Abstract
Background:
Both microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) have been shown to have a critical role in the regulation of cellular processes such as cell growth and apoptosis, as well as cancer progression and metastasis. lncRNAs are aberrantly expressed in many diseases including cancer. Although it is well known that miRNAs can target a large number of protein-coding genes, little is known whether miRNAs can also target lncRNAs. In the present study, we determine whether miR-21 can regulate lncRNA cancer susceptibility candidate 2 (CASC2) in colorectal cancer.
Materials and Methods:
LS174T cells were transfected with locked nucleic acid (LNA)-anti-miR-21 and scrambled LNA for 24, 48 and 72 h. The expression of miR-21 and lncCASC2 was evaluated by quantitative reverse transcriptase polymerase chain reaction.
Results:
However, contrary to what we expected and reported by others, lncCASC2 quantity was significantly reduced in LNA treated LS174T cells compared to the scrambled treated and normal untreated cells (P < 0.05).
Conclusion:
The interaction of miRNA and lncRNA are not as simple as suggested by other reports. Moreover, it could be complex molecular mechanisms underlying the communication of various noncoding RNA elements.
Keywords: Cancer susceptibility candidate 2 gene, colorectal cancer, long noncoding RNAs, microRNA
Introduction
Colorectal cancer (CRC) is one of the most common malignancies worldwide.[1] Unfortunately, the occurrence of cancer trends among adults younger than 50 years.[2] Younger patients diagnosed with higher degree of malignancy.[3] Despite the substantial progress in diagnosis and treatment, we still encounter with high mortality rate of advanced CRC. Data presented an increased level of incidence and mortality among patients with advanced cancer.[4]
This means that identification of molecular basis involved to find novel therapeutic options for treatment of these patients is vital.
It has recently been indicated that noncoding RNAs (ncRNA) play crucial roles in the tumorigenic of CRC.[5] The class of long ncRNA (lncRNA) contains a very heterogeneous subclass of ncRNA.[6]
Despite often being represented as transcriptional noise, it has verified that lncRNAs can be regulating nearly every step within the life cycle of a gene.[7,8,9,10] There are many reports stating lncRNAs involvement in development and progression of various cancers, including breast cancer,[11,12] gastric cancer,[13,14] bladder cancer,[15,16] lung cancer,[17] and CRC[18,19] and glioma.[20] lncRNAs act to regulate gene expression through various mechanisms.[21] A large number of lncRNAs may function as endogenous sponges for other kinds of RNAs such as various microRNAs (miRNAs) which consequently impede miR-mediated functions.[22] Understanding the detailed molecular mechanisms through which lncRNAs function in various diseases might be leading to serve this ncRNA in novel diagnostic or therapeutic strategies. Cancer susceptibility candidate 2 (CASC2), as a novel lncRNA, was originally known as a downregulated gene in endometrial cancer. It has been characterized as a tumor suppressor in several human malignancies, including CRC.[23] Although the CASC2 dysregulated expression points out its tumorigenic properties in CRC patients, precise mechanisms of its mediated process is not clear. miRNAs, another class of ncRNA, function as negative gene regulators through specific complementarily sites at the 3-untranslated region of target messenger RNAs (mRNAs), which result in degradation or translational repression.[24,25] miRNAs have important widely regulatory functions in diverse cellular processes at the transcriptional or posttranscriptional level.[26,27,28] There is a growing body of evidence showing the aberrant expression of certain miRNAs, in particular, miR-21, is associated with carcinogenesis, tumor size, metastasis, and survival rate of patients with CRC.[29,30,31] It is indicated that miR-21 contents increase during CRC progression from precancerous conditions to advanced carcinoma.[32] Among the mechanisms, taking part in CASC2 regulation has been demonstrated being miR-21 action, it has been shown that there is a reciprocal repression between CASC2 and miR-21 in several malignancies including human gliomas. Wang et al. demonstrated that CASC2 expression is dramatically decreased in glioma tissues and it plays a tumor-suppressive function through negative regulation of miR-21, which may be a novel target for therapeutic modalities. Bioinformatics and luciferase reporter assays provided information that miR-21 binds to CASC2 in a sequence-specific manner and they regulate each other reciprocally. In regard to this repression of each of them could upregulate another one as seen in the previous study on glioma cells.[33] In light of vital role of CASC2 and miR-21 in colon carcinogenesis, in the present study, we inhibited miR-21 using locked nucleic acid (LNA) technology in LS174T colorectal cell line.
