Abstract
Background
Chronic inflammatory conditions are associated with higher tumor incidence through epigenetic and genetic alterations. Here, we focused on an association between an inflammation marker, C-reactive-protein (CRP), and global DNA methylation levels of peripheral blood leukocytes.
Methods
The subjects were 384 healthy Japanese women enrolled as the control group of a case-control study for breast cancer conducted from 2001 to 2005. Global DNA methylation was quantified by Luminometric Methylation Assay (LUMA).
Results
With adjustment for lifestyle-related factors, including folate intake, the global DNA methylation level of peripheral blood leukocytes was significantly but weakly increased by 0.43% per quartile category for CRP (P for trend = 0.010). Estimated methylation levels stratified by CRP quartile were 70.0%, 70.8%, 71.4%, and 71.3%, respectively. In addition, interaction between polymorphism of MTHFR (rs1801133, known as C677T) and CRP was significant (P for interaction = 0.046); the global methylation level was significantly increased by 0.61% per quartile category for CRP in the CT/TT group (those with the minor allele T, P for trend = 0.001), whereas no association was observed in the CC group (wild type).
Conclusions
Our study suggests that CRP concentration is weakly associated with global DNA methylation level. However, this association was observed more clearly in individuals with the minor allele of the MTHFR missense SNP rs1801133. By elucidating the complex mechanism of the regulation of DNA methylation by both acquired and genetic factors, our results may be important for cancer prevention.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4089-z) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, C reactive protein, Inflammation, Folate metabolism, Folate intake
Background
Aberrant DNA methylation is known as an essential factor for tumor initiation and promotion. Tumor cell genomes are generally characterized with region-specific hypermethylation and global hypomethylation; typically, the former occurs in promoter regions of tumor suppressor genes, and the latter is represented by hypomethylation in repetitive elements, such as LINE-1 [1–3]. In addition, several studies have reported an association between the level of global methylation of the peripheral blood leukocytes and the development of cancers, which suggests that metabolic conditions in cancer patients may induce aberrant DNA methylation systemically, not only in the primary organ [4–7]. Researchers in the epigenetics field conventionally consider aging and inflammation as initiators of aberrant DNA methylation [2, 8]. Methylation levels of many genes in healthy tissues alter with age [8–12], and aging is obviously related to tumor incidence, as a reflection of the mixture of complex exposures. Moreover, higher tumor incidence is also associated with various chronic inflammatory conditions, such as H. pylori (HP) gastritis and ulcerative colitis (UC), and previous reports suggest that the mechanisms of these associations can be through epigenetic alterations [8, 13]. In a colitis-induced mouse model, DNA hypermethylation was induced in low CpG regions called “DNA methylation valleys” [14].
DNA methylation is also known for its relationship with nutrition. In particular, one-carbon metabolism, which is tightly linked to folate metabolism, involves the step in which the methyl group is added to the cytosine base [15]. Our group previously observed a correlation between folate intake and methylation levels, and assessed interaction between SNPs related to one-carbon metabolism (such as MTHFR etc.) and its intake in Japanese healthy women [15]. In that study, we measured global DNA methylation levels of peripheral blood cells by the LUMA method [16, 17]. Although local tissues can be used to evaluate the effect of local (and mostly direct) exposure to risk factors (such as smoking, alcohol and infection) on DNA methylation, we used peripheral blood cells to assess the influence of systemic exposure such as nutrition levels and the association with serum marker values. Of course, the significance of variation in the DNA methylation of peripheral blood cells for tumorigenesis is not as simple as methylation levels in local tissues, such as gastrointestinal mucosa; nevertheless, it is considered a reasonable method for measuring the systemic effect of an “epimutagen” [4–6, 15, 18, 19].
Here, we focused on one of the most-used quantitative inflammation markers, CRP, and global methylation levels in peripheral blood cells. Since CRP is considered a disease marker for various diseases, including malignant tumors [20–24], the epigenetic effect of high-CRP conditions over the long term is a reasonable research question. In addition, our investigation included statistical adjustment of lifestyle-related characteristics, including folate intake, and examined statistical interaction by five one-carbon metabolism related-SNPs in the MTHFR, MTR, and MTRR genes. The study population was a well-characterized control group for a breast cancer case-control study in Japan, and validated methods were used to measure all variables. We consider that this study will contribute to understanding the mechanism by which systemic inflammation is associated with global DNA methylation, and of effect modification by individual folate-related factors.
