Abstract
Currently, display-based methods are well established and widely used in antibody engineering for affinity maturation and structural stability improvement. We obtained a novel anti-human programmed death 1 (PD-1) antibody using computer-aided design and a mammalian cell display technology platform. We used computer-aided modeling and distance geometry methods to predict and assign the key residues that contributed to the binding of human PD-L1 to PD-1. Then, we analyzed the sequence of nivolumab (an anti-human PD-1 antibody, referred to as MIL75 in the article) to determine the template for antibody design and library construction. We identified a series of potential substitutions on the obtained template and constructed a virtual epitope-targeted antibody library based on the physicochemical properties and each possible location of the assigned key residues. The virtual antibody libraries were displayed on the surface of mammalian cells as the antigen-binding fragments of full-length immunoglobulin G. Then, we used flow cytometry and sequencing approaches to sort and screen the candidates. Finally, we obtained a novel anti-human PD-1 antibody named FV78. FV78 competitively recognized the PD-1 epitopes that interacted with MIL75 and possessed an affinity comparable to MIL75. Our results implied that FV78 possessed equivalent bioactivity in vitro and in vivo compared with MIL75, which highlighted the probability and prospect of FV78 becoming a new potential antibody therapy.
Keywords: antibody, computer-guided modeling, distance geometry, human PD-1, mammalian cell-displayed antibody library
INTRODUCTION
Antibody engineering has been successful at dramatically altering the function and utility of many antibody-based drugs for clinical applications.1 The ability to alter the binding of an antibody to its target (that is, antigen) has substantial effects on its biological function and has led to the creation of numerous antibody therapeutics.2 With the development of structural biology and molecular biology, a series of technologies, including site-directed mutagenesis and the introduction of random mutations representing all possible single amino-acid substitutions in the binding domains (that is, antibody complementarity-determining regions CDRs), have been used to engineer and screen antibody therapeutics with high affinity.3 However, these methods introduce a large number of non-functional antibody fragments by accident, which are comparatively complex and require time and energy.
The antibody–antigen-binding fragment (Fab) contains six CDRs that present a large contiguous surface for antigen recognition.4, 5 The immune system contains a highly diverse population of antibodies, each of which can be distinguished by a unique set of CDRs that confers antigen specificity.6 The database of natural antibody sequences has revealed clear biases for particular amino acids, although the compiled CDR sequences are highly diverse.7, 8, 9 Furthermore, the structural database revealed that these biases were even greater when residues that mediated antigen recognition through direct contact were considered.10, 11 With the development of bioinformatics and computational biology, computational kinetic models have helped set quantitative biophysical goals12 and improve protein modeling and design.13 In our previous study, a series of novel antagonist peptides, antibody-linked molecules and scFvs were designed based on protein–protein interaction models and the orientation of the key binding amino-acid residues.14, 15, 16 A ‘me better’ antibody was obtained based on the key antibody amino-acid residues involved in the interaction with antigens using the 3-D modeling structure of Her2 and its functional antibody MIL5.17 We also obtained the novel anti-vascular endothelial growth factor (VEGF)-neutralizing antibody MIL60 based on the theoretical interaction model between VEGF and its receptor VEGF receptor.18
Over the past two decades, display and screening technologies have been widely used to develop new monoclonal antibodies (mAbs) for human therapy. A series of studies have shown that target-specific and high-affinity antibodies could be rapidly obtained from a large antibody library using in vitro display technologies. Currently, phage display is the most widely used method to display and screen large libraries of antibodies or to engineer selected antibodies.19 However, this display technology has drawbacks in the practical application process. First, the single-chain antibody fragment (scFv) displayed on the phage may contain structural differences compared with the full-length immunoglobulin G (IgG) molecule. Second, differences may exist between prokaryotic expression systems and eukaryotic expression systems in terms of protein post-translational modifications, including glycosylation and methylation; however, eukaryotic expression cells, such as the Chinese hamster ovarian cell line (CHO), are commonly used in the industrial antibody production process. Moreover, the antibody expression levels are unpredictable when the phage library-screened antibody sequences are converted into a mammalian cell expression system owing to the codon usage bias between prokaryotic and eukaryotic expression systems. In comparison, mammalian cells represent a very promising display system for antibody libraries. Recently, Zhou Chen et al.20 displayed small randomly mutagenized IgG libraries on the surfaces of mammalian cells. However, the main drawback of mutant libraries is that most random combination sequences are not converted into functional antibodies. In addition, the capacity of the mammalian cell library is usually limited to 106 owing to the limitations of the transfection approach and site-directed integration, which restricts its application. To avoid these limitations, we made a successful attempt to construct a targeted epitope mammalian cell-displayed antibody library based on computer-aided analysis and design.
The important inhibitory function of the programmed death 1 (PD-1) protein was first discovered owing to the autoimmune-like phenotype of PD-1-/-mice.21 To date, PD-1 has been linked to several autoimmune disorders through genetic analyses of human patients. One of its ligands, PD-L1, has a pivotal role in the ability of tumor cells to evade the host immune system. Many studies have shown that PD-L1 is selectively expressed on many tumors or on cells within the tumor microenvironment in response to inflammatory stimuli.22, 23, 24 Blockade of the interaction between PD-1 and PD-L1 enhances immune functions in vitro and mediates anti-tumor activity in preclinical models. Nivolumab (Opdivo, Bristol-Myers Squibb; BMS) is an anti-human PD-1 antibody used in the clinic. Nivolumab is a high-affinity, fully human IgG4 mAb that inhibits the binding of PD-1 to PD-L1. This promising antibody drug has been approved by the US Food and Drug Administration for the treatment of metastatic melanoma and advanced non-small cell lung cancer. Many clinical indications, such as castration-resistant prostate cancer, renal cell cancer, colorectal cancer and classical Hodgkin’s lymphoma, are clinical trials.25, 26
Herein, a virtual epitope-targeted antibody library was designed using a general template originating from the given anti-PD-1 antibody allele distribution based on the predicted key amino-acid residues involved in human PD-L1 binding to PD-1. Then, the designed antibody library was displayed on mammalian cell membranes for screening. After several rounds of screening and assessment of the candidates, the novel antibody FV78 was selected. FV78 showed good bioactivity in vitro and in vivo. Therefore, we successfully selected a novel anti-human PD-1 antibody using a targeted epitope mammalian cell antibody library.
