial location and low-all-histone deacetylases expression were associated with shorter survival in both univariate and multivariate analysis (axial location, P=0.008 and 0.010; low-all-HDACs, P=0.013 and 0.038, respectively). Low levels of all-histone deacetylases expression were significantly associated with advanced disease status and short survival. These findings may be a guide to future use of histone deacetylase inhibitors in osteosarcoma patients.
Osteosarcoma, a highly aggressive primary malignant bone tumor, accounts for 50% of all bone sarcomas and is frequently found in children and adolescents.1, 2 Although the 5-year disease-free survival of non-metastatic patients has improved to 70%, overall survival of patients with lung metastasis at initial diagnosis (~20% of patients) is only 30%.3, 4 Half the patients without initial metastasis go on to develop lung metastases within 36 months of diagnosis.5 Defining at risk groups is important for improving therapeutic plans for individuals. Clinicopathologic factors, including age, tumor site, tumor size, serum level of alkaline phosphatase, multidrug resistance, and so on, have been reported as prognostic indicators in non-metastatic osteosarcoma of the extremities.6
Histone deacetylases are a group of enzymes that regulate gene expression through removal of an acetyl group from the lysine residue of histones. Four classes of histone deacetylases and 18 histone deacetylase isoforms have been classified in mammalian cells. Class I histone deacetylases consist of histone deacetylase 1, 2, 3, and 8. Class II histone deacetylases are subdivided into class IIA (histone deacetylase 4, 5, 7, and 9) and class IIB (histone deacetylase 6 and 10). Class III histone deacetylases or sirtuins consist of SIRT1–7. The only member of Class IV histone deacetylases is histone deacetylase 11.7 Interactions of histone deacetylases with each other histone deacetylases and with other proteins are varied and diverse.7 Alteration of histone acetylation can induce specific gene activation or repression. Overexpression of histone deacetylases has been found to be an independent negative prognosis indicator in various cancers, including prostate, colorectal, breast, lung, liver, and gastric cancers.8 Some evidence has demonstrated that knockdown of histone deacetylases can induce cell cycle arrest, inhibit cancer cell proliferation and angiogenesis, as well as trigger apoptosis, differentiation and senescence.9
Four U.S. Food and Drug Administration (FDA)-approved histone deacetylase inhibitors, including three non-specific histone deacetylase inhibitors (vorinostat, belinostat, and panobinostat), affect all class I, II, and IV histone deacetylases and one specific Class I histone deacetylase inhibitor (romidepsin).7 Vorinostat and romidepsin are currently being used with T-cell lymphoma patients. Many other inhibitors are undergoing clinical trials in various cancers.8 Growing evidence from in vitro and in vivo animal studies has demonstrated that histone deacetylase inhibitors exert their anti-tumor activity in osteosarcoma both as a single drug10 and acting synergistically with other anticancer agents,11, 12, 13, 14 sensitize cancer cells to radiation,15 increase natural killer cell-mediated cytotoxicity16 as well as inducing cell differentiation.17 These experimental results suggest the probable future use of histone deacetylase inhibitors in osteosarcoma. However, little is known about the mechanisms of histone deacetylases in carcinogenesis and the expression patterns of histone deacetylase isoforms in actual human osteosarcoma tissues.
Understanding the expression profiling of histone deacetylases in osteosarcoma should provide greater insight into the roles of these enzymes in carcinogenesis as well as their association with clinical outcomes. The present study focused on exploring the expression patterns of class I histone deacetylase isoforms 1, 2, and 3 in osteosarcoma cells, and comparing that to benign osteoblasts as well as studying the expression patterns of histone deacetylases and their prognostic significance in a cohort of osteosarcoma patients.
Materials and methods
Patient Characteristics
Eighty-nine osteosarcoma patients diagnosed and treated at Maharaj Nakorn Chiang Mai Hospital, Thailand, between 2000 and 2015, were included in this study. Patients had been treated with a standard neoadjuvant regimen or underwent surgery at Maharaj Nakorn Chiang Mai Hospital and were followed-up for survival analysis until 30 June 2016. The Research Ethics Committee of the Faculty of Medicine, Chiang Mai University, approved the study. Clinicopathological parameters, including date of diagnosis, Enneking staging, location of tumor, tumor size, metastatic status, and percentage of tumor necrosis after chemotherapy were retrieved from hospital records and pathology reports as shown in Table 2. All primary biopsy slides were reviewed by a bone and soft tissue pathologist (JS).
Patient-Derived Osteosarcoma and Osteoblast Cells
Primary osteosarcoma and osteoblast cells were extracted following to previously reported protocol.18 Primary osteosarcoma cells were obtained from chemo-naïve tissues of 6 patients. The tissues were washed in phosphate-buffered saline (PBS), minced into small pieces and incubated in 5 mg/ml collagenase type I solution (Gibco, Boston, MA, USA) at 37°C for 18 h. Cell pellets were isolated by centrifugation at 1200 g for 5 min at room temperature. Cells were cultured in freshly prepared Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Gibco, Boston, MA, USA) at 37 °C in a humidified 5% CO2 incubator.
Primary osteoblast cells were obtained from bone grafts of six healthy participants who had been diagnosed with other non-cancer orthopedic conditions, and who had required autologous bone grafts for substitution procedures. Osteoblastic cells were extracted from bone samples by sequential collagenase type I-trypsin digestion. The cells were isolated by centrifugation at 1200 g for 5 min at room temperature and cultured in freshly prepared Dulbecco’s modified Eagle’s medium with fetal bovine serum at 37 °C in a humidified 5% CO2 incubator. The osteoblastic cells from the 2nd to 4th passages were used for characterization and proteins were extracted for western blotting analysis.
Characterization of Primary Cells
Growth rates of primary cells were measured using a hemocytometer within a specified time period and the doubling time of each cell was calculated. Expression of molecular markers was measured using a quantitative real-time polymerase chain reaction method.18 Osteogenic markers included collagen type I, osteonectin, and bone sialoprotein. Representative cancer markers were MMP-9 and collagen type X. Alkaline phosphatase (ALP) activity assay was conducted kinetically by monitoring the conversion of p-nitrophenyl phosphate (pNPP) to p-nitrophenol. Alizarin red-S histochemical analysis was performed to measure mineralization ability.
