Abstract
We used structured illumination microscopy (SIM) to obtain super-resolution images of muscle attachment structures in C. elegans striated muscle. SIM imaging of M-line components revealed two patterns: PAT-3 (β-integrin) and proteins that interact in a complex with the cytoplasmic tail of β-integrin and localize to the basal muscle cell membrane (UNC-112 (kindlin), PAT-4 (ILK), UNC-97 (PINCH), PAT-6 (α-parvin) and UNC-95), are found in discrete, angled segments with gaps. In contrast, proteins localized throughout the depth of the M-line (UNC-89 (obscurin) and UNC-98) are imaged as continuous lines. Systematic immunostaining of muscle cell boundaries revealed that dense body components close to the basal muscle cell membrane also localize at cell boundaries. SIM imaging of muscle cell boundaries reveal “zipper-like” structures. Electron micrographs reveal electron dense material similar in appearance to dense bodies located adjacent to the basolateral cell membranes of adjacent muscle cells separated by ECM. Moreover, by EM, there are a variety of features of the muscle cell boundaries that help explain the zipper-like pattern of muscle protein localization observed by SIM. Short dense bodies in atn-1 mutants that are null for α-actinin and lack the deeper extensions of dense bodies, showed “zipper-like” structures by SIM similar to cell boundary structures, further indicating that the surface-proximal components of dense bodies form the “zipper-like” structures at cell boundaries. Moreover, mutants in thin and thick filament components do not have “dot-like” dense bodies, suggesting that myofilament tension is required for assembly or maintenance of proper dense body shape.
Keywords: muscle, integrin adhesion sites, super resolution microscopy, electron microscopy, C. elegans
Introduction
Sarcomeres, highly ordered assemblages of several hundred proteins, perform the work of muscle contraction. Despite ever increasing knowledge of the components of sarcomeres and their functions, a clear picture about how sarcomeres are assembled and maintained in the face of muscle contraction is still not available. A number of laboratories are studying the questions of sarcomere assembly and maintenance in the model genetic organism, C. elegans. In addition to being an excellent system to carry out mutational analysis in a whole organism, through both forward and reverse genetics, this nematode offers several advantages for studying muscle. These include its optical transparency, which allows evaluation of myofibrillar structure by polarized light, and localization of GFP tagged proteins in live animals. In addition, its usual self-fertilization allows propagation of muscle mutants that would be unable to mate. The major striated muscle of C. elegans is found in the body wall and used for locomotion, and the myofibrils are restricted to a narrow ~1.5 µm zone adjacent to the cell membrane along the outer side of the muscle cell [Moerman and Fire, 1997]. The thin filaments are attached to the dense bodies (Z-disk analogs, not line but “dot” or “finger” like structures), and the thick filaments are organized around M-lines. Moreover, all the dense bodies and M-lines are anchored to the muscle cell membrane and extracellular matrix (ECM, basement membrane), which is attached to the hypodermis and thus to the cuticle [Waterston, 1988; Moerman and Fire, 1997; Moerman and Williams, 2006; Gieseler et al., 2016;]. This allows the force of muscle contraction to be transmitted directly to the cuticle and allows movement of the whole animal.
Many of the proteins of myofilaments, attachment structures, and regulators of contraction/relaxation have been identified through genetic and molecular biological analyses, and characterized cell biologically by using specific antibodies and GFP fusion proteins [Moerman and Fire, 1997; Qadota and Benian, 2010; Gieseler et al., 2016]. For C. elegans muscle, electron microscopy images are also available [Waterston et al., 1980; Zengel and Epstein, 1980; Waterston, 1988; Gieseler et al., 2016]. However, since conventional fluorescence microscopy has limited resolution (~250 nm in the x-y plane) and EM provides too narrow a view, whole muscle three-dimensional structure is difficult to perceive in single images. In this study, we applied the super resolution microscopy technique, structured illumination microscopy (SIM) with ~120 nm resolution, to observe C. elegans muscle attachment structures, and learned greater detail about the differential localization of components of M-lines and the composition and structure of muscle cell boundaries, and from data using several genetic mutants we propose that muscle tension affects the structure of dense bodies. In addition, we used electron microscopy to further interpret our SIM images of muscle cell boundaries.
Results
The base of M-lines contains multiple proteins localized in discreet separated segments
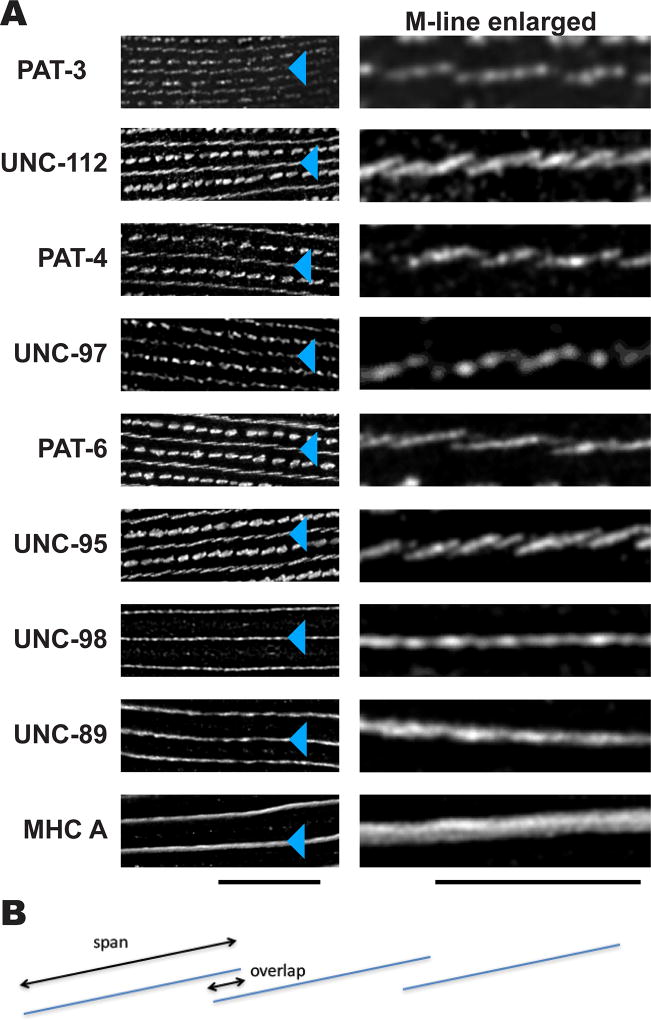
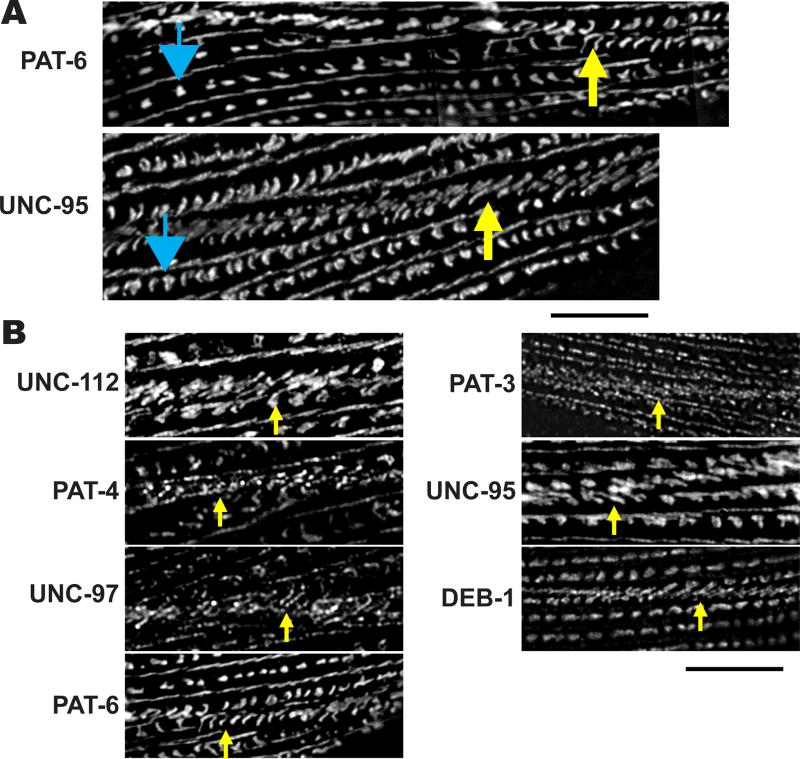
We applied the SIM technique to obtain a higher resolution view of sarcomere structures upon immunostaining with a battery of antibodies to various proteins. Using this method revealed a greater level of complexity and order than could be viewed with conventional widefield or confocal immunofluorescence microscopy. With conventional microscopy, M-lines appear as continuous lines [Moerman and Williams, 2006]. Using SIM, antibodies to multiple proteins show localization to discreet segments (Figure 1A). SIM images of M-lines immunostained with anti-PAT-3 (β-integrin), anti-UNC-112 (kindlin), anti-PAT-4 (ILK), anti-UNC-97 (PINCH), anti-PAT-6 (α-parvin), and anti-UNC-95, showed discontinuous and angled lines (see enlarged images on the right side of Figure 1A). These proteins are localized near the cell membrane [Gieseler et al., 2016]. UNC-112 interacts directly with the cytoplasmic tail of β-integrin (PAT-3) [Qadota et al., 2012]; by genetic criteria, yeast 2-hybrid data, and co-immunoprecipitation experiments, UNC-112/PAT-4/PAT-6/UNC-97 form a four-protein complex [Mackinnon et al., 2002; Lin et al., 2003; Norman et al., 2007; Qadota et al., 2014]. Measurements from Figure 1A indicate that the discontinuous lines of localization of each of the four-protein complex proteins span 1.4—1.9 µm, with maximum overlaps of 200 nm (see interpretative drawing, Figure 1B). In contrast, SIM images of anti-UNC-98, anti-UNC-89 (obscurin), and anti-MHC A (myosin heavy chain A) staining of M-lines showed straight continuous lines (Figure 1A). (It should be noted that MHC A is not just restricted to the M-line, but more broadly distributed in the center of the A-band, as MHC A is a component of the central region of thick filaments spanning a region of 1.8 µm (Miller et al., 1983)).
Figure 1. Differential localization of M-line components.
(A) SIM imaging of M-line components. Wild type worms were immunostained with antibodies to each of the indicated proteins, and imaged by SIM super resolution microscopy. For images in the left column, black bar represents 10 µm; for images in the right column, the black bar represents 5 µm. Blue triangles point to M-lines. PAT-3 (β-integrin), UNC-112 (kindlin), PAT-4 (ILK), UNC-97 (PINCH), PAT-6 (α-parvin), and UNC-95 are known or suspected to be located close to the muscle cell membrane. The images show that antibodies to these proteins localize as discontinuous and angled lines. UNC-98, UNC-89 and MHC A are known or suspected to be located throughout the depth of M-lines. The images show that antibodies to these proteins localize as straight, continuous lines. (B) Diagram depicting the “span” and “overlap” of discontinuous localization of many of the M-line proteins shown on the right side of part (A).
