Version Changes
Revised. Amendments from Version 1
This version addresses all major and most minor criticisms of the reviewers. We simplified the biological models and concentrated mostly on renal metabolism, by adding new experimental data. We also removed 900 MHz data and are now using NMR data acquired at 600 MHz only. We redesigned most figures to exclude spectral overlays to improve clarity and added line shape simulations. We also added a new figure, which is now figure 5, so that the previous figures 5 and 6 are now figures 6 and 7, respectively.
Abstract
Tracing the fate of stable isotopically-enriched nutrients is a sophisticated method of describing and quantifying the activity of metabolic pathways. Nuclear Magnetic Resonance (NMR) spectroscopy offers high resolution data in terms of resolving metabolic pathway utilisation. Despite this, NMR spectroscopy is under-utilised due to length of time required to collect the data, quantification requiring multiple samples and complicated analysis. Here we present two techniques, quantitative spectral filters and enhancement of the splitting of 13C signals due to homonuclear 13C, 13C or heteronuclear 13C, 15N J-coupling in 1H, 13C-HSQC NMR spectra. Together, these allow the rapid collection of NMR spectroscopy data in a quantitative manner on a single sample. The reduced duration of HSQC spectra data acquisition opens up the possibility of real-time tracing of metabolism including the study of metabolic pathways in vivo. We show how these techniques can be used to trace the fate of labelled nutrients in a whole organ model of kidney preservation prior to transplantation using a porcine kidney as a model organ. In addition, we show how the use of multiple nutrients, differentially labelled with 13C and 15N, can be used to provide additional information with which to profile metabolic pathways.
Keywords: NMR, stable-isotope tracing, 13C, 15N, splitting enhancement, metabolism, tracer
Introduction
Investigations of metabolism in health and disease increasingly rely on tracing the use of stable isotope-enriched nutrients through the cell’s metabolic pathways. The most widely utilised technology platform to analyse the resulting complex patterns of labelling in multiple cellular metabolites is mass spectrometry (MS), due to its high sensitivity, short run times and a resulting low-cost operation 1– 12. Conversely, NMR spectroscopy is relatively under-utilised, despite being able to provide higher resolution information on the conversion of synthetically produced stable isotopes of nutrients are incorporated into cellular metabolites 12– 19. This is because NMR spectroscopy has historically suffered from low sensitivity, long acquisition times and the need for complex analytical tools.
NMR spectroscopy is, however, ideally suited to answering some of the more pressing questions about metabolic control in health and disease. We currently have limited knowledge about the compartmentalisation of metabolic pathways in metabolically-active organelles, such as mitochondria, and therefore whether the same metabolite is selectively utilised for distinct purposes in different compartments 20. Given the recent drive to target metabolism in various diseases, understanding the control of metabolism by different tissues is critical to the ability to select specific therapies which target the desired pathways within appropriate cellular compartments. While sample analysis by high-resolution NMR spectroscopy is performed ex vivo or in vitro, the data obtained provide information on metabolic pathways in vivo.
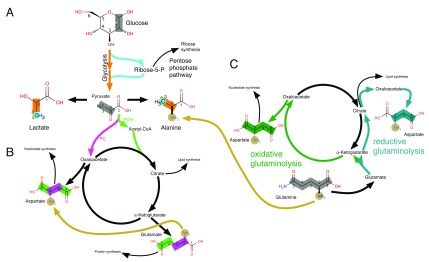
Stable isotope-enriched metabolic precursors, such as glucose or glutamine, are employed as metabolic tracers. These synthetically produced nutrients are enriched in isotopes with a low natural abundance, such as 13C or 15N. Despite the fact that metabolites can arise from multiple sources, the contribution of the different metabolic pathways to the synthesis of this metabolite can be determined through the analysis of the 13C and/or 15N distribution within the metabolite ( Figure 1). The couplings are visualised in the indirect dimension of an HSQC spectrum allowing the determination of the percentage incorporation of isotopic label into adjacent nuclei. While MS data does not need a reference sample to distinguish between labelled and unlabelled metabolites, it is not always possible to derive the exact distribution of labelled nuclei within a molecule. In contrast, NMR spectroscopy data can resolve label distribution at the atomic level, enabling a clearer picture of the label distribution in metabolites.
Figure 1. Tracing of metabolic pathways.
The labelling patterns arising from [1,2- 13C] glucose ( A & B) as well as from [U- 13C, 15N] glutamine ( C for 13C labelling and A– C for 15N labelling) are shown. Metabolism of [1,2- 13C] glucose leads to distinctive labelling patterns in lactate and alanine ([2,3- 13C] lactate/alanine when using glycolysis and [3- 13C] lactate/alanine when using the pentose phosphate shunt, PPP) (Panel A). Similarly, glutamate and aspartate express different labelling patterns from [1,2- 13C] glucose, depending whether they were synthesised via pyruvate dehydrogenase (PDH; resulting in [4,5- 13C] glutamate) or via the pyruvate carboxylase (PC; resulting in [2,3- 13C] aspartate) route (Panel B). Metabolisation of labelled glutamine can reveal other anaplerotic pathway activities such as reductive carboxylation (Panel C).
Our recently published combined NMR spectroscopy and MS approach (CANMS) harnesses the strengths of both modalities to produce highly-resolved metabolism information in the form of metabolite isotopomers 19. The detailed interpretation of MS isotopologue data, when using MS data in isolation, requires use of a pre-defined metabolic model. In contrast, the integrated analysis of NMR spectroscopy and MS data makes fewer assumptions about the metabolic network, providing a more accurate insight into relative pathway contributions than is possible with current established methods or the independent analysis of MS or NMR spectroscopy data alone. For example, proton-less carbon atoms do not give rise to a signal in 2D-HSQC NMR spectra, although incomplete information on those carbons is available via splitting of adjacent carbon nucleus signals. The combination of NMR spectroscopy and MS analysis fills this gap as the MS data provides information on the amount of single carbon labelling into those carbon nuclei via “m+x” isotopologues. [1,2- 13C] glucose is the optimal tracer to assess metabolic flux through glycolysis vs pentose phosphate pathway (PPP) shunting back into glycolysis. While the glycolytically derived isotopomer of lactate is [2,3- 13C] lactate, the PPP derived isotopomers can be [3- 13C], [1- 13C] or [1,3- 13C] lactate. Although the first isotopomer can be assessed with NMR spectroscopy data, the other two isotopomers include labelling in C(1), which HSQC NMR spectroscopy is “blind” to. In these cases, MS data adds new information to the NMR spectroscopy data by contributing the isotopologues NMR spectroscopy is not able to detect, while NMR spectroscopy adds to the MS data by differentiating between [1,3- 13C] and [2,3- 13C] lactate.
A major drawback of utilising 13C- 13C scalar coupling information to derive isotopomer distributions is the time required to acquire spectra. For example, around four hours are required for the acquisition of a 2D-HSQC NMR spectrum with high-resolution in the 13C dimension, even when using fast, state-of-the-art non-uniform sampling (NUS) techniques.
