Abstract
MicroRNAs (miRNAs) function as important regulators in the immune response and inflammation. Several approaches have been reported to computationally predict miRNAs and their potential targets. However, there are still many miRNA–target interactions that are unpredictable by using the current computational algorithms. We established a miRNA in vivo precipitation method (miRIP) to identify unpredictable miRNAs with definite targets in these cells. Because Stat3 is a well-known transcription factor involved in innate immunity and inflammation, we utilized the miRIP method to identify miRNAs that bind Stat3 mRNA in macrophages. Among the captured miRNAs, miR-151-3p was confirmed to interact with Stat3 mRNA 3′-UTR and downregulate the Stat3 protein levels. LPS stimulation decreased miR-151-3p expression, thereby increasing IL-6 production. Therefore, we found that miR-151-3p inhibited LPS-induced IL-6 production by targeting Stat3. These data further confirmed miRIP as an efficient method to identify unpredictable miRNAs and explore miRNAs-mediated regulation in innate immunity and inflammation.
Keywords: innate immunity, IL-6, macrophage, miR-151-3p, Stat3
Introduction
MicroRNAs (miRNAs) are small non-coding RNA molecules that play important roles in various physiological and pathophysiological processes, including immune response and inflammation.1, 2, 3 As the first defense line against invading pathogens, the innate immune system is tightly regulated by a wide range of factors via post-transcriptional and epigenetic modifications.4, 5, 6 Among these factors, miRNAs have been widely demonstrated to be involved in important signaling pathways in innate immune responses.7, 8, 9, 10 Several miRNAs play pro-inflammatory roles, typified by miR-155, and others are negative feedback regulators of inflammatory responses, such as miR-146a and miR-29.11, 12, 13 Although the interaction between innate immune-related genes and miRNAs has been elucidated in many studies, the roles of miRNAs in regulating innate immune responses are far from fully understood. In addition, identifying miRNAs that target specific mRNA in vivo remains a significant challenge, as one miRNA can target multiple mRNAs and vice versa.
Many transcriptional factors have been identified as important initiators and regulators of innate immunity and inflammation. The superfamily of the signal transducer and activator of transcription (STAT) is well known in the field. Among the Stat superfamily, Stat3, predominantly activated by gp130-acting cytokines (for example, IL-6, IL-11, oncostatin-M, LIF), is critical in the signal transduction of cytokine function and the production of pro-inflammatory cytokines.14 These cytokines bind to their receptors and induce the activation of Jak kinases, which activate Stat3. Then, Stat3 is phosphorylated, dimerized and translocated into the nucleus, where it can initiate and regulate the expression of target genes, such as IL-6. In addition to the increase of phosphorylated Stat3 (p-Stat3), total Stat3 protein is upregulated upon the stimulation of LPS and IL-6.15, 16 However, the epigenetic mechanism underlying the upregulation of total Stat3 protein in innate immune responses remain unclear.
Previous studies have focused on the regulation of the Stat3 gene at the transcriptional level; for example, the Stat3 gene itself is activated via IL-6 signaling through an IL-6 response element within the Stat3 gene promoter that contains both a low-affinity Stat3-binding element and a cAMP response element, thus activating the Stat3 gene in cooperation with an unidentified CREB.17 Post-transcriptional control has increasingly emerged as a prominent mechanism of Stat3 regulation, particularly through miRNA targeting. Multiple miRNAs, such as miR-17, miR-20a, miR-93, miR-106b, miR-125b and miR-199, were identified to target Stat3.18, 19, 20 However, further investigation is needed to identify other Stat3-targeting miRNAs under physiological conditions, particularly in innate immunity.
The 3′-untranslated region (3′-UTR) sequence of mouse Stat3 mRNA contains ~1895 nucleotides, suggesting that Stat3 mRNA is targeted by many miRNAs. Identifying the true miRNAs that target Stat3 mRNA is a challenging issue. To characterize the in vivo mRNA:miRNA interactions that cannot be predicted by using the current computational algorithms, we established the miRIP method (miRNA in vivo precipitation) and used this method to identify miR-92a, which targets p21 mRNA.21 In this study, by using biotin-tagged anti-sense oligonucleotides for Stat3 mRNA (Stat3 probe), we identified specific Stat3-interacting miRNAs. Among these miRNAs, miR-151-3p was confirmed to interact with Stat3 mRNA and consequently downregulate the Stat3 protein levels in mouse macrophages in response to LPS stimulation. LPS stimulation decreased the level of miR-151-3p in mouse macrophages, leading to the upregulation of Stat3 protein level and promotion of the production of the pro-inflammatory cytokine IL-6. In this study, miRIP was further demonstrated to be an efficient method for identifying unpredictable interacting miRNAs in the immune system, and furthermore, miR-151-3p was found to be important in amplifying the initial innate immune responses upon pathogen invasion.
Materials and methods
Mice and cell cultures
Male C57BL/6 mice (6–8 weeks) obtained from Joint Ventures Sipper BK Experimental Animal were used to prepare primary mouse peritoneal macrophages. All animal experiments were conducted according to the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Second Military Medical University, Shanghai. The RAW264.7 cell line was obtained from the American Type Culture Collection.
Primary mouse peritoneal macrophages were prepared and cultured, as previously described.22 RAW264.7 cells were cultured and maintained in DMEM containing 10% FBS at 37 °C in 5% CO2. All cell culture media and other reagents were entirely free of endotoxins.
miRNAs in vivo precipitation (miRIP)
miRIP was performed as previously described.21 Briefly, RAW264.7 cells were transfected with a biotin-tagged specific probe or a control probe 24 h before the cells were coll (the probe sequences are listed in Supplementary Table S1). The cells were cross-linked using 1% formaldehyde for 10 min, equilibrated in glycine buffer for 5 min, washed three times with cold PBS, scraped with 1 ml of lysis buffer and incubated for 10 min. The lysis mixtures were sonicated (VCX130, SONICS, and MATERIALS) using the following parameters: 50% amplitude, 30 s of a constant pulse, and a 30-s pause for 10 min. Subsequently, the samples were centrifuged at 10 000g for 10 min. The supernatant lysate was incubated with 1 ml of M-280 beads (Invitrogen, Carlsbad, CA, USA) for 1 h under rotation. The beads sample mixture was washed twice with wash buffer, and finally incubated with 200 μl of lysis buffer for 2 h to reverse the formaldehyde cross-links. Subsequently, the RNA was extracted using TRIzol. After DNase I treatment, the RNA was repurified using TRIzol.