Materials and Methods
Cell culture
The Human colorectal adenocarcinoma cell line LS174T was purchased from the National Cell Bank of Iran (Pasteur Institute, Iran). The cells were maintained in Dulbecco modified Eagle medium (Gibco, USA) in humidified incubator containing 5% CO2 at 37°C.
Cell transfection with locked nucleic acid anti-miR-21
miR-21 inhibitor (LNA-anti-miR-21) transfection was performed according to an established protocol. Briefly, cells were transfected using an X-tremeGENE small interfering RNA (siRNA) Transfection Reagent Kit (Roche, Mannheim, Germany), 24 h after seeding into the well.
LNA-anti-miR-21 was added to the culture media at a final oligonucleotide concentration of 50 pmol. The LNA-anti-miR-21-5P and scrambled LNA molecules were synthesized by Exiqon with fluorescein dye, 6-carboxyfluorescein (6-FAM) moiety at the 5′ end. The following sequences are LNA-anti-miR-21, 5′-FAM-ACATCAGTCTGATAAGCT-3′, and its control oligo, LNA-scramble, 5′-FAM-GTGTAACACGTCTATACGCCCA-3′.
Ten percent fetal bovine serum was added 10 h posttransfection, followed by incubation for an additional 24, 48 and 72 h. Untreated cells as a control and cells transfected with oligo control for LNA-anti-miR-21 (LNA-scramble) were cultured parallel to the LNA-antimiR transfected cells. The evaluation of the transfection was detected by FACSCalibur flow cytometer and fluorescent microscopy.
Cell viability assay
Cell invasion assays were performed using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) assay (Bio-IDEA, Tehran, Iran). In brief, cells were seeded at a density of 2 × 104 cells/well into six-well plates, following 24 h of incubation. The MTT assay was performed 24, 48, and 72 h after transfection. After adding the MTT solution (12 mM) incubate at 37°C in a dark room for 2 h. Afterward, 200 μl dimethyl sulfoxide was added. The plates were capturing at the wavelength of 570 nm.
Measurement of apoptosis
Apoptotic cells were measured by an Annexin-V-FLUOS Staining Kit (Roche, Mannheim, Germany) according to the manufacturer's protocols. 2 × 105 cells/well were seeded in six-well plates. After 24 h incubation, the procedure was performed at 24, 48 and 72 h after transfection. After collection of floating cells, adherent cells were rinsed with Ca2+ and Mg2+-free phosphate-buffered saline and harvested after incubation with 2 mM ethylenediaminetetraacetic acid in phosphate-buffered saline enzyme free for 10 min with gentle movement. After the cells were visibly detached, they were suspended by 5 ml of complete growth medium. Then, both floating and adherent cells were pooled and rinsed twice with phosphate-buffered saline. After centrifuging for 4 min at 190 g at 4°C, the resulting pellet was resuspended in 100 μl incubation buffer with 2 μl Annexin-V and 2 μl propidium iodide for 15 min in a dark room at 4°C. Then, the samples immediately analyzed by using a FACSCalibur flow cytometer with Annexin-V and PI excitation/emission wavelengths (488/525 and 488/675 nm, respectively).
Quantitative reverse transcriptase polymerase chain reaction
Briefly, RNAs from LS174T cells were isolated with a miRCURY RNA Isolation Kit (Exiqon, Copenhagen, Denmark), and the concentration and purity of RNA were measured by spectrophotometry. mRNAs complementary DNA (cDNA) were synthesized by Universal cDNA Synthesis Kit (Exiqon, Copenhagen, Denmark) according to manufacturer's protocol. The amount of miR-21 inhibition determined by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) (SYBR Green Master Mix Kit) with specific hsa-miR-21-5p and hsa-let-7a-5p primers. As an internal control, hsa-let-7a was used for miR-21 template normalization. For evaluation of CASC2 gene expression, one microgram of total RNA was used to synthesize cDNA using Fermentas Kit. Relative levels of lncRNA were carried out using SYBR® Green Master Mix Kit TM (Exiqon, Copenhagen, Denmark) and normalized to levels of endogenous control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), mRNA. 2ΔΔCt method was used for calculation of relative gene expression levels.
The primers’ sequences for CASC2 were: forward 5′-CGGCAGATGGAGATTCAGAAACATAAGG-3′; reverse 5′ GCAACACACTTCTTACTCGGAATAGGG-3′ and GAPDH primers were forward 5′-AGTCCACTGGCGTCTTCAC-3′; reverse 5′-AGGCATTGCTGATGATCTTGAG-3′. All PCR reactions were run in triplicate by The StepOnePlus TM Real-Time PCR (Applied Biosystems, USA) instrument.