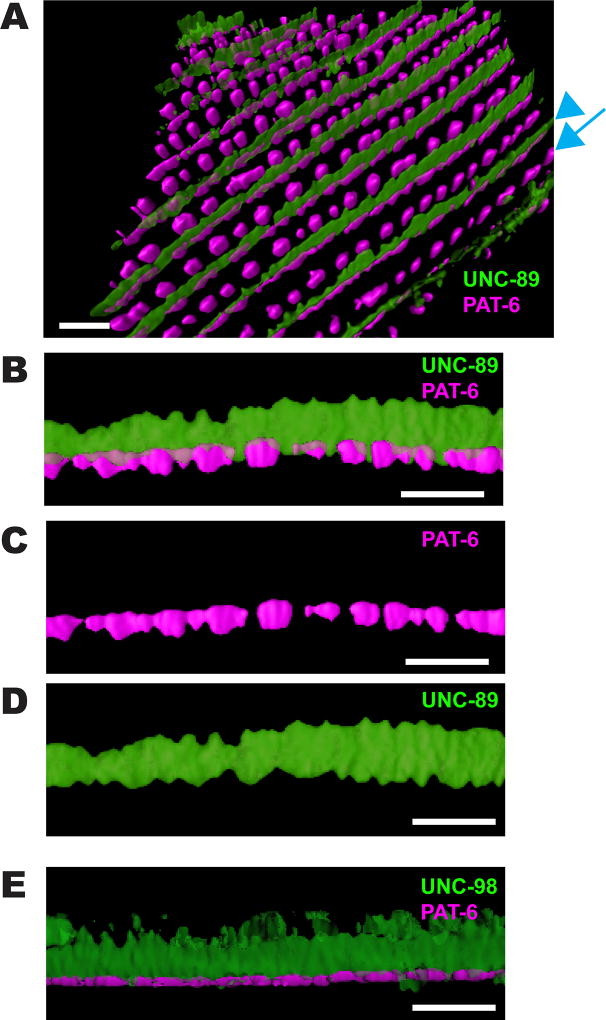
3D reconstructions of SIM images revealed additional information about the M-line. A 3D reconstruction of co-staining with anti-PAT-6 (magenta) and anti-UNC-89 (green) is shown in Figure 2A–D. Viewing a single M-line from the side (Figure 2B) confirms that PAT-6 is localized in discrete segments with gaps (or sometimes more continuous), that UNC-89 is localized in a continuous wall, and this wall fills the gaps between PAT-6 segments. Thus, the base of the M-line is actually continuous with the cell membrane with no gaps. This image also confirms our previous report that UNC-89 is localized throughout the depth of the myofilament lattice [Warner et al., 2013]. As indicated in Figure 2B, the maximum height of the UNC-89 “wall” is ~2.0 µm, which is close to the maximum heights of M-lines observed by electron microscopy (1.5–2 µm). We also performed 3D reconstruction of immunostaining of two additional components of M-lines, the C2H2 Zn finger protein UNC-98 [Mercer et al., 2003], and myosin heavy chain A (MHC A) [Miller et al., 1983]. As shown in Figures 2E and 3A, UNC-98 and MHC A, each localize as continuous walls spanning ~2 µm from near the muscle membrane to deeper inside the sarcomere. The deepest part of the UNC-98 staining, which appears fuzzy, is likely to be artifact due to a weak signal. Co-staining of anti-UNC-89 with anti-MHC A (Figure 3B) shows that UNC-89 (green) does not extend as close to the outer cell membrane, or as deep into the muscle cell as MHC A. To summarize, our SIM observations show that the M-line, near the muscle cell membrane, contains various components (PAT-3, UNC-112, PAT-4, PAT-6, UNC-97, UNC-95), localized in separated segments, but other components of the M-line (UNC-89, UNC-98 and MHC A) are localized throughout the depth of the sarcomere as a continuous wall and fill-in these gaps.
Figure 2. 3D rendering of UNC-89, PAT-6 and UNC-98 localization.
(A) Angled view of localization of UNC-89 (green) and PAT-6 (magenta) in a portion of a single body wall muscle cell. A blue arrowhead points to an M-line, and a blue arrow points to a row of dense bodies. Scale bar, 3 µm. (B) Side view of a portion of a single M-line from (A). Orientation: at the bottom is the outer muscle cell membrane, and at top is deeper into the muscle cell. Note that PAT-6 (magenta) is localized to discrete segments near the muscle cell membrane, and only partially co-localizes deeper with UNC-89 (green). UNC-89 spans from near the muscle cell membrane deep into the sarcomere. Also note that UNC-89 extends close to the muscle cell membrane in the gaps of PAT-6 localization. Scale bar, 3 µm. (C) and (D) are the individual channels for PAT-6 and UNC-89 localization, respectively, from (B). Scale bar, 3 µm. (E) Side view of a single M-line immunostained with anti-UNC-98 (green) and anti-PAT-6 (magenta). Note that UNC-98, like UNC-89, spans ~2 µm from near the muscle cell membrane deep into the sarcomere. The deepest parts of UNC-98 that appear fuzzy are likely to be non-specific staining. Scale bar, 3 µm.
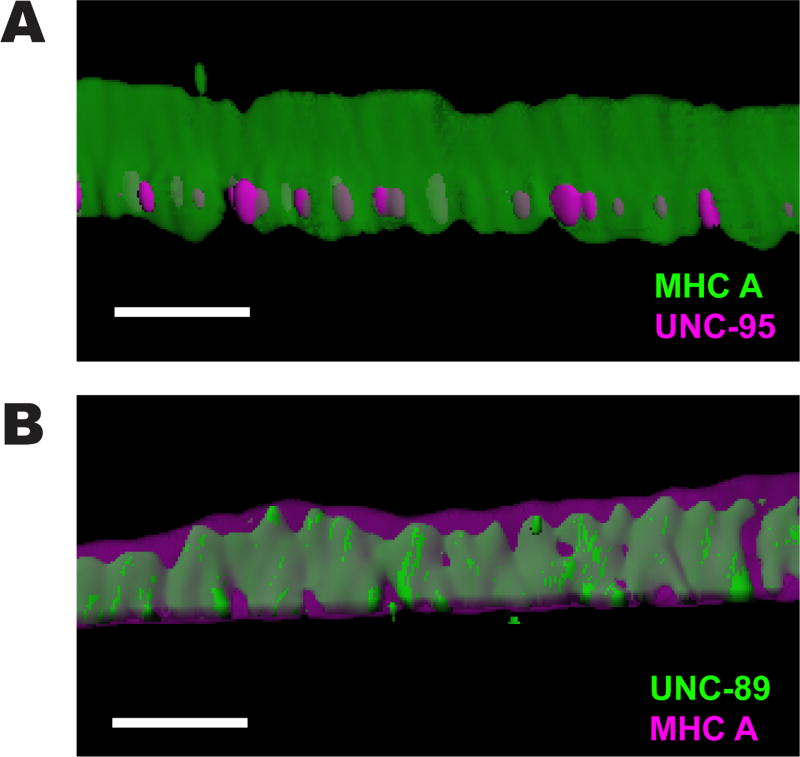
Figure 3. 3D rendering of localization of MHC A with UNC-95, and UNC-89 with MHC A.
(A) Side view of a single M-line immunostained with anti-MHC A (green) and anti-UNC-95 (magenta). MHC A also spans ~2 µm from near the muscle cell membrane deep into the sarcomere. Scale bar, 2 µm. (B) Side view of a single M-line immunostained with anti-UNC-89 (green) and anti-MHC A (magenta). Note that MHC A localizes beyond the localization of UNC-89. Scale bar, 2 µm.
Components of muscle cell boundaries
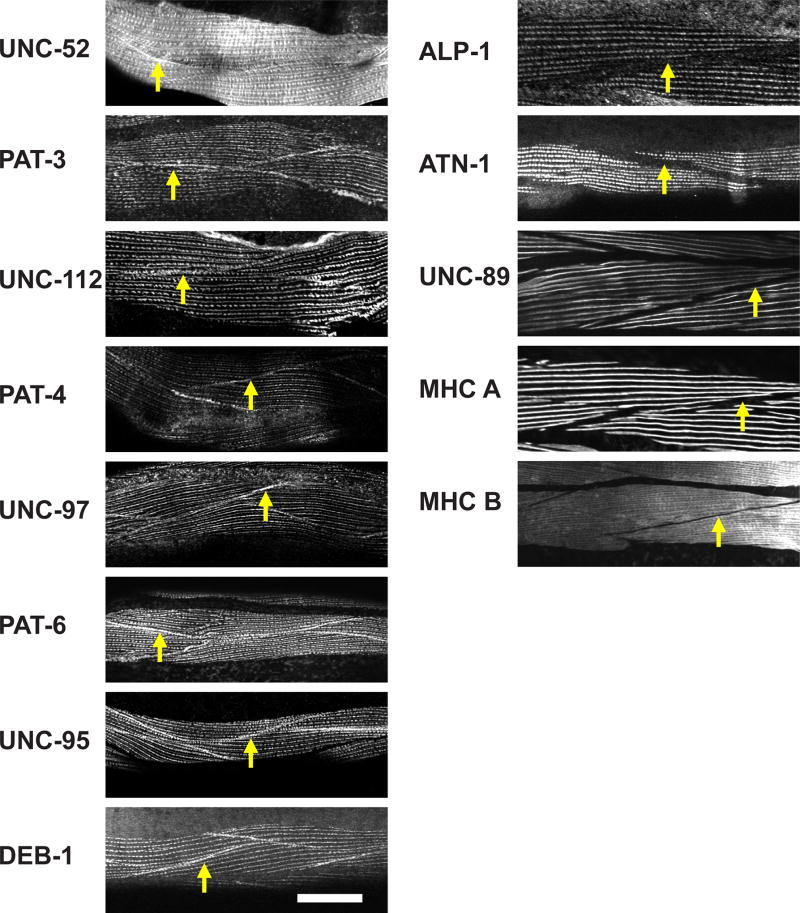
Next, we wanted to examine muscle cell boundary structures. Some muscle cell attachment proteins have been reported to be localized at muscle cell boundaries [Gieseler et al., 2016], but the protein composition of muscle cell boundaries has not been characterized systematically. In some cases, localization has been performed using overexpressed GFP fusions rather than the localization of endogenous proteins using specific antibodies (for example, UNC-97 [Hobert et al. 1999] and UNC-95 [Broday et al. 2004]). We immunostained C. elegans muscle with antibodies to a set of 13 proteins, and examined muscle cell boundaries using confocal microscopy (Figure 4). UNC-52 (perlecan), PAT-3 (β-integrin), UNC-112 (kindlin), PAT-4 (ILK), UNC-97 (PINCH), PAT-6 (α-parvin), UNC-95 and DEB-1 (vinculin) were localized at muscle cell boundaries, but ALP-1 (enigma), ATN-1 (α-actinin), UNC-89 (obscurin), MHC A (myosin heavy chain), and MHC B (myosin heavy chain) were not localized at muscle cell boundaries. The common characteristic of the proteins that localize to muscle cell boundaries is that these proteins also localize close to the membrane or at the base of dense bodies. Moreover, proteins that localize specifically to M-lines, the center of A-bands, or to the deeper parts of dense bodies do not localize at muscle cell boundaries.