Exiqon multiplex miRNA arrays
Expression analysis of miRNAs was performed using the miRCURY LNA Universal RT microRNA PCR System (Exiqon, Vedbaek, Denmark) according to the manufacturer's instructions. Initial data analysis was performed using the software supplied with the real-time PCR instrument to obtain raw Ct values (Ct or Cq, depending on PCR instrument). Briefly, two miRCURY LNA Universal RT microRNA PCRs were performed in this study (GSE77873): one was for the RNA samples identified using the Stat3 specific probe P2, and the other was for the RNA samples identified using the control probe C5. First, the raw Ct value of all miRNAs in P2 and C5 samples were normalized to the Ct value of U6 in P2 and C5 samples, respectively, to yield the ΔCt value. Next, the ΔCt values of all miRNAs from the P2 sample were normalized to ΔCt values from the C5 sample to yield ΔΔCt. Finally, the 2−ΔΔCt value was calculated for each miRNA as the relative level of a certain miR in the P2 probe sample compared with the C5 probe sample. If the P2/C5 ratio exceeded 10, then a specific interaction exists, and the miRNA is ‘enriched’.
Vector construction
The selected region (2723–3279) of Stat3 mRNA 3′-UTR was amplified from normal mouse genomic DNA using the primers listed in Supplementary Table S1 and inserted into the Hind III/Spe I sites of the pMIR-Report construct (Ambion, Austin, TX, USA). The Stat3 mRNA 3′-UTR mutant fragment was constructed and amplified by two PCR rounds. In the first round of PCR, Stat3 3′-UTR F primer and Stat3 3′-UTR Mut R primer were utilized for the clone of the first half of Stat3 mRNA 3′-UTR mutant fragment, and Stat3 3′-UTR Mut F primer and Stat3 3′-UTR R primer were utilized for the second round. Then, the PCR products of the first round were mixed and diluted for use as a template in the second round of PCR. The Stat3 3′-UTR mutant fragment was finally amplified using Stat3 3′-UTR F/R primer and inserted into the Hind III/Spe I sites of the pMIR-Report construct (Ambion). The coding fragment of Stat3 was amplified using the primers listed in Supplementary Table S1 from normal mouse complementary DNA (cDNA) and inserted into the Xho I/Hind III sites of the pBflag construct (Ambion). All constructs were verified by direct sequencing.
Oligonucleotide transfection
RNA oligonucleotides were chemically synthesized and purified at Invitrogen. The sense sequence of mouse miR-151-3p mimics was 5′-CUA GAC UGA GGC UCC UUG AGG-3′ and the antisense sequence was 5′-UCA AGG AGC CUC AGUCUA GUU-3′. Negative control mimics were 5′-UUC UCCGAACGU GUC ACG UdTdT-3′ and 5′-ACG UGA CAC GUUCGG AGA AdTdT-3′. The sequence of the mouse miR-151-3P inhibitor was 5′-CCU CAA GGA GCC UCA GUC UAG-3′ and the sequence of the inhibitor control was 5′-CAG UAC UUUUGU GUA GUA CAA-3′. The Stat3 siRNAs mix was obtained from Genepharma (Shanghai, China). The transfections were performed using INTERFERin reagent (Polyplus-transfection, Illkirch, France) and Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. The final concentrations of Stat3 siRNAs, miR-151-3p mimics and inhibitor used were 20, 50 and 100 nM.
RNA isolation, reverse-transcription, and quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cell lines and mouse primary macrophages using TRIzol reagent (Invitrogen). cDNA synthesis was performed using ReverTra Ace (TOYOBO, Osaka, Japan) and the All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA), respectively, according to the manufacturer's instructions.
Amplification and detection were performed using the Light Cycler 480II/96 and the LightCycler2.0 (Roche, Rotkreuz, Switzerland). The miR-151-3p and Stat3 mRNA levels were normalized to U6 and Actin, respectively, to yield a 2−ΔΔCt value for the relative expression of each transcript. The primers used are shown in Supplementary Table S1.
Luciferase reporter assay
Briefly, the cells plated in a 24-well plate were co-transfected with 50 nM miRNA mimics or 100 nM miRNA inhibitor, 25 ng of firefly luciferase reporter comprising Stat3 3′-UTR, and 6 ng of pRL-TK (Promega, Madison, WI, USA) using jetPRIME and jetPEI Macrophages (Polyplus-transfection, Illkirch, France). The cells were collected at 36 h after the last transfection and analyzed using a Dual-Luciferase Reporter Assay System (Promega).
Western blotting
Total protein was extracted after lysing the cells in RIPA buffer containing protease inhibitors. The protein samples were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes. After blocking with 5% non-fat milk in TBS-T, the membranes were incubated with primary antibody. The following antibodies were used: anti-Erk (1:3000), anti-Phospho-Erk (1:3000), anti-p65 (1:3000), anti-Phospho-p65 (1:3000), anti-IKKβ (1:3000), anti-Phospho-IKKβ (1:3000), anti-IkBα (1:3000), anti-Phospho-IkBα (1:3000), anti-Stat3 (79D7) (1:3000), anti-Phospho-Stat3 (Tyr-705) (1:3000), anti-β-Actin (1:10 000), anti-Gapdh (1:10 000). Goat-anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (1:2000) and goat-anti-mouse IgG conjugated to horseradish peroxidase (HRP) (1:2000) (Cell Signaling Technology, Danvers, MA, USA) were used as the secondary antibodies. Protein was detected with image acquisition using Chemiluminescent Western Blot Scanner (Gene Company, Hong Kong, China).
Statistical analysis
All data are presented as the means±s.d. Student's t test was used to analyze the difference between two experimental groups, and a P-value<0.05 indicates statistical significance.