Here we describe two developments, quantitative spectral filters and signal splitting enhancement, to facilitate and speed-up the acquisition of NMR spectra for tracer-based analysis of metabolism. Such techniques permit high throughput metabolic pathway profiling, increasing access, affordability and sensitivity when using NMR spectroscopy as an investigative modality. Additionally, these developments facilitate fast detection of 15N labelling, especially when combined with 13C tracing, thus providing extra information allowing more accurate metabolic pathway profiling.
Methods and results
Quantitative spectral filters for 13C tracer observation: 1D Spectral filters
Experimental setup. A porcine kidney was procured from a slaughterhouse (FA Gill, Wolverhampton) following approximately 14 minutes warm ischaemic time (WIT) as per previous experimental methodology 18. No animal was killed solely for experimental purposes; all were due for human consumption, therefore no ethical approval was required. After 2 hours cold ischaemic time, kidneys were subject to 18 hours of hypothermic machine perfusion. The perfusate sample was collected after 6 hrs of perfusion and prepared for NMR analysis.
1D NMR spectra were acquired using a Bruker Avance III 600 MHz NMR spectrometer equipped with a 5mm z-PFG TCI Cryoprobe. 128 transients were acquired for each spectrum with a 5 s interscan relaxation delay. A total of 32768 data points with a spectral width of 12 ppm was acquired for each FID using an adiabatic bi-level decoupling scheme to suppress 1H, 13C J-coupling during acquisition 21. While decoupling for this long (2.25 s) was possible because of the cryogenic probe and may potentially work with a room temperature probe, care must be taken as there will be significant sample heating. The sample heating can be significantly reduced, with only very minor reduction in resolution, by acquiring for only 1.125 s. In order to estimate whether a specific spectrometer with a cryoprobe can tolerate the power dissipation originating from the decoupling sequence specific attention should be paid to the cryogas heater current, which should never fall below its system specific lower limit.
The spectra were processed within the MetaboLab software package (version 2018.07182055) 22. A 0.5 Hz line broadening was applied with zero-filling of the data up to 131072 real data points prior to Fourier transformation. The resulting spectra were referenced using DSS and manually phase corrected. Subsequently the spectral baseline was corrected using MetaboLab’s spline baseline correction before the spectra were exported to Bruker format for metabolites to be quantified in the Chenomx software package (version 8.2, http://www.chenomx.com).
NMR methodology. Despite their relative simplicity and limited resolution, 1D-NMR spectra are highly sensitive tools with which to identify and quantify metabolites. Spectral filters enable the acquisition of spectra which filter out certain signals, thereby reducing ambiguity in 1D spectra associated with attributing peaks to nuclei within metabolites. For example, one can acquire 1D 1H NMR spectra of protons bound to 13C only, simplifying signal assignment and analysis of the acquired spectra. The simplest approach to collect such spectra would be to acquire the first increment of a 2D- 1H, 13C-HSQC spectrum. However, signal intensities are not directly comparable with those in standard 1D- 1H NMR spectra. It is therefore not possible to directly derive 13C percentages based on a comparison of those spectra with standard proton 1D spectra unless only a small subset of molecules is labelled with 13C and one accompanying spectrum is scaled so that the majority of signals within the two spectra are of same intensity 23. Spectral filters such as BIRD, TANGO and POCE 24– 28 originated in protein NMR spectroscopy to filter out certain parts of the magnetisation and therefore quantitative data cannot be gained from resultant output spectra. Quantitative comparisons between unfiltered and filtered spectra are usually unnecessary, except for tracer-based analysis. Here we present a novel spectral filter which enables quantitative analysis of resultant spectra from single samples, enriched with 13C tracer.
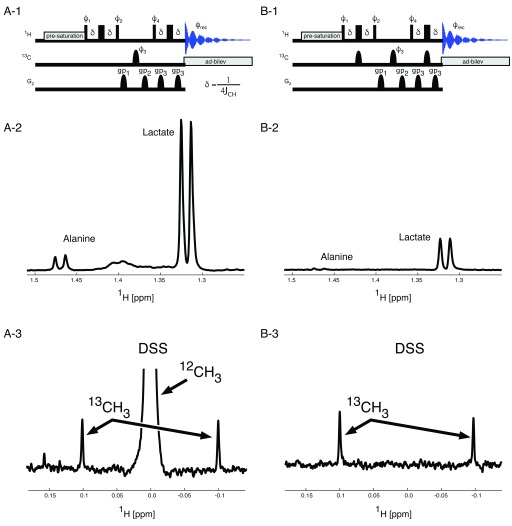
Panels A-1 and B-1 in Figure 2 show the pulse sequences implementing the quantitative spectral filter. While the central 13C π-pulse (phase ϕ 3) is used only in odd numbered transients and replaced with a delay of the same length during even numbered transients, the two other 13C π-pulses are only used in the 12C filtering experiment (B-1), where 1H magnetisation to 12C neighbours is filtered out, so that only 13C bound 1H magnetisation contributes to the signal intensities in the 1D spectrum. The phase cycle ϕ 1 changes as well between the 2 experiments, as indicated in the figure legend.
Figure 2. Spectral Filters in 1D spectroscopy.
To determine percentage 13C incorporation two spectra are acquired per sample. One spectrum ( A) contains 1H signals originating from all protons (all 1H spectrum), while the second spectrum ( B) only contains signals from protons attached to a 13C nucleus. The central 13C π–pulse with phase ϕ 3 is executed every second transient in both experiments. The proton magnetisation in the all 1H pulse sequence is the same for 12C and for 13C bound protons and as a consequence, all 1H magnetisation is longitudinal during the interval between 1H pulses with the phases ϕ 2 and ϕ 4. Because of the additional 13C π–pulses in the 13C bound 1H pulse sequence, the magnetisation for the two different kinds of protons develops differently. Here only the 13C bound 1H magnetisation is longitudinal in the interval between the 1H pulses with the phases ϕ 2 and ϕ 4. Therefore, the 12C bound 1H magnetisation can be destroyed using the two pulse field gradients labelled gp 1 and gp 2. A recovery delay of 200 µs was used after each gradient. The central 13C π–pulse with phase ϕ 4 improves magnetisation selection as it is accompanied by phase changes of the 1H pulses and the receiver. All 13C π-pulses are adiabatic Chirp pulses with γB 1max = 10 kHz. 1H, 13C J-coupling is suppressed during acquisition using adiabatic bilevel decoupling (ad-bilev) 21. The pulse phases are: ϕ 1 = x, x, -x, -x; ϕ 2 = x, x, -x, -x for the all 1H spectrum ( A-1) and y, y, y, y for the 13C bound 1H spectrum ( B-1), ϕ 3 = x, y, -x, -y, ϕ 4 = x, -x, -x, x, y, -y, -y, y for the all 1H spectrum ( A-1) and y, -y, -y, y, -x, x, x, -x for the 13C bound 1H spectrum ( B-1). Panels A-2 and B-2 show the peaks from Alanine and Lactate methyl protons in the all 1H spectrum ( A-2) and the 13C bound 1H spectrum ( B-2). The scaling of the two spectra is identical allowing easy determination of the percentage incorporation of 13C metabolites. Panels A-3 and B-3 demonstrate the complete removal of the 12C bound proton signal from the 13C edited spectrum (B-3) leaving only the natural abundance 13C signals to be observed.
