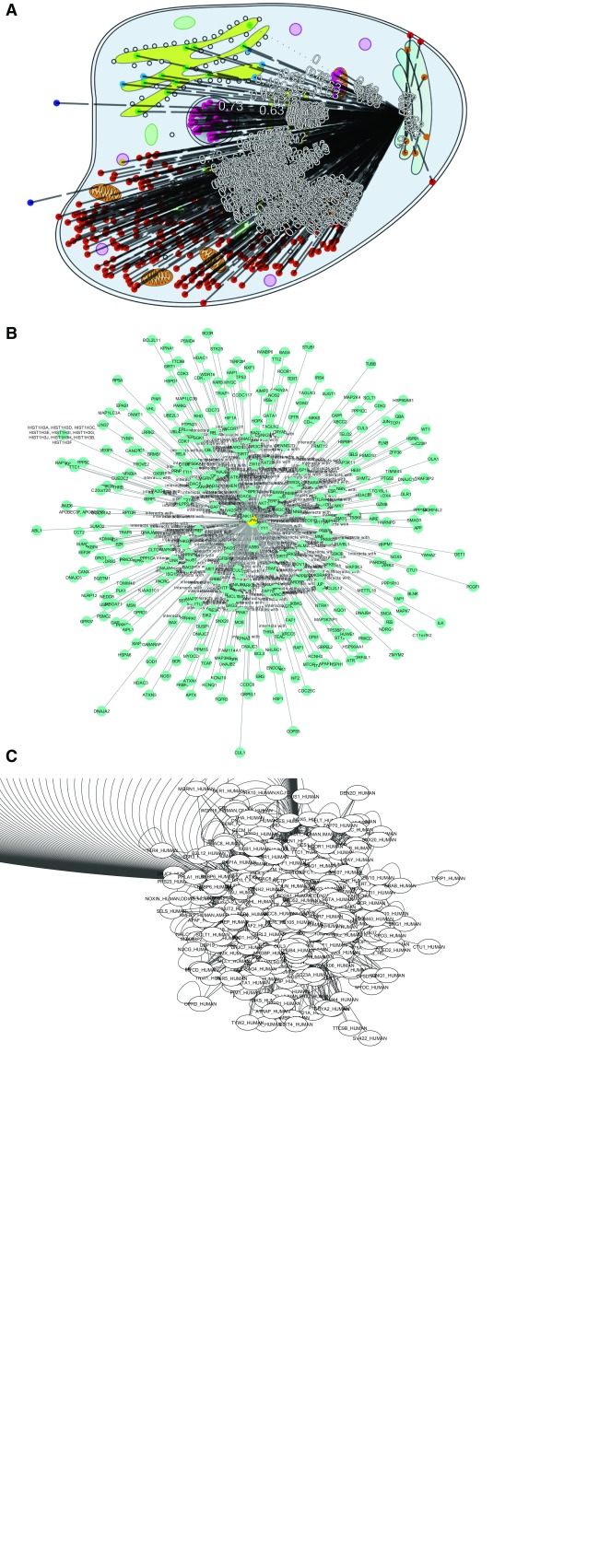
Figure 8.
PPI hub in CellMap ( A), Cytoscape.js ( B) and Cytoscape desktop ( C). For HSPA4 (Heat shock 70 kDa protein 4, UniProt identifier P34932), we show some of the PPIs known (according to HIPPIE HSPA4 has 338 interaction partners). We chose this as one example of a protein with many more PPIs than the average protein (“PPI hub”). The figure compares how three different PPI viewers cope with the HSPA4 network: ( A) CellMap ( http://cell.dallago.us/protein/P34932), ( B) HIPPIE’s Cytoscape.js visualizer and ( C) the desktop version of Cytoscape. Proteins in CellMap are represented as colored dots on the map (image) of the cell, and upon selecting the protein of interest an overlay of edges is drawn. In Cytoscape and Cytoscape.js, proteins are represented as nodes containing a label (protein name as UniProt identifier), and edges are directly inferred from the data. The Cytoscape.js visualization was taken directly from HIPPIE. The Cytoscape network was automatically drawn upon loading the HIPPIE dataset and selecting the protein of interest and it’s direct neighbors.