Figure 4. Components of structures at body wall muscle cell boundaries.
Each panel shows immunostaining with antibodies to the indicated proteins, and imaged with a standard confocal microscope. Yellow arrows point to boundaries between adjacent spindle-shaped body wall muscle cells. As shown in the column of images on the left, UNC-52 (perlecan), PAT-3 (β-integrin), UNC-112 (kindlin), PAT-4 (ILK), UNC-97 (PINCH), PAT-6 (α-parvin), UNC-95, and DEB-1 (vinculin), are localized at the muscle cell boundary. These proteins are also known to localize at the membrane-proximal region of dense bodies. As shown in the column of images on the right, muscle cell boundaries lack the following proteins: ALP-1 (Enigma), ATN-1 (α-actinin), UNC-89 (obscurin), MHC A (myosin heavy chain A), and MHC B (myosin heavy chain B). White bar represents 10 µm.
High-resolution structures of muscle cell boundaries
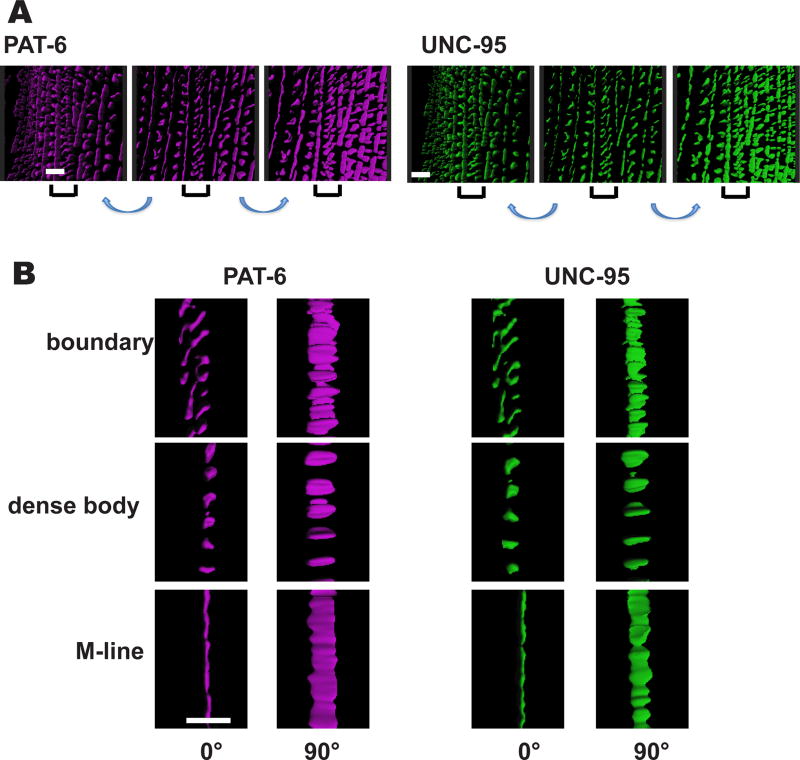
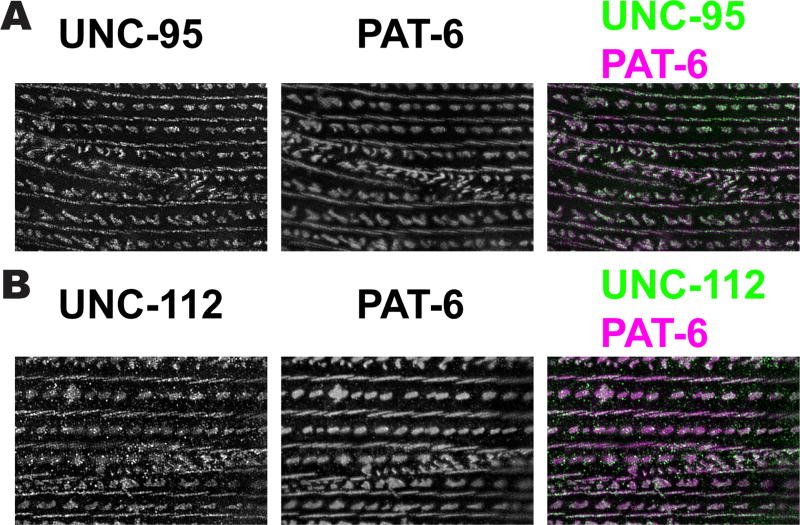
We further examined by SIM imaging the muscle cell boundary structures after immunostaining with anti-PAT-3, anti-UNC-112, anti-PAT-4, anti-UNC-97, anti-PAT-6, anti-UNC-95 and anti-DEB-1. It can be discerned that these cell boundary structures have a “zipper-like” appearance, with half of the structure on each of the opposing muscle cells (Figure 5). As described above, proteins that localize at muscle cell boundaries also localize to the base of dense bodies. SIM imaging of a muscle cell when viewed from above, immunostained with anti-PAT-6 or anti-UNC-95 (Figure 5A), reveals that there is a gradual transition from dense body “dot-like” structures, to “zipper-like” structures at the muscle cell boundary (in Figure 5A, begin with dots indicated by the blue arrow on the left, and proceed to the right until zipper structures appear, indicated by the yellow arrow). All the proteins that localize at muscle cell boundaries viewed by confocal microscopy (Figure 4), localize as “zipper-like” structures by SIM (Figure 5B). 3D reconstruction of SIM images of anti-PAT-6 or anti-UNC-95 staining shows these zipper-like structures in greater detail (Figure 6). The zippers consist of short parallel walls oriented diagonally, approximately 45 degrees from the axis of M-line walls. In Figure 6A, the middle of each set of 3 panels shows part of a muscle cell when viewed directly from above; on each side the plane has been tilted slightly. Figure 6B shows localizations of these proteins at boundaries, dense bodies and M-lines, from above (left panels), and from the side (right panels). From the side views it can be discerned that the heights of the localizations of PAT-6 and UNC-95 are approximately the same at boundaries, dense bodies and M-lines. Although Figure 6 suggests that PAT-6 and UNC-95 localize similarly in these structures, we wanted to determine if the two proteins co-localize. Thus, we co-stained body wall muscle with both antibodies. As shown in Figure 7A, this experiment indicates that PAT-6 and UNC-95 co-localize at M-lines, dense bodies and cell boundaries. A similar experiment shows that PAT-6 also co-localizes with UNC-112 (Figure 7B). We also attempted to determine if by SIM, other members of the four-protein complex (UNC-112, PAT-4, PAT-6 and UNC-97) also co-localized, but the weaker available antibodies to PAT-4 and UNC-97 precluded our ability to do so.
Figure 5. SIM imaging reveals new details about the structure of muscle cell boundaries.
(A) Wild type nematodes were immunostained with anti-PAT-6 and anti-UNC-95 antibodies. Blue arrows point to dense bodies, and yellow arrows point to muscle cell boundary structures. Starting at a dense body indicated with a blue arrow and moving horizontally or slightly diagonally to the right, it appears that the “dot” like dense body structures gradually transition in appearance to the “zipper” like boundary structure indicated with the yellow arrow. Black bar represents 10 µm.
(B) Each panel shows a muscle cell boundary structure indicated with a yellow arrow stained with an antibody to a different protein. Note that all the indicated proteins are part of the “zipper” like structures found at the muscle cell boundary. Scale bar represents 10 µm.
Figure 6. 3D rendering of muscle cell boundaries.
3D rendering of a z-stack of portions of two adjacent body wall muscle cells immunostained with either anti-PAT-6 or anti-UNC-95 are shown. (A) The center panel of each set of 3 images shows a view from the outer muscle cell surface; the panels on either side of the center panel show the same portion of the cell tilted to the left or to the right, respectively. Brackets indicate the muscle cell boundary. Scale bar, 2 µm. (B) Localization of PAT-6 and UNC-95 at boundaries, dense bodies and M-lines, from above (0°), and from the side (90°). From the side views note that the heights of the localizations of PAT-6 and UNC-95 are approximately the same at boundaries, dense bodies and M-lines. Scale bar, 2 µm.
Figure 7. SIM imaging does not reveal any differences in the localization of three integrin adhesion complex proteins, PAT-6, UNC-95 and UNC-112.
(A) Image of two body wall muscle cells co-immunstained with antibodies to UNC-95 and PAT-6. (B) Image of two body wall muscle cells co-immunostained with antibodies to UNC-112 and PAT-6. Scale bar, 10 µm.
