Abstract
Rationale: Quantification of type 2 inflammation provided a molecular basis for heterogeneity in asthma. Non–type 2 pathways that contribute to asthma pathogenesis are not well understood.
Objectives: To identify dysregulated pathways beyond type 2 inflammation.
Methods: We applied RNA sequencing to airway epithelial brushings obtained from subjects with stable mild asthma not on corticosteroids (n = 19) and healthy control subjects (n = 16). Sequencing reads were mapped to human and viral genomes. In the same cohort, and in a separate group with severe asthma (n = 301), we profiled blood gene expression with microarrays.
Measurements and Main Results: In airway brushings from mild asthma on inhaled corticosteroids, RNA sequencing yielded 1,379 differentially expressed genes (false discovery rate < 0.01). Pathway analysis revealed increased expression of type 2 markers, IFN-stimulated genes (ISGs), and endoplasmic reticulum (ER) stress-related genes. Airway epithelial ISG expression was not associated with type 2 inflammation in asthma or with viral transcripts but was associated with reduced lung function by FEV1 (ρ = −0.72; P = 0.0004). ER stress was confirmed by an increase in XBP1 (X-box binding protein 1) splicing in mild asthma and was associated with both type 2 inflammation and ISG expression. ISGs were also the most activated genes in blood cells in asthma and were correlated with airway ISG expression (ρ = 0.55; P = 0.030). High blood ISG expression in severe asthma was similarly unrelated to type 2 inflammation.
Conclusions: ISG activation is prominent in asthma, independent of viral transcripts, orthogonal to type 2 inflammation, and associated with distinct clinical features. ER stress is associated with both type 2 inflammation and ISG expression.
Keywords: asthma, type 2 inflammation, IFNs, endoplasmic reticulum stress, blood gene expression
At a Glance Commentary
Scientific Knowledge on the Subject
Although a subgroup of patients with asthma benefits from blockade of the type 2 cytokines IL-4, IL-5, and IL-13, there is an unmet need to identify non–type 2 mechanisms in asthma. Dysregulation in non–type 2 pathways has been demonstrated; however, the relationship of these pathways to type 2 inflammation and lung function impairment, and the impact of their presence on treatment response, have not been reported.
What This Study Adds to the Field
In this study, IFN-stimulated gene (ISG) expression was increased in mild asthma compared with health as assessed by RNA sequencing of airway epithelial brushings. ISG expression was orthogonal to type 2 inflammation, did not require the presence of virus, and was associated with reduced FEV1 and enhanced endoplasmic reticulum stress. ISG expression was also increased in whole blood, where it was similarly orthogonal to type 2 inflammation in severe asthma. Thus, ISG expression may be a viable target for future therapy. Both type 2 inflammation and ISG expression are independently associated with endoplasmic reticulum stress, which therefore represents an additional potential therapeutic target.
Allergic, type 2 cytokine-driven inflammation is associated with airway obstruction, hyperresponsiveness, remodeling, and response to inhaled corticosteroids (ICS) in asthma (1, 2). Therapeutic targeting of type 2 inflammation has been successful in some but not all patients (3). The subset of patients who respond poorly to therapies targeting type 2 inflammation may have non–type 2 inflammatory mechanisms driving their disease. Furthermore, these non–type 2 pathways likely coexist with type 2 inflammation in some individuals and contribute to severe asthma (3).
The airway epithelium is central to the initiation and maintenance of allergic immune responses, and contributes to asthma pathogenesis, including mucus hypersecretion and airflow obstruction (4). Microarray gene expression profiling of human airway epithelial cells was crucial in establishing the type 2 endotype (1). New technologies incorporating massively parallel RNA sequencing (RNA-seq) of airway samples overcome the sensitivity, specificity, and signal saturation limitations of microarrays, and can offer new insight into airway disease mechanisms (5, 6). In this study, we hypothesized that airway epithelial RNA-seq in asthma would identify non–type 2 pathways of inflammation and epithelial cell dysfunction that are associated with a more severe asthma phenotype. Some of the results of these studies have been previously reported in the form of abstracts (7, 8).
Methods
Additional details are reported in the Methods section of the online supplement.
Subjects and Procedures
Mild asthma study
We recruited 22 healthy control subjects and 32 subjects with mild asthma not on ICS via community-based advertising (NCT00595153). Baseline characterization included medical history and examination, spirometry, measurement of provocative concentration causing a 20% fall in FEV1 (PC20) and fractional exhaled nitric oxide, complete blood count and differential, serum total IgE, and collection of PAXgene blood RNA tubes (Qiagen). A total of 1–2 weeks following the baseline visit 20 healthy subjects and 25 subjects with mild asthma underwent spirometry, bronchoscopy, and blood collection; reasons for dropout of subjects are described in the Methods section of the online supplement. Subjects with asthma were thereafter treated with inhaled budesonide, 180 μg twice daily, for 8 weeks and returned for spirometry 4 weeks after ICS was started, and for repeat bronchoscopy and spirometry 8 weeks after ICS was started.
Severe asthma study
To validate findings in a severe asthma population, we used PAXgene whole-blood RNA samples from a previously reported prospective, 48-week double-blind randomized placebo-controlled trial of omalizumab (anti-IgE) (NCT00314574) (9). Subjects had poorly controlled asthma despite high-dose ICS and long-acting β-agonists. A total of 427 subjects received omalizumab and 423 placebo, with PAXgene tubes collected in a subgroup (Figure 1).
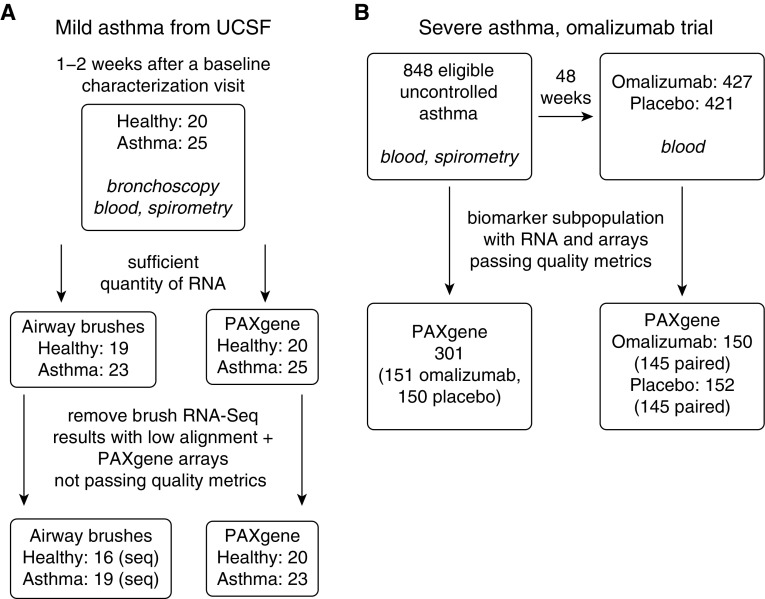
Figure 1.
Cohorts and samples contributing to the results in this publication. (A) Sample numbers from the mild asthma cohort from University of California, San Francisco (UCSF) before treatment with inhaled corticosteroids. Dropout from low RNA quantity was caused by inadequate RNA remaining for sequencing given that microarray and qPCR studies had used RNA from this study population previously. After sequencing, four subjects with mild asthma and three healthy control samples were removed from further analysis because of high mitochondrial RNA and ribosomal RNA abundance leading to insufficient depth of sequencing of protein coding reads. (B) Sample numbers from the randomized placebo-controlled trial of omalizumab in severe asthma. PAXgene = whole blood RNA collection tube; RNA-seq = RNA sequencing.
Gene Expression Measurements
For RNA-seq of airway epithelial brushings, library preparation and multiplexing was done with the Illumina TruSeq Stranded Total RNA with Ribo-zero Human/Mouse/Rat Kit (Illumina Inc.) per manufacturer’s protocol. We performed 100 base pair paired-end sequencing on multiplexed samples via an Illumina HiSeq 2500 at the University of California, San Francisco, Genomics Core. PAXgene RNA samples were run on Affymetrix HGU133A Plus 2.0 arrays as previously described (10).
Differential Expression
RNA-Seq
We performed read count normalization and differential expression analyses using the DESeq2 package (11), and corrected for multiple comparison using the false discovery rate (FDR) (12). Genes with an FDR less than 0.01 were called significant. We used regularized-logarithm transformed counts from DESeq2's rlog function for unsupervised clustering, heatmaps, and multigene metric generation.
Microarray
We performed moderated t tests with the limma package (13, 14) in R. Genes with an FDR less than 0.01 were called significant except in the case of whole-blood samples, which had weaker differential expression and for which an unadjusted P less than 0.05 was used before pathway analysis.
Comparison of Multigene Metrics with Clinical Variables
We performed unadjusted analyses using Spearman rank correlation and Welch’s t test, as appropriate. In adjusted analyses, we developed linear models and sought interactions between multigene metrics in association with lung function. We computed annualized exacerbation rates using zero-intercept generalized linear models, and used a model with an intercept to compute exacerbation rate reduction and perform all statistical tests of significance for exacerbations.