Panels A-2 and B-2 in Figure 2 depict two sample spectra from a perfusate sample where a cadaveric porcine kidney was perfused with modified University of Wisconsin machine perfusion solution (UW MPS) during a period of hypothermic machine perfusion. The standard unlabelled glucose constituent (10 mM) within classical UW MPS was replaced with 10 mM universally labelled glucose, for use as a metabolic tracer during the 18 hour perfusion.
While filters such as BIRD utilise relaxation to minimise the unwanted part of the magnetisation, methods such as TANGO or POCE generate magnetisation where 12C bound 1H atoms possess either the same or the opposite phase compared to the magnetisation of 13C bound 1H atoms, generating two different spectra. By subtraction of these two spectra, the magnetisation of 13C bound 1H atoms can then be calculated. In case of low 13C incorporation, as with any difference technique, subtracting two very large signals in presence of a small signal can lead to substantial artefacts. The quantitative spectral filter works slightly differently compared to TANGO and POCE as the pulse sequence depicted in Figure 2, panel B-1 makes use of two gradient pulses (gp 1 and gp 2) to destroy unwanted magnetisation. As an example, panels A-3 and B3 in Figure 2 show a variant of the pulse sequences where the adiabatic bilevel decoupling (ad-bilev) 21 has been omitted, so that the 12C and the 13C bound 1H signals appear separated in the spectrum. The 12CH 3 signal in A-3 has been truncated to be able to visualise the 13C satellites, which appear with 0.5% of the peak height of the 12CH 3 signal. As can be seen in panel B-3, the 12CH 3 signal is completely suppressed without leaving an artefact, so that even signals from naturally occurring 13C alone are easily detectable in a 13C decoupled spectrum.
The quantitative spectral filter is invariant with respect to differential 1H relaxation rates or signal multiplicities. As with any J-coupling based filtering approach, the 12C filtered spectrum will be scaled with a factor that is proportional to sin(2πδJ CH) 2, where δ is the delay during the first and last spin echo. δ is usually set to ¼J CH with J CH = 145 Hz. Assuming a minimum J CH of 120 Hz and a maximum J CH of 165 Hz, results in a maximum downscaling of 7.2%.
J-Coupling based splitting enhancement in 2D-NMR spectra
Experimental setup. Slaughterhouse porcine kidneys (WIT-15 minutes) were cannulated and flushed with chilled Soltran solution (Baxter) as performed in clinical practice. Kidneys were placed in static cold storage en route to our laboratory, where they were immediately perfused with modified KPS-1 using the Lifeport Kidney Transporter 1.0 (Organ Recovery Systems), which has been modified to include a paediatric oxygenator. Oxygen was supplied at a flow rate of 0.7 L/min for the duration of the 24 hours perfusion period.
At the end of the perfusion period, the kidney was removed from the perfusion circuit and laterally bisected. Sections of cortex and medulla were isolated and snap frozen in liquid nitrogen. These tissues were powdered, also under liquid nitrogen, and 0.5 g was placed in 7 ml homogenisation tube (Precellys, CK28), containing 5.1 ml of HPLC grade methanol (−80°C) to quench metabolism. These were homogenised using the Precellys 24 dual homogeniser (8x 5000 rpm for 15 s). The samples were mixed with 4.65 ml deionised water and 5.1 ml HPLC grade chloroform and vigorously agitated. Biphasic separation of polar and non-polar solvents was performed by centrifugation (1300 g, 15 minutes, 4°C), after which 4.5 ml of the polar layer was aspirated and dried overnight at 35°C.
The dried extracts were resuspended in 60 µl NMR buffer (0.1 M phosphate buffer, 0.5 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid, 2 mM imidazole and 10% D 2O). These suspensions were sonicated to dissolve micro particles and then 35 µl of this solution was added to 1.7 mm NMR tubes.
1H, 13C-HSQC spectra were acquired using a Bruker Avance III 600 MHz NMR spectrometer equipped with a 1.7 mm z-PFG TCI Cryoprobe. The HSQC spectra were acquired using 2 transients per increment with echo/anti-echo gradient coherence selection and an additional pre-saturation for suppressing the water resonance during the 1.5 s interscan relaxation delay. The 1H dimension was acquired with a spectral width of 13 ppm using 512 complex data points. The 13C dimension was acquired with a spectral width of 160 ppm using 25% (2048) of 8192 data points using a non-uniform sampling scheme. The non-uniformly sampled spectra were reconstructed via the compressed sensing algorithm within the MDDNMR (version 2.5) 29 and processed using NMRPipe (version 9.2) 30. All spectra were processed without baseline correction to avoid complications in the multiplet analysis procedure.
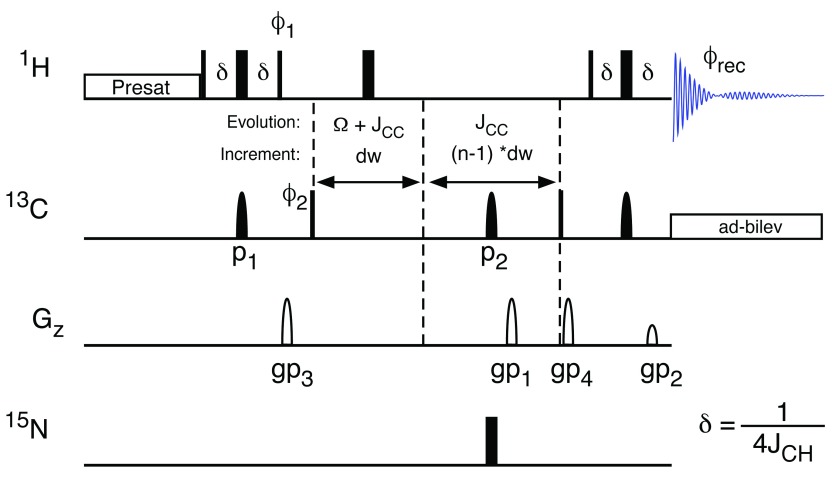
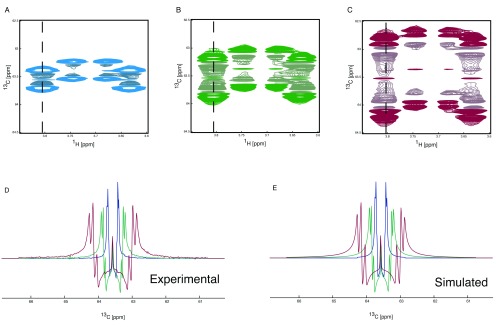
NMR methodology. The relatively long acquisition times of 2D-HSQC spectra are necessary to generate the spectral resolution required to resolve complex multiplet patterns 19. Here we present a technique to manipulate the appearance of NMR multiplets in the indirect dimension of 2D-HSQC spectra. The ability to expand the splitting caused by J-coupling has previously been reported 31, 32. Here we apply this technique in order to negate the requirement for the collection of large number of increments in the 13C dimension, which, together with methods such as non-uniform sampling 29, 33 and variation of the repetition time 34 significantly reduces the time required to acquire 2D-HSQC spectra with sufficient resolution. It also means that at increasingly higher magnetic fields, the advantages of extra sensitivity and increased 1H chemical shift resolution are not negated by the increased 13C increments needed in in order to resolve J-couplings. Enhancement of the splitting due to J-coupling can be achieved by incrementing the spin echo delay after the period where chemical shift of 13C evolves in parallel with the chemical shift evolution ( Figure 3). This spin echo refocuses the 13C chemical shift and the 1H- 13C coupling, but allows the 13C- 13C coupling to evolve further. The delays in the spin echo are proportional to those in the Ω+J CC evolution period with the amount of extra coupling achieved being defined by the stretch of the J CC increment compared to the Ω+J CC increment. Thus, the 13C- 13C J-couplings can be expanded as required ( Figure 4). The ability to scale the signal splittings to varying extents means that the experiment can be tuned to the requirements of the sample and which metabolites are present, and of interest. Figure 4 demonstrates the effect of J-coupling splitting enhancement on 2D HSQC spectra, displaying C(6) of 13C enriched glucose. The 13C trace through the left-most signal ( Figure 4D), demonstrates clearly that while the singlet in the middle of the multiplet does not change, the splitting due to the 1J CC coupling increases and in fact splits into multiple signals as the splittings of previously unresolved long-range couplings are amplified so that they are large enough to become resolved in the spectrum. These splittings can easily be simulated ( Figure 4E), thereby providing additional information with which to model metabolic pathways.