ECM proteins localize between muscle cells
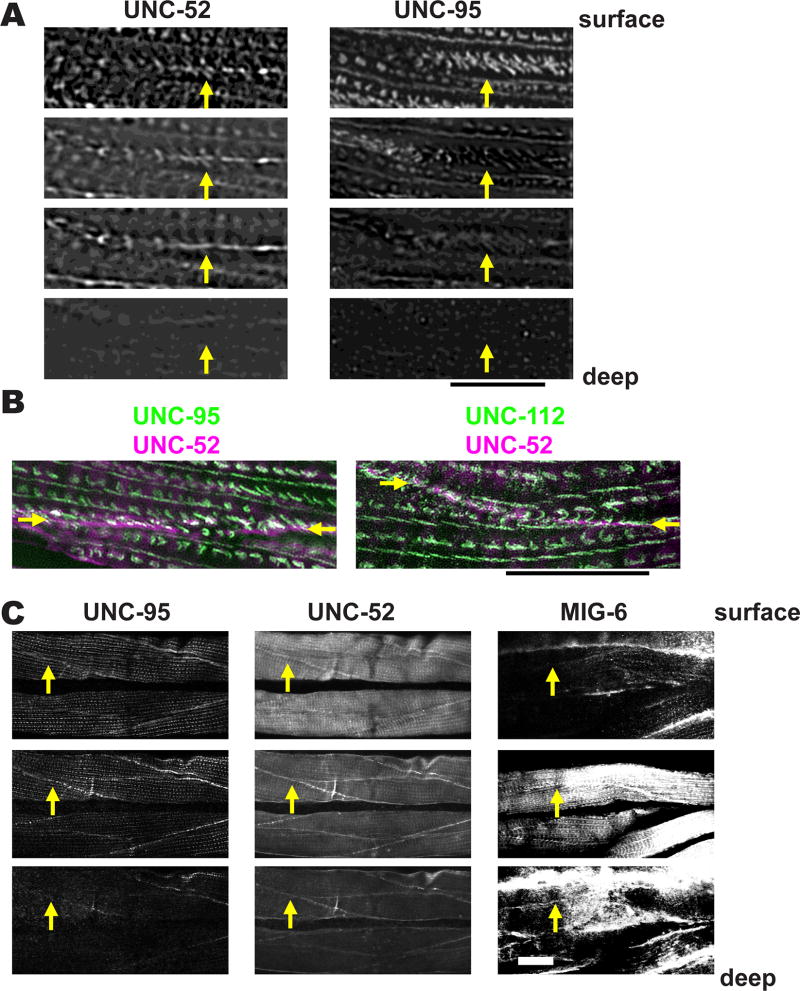
UNC-52 (perlecan) is a basement membrane protein that concentrates at the base of M-lines and dense bodies and muscle cell boundaries [Mullen et al. 1999]. We co-stained worm muscles with anti-UNC-95 and anti-UNC-52 (perlecan), and observed muscle cell boundary structures using SIM. Near the outer cell membrane, both proteins show “zipper-like” structures as shown in Figure 8A (top panel). Traveling deeper into the muscle cell, UNC-95 localizes to “zipper-like” structures, but UNC-52 localizes to a straight line (Figure 8A). This observation suggests that ECM proteins like UNC-52 exist between muscle cells. To obtain more evidence for this hypothesis, we used SIM to observe muscle cells co-stained with either anti-UNC-52 and anti-UNC-95, or with anti-UNC-52 and andi-UNC-112. As shown in Figure 8B, UNC-52 (magenta) mostly localizes between the zipper-like structures containing UNC-95 or UNC-112 (green).
Figure 8. Muscle cell boundaries include extra cellular matrix proteins.
(A) Worms were co-immunostained with antibodies to UNC-95 and the ECM component UNC-52 (perlecan). Images were obtained by SIM, and each image was taken at 0.4 µm intervals. The yellow arrows point to muscle cell boundaries. Scale bar represents 10 µm.
(B) SIM images of a portion of two adjacent body wall muscle cells co-stained with either antibodies to UNC-52 and UNC-95 (left), or UNC-52 and UNC-112 (right). Arrows mark the muscle cell boundary. Note that most of the ECM protein UNC-52 lies between the zipper-like structures labeled with either UNC-95 or UNC-112. Scale bar, 10 µm.
(C) UNC-52 (perlecan) and MIG-6 (papilin, lacunin) localize at muscle cell boundaries both at “surface” (close to membrane adjacent to hypodermis) and “inside” (away from this surface) focal planes. Animals were immunostained with antibodies to UNC-95, UNC-52 and MIG-6, and images were obtained by confocal microscopy, and show three consecutive 1.0 µm optical sections. The yellow arrows point to muscle cell boundaries. White bar represents 10 µm.
We next wondered if other known ECM proteins could be detected at muscle cell boundaries. Thus, we immunostained worms with antibodies to UNC-95, UNC-52 and MIG-6 (Figure 8C). MIG-6 is homologous to the mammalian ECM proteins papilin and lacunin, and is essential for embryonic hypodermal cell closure, proper distal tip cell migration, and neuronal cell migration [Hedgecock et al. 1987; Kramerova et al., 2000; Kawano et al., 2009]. Kawano et al. (2009) report that antibodies to MIG-6 stain muscle, but no details were given. At the surface focal planes, UNC-95, UNC-52 and MIG-6 are localized at dense bodies, M-lines, and cell boundaries. At the deeper focal planes, nearly all UNC-95 has disappeared, whereas for UNC-52 and MIG-6, although M-line and dense body staining is weak, cell boundary staining remains strong. (Figure 8C). Overall, our results demonstrate that muscle cell boundaries are not “cell-to-cell” contacts, but “cell-ECM-cell” contacts (Figure 9).
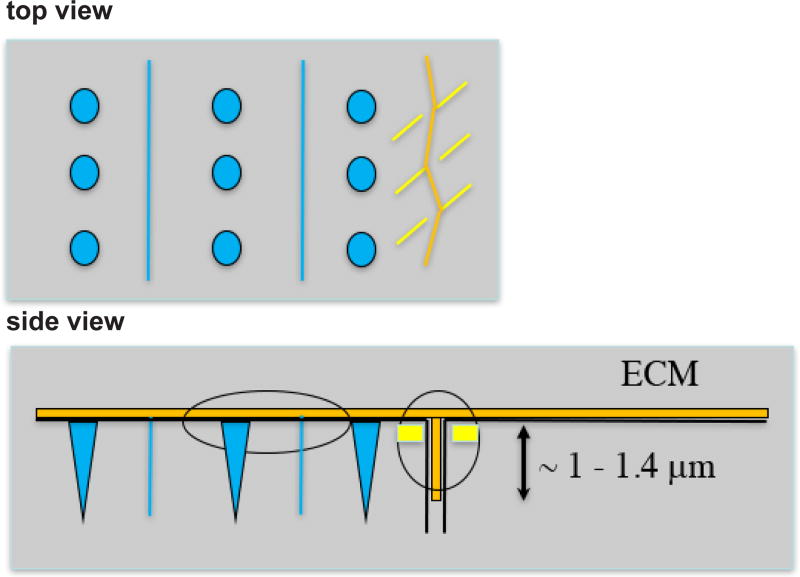
Figure 9. Drawing depicting ECM (orange), M-lines and dense bodies (blue) and attachment plaques at a muscle cell boundary (yellow).
The ECM lies at the base of the cells, and extends between cells for a distance of approximately 1–1.4 µm, based on Figure 8A. The attachment plaques do not extend as deeply as ECM between the muscle cells.
Absence of deeper components of dense bodies results in alteration in the shape of dense bodies
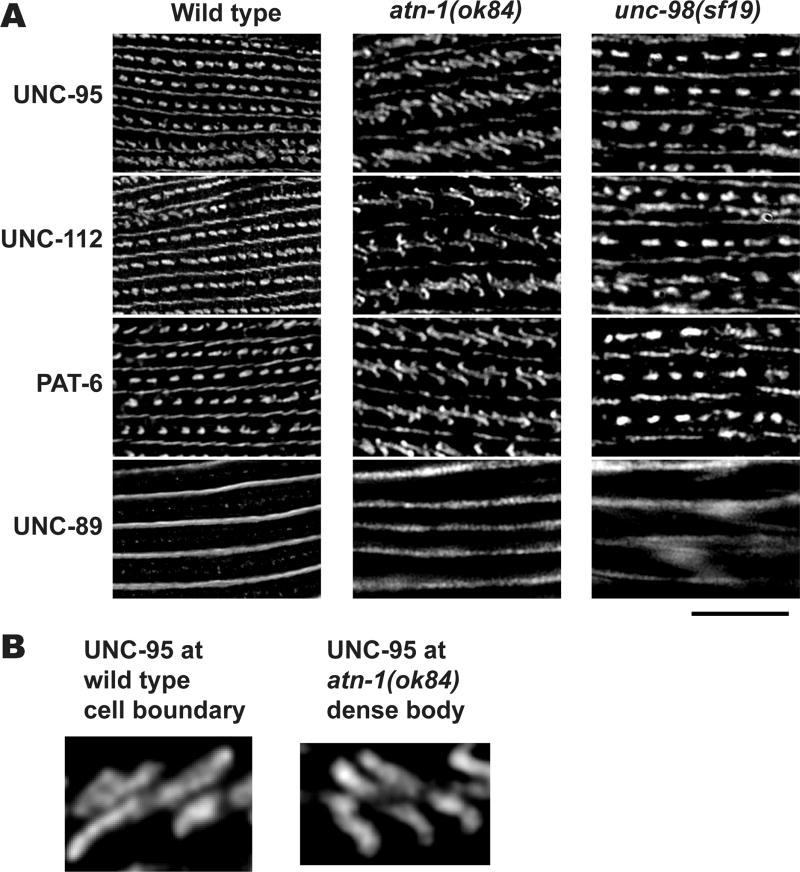
Next we characterized muscle attachment structures in various mutants with the high resolution SIM imaging technique. We examined the effects of loss of function mutations in atn-1 and unc-98. atn-1 encodes the sole α-actinin in nematodes that is localized to the major deeper portion of the dense body but not at its membrane-proximal base [Barstead et al. 1991]. unc-98 encodes a C2H2 Zn finger protein that localizes to M-lines [Mercer et al. 2003]. Deletion of atn-1 results in short and wide dense bodies [Moulder et al., 2010]. Mutation in unc-98 results in abolishment of M-lines [Mercer et al., 2003]. We stained each of the mutant strains with anti-UNC-112, anti-UNC-95, anti-PAT-6 and anti-UNC-89, and obtained SIM images. The unc-98 mutant shows normal dense bodies and normal M-lines at their bases (stained with UNC-95, UNC-112 or PAT-6), but abnormal M-lines at their deeper portions (stained with UNC-89). However, the atn-1 deletion mutant, ok84, shows “zipper-like” dense bodies and normal M-lines (Figure 10). The shape of dense bodies in the atn-1 mutant is similar to that of normal muscle cell boundary structures (compare Figure 5 and Figure 10). In atn-1(ok84), M-lines visualized by anti-UNC-89 staining are normal. As described above, the difference between normal dense bodies (“dots”) and normal cell boundaries (“zippers”) is the presence of the deeper protein components of dense bodies, like ATN-1. Thus, by SIM, the similarity in appearance between dense bodies of atn-1 mutants, which lack the deeper components of dense bodies (e.g. ATN-1, ALP-1, ZYX-1), and the appearance of normal cell boundary structures, confirms this model.
Figure 10. Lack of a deeper component of dense bodies changes their shape from “dot” like to “zipper” like.
(A) Wild type, atn-1 (ok84), and unc-98 (sf19) worms were immunostained with antibodies against UNC-95, UNC-112, PAT-6, and UNC-89. Anti-UNC-95, anti-UNC-112, and anti-PAT-6, localize to dense bodies and M-lines. Anti-UNC-89, localize only to M-lines. Images were captured by SIM. In the atn-1 (ok84) mutant, the dense bodies all appear like the “zipper” like structures normally found at muscle cell boundaries (compare with Figure 5). In contrast, in the unc-98(sf19) mutant, dense bodies mostly appear normal. Black scale bar represents 10 µm. (B) Enlargement of UNC-95 localization at a wild type cell boundary (left) and at an atn-1(ok84) dense body (right). Note the similarity in these two structures. Scale bar, 1 µm.