Results
Identification of miR-151-3p as an unpredictable Stat3 mRNA-targeting miRNA
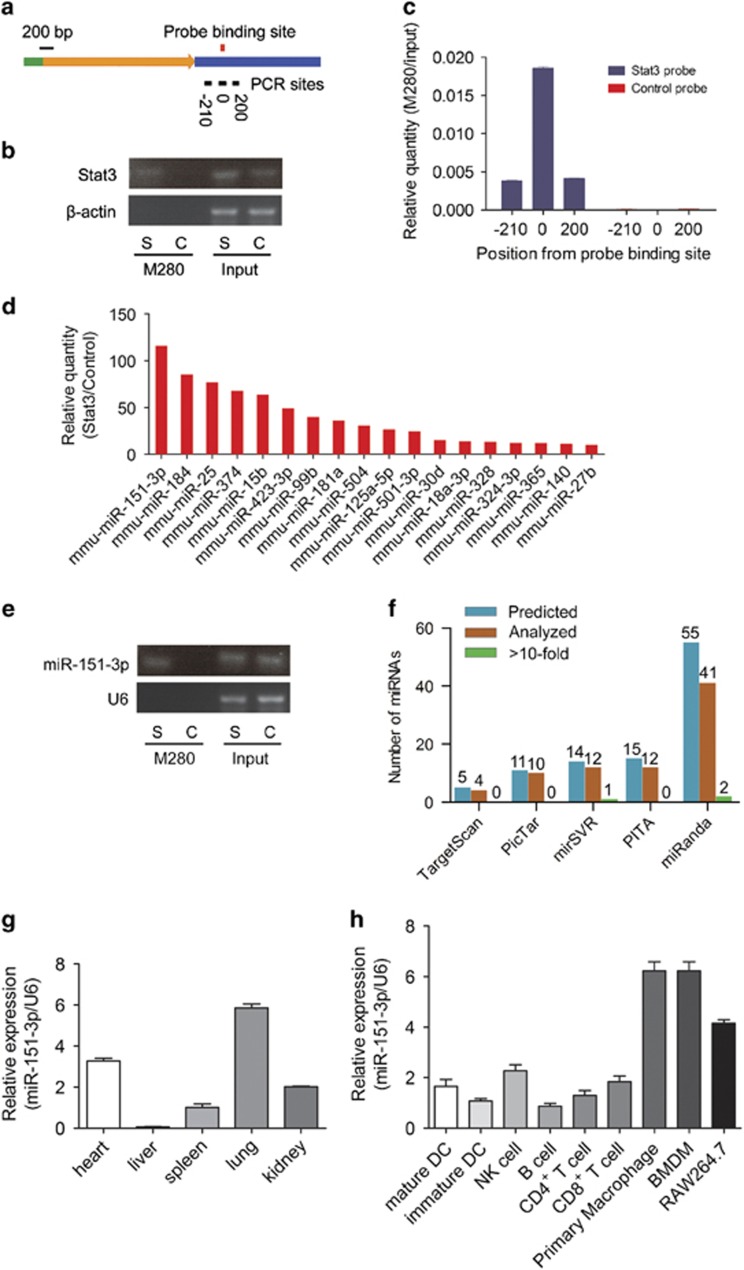
The 3′-UTR of Stat3 mRNA is relatively long with ~1895 nucleotides, indicating that Stat3 mRNA is a potential target for miRNAs (Figure 1a). To explore whether miRNAs are involved in the regulation of Stat3 in vivo, miRIP was performed to purify miRNAs binding to Stat3 mRNA. The Stat3 mRNA probe was transfected into RAW264.7 cells to hybridize Stat3 mRNA for 24 h. The interacting region of Stat3 mRNA was purified using Streptavidin Dynabeads (M-280) and isolated using TRIzol reagent. The enriched region of Stat3 mRNA was confirmed by RT-PCR (Figure 1b), confirming the specificity of miRIP. The scope analysis of purified mRNA was also conducted by RT-PCR using multiple primers in different positions. As shown in Figure 1c, the majority segments purified by miRIP in RAW264.7 cells were ~500 bp around the probe binding site.
Figure 1.
Identification of miR-151-3p as an unexpected Stat3 mRNA-targeting miRNA. (a) Stat3 mRNA and probe-binding sites. The green bar, 5′-UTR; the yellow arrow, coding sequence; the blue bar, 3′-UTR; the black bars and the numbers at the bottom denote regions analyzed using RT-qPCR; red bar, the probe binding sites. (b) RT-qPCR analysis of region '0' of Stat3 mRNAs from Stat3 probe and control probe affinity purified complex in RAW264.7 cells. S, Stat3 probe; C, Control probe. (c) RT-qPCR analysis of regions of Stat3 mRNAs from Stat3 probe and control probe affinity purified complex in RAW264.7 cells. (d) Multiple miRNA array data of Stat3 probe and control probe affinity purified complex in RAW264.7 cells. The data are shown as normalized values for the enriched miRNAs (>10-fold). (e) RT-qPCR analysis of the most enriched miR-151-3p. S, Stat3 probe; C, control probe. (f) The comparisons of miRNAs identified by miRIP and predicted using TargetScan, PicTar, PITA, miRanda and mirSVR in the 500-bp region around probe-binding sites for Stat3 mRNAs. (g) The expression of miR-151-3p in different organs was measured using RT-qPCR and normalized to the expression of U6. (h) The expression of miR-151-3p in different immune cells was measured using RT-qPCR and normalized to the expression of U6. Data are shown as the means±s.d. (n=3) of one representative experiment. Similar results were obtained in at least three independent experiments.
Exiqon multiplex miRNA array was performed to simultaneously analyze several hundred miRNAs (GSE77873). The top 18 enriched miRNAs are shown in Figure 1d, and miR-151-3p, the most robustly interacting miRNA, was confirmed by RT-PCR (Figure 1e). To examine whether the purified miRNAs could be predicted by computational algorithms, TargetScan, PicTar, PITA, miRanda and mirSVR were utilized to predict all potential miRNAs targeting a given mRNA region isolated using the Stat3-specific probe. Surprisingly, several robustly interacting miRNAs (>10-fold) identified using this technology could not be predicted by computational prediction algorithms, and several of the predicted miRNAs showed a weak interaction with Stat3 mRNA in RAW264.7 cells (Figure 1f and Supplementary Table S2). These data reveal that previous computational algorithms are defective to predict the truly enriched Stat3 mRNA binding miRNAs, and our in vivo miRIP assay can identify miR-151-3p as an unpredictable microRNA that binds Stat3 mRNA.
The distribution of miR-151-3p was further examined among different organs and different immune cells. The results showed that miR-151-3p was highly expressed in the lungs and heart and enriched in macrophages compared with other hematopoietic cells (Figures 1g and h). In addition, CD4+ T cells, CD8+ T cells and B cells were isolated from spleens of wild-type mice. The expression of miR-151-3p remained unchanged during the activation of CD4+ T cells (Supplementary Figure 1a), CD8+ T cells (Supplementary Figure 1b) and B cells (Supplementary Figures 1c and d). These results strongly indicated that miR-151-3p is more likely to participate in the function of macrophages instead of T/B cells.