Metatranscriptome Preprocessing and Differential Expression
We used a recently described custom bioinformatics pipeline to detect viral transcripts (15). Briefly, sequencing reads underwent quality and complexity filtering (16) followed by subtractive alignment to the human genome (ENSEMBL GRCh38) using STAR (17). Remaining reads underwent additional subtractive alignment against phage and nonhuman tropic viruses using Bowtie2 (18). Identical reads were compressed (19) and remaining sequences were aligned to the National Center for Biotechnology Information nucleotide database using GSNAPL to identify viral transcripts (20).
Statistics and Plots
All analyses were done in the R statistical environment. Correlations were reported either as Spearman ρ or Pearson r as indicated. Heatmaps were created using the gplots package. All other plots were created with the ggplot2 package (21). P less than 0.05 was taken as statistically significant without correction for multiple testing unless otherwise indicated. Hierarchical clustering was performed with Euclidean distance and complete linkage.
Study Approval
All study sites obtained institutional review board approval and written informed consent from study participants before enrollment.
Results
Differential Airway Epithelial Gene Expression by RNA-Seq
RNA-seq data from airway epithelial brushings passed quality control in 16 healthy control subjects and 19 subjects with mild asthma not on ICS (Figure 1). Subject demographics are described in Table 1. A total of 1,379 transcripts were differentially expressed between asthma and health (FDR < 0.01) (see Table E1 in the online supplement). The most highly differentially expressed genes included those known to be induced by type 2 cytokine exposure, such as POSTN (periostin), SERPINB2 (serpin family B member 2), and CLCA1 (chloride channel accessory 1) (10).
Table 1.
Baseline Subject Characteristics
| Healthy Control Subjects | Subjects with Asthma | P Value | |
|---|---|---|---|
| Sample size of RNA-seq data | 16 | 19 | |
| Age, yr | 34 ± 10 | 34 ± 13 | 0.94 |
| BMI, kg/m2 | 26 ± 4 | 29 ± 6 | 0.14 |
| Female sex, % | 38 | 58 | 0.39 |
| Ethnicity* | |||
| White | 9 | 9 | |
| Black or African American | 1 | 1 | |
| Hispanic | 3 | 4 | |
| Asian/Pacific Islander | 2 | 5 | |
| Native American | 1 | 0 | |
| FEV1, % predicted | 98 ± 13 | 84 ± 14 | 0.0042 |
| Δ FEV1 with albuterol, % of baseline | 5.2 ± 0.4.5 | 13.8 ± 10.2 | 0.0027 |
| FEV1/FVC | 0.78 ± 0.061 | 0.74 ± 0.077 | 0.10 |
| Methacholine PC20, mg/ml | >10 (n = 15) | 0.57 (0.34–1.55) | <0.0001 |
| IgE, IU/ml | 17 (11–29) (n = 15) | 247 (129–361) | 0.00040 |
| Blood eosinophils, ×109/L | 0.11 (0.06–0.19) | 0.36 (0.13–0.50) | 0.0011 |
| FeNO, ppb | 16 (12–23) | 66 (20–85) (n = 14) | 0.010 |
| Epithelial brushings | |||
| Nonsquamous epithelial, % | 96 (95–98) | 98 (97–99) | 0.051 |
| Neutrophil | 0.2 (0–0.4) | 0 (0–0.2) | 0.034 |
| Lymphocyte | 0.10 (0–0.45) | 0 (0–0) | 0.055 |
| Eosinophil | 0 (0–0) | 0 (0–0.10) | 0.29 |
| Macrophage | 3 (1.3–4.7) | 1.4 (0.9–2.7) | 0.066 |
Definition of abbreviations: BMI = body mass index; FeNO = fractional exhaled nitric oxide; PC20 = provocative concentration causing a 20% fall in FEV1; RNA-seq = RNA sequencing.
Values are reported as mean ± SD or median (25–75th percentile). All data are from the baseline visit except spirometry, which are reported from visit 2 (immediately before initiation of RNA sequencing). P values are based on t test (where means reported), Wilcoxon rank sum (where medians reported), or chi-square test for proportions.
In cases of multiple ethnicities, the lowest-numbered ethnicity from this list is reported: 1) Hispanic, 2) black or African American, 3) Pacific Islander, 4) Native American, 5) Asian, and 6) white.
We used ingenuity pathway analysis (IPA) to predict upstream regulators of overrepresented genes (Table 2). As expected, regulators of type 2 inflammation including IL-5 and IL-13 scored well. In addition, type I (IFN-α, β) and III (IFN-λ) IFN and their downstream mediators (IRF [IFN regulatory factor]-1, IRF7, IFNAR2 [IFN α and β receptor subunit 2], STAT1 [signal transducer and activator of transcription 1]) were among the pathways most significantly associated with asthma (Table 2). Unlike the IFN-stimulated genes (ISGs), IFNs themselves and most other upstream regulators involved in IFN signaling as identified by IPA were not differentially expressed in brushings from subjects with asthma. For example, STAT1, an upstream regulator previously reported to be dysregulated in asthma (22), and secretory leukocyte protease inhibitor, previously reported to be repressed by and inversely related to IFN-γ in severe asthma (23), were not significantly altered in our asthma group (see Table E2).
Table 2.
IPA Upstream Regulator Analysis of Epithelial Brushing RNA-Seq–based Gene Expression
| Upstream Regulator | Activation z-Score | P Value of Overlap |
|---|---|---|
| IL5* | 4.93 | 2.71 × 10−4 |
| EIF4E† | 4.80 | 4.56 × 10−6 |
| Interferon-α group‡ | 4.79 | 1.23 × 10−3 |
| CD38 | 4.64 | 1.28 × 10−4 |
| IFNA2‡ | 4.41 | 9.97 × 10−3 |
| IL13* | 4.31 | 1.40 × 10−2 |
| XBP1† | 4.22 | 1.26 × 10−7 |
| MYC | 4.16 | 7.30 × 10−3 |
| STAT1‡ | 4.15 | 3.12 × 10−2 |
| IRF7‡ | 4.14 | 7.77 × 10−3 |
| IFNB1‡ | 3.79 | 4.17 × 10−2 |
| AGT | 3.74 | 2.44 × 10−2 |
| IFNL1‡ | 3.45 | 1.24 × 10−3 |
| TP53 | 3.39 | 4.61 × 10−3 |
| IRF1‡ | 3.37 | 6.91 × 10−4 |
| PRL | 2.89 | 1.21 × 10−4 |
| VEGFA | 2.83 | 1.10 × 10−2 |
| EIF2AK2† | 2.71 | 4.97 × 10−3 |
| HRAS | 2.71 | 1.03 × 10−2 |
| PAF1 | 2.65 | 1.95 × 10−2 |
| IFNA1/IFNA13‡ | 2.60 | 1.27 × 10−3 |
| Ifn group‡ | 2.49 | 4.59 × 10−2 |
| IFNAR2‡ | 2.45 | 1.37 × 10−3 |
| MYCN | 2.28 | 5.21 × 10−4 |
| THBS4 | 2.20 | 1.93 × 10−2 |
| APP | 2.02 | 4.28 × 10−3 |
Definition of abbreviations: IPA = ingenuity pathway analysis; RNA-seq = RNA sequencing.
Regulators are filtered by 1) genes, proteins, or RNA; 2) activated predicted state; 3) activation z-score ≥ 2; and 4) P < 0.05. The list is sorted by activation z-score. P value of overlap is the probability that the observed overlap between the list of genes differentially expressed between asthma and health, and the set of genes assigned by IPA as downstream of the listed upstream regulator, would occur by chance (Fisher’s exact test). The strength of activity of the regulator in disease, as determined by the net observed direction of changes in the overlapping downstream genes, is represented by an activation score (a more positive/negative value indicates that a larger proportion of genes changed in a direction consistent with activation/inhibition of the regulator in asthma).
Genes demonstrating presence of airway type 2 inflammation.
Genes/groups demonstrating presence of epithelial endoplasmic reticulum stress.
Genes/groups demonstrating presence of airway IFNs leading to epithelial IFN-stimulated gene expression.
Comparison of RNA-Seq and Microarrays
The presence of strong signals for non–type 2 inflammation in airway brushings analyzed by RNA-seq is a new finding in comparison with our previous microarray performed with similar subjects (1, 10). Whereas our RNA-seq study found 1,379 differentially expressed genes at an FDR less than 0.01, the earlier microarray study reported only 22 genes as differentially expressed at P less than 0.05 using a Bonferroni correction, and this increased to only 36 genes on reanalysis with a FDR less than 0.01 cutoff (10). However, although the prior microarray study had fewer statistically significant genes, the overall pattern of expression differences between groups was quite similar between the two studies as assessed using the Camera competitive gene set test (24), with significant overlap of both upregulated genes (n = 915; P = 3.46 × 10−28) and downregulated genes (n = 284; P = 4.12 × 10−15) in subjects with asthma compared with healthy control subjects (see Figure E1A for enrichment plots). This current paper presents RNA-seq generated from samples collected in one stand-alone clinical study, whereas our prior report was derived from samples from four different banked clinical studies that used comparable sample collection methods. This design may have avoided several clinical and technical sources of variation and improved our statistical power to find differences.