Ultrastructural appearance of cell-to-cell boundaries is consistent with SIM observations
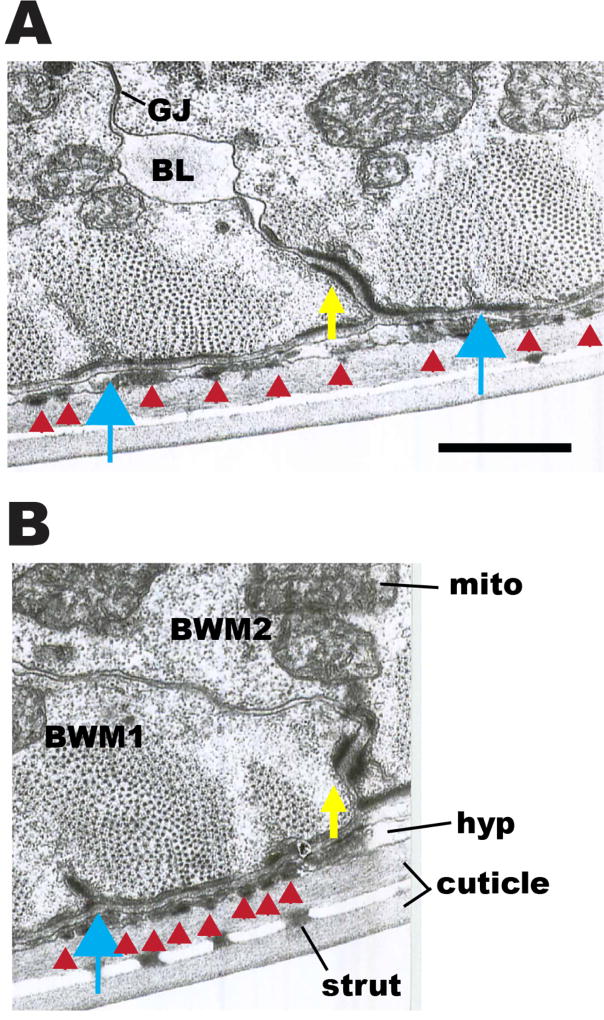
Transmission EM images of the boundaries between adjacent body wall muscle cells confirm and extend our SIM observations. As shown in cross sections in Figure 11A and B, electron dense material similar in appearance to material that comprises dense bodies is located adjacent to the basolateral cell membranes of adjacent muscle cells, separated by ECM-appearing material (indicated by yellow arrows in Figure 11A and B). The structures observed by EM likely correspond to the zipper-like structures observed by SIM. Indeed, by examining serial thin sections, one can follow the gradual transition from the punctate (“dot”) organization of the standard dense body as it moves along the base of a muscle membrane from section to section until reaching the parallel spaced (“zipper”) dense body domains when the dense body shifts to a cell-cell border, and then proceeds slowly back to a dot organization in the next muscle on the opposite side of the zippered region (data not shown).
Figure 11. Transmission EM images of boundaries between adjacent body wall muscle cells.
(A) and (B): Two cross-sectional views of body wall muscle, showing portions of two adjacent muscle cells. Yellow arrows point to electron dense material at basolateral membranes between two adjoining muscle cells (BWM1, BWM2). Discontinuous patches of this dense material can be followed along cell borders in serial thin sections to lead up to dense bodies (blue arrows), and for the most part the same proteins localize to this zone as for dense bodies. Flat black bars of electron dense material (red arrowheads) are also seen on the basal membrane of the muscles beneath each sarcomere, and filling the thin hypodermal layer (hyp) coincident with the sarcomeres; these represent attachment points between muscle, hypodermis and cuticle. Some basal lamina material (BL) projects inward from the basal zone between muscle cells and can collect in spaces along their lateral borders. Cuticle has an inner and outer layer, connected by struts. Mito, mitochondria. GJ, gap junction. Scale bar, 1 µm.
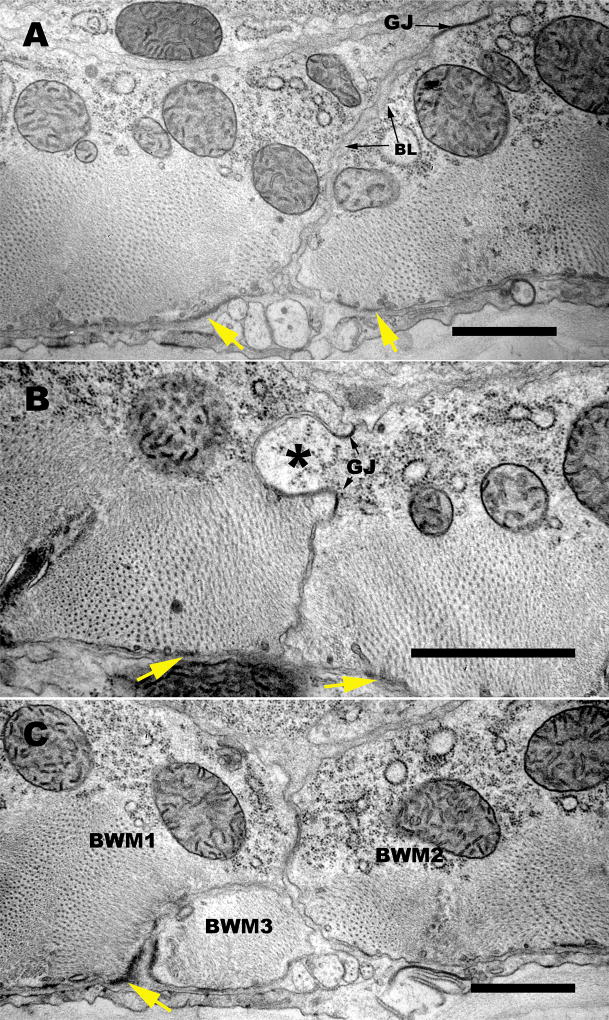
One can also detect the presence of basement membrane material projecting into the extracellular spaces between neighboring muscles, extending from the basal edge along the lateral muscle surfaces, and sometimes accumulating in local spaces (labeled “BL” in Figure 11A). Sometimes this basal lamina projects all the way to the inward facing surface of the body wall muscle, but it is often interrupted by the presence of gap junctions on these lateral membranes, particularly at the most distal zones, away from basal structures. Furthermore, EM views of the muscle cell-muscle cell border in transverse thin sections display a range of different deflections of this lateral border that help to explain the zig zag zipper-like nature seen by SIM (Figure 12). In many cases, the lateral borders are seen to slant almost diagonally away from the dense body contact zones, with several minor changes in orientation along the way. Sometimes a third thin wedge of muscle intervenes at this basal border, coming from a neighboring muscle in the same quadrant (Figure 12C) to cause a sharp deflection of the lateral border. In other places there are distinctive interlocking fingers where one muscle cell pushes deeply into its neighbor. Such fingers feature dense body-like material in some cases (more basally) but often feature one or more gap junctions where the fingers interlock more apically (Figure 12B). All of the above geometries are likely to contribute in part to the irregular zippering along lateral borders seen by SIM.
Figure 12. Several geometries along the lateral border contribute to the zipper-like structure there.
(A,B,C). Three transverse electron micrographs display some of the variety of features along the muscle cell-muscle cell apposition which help to produce a zig-zag pattern of the muscle protein localization observed by SIM. (A) Here the two adjoining muscle cells feature an overall slanting interface, from the region of the dense body at the basal surface to a large gap junction (GJ) near the apical limit of the border zone. Some basement membrane (BL) material fills the extracellular spaces here all the way to the gap junction. (B) Here a large “finger” of muscle tissue (asterisk) protrudes from one muscle cell into its neighbor, and two gap junctions (GJ) mark the base of this finger, connecting the two cells more intimately. (C) Here the border between two muscle cells (BWM1, BWM2) is invaded by the extreme distal portion of a third muscle cell (BWM3), so that the dense body material (marked with a yellow arrow) connects muscle cells 1 and 3, while the lateral border deviates sharply to allow muscle cells 1 and 2 to meet further in; note that BWM cells 1 and 2 are also connected by a gap junction (GJ). All of these geometries are common, and large enough to contribute to the flexible zig-zag shapes detectable by SIM. Scale bars, 1 µm.
As seen by polarized light, the sarcomere lattice projects along the spindle-shaped muscle at a shallow angle with respect to the central axis of the muscle spindle itself. This shallow orientation difference accounts for the steady left/right progression of dense body structures along the surface of the muscle membrane as one moves through serial sections. Polarized light also helps to illustrate the long spindle-like shapes of these muscles, whose extreme distal extensions lie in proximity to near neighbors (as shown in Figure 12C; see also MusFIG 7 in WormAtlas (Altun and Hall, 2009) for a significant portion of total muscle length. These situations contribute to slowly progressing diagonal interfaces when two muscles are more similar in dimension, much like that shown in Figure 12A, and sharp changes in the lateral borders when one muscle overrides the distal extreme of a neighbor.