Immunohistochemistry and Scoring
Formalin-fixed paraffin-embedded tissues were obtained from archival paraffin blocks at the Department of Pathology, Faculty of Medicine, Chiang Mai University. Formalin-fixed paraffin-embedded tissues were immunostained using the Ventana automated straining system (Ventana Medical Systems, Tucson, AZ, USA). An Ultraview Universal DAB Detection Kit (Ventana Medical Systems, Tucson, AZ, USA), an indirect biotin-free system, was used to detect primary antibodies. Optimized conditions, including dilution, incubation time, and positive control tissues for each antibody, are shown in Table 1.19, 20, 21, 22
Table 1. List of antibodies and positive controls.
| Antibody | Immunoreactivity | Cat./ company | Dilution | Incubation (min) | Positive control tissue | Reference |
|---|---|---|---|---|---|---|
| Histone deacetylase 1 | Nucleus | ab19845/ Abcam | 1:10000 | 32 | Histone deacetylase 1,2: | 19 |
| Histone deacetylase 2 | Nucleus | ab39669/ Abcam | 1:1500 | 32 | Normal colon/ spleen/ liver | |
| Histone deacetylase 3 | Nucleus | ab32369/ Abcam | 1:1000 | 32 | Histone deacetylase 1,2,3: Normal kidney, pancreas Histone deacetylase 1: Tonsil Manufacturer | 20, 21, 22 |
Nuclear staining of each histone deacetylase isoform was evaluated (by PC and JS) without prior knowledge of clinical data using a semi-quantitative immunoreactive scoring system.23 The percentage of immunoreactive cells was estimated and scored as follows: negative=0, positive staining <10%=1, positive staining ≥10 and <33%=2, positive staining ≥33% and <66%=3, positive staining ≥66%=4. Intensity of staining was scored on a scale of 0–3: no color reaction=0, mild reaction=1, moderate reaction=2, and intense reaction=3. Immunoreactive score (IRS) was derived by multiplying immunoreactive cell scores and intensity of staining scores to compute an immunoreactive score ranging from 0 to 12.
Statistical Analysis
Statistical analyses were performed using STATA version 11. The significance of correlation between staining patterns of histone deacetylase isoforms and clinicopathologic data was tested using Student’s t-test or one-way ANOVA for parametric tests and the Mann–Whitney U test or the Kruskal–Wallis test for nonparametric tests. Association between histone deacetylase expression and overall survival of osteosarcoma patients was evaluated by univariate survival analysis using the Kaplan–Meier method together with the log-rank test. Multivariate survival analysis was carried out using Cox regression of proportional hazards to probe for significance at the 95% confidence interval (CI). P-values<0.05 were considered to be statistically significant.
Results
Expression of Histone Deacetylases in Primary Osteosarcoma and Osteoblast Cells
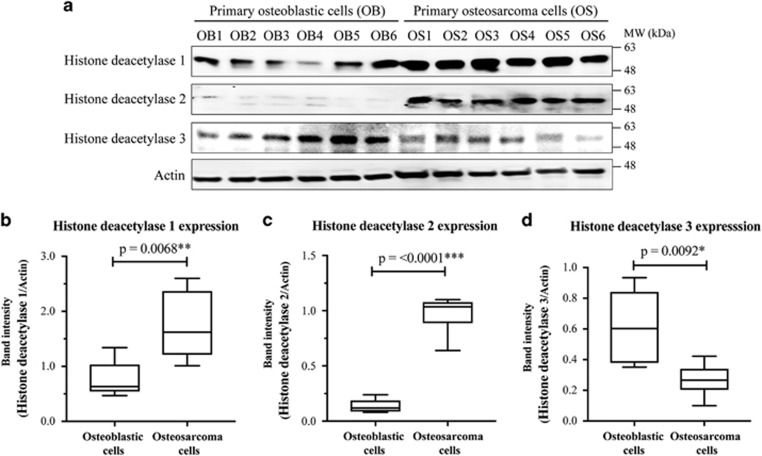
All isolated cells were characterized using specific markers and biological analysis to confirm their osteogenic and oncogenic properties (Supplementary data). Results using the western blotting technique showed that histone deacetylase isoforms 1 and 2 were statistically significantly upregulated in primary osteosarcoma cells compared to osteoblastic cells, whereas the expression levels of histone deacetylase 3 were the reverse (Figure 1).
Figure 1.
Expression of histone deacetylase isoforms in primary osteosarcoma and osteoblastic cells. (a) Representative western blots of histone deacetylase isoforms 1, 2, and 3 of primary osteoblastic (OB) and osteosarcoma (OS) cells. (b–d): Boxplots showing band intensities of histone deacetylase isoforms of individual samples relative to actin.
Expression Patterns of Histone Deacetylases in the Osteosarcoma Cohort
There were 89 osteosarcoma cases available with the clinicopathologic data and paraffin blocks. Overall, patient survival ranged from 2 to 166 months after initial diagnosis with a median of 18 months. Sixty patients died at 2–86 months. Median follow-up of surviving patients (n=29) was 52 months (range: 13–166 months). Their 1-year and 5-year survival rates were 70 and 32%, respectively.