To directly assess how the use of RNA-seq versus microarrays affected the ability to detect gene expression changes and the ISG signature, we compared our RNA-seq data with microarray data generated using the same RNA samples. These microarray data represent a subset of the data previously reported in our comparison of subjects with asthma and chronic obstructive pulmonary disease (25). At an FDR less than 0.01, a total of 1,734 genes were differentially expressed by microarray between subjects with asthma and healthy control subjects. Comparison of RNA-seq and microarray data from the same samples using the Camera competitive gene set test showed significant overlap of both upregulated genes (P = 1.11 × 10−59) and downregulated genes (P = 3.50 × 10−42) (see Figure E1B). Across ISGs, microarray and sequencing showed concordant differences between asthma and health, but the magnitude of the fold differences were greater in the sequencing results (see Figure E2A). The finding that some ISGs were significantly increased in asthma by RNA-seq but not by microarrays could not be explained consistently by low microarray signal intensity (see Figure E2B).
Development of a Metric of Airway ISG Expression
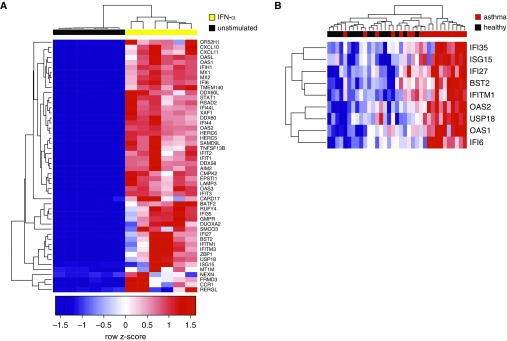
We first focused on ISGs. To generate a gene expression signature specific to airway epithelial cells, we cultured human bronchial epithelial cells at an air–liquid interface and stimulated them with IFN-α (10 ng/ml for 24 h). We chose IFN-α given its significance in the IPA list and the largely overlapping gene expression responses to type I and type III IFNs (26). RNA-seq of IFN-α-stimulated epithelial cells yielded 1,031 IFN-α-induced genes (FDR < 0.01; top 50 in Figure 2A). We restricted this top 50 list to those differentially expressed in asthmatic airway brushings (FDR < 0.01), yielding nine genes: IFI6 (IFNα-inducible protein 6), OAS1 (2′-5′-oligoadenylate synthetase 1), USP18 (ubiquitin specific peptidase 18), OSA2, IFITM1 (IFN-induced transmembrane protein 1), BST2 (bone marrow stromal cell antigen 2), IFI27 (IFNα-inducible protein 27), and IFI35 (IFN-induced protein 35). This degree of overlap is significant because the probability of finding nine or more genes overlapping with the set of 1,379 genes differentially expressed in human airway brushings, when drawing 50 genes randomly without replacement from 19,938 protein-coding genes in the epithelial culture sequencing data, is P equal to 0.0067 by a Fisher’s exact test.
Figure 2.
Development of an airway epithelial brushing IFN-stimulated gene expression metric in mild asthma. (A) Hierarchical clustering of primary human epithelial cells cultured at an air–liquid interface (columns) using the top 50 (by fold-difference) genes (rows) induced by IFN-α (yellow, n = 6) relative to unstimulated (black, n = 6). All gene expression measurements are from RNA sequencing, with blue indicating low relative expression, and red indicating high relative expression. (B) Visualization of intersection of genes upregulated in asthma relative to health (false discovery rate < 0.01) and IFN-α–stimulated genes from A by supervised hierarchical clustering. n = 19 asthma (red); n = 16 healthy (black).
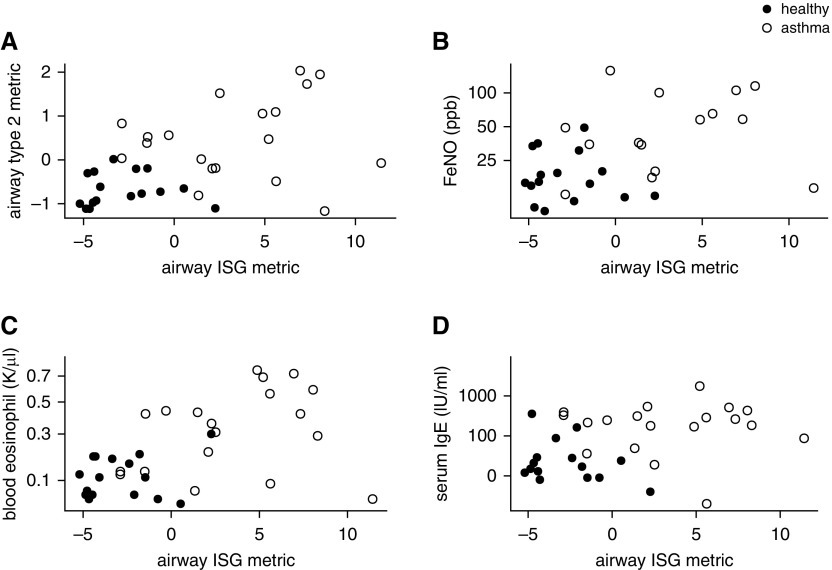
Grouping subjects by the expression of these nine IFN-induced genes shows that 12 of 19 subjects with asthma have coordinately elevated expression for almost all of these genes (Figure 2B). The centered, regularized read counts for these nine genes were added to form an airway ISG metric. We found no association between the epithelial ISG metric and our previously published type 2 inflammatory metric (ρ = 0.12; P = 0.62) (2), nor other markers of allergic inflammation including blood eosinophils, serum IgE, and fractional exhaled nitric oxide (Figure 3).
Figure 3.
Airway IFN-stimulated gene expression is not associated with airway type 2 inflammation. The airway IFN-stimulated gene metric was calculated from the expression of the nine signature genes in airway epithelial brushings acquired at bronchoscopy. Covariates were measured at baseline. Scatterplots of covariates versus the airway IFN-stimulated gene metric are shown for the following variables. (A) Airway epithelial type 2 gene expression metric (n = 16 healthy [solid circles] and n = 19 mild asthma [open circles]; ρ = 0.12; P = 0.62 within asthma). (B) Exhaled nitric oxide on a log10 axis (n = 16 healthy [solid circles] and n = 15 mild asthma [open circles]; ρ = 0.30; P = 0.28 within asthma). (C) Blood eosinophils on a square root axis (n = 16 healthy [solid circles] and n = 19 mild asthma [open circles]; ρ = 0.22; P = 0.36 within asthma). (D) Total serum IgE (n = 15 healthy [solid circles] and n = 19 mild asthma [open circles]; ρ = −0.023; P = 0.93 within asthma). Spearman rank correlation used in all cases. FeNO = fractional exhaled nitric oxide; ISG = IFN-stimulated gene.
