Abstract
Background:
Extended-spectrum β-lactamase (ESBL)-producing is a significant resistant mechanism to β-lactams in Enterobacteriaceae, especially in Klebsiella pneumoniae. The main objectives of this study were to genetically characterize urinary clinical isolates of K. pneumoniae through the investigating of blaTEM, blaCTX-M and using molecular typing by Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) method. We also determined the frequency of antibiotic resistance of K. pneumoniae strains to characterize the β-lactamases included.
Materials and Methods:
A cross-sectional study was carried out to evaluate 98 strains of K. pneumoniae isolated from urine culture of outpatients referred to Al-Zahra Hospital, Isfahan, Iran. Antibiotic susceptibility testing was performed using Kirby–Bauer's method. Screening of ESBLs was carried out using double-disk screening test. PCR technique was performed to detect TEM and CTX-M genes. The total DNA of each strain was tested by ERIC-PCR.
Results:
In 98 K. pneumoniae studied clinical isolates, 25.5% were ESBL producing and 44.9% multidrug-resistant (MDR). From 25 ESBL isolates, 23 (92%) cases showed MDR phenotype. In ESBL producing isolates, 23 (92%) were blaCTX-M and 19 (76%) blaTEM positive. The antimicrobial drug susceptibilities of ESBL isolates indicated high resistant rates for cefotaxime and ceftazidime. All 25 ESBL producing isolates were resistant to cefotaxime. Complex patterns of fingerprints isolates showed that 36% of the isolates were belonged to the cluster no 5.
Conclusion:
This study revealed high antimicrobial resistance rates among ESBL isolates which can lead to various health difficulties. Epidemiological data collection from patients is recommended to develop the strategies to manage antibiotic resistance.
Keywords: Antimicrobial resistance, blaCTX-M, blaTEM, Klebsiella pneumoniae, multidrug-resistant
Introduction
Klebsiella pneumoniae is one of the most important human bacterial pathogens, result to the extensive range of community and hospital acquired infections that may lead to morbidity and mortality, particularly in immunocompromised patients.[1,2] Urinary tract infections (UTIs)-induced by multidrug-resistant (MDR) K. pneumoniae isolates are a serious public health problem. The potency of many antimicrobial agents have been limited, also these bacteria are hard and expensive to cure.[3]
During the past decade, drug companies improved many novel Gram-positive antimicrobial agents to fight MDR bacteria, but unfortunately, the growing problem of MDR in Gram-negative bacteria was not paralleled by the developing of novel antimicrobials.[4] Estimation of epidemiological data caused by MDR K. pneumoniae in various parts of the world indicated to increased prevalence with distinct variety in many regions. These divergences require geographical surveys due to their resistance patterns and risk factors.[2]
Recently, the report of the Infectious Diseases Society of America particularly referred extended-spectrum cephalosporin-resistant Escherichia coli and Klebsiella spp. as a category of MDR Gram-negative bacilli.[5]
β-lactams are the most usual drugs against bacterial infections and extended spectrum β-lactamase (ESBL) producing is a significant resistant mechanism to β-lactams in Enterobacteriaceae, especially in K. pneumoniae.[6,7]
ESBLs producing isolates mainly exists in hospitals and frequently in Intensive Care Units (ICUs), but recently have been developed in the community.[8]
ESBLs belong to the Ambler class A, display resistance to β-lactam antibiotics except cephamycins and carbapenems, and are inhibited by clavulanic acid.[9] A previous similar study in China was shown high rates of MDR, extensively drug-resistant and pandrug-resistant strains among K. pneumoniae isolates, also most of these antibiotic resistance genes were transferable.[1]