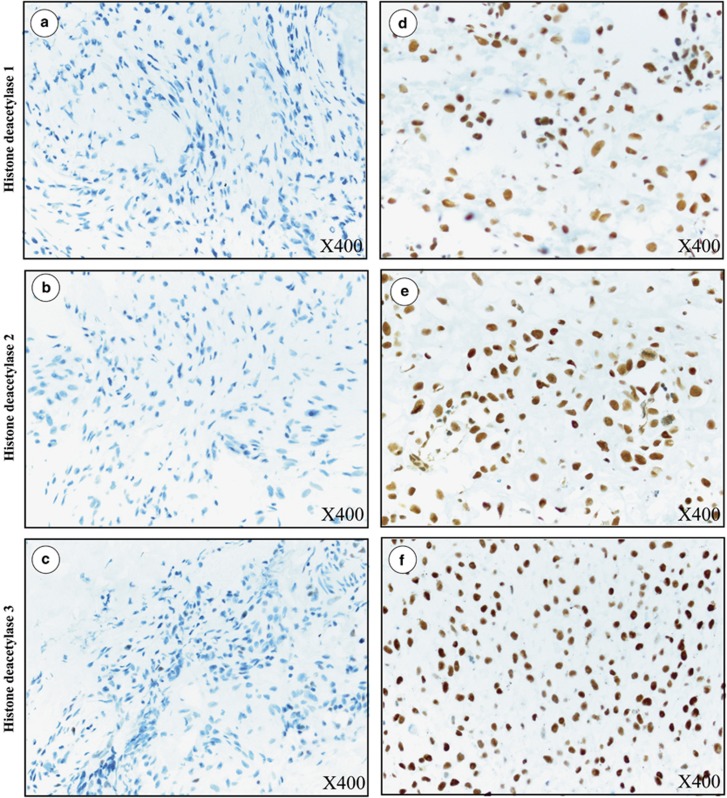
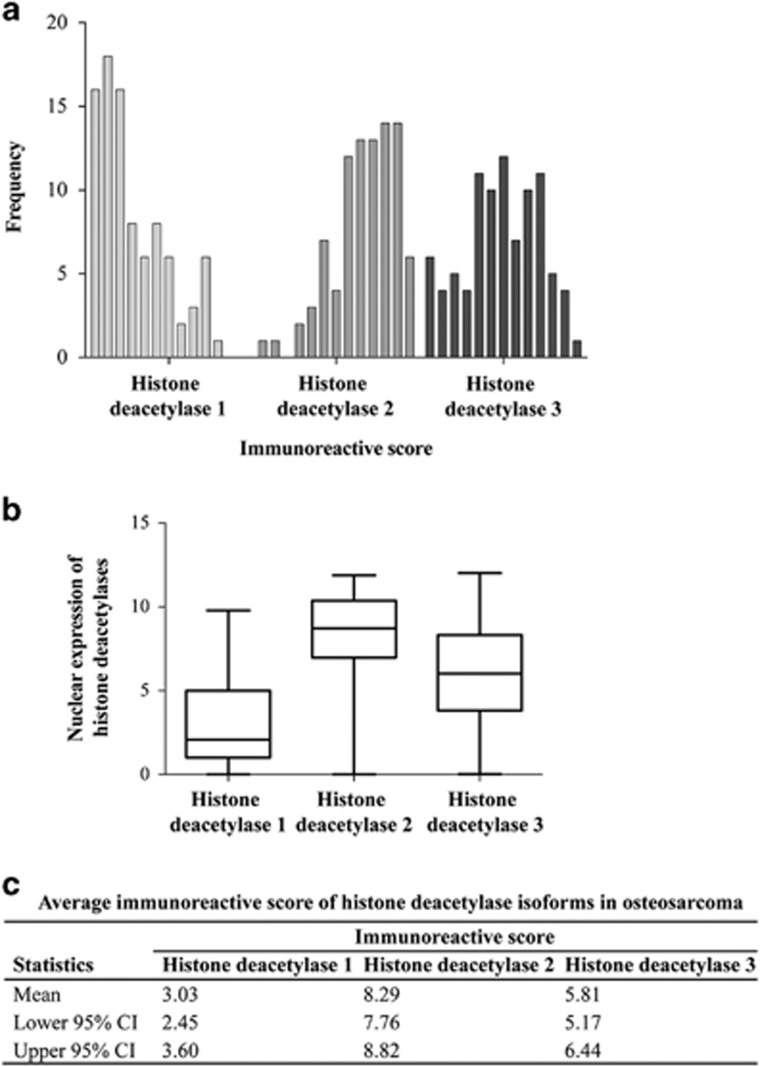
All three histone deacetylase isoforms, histone deacetylase 1, histone deacetylase 2, and histone deacetylase 3, were expressed predominantly in the nuclei of osteosarcoma cells (Figure 2). Cytoplasmic staining of histone deacetylase 2 was found to be partial in one case and strongly diffused in another (data not shown). Positive nuclear staining (Immunoreactive score>0) of histone deacetylase isoforms of all cases combined were 82% (histone deacetylase 1), 99% (histone deacetylase 2) and 93% (histone deacetylase 3). Distribution histone deacetylase expression is shown in Figure 3. Histone deacetylase 2 was the most highly expressed isoform in osteosarcoma, with 55% of all osteosarcoma tissues strongly stained (Immunoreactive score >8). Nuclear expression of histone deacetylase 1, on the other hand, was weak (Immunoreactive score 0 to 4) in 70% of cases.
Figure 2.
Immunohistochemical staining of histone deacetylase isoforms in osteosarcoma tissues (X400). (a–c) Negative histone deacetylase 1, histone deacetylase 2, histone deacetylase 3 (Immunoreactive score=0). (d–f) strong histone deacetylase 1 (Immunoreactive score=10), histone deacetylase 2 (Immunoreactive score=12) and histone deacetylase 3 (Immunoreactive score=12) nuclear staining.
Figure 3.
Nuclear staining scores of each histone deacetylase isoforms in the osteosarcoma cohort. (a) Frequency of osteosarcoma cases with differential immunoreactive score. (b) Levels of nuclear expression of histone deacetylase isoform 1, 2, and 3 in osteosarcoma. (c) Table summarizes average immunoreactive score of histone deacetylase isoforms in this studied cohort.
Correlation of Histone Deacetylase Expression and Clinicopathological Factors
Characteristics of osteosarcoma patients in this cohort are shown in Table 2. Significant variation of each histone deacetylase isoform was observed among individual histone deacetylase isoforms and clinicopathological factors. Expression levels of histone deacetylase 1 were significantly lower in the late Enneking stages (P=0.014) and with an initial metastatic phenotype (P=0.040). Expression of histone deacetylase 3 was significantly lower in adults (age >15 years) than in children (age ≤15 years) (P=0.0259).