MATERIALS AND METHODS
Reagents and cell lines
The restriction enzymes were purchased from NEB (Ipswich, MA, USA). The 2xTaq PCR MasterMix was purchased from TIANGEN BioTECH (Bejing, China) Co., Ltd. The Lipofectamine 2000 transfection reagent and Opti-MEM medium were obtained from Invitrogen Life Technologies (Carlsbad, CA, USA). Hygromycin B was purchased from Roche (Basel, Switzerland). Fetal bovine serum (FBS) was purchased from Life Technologies (Tarrytown, NY, USA), the DNA Mini Kit was purchased from Qiagen (Hilden, Germany), and the pGEM-T Easy Vector System was purchased from Promega (Madison, WI, USA). The FACSCalibur machine was obtained from BD (Franklin Lakes, NJ, USA). The glass bottom dish was obtained from In Vitro Scientific (Mountain View, CA, USA). The Biacore T200, Fab capture kit and the related evaluation software were purchased from GE Healthcare (Little Chalfont, Buckinghamshire, UK). The monocyte purification kit and CD4+ T-cell isolation kit were obtained from Miltenyi Biotec (Bergisch Gladbach, Germany), interleukin-4 (IL-4) and granulocyte-macrophage colony-stimulating factor (GM-CSF) were obtained from R&D Systems (Minneapolis, MN, USA) and the human IFN-γ ELISA MAX Deluxe kit was obtained from BioLegend (San Diego, CA, USA).
PEGFP-N1 and pDsRed2-1 were purchased from Clontech Laboratories, Inc (Madison, WI, USA). The CD274 (NM_014143) Human cDNA ORF Clone was purchased from OriGene Technologies (Rockville, MD, USA). The pcDNA5/FRT and POG44 vectors and the Flp-In-CHO cells were purchased from Invitrogen Life Technologies.
MIL75, which is a full-length IgG1 antibody containing the same variable region sequences as nivolumab (BMS), was expressed in our laboratory. The PD-1/Fc, PD-L1/Fc and PD-L2/Fc fusion proteins were prepared in our laboratory. MIL75-Biotin, PD-1/Fc-Biotin and PD-L2/Fc-Biotin were labeled by Jiaxuan Biotech (Beijing, China). The fluorescein isothiocyanate (FITC)-conjugated anti-human IgG (Kappa) (anti-human IgG (Kappa)-FITC) and PE-conjugated anti-human IgG (Kappa) (anti-human IgG (Kappa)-PE) were purchased from BD Biosciences. The FITC-conjugated PD-1 was purchased from Jiaxuan Biotech.
The NPG mice were purchased from Beijing Vitalstar Biotechnology Co., Ltd (Beijing, China).
Computer-guided modeling
The protein 3-D structure computer-guided homology modeling program InsightII 2005 (MSI) was used to generate the homology model of the protein and its receptor. The original theoretical structures of relative proteins were established using the crystal structures of PD-1, PD-L1 and a homologous antibody as the structural templates.
Molecular dynamics simulations were performed for the 3-D theoretical structures of the proteins (human PD-1 and PD-L1) using an all-atom model with an explicit solvent. The starting structures for the simulations were obtained from X-ray structures (human PD-1 PDB code: 3RRQ; human PD-L1 PDB code: 4Z18 and the PD-1 and PD-L1 complex structure PDB code: 4ZQK). Structures of the whole proteins (PD-1, PD-L1 and the PD-1 and PD-L1 complex) were obtained using explicit-atom simulations for 100 ns using CVFF and the CHARMM force field. The simulations were performed at 298 K and 1 atm in the NPT ensemble. During the simulation procedure, the energy was initially minimized with the steepest descent method by fixing the protein to allow the water to relax around the protein. Then, the restraints were removed and the structure was further minimized with the steepest descents adopted based on the Newton–Raphson methods. The system was slowly heated to room temperature with a 5 °C increment every 1 ps using a 1-fs time step. The system was equilibrated for 1 ns before starting the computation of the various properties from the simulation. The configurations were saved every 1 ps during the simulation for statistical analysis.
Rules for selecting key residues in PD-L1
The theoretical 3-D complex structures of PD-1 and PD-L1 were analyzed using computer graphics and geometry distance methods. The key amino-acid residues involved in PD-L1 binding to PD-1 were predicted using the solvent accessible surface area calculation method according to the Van der Waals interactions and intra-molecular hydrogen bond formation.
Rules for choosing the suitable antibody template
The IMGT V domain gene and allele of nivolumab (anti-human PD-1 antibody) were fully studied using the antibody sequence alignment method based on the human antibody variable domain database27, 28 (http://www.imgt.org). The 3-D structure of nivolumab was modeled according to the determined amino-acid sequence of the human antibody variable domain allele and its physicochemical properties were theoretically analyzed. The complementarity-determining region of an antibody and its adjacent position are pivotal for antigen-binding; thus, when the ratio of the accessible surface area of an amino acid in the protein is >60%, typically the amino-acid residue is considered to be exposed in solution. In this case, the CDR amino-acid residues that contributed to the interaction between the antibodies and antigens and possessed better solvent accessibility (that is, were regarded as exposed) were assigned first. Then, the amino acids that possessed polar and similar physicochemical properties replaced the assigned residues in the parent model. Because HCDR3 had the highest variation of the six CDRs in the antibody, in this study the parent model of HCDR3 in the library was chosen from MIL75.
Cell culture
Flp-In-CHO cells were cultivated in DMEM/F12 medium supplemented with 100 units/ml of penicillin, 100 units/ml of streptomycin and 10% FBS. The cells were incubated in a humidified incubator (Thermo, Waltham, MA, USA) at 37 °C with 5% CO2.
Construction of the Fab vector for antibody display on mammalian cells
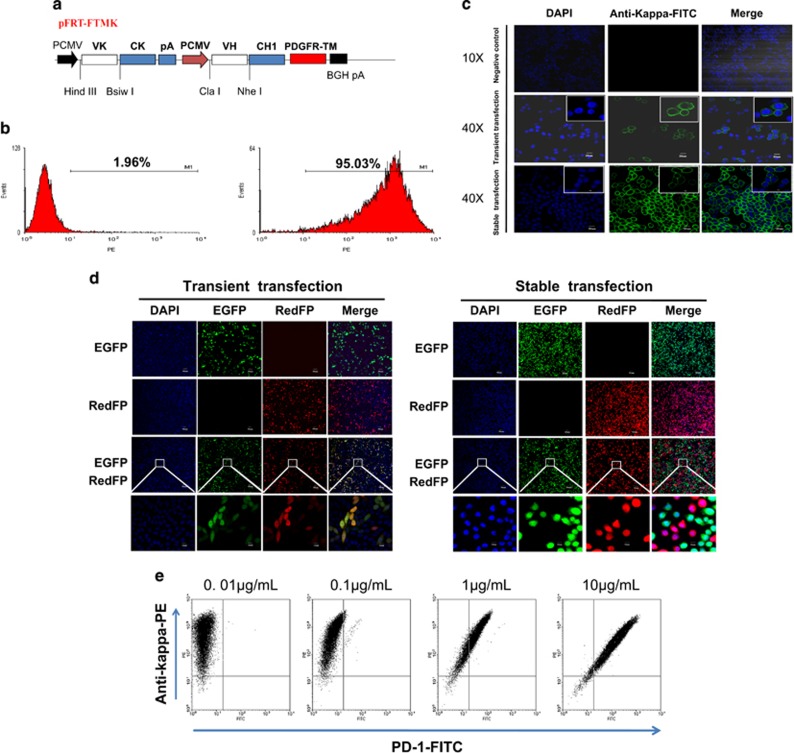
To implement candidate antibody screening, a novel mammalian cell-displayed antibody vector (pFRT-FTMK for short) was constructed as shown in Figure 2a. The Flp-In System (Invitrogen) was used to perform the integration and expression of the gene of interest in mammalian cells at a specific genomic location in this work. Using the pcDNA5/FRT vector, two parallel expression cassettes were cloned into the parent vector to express an antibody with both a light chain (VL+CL) and part of a heavy chain (VH+CH1) under the control of each PCMV promoter. Two sets of restriction enzyme-cutting sites were selected for the insertion of the heavy chain (HindIII and BsiwI) and light chain (ClaI and NheI) of the antibody variable regions. The transmembrane region of the platelet-derived growth factor receptor was used to ensure Fab antibody membrane expression.29
Figure 2.