miR-151-3p targets Stat3 mRNA and regulates Stat3 expression in macrophages
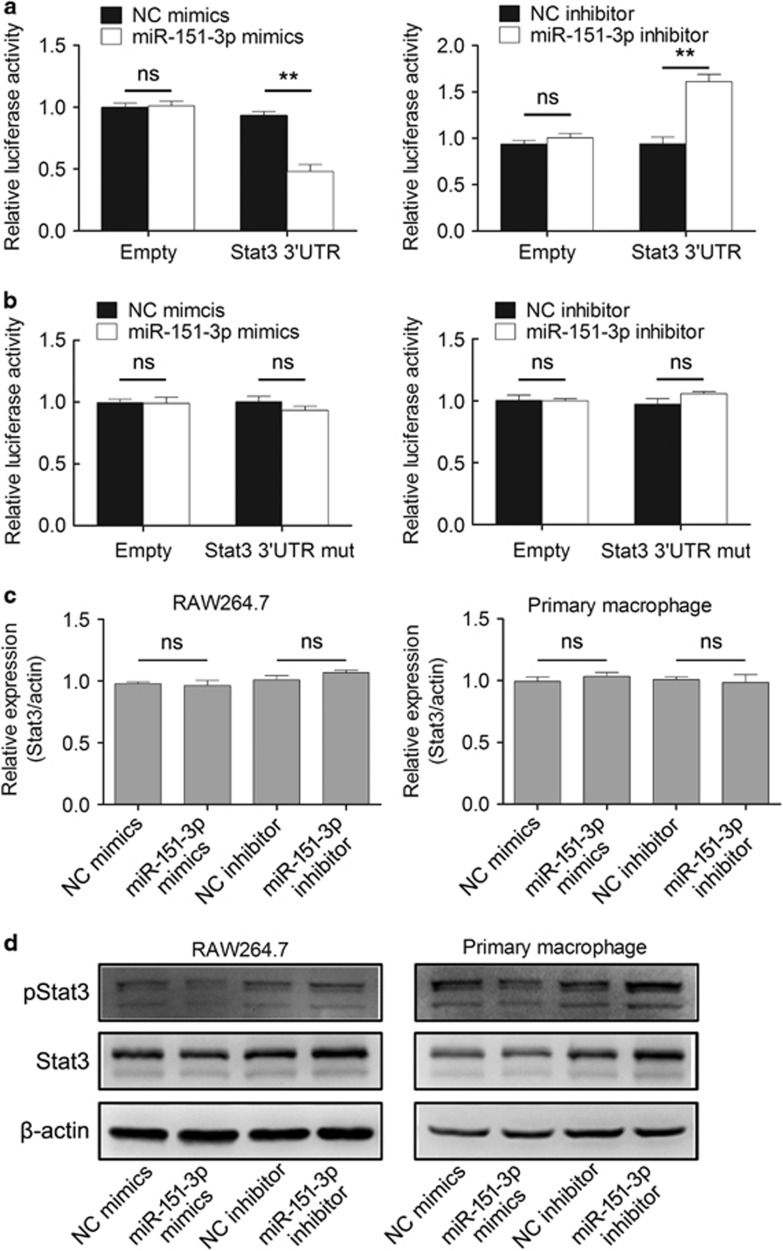
Several experiments were performed to confirm whether miR-151-3p regulates the expression of the target gene Stat3. Luciferase assay was conducted to confirm the interaction between miR-151-3p and the 3′-UTR of Stat3 mRNA. Luciferase reporter plasmids with a selected region (2723–3279) of Stat3 3′-UTR or Stat3 3′-UTR mutant (fragment 474–537 of the selected region were deleted) were constructed, and a dual-luciferase reporter assay was employed in RAW264.7 cells. The results indicated that the expression of luciferase reporter plasmid containing 3′-UTR of Stat3 was inhibited by co-transfection with miR-151-3p mimics, and the opposite result was obtained when transfected with miR-151-3p inhibitor (Figure 2a). In addition, no difference was observed between scrambled negative control mimics and miR-151-3p mimics in cells co-transfected with Stat3 3′-UTR mutant luciferase reporter plasmid, and a similar result was obtained after transfection with a miR-151-3p inhibitor (Figure 2b).
Figure 2.
miR-151-3p targets Stat3 mRNA and regulates Stat3 expression in macrophages. (a and b) Analysis of luciferase activity. RAW264.7 cells were co-transfected with pMIRfirefly luciferase reporter plasmids, pTK-Renilla luciferase plasmids, together with miR-151-3p inhibitor or mimics. After 36 h, firefly luciferase activity was measured and normalized to Renilla luciferase activity. (c) Effects of miR-151-3p on the endogenous Stat3 mRNA level were analyzed using RT-qPCR in RAW264.7 cells and primary peritoneal macrophages. (d) Effects of miR-151-3p on the endogenous Stat3 protein and p-Stat3 levels after LPS stimulation for 30 min were analyzed by western blotting in RAW264.7 cells and primary peritoneal macrophages. The data are shown as the means±s.d. (n=3) of one representative experiment. Similar results were obtained in at least three independent experiments. **P<0.01, unpaired Student's t-test. NC, negative control oligonucleotides; ns, not significant.
Furthermore, RAW264.7 cells were transfected with miR-151-3p inhibitor or mimics. RT-PCR and western blot results showed that the miR-151-3p inhibitor or mimics did not affect the level of Stat3 mRNA and that the Stat3 protein level was increased in the presence of the miR-151-3p inhibitor and decreased by miR-151-3p mimics in LPS stimulated macrophages (Figures 2c and d,Supplementary Figure 2a). Similar results were observed in primary peritoneal macrophages (Figures 2c and d,Supplementary Figure 2a). Therefore, miR-151-3p may target the Stat3 mRNA 3′-UTR and then downregulate the expression of Stat3 at the post-transcriptional level in macrophages.
LPS significantly decreases miR-151-3p and upregulates Stat3 protein levels in macrophages
The expression of miRNAs, such as miR-146a and miR-155, is downregulated after LPS stimulation. To confirm whether miR-151-3p is regulated by LPS stimulation in macrophages, RAW264.7 cells were stimulated with LPS, and the expression of miR-151-3p was analyzed using RT-PCR. miR-151-3p was rapidly downregulated in RAW264.7 cells within 30 min after LPS stimulation (Figure 3a). A similar downregulation pattern of miR-151-3p was observed in primary peritoneal macrophages and bone marrow-derived macrophages in response to LPS stimulation (Figures 3a and b). However, the expression of miR-151-3p in bone marrow-derived dendritic cells (BMDC), mouse embryo fibroblasts (MEF) and colon cancer cell line CT-26 did not fit this pattern (Figure 3b).
Figure 3.
LPS stimulation downregulates miR-151-3p expression and upregulates Stat3 protein levels in macrophages. (a) RAW264.7 cells and primary peritoneal macrophages were stimulated with or without 100 ng/ml LPS for the indicated time points. The expression of miR-151-3p was measured using RT-qPCR and normalized to the expression of U6. (b) Bone marrow-derived macrophages (BMDM), bone marrow-derived dendritic cells (BMDC), mouse embryo fibroblasts (MEF) and the colon cancer cell line CT-26 were stimulated with or without 100 ng/ml LPS for the indicated time points. The expression of miR-151-3p was measured using RT-qPCR and normalized to the expression of U6. (c and d) The levels of p-Stat3 and Stat3 protein (c) and Stat3 mRNA (d) were analyzed by western blotting and RT-qPCR in macrophages stimulated with or without LPS (100 ng/ml) as indicated. Data are shown as the means±s.d. (n=3) of one representative experiment. Similar results were obtained in at least three independent experiments. *P<0.05; **P<0.01, unpaired Student's t-test.