Table 2. Characteristics of osteosarcoma patients in study cohort.
| Factor |
Nuclear expression (Immunoreactive scores, mean±SD) |
||||||
|---|---|---|---|---|---|---|---|
| All patients | Histone deacetylase 1 | P-value | Histone deacetylase 2 | P-value | Histone deacetylase 3 | P-value | |
| Age at diagnosis, years [mean=18.64±11.98, median=15 (range 5-73)] | |||||||
| ≤15 | 46 | 3.59±2.94 | 0.053 | 8.38±2.42 | 0.763 | 6.45±2.92 | 0.026* |
| >15 | 43 | 2.43±2.46 | 8.22±2.70 | 5.06±3.06 | |||
| Gender | |||||||
| Male | 50 | 3.07±2.91 | 0.860 | 8.14±2.58 | 0.493 | 5.95±3.06 | 0.597 |
| Female | 39 | 2.98±2.60 | 8.51±2.52 | 5.55±3.08 | |||
| Enneking stage | |||||||
| IIB | 45 | 3.91±3.04 | 0.014* | 8.66±2.27 | 0.125 | 5.63±2.90 | 0.406 |
| III | 34 | 2.29±2.22 | 7.78±2.78 | 5.97±3.20 | |||
| Site | |||||||
| Extremities | 78 | 3.30±2.83 | 0.050 | 8.37±2.50 | 0.833 | 5.92±3.05 | 0.581 |
| Axial | 8 | 1.32±1.17 | 8.17±2.18 | 5.37±3.31 | |||
| Tumor size, cm [mean=8.79±3.92, median=7.9 (range 1.1-21.4)] | |||||||
| ≤10 cm [Median=7.5 (range 1.1-10.0)] | 59 | 3.01±2.81 | 0.416 | 8.04±2.63 | 0.206 | 6.04±2.92 | 0.391 |
| >10 cm [Median=13.7 (range 10.2-21.4)] | 18 | 3.66±2.94 | 8.91±2.11 | 5.28±3.19 | |||
| Metastasis | |||||||
| At diagnosis | 34 | 2.29±2.22 | 0.040# | 7.78±2.78 | 0.270 | 5.97±3.20 | 0.627 |
| At follow-up | 21 | 3.80±3.36 | 8.44±2.48 | 5.41±2.45 | |||
| No | 24 | 4.01±2.81 | 8.84±2.10 | 5.82±3.28 | |||
| Chemoresistance | |||||||
| Good responders (Tumor necrosis≥90%) | 8 | 3.20±3.35 | 0.735 | 9.54±2.67 | 0.233 | 5.22±2.81 | 0.654 |
| Poor responders (Tumor necrosis<90%) | 50 | 3.62±3.02 | 8.44±2.34 | 5.78±2.93 | |||
P-values were calculated with *Mann–Whitney U test, and #Kruskal–Wallis test, P-value<0.05 shown in bold.
Survival Analysis of Expression Patterns of Class I Histone Deacetylases in High-grade Osteosarcoma
Immunoreactive score cutoff points for each histone deacetylase isoform were defined by receiver operating characteristic (ROC) curves: histone deacetylase 1, immunoreactive score=1; histone deacetylase 2, immunoreactive score=10; histone deacetylase 3, immunoreactive score=6. Combined expression profiles of three isoforms of histone deacetylase were combined into two groups: Group 1—none or some histone deacetylase isoforms expressed at immunoreactive score lower than the cu-off (71 cases, 80%), and Group2—all-histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff in (18 cases, 20%).
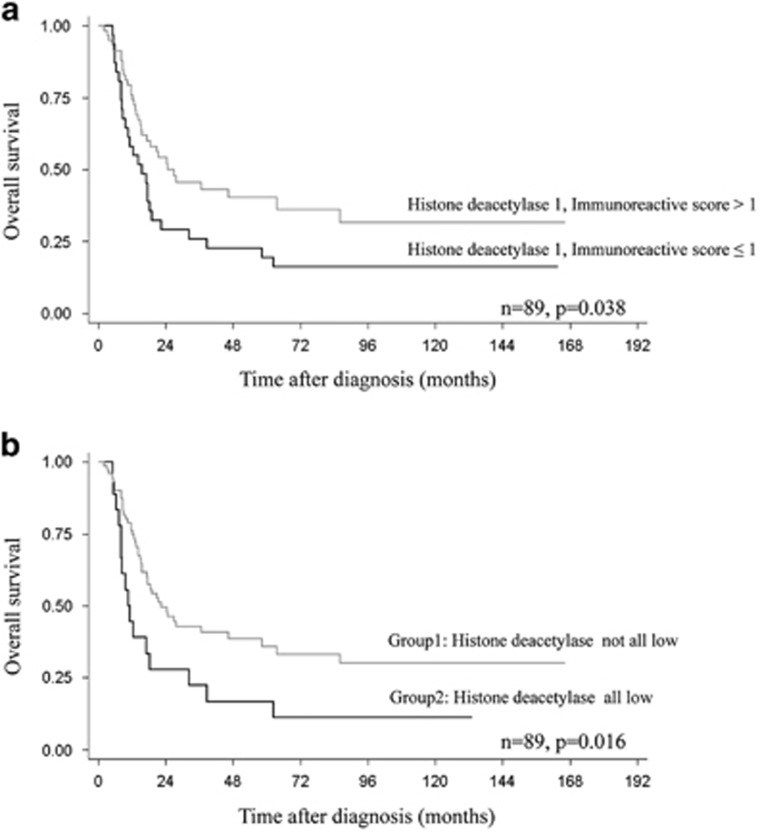
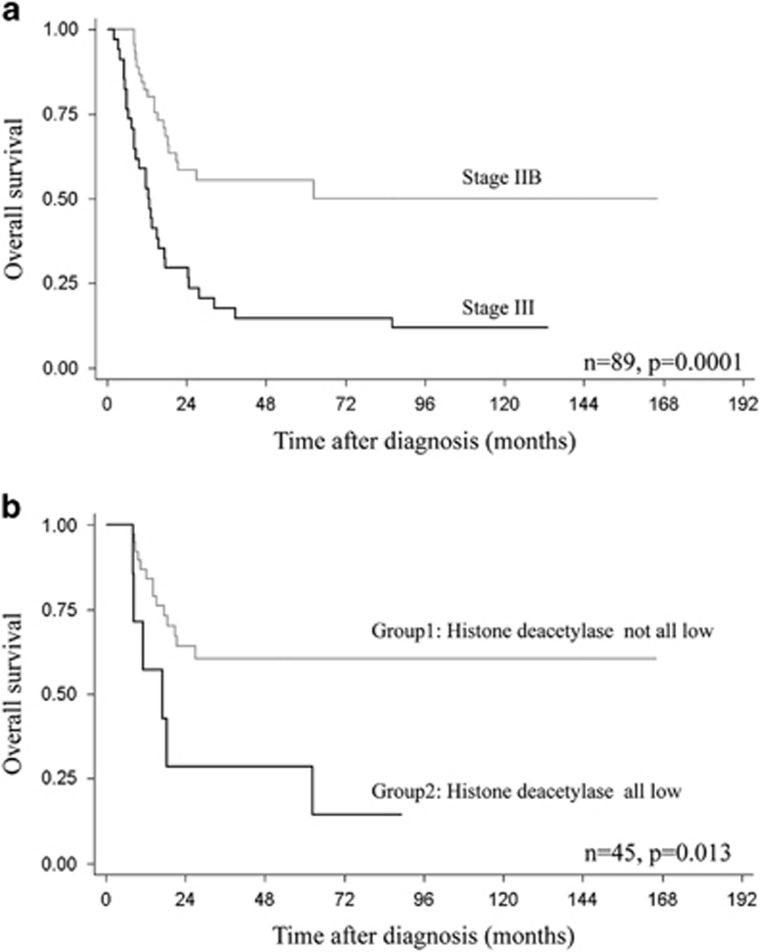
Univariate survival analysis indicated that low levels of nuclear expression of histone deacetylase 1 (Immunoreactive score≤1) was a significant negative predictor of high-grade osteosarcoma (P=0.038) (Table 3 and Figure 4a). Five-year survival rates were 19% in patients with low-histone deacetylase 1 expression (Immunoreactive score≤1), but increased to 40% in patients with higher histone deacetylase 1 (Immunoreactive score>1). Combined histone deacetylases were also significantly associated with shorter survival (P=0.016). Figure 4b shows the 5-year survival rates of 36% and 17% in Group1 and Group2 patients, respectively. Other significant negative prognosticators identified in this study were advanced stage (P<0.001), location of the tumor at the axial site (P=0.009), and the presence of metastases at initial evaluation (P<0.001).
Table 3. Median survival analyses of patients with high-grade osteosarcoma.