Construction and verification of the ability of the pFRT-FTMK vector to display a Fab antibody on mammalian cells. (a) Schematic illustration of the Fab antibody vector used for display on the mammalian cell membrane. Two parallel pCMV/T7 promoters were chosen for light chain (VK+CK) and partial heavy chain (VH+CH1) transcription followed by the transmembrane sequence (PDGFR-TM). (b) Flow cytometry analysis of the Fab antibody displayed on the cell surface. The MIL75-pFRT-FTMK vector was stably transfected into Flp-In-CHO cells. Null cells (left) and transfected cells (right) were stained with anti-human IgG (Kappa)-PE. (c) Laser confocal scanning microscopy analysis of the Fab antibody displayed on the cell surface following transient and stable transfection of MIL75-pFRT-FTMK into Flp-In-CHO cells. The cells were stained with anti-human IgG(Kappa)-FITC; null cells without any transfection was used as the negative control. (d) Laser confocal scanning microscopy analysis of the expression and location of the red and green fluorescent proteins. The pFRT-EGFP and pFRT-redFP vectors were independently transfected or co-transfected into Flp-In-CHO cells. Under transient transfection, the red and green fluorescent proteins were located in the transfected cells nearly simultaneously. By contrast, the red and green fluorescent proteins were located separately after stable transfection. (e) MIL75-pFRT-FTMK stably transfected cells (5 × 105) were stained with serial dilutions of FITC-conjugated PD-1 (0.01, 0.1, 1 or 10 μg/ml) and a fixed concentration of anti-human IgG(Kappa)-PE.
Transient and stable transfection of the MIL75-pFRT-FTMK, pFRT-EGFP and pFRT-redFP vectors
The MIL75 VH and VL genes were amplified and cloned into pFRT-FTMK. Then, the constructed vector was transiently transfected into Flp-In-CHO cells. After 48 h of transfection, the cells were collected for analysis. MIL75-pFRT-FTMK and POG44 were co-transfected (1:9) into Flp-In-CHO cells. After 48 h of transfection, the cells were cultured under selection pressure by hygromycin B (1 mg/ml). Two weeks later, the surviving cells were pooled together for analysis. The green and red fluorescent protein genes were amplified from the PEGFP-N1 and pDsRed2-1 vectors, cloned into pcDNA5/FRT, and named pFRT-EGFP and pFRT-redFP, respectively. The two vectors were transfected alone or co-transfected (1:1) into Flp-In-CHO cells. Similar to the above experimental method, the transiently transfected and stably transfected cells were collected for analysis.
Laser confocal scanning microscopy
A total of 2 × 105 cells were seeded into a glass bottom dish and incubated in a humidified incubator at 37 °C with 5% CO2. After 24 h, the cells were stained with anti-human IgG (Kappa)-FITC and incubated for 30 min at 4 °C. Then, the cells were washed twice with 500 μl of phosphate-buffered salin (PBS) and immobilized with 4% paraformaldehyde. Prior to the experiment, 4′,6-diamidino-2-phenylindole was added and incubated with the cells for >5 min to stain the nuclei. Similar experimental conditions were used in addition to fluorescence labeling for the pFRT-EGFP and pFRT-redFP vector-transfected cells.
Flow cytometry analysis
Cells (5 × 105) were suspended in assay buffer (PBS containing 2% fetal calf serum) and stained with serial dilutions of FITC-conjugated PD-1. After incubation for 30 min at 4 °C, the cells were washed twice with assay buffer by centrifugation. The cell fluorescence was analyzed using the FACSCalibur flow cytometer.
To evaluate the competitive activity of the antibody on the PD-1/PD-L1 interaction, we constructed a cell line that highly expressed PD-L1 (CHO-K1/PD-L1) using the stable transfection method and determined the saturation concentration of FITC-PD-1 (20 μg/ml). Using this concentration, a gradient concentration of antibody (400, 100, 25, 6.25, 1.625 and 0.4065 μg/ml) was added to the stained CHO-K1/PD-L1 cells (5 × 105/sample). Cell fluorescence was analyzed with a FACSCalibur flow cytometer.
Fluorescence-activated cell sorting
First, 5 × 107 cells from the antibody cell libraries were collected and resuspended in 2 ml of PBS. Then, the cells were stained with anti-human IgG (Kappa)-PE (20 μl/2.5 × 106 cells) and FITC-conjugated PD-1 (10 μg/ml). The double-color fluorescence was analyzed, and the cells were sorted using the FACSAriaIII flow cytometer.
Mammalian cell-displayed antibody library construction and screening
Based on the theoretical analysis, the mammalian cell-displayed antibody library was constructed to introduce the mutant sites. Using overlap PCR, the determined parental antibody sequences (human IgHV3-33*01 and IgKV3-11*01) and the involved important mutant sites were inserted into the pFRT-FTMK vector. TOP10 cells competent for electrotransformation were prepared and used for antibody library plasmid transformation. Finally, the corresponding transformed clones were counted. To test the quality and diversity of the constructed antibody library, a portion of the VH (278 base pairs) and VL (183 base pairs) fragments were obtained separately by PCR. The specific primers designed to amplify the corresponding light chain and heavy chain fragments were as follows: light chain forward 5′-AAGCCTGGCCAGGCCCCTAGACT and reverse 5′-GCCGAAGGTTCTAGGCCAGTT and heavy chain forward 5′-CTGGCAGAAGCCTGAGACTG and reverse 3′-GCCCTGGCCCCAGTAGTC. DNA sequences reflecting the diversity of the constructed plasmids were analyzed using second-generation sequencing. Using the constructed antibody plasmid library, the plasmid with POG44 was co-transfected into the Flp-In System with a 1:9 ratio. The optimal selection pressure was 1000 mg/l with hygromycin as the marker. Two weeks later, the original targeted epitope mammalian cell-displayed antibody library was obtained. Then, the antibodies targeted to human PD-1 with high affinity and expression levels were enriched by fluorescence-activated cell sorting (FACS).