To examine the role of miR-151-3p, a Stat3 mRNA binding miRNA, in innate response and inflammation, we first analyzed pStat3 activation in macrophages (RAW264.7 cells and primary peritoneal macrophages) in response to LPS stimulation (Figure 3c and Supplementary Figure 2b). The results showed that both p-Stat3 and total Stat3 levels were increased upon LPS stimulation in macrophages, consistent with the results of previous studies.13, 14 The levels of p-Stat3 rapidly increased, peaking at 30 min and returned to relatively low levels by 60 min. An analysis of Stat3 mRNA in cells stimulated with LPS was also performed. The results showed Stat3 mRNA levels remained unchanged during the same period (Figure 3d), suggesting the posttranscriptional regulation of Stat3. Collectively, these results suggest that LPS stimulation rapidly decreases miR-151-3p, consequently leading to an increase in the Stat3 levels in macrophages posttranscriptionally, which eventually enhances Stat3 phosphorylation.
LPS-induced downregulation of miR-151-3p promotes Stat3-dependent gene expression in macrophages
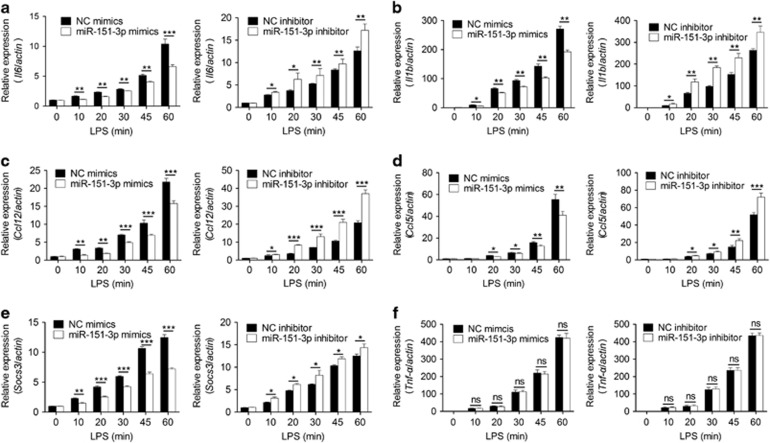
To explore the physiological significance of the LPS-induced downregulation of miR-151-3p, we examined the expression of many Stat3-dependent genes and used Tnf as a Stat3 independent control gene. The results showed that many Stat3-dependent genes, such as IL-6 (Figure 4a), IL-1b (Figure 4b), Ccl12 (Figure 4c), Ccl5 (Figure 4d) and Socs3 (Figure 4e), were upregulated after the transfection of miR-151-3p mimics in LPS-stimulated macrophages compared with scrambled negative control mimics. Similar results were not observed for the expression of Tnf-α (Figure 4f). Consistently, miR-151-3p inhibitor-transfected macrophages decreased Stat3-dependent gene expression compared with scrambled negative control inhibitor (Figures 4a–e). On the basis of these results, the LPS-induced downregulation of miR-151-3p could play important roles in innate immunity by regulating Stat3.
Figure 4.
miR-151-3p downregulates Stat3 dependent genes in macrophages. Macrophages were stimulated with or without 100 ng/ml LPS for the indicated time points. The expression of IL-6 (a), IL-1b (b), Ccl5 (c), Ccl12 (d), Socs3 (e) and Tnf-α (f) was measured using RT-qPCR. Data are shown as the mean±s.d. (n=3) of one representative experiment. Similar results were obtained in at least three independent experiments. *P<0.05; **P<0.01, unpaired Student's t-test.
IL-6 is not involved in instant Stat3 activation after LPS stimulation
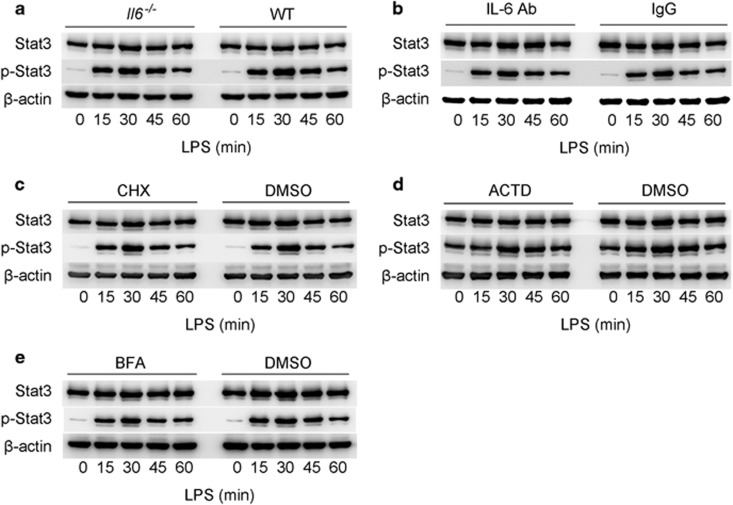
As IL-6 is a strong stimulator of Stat3 activation, and LPS triggers IL-6 via the NF-κB pathway, the activation of Stat3 after LPS stimulation may be a subsequent event of IL-6 secretion. Thus, we examined whether the activation of Stat3 upon LPS stimulation resulted from autocrine or paracrine IL-6. Primary macrophages from IL-6−/− mice were stimulated with LPS and analyzed using western blotting. The results showed that the lack of IL-6 did not affect the phosphorylation of Stat3 within one hour of LPS stimulation (Figure 5a and Supplementary Figure 2c). In addition, we neutralized IL-6 in LPS-stimulated macrophages and found that Stat3 could still be phosphorylated by LPS stimulation within one hour (Figure 5b and Supplementary Figure 2d). Moreover, we inhibited the transcription, translation of IL-6 mRNA and secretion of IL-6 with cyclohexane (CHX), actinomycin D (ACTD) and brefeldin A (BFA), respectively. Western blot results showed instant Stat3 phosphorylation in these cells upon LPS simulation (Figures 5c–e and Supplementary Figures 2e–g). In addition, the observed phenomenon exists within one hour after LPS stimulation, while the standard LPS/IL-6/Stat3 loop requires more than one hour,23 suggesting that LPS may trigger instant Stat3 phosphorylation aside from the canonical LPS/NF-κB/IL-6/Stat3 loop. Taken together, these results convincingly demonstrated that the phosphorylation of Stat3 within one hour of LPS stimulation was not driven by IL-6. However, the detailed mechanism needs further elucidation.
Figure 5.