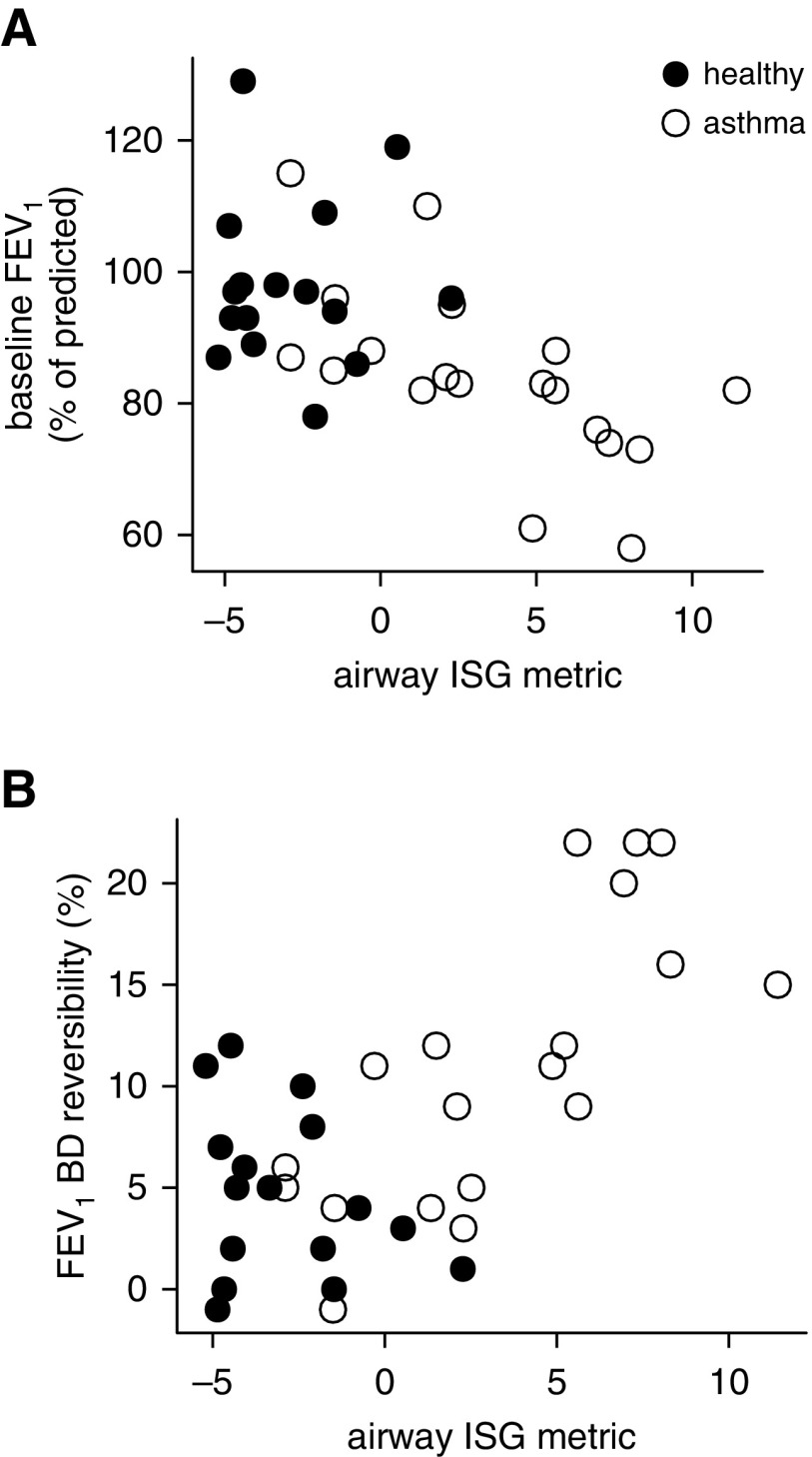
Airway ISG Expression Is Associated with Reduced Lung Function
Higher airway ISG expression was associated with reduced FEV1 (ρ = −0.72; P = 0.0004) and increased bronchodilator response among subjects with asthma (ρ = 0.75; P = 0.0002) (Figures 4A and 4B). To validate this on a genome-wide scale, we performed IPA of the pathways associated with lower FEV1 in asthma using all the RNA-seq data and found that IFN-related pathways represented 5 of the top 10 pathways (see Table E3). We found no relationship between the ISG metric and age, sex, body mass index, or PC20. Weaker relationships were seen between type 2 inflammation and lung function (ρ = −0.40, P = 0.050 for FEV1; ρ = 0.44, P = 0.032 for bronchodilator response). The association between ISG expression and FEV1 remained significant when adjusted for the type 2 metric (P = 0.0056), and there was no significant interaction between the type 2 and ISG metrics (P = 0.75 for interaction term).
Figure 4.
Relationship of airway IFN-stimulated gene expression to lung function in mild asthma. (A) Baseline FEV1 as a percent of the predicted value, measured immediately before and on the day of bronchoscopy before bronchodilator (BD) (ρ = −0.72; P = 0.0004 within asthma). (B) Change in FEV1 as a percentage of predicted after administration of BD (ρ = 0.75; P = 0.0002 within asthma). Spearman rank correlation used and n = 16 healthy (solid circles) and n = 19 asthma (open circles) in all cases. ISG = IFN-stimulated gene.
Effect of ICS on Airway ISG Expression and Lung Function
Airway epithelial brush ISG activity was significantly attenuated by 8 weeks of ICS (see Figure E3A; P = 0.007 before vs. after ICS). After ICS treatment, airway ISG activity was not associated with FEV1 or bronchodilator reversibility (see Figures E3B and E3C). The relationship between baseline ISG expression and the change in FEV1 was significant after 2 but not after 4 and 8 weeks of ICS treatment (see Table E4). Thus, ISG expression at baseline had a mixed effect on ICS response, unlike the more consistent relationship seen for type 2 inflammation (2) (see Table E4). Similarly, compared with type 2 inflammation, ISG expression had lower area under the curve by receiver operating characteristic analysis in predicting those with a positive ICS response (see Figure E3D).
Airway ISG Expression Is Not Associated with a Distinct Viral Signature
Given the well-established relationship between IFNs and viral infection, we screened the airway epithelial RNA-seq for viral transcripts using our metagenomics pipeline. Potential viral pathogens were identified in low abundance in a small subset of subjects. A virus had to be present at more than 15 reads to be considered present. We identified 35 human rhinovirus A reads in one subject with asthma with high ISG levels (see Figure E4 for the alignment of viral reads from this subject against the human rhinovirus A genome). Human herpes virus was detected in four healthy control subjects and one asthma subject with low ISG levels. To confirm the overall negative findings, we assessed the presence of three common respiratory viruses (respiratory syncytial virus, human rhinovirus, and parainfluenza virus type 3) by validated qPCR assays. All three viruses were undetectable in the human airway brush RNA samples, but were detected in positive control samples. The discordance between qPCR and RNA-Seq for human rhinovirus in the one subject previously mentioned suggests that the limit of detection for the qPCR assay is near 35 RNA-Seq reads in this dataset.
Whole-Blood Gene Expression Reflects Airway ISG Expression
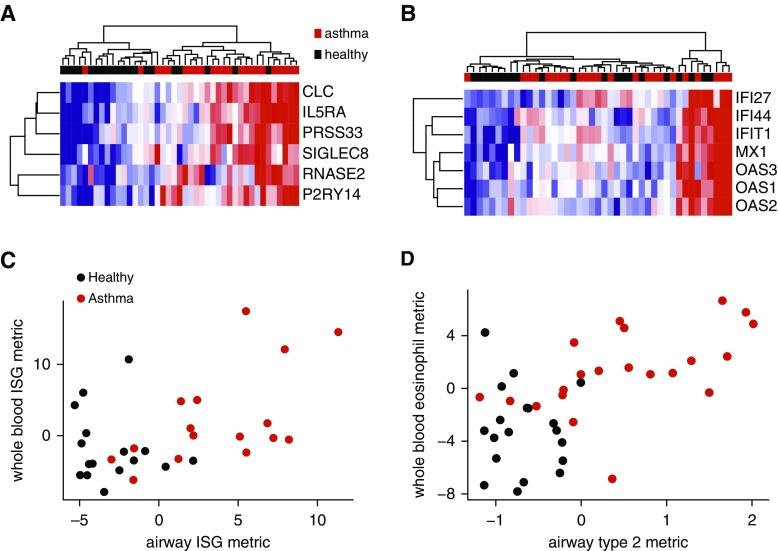
Whole-blood PAXgene profiling by microarrays in 23 subjects with asthma not on ICS and 20 healthy control subjects identified 1,372 differentially expressed transcripts (P < 0.05, see Table E5). Eosinophil-associated genes (SIGLEC8 [sialic acid binding Ig-like lectin 8], CLC [Charcot-Leyden crystal galectin], IL5RA [IL5 receptor subunit α], PRSS33 [protease, serine 33], P2RY14 [purinergic receptor P2Y14], RNASE2 [ribonuclease A family member 2]) topped this list (Figure 5A) (27). The high degree of correlation between these genes and with the percentage of blood eosinophils, but not neutrophils (see Figure E5 and Table E6), suggests that the differential expression is caused by the differences in eosinophil numbers between subjects with asthma and healthy control subjects; other cell types were not significantly different between groups (see Figure E6).
Figure 5.
Whole-blood gene expression reflects airway IFN-stimulated gene expression in mild asthma. (A) Unsupervised hierarchical clustering of whole-blood gene expression by six eosinophil-associated genes differentially expressed in asthma (P < 0.05). Samples (columns) were collected on the day of and immediately preceding bronchoscopy. Gene expression measurements are from microarrays, with blue indicating low relative expression, and red indicating high relative expression. n = 23 asthma (red) and n = 20 healthy (black). (B) Unsupervised hierarchical clustering of whole-blood gene expression by seven IFN-stimulated genes previously identified in whole blood. (C) Whole-blood IFN-stimulated gene metric versus airway (epithelial brushing) IFN-stimulated gene metric (n = 16 asthma; ρ = 0.55; P = 0.030). (D) Whole-blood eosinophil-associated gene metric versus blood eosinophils (n = 21 asthma; ρ = 0.68; P = 0.00094). Whole-blood gene expression metrics were made in the same manner as they were for the airway samples and related across tissues. Spearman rank correlation used in all cases. ISG = IFN-stimulated gene.