| Factor | Patients | Events (Death) | Median survival, months | P-value |
|---|---|---|---|---|
| Age at diagnosis, years | ||||
| ≤15 | 46 | 28 | 17.7 | 0.428 |
| >15 | 43 | 32 | 19.3 | |
| Gender | ||||
| Male | 50 | 40 | 18.1 | 0.067 |
| Female | 39 | 21 | 32.5 | |
| Enneking stage | ||||
| IIB | 45 | 20 | 62.3 | <0.001 |
| III | 34 | 30 | 12.6 | |
| Site | ||||
| Extremities | 78 | 49 | 24.4 | 0.009 |
| Axial | 8 | 8 | 11.4 | |
| Tumor size | ||||
| ≤10 cm | 59 | 36 | 24.4 | 0.171 |
| >10 cm | 18 | 13 | 12.8 | |
| Metastasis | ||||
| At diagnosis | 34 | 30 | 12.6 | <0.001 |
| At follow-up | 21 | 13 | 20.9 | |
| No | 24 | 7 | Undefined | |
| Chemoresistance | ||||
| Good responders | 8 | 3 | Undefined | 0.245 |
| Poor responders | 50 | 32 | 21.4 | |
| Histone deacetylase 1 expression | ||||
| Low (Immunoreactive score≤1) | 31 | 26 | 15.5 | 0.038 |
| High (Immunoreactive score>1) | 58 | 34 | 24.8 | |
| Histone deacetylase 2 expression | ||||
| Low (Immunoreactive score≤10) | 63 | 44 | 18.5 | 0.777 |
| High (Immunoreactive score>10) | 26 | 16 | 20.9 | |
| Histone deacetylase 3 expression | ||||
| Low (Immunoreactive score≤6) | 50 | 35 | 19.3 | 0.571 |
| High (Immunoreactive score>6) | 39 | 25 | 22.6 | |
| Histone deacetylases | ||||
| Group1a: Not all low | 71 | 44 | 22.6 | 0.016 |
| Group2b: All low | 18 | 16 | 10.7 |
P-values were calculated with the log-rank test, P-value<0.05 shown in bold.
Group1—none or some histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Group2—all-histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Figure 4.
Kaplan–Meier curves showing overall survival according to (a) histone deacetylase 1 expression and (b) combined histone deacetylase expression in all high-grade osteosarcoma cases; Group1—none or some histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff; Group2—all-histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Only low expression of combined histone deacetylases (HR: 2.29 (1.16–4.51), P=0.017) and axial location (HR: 3.12 (1.30–7.52)) continued to be independent poor prognostic factors in the multivariate survival analysis (Table 4).
Table 4. Multivariate analysis of overall survival in high-grade osteosarcoma with inclusion of class I histone deacetylase expression.
| Factor |
Overall survival |
|
|---|---|---|
| HR (95% CI) | P-value | |
| Age at diagnosis, years | ||
| ≤15 | 1.00 | — |
| >15 | 0.91 (0.50–1.67) | 0.771 |
| Gender | ||
| Male | 1.00 | — |
| Female | 0.73 (0.39–1.36) | 0.321 |
| Site | ||
| Extremities | 1.00 | — |
| Axial | 3.12 (1.30–7.52) | 0.011 |
| Tumor size | ||
| ≤10 cm | 1.00 | — |
| >10 cm | 1.70 (0.88–3.25) | 0.112 |
| Histone deacetylases | ||
| Group1a: Not all low | 1.00 | — |
| Group2b: All low | 1.96 (1.04–3.73) | 0.039 |
P-values<0.05 were shown in bold.
Group1—none or some histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Group2—all-histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Survival Analysis of Expression Patterns of Class I Histone Deacetylases in Stage IIB Osteosarcoma
Stage was a strong indicator of a poor prognosis (Figure 5a). Half the stage III patients died within 13 months and only 12% (4/34) survived until the final follow-up (Table 3). Subgroup survival analysis was performed with stage IIB cases (Table 5). The 1-year and 5-year survival rates of patients with stage IIB osteosarcoma were 82 and 55%, respectively.