Figure 3. Splitting Enhanced HSQC Spectroscopy.
The splitting enhancement due to J-coupling is achieved using an additional spin echo subsequent to the 13C evolution period. The delays in the spin-echo for the J-coupling enhancement are multiples of the dwell time (dw). While the chemical shift evolves with dw, which is determined by setting the spectral width of the spectrum, splittings are enhanced depending on the increment of the 13C gradient selection spin-echo. The HSQC spectrum is acquired using echo/anti-echo for quadrature detection to allow for efficient removal of artefacts in only two scans per increment. Optionally, the 13C, 15N-couplings can be scaled by the introduction of the 15N π-pulse simultaneously with the 13C π-pulse (labelled p2). 1H, 13C J-coupling is suppressed during acquisition using adiabatic bilevel decoupling (ad-bilev) 21. The pulse phases are: ϕ 1 = y; ϕ 2 = x, -x; ϕ rec = x, -x.
Figure 4. J-coupling splitting enhancement HSQC spectroscopy.
1H, 13C HSQC spectra showing the C(6) of glucose are shown ( A, B and C). The spectrum with no J-coupling splitting enhancement is shown in blue ( A), with an enhancement of two in green ( B) and with an enhancement of four in red ( C). The 13C trace of the HSQC spectra ( D), taken from the 1H frequency as depicted by the dashed line, clearly shows the increase in observed splitting. The J-coupling splitting enhancement is achieved using an additional spin echo subsequent to the 13C evolution period. The delays to achieve the scaling of the splittings are multiples of dw such the use of a delay of 3*dw will result in a J-coupling splitting enhancement of 4 (one from the t 1 evolution and three from the J-coupling splitting enhancement spin echo). The observed splitting can be simulated ( E) giving the following incorporation percentages. From the no J-coupling splitting enhancement spectrum 6.8% / 41.2% / 52 % for [6- 13C] / [5,6- 13C] / [U- 13C], from the two-fold J-coupling splitting enhancement 6.3% / 41.4% / 52.3 % for [6- 13C] / [5,6- 13C] / [U- 13C] and from the four-fold J-coupling splitting enhancement 5.9% / 41.6% / 52.5 % for [6- 13C] / [5,6- 13C] / [U- 13C].
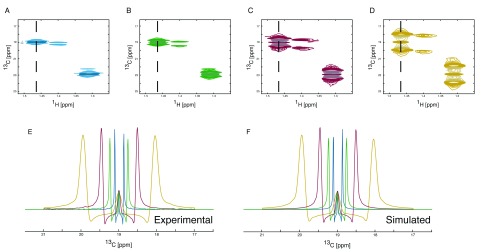
Large expansion of J-coupling also allows for rapid collection of data, as the resolution required to resolve them becomes diminished ( Figure 5). However, this should be tempered by the need to avoid unnecessary overlap of signals. To date, the authors have acquired 2D spectra with up to eight fold enhanced 13C J-couplings, combined with shortening the acquisition by using variable pulse sequence repetition times 34, leading to an overall decrease in acquisition time by a factor larger than 10 ( Figure 5). Panels A to D show the spectral region of the methyl groups of lactate and alanine. Panel E shows a cross section, as marked in the 2D spectra, from alanine, whereas panel F shows the corresponding simulations of those multiplets. The acquisition times for the different spectra were 233, 110, 51 and 24 minutes ( Table 1). Whilst the lines in the spectrum become broader due to the shorter acquisition times, this is negated by the increase in splittings, allowing the analysis of the multiplets with similar precision. Shorter acquisition times may be achieved by including spectral folding in the acquisition protocol or by incorporating new fast acquisition schemes such as ASAP- or ALSOFAST-HSQC 35.
Figure 5. Simulation of J-coupling splitting enhanced spectra.
1H, 13C HSQC spectra showing the C(3) of alanine are shown ( A, B, C and D). The enhancement of J-coupling in the 1H, 13C-HSQC spectra leads to increased separation of coupled peaks and allows the collection of data with reduced numbers of increments in the 13C dimension resulting in shorter acquisition times. The number of points collected in the 13C dimension can be reduced to match the increase in J-coupling enhancement as the loss of resolution will be counteracted by the increase in separation of the coupled peaks. Spectra with no enhancement ( A), two-fold enhancement ( B), four-fold enhancement ( C) and eight-fold enhancement ( D) were collected. The overlay of the differently enhanced spectra clearly shows the effect of the enhancement ( E). The enhancement can be tailored to meet the need in order to maximise separation without significantly increasing signal overlap whilst achieving the maximum reduction in acquisition time possible. The splitting enhancement can easily be included in the simulation parameters ( F) resulting in no adverse effects on the simulated spectra. The simulation gives the following incorporation percentages. From the no J-coupling splitting enhancement spectrum 12.1% / 87.9% for [3- 13C] / [2,3- 13C], from the two-fold J-coupling splitting enhancement 12.0% / 88.0% for [3- 13C] / [2,3- 13C], from the four-fold J-coupling splitting enhancement 12.0% / 88.0% for [3- 13C] / [2,3- 13C] and from the eight-fold J-coupling splitting enhancement 12.1% / 87.9% for [3- 13C] / [2,3- 13C].
Table 1. Comparison of spectroscopic techniques.