We used IPA on the differentially expressed gene list to identify non-eosinophil-associated pathways and found that IFN-associated genes were the top-scoring upstream regulators in asthma (e.g., IRF7 [IFN regulatory factor 7], STAT1, IFNB1 [IFNβ 1], IFNA2 [IFNα 2]) (Table 3). We developed a blood ISG metric by combining the expression of seven intercorrelated genes known to be responsive to IFN signaling in peripheral blood mononuclear cells: IFI27, IFI44, MX1 (MX dynamin like GTPase 1), IFIT1 (IFN-induced protein with tetratricopeptide repeats 1), OAS1, OAS2, and OAS3 (Figure 5B) (28). We found significant correlation between airway and blood ISG metrics across subjects with asthma (ρ = 0.55; P = 0.03) (Figure 5C). A similarly robust relationship was seen between a blood eosinophil gene expression metric and blood eosinophils (ρ = 0.68; P = 00094, within asthma) (Figure 5D). Unlike in the airway, blood ISG expression was not correlated with FEV1 in asthma (n = 23; ρ = −0.29; P = 0.19) or affected by ICS (see Figure E3). There was a modest negative correlation between blood ISG expression and basophils but no other cell types (see Table E5). The expression of a subset of differentially expressed genes was validated by qPCR, with good correlation between platforms and consistency of differential expression (see Figure E7 and Table E7).
Table 3.
IPA Upstream Regulator Analysis of Whole-Blood Array–based Gene Expression
| Upstream Regulator | Activation z-Score | P Value of Overlap |
|---|---|---|
| IRF7* | 4.78 | 5.68 × 10−6 |
| STAT1* | 3.72 | 1.32 × 10−3 |
| Cg | 3.68 | 3.01 × 10−2 |
| Interferon-α group* | 3.57 | 3.91 × 10−4 |
| PRL | 3.31 | 2.90 × 10−2 |
| IFNB1* | 3.29 | 7.80 × 10−3 |
| IFNA2* | 3.18 | 5.06 × 10−6 |
| IRF1* | 3.04 | 1.30 × 10−3 |
| IFN-β group* | 3.01 | 8.66 × 10−3 |
| TGM2 | 3.00 | 3.15 × 10−2 |
| Ifnar group* | 2.87 | 1.57 × 10−3 |
| Ifn group* | 2.78 | 9.94 × 10−3 |
| IFNG* | 2.71 | 2.26 × 10−4 |
| GATA2 | 2.58 | 1.14 × 10−2 |
| SMARCA4 | 2.58 | 1.28 × 10−3 |
| JUN | 2.45 | 4.57 × 10−2 |
| IL4† | 2.40 | 2.53 × 10−6 |
| IL5† | 2.36 | 6.29 × 10−6 |
| IL2 | 2.30 | 2.00 × 10−4 |
| STAT3* | 2.20 | 3.57 × 10−5 |
| RNASE2 | 2.17 | 4.03 × 10−2 |
| IL27 | 2.16 | 1.56 × 10−2 |
| IFNL1* | 2.04 | 1.28 × 10−4 |
| IFNAR2* | 2.00 | 2.70 × 10−2 |
Definition of abbreviation: IPA = ingenuity pathway analysis.
Regulators are filtered by 1) genes, proteins, or RNA; 2) activated predicted state; 3) activation z-score ≥ 2; and 4) P < 0.05. The list is sorted by activation z-score.
Genes/groups demonstrating presence of airway IFN leading to epithelial IFN-stimulated gene expression.
Genes demonstrating presence of airway type 2 inflammation.
ISG Expression Is Increased in Severe Asthma
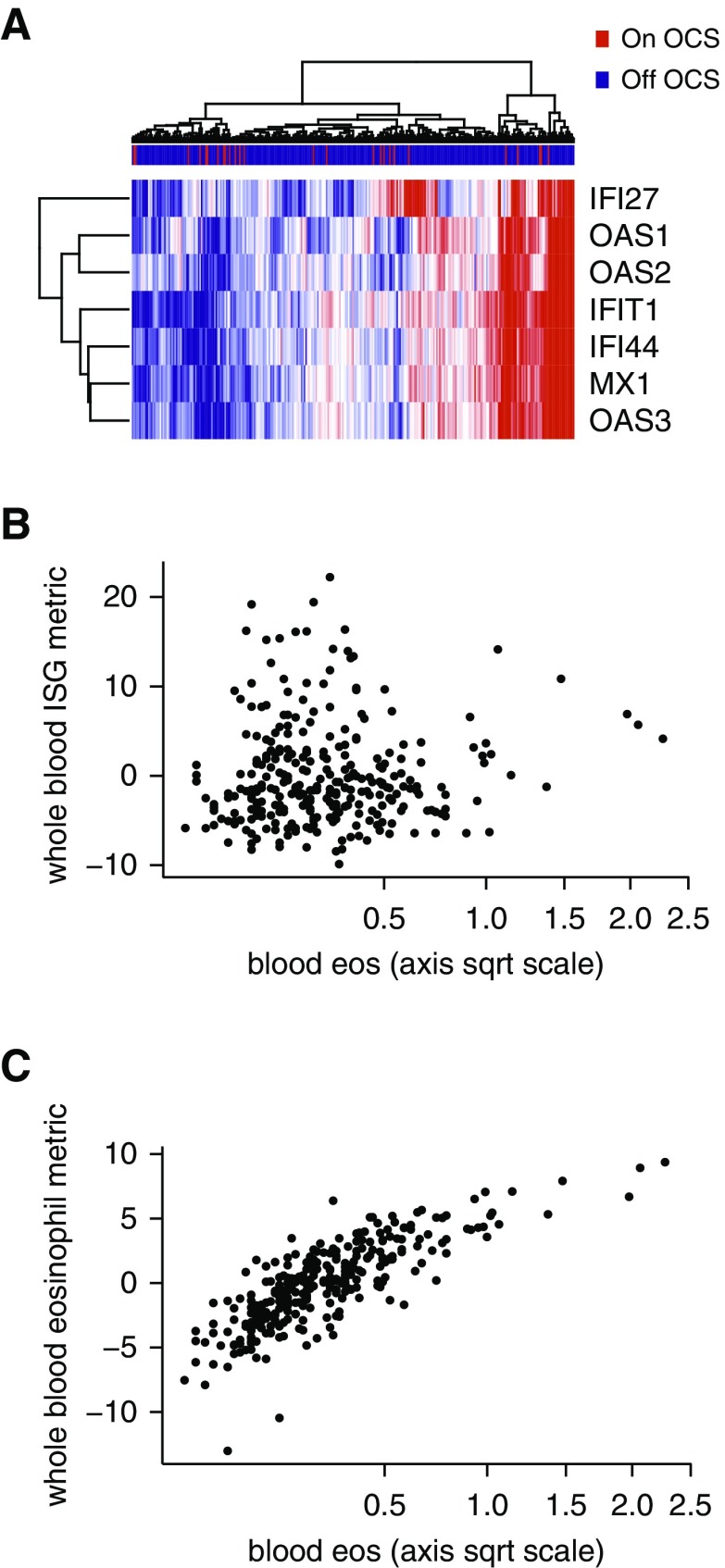
Demographic data for the subjects from the severe asthma study, in which they were randomized to placebo or omalizumab, are available in Table E8 (9). Unsupervised clustering of the blood ISG genes at baseline using microarrays showed a substantial fraction of subjects with elevated expression (Figure 6A and Table 4). There was no relationship between the blood ISG metric and type 2 inflammation as assessed by blood eosinophils (Figure 6B) (2); whereas, as expected, the blood eosinophil gene metric was significantly associated with blood eosinophils (Figure 6C). Consistent with the effects of ICS in mild asthma, blood ISG expression and FEV1 were not correlated in severe asthma (n = 301; ρ = 0.0030; P = 0.96). The lack of associations agrees with a recent description of “T1”-high severe asthma (29). Blood ISG expression in severe asthma was also unaffected by oral corticosteroid use (Figure 6A ; see Figure E3). High compared with low ISG expression was not associated with a different exacerbation rate in the placebo arm (Table 4). In this trial, treatment with omalizumab reduced the exacerbation rate over 48 weeks relative to placebo. Markers of type 2 inflammation identified individuals more likely to respond (30). In contrast, a high blood ISG metric was not associated with a reduction in exacerbations in response to omalizumab (exacerbation rate reduction = 1.27%; 95% confidence interval, −60.13 to 39.20; P = 0.96 for effect of treatment on number of exacerbations ) (Table 4). Although there was a trend for a response to omamlizumab for the group with a low blood ISG metric, the effect of ISG status on treatment response was not significant (P = 0.3 for interaction term) (Table 4).
Figure 6.
IFN-stimulated gene (ISG) expression is increased in severe asthma and unrelated to type 2 inflammation. (A) Unsupervised hierarchical clustering of whole-blood gene expression by seven ISGs. Severe asthma samples (columns) were collected at baseline before change in treatment. Gene expression measurements are from microarrays, with blue indicating low relative expression, and red indicating high relative expression, n = 301 severe asthma (color-coded red if on oral corticosteroids). (B) Whole-blood ISG expression is not related to blood eosinophil count, a measure of type 2 inflammation (n = 288 severe asthma; ρ = 0.057; P = 0.34). (C) Whole-blood eosinophil-associated gene expression is associated with blood eosinophil count (n = 288 severe asthma; ρ = 0.82; P < 2.2 × 10−16). Spearman rank correlation was used in both B and C. eos = eosinophils; OCS = oral corticosteroids; sqrt = square root.