Methods
Study subjects and data collection
Subjects were the control group of a hospital-based case-control study for breast cancer conducted from May 2001 to September 2005 at four hospitals in Nagano Prefecture, Japan. Details of this study have been described previously [25, 26]. In this study, healthy female individuals were selected by medical check-up in either of four hospitals and confirmed to not have any cancer. Each subject was recruited as a control for a case matched by age (within 3 years) and residential area. After exclusion of subjects with extremely low or high total energy intake (< 500 or 4000 kcal, respectively) or those without available DNA samples, 384 healthy Japanese women were included in the study.
Participants answered a self-administered questionnaire that included questions on the following: demographic and anthropometric characteristics, smoking habit, family history of cancer, physical activity, medical history, and menstrual and reproductive history. Dietary habits were investigated using a 136-item semiquantitative food frequency questionnaire (FFQ) that was developed and validated in a Japanese population [27, 28]. Spearman’s correlation coefficients between energy-adjusted folate intakes estimated from the FFQ and from dietary records were 0.35–0.50. Participants donated blood samples at the time they returned the questionnaire. Details of these protocols have been described previously [15].
Laboratory analysis
Plasma CRP concentrations were measured by a commercial laboratory (LSI Medience Corporation, Tokyo, Japan) using a latex-enhanced high-sensitivity assay on a BN II nephelometer (Dade Behring Marburg GmbH, Marburg, Germany). The intra-assay coefficient of variation from the quality control samples was 4.8% at 0.036 mg/dL.
Genomic DNA was extracted from the whole blood using a Qiagen FlexiGene DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Global DNA methylation was quantified by Luminometric Methylation Assay (LUMA) [16, 17], which was described and validated for assessing small differences in our in-house testing [15]. Briefly, after cleaving 300 nanograms of genomic DNA with either HapII + EcoRI or MspI + EcoRI in two separate reactions, the HpaII/EcoRI and MspI/EcoRI ratios were calculated by pyrosequencing-based quantification of (dGTP + dCTP)/dATP for each reaction. The HpaII/MspI ratio was then calculated as (HpaII/EcoRI)/(MspI/EcoRI), which corresponds to the proportion of unmethylated cytosine in CCGG sequences in the genome [15]. Reproducibility of the assay for global methylation level was considered high (Total assay CV, 6.4%, which is always higher than intra- and inter- assay CV), suggesting that measurement errors were minimal.
In addition we focused on three genes, MTHFR, MTR, and MTRR, which are known to be involved in DNA methylation in one-carbon metabolism, and selected SNPs in consideration of the availability of functional information. Five polymorphisms in MTHFR (rs1801133 and rs1801131), MTR (rs1805087), and MTRR (rs10380 and rs162049) genes were genotyped by TaqMan SNP Genotyping Assay (Applied Biosystems, USA). These protocols have been described in detail elsewhere [15].
Statistical analysis
Pre-analysis data processing
Folate intake was adjusted for total energy intake using the residual method. Serum CRP concentration and folate intake was divided into quartile categories because inappropriate influence of outlier should be avoided and linear correlation could not be assumed for untransformed value.
Adjustment for potential confounders
The following variables were used for adjustment: age (continuous); body mass index (BMI, continuous); smoking (never smokers, past smokers, current smokers); alcohol drinking (non-drinkers, occasional drinkers, regular drinkers of < 150 g ethanol/week, regular drinkers of ≥150 g ethanol/week); and physical activity in the past 5 years (no, 2 days/week, 3 days/week). Adjusted mean global methylation levels were calculated according to CRP categories and SNPs related to one-carbon metabolism using a multivariate linear regression model with covariates above. Subjects missing measurement data for at least one covariate were excluded from the multivariate analyses.
Testing for linear trend and statistical interaction
To test linear trends for mean CRP levels, regression coefficients (β) were calculated in the multivariable linear regression model using categories of each CRP level as ordinal variables. Log-transformed CRP was used in the model as a sensitivity analysis of linearity. To investigate statistical interaction by effect modifiers, subgroup analyses and tests for interaction (Wald test) were carried out with SNPs related to one-carbon metabolism and folate intake.
General considerations
Since the analyzed results are highly correlated with each other and considered to be too strict for exploratory research, statistical multiplicity was not adjusted. All P-values are two-sided, and those under 0.05 were considered significant. All statistical analyses were performed with SAS version 9.3 (SAS Institute, USA).