Statistical analysis
Data are presented as mean ± standard deviation (SD) and analyzed using SPSS v. 22 statistical software (IBM, Chicago, IL, USA) by the two-way analysis of variance test with Fisher's post hoc followed by Scheffé multiple comparison test. Statistical significance was accepted at the P < 0.05 level.
Results
Locked nucleic acid anti-miR-21 reduced the expression of miR-21
For inhibition of miR-21, the miRCURY LNA miRNA Inhibitor™ was transfected to LS174T cells with the X-tremeGENE siRNA Transfection Reagent™. On the basis of the initial optimization experiments, transfection was performed with 50 pM of LNA-anti-miR in 5 μl transfection reagent in a total volume of 200 μl Opti-MEMI medium (Gibco, USA). The transfection efficacy was >85% in the optimized situation. The expression of miR-21 was evaluated by qRT-PCR after transfection for 24, 48, and 72 h. The results showed downregulated level of miR-21 in LNA-anti-miR treated group. No statistically significant difference was found between values of miR-21 in cells transfected with scrambled LNA and control groups during the incubation (P > 0.05). In all three time points, the expression of miR-21 was considerably lower in the LNA-anti-miR group compared to scramble LNA and untreated groups (P < 0.01) [Figure 1].
Figure 1.
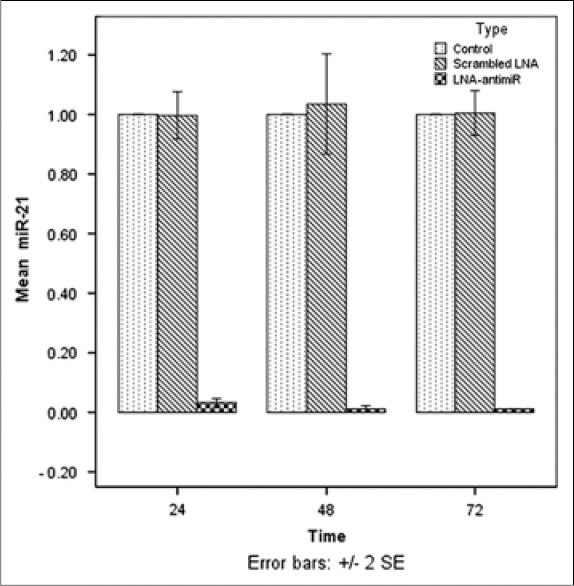
Assessment of the miR-21 level by real-time polymerase chain reaction, 24, 48, and 72 h after transfection. The expression of miR-21 was normalized to let-7a expression using the ΔΔCt method. The untreated group was considered as a reference for each time point. Data were mean ± standard deviation of three independent experiments
Modulation of cancer susceptibility candidate 2 by locked nucleic acid-anti-miR-21
To assess lncRNA CASC2 as a target gene of miR-21 in LS174T, we evaluated the expression levels of lncRNA CASC2 in 3 groups: LS174T cells pretreated with control oligo (LNA-scramble) or LNA-anti-miR-21 and untreated group by qRT-PCR. As shown in Figure 2, CASC2 expression levels were significantly down-regulated in group transfected with LNA-anti-miR (P < 0.001).
Figure 2.
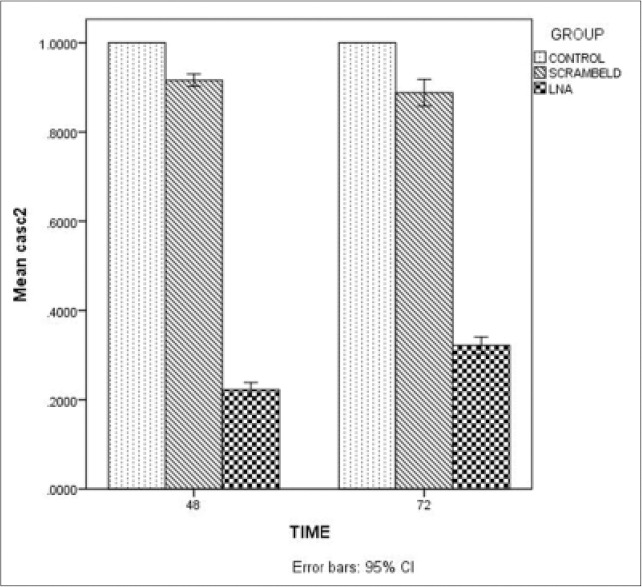
The expression of cancer susceptibility candidate 2 long noncoding RNA in locked nucleic acid-anti-miR, scramble locked nucleic acid and untreated groups in LS174T colon adenocarcinoma cells. The long noncoding cancer susceptibility candidate 2 levels, 48 and 72 h after transfection was analyzed by quantitative reverse transcriptase-polymerase chain reaction. The expression was normalized to glyceraldehyde-3-phosphate dehydrogenase
Locked nucleic acid-anti-miR-21 reduced viability of LS174T cells
To further examine the functional effect of anti-miR-21 on LS174T cells, we measured cell viability by MTT assay. Treatment of cells with LNA anti-miR-21 showed a significant reduction in cell viability in all their time points compared with the control and scrambled LNA groups, especially after 72 h, with >50% reduction (P < 0.01).