ELISA
An enzyme-labeled plate was coated with 1 μg/ml of PD-1 and the CD28 family fusion protein at 4 °C overnight. Then, the plate was blocked with 1.5% casein for 1.5 h at 37 °C. Serial antibody concentration gradients were added to the appropriate wells. The antigen–antibody complexes were formed at 37 °C for 2 h. After washing, horse radish peroxidase (HRP)-conjugated goat anti-human IgG was added to the plate and incubated for 1 h at room temperature. The binding signals were visualized using the TMB substrate and the light absorbance was measured at 450 nm.
Competitive ELISA
The epitope competition enzyme-linked immunosorbent assay (ELISA) experiments were performed to evaluate the degree of epitope overlap between MIL75 and the candidate antibody. MIL75 were labeled with biotin and serial antibody concentrations were added to compete with the binding activity between MIL75-biotin and PD-1.
The activity of the designed antibody that inhibited the interaction between PD-1 and its ligands PDL-1 and PDL-2 was evaluated by the competitive ELISA. To test the antibody-blocking activity between PD-1 and PD-L1, enzyme-labeled plates were coated with PD-L1/Fc. Biotin-labeled PD-1/Fc was diluted to an appropriate concentration (5 μg/ml) and the antibody candidates were serially diluted with the configured PD-1/Fc-Biotin solution. TMB and avidin-biotin-HRP were used to test the blocking signals. Similarly, an enzyme-labeled plate was coated with PD-L2/Fc. Biotin-labeled PD-1/Fc was diluted to an appropriate concentration (10 μg/ml) and the antibody candidates were serially diluted with the configured PD-2/Fc-Biotin solution. The blocking signals were tested using the substrate and enzyme as described above.
BIaCore analysis
Surface plasmon resonance experiments were performed at 25 °C using a Biacore T200 machine with HBS-EP as the running buffer. For these measurements, the anti-human Fab antibody was coupled on a CM5 chip at pH 5.0 using an amine coupling kit (GE Healthcare). Samples at a concentration of 1–2 μg/ml were captured on the second flow cell at a flow rate of 30 ml/min. A dilution series of the PD-1/Fc fusion protein (from 3.125–50 nM) was passed through both flow cells at 30 μl/min to record the association phase (120 s). The dissociation phase was monitored for 1200 s and triggered by replacing the sample solution with HBS-EP. Bulk refractive index differences were corrected by subtracting the response obtained on the reference flow cell (first flow cell). After each cycle, the sensor surface was regenerated via a short treatment with 10 mM glycine-HCl (pH 2.1). The binding kinetics were recorded and analyzed with the Biacore T200 evaluation software (GE Healthcare) using the 1:1 binding model.
Biological activity evaluation in vitro
The in vitro activity was evaluated in a monocyte-induced dendritic cell (DC) and purified allogeneic CD4+ T-cell system. Whole blood samples were obtained from two healthy human donors at different experimental times. First, monocytes were isolated from one donor’s peripheral blood mononuclear cells (PBMCs) using a monocyte purification kit. The monocytes were cultured in vitro at a concentration of 2 × 104 cells per well for 7 days with interleukin-4 (IL-4, 50 ng/ml) and GM-CSF (25 ng/ml). The cell growth medium was exchanged every 3 days. On the 7th day, CD4+ T cells were isolated from another PBMC donor using a CD4+ T-cell isolation kit. In vitro differentiated human DCs were mixed with purified allogeneic CD4+ T cells at a 10:1 (T:DC) ratio. The cells were cultured with or without dose titrations of MIL75 (50 μg/ml; fivefold dilutions; six gradients), the samples (50 μg/ml; fivefold dilutions; six gradients) or the control MIL50 antibody (50 μg/ml; a humanized anti-ricin IgG antibody from our laboratory). After 5 days, IFN-γ secretion in the supernatants was analyzed using the Human IFN-γ ELISA MAX Deluxe kit.
Biological activity analysis in vivo
Twenty-nine NPG mice were fed in the specific pathogen free animal feeding room for 1 week. Whole blood samples were obtained from one donor. PBMCs were isolated using lymphocyte separation medium. A total of 1 × 106 cells were injected into each mouse by the intravaneous route. One week later, a 10 mg/kg dose of the antibody drug (MIL75 or the screening antibody FV78) or isometric normal saline (NS) was injected two times per week by intravenous injection. The mice were killed when their body weights decreased to 15–20% of the baseline. The median survival times were analyzed.
Statistical analysis
The equation log(agonist) vs. response–variable slope was used to analyze the binding activity and competitive activity of the antibody. The in vitro biological activity evaluation was analyzed by two-way analysis of variance. The log-rank (Mantel–Cox) test was used to analyze the median survival of the mice treated with the anti-PD-1 antibody or NS therapy. Data with P-values ⩽0.05 were considered statistically significant.
RESULTS
Determination of the binding model of human PD-L1 to PD-1
The key amino-acid residues involved in human PD-L1 binding to PD-1 were determined based on the 3-D structures of human PD-1, PD-L1 and the PD-1 and PD-L1 complex using computer graphics technology and the distance geometry method. The RMSDs (root means square distances) of the main chain carbon atoms between the crystal and theoretical structures were 0.012 nm (PD-1), 0.014 nm (PD-L1) and 0.023 nm (PD-1/PD-L1) compared with the crystal structures of the human PD-1, PD-L1 and PD-1/PD-L1 complex, respectively.30 The results indicated that the theoretical structures were rational. The chosen force field and minimization method were suitable for the analysis of the structural properties and interaction model of PD-1/PD-L1.
As shown in Figure 1a, the interaction mode between human PD-1 and PD-L1 fit together effectively. The key amino-acid residues in PD-1 and PD-L1 were marked as shown in Figure 1b. Considering the radius of the amino-acid side chain (2.5 Å) and the intermolecular hydrogen bonding, the important amino-acid residues in PD-L1 were Ile54, Glu58, Gln66, Val68, Lys75, Arg113, Gly119, Gly120, Ala121, Asp122, Tyr123, Lys124 and Arg125 and those in PD-1 were Tyr48, Asn54, Gln55, Lys58, Asp65, Arg66, Ser67, Gln68, Lys111, Gln113, Ile114 and Glu116. The determined key binding amino-acid residues were consistent with the PD-1 and PD-L1 crystal structures.30 The results showed that the theoretical minimized PD-1/PD-L1 complex structure was reasonable. Then, the binding energies between the key residues were calculated based on the theoretical PD-1/PD-L1 complex structure and the suitable force field. The results showed that the amino-acid residues Glu58, Gln66, Asp122 and Ala121 were the most important factors in PD-L1. Although four basic amino-acid residues (Lys75, Arg113, Lys124 and Arg125) in PD-L1 were involved in PD-1 binding, electrostatic binding was not detected.