The acquisition time and signal to noise ratios of various experiments used in this study. A good signal to noise ratio can be achieved using the spectral filtering allowing rapid measurement of quantitative spectra. The signal to noise benefit of the HSQC over the 1D 13C acquisition is clearly seen. The effect of increasing the J-coupling splitting enhancement whilst simultaneously reducing the number of increments on the acquisition time of 1H, 13C-HSQC spectra is also shown.
| Experiment | Acquisition
time [mins] |
Signal to noise
ratio (CH 3 of lactate) |
Transients | TD | SW
[ppm] |
Splitting
enhancement |
|---|---|---|---|---|---|---|
| 1H 1D (all 1H) | 2 | 327.31 | 8 | 16384 | 12 | 1 |
| 1H 1D ( 13C bound 1H) | 2 | 66.13 | 8 | 16384 | 12 | 1 |
| 1H, 13C-HSQC | 233 | 823.86 | 2 | 1024/8192 | 12/160 | 1 |
| 1H, 13C-HSQC | 110 | 770.98 | 2 | 1024/4096 | 12/160 | 2 |
| 1H, 13C-HSQC | 51 | 426.41 | 2 | 1024/2048 | 12/160 | 4 |
| 1H, 13C-HSQC | 24 | 283.38 | 2 | 1024/1024 | 12/160 | 8 |
| 13C 1D (30° excitation) | 1414 | 59.95 | 16384 | 65538 | 239 | 1 |
15N tracing
Experimental setup. 2D 14N spectral filter - Sample preparation is described elsewhere 16. 2D 1H 13C-HSQC NMR spectra with and without 14N filtering were acquired using a Bruker Avance III 600 MHz NMR spectrometer equipped with a 1.7 mm z-PFG TCI Cryoprobe. The HSQC spectra were acquired using 2 transients per increment with echo/anti-echo gradient coherence selection and an additional pre-saturation during the 1.5 s interscan relaxation delay to suppress the water resonance. The 1H dimension was acquired with a spectral width of 13 ppm using 512 complex data points. The 13C dimension was acquired with a spectral width of 160 ppm using 2048 data points. The spectra were processed with quadratic cosine window functions and without baseline correction to avoid complications in the multiplet analysis procedure.
13C, 15N J-coupling splitting enhancement. The human Renal Proximal tubule cell line (RPTEC/TERT1, supplied by Evercyte GmBH, Austria) was used to investigate the metabolic fates of both carbon from glucose, and carbon and nitrogen from glutamine. Cells were expanded as described elsewhere 36, with population doubling level (PDL) routinely tracked using in-house software (PDL calculator, EcoCyto). Cells with PDL between 43 and 45 were collated and seeded at a density of 4×10 4/cm 2 in 75 cm 2 flasks containing 240 µl/cm 2 flux media (Zenbio, cat DMEMf12-NGG002), supplemented as above with the addition of 17.5 mM [1,2- 13C] D-Glucose (sigma 453188) and 2 mM [U- 13C,U- 15N] L-Glutamine (sigma, 607983). Cell culture was continued for 48 hours to allow isotopic labelling, after which cells were washed with ice-cold saline solution (0.9%) and collected by scraping into 2 ml pre-chilled methanol (-20°C), 2 ml water (4°C) and 2 ml chloroform (-20°C). The solution was vigorously mixed for 10 minutes, after which lysates were centrifuged at 15,000 g for 15 min at 4°C. 1 ml of the sample was aspirated for NMR analysis. Samples were dried using a Savant (SPD1010) speedvac concentrator and then resuspended in 60 µL of 100 mM sodium phosphate buffer, containing 0.5 mM DSS, 2 mM Imidazole in D 2O, pH 7.0. The samples were vortexed and subsequently sonicated for 10 min and then centrifuged at 15000 g for 30 seconds to collate the fluid. Finally 35 µl of the samples were transferred to 1.7 mm NMR tubes.
2D- 1H, 13C-HSQC and 2D- 1H, 15N-HSQC NMR spectra were acquired using a Bruker Avance III 600 MHz NMR spectrometer equipped with a 1.7 mm z-PFG TCI Cryoprobe. The HSQC spectra were acquired using 2 transients per increment with echo/anti-echo gradient coherence selection with an additional pre-saturation to suppress the water resonance during the 1.5 s interscan relaxation delay. The 1H dimension of the 1H, 13C-HSQC spectra was acquired with a spectral width of 12 ppm using 512 complex data points. The 13C dimension was acquired with a spectral width of 160 ppm using 25% (2048) of 8192 data points using a non-uniform sampling scheme. The 1H dimension of the 1H, 15N-HSQC spectra was acquired with a spectral width of 12 ppm using 1024 complex data points. The 15N dimension was acquired with a spectral width of 40 ppm using 256 data. All non-uniformly sampled spectra were reconstructed via the compressed sensing algorithm within MDDNMR (version 2.5) 29 and processed using NMRpipe (version 9.2) 30. All spectra were processed without baseline correction to avoid complications in the multiplet analysis procedure especially with regards to the negative peaks caused by the echo/anti-echo coherence selection with gradients.
All NMR spectra in this article were processed within the MetaboLab software package (version 2018. 07182055; http://metabolab.uk) 22.
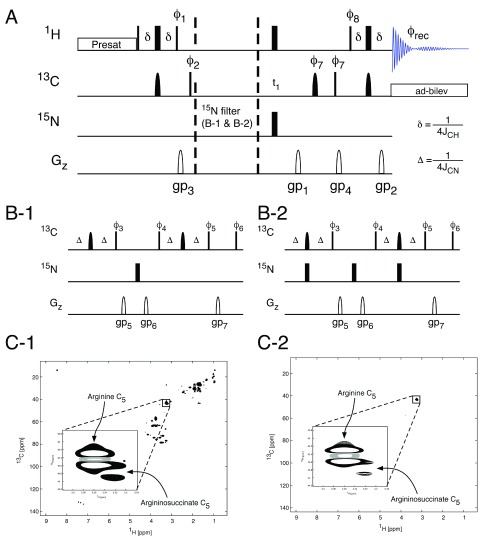
NMR methodology. Both aforementioned methods can be used to detect 15N labelling in metabolites, which alongside 13C isotope incorporation can provide additional much-needed information on the overlapping activity of multiple metabolic pathways. 2D spectroscopic filters are an extension of the 1D concept and as such can be used to simplify increasingly complex 2D spectra by selectively observing a subset of metabolites in which nuclei of interest have been incorporated. For example, the analysis of the 13C nuclei that are adjacent to 15N nuclei using 2D spectra permits a simplified unequivocal description of the nature in which two metabolic pathways converge.
Similar to the 1D method, the acquisition of two spectra is required in order to enable a quantitative analysis of the amount of 15N labelling in the presence of 13C labelling within the metabolite. If spectral simplification is the goal, a single spectrum is sufficient 16. The pulse sequence ( Figure 6) is a gradient selected 1H, 13C-HSQC spectrum with the spectral filter added once the 1H magnetisation has been transferred to the 13C nucleus. The spectrum collected with the 14N spectral filter (Panel C-2, Figure 6) contains only two visible NMR signals, corresponding to arginine and arginosuccinate, clearly showing how the filter can simplify complex spectra for easier analysis.