LPS activates Stat3 in an IL-6 independent manner. (a) Primary macrophages from IL-6−/− and WT mice were treated with LPS (100 ng/ml) for the indicated time points. p-Stat3 and Stat3 were analyzed using immunoblotting. (b) Primary macrophages were treated with IL-6 antibody or mouse normal IgG and subsequently treated with LPS for the indicated time points. p-Stat3 and Stat3 were analyzed through immunoblotting. (c–e) Primary macrophages were treated with CHX, ACTD or BFA for eight hours and then stimulated with LPS (100 ng/ml) for the indicated time points. p-Stat3 and Stat3 levels were analyzed using immunoblotting.
miR-151-3p suppresses IL-6 production in LPS-stimulated macrophages by reducing Stat3 expression
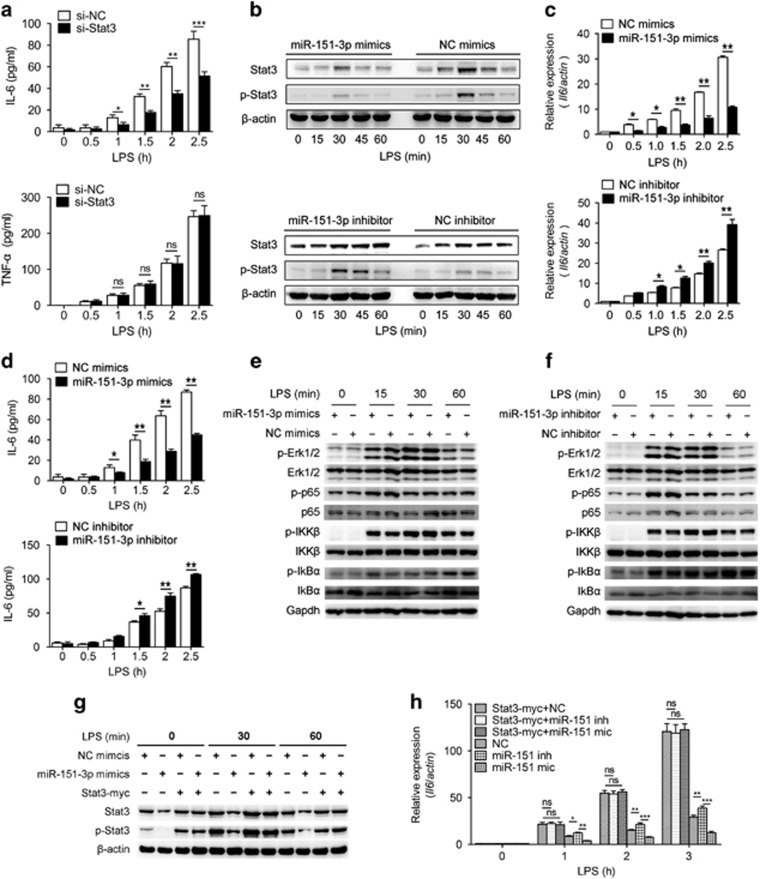
As the upregulation of Stat3 post LPS stimulation was not induced by the secretion of IL-6, we further examined the role of Stat3 in LPS-induced cytokine production. We silenced Stat3 by using Stat3 siRNA, and the results indicated decreased IL-6 production upon LPS stimulation but had no effect on Tnf-α production (Figure 6a).
Figure 6.
miR-151-3p negatively regulates LPS-induced IL-6 production in primary macrophages by targeting Stat3. (a) Primary macrophages were transfected with Stat3 siRNA or a scrambled negative control. Quantification of IL-6 and Tnf-α in cell culture supernatant were determined using ELISA after stimulation with LPS (100 ng/ml) for the indicated time. (b) Primary macrophages were transfected with mimics or inhibitors of miR-151-3p, then stimulated with LPS (100 ng/ml) for the indicated time points. p-Stat3 and Stat3 levels were analyzed by immunoblotting. (c and d) Primary macrophages were transfected with miR-151-3p mimics or inhibitors. After 36 h, the cells were stimulated with or without LPS (100 ng/ml). IL-6 was measured at the indicated time points using RT-qPCR (c) and ELISA assay (d). (e and f) Representative immunoblot analysis of phosphorylated Erk, total Erk, phosphorylated p65, total p65, phosphorylated IKKβ, total IKKβ, phosphorylated IkBα, total IkBα and GAPDH from primary macrophages transfected with miR-151-3p mimics (e) or inhibitors (f) and then stimulated with 100 ng/ml LPS for the indicated time points. (g and h) Primary macrophages were transfected with negative control, miR-151-3p mimics, negative control plus the Stat3-myc overexpression plasmid as indicated. After 48 h, the cells were stimulated with LPS (100 ng/ml) for the indicated time points. The levels of Stat3 and p-Stat3 were analyzed by immunoblotting (g), and the levels of IL-6 mRNA were measured using RT-qPCR (h). The data are shown as the means±s.d. (n=3) of one representative experiment. Similar results were obtained in at least three independent experiments. NC, scrambled negative control oligonucleotides. *P<0.05; **P<0.01; ***P<0.001, unpaired Student's t-test.
We further investigated whether the LPS-induced downregulation of miR-151-3p could influence IL-6 expression by targeting Stat3. The transfection of miR-151-3p mimics markedly decreased the levels of Stat3 protein and p-Stat3 in primary macrophages stimulated with LPS (Figure 6b and Supplementary Figure 2h). Conversely, increased levels of Stat3 and p-Stat3 expression were observed in macrophages treated with miR-151-3p inhibitor (Figure 6b and Supplementary Figure 2i). The upregulation of IL-6 upon LPS stimulation was markedly reduced in macrophages treated with miR-151-3p mimics, but the opposite phenomenon was observed in cells transfected with the miR-151-3p inhibitor (Figures 6c and d).
To further investigate whether the changes in IL-6 were due to the differences in the NF-κB signaling pathway between miR-151-3p and scrambled negative control mimic-transfected macrophages, we examined the activation of the NF-κB signaling pathway. No significant difference in the level of p-IKKβ, p-IkBα and p-p65 was observed between miR-151-3p-treated macrophages and scrambled negative control mimic-treated macrophages (Figure 6e). Similar results were obtained in inhibitor-transfected macrophages (Figure 6f). These results suggest that the changes in IL-6 are not induced by the classical NF-κB signaling pathway.
Next, both gain-of-function and rescue studies were performed to convincingly demonstrate the role of miR-151-3p in the regulation of IL-6 via Stat3 in macrophages. Indeed, miR-151-3p could downregulate Stat3 phosphorylation and activation. However, when Stat3 was overexpressed without 3′-UTR, miR-151-3p could no longer regulate Stat3 expression (Figure 6g and Supplementary Figure 2j) or IL-6 transcription (Figure 6h). The overexpression of Stat3 by transfecting Stat3 expression plasmid in miR-151-3p mimic-transfected macrophages could rescue the suppression of IL-6 expression by miR-151-3p (Figure 6h). Therefore, miR-151-3p reduces IL-6 production upon LPS stimulation by suppressing Stat3 activation. The LPS-induced downregulation of miR-151-3p in macrophages contributes to the production of IL-6 by increasing Stat3 protein levels and consequently amplifying the Stat3 pathway in inflammatory responses.
Discussion
Exploring miRNA–mRNA interactions remains as a significant challenge because of the limited knowledge of the rules governing these processes and the high false-positive rate of computational algorithms.24 Technologies have been developed to identify the targets of certain miRNA and extensively explored over the past decade.25, 26, 27, 28, 29, 30 However, there are few experimental methods for purifying miRNA regulomes. For example, microRNA capture affinity technology (miR-CATCH)31 and the identification of miRNAs targeting a single gene by applying short biotinylated DNA anti-sense oligonucleotide mix32 cannot reflect the physiological state in vivo. Here, in continuation of a previous work concerning identification of unexpected miRNAs in human cancer cells,21 several unexpected miRNAs targeting mouse Stat3 mRNA were identified using a miRIP approach. Among these miRNAs, miR-151-3p was confirmed to directly target Stat3 mRNA.