Table 4.
Association of ISG Expression with Subject Characteristics and Exacerbation Rates in Response to Omalizumab in Severe Asthma
| ISG-High* | ISG-Low* | P Value | |
|---|---|---|---|
| Age, yr† | 45.5 ± 13.8 | 46.0 ± 13.8 | 0.72 |
| Female, %† | 77 | 57 | 0.0002 |
| BMI, kg/m2† | 33.2 ± 7.9 | 31.0 ± 6.9 | 0.01 |
| Number of exacerbations in year before enrollment† | 2 (0 to 11) | 2 (0 to 8) | 0.66 |
| N, placebo/omalizumab | 76/75 | 74/76 | — |
| Annualized exacerbation rate | — | ||
| Placebo (95% CI) | 1.0 (0.74 to 1.36) | 0.80 (0.56 to 1.11) | |
| Omalizumab (95% CI) | 1.0 (0.73 to 1.34) | 0.55 (0.36 to 0.81) | |
| P value for effect of treatment‡ | 0.96 | 0.11 | |
| Exacerbation rate reduction % (95% CI) | 1.3 (−60.3 to 39.2) | 31.1 (−7.9 to 56.0) | 0.3§ |
Definition of abbreviations: BMI = body mass index; CI = confidence interval; ISG = IFN-stimulated gene.
ISG-high and -low are determined as ISG expression metrics above or below the median value for all subjects at baseline.
Baseline traits are presented as mean ± SD or median (range), and statistical differences were assessed by a two-sided Welch two-sample t test for the former and a generalized linear model with quasi-Poisson error distribution for the latter, as described in the Methods. For female percentage, a chi-square test for equality of proportions was performed.
P value for treatment term with ISG-low and -high modeled separately as described in the Methods.
P value for interaction term in a model of exacerbation rate including treatment and ISG status as predictors.
Airway Epithelial Endoplasmic Reticulum Stress Is Common to ISG and Type 2 Inflammation in Asthma
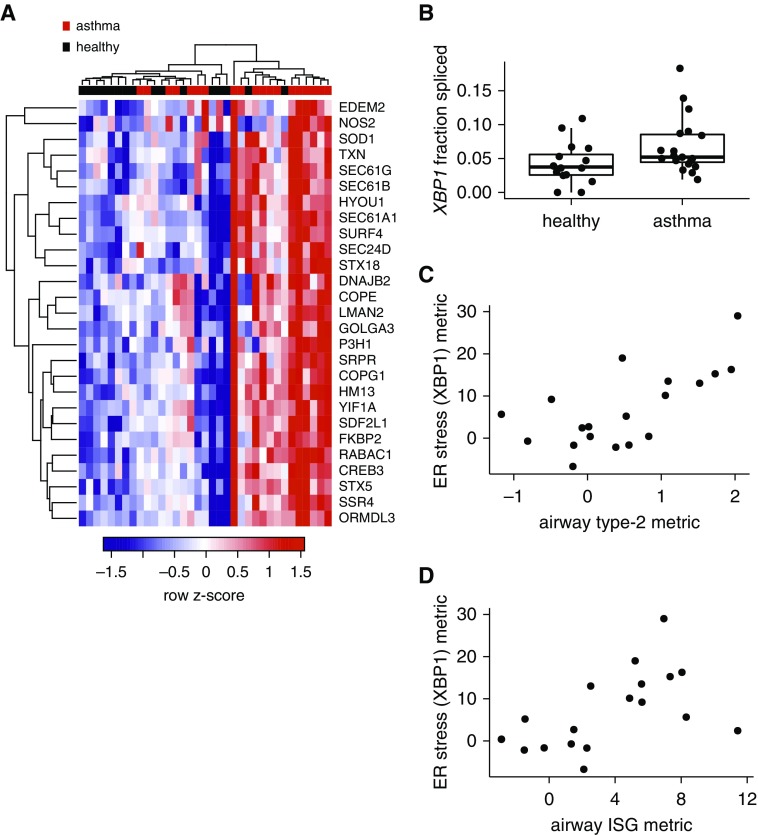
In response to endoplasmic reticulum (ER) stress, cells activate an unfolded-protein response (UPR) initiated by one of three signal transduction pathways: 1) IRE1α (inositol-requiring enzyme 1 α)–XBP1 (X-box binding protein 1), 2) PERK (protein kinase R [PKR]-like endoplasmic reticulum kinase), and 3) ATF6 (activation transcription factor 6). Our IPA analysis showed prominent activation of the IRE1α–XPB1 (IPA regulator XBP1) pathway in mild asthmatic airway brushings (Table 2 and Figure 7A). Activation of IRE1α leads to splicing of XBP1 mRNA to an activated form, which we confirmed in epithelial brushings through genome-wide splicing analysis of the airway brushing RNA-seq data (FDR = 0.0001) (Figure 7B). Notably, IPA upstream regulator analysis did not reveal significant activation of the XBP1 pathway in whole-blood microarray gene expression (Table 2). There was significant attenuation of ER stress after 8 weeks of ICS (see Figure E3).
Figure 7.
Epithelial endoplasmic reticulum (ER) stress markers are elevated in mild asthma and associated with both high type 2 inflammation and IFN-stimulated gene expression. (A) Hierarchical clustering of airway epithelial brushing gene expression samples (columns) using genes differentially expressed (false discovery rate < 0.01) in asthma versus health and downstream of XBP1 (X-box binding protein 1) in IPA. (B) XBP1 splicing assessed bioinformatically from RNA sequencing data through use of the rMATS package in R (false discovery rate = 0.0001 for genome-wide splicing analysis). (C) Relationship of an ER stress gene expression metric consisting of genes downstream of XBP1 per IPA, to airway type 2 inflammation (n = 19 mild asthma; ρ = 0.62; P = 0.0058). (D) ER stress gene expression metric versus the airway IFN-stimulated gene metric (n = 19 mild asthma; ρ = 0.62; P = 0.0052). Only asthma shown. IPA = ingenuity pathway analysis; ISG = IFN-stimulated gene.
We examined the relationship between XBP1 activation and airway ISG expression and type 2 inflammation. Genes downstream of XBP1 were combined into a single metric of ER stress that was associated with heightened type 2 inflammation and ISG expression (Figures 7C and 7D). Individuals with asthma and low levels of type 2 inflammation and ISG expression did not have activation of the XBP1 arm of the UPR. Type 2 inflammation and ISG expression remained significantly associated with ER stress when added in a linear model (P = 0.00076 and 0.0059, respectively), without a significant interaction (P = 0.85). Consistent with the association between XBP1 activation and both type 2 inflammation and ISG expression, stimulation of epithelial cell cultures by IL-13 or IFN-α at an air–liquid interface showed activation of all arms of the UPR (see Table E9).
Discussion
We found that a clinically distinct subset of asthma has increased ISG expression in the airway and in peripheral blood. In subjects with mild asthma not on ICS, this ISG-high subgroup had decreased lung function and increased bronchodilator reversibility. In both mild and severe asthma, ISG expression was independent of type 2 inflammation. These data extend prior reports that support the presence of airway IFNs in stable asthma (23, 29, 31–34) by showing that IFN-driven inflammation is orthogonal to type 2 inflammation and is not associated with the presence of viral infection. Furthermore, we found that epithelial ER stress is related to both ISG expression and type 2 inflammation, making it a potential common downstream therapeutic target.
We did not find an association between the presence of viral RNA as detected by RNA-seq or qPCR and the IFN response in this immunocompetent and clinically stable group. One recent report found evidence for IFN responses associated with occult viral transcripts in nasal brushings in some asymptomatic children with asthma (35). Our experiments used similar PCR methods and did not detect viral transcripts in the lower airway in this subgroup, with the exception of one subject. Our findings may differ because of differences in location (upper vs. lower airway) and age. In contrast to stable asthma, a large percentage of asthma exacerbations are induced by viral infection, particularly with rhinovirus, and subjects with asthma have increased lower respiratory tract symptoms after viral infection (36). Deficient type I and type III IFN production in response to viral infection has been observed in epithelial cells recovered from asthmatic airways (37, 38). These findings in an ex vivo infection model are nonetheless potentially compatible with our results of heightened responses during stable disease. In fact, baseline elevated IFN responses can lead to desensitization to type I IFN stimulation through the ISG USP18, which we found induced in airway brushings (39).
Although ISGs have been most studied in association with viral infection, host expression of ISGs can also occur in response to LPS-containing gram-negative bacteria and some intracellular bacteria and fungi (40, 41). Future studies will address the relationship between airway ISG expression and the microbiome, which is dysregulated in asthma (42–47).