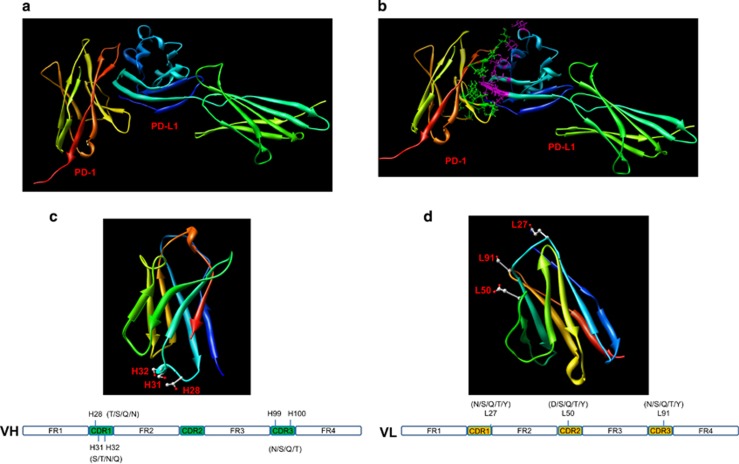
Figure 1.
Computer-aided homology modeling. (a) The ribbon structure of the human PD-1 and PD-L1 complex based on the computer-guided modeling and docking methods. (b) The fine structure of the binding mode between human PD-1 and PD-L1. The purple sticks denote the key amino-acid residues in human PD-L1, whereas the green balls and sticks denote the key amino-acid residues in human PD-1. (c) The 3-D theoretical structure of IgHV3-33*01, where the balls and sticks denote H28, H31 and H32. (d) The 3-D theoretical structure of IgHK1-11*01, where the balls and sticks denote L27, L50 and L91.
Generation of the virtual epitope-targeted library
The closet gene and allele of the variable domains of the anti-human PD-1 clinical antibody nivolumab were analyzed using the Domain Gap Align method in IMGT (http://www.imgt.org). The results showed that the heavy chain variable domain belonged to human IgHV3-33*01, whereas the light chain belonged to human IgKV3-11*01. The 3-D structures of IgHV3-33*01 and IgKV3-11*01 were constructed using the computer-guided homology modeling method. As shown in Figure 1c, the polar positions H28, H30 and H31 were the most accessible. Positions H99 and H100 were also polar residues. Using the rules for choosing the suitable antibody template, positions H28, H30, H31, H99 and H100 in the variable domain of the IgHV3-33*01 heavy chain were assigned as the key locations and the mutant was designed as shown in Figure 1c. Similarly, positions L27, L50 and L91 in the variable domain of the IgKV3-11*01 light chain were assigned as shown in Figure 1d.
Verification of the pFRT-FTMK vector for cell surface display and site-directed integration
To verify that the constructed pFRT-FTMK vector could display Fab-scaffolding proteins on the cell surface, MIL75-pFRT-FTMK was constructed as described in the Materials and Methods. The flow cytometry analysis results indicated that ~95% of the cells displayed the MIL75-Fab antibody (Figure 2b). The antibody expression levels were further examined by laser scanning confocal microscopy. Using this method, the Fab antibody was detectable on the cell surface whether it was transiently or stably transfected. Almost all of the stably transfected cells were labeled with the green fluorescent protein (Figure 2c).
To ensure that a single copy of the plasmid was integrated into every cell via the Flp-In system, pFRT-EGFP and pFRT -redFP were constructed as described in the Materials and Methods section. The data shown in Figure 2d demonstrated that the red and green fluorescent plasmids were randomly transfected into the cells by transient co-transfection. Most of the cells showed green and red fluorescence colocalized into a single cell. However, a single fluorescent protein gene was expressed in individual cells following stable transfection, which indicated that only a single pFRT-FTMK Fab antibody expression vector could be integrated into the Flp-In-CHO cell genomes.
Construction and screening of the epitope-target anti-PD-1 antibody library
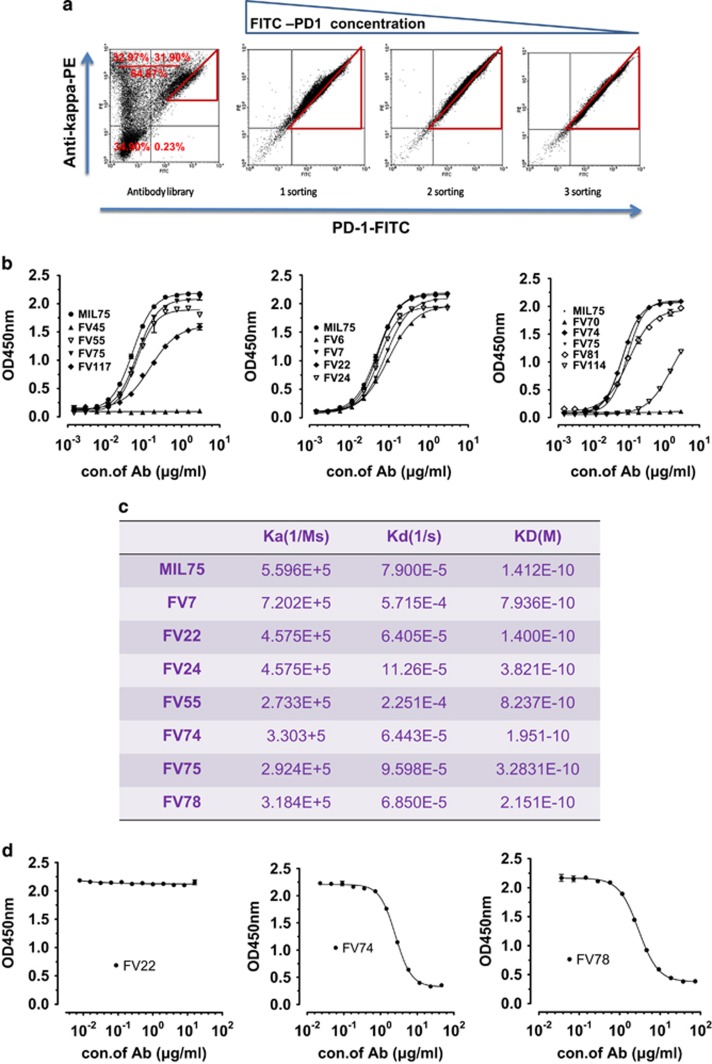
Using the constructed Fab membrane expression vector pFRT-FTMK (Figure 2a) described above, the designed human IgHV3-33*01 and IgKV3-11*01 chains were chosen as the parent sequences. The potential key sites determined above were designed (Figures 1c and d) and an antibody plasmid library was constructed. The corresponding bacterial clones numbered ~108. The DNA sequence diversity of the constructed antibody library was analyzed using second-generation sequencing analysis. A total of 30 497 VH sequences and 72016 VL sequences were obtained separately, representing 88.92% and 92.57% of the theoretical sequences, respectively, created by designing the degenerate primers. In summary, we successfully constructed the antibody plasmid library, which paved the way for the construction and screening of the cell library. As shown in Figure 3a, ~64.87% of the subpopulation among the total cell library displayed a complete Fab antibody and 31.90% of the subpopulation had specific antigen-binding activity. After three rounds of screening and enrichment with the FITC-labeled human PD-1 antigen concentration gradient (10, 5 and 1 μg/ml), we obtained enriched positive cell clones with high affinity and expression levels (Figure 3a).