Figure 6. Filtered HSQC spectroscopy.
The application of a 15N filtering block in the 1H, 13C-HSQC pulse sequence ( A) allows the observation of 1H, 13C groups directly coupled to 15N nuclei. In the sequence in panel B-1 no filtering will be observed and the resulting spectrum ( C-1) will contain all 1H, 13C groups adjacent to either 14N or 15N nuclei. The use of a 15N filter ( B-2) will result in only those 1H, 13C groups adjacent to a 15N nuclei being observed in the resulting 1H, 13C HSQC spectrum ( C2). 1H, 13C J-coupling is suppressed during acquisition using adiabatic bilevel decoupling (ad-bilev) 21. The pulse phases are: ϕ 1 = y; ϕ 2 = x, -x; ϕ 3 = x for the no filter sequence (panel B-1) and y for the 15N filtered sequence (panel B-2); ϕ 4 = y, –y for the no filter sequence and y, y for the 15N filtered sequence; ϕ 5 = y, -y for the no filter sequence x, -x for the 15N filtered sequence; ϕ 6 = x, x, -x, -x; ϕ 7 = x, x, x, x, -x, -x, -x, -x; ϕ 8 = x, x, x, x, y, y, y, y; ϕ rec = x, -x, -x, x, y, -y, -y, y.
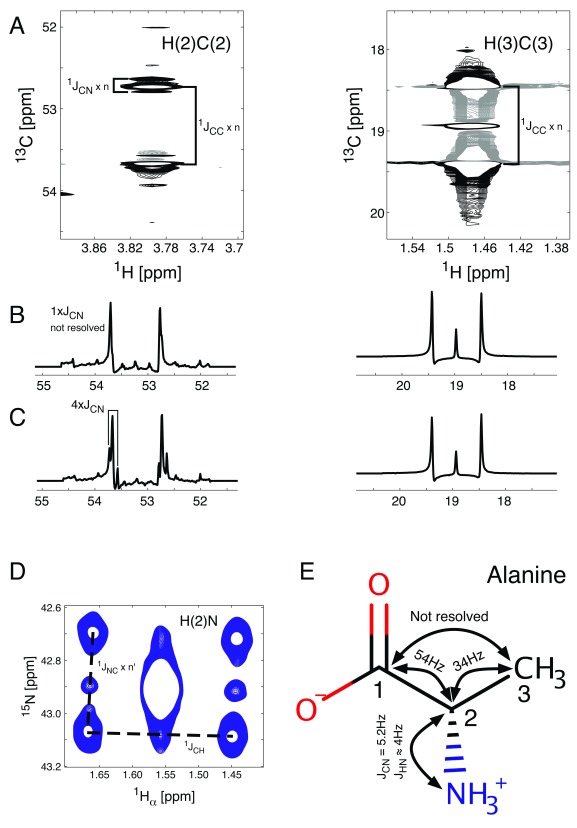
While 2D spectral filters serve a purpose, their quantitative usage is limited by the variability of the 1J CN constant. J-coupling splitting enhancement on the other hand can be easily extended to include 13C- 15N J- coupling splitting enhancement. Indeed, the addition of a single 15N π–pulse simultaneous with the central 13C π–pulse ( Figure 3) is sufficient to enhance the apparent 13C- 15N J-coupling splitting in 1H, 13C-HSQC spectra, an example of which is given in Figure 7. The 2D signals for the J CN splitting scaled spectrum are shown in panel A. Panels B and C show traces of the 13C multiplets for carbon atoms 2 and 3 of alanine. While the J CC splittings are enhanced by a factor of 4 in both spectra, the apparent J CN splittings are unchanged in the spectrum in panel B, whereas they are enhanced by a factor of 4 in the spectra in panel C. Because the 2J CN coupling between C(3) and N is negligible, both traces for C(3) overlap perfectly. C(2) on the other hand experiences a 1J CN coupling, which is too small to be resolved when the J CN splitting is not enhanced and is only detectable in panel C.
Figure 7. Splitting due to 15N and 13C incorporation.
Regions of the 1H, 13C-HSQC spectrum containing signals from alanine (panels A & B). The 13C traces of the alanine signals are shown with either no J CN-coupling splitting enhancement ( B) or four-fold J CN-coupling splitting enhancement ( C). The signals are split by either 1J CC or 1J CN coupling contributions. The long range 1H, 15N-HSQC spectrum ( D) is composed of unlabelled alanine (central peak) and 13C/ 15N labelled alanine (6 outer signals, 4 split by 2J HC and 1J NC couplings and 2 only split by 1J HC (values for the coupling constants are shown in panel E)).
J CN coupling, as any J-coupling, works in two directions, therefore a similar approach can be followed from the opposite direction. While amine groups of small molecules are notoriously difficult to observe due to chemical exchange of amine protons with solvent molecules, a long-range HSQC spectrum can be acquired. In such a spectrum proton magnetisation is transferred from H α (the proton bound to C(2)) via the 2J HN coupling. The splitting due to the J NC coupling then can be enhanced to show the appearance of 13C labelling in molecules which contain 15N next to those labelled carbon nuclei. As in this case, where the 13C nucleus is also bound to the proton determining the chemical shift on the horizontal axis, that same proton signal will be split by the 1J CH coupling constant. The result in this case is therefore a signal split into 7 2D components ( Figure 7, panel D), demonstrating that alanine was either recycled from unlabelled alanine which was incorporated into proteins, synthesised de-novo from [U- 13C] glucose and 15N labelled glutamate which originated from [U- 13C, U- 15N] glutamine that was added to the growth medium in addition to the [U- 13C] glucose or just synthesised de novo from [U- 13C] glucose with an unlabelled amino group transferred to form alanine. In conjunction with MS data, this complementarity between the 2D- 1H, 13C- and the 2D- 1H, 15N-HSQC spectra enables a model-free metabolism analysis using multiple nutrients as tracer sources in a single sample.
Discussion
Changes in metabolism are increasingly being recognised as central to the pathogenesis of a number of different diseases. Although metabolomic studies have helped determine aspects of disease phenotype, tracing the changing use of specific metabolic pathways using stable isotope-enriched nutrients provides higher resolution information on altered metabolic pathway activity that may lead to the identification of specific novel therapeutic targets. Over the last few years, development of magnet and probe technology, including innovative ultra-sensitive microprobes, has enabled the study of systems that were not previously amenable to NMR spectroscopy. Parallel advancement in the methods used to acquire and analyse data from samples will increase the amount of information we can gain from such samples.