TEM and SHV types are the first ESBLs have developed by genetic mutation from native β-lactamases. New classes of ESBLs have emerged such as CTX-M, PER, VEB, TLA-1, GES/IBC, SFO-1, and BES-1. However, CTX-M enzymes have been widely detected worldwide. The prevalence of the different types of ESBLs varies by region and the clinical state and is changing over time.[6]
Recently, the CTX-M enzymes seem to be more prevalent throughout the world, whereas other ESBLs of the SHV and TEM subgroups appear to decrease. Several nosocomial outbreaks caused by CTX-M producing K. pneumoniaee because of misuse of broad-spectrum antibiotics, especially in ICU or immunodeficient patients.[7,8,9]
The CTX-M family was grouped based on sequence similarity into four distinct subtypes included CTX-M-1, CTX-M-2, CTX-M-8, and CTX-M-9.[10]
ESBLs derived from TEM-and SHV-type penicillinases, have been extending in Spain, the United Kingdom, Canada, China, and Korea, especially in the Enterobacteriaceae including K. pneumoniae and E. coli.[7,10] In contrast with TEM and SHV type ESBLs, most of the CTX-M producers show degrees of resistance to cefotaxime and ceftriaxone.[10]
The phenotype of β-lactams resistance may propose the existence of CTX-M enzymes; therefore, polymerase chain reaction (PCR) has been performed greatly to identify bla genes.[10]
Enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) produce specific strain patterns by amplification of repetitive DNA elements. ERIC-PCR has demonstrated the microbial variety and functional evolutionary of microorganisms.[11]
The main objectives of this study were to genetically characterize urinary clinical isolates of K. pneumoniae through the investigating of blaTEM, blaCTX-M (the two most common genes) and using molecular typing by ERIC-PCR. We also determined the frequency of antibiotic resistance of K. pneumoniae strains to characterize the β-lactamases included and to highlight the genetic variety of these isolates.
Materials and Methods
Study design and population
This retrospective cross-sectional study was carried out from February 2015 to December 2015 in Al-Zahra Hospital, affiliated to Isfahan University of Medical Sciences.
Subjects and methods
During the study, a total of 98 nonduplicate clinical isolates of K. pneumoniae were recovered from urine specimens of patients that were referred to different wards of Al-Zahra Hospital, Isfahan, Iran.
Subsequent taking informed permission from the patients, their demographic information, including age and gender were completed in question sheets. All the parts of the study were supervised and approved by the Ethical Committee of Isfahan University of Medical Sciences (code 394573).
Sample collection and microbial detection
Urine specimens were cultured on blood agar and MacConkey agar plates (Hi-Media, India) at 37°C for 24 h. The grown isolates were recognized by the morphology of colonies, Gram-staining, and biochemical tests properties. K. pneumoniae colonies on blood agar medium were large, cupola shape, and mucoid that move to merge. On MacConkey medium, K. pneumoniae colonies were large, mucoid dark pink, which specifies the lactose fermentation. The biochemical tests for K. pneumoniae included of negative indole, positive urease, positive VP, positive Simmons’ citrate agar, variable magnetic resonance, and no motile. Bacterial isolates were refrigerated on nutrient agar plates or frozen (−70°C) in microtubes containing 15% glycerol and tryptic soy broth medium.[2] Afterward, molecular confirmation was performed by PCR of the ureD gene. Analysis for the presence of the ureD gene demonstrated that all the isolates confirmed as K. pneumoniae were positive for the ureD gene.