Figure 3.
Screening and characterization of the mammalian cell antibody library for functional antibody candidates. (a) Sorting of the mammalian cell antibody library to identify clones with high affinity and expression levels. The cell library were stained with serial dilutions of FITC-conjugated PD-1 (10, 10, 5 or 1 μg/ml) and a fixed concentration of anti-human IgG(Kappa)-PE. Fluorescence-activated cell sorting were performed three times with a declining PD-1 antigen concentration. (b) ELSIA detection of the binding activity of the antibody candidates to PD-1/Fc. (c) Biacore analysis of the antibody candidates’ affinities. (d) Competitive ELISA for epitope assessment. The initial antibody concentrations of FV22, FV74 and FV78 were 16, 46 and 74 μg/ml, respectively.
Sequencing and binding activity of selected candidates
Total DNA was isolated from the cell library screened above and the VK+CK+PCMV+VH fragments (Figure 1a) were amplified using PCR. Then, the PCR products were inserted into the PGEM-T vector for transformation. A total of 120 clones were selected for the sequence homology analysis. Thirteen representative antibody sequences out of 120 were chosen and used to prepare full-length IgG1 antibodies. Most of the clones (with the exception of FV6, FV45, FV70, FV81, FV114 and FV117) showed binding activity similar to MIL75 in the ELISA results shown in Figure 3b. Then, the remaining seven antibody clones were analyzed by Biacore. The affinities of all detected antibodies are shown in the same order of magnitude. FV22 (KD=0.14 nM), FV74 (KD=0.19 nM) and FV78 (KD=0.21 nM) showed more similar affinities compared with the MIL75 positive control (KD=0.14 nM) (Figure 3c). According to Figure 3d, FV74 and FV78 had a large epitope overlap with MIL75, whereas FV22 had no epitope overlap with MIL75. The antibody transient expression level analysis indicated that the FV78 expression level was the highest among the three antibodies. Therefore, FV78 was chosen for the functional assessment.
Preliminary functional assessment of FV78
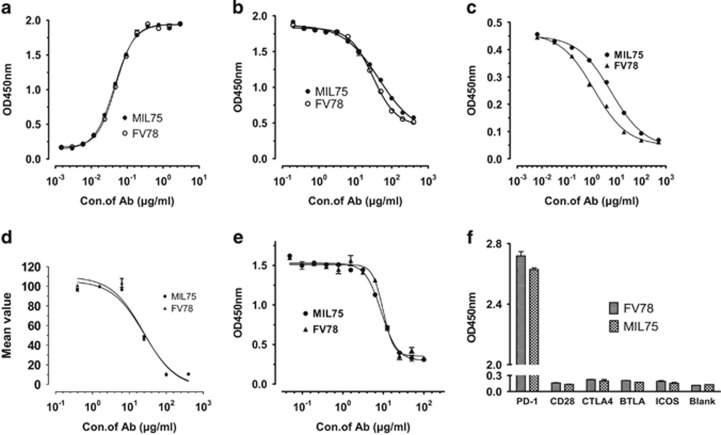
The binding activities of FV78 and MIL75 in the PD-1-coated plates are shown in Figure 4a. The EC50 values for FV78 and MIL75 were 0.048 and 0.044 μg/ml, respectively, suggesting that the binding activity of FV78 was similar to MIL75. The analysis of the binding activity of FV78 to the CD28 family indicated that FV78 had no cross-reactivity with the other members of CD28 family (Figure 4e). As shown in Figure 4b, the binding activity of biotin-MIL75 to the PD-1-coated plate was inhibited by FV78 in a dose-dependent manner. The EC50 values for FV78 and MIL75 were 30 and 45 μg/ml, respectively. This result implied that the two mAbs (MIL75 and FV78) might recognize an overlapping epitope region of PD-1. Therefore, the competitive activity of FV78 and MIL75 against the binding of PD-L1 to PD-1 was analyzed (Figure 4c). The binding of biotin-PD-1 to the PD-L1-coated plate was inhibited by FV78 in a dose-dependent manner. The EC50 values of FV78 and MIL75 for the inhibition of the binding between PD-L1 and PD-L1 were 1.230 and 6.059 μg/ml, respectively, which indicated that FV78 was able to inhibit the binding of PD-L1 to PD-1 more effectively. By contrast, no difference in the competitive activity of the antibody against the binding of PD-L1 to PD-1 was detected in the FACS analysis, where the EC50 values of FV78 and MIL75 were 26.61 and 24.61 μg/ml, respectively. Next, the competitive activity of FV78 and MIL75 with PD-L2 bound to PD-1 was compared as shown in Figure 4d. FV78 inhibited the binding of biotin-PD-1 to the PD-L2-coated plate in a dose-dependent manner. The EC50 values of FV78 and MIL75 for the inhibition of the interaction between PDL2 and PD-1 were 9.86 and 8.91 μg/ml, respectively, indicating that FV78 effectively inhibited the interaction between PD-1 and PD-L2 (Figure 4d).
Figure 4.
Preliminary functional assessment of FV78. (a) Binding activity of FV78 to PD-1/Fc. (b) Epitope comparability of FV78 and MIL75 binding to PD-1/Fc by ELISA assay. (c) Analysis of the competitive activity of FV78 and MIL75 against the binding of PD-L1 to PD-1 by ELISA assay. (d) Analysis of the competitive activity of FV78 and MIL75 against the binding of PD-L1 to PD-1 by FACS assay. (e) Analysis of the competitive activity of FV78 and MIL75 against the binding of PD-L2 to PD-1 by ELISA assay. (f) Evaluation of the cross-reactive binding activity of FV78 to other CD28 family members.