Results
The study population is the same as in our previous study of DNA methylation and folate-related factors [15]. Briefly, mean age was 54.1 years (SD 10.3); mean body mass index was 23.0 (SD 3.2); smoking status was never 92.7%, former 2.1%, and current 5.2%; and drinking status was non-drinker 60.4%, sometimes 10.2%, < 150 g/week 22.7%, and ≥150 g/week 6.8%. After the evaluation of CRP distribution, we excluded four subjects with CRP > 0.5 mg/dL, which is an upper measurement limit of hs-CRP, because the possibility of acute inflammation could not be excluded. A total of 380 subjects was then used for analysis.
First, we investigated the crude association between serum CRP concentration and global DNA methylation (Table 1), but did not observe significant association (P for trend = 0.071). Next, we included potential confounders into the model. On adjustment for conventional lifestyle-related factors such as age, BMI, smoking, exercise, and drinking status, global methylation level was shown to be significantly increased by 0.43% per quartile category for CRP, and a trend test was statistically significant (adjusted model 1, P for trend = 0.011). Adjustment for BMI was considered to have the greatest influence on the coefficient of CRP (the coefficient changed from 0.28% to 0.42%, with P = 0.010 before/after BMI adjustment only). In addition, since we observed an association between folate intake and global methylation level [15], we included folate intake into the model (Adjusted Model 2), but the CRP trend did not remarkably change from the previous model (P for trend = 0.010). As shown in Table 1, log-transformed CRP (a numerical, not a categorical variable) was associated with global methylation level, and total energy intake did not influence the association (P for trend = 0.011). The association was still observed (P for trend = 0.019) and at the upper limit of total energy intake of 3000 kcal. Details of models are shown in Additional file 1: Table S1.
Table 1.
Assessment of the association between serum CRP concentration and global methylation level with and without adjustment for life-style factors using the general linear model
| 95% confidence interval | 95% confidence interval | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | CRP (mg/dL) | N | Estimated mean methylation (%) | Lower | Upper | βc | Lower | Upper | P value for trend |
| Crude model | 0.002–0.011 | 101 | 69.7 | 69.0 | 70.3 | ||||
| 0.012–0.023 | 89 | 70.2 | 69.5 | 70.9 | |||||
| 0.024–0.048 | 93 | 70.7 | 70.0 | 71.3 | |||||
| 0.049–0.491 | 93 | 70.4 | 69.8 | 71.1 | |||||
| 0.28 | −0.02 | 0.58 | 0.071 | ||||||
| Log-transformed CRP | 0.25 | −0.07 | 0.56 | 0.123 | |||||
| Adjusted model 1a | 0.002–0.011 | 100 | 70.1 | 69.0 | 71.2 | ||||
| 0.012–0.023 | 89 | 70.7 | 69.6 | 71.9 | |||||
| 0.024–0.048 | 92 | 71.4 | 70.3 | 72.6 | |||||
| 0.049–0.491 | 92 | 71.3 | 70.1 | 72.5 | |||||
| 0.43 | 0.10 | 0.76 | 0.011 | ||||||
| Log-transformed CRP | 0.42 | 0.07 | 0.78 | 0.019 | |||||
| Adjusted model 2b | 0.002–0.011 | 100 | 70.0 | 68.9 | 71.1 | ||||
| 0.012–0.023 | 89 | 70.8 | 69.6 | 71.9 | |||||
| 0.024–0.048 | 92 | 71.4 | 70.2 | 72.6 | |||||
| 0.049–0.491 | 92 | 71.3 | 70.1 | 72.4 | |||||
| 0.43 | 0.10 | 0.76 | 0.010 | ||||||
| Log-transformed CRP | 0.42 | 0.06 | 0.79 | 0.021 | |||||
aAdjusted by age, BMI, smoking (never, former, current), exercise (none/week, 1–2 times/week, ≥3 times/week), and drinking (non-drinker, sometimes, < 150 g/week, ≥150 g/week). Estimates were calculated with average values of continuous variables
bAdjusted by the factors in adjusted model 1 with folate intake
cAverage relative methylation difference per 1-category increase (included as a continuous variable)
Subsequently, we examined the statistical interaction by one-carbon metabolism related-SNP, folate intake and drinking status. As shown in Table 2, interaction between a polymorphism of MTHFR (rs1801133, known as C677T) and CRP was statistically significant (P for interaction = 0.046): the effect of CRP trend was significant in the CT/TT group (individuals with the minor allele T, P = 0.001), but no association was observed in the CC group (wild type). Although not presented in the table, the interaction was not significant (P = 0.109) when log-transformed CRP values were used instead of the CRP category, but the association was similar in the stratified analyses (β = 0.567, P = 0.005 in the CT/TT group; β = − 0.028, P = 0.932 in the CC group). Although no significant interaction was observed between the other SNPs and CRP status, one side of the divided subgroups showed significant P values in every analysis. Using Adjusted Model 2 in Table 1, we calculated adjusted estimates of global methylation levels stratified by CRP category. As shown in Table 3, a monotonous increase with CRP category was clearly observed in the MTHFR CT/TT group, but not in the CC group. In addition, on stratification all of subgroups by the single factors listed in Table 3, alcohol drinkers in the CT/TT group showed the highest regression coefficient (β = 0.83, P = 0.007) for the association between DNA methylation and CRP level (Table 4). A similar tendency was observed when using log-transformed CRP values.