Antibiotic susceptibility
The estimate of individuals for antibiotic susceptibilities was performed using disk diffusion method (Kirby–Bauer's) as stated by the Clinical and Laboratory Standards Institute (CLSI) guidelines.[12] The following antibiotics were tested: piperacillin/tazobactam (100/10 μg), cephalothin (30 μg), cefotaxime (30 μg), imipenem (10 μg), cefoxitin (30 μg), ceftazidime (30 μg), gentamicin (10 μg), tetracycline (30 μg), tigecycline (15 μg), ciprofloxacin (5 μg), colistin (50 μg), trimethoprim-sulfamethoxazole (TS) (1.25/23.75 μg), and nitrofurantoin (300 μg). The antibiotic disks were obtained from MAST, UK. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as negative and positive control strains, respectively.[12]
Phenotypic extended spectrum β-lactamase recognition
K. pneumoniae isolates were screened for possible ESBL production using ceftazidime and cefoxitin disks. These strains were further tested for ESBL producing by double-disk synergy test (DDST). This phenotypic assay is based on the exhibition of a synergy image between disks with cefotaxime (30 μg) and ceftazidime (30 μg), which were placed about 20–30 mm from a disk with clavulanic acid (10 μg). E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as quality control strains.[10]
Molecular characterization of resistance genes
Bacterial genomic DNA of suspected ESBLs isolates was extracted directly from 24 h colonies on 5% sheep blood agar plates (Hi-Media, India) by boiling a dense suspension in sterile distilled water for 10 min and centrifugation for 2 min at 13,000 ×g.[13] ESBL-encoding genes were recognized using specific primers for the blaTEM gene (forward primer: 5’AGTATTCAACATTTCCGTGTCG3’ and reverse primer 5’GCTTAATCAGTGAGGCACCTATC3’) and blaCTX-M gene (forward primer: 5’CGTGCTGATGAGCGCTTTGC 3’ and reverse primer 5’AGATCACCGCGATATCGTTG 3’). DNA amplification was performed using the following thermal cycling conditions: Initial denaturation at 95°C for 5 min, followed by 25 cycles of denaturation at 95°C for 30 s, annealing temperature of 59°C for bla CTX-M and 61°C for blaTEM for 25 s and extension at 72°C for 45 s followed by a final extension at 72°C for 10 min. PCR products were analyzed by agarose gel electrophoresis for 1 h at 80 V in 1.5% agarose gel containing 0.05 mg/L safe view to detect specific bands in 0.5X TAE buffer. Primers were purchased from Metabion (Munich, Germany), Master Mix from Amplicon and other molecular materials were bought from Cinnagen, Iran. K. pneumoniae ATCC 7881 was used as a positive control strain.
Enterobacterial repetitive intergenic consensus polymerase chain reaction
The total DNA of each strain was tested by ERIC-PCR. DNA was amplified using the primers ERIC-R: 5’-AAGCTCCTGGGGATTCA-3’and ERIC-F: 5’AGTAAGTGACTGGGGTGAGCG-3’ with the following program: Initial denaturation at 95°C for 5 min, with the next forty cycles includes a denaturation step at 92°C for 30 s; annealing at 60°C for 45 s; extension at 72°C for 30 s; followed by a final extension step at 72°C for 10 min and final storage at 4°C. The ERIC-PCR reaction was performed in a 20 μL volumes containing 8 μL deionized water, 10 μL Master Mix (Amplicon), 0.5 μL of each Primer, and 1 μL of the crude extract DNA.
The ERIC-PCR products were screened by electrophoresis for 45 min in electric field of 100 V on 1.5% agarose gel. The 100 base pair DNA marker (Cinnagen) was used and the gels were subsequently stained for 30 min in a solution containing 0.5 mg of safe view per milliliter.[14]
Computer-assisted enterobacterial repetitive intergenic consensus polymerase chain reaction DNA fingerprint analysis
ERIC-PCR fingerprints of amplified DNA fragments were visually compared. The difference of the patterns by at least one band was classified as different strains. The 1.0 kb DNA marker was used as an external reference standard to normalize positions of the bands on the different lanes and gels. The existence of a given band was coded as 1 and the lack of a given band was coded as 0 in an information matrix and analyzed using the PyElph 1.4 software (BMC Bioinformatics U.S). Dendrograms of differences were set up for each case.
Definitions
MDR was explained as obtained insusceptibility to at least one agent in three or more antimicrobial classes.[1]
Statistical analyses
Data were entered and analyzed using SPSS software version 20 (IBM U.S 2009). The description of the results was carried out by frequencies. For categorical variables, different groups were compared using the Chi-square test or Fisher's exact test. Logistic regression was used for analyzing independent variables that determine an outcome.
All probabilities were two-tailed and the value of P = 0.05 was considered to be statistically significant.