The biological activity of FV78 in vitro and in vivo
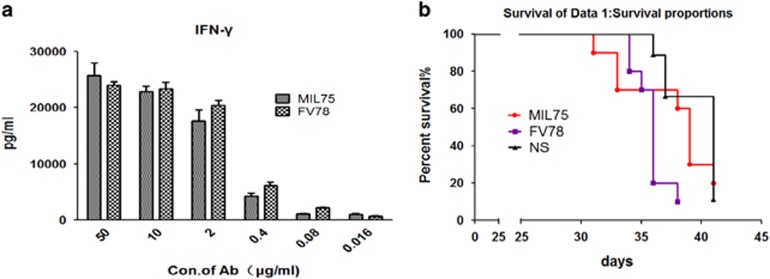
The ability of FV78 to stimulate T-cell responses was evaluated in vitro. Figure 5a showed that IFN-γ release was titratably enhanced because the PD-1/PDL1 suppression signal was blocked once FV78 bound to PD-1. This finding implied that FV78 was able to reverse T-cell activity in a dose-dependent manner. As shown in Figure 5b, when 106 freshly isolated human PBMCs were injected intravenously via the tail vein into adult 6–10-week-old NPG mice, the FV78 antibody effectively activated the immune system, aggravated the graft versus host disease (GVHD) reaction, and accelerated the death of the mice (MST and FV78 vs N.S.=41 days vs 36 days, P<0.05). The effect of the FV78 antibody was not significantly different compared with the positive control MIL75 antibody (MST, FV78 vs MIL75=39 days vs 36, P=0.08). These results indicated that the new FV78 antibody was not weaker than the positive control MIL75 antibody.
Figure 5.
Thein vitro and in vivo biological activity of FV78. (a) In vitro activity analysis of the ability of FV78 to stimulate T cells. Serial concentrations of FV78 and the corresponding MIL75 concentration were added in a concentration gradient. IFN-γ release was detected by ELISA. (b) In vivo biological activity analysis of FV78. FV78 accelerated the death of humanized NPG mice compared to the N.S. condition (P=0.044).
DISCUSSION
Engineering antibodies that target biologically relevant epitopes, whether they are active sites of enzymes or ligand-activated domains of cell surface receptors, are critical for therapeutic lead identification. However, engineering the given antibody using the random mutation method may cause the large majority of the mutant antibodies to lose their biological activity. Furthermore, antibody epitopes that recognize the antigen may drift if some key amino-acid residues are replaced owing to the complexity of the special interaction between the antigen and antibody. Therefore, the substitution of specific amino acids based on structural analysis is a good method. In this study, three antibody candidates (FV22, FV74 and FV78) were selected from a targeted epitope mammalian cell-displayed antibody library. FV74 and FV78 showed large epitope overlaps with MIL75, whereas the FV22 epitope recognized a completely different antigen compared with MIL75.
The CDR regions were compared among the Fv22, Fv78 and MIL75 antibodies using the multiple sequence alignment method (ClustalW, www.ebi.ac.uk). The results showed that Fv22 possessed phenylalanine (F) in the heavy chain Fv fragment at position 58 of the CDR2 region, whereas the MIL75 and Fv78 antibodies possessed arginine (R). Importantly, Fv22 possessed the residues A97T98E99G100D101Y102, Fv78 possessed A97T98N99N100D101Y102 and MIL75 possessed A97T98N99D100D101Y102 in the key CDR3 region and Fv22 possessed the residues D50S51S52N53, Fv78 possessed T50A51S52N53 and MIL75 possessed D50A51S52N53 in the light chain FV fragment of the important CDR2 region. Further analysis of the FV22 sequence showed that the amino acids F in CDR2_H58 and G in CDR3_H100 were introduced unexpectedly by degenerate primer design in the overlap PCR, which caused minor changes in the antibody’s spatial structure and antigen epitope drift from the theoretical analysis. Then, theoretical 3-D structures of the three antibodies’ (FV22, FV78 and MIL75) variable regions were constructed using the computer-guided homology modeling method. The spatial structure of the antibody and human PD-1 complex was modeled based on the molecular docking and dynamics methods results (supplement). The theoretical results showed that the key amino-acid residues in human PD-1 identified by Fv22 were almost completely different from those of FV78 and MIL75. Thus, the predicted key amino-acid sites within the antibody’s CDR region may be crucial for the antibody function. Therefore, site-specific mutations introduced in accordance with the physical and chemical properties of the key amino acids rather than random mutations could better guarantee the quality and proportion of the functional antibodies in the antibody library.
Kirkham31 created an anti-barstar antibody repertoire based on structural information of the interaction between the barnase enzyme and the corresponding inhibitor barstar. Similarly, in this study we circumvented the complexity of the natural immune system using precisely defined synthetic libraries. As a result, we were able to investigate the special role that the key amino-acid residues played in the protein–protein-binding sites. We generated libraries with restricted diversities and displayed the diverse surfaces on a fixed scaffold (that is, human IgHV3-33 for the heavy chain variable domain and human IgKV3-11 for the light chain variable domain) that was formed by the framework regions and buried CDR residues. Our results demonstrated that the key amino-acid residue (that is, those belonging to the ligand–receptor interaction) side chains were capable of mediating most of the contacts necessary for high-affinity antigen recognition in the context of a suitable scaffold. Thus, it seems very likely that the overabundance of key amino-acid residues in natural ligand–receptor-binding sites is a consequence of the side chains being particularly well suited to make productive contacts with the antibody–antigen interaction.
The mammalian cell antibody library is a very promising technology platform compared with the traditional antibody library technology (that is, the phage antibody library or ribosome antibody library). Full-length IgG or Fab displayed on the mammalian cell adopts a natural structure and subtle protein post-translational modifications. The FACS high-throughput screening method can be used to rapidly screen candidates with high affinity and high expression levels. The antibodies converted from the candidates screened from the mammalian cell antibody library could fully maintain the desired properties (that is, affinity and expression level). However, the diversity of the constructed cell library is only ~106 owing to the limitations of the transfection approach and efficiency shortages of site-directed integration, which limits its applications. In this work, we constructed a targeted epitope mammalian cell-displayed antibody library based on computer-aided analysis and design, which could contribute to the construction of an improved library. The use of computer-aided molecular design for the virtual antibody library could increase the proportions of functional antibodies within the antibody library to a certain extent compared with the random mutation antibody library proposed in Zhou’s20 article (the functional antibodies in this article vs Zhou’s article were 31.90 vs 7%).