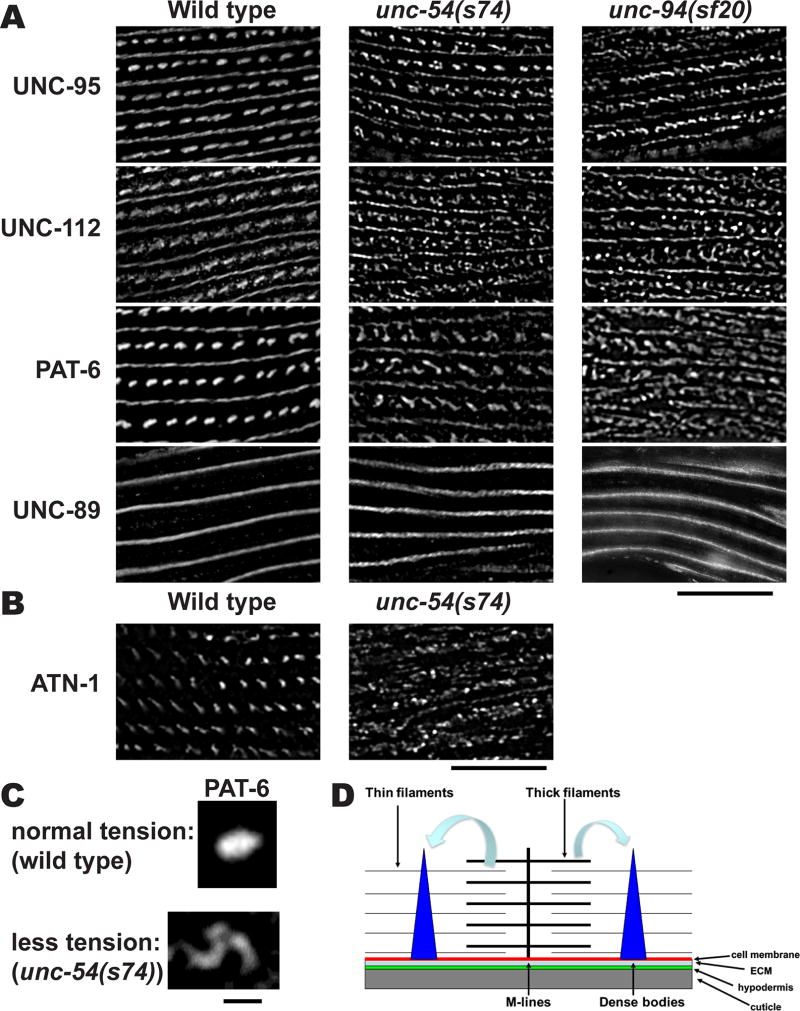
Mutations in components of thin filaments and thick filaments affect the shape of dense bodies
Since the atn-1 null mutant displays a reduced ability to transmit the force of muscle contraction [Moulder et al., 2010], we hypothesized that tension normally exerted by myofilaments might change the shape of dense bodies from “zipper” like to “dot” like. To obtain evidence for this idea, we immunostained unc-54(s74) and unc-94(sf20) mutants with anti-UNC-112, anti-UNC-95, anti-PAT-6 and anti-UNC-89 antibodies and examined their structures by SIM (Figure 13A). The unc-54 gene encodes myosin heavy chain B, and localizes to the outer, major portions of muscle thick filaments, and not the inner portion of thick filaments that are cross-linked at the M-line [Epstein et al., 1974; Miller et al., 1983]. unc-54(s74) contains an arginine to cysteine change near the ATP-binding site in the head region of myosin heavy chain and shows nearly normal muscle structure by polarized light microscopy and EM, but slow and stiff movement [Moerman et al., 1982; Dibb et al., 1985]. unc-94 encodes tropomodulin and is localized to the minus ends of thin filaments [Stevenson et al., 2007], whereas the plus ends of thin filaments are located near dense bodies. The unc-94(sf20) mutant has slow movement and disorganization of sarcomeres, including abnormal accumulation of F-actin outside of normal thin filaments [Stevenson et al., 2007]. In both mutants, basal components of dense bodies, visualized by immunostaining of anti-UNC-112, anti-UNC-95, and anti-PAT-6 showed no “dot” like structures. Similarly, the deeper component of dense bodies, ATN-1, also does not have a dot-like appearance in unc-54(s74) (Figure 13B). M-lines that are visualized by anti-UNC-89 staining were normal. Since neither UNC-54 nor UNC-94 proteins are localized at dense bodies, our interpretation is that reduced tension caused by defects in thick filaments or thin filaments changes the shape of dense bodies (Figure 13C). These results and interpretation support our hypothesis that myofilament tension is required for maintenance of the shape of dense bodies (Figure 13D).
Figure 13. Reduced contractile force affects the shape of dense bodies.
(A) Wild type, unc-54(s74), and unc-94(sf20) worms were immunostained with antibodies against UNC-95, UNC-112, PAT-6, and UNC-89. Images were captured by SIM. The unc-54 gene encodes myosin heavy chain B, one of the components of thick filaments, and the s74 allele shows nearly normal sarcomere structure, but the animals move slowly and stiffly, as compared to wild type. The unc-94 gene encodes a tropomodulin, localized at the pointed ends of thin filaments. Both in the unc-54(s74) and unc-94(sf20) mutants, the membrane-proximal components of dense bodies do not appear “dot” like, but appear closer to “zipper” like. Black bar represents 10 µm. (B) Wild type and unc-54(s74) muscle stained with antibodies to ATN-1 (α-actinin), a deeper component of dense bodies. Note that the deeper portions of dense bodies appear disorganized. Scale bar, 10 µm. (C) Enlargement of single dense bodies stained with anti-PAT-6 from wild type (top) and from unc-54(s74) muscle (bottom). Note the that under normal tension conditions (wild type muscle) the dense body appears as a dot, but that under reduced tension (unc-54(s74)), the dense body appears fragmented. Scale bar, 1 µm. (D) Model for determination of the shape of a type of muscle attachment structure, the dense body. Lack of deeper part of dense bodies (atn-1 mutant), reduction of contractile force (unc-54 mutant), or defects in actin thin filaments (unc-94 mutant) causes alteration of dense body shape, suggesting that the maintenance of normal “dot” like dense bodies requires normal myofilament tension.
Discussion
Differential localization of M-line and dense body components
Our SIM observations revealed two different patterns of localization for nine components of M-lines: PAT-3, UNC-112, PAT-4, UNC-97, PAT-6 and UNC-95 localize to discrete angled segments with gaps, and UNC-98, UNC-89 and MHC A localize to more continuous lines (Figure 1A). Although SIM has allowed us for the first time to clearly observe these discontinuities, previous reports have hinted at such discontinuities by conventional microscopy. The best example was reported using antibodies to PAT-3 (β-integrin) by Francis and Waterston (1985). Other examples include the localization of UNC-95, using a GFP fusion (Broday et al., 2004), or an antibody (Qadota et al., 2007). The proteins found in discrete segments with gaps are proteins that interact directly (UNC-112), or in complexes with the cytoplasmic tail of β-integrin (PAT-3), and thus are likely to lie close to the muscle cell membrane. Our previous confocal z-series [supplementary data in Warner et al., 2013], showed that indeed UNC-112 and PAT-6 localize close to the muscle cell membrane. In addition, the 3D reconstruction of a z-series taken by SIM (Figure 2), also shows that PAT-6 localizes close to the muscle cell membrane. Two proteins, UNC-89 and UNC-98, found in continuous lines localize throughout the depth of the M-line, from near the muscle cell membrane to deep into the myofilament lattice: We had reported earlier that UNC-89 localizes throughout the depth of the M-line based on a z-series taken by confocal microscopy [supplementary data in Warner et al., 2013]. By a similar method, UNC-98 is also located throughout the depth of the M-line [Miller, 2006]. That UNC-89 is full depth is further confirmed by examining the 3D reconstruction of a z series taken by SIM shown in Figure 2. Moreover, this reconstruction shows that UNC-89 localizes as a continuous wall, and that gaps of PAT-6 localization near the muscle cell membrane are “filled” with UNC-89. Finally, MHC A also localizes as a continuous wall (Figure 3), and MHC A extends somewhat more deeply than does UNC-89. This relationship between UNC-89 and MHC A localization is consistent with transmission EM images which show that in cross sections of body wall muscle, thick filaments are found more deeply into the muscle cell than the limit of the M-line (e.g. see EM images in Gieseler et al. 2016), which is the location of UNC-89.
Dense bodies also have differential localization of proteins depending on distance from the muscle cell basal cell membrane [Gieseler et al. 2016]. The surface portion of dense bodies contains integrins (PAT-2 and PAT-3), the integrin-associated four-protein complex (UNC-112, PAT-4, UNC-97, and PAT-6), UNC-95, CPNA-1, and DEB-1. Middle and deepest parts of dense bodies consist of ATN-1 and ALP-1. ZYX-1 is localized only at the middle part of dense bodies. The differential localization of protein components of dense bodies also might affect the assembly or maintenance of the shape of dense bodies (see below).
Contact site structures for adjacent muscle cells contain a subset of integrin associated proteins
M-lines and dense bodies consist of different sets of proteins. Some proteins are localized both at M-lines and dense bodies (integrins (PAT-2 and PAT-3), the integrin-associated four-protein complex (UNC-112, PAT-4, UNC-97, and PAT-6), UNC-95, and CPNA-1 [Gieseler et al., 2016]. M-lines contain M-lines specific proteins (UNC-96, UNC-98, UNC-89, LIM-8, and LIM-9). Dense bodies also contain dense body specific proteins (DEB-1, ATN-1, and ALP-1). Proteins located at specific sites are likely to be required for site-specific functions, such as the linkage from cell membrane to thin or to thick filaments (dense bodies and M-lines, respectively). The muscle cell boundary involves a third set of proteins containing integrins and integrin associated proteins. As shown in Figure 4, only those proteins that localize to the base of dense bodies also localize to muscle cell boundaries. However, proteins that localize only at cell boundaries have not yet been identified, other than the innexins (Liu et al., 2013). Such proteins are likely to be important for the function of cell boundary structures, for example essential for lateral force transmission, and/or the integrity of each body wall muscle quadrant, as well as for cell-cell signaling. Further genetic and molecular biological studies should identify such proteins.
When viewed from above, muscle cell boundary structures appear “zipper-like”, whereas dense bodies appear “dot or dash-like”. We showed that muscle cell boundaries lack ATN-1 and ALP-1, which are components of the deeper parts of dense bodies, suggesting that the cell boundary only contains the surface components of dense bodies. A component that is crucial for the characteristic shape of dense bodies is α-actinin, since in the atn-1 null mutant, the dense bodies appear short and broad when viewed by EM [Moulder et al., 2010]. Similarly, in the atn-1 null mutant, when immuno-stained with antibodies to UNC-95, UNC-112 and PAT-6, the dense bodies appear “zipper-like” (Figure 10), identical to the normal boundary structures. EM observations (Figure 11) further support the concept that cell boundary structures or adhesion plaques are similar to the short dense bodies that are found in atn-1 mutants. 3D reconstruction of SIM images of anti-PAT-6 or anti-UNC-95 shows these zipper-like structures consist of short parallel walls oriented diagonally, approximately 45 degrees from the axis of M-line walls, and these structures extend somewhat less deeply than M-lines and dense bodies (Figure 6).
The muscle cell boundary attachment sites consist of cell-ECM-cell contacts
We showed that two ECM proteins (UNC-52 (perlecan), MIG-6 (papilin, lacunin)) localize between adjacent muscle cells (Figure 8), suggesting that the lateral muscle cell attachment site is a cell-ECM-cell structure. Supporting this finding, no cadherins or catenins have been reported to localize at the muscle cell boundary, and loss of function mutations in either of those proteins indicate involvement in hypodermal cell attachments, but not muscle cell attachments [Costa et al., 1998; Pettitt et al., 2003]. We localized anti-UNC-52 and anti-MIG-6 antibodies between adjacent muscle cells (Figure 8C). Using GFP fusion proteins, Muriel et al. [2005] reported that FBL-1 (C. elegans fibulin) also localizes at the muscle cell boundary. Furthermore, nematodes having mutations in fbl-1 showed gaps at the muscle cell boundary, suggesting that FBL-1 is crucial for maintaining contacts between adjacent muscle cells [Muriel et al., 2005]. FBL-1 localization at the cell boundary has not yet been examined for depth of localization, and the localization of other ECM proteins (for example, type IV collagens (LET-2 and EMB-9)) that comprise the basement membrane have not yet been examined at muscle cell boundaries. Further examination of ECM proteins at muscle cell boundaries and their loss of function mutants should clarify which ECM proteins, in addition to FBL-1, are essential components of muscle cell boundaries.