Results
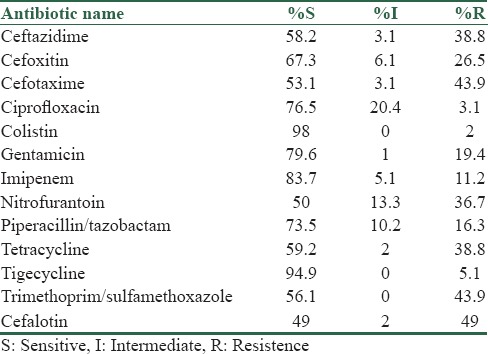
During the period of our study, 98 strains of K. pneumoniae were isolated. Thirty-five (35.7%) cases were male and 63 (64.3%) were female. The age range of patients was 11–89 (42.41 ± 18.36) years. The antimicrobial susceptibility profile showed that colistin had the highest level of activity in these isolates, also susceptibility to imipenem was 80%. Carbapenems presented the highest levels of activity; imipenem inhibited from more than 80% of the isolates. More than 50% of the strains were susceptible to piperacillin-tazobactam and TS [Table 1].
Table 1.
Percentage of antimicrobial susceptibility of 98 strains of Klebsiella pneumoniae
From the 98 studied isolates, 25 (25.5%) were ESBL producers by the DDST, and 44 (44.9%) indicated MDR phenotype. Among ESBL producing isolates, 23 (92%) cases examined MDR (P = 0.001, odds ratio (OR) =28.47, confidence interval: 6.16–131.65). The prevalence of blaCTX-M and blaTEM producing K. pneumoniae isolates were assessed 24 (24.5%) and 18 (18.4%), respectively. In 25 ESBL producing strains, 23 (92%) blaCTX-M and 19 (76%) blaTEM were positive [Figures 1 and 2].
Figure 1.
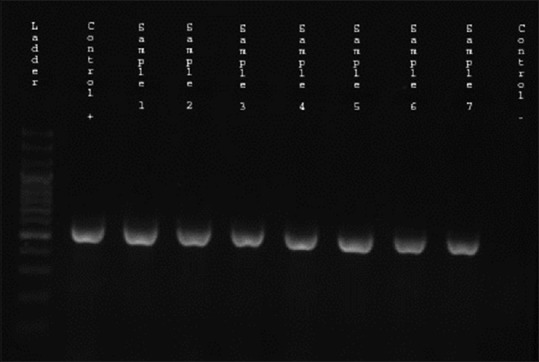
Gel electrophoresis of polymerase chain reaction products following amplification with specific primer for blaCTX-M gene (568 bp) Lane 1: The 100 bp ladder, Lane 2: blaCTX-M positive control, Lane 3–9: Positive clinical isolates for blaCTX-M gene, Lane 10: Negative control
Figure 2.
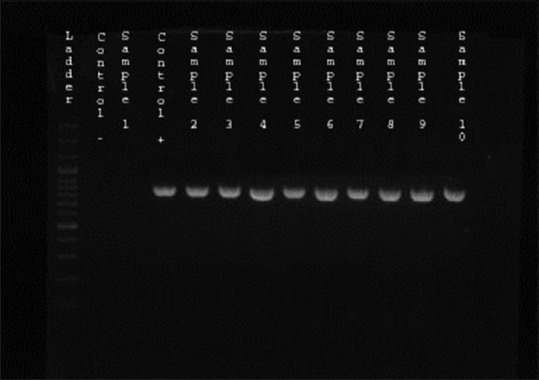
Gel electrophoresis of polymerase chain reaction products following amplification with specific primer for blaTEM gene (850 bp) Lane 1: The 100 bp ladder, Lane 2: blaTEM negative control, Lane 3: Negative clinical isolate, Lane 4: Positive control, Lane 5–13: Positive clinical isolates for blaTEM gene
The antimicrobial drug susceptibilities of ESBL producing K. pneumoniae isolates showed the high resistant rates for cefotaxime and ceftazidime (100% and 88%), respectively, and the high sensitivity rates for colistin and tigecycline (100% and 92%), respectively [Table 2].
Table 2.