Figure 5.
Kaplan–Meier curves showing overall survival according to (a) enneking stage and (b) combined histone deacetylase expression in stage IIB; Group1—none or some histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff; Group2—all-histone deacetylase isoforms expressed at immunoreactive score lower than the cut-off.
Table 5. Median survival analyses of patients with stage IIB osteosarcoma.
| Factor | Patients | Events (Death) | Median survival, months | P-value |
|---|---|---|---|---|
| Age at diagnosis, years | ||||
| ≤15 | 24 | 8 | Undefined | 0.222 |
| >15 | 21 | 12 | 21.4 | |
| Gender | ||||
| Male | 22 | 13 | 21.4 | 0.145 |
| Female | 23 | 7 | Undefined | |
| Site | ||||
| Extremities | 42 | 17 | Undefined | 0.008 |
| Axial | 3 | 3 | 12.2 | |
| Tumor size | ||||
| ≤10 cm | 34 | 15 | Undefined | 0.936 |
| >10 cm | 9 | 4 | Undefined | |
| Metastasis | ||||
| Yes | 21 | 13 | 20.9 | 0.057 |
| No | 24 | 7 | Undefined | |
| Chemoresistance | ||||
| Good responders (Tumor necrosis≥90%) | 5 | 0 | Undefined | 0.064 |
| Poor responders (Tumor necrosis<90%) | 32 | 16 | 27.1 | |
| Histone deacetylase 1 expression | ||||
| Low (Immunoreactive score≤1) | 11 | 8 | 17.7 | 0.051 |
| High (Immunoreactive score>1) | 34 | 12 | Undefined | |
| Histone deacetylase 2 expression | ||||
| Low (Immunoreactive score≤10) | 31 | 15 | 62.3 | 0.341 |
| High (Immunoreactive score>10) | 14 | 5 | Undefined | |
| Histone deacetylase 3 expression | ||||
| Low (Immunoreactive score≤6) | 28 | 15 | 27.1 | 0.171 |
| High (Immunoreactive score>6) | 17 | 5 | Undefined | |
| Histone deacetylases | ||||
| Group1a: Not all low | 38 | 14 | Undefined | 0.013 |
| Group2b: All low | 7 | 6 | 17.1 | |
P-values were calculated with the log-rank test, P-value<0.05 shown in bold.
Undefined; the estimated median survival was not reached.
Group1—none or some histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Group2—all-histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Univariate survival analysis showed that all-low -histone deacetylases were significantly correlated with poor prognosis in stage IIB (P=0.013) (Figure 5b). The 5-year survival rate was 61% in the not all-low-histone deacetylases patients, whereas it decreased to 29% in the all-low-histone deacetylases patients. Another significant negative prognosticator observed in stage IIB osteosarcoma was axial location (P=0.008). Initial metastasis (P=0.057) and low-histone deacetylase 1 expression (P=0.051) were marginally associated with shorter survival. (Table 5)
In multivariate analysis, all-low-histone deacetylases (HR: 3.61 (1.08–12.10), P=0.037) along with axial location (HR: 6.34 [1.55–26.04), P=0.10) continued as significant poor prognostic factors in stage IIB (Table 6).
Table 6. Multivariate analysis of overall survival in stage IIB osteosarcoma with inclusion of class I histone deacetylase expression.
| Factor |
Overall survival |
|
|---|---|---|
| HR (95% CI) | P-value | |
| Age at diagnosis, years | ||
| ≤15 | 1.00 | — |
| >15 | 1.47 (0.56–3.90) | 0.436 |
| Gender | ||
| Male | 1.00 | — |
| Female | 0.93 (0.33–2.67) | 0.898 |
| Site | ||
| Extremities | 1.00 | — |
| Axial | 6.34 (1.55–26.04) | 0.010 |
| Tumor size | ||
| ≤10 cm | 1.00 | — |
| >10 cm | 0.86 (0.27–2.70) | 0.791 |
| Histone deacetylases | ||
| Group1a: Not all low | 1.00 | — |
| Group2b: All low | 3.44 (1.07–11.09) | 0.038 |
P-values <0.05 were shown in bold.
Group1—none or some histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Group2—all-histone deacetylase isoforms expressed at immunoreactive score lower than the cutoff.
Discussion
To our knowledge, this is the first report on the association of expression patterns of histone deacetylases in osteosarcoma with clinicopathological parameters and survival of patients. In this study, class I histone deacetylase isoforms (histone deacetylase 1, histone deacetylase 2, and histone deacetylase 3) were differentially expressed in the majority of cases. We found that low expression of histone deacetylase 1 was correlated with poor outcomes such as metastasis and advanced stages. These findings are consistent with in vitro experiments in demonstrating the important roles of histone deacetylases in pulmonary metastases of osteosarcoma.24, 25, 26, 27 The most studied metastatic mechanism in this bone tumor was the Fas/FasL signaling pathway. Fas ligand (FasL) was constitutively expressed in some organs, including lungs. When tumor cells carrying Fas receptors migrate to the lungs, the receptor binds to its ligand to form a ‘‘death-inducing signaling complex’’.24 This assembly further triggers apoptosis via Caspase 8/Caspase 3 activation. Thus, only Fas-negative osteosarcoma cells can form lung metastases. Surprisingly, after treatment with the histone deacetylase 1/3-specific inhibitor, the Fas gene was upregulated in LM7, CCH-OS-D, and U2OS osteosarcoma cells, eventually resulting in cell death.25, 26, 27 This significant function of histone deacetylases was confirmed in an in vivo study that demonstrated an anti-metastatic effect of MS-275, an histone deacetylase inhibitor, in nu/nu-mice with osteosarcoma lung metastases.28 This evidence illustrates the pivotal roles of histone deacetylases in lung metastases, particularly, through Fas/FasL complex modulation.