Until recently, miR-151-3p could not be predicted using bioinformatics tools, even with the loosest parameters. Such results indicated that miR-151-3p was unpredictable using most bioinformatics tools. However, with more evolving bioinformatics tools, the prediction of miRNA:mRNA interactions would become increasingly precise. Other than miR-151-3p, miRNAs such as miR-27b and miR-184, also impact the expression of mouse Stat3, although not as significantly as miR-151-3p, thus confirming the accuracy and efficiency of the method.
MicroRNA-151-3p was primarily expressed in the lungs and enriched in macrophages compared with its expression in other hematopoietic cells. The expression levels of miR-151-3p in T cells and B cells were relatively lower than that in macrophages and almost remained unchanged during the activation of CD4+ T cells, CD8+ T cells, and B cells; however, we cannot conclude that miR-151-3p has no function in T/B cells. The influence of miR-151-3p on T/B cells function needs further investigation. As an organ susceptible to various potential pathogens, it is reasonable that the lung possesses the highest miR-151 expression to maintain homeostasis.
MicroRNA −151-3p was also markedly decreased in LPS-induced macrophages, and the down-regulation of miR-151-3p led to the up-regulation of Stat3, which further promoted the subsequent production of many Stat3-dependent genes, such as the pro-inflammatory cytokine IL-6. The level of miR-151-3p after LPS stimulation diminished at 30 min, began to recover at 45 min, and finally returned to the baseline levels at 60 min. We assume that this finding may have biological significance, as the overactivation of STAT3/IL6 signaling could be avoided in this manner. The recovery of miR-151-3p restrains IL-6 production, and thus the risk of the harm resulting from uncontrolled immune responses may be minimized. In this case, we speculated that miR-151-3p maintains Stat3-dependent pro-inflammatory cytokines from activation, thus avoids casual inflammation in a healthy state, and that during infection, miR-151-3p enables the efficient clearance of pathogens, prevents the over-production of pro-inflammatory response and may participate in inflammation resolution. In general, miR-151-3p functions in maintaining macrophage homeostasis in a basal state and restricts harmful inflammatory responses after infection or injury.
Stat3 acts as a hinge in various signaling pathways in immune cells and inflammation responses and to some extent maintains the immune homeostasis. The upstream signal sources of Stat3 (IL-6, IL-21, IL-23, IL-10 and so on) and the downstream targets of Stat3 (IL-6, IL-17, IL-10 and so on) confer Stat3 a dual role in anti-inflammatory and pro-inflammatory responses. Furthermore, Stat3 is also pivotal in different immune cell type maintenance functions.33 Stat3 plays an important role in Th2 cell responses34 and functions in Treg cell differentiation.35 In addition, Stat3 was crucial for Th17 differentiation through IL-23 and IL-6 signals.36
In addition to T cells, Stat3 plays important roles in myeloid cells, particularly in DC and macrophage differentiation and function.37, 38 Most of the functions of Stat3 were realized by phosphorylation, methylation or acetylation modifications at different positions, but total Stat3 protein is rarely observed in innate immune responses.15, 16 Previous data have revealed relatively high levels of Stat3 phosphorylation in RAW264.7 cells at 30 min after LPS stimulation,39 similar to our results. Our current study showed that total Stat3 continued to increase at the protein level, but the Stat3 mRNA level did not change in macrophages upon LPS treatment, implying the post-transcriptional regulation of total Stat3 protein during this process. In addition, we investigated the roles of both IL-6 in LPS-induced instant Stat3 phosphorylation and Stat3 in LPS-induced cytokine production. We convincingly demonstrated that the phosphorylation of Stat3 within one hour of LPS stimulation was not driven by IL-6 secretion and that Stat3 does play an important role in LPS-induced IL-6 secretion aside from the canonical LPS/NF-κB/IL-6 signaling pathway. Although multiple miRNAs, including miR-17, miR-20a, miR-93, miR-106b, miR-125b, miR-199 and miR-223, target Stat3 mRNA,18, 19, 20 it is likely that Stat3 mRNA is targeted by many other miRNAs based on the long 3′-UTR of Stat3. In this study, several unexpected miRNAs targeting Stat3 mRNA were identified, and a new Stat3 targeting miRNA (miR-151-3p) was verified to directly interact with Stat3 mRNA and function in inflammatory responses.
The fine regulation of pattern recognition receptors is indispensable for innate immunity.40 In recent years, both post-transcriptional and epigenetic modifications participate in the sophisticated regulatory network of the immune system.5, 6 In addition to these modifications, non-coding RNAs and microRNAs have important functions in innate immune responses and inflammation.3, 41 Unlike regulators, such as cytokines or other noncoding RNAs, miRNAs function primarily as a buffer to maintain the balance of immune responses. For example, the downregulation of miR-497 by IL-17 results in high HIF-1α expression and the increased production of IL-1β and IL-6 by astrocytes in EAE mice.42 In addition, microRNAs might be used by pathogens to attenuate human immunity in chronic viral infections.43, 44 For miR-151-3p, the miRIP fished miRNA examined in human cancers 45 and early mouse embryo development,46 the function in innate immunity is far from clear, reflecting a lack of target gene information. Although the detailed mechanism of the novel fast Stat3 phosphorylation pathway in our study remains unclear, the enhanced production of pro-inflammatory cytokine IL-6 was NF-κB independent.
In summary, using the miRIP method, we identified computationally unpredictable miRNAs targeting mouse Stat3 mRNA and showed that miR-151-3p is a negative regulator of innate immune responses and inflammation by inhibiting IL-6 expression by downregulating the Stat3 and p-Stat3 levels. Thus, we speculated that at steady state, miR-151-3p restricts Stat3 and maintains macrophages in a tolerant state; under infection, miR-151-3p participates in preventing the innate immune system from over-activation. The findings of our current study in immune cells further confirmed that miRIP is an efficient method for identifying miRNAs that bind target mRNAs in the immune system and expands its application in the identification of previously unexpected miRNAs in cancer.
Acknowledgments
We thank Dr Taoyong Chen and Dr Mingjin Yang for helpful discussion and Ms Mei Jin for technical assistance. This work was supported by Grants from the National Key Basic Research Program of China (2013CB530502, 2015CB964403) and the National Natural Science Foundation of China (31470849, 31390431 and 81471569), and the Shanghai Committee of Science and Technology (2015QA1404700).
Footnotes
Supplementary Information for this article can be found on the Cellular & Molecular Immunology website (http://www.nature.com/cmi)
There were no financial disclosures from any authors.