Antibiotic susceptibility pattern of Klebsiella pneumoniae extended spectrum β-lactamase producing strains with disk diffusion method
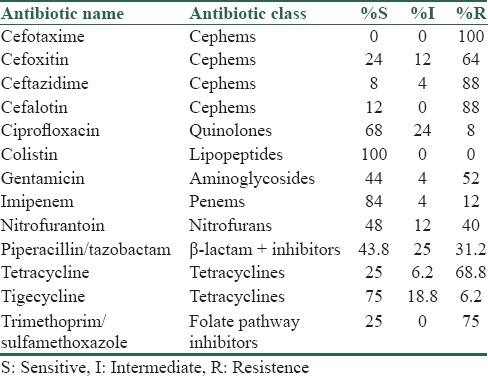
All 23 blaCTX-M producing K. pneumoniae isolates examined were resistant to cefotaxime, and all but one isolate of blaTEM were resistant to cefotaxime.
Significantly, the rates of resistance to most antibiotics were much higher among ESBL positive isolates than ESBL negative isolates (P = 0.038). No significant statistical differences were found between gender or age group and MDR, ESBL, and anti-microbial resistant (P > 0.05).
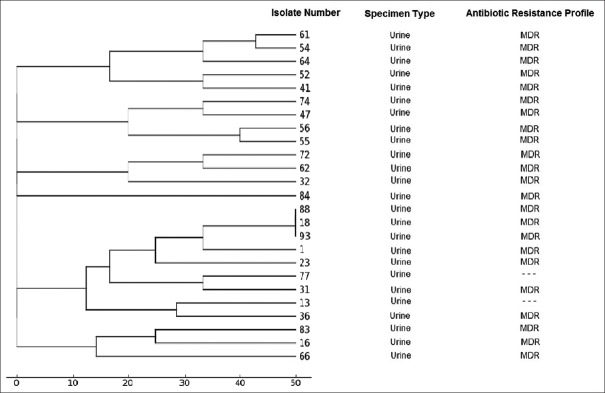
The genomic diversity analysis of 25 strains of ESBL strains of K. pneumoniae has been carried out using the ERIC-PCR fingerprinting method with ERIC-type primer.
The ERIC-PCR profiles revealed differentiation of the 23 band pattern which grouped into six main cluster with a maximum of 24% similarity. Complex patterns of fingerprints isolates showed that 36% of the isolates were belonged to the five cluster [Figure 3].
Figure 3.
Dendrogram based on the cluster analysis (UPGMA) of the estimate of genetic similarity by ERIC - polymerase chain reaction drawn with PyElph 1.4 software
Discussion
UTIs are one of the most common infectious diseases, and K. pneumoniae is one of the most prevalent organisms causing MDR UTI. Most of the epidemiologic studies focus on antibiotic resistance patterns in different regions.[3] Therefore, in this study, we determined the frequency of antibiotic resistance of K. pneumoniae strains and the genetic diversity of these isolates in outpatient referred to Al-Zahra Hospital, Isfahan, Iran.
In general, urinary K. pneumoniae isolates have high rates of resistance to the commonly used antimicrobial agents.[15] Our findings showed that colistin had the highest level of activity in these isolates, also susceptibility to imipenem was 80%. Carbapenems presented the highest levels of activity; More than 50% of the strains were susceptible to piperacillin-tazobactam and trimetoprim/sulfamethoxazole. Thus, the cross-resistance with cephems could be related to the misusing of broad-spectrum antibiotics such as penicillins, cephalosporins, chloramphenicol, tetracyclines, and aminoglycosides.
Other studies that performed in Iran (Tehran, Zahedan) and also in Algeria had shown high rates of resistance to carbapenems, β-lactams, ciprofloxacin, aminoglycosides, but intermediately susceptible to tigecycline and susceptible to gentamicin. In spite of our study, high rates of colistin-resistant strains reported from Algeria.[5,6,16,17,18]
The emergence of MDR K. pneumoniae is a worldwide important problem and many studies with different results has been conducted in recent years.[19] In this study, the prevalence of MDR was 44.9% among the K. pneumoniae isolates and. In another study that performed in Iran, the prevalence of MDR K. pneumoniae isolates was reported 74%, which is more than our results. Furthermore, in other study that was conducted in Kashan, MDR K. pneumoniae rate was 46.6% and no imipenem resistance was reported.[2] In others studies that were conducted on positive urine cultures for K. pneumoniae in China, Pakistan, and Bangladesh the percentage of MDR isolates were 63.8%, 71%, and 85%, respectively.