In summary, our results demonstrated that synthetic antibody libraries could be used to investigate the relative suitability of different amino acids for antigen recognition. By combining the usage of theoretical predictions with the mammalian cell antibody library-screening platform, Fv78 was screened and selected as a novel anti-human PD-1 antibody. FV78 possessed equivalent affinity and a highly overlapping epitope with MIL75. Furthermore, FV78 could efficiently block the interaction between PD-1 and PD-L1/PD-L2. FV78 was more efficient in blocking the PD-L1 and PD-1 interaction than MIL75 in the competitive ELISA, although no difference was detected in the FACS assay. The in vitro IFN-γ secretion experiment implied that FV78 could stimulate T cells in a dose-dependent manner. We evaluated the antibody’s preliminary activity in vivo using on the GVHD mouse model. Although the GVHD animal model is not an ideal animal model for PD-1 antibody evaluation, the current results indicated that FV78 could accelerate the death of PBMC-grafted NPG mice. Additional animal models, such as Homo sapiens PD-1 transgenic mice, are needed for in vivo pharmacodynamics evaluations. The abovementioned results highlight the probability and prospect of FV78 becoming a new potential antibody drug.
Acknowledgments
The work was supported by the National Natural Sciences Foundation of China grant (No. 81272528 and No.31370938), National High Technology Research and Development Program (863 Program, No. 2012AA02A302), National Science and Technology Major Projects for 'Major New Drugs Innovation and Development' (2014ZX09304311-001-002-004) and the Beijing Natural Science Foundation (No. 5152022).
Footnotes
Supplementary Information for this article can be found on the Cellular & Molecular Immunology website (http://www.nature.com/cmi)
The authors declare no conflict of interest.
Supplementary Material
References
- Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol 2006; 6: 343–357. [DOI] [PubMed] [Google Scholar]
- Presta LG. Molecular engineering and design of therapeutic antibodies. Curr Opin Immunol 2008; 20: 460–470. [DOI] [PubMed] [Google Scholar]
- Forsyth CM, Juan V, Akamatsu Y, DuBridge RB, Doan M, Ivanov AV et al. Deep mutational scanning of an antibody against epidermal growth factor receptor using mammalian cell display and massively parallel pyrosequencing. mAbs 2013; 5: 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit AG, Mariuzza RA, Phillips SE, Poljak RJ. Three-dimensional structure of an antigen-antibody complex at 2.8A resolution. Science 1986; 233: 747–753. [DOI] [PubMed] [Google Scholar]
- Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 1986; 321: 522–525. [DOI] [PubMed] [Google Scholar]
- Berek C, Griffiths GM, Milstein C. Molecular events during maturation of the immune response to oxazolone. Nature 1985; 316: 412–418. [DOI] [PubMed] [Google Scholar]
- Lea S, Stuart D. Analysis of antigenic surfaces of proteins. FASEB J 1995; 9: 87–93. [DOI] [PubMed] [Google Scholar]
- Padlan EA. On the nature of antibody combining sites: unusual structural features that may confer on these sites an enhanced capacity for binding ligands. Proteins 1990; 7: 112–124. [DOI] [PubMed] [Google Scholar]
- Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA et al. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol 2003; 334: 733–749. [DOI] [PubMed] [Google Scholar]
- Davies DR, Cohen GH. Interactions of protein antigens with antibodies. Proc Natl Acad Sci USA 1996; 93: 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padlan EA. Anatomy of the antibody molecule. Mol Immunol 1994; 31: 169–217. [DOI] [PubMed] [Google Scholar]
- Rao BM, Lauffenburger DA, Wittrup KD. Integrating cell-level kinetic modeling into the design of engineered protein therapeutics. Nat Biotechnol 2005; 23: 191–194. [DOI] [PubMed] [Google Scholar]
- De Genst E, Areskoug D, Decanniere K, Muyldermans S, Andersson K. Kinetic and affinity predictions of a protein-protein interaction using multivariate experimental design. J Biol Chem 2002; 277: 29897–29907. [DOI] [PubMed] [Google Scholar]
- Zhang W, Feng J, Li Y, Guo N, Shen B. Humanization of an anti-human TNF-α antibody by variable region resurfacing with the aid of molecular modeling. Mol Immunol 2005; 42: 1445–1451. [DOI] [PubMed] [Google Scholar]
- Qin W, Feng J, Zhang W, Li Y, Shen B. A novel TNFalpha antagonizing peptide-Fc fusion protein designed based on CDRs of TNFalpha neutralizing monoclonal antibody. Biochem Biophys Res Commun 2004; 322: 1024–1028. [DOI] [PubMed] [Google Scholar]
- Chang H, Qin W, Li Y, Zhang J, Lin Z, Lv M et al. A novel human scFv fragment against TNF-alpha from de novo design method. Mol Immunol 2007; 44: 3789–3796. [DOI] [PubMed] [Google Scholar]
- Qiao C, Lv M, Li X, Geng J, Li Y, Zhang J et al. Affinity maturation of antiHER2 monoclonal antibody MIL5 using an epitope-specific synthetic phage library by computational design. J Biomol Struct Dyn 2013; 31: 511–521. [DOI] [PubMed] [Google Scholar]
- Yang J, Wang Q, Qiao C, Lin Z, Li X, Huang Y et al. Potent anti-angiogenesis and anti-tumor activity of a novel human anti-VEGF antibody, MIL60. Cell Mol Immunol 2014; 11: 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol 2005; 23: 1105–1116. [DOI] [PubMed] [Google Scholar]
- Zhou C, Jacobsen FW, Cai L, Chen Q, Shen WD. Development of a novel mammalian cell surface antibody display platform. mAbs 2010; 2: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Honjo T. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol 2006; 27: 195–201. [DOI] [PubMed] [Google Scholar]
- Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 2002; 8: 793–800. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci USA 2002; 99: 12293–12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol 2008; 8: 467–477. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366: 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasboas JC, Ansell SM. Nivolumab for the treatment of classical Hodgkin lymphoma after failure of autologous stem cell transplant and brentuximab. Expert Rev Anticancer Ther 2016; 16: 5–12. [DOI] [PubMed] [Google Scholar]
- Ehrenmann F, Kaas Q, Lefranc M-P. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res 2010; 38: D301–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenmann F, Lefranc M-P. IMGT/DomainGapAlign: IMGT standardized analysis of amino acid sequences of variable, constant, and groove domains (IG, TR, MH, IgSF, MhSF). Cold Spring Harb Protoc 2011; 2011: 737–749. [DOI] [PubMed] [Google Scholar]
- Gronwald RG, Grant FJ, Haldeman BA, Hart CE, O’Hara PJ, Hagen FS et al. Cloning and expression of a cDNA coding for the human platelet-derived growth factor receptor: evidence for more than one receptor class. Proc Natl Acad Sci USA 1988; 85: 3435–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak KM, Kitel R, Przetocka S, Golik P, Guzik K, Musielak B et al. Structure of the complex of human programmed death 1, PD-1, and Its ligand PD-L1. Struct Lond Engl 1993 2015; 23: 2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham PM, Neri D, Winter G. Towards the design of an antibody that recognises a given protein epitope. J Mol Biol 1999; 285: 909–915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.