In our study, type 2 inflammation and ISG expression were not correlated. Previous work found that T-helper cell type 1 and IFN responses have suppressive effects on type 2 responses (48), suggesting that the two should be inversely related. Our results do not exclude the possibility that ISG expression by epithelial cells represents a regulatory mechanism aimed at dampening excessive type 2 inflammation. If true, this could lead to dual responses in some individuals, whereas in others ISG expression may persist after type 2 responses recede.
Although type 2 and IFN-driven inflammation were unrelated in airway epithelial brushings, both were independently associated with markers of airway epithelial ER stress. Thus, IRE1-XBP1–regulated airway epithelial ER stress may be a final common pathway for both types of inflammation. ER stress is a pathway of established importance in many diseases and airway epithelial ER stress is emerging as potentially important in asthma. The inflammatory environment of the airway in asthma offers multiple ways in which ER stress, or the UPR, can be induced. In mice, house-dust mite allergen challenge is a potent activator of epithelial ER stress, and knockdown of two ER stress-associated proteins, ATF6 and ERp57, decreased airway inflammation, hyperresponsiveness, and fibrosis (49, 50). In another study, IRE1β-initiated ER stress was required for epithelial mucin production (51). These studies support the notion that type 2 inflammation, which is associated with increased epithelial production and secretion of mucus and innate cytokines, can lead to ER stress. How IFN-driven inflammation might induce ER stress is less well understood. In our data, there was evidence of heightened ER stress-related gene expression in airway brushings but not in blood samples in mild asthma, and therefore we were unable to assess this pathway in severe asthma. Further studies in severe asthma will be important because inhibition of ER stress may be a novel therapeutic strategy for asthma based on preclinical results from animal models (52).
The largely overlapping responses of type I (IFN-α/β), II (IFN-γ), and III (IFN-λ) IFNs preclude distinguishing these pathways using transcript profiling (26). The cellular source of airway IFNs in our study is also unknown. Although type I and type III IFNs are secreted by many cell types including epithelial and hematopoietic cells, type II IFN secretion is limited to specific immune cells including T-helper cell type 1, natural killer T, and natural killer cells. In our airway brushing RNA-seq data, IFNs were not differentially expressed, suggesting that epithelial cells or luminal hematopoietic cells are not a likely source. Uncollected airway cells, such as inflammatory cells in the lamina propria, may contribute to airway IFNs.
Recently, approximately 70% of humans with severe asthma were found to have levels of IFN-γ-producing BAL fluid CD4+ T cells higher than the upper limit seen in mild-moderate asthma (23). The relationships between these IFN-γ-producing T cells, type 2 inflammation, and lung function were not presented, which combined with the lack of healthy control subjects precludes direct comparison with our findings. Additional data from that study suggest that IFN-γ-induced downregulation of SLPI (secretory leukocyte protease inhibitor) in airway epithelial cells leads to increased airway hyperresponsiveness in severe asthma. We did not find differential expression of the SLPI gene in mild asthma. Furthermore, the production of type I and type III IFNs was not reported in that study, leaving uncertain which IFNs drive airway ISG expression in asthma.
Other potential explanations for the increased ISG expression that we observed include impaired resolution of IFN-driven inflammation, a higher gain in the IFN response pathways (22), or expression of endogenous retroelements. An abnormally high gain has the most support based on the previously reported finding of constitutive activation of the STAT1 transcription factor, a key mediator of intracellular IFN responses, in the airway epithelium from subjects with mild asthma (22). In the current study, we were limited to studying gene expression and therefore were unable to address the activation of STAT1. Although STAT1 is known to be downstream of IFN stimulation and was induced by IFN-α in our epithelial cultures, it was not differentially upregulated in our airway brushings from asthma relative to health. Possible explanations for this difference are that there are stimuli acting on the epithelium beyond IFNs in vivo (e.g., type 2 inflammation, microbiome), and that the level and duration of IFN stimulation differs between in vivo and culture conditions.
Our paired whole-blood gene expression analysis showed that blood cell ISG expression is elevated in stable asthma, not just with asthma exacerbations and suboptimal control (53, 54). The ISG signature in blood is not as prominent as it is in the airway, and therefore we used a significance threshold of an unadjusted P less than 0.05 rather than a more stringent FDR cutoff to identify a sufficient number of genes for optimal pathway analysis. Our data further show that a subgroup of severe subjects with asthma has elevated whole-blood ISG expression and that as in mild asthma, ISG expression and type 2 inflammation are orthogonal. Our data also suggest that elevated ISG expression in severe asthma may be associated with decreased exacerbation reduction benefit from anti-IgE therapy. This is an intriguing trend identified in an exploratory study and can be tested in adequately powered trials in the future. The finding raises the possibility that incorporation of blood gene expression biomarkers could complement type 2 inflammatory markers and support broader clinical detection of ISG expression in asthma.
Notably, the presence of subgroups with elevated ISG expression in both mild and severe asthma parallels what has been found for the established endotype of type 2 inflammation (1, 55). In addition, similar to characteristics of type 2 inflammation, we found that ICS treatment of subjects with mild asthma led to reduced ISG expression and loss of the relationship of ISG expression to lung function, and that ISG expression is unrelated to FEV1 in severe asthma (2, 55). Thus, the presence of elevated ISG expression in mild asthma, and the effects of ICS on ISG expression and its relationship to FEV1, does not diminish the potential importance of IFN-driven inflammation in severe asthma.
This study adds to an expanding literature on asthma phenotyping and demonstrates that differential ISG expression defines a clinically relevant subgroup of subjects with asthma. This work informs further studies in larger cohorts designed to delineate the diversity of ISG responses. Additional evaluation of ISG expression in severe asthma, including associations with responses to existing and emerging therapeutics, will be an important future direction. ER stress may be a common pathway for both type 2 and IFN-driven inflammation, and may offer a new target for the treatment of asthma.
Acknowledgments
Acknowledgment
The authors thank the University of California, San Francisco, Sandler Genomics Core and the Parnassus Genomics Core for providing high-quality RNA sequencing data. Walter E. Finkbeiner gave valuable support for the cell culture experiments and was funded by the Cystic Fibrosis Foundation (RDP-R613) and NIH DK072517.
Footnotes
Supported by grants from the NIH: K23HL116657 (N.R.B.), K23HL123778 (S.A.C.), K12 HL11999 (S.A.C. and D.J.E.), P01HL107202 (J.V.F.), AI077439 (D.J.E. and P.G.W.), and HL137013, AI077439, and HL107202 (P.G.W.); from the Sandler Asthma Basic Research Center; and from Genentech.
Author Contributions: N.R.B., S.A.C., and P.G.W. conceived the hypothesis, interpreted data, and drafted the manuscript. N.R.B. and S.A.C. designed and performed analysis of epithelial brushing and blood gene expression and clinical data. D.J.E. gave guidance on gene expression analyses and revised the manuscript critically for intellectual content. S.N. and O.D.S. provided computational support. C.P.N. coordinated the bronchoscopy study and participated in collection of samples and clinical data. D.F.C. performed analysis of blood microarray data and exacerbations in the severe asthma study. K.L.J. performed qPCR and assisted in analysis. S.G. assessed RNA quality and quantity and contributed to library generation. L.R.B., L.T.Z., J.L.P., and D.J.E. designed, performed, and analyzed data from the epithelial cultures. C.L. and J.L.D. provided S.A.C. with assistance in the analysis of nonhuman reads. J.R.A. designed the study in mild asthma. J.V.F. and P.G.W. designed the study in mild asthma and with N.R.B. participated in the collection of samples and clinical data. All authors read and approved the final manuscript.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201706-1070OC on October 24, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhakta NR, Solberg OD, Nguyen CP, Nguyen CN, Arron JR, Fahy JV, et al. A qPCR-based metric of Th2 airway inflammation in asthma. Clin Transl Allergy. 2013;3:1–9. doi: 10.1186/2045-7022-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray A, Raundhal M, Oriss TB, Ray P, Wenzel SE. Current concepts of severe asthma. J Clin Invest. 2016;126:2394–2403. doi: 10.1172/JCI84144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fahy JV. Type 2 inflammation in asthma: present in most, absent in many. Nat Rev Immunol. 2015;15:57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.’t Hoen PA, Friedländer MR, Almlöf J, Sammeth M, Pulyakhina I, Anvar SY, et al. GEUVADIS Consortium. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31:1015–1022. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 6.SEQC/MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014;32:903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christenson SA, Bhakta NR, Nerella S, Pollack JL, Barbeau R, Barczak AJ, et al. RNA-sequencing of airway epithelial brushings identifies interferon-driven gene expression changes in asthma [abstract] Am J Respir Crit Care Med. 2015;191:A3626. [Google Scholar]
- 8.Bonser LR, Christenson SA, Pollack JL, Barbeau R, Barczak AJ, Zlock L, et al. Investigating mRNA and miRNA alterations induced by asthma-associated cytokine stimulation in a human airway model using RNASeq [abstract] Am J Respir Crit Care Med. 2016;193:A4624. [Google Scholar]
- 9.Hanania NA, Alpan O, Hamilos DL, Condemi JJ, Reyes-Rivera I, Zhu J, et al. Omalizumab in severe allergic asthma inadequately controlled with standard therapy: a randomized trial. Ann Intern Med. 2011;154:573–582. doi: 10.7326/0003-4819-154-9-201105030-00002. [DOI] [PubMed] [Google Scholar]
- 10.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289. [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 15.Doan T, Wilson MR, Crawford ED, Chow ED, Khan LM, Knopp KA, et al. Illuminating uveitis: metagenomic deep sequencing identifies common and rare pathogens Genome Med201681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruby JG, Bellare P, Derisi JL. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3 (Bethesda) 2013;3:865–880. doi: 10.1534/g3.113.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2009.