Supplementary Material
References
- Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol 2016; 16: 279–294. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012; 148: 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Dai Y, Yang Y, Huang C, Meng X, Wu B et al. Emerging role of microRNAs in regulating macrophage activation and polarization in immune response and inflammation. Immunology 2016; 148: 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cao X. Cellular and molecular regulation of innate inflammatory responses. Cell Mol Immunol 2016; 13: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Qian C, Cao X. Post-Translational Modification Control of Innate Immunity. Immunity 2016; 45: 15–30. [DOI] [PubMed] [Google Scholar]
- Alvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol 2015; 15: 7–17. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol 2012; 30: 295–312. [DOI] [PubMed] [Google Scholar]
- Forster SC, Tate MD, Hertzog PJ. MicroRNA as type I interferon-regulated transcripts and modulators of the innate immune response. Front Immunol 2015; 6: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth LA, Boardman DA, Tung SL, Lechler R, Lombardi G. MicroRNAs affect dendritic cell function and phenotype. Immunology 2015; 144: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shi X. MicroRNAs in the regulation of TLR and RIG-I pathways. Cell Mol Immunol 2013; 10: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating MicroRNAs. Immunity 2009; 31: 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 2006; 103: 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol 2011; 12: 861–869. [DOI] [PubMed] [Google Scholar]
- Garbers C, Aparicio-Siegmund S, Rose-John S. The IL-6/gp130/STAT3 signaling axis: recent advances towards specific inhibition. Curr Opin Immunol 2015; 34: 75–82. [DOI] [PubMed] [Google Scholar]
- Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NF kappa B. Genes Dev 2007; 21: 1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Wang H, Liu Y, Song Y, Lai L, Han Q et al. Inducible MicroRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1 beta production in macrophages by targeting STAT3. PLoS One 2012; 7: e42971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiba M, Nakajima K, Yamanaka Y, Kiuchi N, Hirano T. Autoregulation of the Stat3 gene through cooperation with a cAMP-responsive element-binding protein. J Biol Chem 1998; 273: 6132–6138. [DOI] [PubMed] [Google Scholar]
- Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol 2009; 333: 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A et al. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood 2011; 117: 4338–4348. [DOI] [PubMed] [Google Scholar]
- Haghikia A, Missol-Kolka E, Tsikas D, Venturini L, Brundiers S, Castoldi M et al. Signal transducer and activator of transcription 3-mediated regulation of miR-199a-5p links cardiomyocyte and endothelial cell function in the heart: a key role for ubiquitin-conjugating enzymes. Eur Heart J 2011; 32: 1287–1297. [DOI] [PubMed] [Google Scholar]
- Su X, Wang H, Ge W, Yang M, Hou J, Chen T et al. An in vivo method to identify microRNA targets not predicted by computation algorithms: p21 targeting by miR-92a in cancer. Cancer Res 2015; 75: 2875–2885. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Q, Ding Y, Liu Y, Zhao D, Zhao K et al. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat Immunol 2016; 17: 806–815. [DOI] [PubMed] [Google Scholar]
- Bode JG, Ehlting C, Haeussinger D. The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis. Cell Signal 2012; 24: 1185–1194. [DOI] [PubMed] [Google Scholar]
- Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med 2011; 11: 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA 2007; 13: 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA 2007; 104: 19291–19296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andachi Y. A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target. RNA 2008; 14: 2440–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu RJ, Yang HJ, Tsai HJ. Labeled microRNA pull-down assay system: an experimental approach for high-throughput identification of microRNA-target mRNAs. Nucleic Acids Res 2009; 37: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LP, Seinen E, Duns G, de Jong D, Sibon OCM, Poppema S et al. A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res 2009; 37: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz L, Bilen B, Hausser J, Zavolan M. Argonaute CLIP—A method to identify in vivo targets of miRINAs. Methods 2012; 58: 106–112. [DOI] [PubMed] [Google Scholar]
- Vencken S, Hassan T, McElvaney NG, Smith SGJ, Greene CM. miR-CATCH: microRNA capture affinity technology. Methods Mol Biol 2015; 1218: 365–373. [DOI] [PubMed] [Google Scholar]
- Wei K, Yan F, Xiao H, Yang X, Xie G, Xiao Y et al. Affinity purification of binding miRNAs for messenger RNA fused with a common tag. Int J Mol Sci 2014; 15: 14753–14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura H, Murakami M, Okuyama Y, Tsuruoka M, Kitabayashi C, Kanamoto M et al. Interleukin-17 promotes autoimmunity by triggering a positive-feedback loop via interleukin-6 induction. Immunity 2008; 29: 628–636. [DOI] [PubMed] [Google Scholar]
- Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J et al. The transcription factor STAT3 is required for T helper 2 cell development. Immunity 2011; 34: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Amarnath S, Mariotti J, Kim YC, Foley J, Eckhaus M et al. STAT3 transcription factor promotes instability of nTreg cells and limits generation of iTreg cells during acute murine graft-versus-host disease. Immunity 2012; 37: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 2008; 452: 773–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014; 344: 310–313. [DOI] [PubMed] [Google Scholar]
- Kumar V, Cheng P, Condamine T, Mony S, Languino LR, McCaffrey JC et al. CD45 phosphatase inhibits STAT3 transcription factor activity in myeloid cells and promotes tumor-associated macrophage differentiation. Immunity 2016; 44: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YJ, Jeong M, Lee KT, Jang DS, Choi JH. Isocyperol, isolated from the rhizomes of Cyperus rotundus, inhibits LPS-induced inflammatory responses via suppression of the NF-kappa B and STAT3 pathways and ROS stress in LPS-stimulated RAW 264.7 cells. Int Immunopharmacol 2016; 38: 61–69. [DOI] [PubMed] [Google Scholar]
- Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol 2016; 16: 35–50. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cao X. Long noncoding RNAs in innate immunity. Cell Mol Immunol 2016; 13: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan K, Pang R, Zhao C, Liu X, Gao W, Zhang J et al. IL-17-triggered downregulation of miR-497 results in high HIF-1alpha expression and consequent IL-1beta and IL-6 production by astrocytes in EAE mice. Cell Mol Immunol 2017; 14: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M, Koster S, Sakowski E, Ramkhelawon B, van Solingen C, Oldebeken S et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol 2016; 17: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett HF, Cartwright ANR, Kim H-J, Godec J, Pyrdol J, Aijo T et al. The microRNA miR-31 inhibits CD8(+) T cell function in chronic viral infection. Nat Immunol 2017; 18: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde T. MicroRNA-151 and its hosting gene FAK (focal adhesion kinase) regulate tumor cell migration and spreading of hepatocellular carcinoma. Hepatology 2010; 52: 1164–1166. [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez J, de Dios Hourcade J, del Mazo J. Reprogramming of microRNAs by adenosine-to-inosine editing and the selective elimination of edited microRNA precursors in mouse oocytes and preimplantation embryos. Nucleic Acids Res 2013; 41: 5483–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.