In this paper, we describe how spectral filters and J-splitting enhancement can be used in tracer-based metabolism studies. These techniques overcome some of the major hurdles in the use of NMR spectroscopy. A challenge in the analysis of NMR HSQC spectroscopy data has been the need for an additional “unlabelled” sample in order to determine absolute per carbon 13C incorporation percentages. However, samples cannot be assumed to be biologically identical, thus making analyses problematic due to the inability to determine accurate 13C isotope incorporation values. Systems that demonstrate greater inter-sample variation, such as in vivo tracer studies, are even more prone to these analytical issues. The use of spectral filters negates the requirement for two samples and instead a single sample can be used to determine absolute percentage 13C incorporation and thus allow the scaling of multiplets.
The 2D HSQC spectrum is a powerful tool in the study of metabolism as it takes advantage of the increased sensitivity of the 1H nucleus over 13C and using the splitting due to J-coupling in the 13C dimension allows the indirect visualisation of the 13C incorporation into quaternary carbons. However, long acquisitions times, even when using the latest NUS techniques, limits the number of samples that can be acquired. Reducing the experimental time makes the use of HSQC spectra a much more attractive method in the study of tracer-based metabolism. The use of echo/anti-echo for quadrature detection ensures efficient elimination of unwanted artefacts, whilst using only two scans per increment in the indirect dimension. The changes observed in line shape due to the quadrature detection are predictable and can be easily incorporated into line fitting analysis. As described elsewhere 19, the simulation procedure assumes weak coupling between the different carbon nuclei. The simulation is implemented as a spin echo before the acquisition of a 13C-FID to allow the evolution of 13C, 13C spin coupling prior to the first increment.
The ability to scale the visualised splittings due to J-coupling allows HSQC spectra to be acquired in time equivalent to that of a 1D 1H spectrum, but the HSQC spectrum contains significantly more information. The reduced time required to acquire HSQC spectra means that it is feasible to apply 2D methods to in vivo tracer-based metabolism studies, as well as allowing the use of greater sensitivity of higher field spectrometers while avoiding longer experiment times ( Table 1). Expansion of the splitting due to J-coupling can also bring out smaller long-range couplings that were not apparent in a normal HSQC spectrum. Thus, the scaling of splittings can either be used to decrease acquisition times by allowing data collection at lower resolution or to bring out smaller couplings not previously visible. These small couplings include the 1J CN couplings that are found in many metabolites after the addition of metabolites labelled 15N in conjugation with 13C. This increases the information available and allows more in-depth analysis of complex metabolic pathways. In the example shown ( Figure 6), the cells used for this experiment were deficient in the expression of fumarate hydratase 16 and therefore contained high fumarate levels. One hypothesis was that argininosuccinate is synthesised from fumarate and arginine to minimise intracellular fumarate. In order to ascertain the signal assignment, we used [U- 13C, U- 15N] arginine and were able to show that 15N labelled argininosuccinate was being produced in the cells containing the knock out, but not in wild-type cells 16. This shows the utility of using multiple labelled nutrients to answer fundamental questions in metabolism.
In summary, the spectroscopic tools presented here open up new avenues for tracer-based metabolism studies. Scaling of signal splittings due to J-coupling leads to faster data collection of samples supplemented with nutrients enriched in stable isotopes, such as 13C and 15N. This enables profiling of metabolic pathways and can also be used to enhance sensitivity beyond current technical developments whilst maintaining reasonable data acquisition times. Ultimately, the use of 1D spectral filters as well as the fast acquisition of HSQC spectra leads to the possibility of tracing metabolism in real-time. In addition, simultaneous tracing with multiple nutrients will lead to unprecedented insight into the interplay of converging and intersecting metabolic pathways, both in vitro and in vivo 37.
Data availability
All experimental data for this article is available at: http://doi.org/10.17605/OSF.IO/EQHN3 38.
Experimental NMR datasets for HS-TrAM: 12C filtered (subdirectory 2) 1H spectrum, both of which are 13C decoupled during acquisition. Subdirectories 3 and 4 contain an unfiltered (3) and a 12C filtered (4) spectrum, both without 13C decoupling during acquisition. Subdirectories 5 and 6 contain POCE spectra, either with 12C- and 13C-bound 1H with same phase (subdirectory 5), or with opposite phase (subdirectory 6), both without 13C decoupling during acquisition. Subdirectory 7 contains a 30 degree excitation 1D 13C spectrum, acquired using a TXO Cryoprobe. The file jEnhanced_13C_HSQC.zip contains the 1H, 13C-HSQC spectra with different J-coupling splitting enhancements.
The file 13C_HSQC_14N_filter_and_15N_HSQC.zip contains an unfiltered (subdirectory 1) and a 14N filtered (subdirectory 2) 1H, 13C-HSQC spectrum. The file jEnhanced_13C_15N_HSQC.zip contains the following spectra: 4 x 13C, 13C splitting enhancement 1H, 13C-HSQC in subdirectory 1, 4 x 13C, 13C splitting enhancement and 4 x 13C, 15N splitting enhancement 1H, 13C-HSQC in subdirectory 2 and 4 x 13C, 15N splitting enhancement 1H, 15N-HSQC in subdirectory 3.
License: CC0 1.0 Universal
Acknowledgements
We thank HWB-NMR at the University of Birmingham for providing open access to their Wellcome Trust-funded spectrometers. Organ Recovery Systems donated perfusion equipment.