The probable explanation for this higher rates of MDR and antibiotic resistance, especially in developing countries may be attributable to irregular antibiotic prescription, genetic, geographic, social behaviors and sampling biases and different patients’ characteristics.[2] In this study, the rate of antibiotic resistance in unhospitalized is lower than hospitalized patients that mentioned better condition.
According to the present study, gender or age group were not known as risk factors for MDR strains (P > 0.05), despite findings of a similar study that was performed in Sudan showed the male gender as the MDR risk factor, which is inconsistent with our investigation. Moreover, no significant difference was found between MDR rate and age group according to the previous studies in Kashan, Iran, and Sudan, which is compatible with our research.[2,20]
Recently, many studies have reported epidemic outbreaks of ESBL-producing Enterobacteriaceae.[21,22]
The overall prevalence of ESBL-producing K. pneumoniae isolates altered significantly according to geographic districts of countries and the different hospital conditions. ESBL prevalence rate in our study is 25.5% that is upper compared to those reported in Northern Algeria (19.9%), Spain (20.8%), and Tunisia (20.2%). However, higher prevalence rates of ESBL-producing K. pneumoniae isolates were recognized in Tehran, Iran (69.7%),[17] Zahedan, Iran (66.7%),[16] Ahvaz, Iran (27%),[23] Isfahan, Iran (40%),[24] South America (45.4%–51.9%), France (26%) and Saudi Arabia (55%) and 70% in Pakistan.[3,6] The reason for lower rate of antibiotic, antimicrobial resistance was that isolates were taking only from outpatients referred to hospital.
Antimicrobial susceptibility analysis of the ESBL-producing isolates found highly prevalent resistances to third generation cephalosporins and about 50% to gentamicins. Our results are similar to other study that was performed in Madagascar (100%) to third generation cephalosporins, 87.7% to gentamicins.[9] It is now verified that the consumption of antibiotics, especially third generation cephalosporins, is the most major risk factor in the development of bacterial resistance.[6]
From the 25 ESBL studied isolates, 23 (92%) cases examined MDR (P = 0.001, OR = 28.47) and this finding was according to many other similar studies. These results indicated that there is a strong association between infection with ESBL-producing K. pneumoniae and the use of several different antibiotics. For controlling of outbreaks of infection emphasis must be placed on the logical and sensible use of all antimicrobial agents.[24]
The prevalence of blaCTX-M and blaTEM genes in ESBL strains were 92% and 76%, respectively. These results demonstrated a high proportion of CTX-M and TEM enzymes among ESBL-producing strains in outpatients referred to Al-Zahra Hospital. Our data were in accordance with other studies that performed in different parts of our country and showed the most prevalent genes were CTX-M types, SHV and TEM.[17] In Morocco, ESBL-producing Enterobacteriaceae have been detected in different hospitals and the ESBL genes isolated in Morocco were blaTEM, blaSHV, blaDHA, and blaOXA types.[25] ESBL-producing K. pneumoniae isolates are a serious problem and describes the high resistant rates to common antibiotics.[3] CTX-M-producing K. pneumoniae isolates demonstrated rapid emergence and spread mid-to late-2000s in the United States.[26]
The coexistence of CTX-M ESBL and TEM type β-lactamases in these isolates may have also contributed to the observed high rate of antimicrobial drug resistance.[26] Most of the CTX-M ESBL positive strains in the current study were detected TEM positive (18/24). These data have clinical suggestion for selecting empiric antibiotic treatment when infection caused by ESBL-producing K. pneumoniae is suspected.[26] Although pulsed-field gel electrophoresis is evaluated as the gold standard method for genotyping, ERIC-PCR is an alternative method for producing fingerprints of bacterial genomes, especially in Enterobacteriaceae.[18,27] Our findings showed that 92% of the isolates were belonged to five independent clusters, therefore, there is considerable diversity of genetic types of CTX-M producers and complexity of the resistance phenotype was found.