We also identified a significant association between low-histone deacetylase 1 expression as well as low-all-histone deacetylases expression and short overall survival in high-grade osteosarcoma patients. These results suggest that the expression patterns of class I histone deacetylases have an effect on the behavior of osteosarcoma which may have clinical implications. Our data point out the possibility that dysregulation of histone deacetylases in osteosarcoma might contribute to disease progression. This finding can be explained by considering molecular functions of histone deacetylases in regulating the activation and/or repression of diverse oncogenic and tumor suppressor genes. That is, an alteration of histone deacetylase expression during the progression of osteosarcoma cells might critically affect a wide range of downstream signaling pathways. Several studies have demonstrated significant changes in the expression levels of resultant proteins upon the inhibition of histone deacetylases. For example, the MDM2-p53 feedback loop was re-activated as a result of an increase in p53 expression and a decrease in MDM2 expression which subsequently resulted in suppression of proliferation of osteosarcoma cells.10 Another study suggested that histone deacetylase inhibitor might enhance natural killer cell recognition of cancer cells by inducing an up-regulation of NKG2D ligands, MICA/B, on osteosarcoma cells and by increasing the expression of NKG2D receptors on natural killer cells.29
In addition, low-all-histone deacetylases expression was found to be independently correlated with short survival, not only in the entire osteosarcoma cohort but also in the subgroup of stage IIB patients. This finding indicates a benefit to defining a high-risk group among osteosarcoma patients, especially in patients without initial metastasis. Similar correlations have been observed with other cancers. Bernard et al.19 reported that lower expression of histone deacetylases and the histone modifications correlated with the lowest patient survival in colorectal cancer. High expression of histone deacetylase 1 and histone deacetylase 6 correlated with better patient survival in ER-positive breast cancers, while high histone deacetylase 2 expression was associated with improved overall survival in ER-negative groups.30
Expression levels of histone deacetylases may not necessarily indicate clinical sensitivity to histone deacetylase inhibitor. The present study demonstrated that histone deacetylase overexpression was not a negative prognostic marker in osteosarcoma; indeed, a decrease in the expression levels of combined class I histone deacetylases isoforms was independently associated with poor prognosis. Interestingly, it was found that histone deacetylase 1 and 2 were upregulated but histone deacetylase 3 was downregulated in primary osteosarcoma cells compared to osteoblastic cells (Figure 1). Considering histone deacetylase 1 and histone deacetylase 2 patterns in normal osteoblast differentiation, expression of protein and mRNA of both histone deacetylases decreased during the process.31 Low levels of histone deacetylase 1 were observed in differentiated osteoblasts. With RNAi knockdown of histone deacetylase 1, it was found that osteoblast differentiation was re-activated, suggesting that histone deacetylase 1 has a role in suppressing osteoblast differentiation.32 Unlike histone deacetylase 1 and histone deacetylase 2, histone deacetylase 3 works as a transcriptional co-repressor of transcription factors involved in osteoblast differentiation.31 Expression of histone deacetylase 3 is maintained throughout the normal differentiation process. Our findings confirm that histone deacetylase expression patterns in osteoblasts and osteosarcoma cells are different. Epigenetic regulation might contribute to transformation of osteoblastic cells to a malignant phenotype, thereby warranting targeted osteosarcoma therapy.
Much evidence has demonstrated the anticancer activity of knockdown of histone deacetylases, including an induction of cell cycle arrest, apoptosis, differentiation and senescence, as well as an inhibition of cell proliferation and angiogenesis.9, 33 Also, the results from in vitro cytotoxicity testing confirm that an alteration of epigenetic regulators, particularly histone deacetylases, was a key event in tumorigenesis of osteosarcoma.13, 34 This study suggests a rationale for targeting histone deacetylases in osteosarcoma, which is potentially useful in the treatment of the majority of patients.
A strength of this study is that it included the primary biopsy formalin-fixed paraffin-embedded tissue of all 89 osteosarcoma cases, including complete clinicopathologic data and survival data. This is the first study of the association between histone deacetylase expression patterns and patient survival. Two potential weaknesses are that there may have been changes to or differences in treatment protocols during the long study period (2000–2015) and the rarity of this type of tumor.
In conclusion, this study demonstrates that expression patterns of class I histone deacetylase isoforms are correlated with clinical outcomes in osteosarcoma. Low levels of all-histone deacetylases are potentially poor prognostic markers for patients who were metastasis-free at the time of diagnosis. This finding also suggests using histone deacetylase inhibitor in treatment of osteosarcoma. Further studies could address potential mechanisms of epigenetic regulation through histone acetylation in tumorigenesis and progression of osteosarcoma, particularly the metastatic phenotype.
Acknowledgments
This study was supported by the National Science and Technology Development Agency (NSTDA), code P-15-50265, Faculty of Medicine, Chiang Mai University, National Research University (NRU) fund, and the Excellence Center in Osteology Research and Training Center (ORTC). We also express our sincere thanks to Dr. G. Lamar Robert, PhD and Associate Professor Dr. Chongchit Sripun Robert, PhD, for editing the English manuscript.
Footnotes
Supplementary Information accompanies the paper on Modern Pathology website (http://www.nature.com/modpathol)
The authors declare no conflict of interest.