Tension is required for maintenance of the shape of dense bodies
We showed that mutations in a thin-filament specific protein, UNC-94 (tropomodulin) or a thick filament-specific protein, UNC-54 (MHC B) result in a change in shape of dense bodies from the normal “dot-like” to “zipper-like” (Figure 13A). Similar results were also obtained using other unc-54 alleles (s95 and st134, data not shown) and unc-94 allele (su177, data not shown). The mutations in these three unc-54 alleles are found in myosin head region, and although these animals move slowly and stiffly, their muscle structure is either normal or close to normal [Moerman et al., 1982; Moerman and Fire, 1997]. unc-94 encodes the F-actin minus end capping protein tropomodulin and two unc-94 mutant alleles have basically the same phenotype [Stevenson et al., 2007]. It is noteworthy that neither UNC-54 nor UNC-94 are located at dense bodies, but are located in thick filaments, or at the minus ends of thin filaments, respectively (the plus ends of thin filaments are located near dense bodies). These results with mutants suggest that defects in non-dense body components can still result in disorganization of dense bodies. Mutations in other non-structural components also result in disorganization of overall muscle structure. unc-27 encodes a troponin I, a component of the thin filaments, and regulator of muscle contraction, at least in vertebrate striated muscle. Mutations in unc-27 show severe muscle disorganization [Burkeen et al., 2004]. Muscle contraction and relaxation are required for dynamic turnover of structural components [Ghosh and Hope, 2010] and defects in this turnover cause muscle disease, such as muscle atrophy [Benian and Epstein, 2011; Epstein and Benian, 2012]. In addition to turnover functions, we propose that normal muscle activity, that produces muscle tension, might be essential for maintaining the structure of one type of muscle attachment site, the dense body (Figure 13C and D). In other words, structural and attachment components are essential for maintenance of overall structure, but also normal contraction and relaxation activities themselves contribute to maintain overall structure. This “bidirectional regulation” is conceptually similar to integrin outside-in and inside-out signaling [Hu and Luo, 2013; Shen et al., 2012]. The molecular mechanisms by which a change in tension translates into regulation of muscle attachment structures is unclear, but further genetic and molecular biological studies may clarify these mechanisms.
EM observations help to explain the zipper-like structures observed by SIM
Electron micrographs (Figure 11) reveal electron dense material similar in appearance to dense bodies located adjacent to the basolateral cell membranes of adjacent muscle cells separated by ECM. These structures are thus compatible with the zipper-like structures filled with ECM components that we observe by SIM immunostaining (Figures 5–9). In addition, by EM, there are a variety of features of the muscle cell boundaries that help explain the zig-zag pattern of muscle protein localization observed by SIM, including diagonal orientation of lateral borders (Figure 11A, B, Figure 12A), and finger-like projections of one cell into another cell (Figure 12B). These angled deviations of the borders and finger-like projections are likely contribute to: (1) the angle of the zipper lines, and (2) the fact that the ECM component, UNC-52, does not localize to a straight or continuous line (Figure 8A), and also that there is actually some overlap between the zipper-like intracellular components UNC-95 and UNC-112, with the ECM component UNC-52 (Figure 8B; this can be seen as “white” overlap between the magenta UNC-52 and green UNC-95 or UNC-112 signals). Moreover, although ours is the first report of finger-like projections between body wall muscle cells, interlocking fingers have been reported between pharyngeal muscle cells and between marginal cells within each pharyngeal segment (Albertson and Thomson, 1976; Altun and Hall, 2009). These fingers in both pharyngeal and body wall muscle cells are likely to provide structural integrity and resistance to breakdown of the muscle tissue during the stress of repeated contraction and relaxation. Finally, this is one of the first reports of gap junctions connecting body wall muscle cells away from the muscle arms (Figure 11A, Figure 12)(an earlier indication of these same appositions was reported by Chen et al., 2007). These are likely to provide a route for signaling between body wall muscle cells, and to permit them to act as larger functional units in C. elegans in which the mononuclear striated muscle cells are not fused into a syncytium. Overall, the finger-like projections and gap junctions that provide structural integrity and signaling between nematode body wall muscle cells, is reminiscent of the intercalated disks of vertebrate cardiac muscle, which also allow smaller mononuclear cells to function in larger units.
Summary
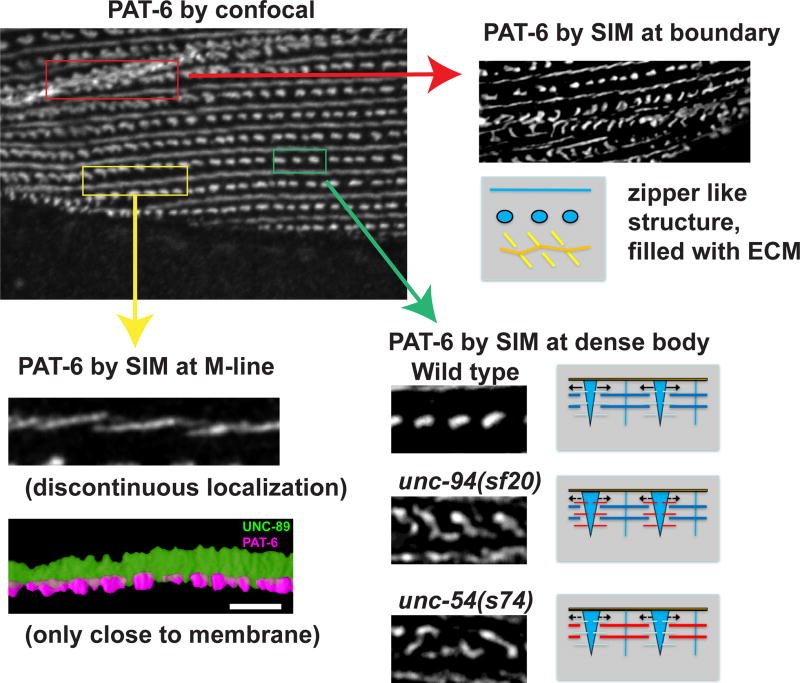
Our SIM imaging as compared with confocal imaging, has revealed new details about integrin adhesion sites in nematode striated muscle (Figure 14): The base components of M-lines (e.g. PAT-6) have a discontinuous localization; some components used at the base of dense bodies are also found at muscle cell boundaries, where they localize in a unique zipper-like structure, filled with ECM components (e.g. UNC-52, MIG-6); and although our view of dense body structure by SIM has not changed our previous views of this structure, nevertheless, mutants in components of the sarcomere other than the dense body, that are involved in generating contractile force (myosin heavy chain or tropomodulin), result in disorganization of dense bodies, suggesting that tension is required to maintain dense body structure. Finally, in regards to the muscle cell boundaries, our EM observations are compatible with our SIM images, and the complexity of muscle cell to muscle cell boundaries are more complex than previously appreciated. This complexity includes diagonal geometry, finger-like projections from one cell to another and numerous gap junctions. These intercellular specializations are all likely to contribute to structural integrity and multiple spindle-shaped body wall muscle cells acting as larger functional units.
Figure 14. Summary of key findings revealed by SIM.
The upper left image shows PAT-6 localized by confocal microscopy. As shown in the red box, by confocal PAT-6 at the muscle cell boundary appears in irregular clusters, but is revealed by SIM imaging (pointed to by a red arrow at the upper right) to exist in distinct zipper like structures, separated by ECM (and depicted in drawing). As shown in the yellow box, by confocal PAT-6 at the M-line appears as an irregular continuous line, but is revealed by SIM (pointed to by a yellow arrow at the bottom left) to localize to distinct slightly diagonal lines. This discontinuity of PAT-6 at the base of the M-line is also clear from 3D rendering (very bottom left). As shown in the green box, by confocal PAT-6 at dense bodies appear as a row of solid ovals. As shown by SIM, in wild type muscle, PAT-6 at dense bodies has the same appearance as revealed by confocal. However, in mutants affecting thin filament function (unc-94 (encodes tropomodulin)) or thick filament function (unc-54 (encodes a myosin heavy chain)) but not appreciably sarcomere structure, dense bodies are mis-shapen—elongated “dumb-bells”, very similar to the appearance of the zipper-like structures at the normal muscle cell boundary. The drawing to the right of the images convey the model that normal tension generated by the thin and thick filaments is required to maintain the proper structure of dense bodies, the main structures in nematode muscle upon which contractile force is applied.
Materials and Methods
C. elegans strains
Standard growth conditions for C. elegans were used [Brenner, 1974]. We used Bristol N2 as the wild-type strain and the following mutant strains: atn-1(ok84) (obtained from Dr. Bob Barstead; (Moulder et al., 2010)), unc-98(sf19) [Mercer et al., 2003], unc-54(s74) [Moerman et al., 1982] and unc-94(sf20) [Stevenson et al., 2007]. N2 (Bristol) and unc-54(s74) were obtained from the Caenorhabditis Genetics Center.
Immunolocalization in adult body-wall muscle
Adult nematodes were fixed and immunostained according to the method described [Nonet et al., 1993], and described in further detail [Wilson et al., 2012]. The following primary antibodies were used at 1:200 dilution: anti-UNC-89 (mouse monoclonal MH42 [Benian et al., 1996; Hresko et al., 1994]); anti-UNC-89 (rabbit polyclonal EU30 [Benian et al., 1996]); anti-myosin heavy chain A (MHC A) (mouse monoclonal 5–6; [Miller et al., 1983]), anti-myosin heavy chain B (MHC B) (mouse monoclonal 5–8; [Miller et al., 1983]), anti-UNC-52 (mouse monoclonal MH2; [Mullen et al., 1999]), anti-DEB-1 (mouse monoclonal MH24; [Francis and Waterston, 1991]), anti-ATN-1 (mouse monoclonal MH35; [Francis and Waterston, 1991]), and at 1:100 dilution: anti-UNC-95 (rabbit polyclonal Benian-13; [Qadota et al., 2007]), anti-UNC-112 (rabbit polyclonal; [Hikita et al., 2005]), anti-PAT-4 (rabbit polyclonal; [Qadota et al., 2012]), anti-UNC-97 (rabbit polyclonal; [Miller et al., 2006]), anti-PAT-6 (rat polyclonal; [Warner et al., 2013]), anti-UNC-98 (rabbit polyclonal; [Mercer et al., 2003]), anti-ALP-1 (rabbit polyclonal; [Han and Beckerle, 2009]), anti-MIG-6 (rabbit polyclonal; [Kawano et al., 2009]). 5–6, MH2, and MH24 were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Secondary antibodies, also used at 1:200 dilution, included anti-rabbit Alexa 488 (Invitrogen), anti-rat Alexa 594 (Invitrogen), and anti-mouse Alexa 594 (Invitrogen).
Confocal microscopy
Images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope and an Apochromat x63/1.4 numerical aperture oil immersion objective, in x2.5 zoom mode. The color balances of the images were adjusted by using Adobe Photoshop (Adobe, San Jose, CA).