There are several potential limitations to our study included failure to distinguish colonization from true infection, lack of a comparison group, small numbers of patients, the inclusion of only specific patient populations, and the limiting of more investigation of patients. It is necessary to mention that we did not perform sequencing as a preferred complementary procedure then we could not see the probable mutations and the mechanism of resistance.
Conclusion
The current study revealed relatively high antimicrobial resistance rates among ESBL isolates. About 11% of imipenem-resistant strains were found. Furthermore, the MDR rate compared with developed countries was high. We found a diversity of the genotypes and the complexity of the resistance phenotypes. Epidemiological data collection from outpatients is recommended to develop the strategies to manage antibiotic resistance.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Wang Q, Li B, Tsang AK, Yi Y, Woo PC, Liu CH. Genotypic analysis of Klebsiella pneumoniae isolates in a Beijing Hospital reveals high genetic diversity and clonal population structure of drug-resistant isolates. PLoS One. 2013;8:e57091. doi: 10.1371/journal.pone.0057091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moini AS, Soltani B, Taghavi Ardakani A, Moravveji A, Erami M, Haji Rezaei M, et al. Multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolated from patients in Kashan, Iran. Jundishapur J Microbiol. 2015;8:e27517. doi: 10.5812/jjm.27517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Bouamri MC, Arsalane L, El Kamouni Y, Zouhair S. Antimicrobial susceptibility of urinary Klebsiella pneumoniae and the emergence of carbapenem-resistant strains: A retrospective study from a university hospital in Morocco, North Africa. Afr J Urol. 2015;21:36–40. [Google Scholar]
- 4.Giamarellou H. Multidrug-resistant Gram-negative bacteria: How to treat and for how long. Int J Antimicrob Agents. 2010;36(Suppl 2):S50–4. doi: 10.1016/j.ijantimicag.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Giske CG, Monnet DL, Cars O, Carmeli Y ReAct-Action on Antibiotic Resistance. Clinical and economic impact of common multidrug-resistant Gram-negative bacilli. Antimicrob Agents Chemother. 2008;52:813–21. doi: 10.1128/AAC.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagha N, Abdelouahid DE, Hassaine H, Robin F, Bonnet R. First characterization of CTX-M-15 and DHA-1 β-lactamases among clinical isolates of Klebsiella pneumoniae in Laghouat Hospital, Algeria. Afr J Microbiol Res. 2014;8:1221–7. [Google Scholar]
- 7.Wachino J, Doi Y, Yamane K, Shibata N, Yagi T, Kubota T, et al. Nosocomial spread of ceftazidime-resistant Klebsiella pneumoniae strains producing a novel class a beta-lactamase, GES-3, in a neonatal intensive care unit in Japan. Antimicrob Agents Chemother. 2004;48:1960–7. doi: 10.1128/AAC.48.6.1960-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed ZB, Ayad A, Mesli E, Messai Y, Bakour R, Drissi M. CTX-M-15 extended-spectrum beta-lactamases in Enterobacteriaceae in the Intensive Care Unit of Tlemcen Hospital, Algeria. East Mediterr Health J. 2012;18:382–6. doi: 10.26719/2012.18.4.382. [DOI] [PubMed] [Google Scholar]
- 9.Rakotonirina HC, Garin B, Randrianirina F, Richard V, Talarmin A, Arlet G. Molecular characterization of multidrug-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiol. 2013;17:13–85. doi: 10.1186/1471-2180-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47:3724–32. doi: 10.1128/AAC.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramazanzadeh R, Zamani S, Zamani S. Genetic diversity in clinical isolates of Escherichia coli by enterobacterial repetitive intergenic consensus (ERIC)-PCR technique in Sanandaj hospitals. Iran J Microbiol. 2013;5:126–31. [PMC free article] [PubMed] [Google Scholar]