Locked nucleic acid anti-miR-21 induces apoptosis in LS174T cells
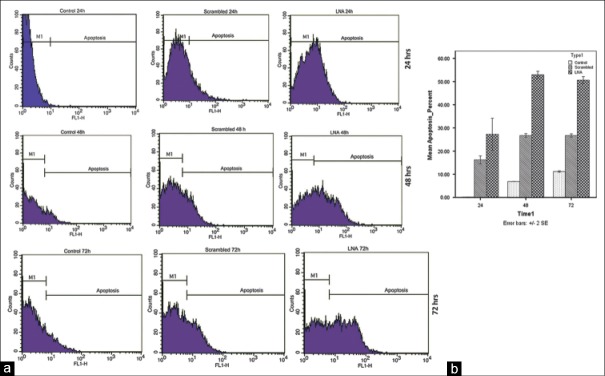
Because the LNA anti-miR-21 suppressed cell viability, we next sought to determine whether the LNA anti-miR-21 also induced apoptosis. To measure the effect of this treatment on apoptosis we used Annexin-V binding assay, which allows quantification of the level of phosphatidylserine exposure at the outer membrane side, a well-characterized event of early apoptosis.[34] At all three time points, The population of Annexin-V-positive cells increased significantly in LNA anti-miR-21 transfected cells (P < 0.01). In addition, we observed the highest amount of apoptosis at 48 h after LNA-anti-miR-21 transfection. LNA-transfected cells show higher percent of apoptosis compared with scrambled LNA-transfected cells and untreated cells (P < 0.01) [Figure 3].
Figure 3.
Assessment of apoptosis by Annexin-V-propidium iodide staining performed 24, 48, and 72 h after transfection. Representative cytofluorometric graphs are shown (a); data shown in the graph are mean ± standard deviation of triplicate experiments (b)
Discussion
In recent years, the prevalence of CRC has increased markedly among individuals between ages 20 and 49 years. A significant proportion of young patients present with advanced disease.[3] Even with great development in diagnostic and therapeutic methods, the overall survival rate is still relatively poor and remains unsatisfactory.[35] Hence, it is imperative to comprehend the pathogenic mechanism of CRC. Nowadays, lncRNAs have gained wide attention as new mediators in tumor biology.[36] Thus, learning the processes by which lncRNA expression is controlled is the first stage toward understanding the basic mechanisms by which they exert their functions in human cancers.
miR-21 is one of the oncogenic miRNAs that are significantly overexpressed in tumor tissue of CRC, which is responsible for the malignant progression of colon cancer.[37] The basic role of miR-21 action may be the consequence of its ability to regulate numerous coding genes[38,39] but growing body of evidence suggested that lncRNA might regulate by similar mechanisms as for protein coding genes.[40]
In this study, we determine whether miR-21 can directly regulate lncRNA CASC2 in CRC and could not find any association. Tumor-suppressor features for the lncRNA CASC2 have been recognized in several cancers. CASC2 was expressed in reduced level in endometrial cancer. Exogenous expression of CASC2 remarkably inhibited the clonal growth in AN3CA endometrial cancer cell line.[23] In the distinct study, lower expression of CASC2 was found in nonsmall cell lung cancer (NSCLC) and significantly associated with advanced tumor node metastasis stage and tumor size. In vitro and in vivo assays showed inhibited NSCLC cell proliferation through CASC2 overexpression.[41] Cao et al. dual-luciferase reporter assays revealed that CASC2 targeted by miR-21 and miR-21 was able to decrease the expression of CASC2 in renal cell carcinoma.[42] In human gliomas, miR-21 could negatively regulate CASC2 function in a sequence-specific manner and therefore destroyed the CASC2-mediated inhibition of glioma cell proliferation and invasion. Moreover, overexpression of miR-21 dramatically suppressed the cell apoptosis caused by CASC2.[33] Despite all these evidence verified the negative regulation of CASC2 by mir-21, surprisingly, our results could not provide any proofs to claim that they are biologically relevant. Indeed, (i) the intricacy of the regulation of this tumor suppressor gene, which may be distinctly, affected depending on the type of cancer and (ii) there are more miRNAs that have a significant negative effect on considered gene. It remains possible that it might be a trans-inhibition of CASC2 results from the synergistic action of multiple miRNAs. Furthermore, extra studies will be needed to gain a full understanding of the molecular mechanisms underlying the communication of various ncRNA elements in a network.