Funding Statement
This work was supported by the Wellcome Trust [099185] and through an Institutional Strategic Support Award given to the University of Birmingham; the National Institute for Health Research [13-0053]; Help Harry Help Others [HelpCU09]; UHB Charitable Funds [17-3-846] and the metabolic tracer analysis core (MTAC) at the University of Birmingham.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 4 approved]
References
- 1. Frezza C, Zheng L, Folger O, et al. : Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477(7363):225–228. 10.1038/nature10363 [DOI] [PubMed] [Google Scholar]
- 2. Walther JL, Metallo CM, Zhang J, et al. : Optimization of 13C isotopic tracers for metabolic flux analysis in mammalian cells. Metab Eng. 2012;14(2):162–171. 10.1016/j.ymben.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hiller K, Metallo CM: Profiling metabolic networks to study cancer metabolism. Curr Opin Biotechnol. 2013;24(1):60–68. 10.1016/j.copbio.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 4. Gravel SP, Andrzejewski S, Avizonis D, et al. : Stable isotope tracer analysis in isolated mitochondria from mammalian systems. Metabolites. 2014;4(2):166–183. 10.3390/metabo4020166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang X, Chen YJ, Cho K, et al. : X 13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal Chem. 2014;86(3):1632–1639. 10.1021/ac403384n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chouchani ET, Pell VR, Gaude E, et al. : Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. 10.1038/nature13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buescher JM, Antoniewicz MR, Boros LG, et al. : A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. 10.1016/j.copbio.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mackay GM, Zheng L, van den Broek NJ, et al. : Analysis of Cell Metabolism Using LC-MS and Isotope Tracers. Methods Enzymol.Elsevier,2015;561:171–196. 10.1016/bs.mie.2015.05.016 [DOI] [PubMed] [Google Scholar]
- 9. Jiang L, Shestov AA, Swain P, et al. : Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532(7598):255–258. 10.1038/nature17393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quek LE, Liu M, Joshi S, et al. : Fast exchange fluxes around the pyruvate node: a leaky cell model to explain the gain and loss of unlabelled and labelled metabolites in a tracer experiment. Cancer Metab. 2016;4:13. 10.1186/s40170-016-0153-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H, Badur MG, Divakaruni AS, et al. : Distinct Metabolic States Can Support Self-Renewal and Lipogenesis in Human Pluripotent Stem Cells under Different Culture Conditions. Cell Rep. 2016;16(6):1536–1547. 10.1016/j.celrep.2016.06.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sauer U: Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol. 2006;2(1):62. 10.1038/msb4100109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lane AN, Fan TW, Higashi RM: Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Methods Cell Biol. 2008;84:541–588. 10.1016/S0091-679X(07)84018-0 [DOI] [PubMed] [Google Scholar]
- 14. Lane AN, Fan TW, Bousamra M, 2nd, et al. : Stable isotope-resolved metabolomics (SIRM) in cancer research with clinical application to nonsmall cell lung cancer. OMICS. 2011;15(3):173–182. 10.1089/omi.2010.0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang Y, Lane AN, Ricketts CJ, et al. : Metabolic reprogramming for producing energy and reducing power in fumarate hydratase null cells from hereditary leiomyomatosis renal cell carcinoma. PLoS One. 2013;8(8):e72179. 10.1371/journal.pone.0072179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adam J, Yang M, Bauerschmidt C, et al. : A role for cytosolic fumarate hydratase in urea cycle metabolism and renal neoplasia. Cell Rep. 2013;3(5):1440–1448. 10.1016/j.celrep.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lussey-Lepoutre C, Hollinshead KE, Ludwig C, et al. : Loss of succinate dehydrogenase activity results in dependency on pyruvate carboxylation for cellular anabolism. Nat Commun. 2015;6:8784. 10.1038/ncomms9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nath J, Smith T, Hollis A, et al. : 13C glucose labelling studies using 2D NMR are a useful tool for determining ex vivo whole organ metabolism during hypothermic machine perfusion of kidneys. Transpl Res. 2016;5:7. 10.1186/s13737-016-0037-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chong M, Jayaraman A, Marin S, et al. : Combined Analysis of NMR and MS Spectra (CANMS). Angew Chem Int Ed Engl. 2017;56(15):4140–4144. 10.1002/anie.201611634 [DOI] [PubMed] [Google Scholar]
- 20. Banke NH, Lewandowski ED: Impaired cytosolic NADH shuttling and elevated UCP3 contribute to inefficient citric acid cycle flux support of postischemic cardiac work in diabetic hearts. J Mol Cell Cardiol. 2015;79:13–20. 10.1016/j.yjmcc.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kupce Ē, Freeman R, Wider G, et al. : Suppression of Cycling Sidebands Using Bi-level Adiabatic Decoupling. J Magn Reson A. 1996;122:81–84. Reference Source [Google Scholar]
- 22. Ludwig C, Günther UL: MetaboLab--advanced NMR data processing and analysis for metabolomics. BMC Bioinformatics. 2011;12:366. 10.1186/1471-2105-12-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howe PW, Ament Z, Knowles K, et al. : Combined use of filtered and edited 1H NMR spectroscopy to detect 13C-enriched compounds in complex mixtures. NMR Biomed. 2012;25(11):1217–1223. 10.1002/nbm.2791 [DOI] [PubMed] [Google Scholar]
- 24. Garbow JR, Weitekamp DP, Pines A: Bilinear rotation decoupling of homonuclear scalar interactions. Chem Phys Lett. 1982;93(5):504–509. 10.1016/0009-2614(82)83229-6 [DOI] [Google Scholar]
- 25. Wimperis S, Freeman R: An excitation sequence which discriminates between direct and long-range CH coupling. J Magn Reson 1969. 1984;58(2):348–353. 10.1016/0022-2364(84)90227-0 [DOI] [Google Scholar]
- 26. Uhrin D, Liptaj T, Kover KE: Modified BIRD Pulses and Design of Heteronuclear Pulse Sequences. J Magn Reson A. 1993;101(1):41–46. 10.1006/jmra.1993.1005 [DOI] [Google Scholar]
- 27. Briand J, Sørensen OW: Simultaneous and independent rotations with arbitrary flip angles and phases for I, ISalpha, and ISbeta spin systems. J Magn Reson. 1998;135(1):44–49. 10.1006/jmre.1998.1556 [DOI] [PubMed] [Google Scholar]
- 28. Henry PG, Marjanska M, Walls JD, et al. : Proton-observed carbon-edited NMR spectroscopy in strongly coupled second-order spin systems. Magn Reson Med. 2006;55(2):250–257. 10.1002/mrm.20764 [DOI] [PubMed] [Google Scholar]
- 29. Kazimierczuk K, Orekhov VY: Accelerated NMR spectroscopy by using compressed sensing. Angew Chem Int Ed. 2011;50(24):5556–5559. 10.1002/anie.201100370 [DOI] [PubMed] [Google Scholar]
- 30. Delaglio F, Grzesiek S, Vuister GW, et al. : NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–93. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- 31. Willker W, Flögel U, Leibfritz D, et al. : Ultra-high-resolved HSQC spectra of multiple- 13C-labeled biofluids. J Magn Reson. 1997;125(1):216–219. 10.1006/jmre.1996.1101 [DOI] [PubMed] [Google Scholar]
- 32. Furihata K, Tashiro M: BASHD-J-resolved-HMBC, an efficient method for measuring proton-proton and heteronuclear long-range coupling constants. Magn Reson Chem. 2014;52(1–2):27–31. 10.1002/mrc.4032 [DOI] [PubMed] [Google Scholar]
- 33. Kazimierczuk K, Zawadzka A, Koźmiński W: Optimization of random time domain sampling in multidimensional NMR. J Magn Reson. 2008;192(1):123–30. 10.1016/j.jmr.2008.02.003 [DOI] [PubMed] [Google Scholar]
- 34. Macura S: Accelerated multidimensional NMR data acquisition by varying the pulse sequence repetition time. J Am Chem Soc. 2009;131(28):9606–9607. 10.1021/ja9011063 [DOI] [PubMed] [Google Scholar]
- 35. Schulze-Sünninghausen D, Becker J, Koos MRM, et al. : Improvements, extensions, and practical aspects of rapid ASAP-HSQC and ALSOFAST-HSQC pulse sequences for studying small molecules at natural abundance. J Magn Reson. 2017;281:151–161. 10.1016/j.jmr.2017.05.012 [DOI] [PubMed] [Google Scholar]
- 36. Aschauer L, Gruber LN, Pfaller W, et al. : Delineation of the key aspects in the regulation of epithelial monolayer formation. Mol Cell Biol. 2013;33(13):2535–2550. 10.1128/MCB.01435-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nilsson R, Jain M: Simultaneous tracing of carbon and nitrogen isotopes in human cells. Mol Biosyst. 2016;12(6):1929–1937. 10.1039/c6mb00009f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ludwig C: HS-TrAM.2017. http://www.doi.org/10.17605/OSF.IO/EQHN3 [Google Scholar]