- 12.Cockerill FR. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Twenty Second International Supplement M100. S22. USA Clinical and Laboratory Standards Institute. 2012 [Google Scholar]
- 13.Sedighi M, Vaez H, Moghoofeie M, Hadifar S, Oryan G, Faghri J. Molecular detection of metallo-ß-lactamase gene blaVIM-1 in imipenem-resistant Pseudomonas aeruginosa strains isolated from hospitalized patients in the hospitals of Isfahan. Adv Biomed Res. 2015;4:57. doi: 10.4103/2277-9175.151872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norouzi A, Azizi O, Nave HH, Shakibaie MR. Analysis of amino acid substitution mutations of gyrA and parC genes in clonal lineage of Klebsilla pneumoniae conferring high-level quinolone resistance. J Med Microbiol Infect Dis. 2014;2:109–17. [Google Scholar]
- 15.Livermore DM. Beta-lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–84. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feizabadi MM, Delfani S, Raji N, Majnooni A, Aligholi M, Shahcheraghi F, et al. Distribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran. Microb Drug Resist. 2010;16:49–53. doi: 10.1089/mdr.2009.0096. [DOI] [PubMed] [Google Scholar]
- 17.Saeidi S, Alavi-Naini R, Shayan S. Antimicrobial susceptibility and distribution of TEM and CTX-M genes among ESBL-producing Klebsiella pneumoniae and Pseudomonas aeruginosa causing urinary tract infections. Zahedan J Res Med Sci. 2014;16:1–5. [Google Scholar]
- 18.Kontopoulou K, Protonotariou E, Vasilakos K, Kriti M, Koteli A, Antoniadou E, et al. Hospital outbreak caused by Klebsiella pneumoniae producing KPC-2 beta-lactamase resistant to colistin. J Hosp Infect. 2010;76:70–3. doi: 10.1016/j.jhin.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Rezaee MA, Sheikhalizadeh V, Hasani A. Detection of integrons among multi-drug resistant (MDR) Escherichia coli strains isolated from clinical specimens in Northern West of Iran. Braz J Microbiol. 2011;42:1308–13. doi: 10.1590/S1517-838220110004000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibrahim ME, Bilal NE, Hamid ME. Increased multi-drug resistant Escherichia coli from hospitals in Khartoum state, Sudan. Afr Health Sci. 2012;12:368–75. doi: 10.4314/ahs.v12i3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–7. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 22.Jones RN. Global epidemiology of antimicrobial resistance among community-acquired and nosocomial pathogens: A five-year summary from the SENTRY Antimicrobial Surveillance Program (1997-2001) Semin Respir Crit Care Med. 2003;24:121–34. doi: 10.1055/s-2003-37923. [DOI] [PubMed] [Google Scholar]
- 23.Khosravi AD, Hoveizavi H, Mehdinejad M. Prevalence of Klebsiella pneumoniae encoding genes for Ctx-M-1, Tem-1 and Shv-1 extended-spectrum beta lactamases (ESBL) enzymes in clinical specimens. Jundishapur J Microbiol. 2013;6:e8256. [Google Scholar]
- 24.Moayednia R, Shokri D, Mobasherizadeh S, Baradaran A, Fatemi SM, Merrikhi A. Frequency assessment of ß-lactamase enzymes in Escherichia coli and Klebsiella isolates in patients with urinary tract infection. J Res Med Sci. 2014;19(Suppl 1):S41–5. [PMC free article] [PubMed] [Google Scholar]
- 25.Barguigua A, El Otmani F, Talmi M, Bourjilat F, Haouzane F, Zerouali K, et al. Characterization of extended-spectrum ß-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates from the community in Morocco. J Med Microbiol. 2011;60(Pt 9):1344–52. doi: 10.1099/jmm.0.032482-0. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Huang T, Surendraiah PK, Wang K, Komal R, Zhuge J, et al. CTX-M ß-lactamase-producing Klebsiella pneumoniae in suburban New York City, New York, USA. Emerg Infect Dis. 2013;19:1803–10. doi: 10.3201/eid1911.121470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: Risk factors for infection and impact of resistance on outcomes. Clin Infect Dis. 2001;32:1162–71. doi: 10.1086/319757. [DOI] [PubMed] [Google Scholar]