Conclusion
Although an inverse correlation between CASC2 and mir-21 levels has been reported in several types of tumors, it had never been investigated in CRC. Thus, further investigation and validation in a larger number of CRC cell models are required to better declare the function and critical mechanisms of lncRNA CASC2 in the progression of colon cancer.
Financial support and sponsorship
We would like to appreciate financial support provided by Isfahan University of Medical Sciences.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell JB, Maggard MA, Liu JH, Etzioni DA, Livingston EH, Ko CY. Rates of colon and rectal cancers are increasing in young adults. Am Surg. 2003;69:866–72. [PubMed] [Google Scholar]
- 3.Siegel RL, Jemal A, Ward EM. Increase in incidence of colorectal cancer among young men and women in the United States. Cancer Epidemiol Biomarkers Prev. 2009;18:1695–8. doi: 10.1158/1055-9965.EPI-09-0186. [DOI] [PubMed] [Google Scholar]
- 4.Sun BO, Li W, Liu N. Curative effect of the recent photofrin photodynamic adjuvant treatment on young patients with advanced colorectal cancer. Oncol Lett. 2016;11:2071–4. doi: 10.3892/ol.2016.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slaby O. Non-coding RNAs in Colorectal Cancer. Springer; 2016. Non-coding RNAs as biomarkers for colorectal cancer screening and early detection; pp. 153–70. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 7.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammad F, Pandey GK, Mondal T, Enroth S, Redrup L, Gyllensten U, et al. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development. 2012;139:2792–803. doi: 10.1242/dev.079566. [DOI] [PubMed] [Google Scholar]
- 10.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–33. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piao HL, Ma L. Non-coding RNAs as regulators of mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2012;17:33–42. doi: 10.1007/s10911-012-9245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8:496–505. doi: 10.4161/rna.8.3.14800. [DOI] [PubMed] [Google Scholar]
- 13.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–73. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–29. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–21. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 16.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–94. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong PR. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–5. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 20.Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, et al. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40:2004–12. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 21.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldinu P, Cossu A, Manca A, Satta MP, Sini MC, Rozzo C, et al. Identification of a novel candidate gene, CASC2, in a region of common allelic loss at chromosome 10q26 in human endometrial cancer. Hum Mutat. 2004;23:318–26. doi: 10.1002/humu.20015. [DOI] [PubMed] [Google Scholar]
- 24.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–98. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 26.Cheng LC, Tavazoie M, Doetsch F. Stem cells: From epigenetics to microRNAs. Neuron. 2005;46:363–7. doi: 10.1016/j.neuron.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 27.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 28.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S, Liang Z, Xu L, Zou F. MicroRNA-21: A ubiquitously expressed pro-survival factor in cancer and other diseases. Mol Cell Biochem. 2012;360:147–58. doi: 10.1007/s11010-011-1052-6. [DOI] [PubMed] [Google Scholar]
- 31.Schetter AJ, Harris CC. Alterations of microRNAs contribute to colon carcinogenesis. Semin Oncol. 2011;38:734–42. doi: 10.1053/j.seminoncol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamichi N, Shimomura R, Inada K, Sakurai K, Haraguchi T, Ozaki Y, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15:4009–16. doi: 10.1158/1078-0432.CCR-08-3257. [DOI] [PubMed] [Google Scholar]
- 33.Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27:275–82. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84:1415–20. [PubMed] [Google Scholar]
- 35.Carpenter B, McKay M, Dundas SR, Lawrie LC, Telfer C, Murray GI. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br J Cancer. 2006;95:921–7. doi: 10.1038/sj.bjc.6603349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 38.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 39.Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–75. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 40.Jeggari A, Marks DS, Larsson E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–3. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He X, Liu Z, Su J, Yang J, Yin D, Han L, et al. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumour Biol. 2016;37:9503–10. doi: 10.1007/s13277-016-4787-6. [DOI] [PubMed] [Google Scholar]
- 42.Cao Y, Xu R, Xu X, Zhou Y, Cui L, He X. Downregulation of lncRNA CASC2 by microRNA-21 increases the proliferation and migration of renal cell carcinoma cells. Mol Med Rep. 2016;14:1019–25. doi: 10.3892/mmr.2016.5337. [DOI] [PubMed] [Google Scholar]