N-SIM microscopy
Super-resolution microscopy was performed with a Nikon N-SIM system in 3D structured illumination mode on an Eclipse Ti-E microscope equipped with a 100×/1.49 NA oil immersion objective, 488- and 561-nm solid-state lasers, and an EM-CCD camera (DU-897, Andor Technology). Super-resolution images were reconstructed using the N-SIM module in NIS-Elements software. The color balances of the images were adjusted by using Adobe Photoshop (Adobe, San Jose, CA). A Z-series every 0.2 µm was taken from the outer muscle cell membrane deeper into the muscle cell. This Z stack was then reconstructed into a 3D image using software on the N-SIM microscope. The reconstructed 3D z-stacks were analyzed using Imaris 8.4 (Bitplane), to visualize the M-lines and rows of dense bodies. 3D surfaces were manually thresholded to best represent the observed breaks in PAT-6 or UNC-95 localization in the M-lines, and individual sections were displayed using clipping planes to include only one M-line.
Electron microscopy
Adult animals were prepared for electron microscopy by standard methods [Hall, 1995]. Briefly, live animals were rinsed in M9 buffer and cut open using a razor blade in buffered aldehydes, rinsed in buffer and re-fixed in buffered osmium tetroxide, then rinsed in buffer and en bloc stained in uranyl acetate before dehydration and embedding in plastic resin. Thin sections were collected onto Formvar-coated slot grids, re-stained with uranyl acetate and/or lead citrate, then viewed with a Philips CM10 electron microscope. Images were collected on film, or using an Olympus Morada digital camera system.
Acknowledgments
We thank Laura Fox-Goharioon and Neil Anthony of Emory University’s Integrated Cellular Imaging (ICI) Core for help in using the N-SIM microscope, and for 3D rendering, respectively. We thank the following individuals for various nematode strains and/or antibodies: Dr. Robert Barstead, Dr. Mary Beckerle, Dr. Joe Culotti, Dr. Henry Epstein (deceased), Dr. Pam Hoppe, and Dr. Kozo Kaibuchi. This study was supported in part by National Institutes of Health Grant RO1AR064307 and a pilot grant from Emory’s ICI Core to G.M.B. Electron microscopy performed by D.H.H. and K.C.Q.N. was supported by NIH OD 010943. Some of the nematode strains used in this work were provided by the Caenorhabditis Genetics Center (funded by the National Institutes of Health, Center for Research Resources).
Abbreviations used
- SIM
structured illumination microscopy
- Unc
uncoordinated
References
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Altun ZF, Hall DH. Alimentary System, Pharynx. WormAtlas. 2009 doi: 10.3908/wormatlas.1.3. [DOI] [Google Scholar]
- Benian GM, Epstein HF. Caenorhabditis elegans muscle: a genetic and molecular model for protein interactions in the heart. Circ Res. 2011;109:1082–1095. doi: 10.1161/CIRCRESAHA.110.237685. [DOI] [PubMed] [Google Scholar]
- Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89 required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L, Kolotuev I, Didier C, Bhoumik A, Podbilewicz B, Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger preotein RNF-5 in C. elegans. J Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkeen AK, Maday SL, Rybicka KK, Sulcove JA, Ward J, Huang MM, Barstead R, Franzini-Armstrong C, Allen TS. Disruption of Caenorhabditis elegans muscle structure and function caused by mutation of troponin I. Biophys J. 2004;86:991–1001. doi: 10.1016/S0006-3495(04)74174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Liu Q, Ge Q, Xie J, Wang ZW. UNC-1 regulates gap junctions important to locomotion in C. elegans. Curr Biol. 2007;17:1334–1339. doi: 10.1016/j.cub.2007.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibb NJ, Brown DM, Karn J, Moerman DG, Bolten SL, Waterston RH. Sequence analysis of mutations that affect the synthesis, assembly and enzymatic activity of unc-54 myosin heavy chain of C. elegans. J Mol Biol. 1985;183:543–551. doi: 10.1016/0022-2836(85)90170-6. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Benian GM. Paradigm shifts in cardiovascular research from Caenorhabditis elegans muscle. Trends Cardiovasc Med. 2012;22:201–209. doi: 10.1016/j.tcm.2012.07.021. [DOI] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseler K, Qadota H, Benian GM. Development, structure, and maintenance of C. elegans body wall muscle. WormBook. 2016:1–59. doi: 10.1895/wormbook.1.81.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SR, Hope IA. Determination of the mobility of novel and established Caenorhabditis elegans sarcomeric proteins in vivo. Eur. J. Cell Biol. 2010;89:437–448. doi: 10.1016/j.ejcb.2009.11.027. [DOI] [PubMed] [Google Scholar]
- Hall DH. Electron microscopy and three-dimensional image reconstruction. Methods Cell Biol. 1995;48:395–436. doi: 10.1016/s0091-679x(08)61397-7. [DOI] [PubMed] [Google Scholar]
- Han HF, Beckerle MC. The ALP-Enigma protein ALP-1 functions in actin filament organization to promote muscle structural integrity in Caenorhabditis elegans. Mol Biol Cell. 2009;20:2361–2370. doi: 10.1091/mbc.E08-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH, Stern BD. Genetics of cell and axon migrations in Caenorhabditis elegans. Development. 1987;100:365–382. doi: 10.1242/dev.100.3.365. [DOI] [PubMed] [Google Scholar]
- Hikita T, Qadota H, Tsuboi D, Taya S, Moerman DG, Kaibuchi K. Identification of a novel Cdc42 GEF that is localized to the PAT-3-mediated adhesive structure. Biochem Biophys Res Commun. 2005;335:139–145. doi: 10.1016/j.bbrc.2005.07.068. [DOI] [PubMed] [Google Scholar]
- Hresko MC, Williams BD, Waterston RH. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Luo BH. Integrin bi-directional signaling across the plasma membrane. J Cell Physiol. 2013;228:306–312. doi: 10.1002/jcp.24154. [DOI] [PubMed] [Google Scholar]
- Kawano T, Zheng H, Merz DC, Kohara Y, Tamai KK, Nishiwaki K, Culotti JG. C. elegans mig-6 encodes papilin isoforms that affect distinct aspects of DTC migration, and interacts genetically with mig-17 and collagen IV. Development. 2009;136:1433–1442. doi: 10.1242/dev.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerova IA, Kawaguchi N, Fessler LI, Nelson RE, Chen Y, Kramerov AA, Kusche-Gullberg M, Kramer JM, Ackley BD, Sieron AL, Prockop DJ, Fessler JH. Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development. 2000;127:5475–5485. doi: 10.1242/dev.127.24.5475. [DOI] [PubMed] [Google Scholar]
- Liu P, Chen B, Altun ZF, Gross MJ, Shan A, Schuman B, Hall DH, Wang ZW. Six innexins contribute to electrical coupling of C. elegans body-wall muscle. PLoS One. 2013;8:e76877. doi: 10.1371/journal.pone.0076877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol Biol Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Miller RK, Qadota H, Landsverk ML, Mercer KB, Epstein HF, Benian GM. UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK. PhD thesis. Emory University; 2006. UNC-98 links an integrin-associated complex to thick filaments in C. elegans muscle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Fire A. Muscle: Structure, Function, and Development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor (NY); 1997. pp. 417–470. [PubMed] [Google Scholar]
- Moerman DG, Plurad S, Waterston RH, Baillie DL. Mutations in the unc-54 myosin heavy chain gene of Caenorhabditis elegans that alter contractility but not muscle structure. Cell. 1982;29:773–781. doi: 10.1016/0092-8674(82)90439-1. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Fire A. Muscle Structure, Function and Development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 417–470. [PubMed] [Google Scholar]
- Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. WormBook. 2006:1–16. doi: 10.1895/wormbook.1.81.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder GL, Cremona GH, Duerr J, Stirman JN, Fields SD, Martin W, Qadota H, Benian GM, Lu H, Barstead RJ. alpha-actinin is required for the proper assembly of Z-disk/focal-adhesion-like structures and for efficient locomotion in Caenorhabditis elegans. J Mol Biol. 2010;403:516–528. doi: 10.1016/j.jmb.2010.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen GP, Rogalski TM, Bush JA, Gorji PR, Moerman DG. Complex patterns of alternative splicing mediate the spatial and temporal distribution of perlecan/UNC-52 in Caenorhabditis elegans. Mol Biol Cell. 1999;10:3205–3221. doi: 10.1091/mbc.10.10.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel JM, Dong C, Hutter H, Vogel BE. Fibulin-1C and Fibulin-1D splice variants have distinct functions and assemble in a hemicentin-dependent manner. Development. 2005;132:4223–4234. doi: 10.1242/dev.02007. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Pettitt J, Cox EA, Broadbent ID, Flett A, Hardin J. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J Cell Biol. 2003;162:15–22. doi: 10.1083/jcb.200212136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Benian GM. Molecular structure of sarcomere-to-membrane attachment at M-Lines in C. elegans muscle. J Biomed Biotechnol. 2010 doi: 10.1155/2010/864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Moerman DG, Benian GM. A molecular mechanism for the requirement of PAT-4 (integrin-linked kinase (ILK)) for the localization of UNC-112 (Kindlin) to integrin adhesion sites. J Biol Chem. 2012;287:28537–28551. doi: 10.1074/jbc.M112.354852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Luo Y, Matsunaga Y, Park AS, Gernert KM, Benian GM. Suppressor mutations suggest a surface on PAT-4 (integrin-linked kinase) that interacts with UNC-112 (kindlin) J Biol Chem. 2014;289:14252–14262. doi: 10.1074/jbc.M114.556308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Delaney MK, Du X. Inside-out, outside-in, and inside-outside-in: G protein signaling in integrin-mediated cell adhesion, spreading, and retraction. Curr Opin Cell Biol. 2012;24:600–606. doi: 10.1016/j.ceb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TO, Mercer KB, Cox EA, Szewczyk NJ, Conley CA, Hardin JD, Benian GM. unc-94 encodes a tropomodulin in Caenorhabditis elegans. J Mol Biol. 2007;374:936–950. doi: 10.1016/j.jmb.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner A, Xiong G, Qadota H, Rogalski T, Vogl AW, Moerman DG, Benian GM. CPNA-1, a copine domain protein, is located at integrin adhesion sites and is required for myofilament stability in Caenorhabditis elegans. Mol Biol Cell. 2013;24:601–616. doi: 10.1091/mbc.E12-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure in C. elegans. Dev Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Waterston RH. Muscle. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; 1988. pp. 281–335. [Google Scholar]
- Wilson KJ, Qadota H, Benian GM. Immunofluorescent localization of proteins in Caenorhabditis elegans muscle. Meth Mol Biol. 2012;798:171–181. doi: 10.1007/978-1-61779-343-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel JM, Epstein HF. Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]