- 22.Sampath D, Castro M, Look DC, Holtzman MJ. Constitutive activation of an epithelial signal transducer and activator of transcription (STAT) pathway in asthma. J Clin Invest. 1999;103:1353–1361. doi: 10.1172/JCI6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raundhal M, Morse C, Khare A, Oriss TB, Milosevic J, Trudeau J, et al. High IFN-γ and low SLPI mark severe asthma in mice and humans. J Clin Invest. 2015;125:3037–3050. doi: 10.1172/JCI80911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christenson SA, Steiling K, van den Berge M, Hijazi K, Hiemstra PS, Postma DS, et al. Asthma-COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;191:758–766. doi: 10.1164/rccm.201408-1458OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esnault S, Kelly EA, Schwantes EA, Liu LY, DeLain LP, Hauer JA, et al. Identification of genes expressed by human airway eosinophils after an in vivo allergen challenge. PLoS One. 2013;8:e67560. doi: 10.1371/journal.pone.0067560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride JM, Jiang J, Abbas AR, Morimoto A, Li J, Maciuca R, et al. Safety and pharmacodynamics of rontalizumab in patients with systemic lupus erythematosus: results of a phase I, placebo-controlled, double-blind, dose-escalation study. Arthritis Rheum. 2012;64:3666–3676. doi: 10.1002/art.34632. [DOI] [PubMed] [Google Scholar]
- 29.Modena BD, Bleecker ER, Busse WW, Erzurum SC, Gaston BM, Jarjour NN, et al. Gene expression correlated with severe asthma characteristics reveals heterogeneous mechanisms of severe disease. Am J Respir Crit Care Med. 2017;195:1449–1463. doi: 10.1164/rccm.201607-1407OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanania NA, Wenzel S, Rosén K, Hsieh HJ, Mosesova S, Choy DF, et al. Exploring the effects of omalizumab in allergic asthma: an analysis of biomarkers in the EXTRA study. Am J Respir Crit Care Med. 2013;187:804–811. doi: 10.1164/rccm.201208-1414OC. [DOI] [PubMed] [Google Scholar]
- 31.Cho SH, Stanciu LA, Holgate ST, Johnston SL. Increased interleukin-4, interleukin-5, and interferon-gamma in airway CD4+ and CD8+ T cells in atopic asthma. Am J Respir Crit Care Med. 2005;171:224–230. doi: 10.1164/rccm.200310-1416OC. [DOI] [PubMed] [Google Scholar]
- 32.Krug N, Madden J, Redington AE, Lackie P, Djukanovic R, Schauer U, et al. T-cell cytokine profile evaluated at the single cell level in BAL and blood in allergic asthma. Am J Respir Cell Mol Biol. 1996;14:319–326. doi: 10.1165/ajrcmb.14.4.8600935. [DOI] [PubMed] [Google Scholar]
- 33.Bullens DM, Decraene A, Dilissen E, Meyts I, De Boeck K, Dupont LJ, et al. Type III IFN-lambda mRNA expression in sputum of adult and school-aged asthmatics. Clin Exp Allergy. 2008;38:1459–1467. doi: 10.1111/j.1365-2222.2008.03045.x. [DOI] [PubMed] [Google Scholar]
- 34.da Silva J, Hilzendeger C, Moermans C, Schleich F, Henket M, Kebadze T, et al. Raised interferon beta, type 3 interferon and interferon stimulated genes: evidence of innate immune activation in neutrophilic asthma. Clin Exp Allergy. 2016;47:313–323. doi: 10.1111/cea.12809. [DOI] [PubMed] [Google Scholar]
- 35.Wesolowska-Andersen A, Everman JL, Davidson R, Rios C, Herrin R, Eng C, et al. Dual RNA-seq reveals viral infections in asthmatic children without respiratory illness which are associated with changes in the airway transcriptome Genome Biol2017181–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corne JM, Marshall C, Smith S, Schreiber J, Sanderson G, Holgate ST, et al. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359:831–834. doi: 10.1016/S0140-6736(02)07953-9. [DOI] [PubMed] [Google Scholar]
- 37.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201:937–947. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med. 2006;12:1023–1026. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- 39.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monroe KM, McWhirter SM, Vance RE. Induction of type I interferons by bacteria. Cell Microbiol. 2010;12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morton CO, Bouzani M, Loeffler J, Rogers TR. Direct interaction studies between Aspergillus fumigatus and human immune cells; what have we learned about pathogenicity and host immunity? Front Microbiol. 2012;3:413. doi: 10.3389/fmicb.2012.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131:346–352. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. National Heart, Lung, and Blood Institute's Asthma Clinical Research Network. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bundy KW, Gent JF, Beckett W, Bracken MB, Belanger K, Triche E, et al. Household airborne Penicillium associated with peak expiratory flow variability in asthmatic children. Ann Allergy Asthma Immunol. 2009;103:26–30. doi: 10.1016/S1081-1206(10)60139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharpe RA, Bearman N, Thornton CR, Husk K, Osborne NJ. Indoor fungal diversity and asthma: a meta-analysis and systematic review of risk factors. J Allergy Clin Immunol. 2015;135:110–122. doi: 10.1016/j.jaci.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Chou H, Wu KG, Yeh CC, Tai HY, Tam MF, Chen YS, et al. The transaldolase, a novel allergen of Fusarium proliferatum, demonstrates IgE cross-reactivity with its human analogue. PLoS One. 2014;9:e103488. doi: 10.1371/journal.pone.0103488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agbetile J, Fairs A, Desai D, Hargadon B, Bourne M, Mutalithas K, et al. Isolation of filamentous fungi from sputum in asthma is associated with reduced post-bronchodilator FEV1. Clin Exp Allergy. 2012;42:782–791. doi: 10.1111/j.1365-2222.2012.03987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzales-van Horn SR, Farrar JD. Interferon at the crossroads of allergy and viral infections. J Leukoc Biol. 2015;98:185–194. doi: 10.1189/jlb.3RU0315-099R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffman SM, Chapman DG, Lahue KG, Cahoon JM, Rattu GK, Daphtary N, et al. Protein disulfide isomerase-endoplasmic reticulum resident protein 57 regulates allergen-induced airways inflammation, fibrosis, and hyperresponsiveness. J Allergy Clin Immunol. 2016;137:822–832.e7. doi: 10.1016/j.jaci.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffman SM, Tully JE, Nolin JD, Lahue KG, Goldman DH, Daphtary N, et al. Endoplasmic reticulum stress mediates house dust mite-induced airway epithelial apoptosis and fibrosis. Respir Res. 2013;14:1–12. doi: 10.1186/1465-9921-14-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martino MB, Jones L, Brighton B, Ehre C, Abdulah L, Davis CW, et al. The ER stress transducer IRE1β is required for airway epithelial mucin production. Mucosal Immunol. 2013;6:639–654. doi: 10.1038/mi.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siddesha JM, Nakada EM, Mihavics BR, Hoffman SM, Rattu GK, Chamberlain N, et al. Effect of a chemical chaperone, tauroursodeoxycholic acid, on HDM-induced allergic airway disease. Am J Physiol Lung Cell Mol Physiol. 2016;310:L1243–L1259. doi: 10.1152/ajplung.00396.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bjornsdottir US, Holgate ST, Reddy PS, Hill AA, McKee CM, Csimma CI, et al. Pathways activated during human asthma exacerbation as revealed by gene expression patterns in blood. PLoS One. 2011;6:e21902. doi: 10.1371/journal.pone.0021902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF, Jr, Liu AH, et al. Asthma BioRepository for Integrative Genomic Exploration (Asthma BRIDGE) Consortium. Gene expression profiling in blood provides reproducible molecular insights into asthma control. Am J Respir Crit Care Med. 2017;195:179–188. doi: 10.1164/rccm.201601-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia G, Erickson RW, Choy DF, Mosesova S, Wu LC, Solberg OD, et al. Bronchoscopic Exploratory Research Study of Biomarkers in Corticosteroid-refractory Asthma (BOBCAT) Study Group. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J Allergy Clin Immunol. 2012;130:647–654.e10. doi: 10.1016/j.jaci.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]