Supplementary Material
References
- Settakorn J, Lekawanvijit S, Arpornchayanon O et al. Spectrum of bone tumors in Chiang Mai University Hospital, Thailand according to WHO classification 2002: A study of 1,001 cases. J Med Assoc Thai 2006;89:780–787. [PubMed] [Google Scholar]
- Heymann MF, Brown HK, Heymann D. Drugs in early clinical development for the treatment of osteosarcoma. Expert Opin Investig Drugs 2016;1–16. [DOI] [PubMed]
- Friebele JC, Peck J, Pan X et al. Osteosarcoma: A Meta-Analysis and Review of the Literature. Am J Orthop (Belle Mead NJ) 2015;44:547–553. [PubMed] [Google Scholar]
- Allison DC, Carney SC, Ahlmann ER et al. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma 20122012;704872. [DOI] [PMC free article] [PubMed]
- Clark JC, Dass CR, Choong PF. A review of clinical and molecular prognostic factors in osteosarcoma. J Cancer Res Clin Oncol 2008;134:281–297. [DOI] [PubMed] [Google Scholar]
- Bacci G, Longhi A, Versari M et al. Prognostic factors for osteosarcoma of the extremity treated with neoadjuvant chemotherapy: 15-year experience in 789 patients treated at a single institution. Cancer 2006;106:1154–1161. [DOI] [PubMed] [Google Scholar]
- Hull EE, Montgomery MR, Leyva KJ. HDAC inhibitors as epigenetic regulators of the immune system: impacts on cancer therapy and inflammatory diseases. Biomed Res Int 2016;2016:8797206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014;124:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 2014;13:673–691. [DOI] [PubMed] [Google Scholar]
- Xie C, Wu B, Chen B et al. Histone deacetylase inhibitor sodium butyrate suppresses proliferation and promotes apoptosis in osteosarcoma cells by regulation of the MDM2-p53 signaling. Onco Targets Ther 2016;9:4005–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Tanaka K, Nakatani F et al. Involvement of P-glycoprotein and MRP1 in resistance to cyclic tetrapeptide subfamily of histone deacetylase inhibitors in the drug-resistant osteosarcoma and Ewing's sarcoma cells. Int J Cancer 2006;118:90–97. [DOI] [PubMed] [Google Scholar]
- Sonnemann J, Trommer N, Becker S et al. Histone deacetylase inhibitor-mediated sensitization to TRAIL-induced apoptosis in childhood malignancies is not associated with upregulation of TRAIL receptor expression, but with potentiated caspase-8 activation. Cancer Biol Ther 2012;13:417–424. [DOI] [PubMed] [Google Scholar]
- Yu D, Kahen E, Cubitt CL et al. Identification of synergistic, clinically achievable, combination therapies for osteosarcoma. Sci Rep 2015;5:16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh MS, Kim CW, Park BS et al. Mechanism of histone deacetylase inhibitor Trichostatin A induced apoptosis in human osteosarcoma cells. Apoptosis 2004;9:583–589. [DOI] [PubMed] [Google Scholar]
- Blattmann C, Thiemann M, Stenzinger A et al. Radiosensitization by histone deacetylase inhibition in an osteosarcoma mouse model. Strahlenther Onkol 2013;189:957–966. [DOI] [PubMed] [Google Scholar]
- Yamanegi K, Yamane J, Kobayashi K et al. Sodium valproate, a histone deacetylase inhibitor, augments the expression of cell-surface NKG2D ligands, MICA/B, without increasing their soluble forms to enhance susceptibility of human osteosarcoma cells to NK cell-mediated cytotoxicity. Oncol Rep 2010;24:1621–1627. [DOI] [PubMed] [Google Scholar]
- Cain JE, McCaw A, Jayasekara WS et al. Sustained low-dose treatment with the histone deacetylase inhibitor LBH589 induces terminal differentiation of osteosarcoma cells. Sarcoma 20132013;608964. [DOI] [PMC free article] [PubMed]
- Pruksakorn D, Teeyakasem P, Klangjorhor J et al. Overexpression of KH-type splicing regulatory protein regulates proliferation, migration, and implantation ability of osteosarcoma. Int J Oncol 2016;49:903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard A, Goossens-Beumer IJ, van Hoesel AQ et al. Nuclear expression of histone deacetylases and their histone modifications predicts clinical outcome in colorectal cancer. Histopathology 2015;66:270–282. [DOI] [PubMed] [Google Scholar]
- Weichert W, Roske A, Gekeler V et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol 2008;9:139–148. [DOI] [PubMed] [Google Scholar]
- Lehmann A, Denkert C, Budczies J et al. High class I HDAC activity and expression are associated with RelA/p65 activation in pancreatic cancer in vitro and in vivo. BMC Cancer 2009;9:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche FR, Weichert W, Roske A et al. Class I histone deacetylases 1, 2 and 3 are highly expressed in renal cell cancer. BMC Cancer 2008;8:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmott JS, Colebatch AJ, Kakavand H et al. Expression of the class 1 histone deacetylases HDAC8 and 3 are associated with improved survival of patients with metastatic melanoma. Mod Pathol 2015;28:884–894. [DOI] [PubMed] [Google Scholar]
- Huang G, Nishimoto K, Yang Y et al. Participation of the Fas/FasL signaling pathway and the lung microenvironment in the development of osteosarcoma lung metastases. Adv Exp Med Biol 2014;804:203–217. [DOI] [PubMed] [Google Scholar]
- Rao-Bindal K, Zhou Z, Kleinerman ES. MS-275 sensitizes osteosarcoma cells to Fas ligand-induced cell death by increasing the localization of Fas in membrane lipid rafts. Cell Death Dis 2012;3:e369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshkina NV, Rao-Bindal K, Kleinerman ES. Effect of the histone deacetylase inhibitor SNDX-275 on Fas signaling in osteosarcoma cells and the feasibility of its topical application for the treatment of osteosarcoma lung metastases. Cancer 2011;117:3457–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaffit F, Vaute O, Chevillard-Briet M et al. Cleavage and cytoplasmic relocalization of histone deacetylase 3 are important for apoptosis progression. Mol Cell Biol 2007;27:554–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao-Bindal K, Koshkina NV, Stewart J et al. The histone deacetylase inhibitor, MS-275 (entinostat), downregulates c-FLIP, sensitizes osteosarcoma cells to FasL, and induces the regression of osteosarcoma lung metastases. Curr Cancer Drug Targets 2013;13:411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Denman CJ, Cobanoglu ZS et al. The narrow-spectrum HDAC inhibitor entinostat enhances NKG2D expression without NK cell toxicity, leading to enhanced recognition of cancer cells. Pharm Res 2015;32:779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Min SK, Park HR et al. Expression of histone deacetylases HDAC1, HDAC2, HDAC3, and HDAC6 in invasive ductal carcinomas of the breast. J Breast Cancer 2014;17:323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee-Lawrence ME, Westendorf JJ. Histone deacetylases in skeletal development and bone mass maintenance. Gene 2011;474:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Suh JH, Kim AY et al. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol Endocrinol 2006;20:2432–2443. [DOI] [PubMed] [Google Scholar]
- Bots M, Verbrugge I, Martin BP et al. Differentiation therapy for the treatment of t(8;21) acute myeloid leukemia using histone deacetylase inhibitors. Blood 2014;123:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayanithy V, Park C, Sarver AL et al. Combinatorial treatment of DNA and chromatin-modifying drugs cause cell death in human and canine osteosarcoma cell lines. PLoS ONE 2012;7:e43720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.