Table 2.
Assessment of interaction between serum CRP concentration and folate-related SNP/factors for global methylation levels
| 95% confidence interval | P value for trend* | P for interaction* | ||||
|---|---|---|---|---|---|---|
| N | β*c | Lower | Upper | |||
| MTHFR (rs1801133) | 0.046 | |||||
| CT/TT | 264 | 0.61 | 0.24 | 0.99 | 0.001 | |
| CC | 109 | −0.09 | −0.69 | 0.52 | 0.778 | |
| MTHFR (rs1801131) | 0.735 | |||||
| CC/CA | 127 | 0.36 | −0.17 | 0.89 | 0.186 | |
| AA | 246 | 0.47 | 0.07 | 0.87 | 0.022 | |
| MTR (rs1805087) | 0.627 | |||||
| GG/GA | 120 | 0.34 | −0.19 | 0.86 | 0.206 | |
| AA | 250 | 0.49 | 0.09 | 0.89 | 0.016 | |
| MTRR (rs162049) | 0.599 | |||||
| AA/AG | 261 | 0.48 | 0.10 | 0.86 | 0.013 | |
| GG | 110 | 0.30 | −0.30 | 0.89 | 0.328 | |
| MTRR (rs10380) | 0.423 | |||||
| TT/CT | 79 | 0.66 | 0.02 | 1.30 | 0.043 | |
| CC | 292 | 0.37 | −0.01 | 0.74 | 0.054 | |
| Drinking statusa | 0.473 | |||||
| Non-drinker | 223 | 0.33 | −0.08 | 0.74 | 0.118 | |
| Former/Current | 150 | 0.56 | 0.06 | 1.06 | 0.028 | |
| Folate intakeb | 0.736 | |||||
| < 340 μg/day | 188 | 0.49 | 0.03 | 0.94 | 0.036 | |
| ≥340 μg/day | 185 | 0.38 | −0.06 | 0.83 | 0.092 | |
*Adjusted by age, BMI, smoking (never, former, current), exercise (none/week, 1–2 times/week, ≥3 times/week), drinking (non-drinker, sometimes, < 150 g/week, ≥150 g/week), and folate intake (quartile)
a Adjusted by age, BMI, smoking (never, former, current), exercise (none/week, 1–2 times/week, ≥3 times/week), and folate intake (quartile)
b Adjusted by age, BMI, smoking (never, former, current), exercise (none/week, 1–2 times/week, ≥3 times/week), and drinking (non-drinker, sometimes, < 150 g/week, ≥150 g/week)
c Average relative methylation difference per 1-category increase (included as continuous variable)
Table 3.
Adjusted estimates for mean methylation level stratified by quartile of serum CRP concentration
| 95% confidence interval | |||||
|---|---|---|---|---|---|
| MTHFR (rs1801133) | CRP (mg/dL) | N | Estimated mean methylation (%)a | Lower | Upper |
| CT/TT | 0.002–0.011 | 73 | 69.8 | 68.7 | 71.0 |
| 0.012–0.023 | 59 | 70.5 | 69.2 | 71.7 | |
| 0.024–0.048 | 63 | 71.4 | 70.1 | 72.7 | |
| 0.049–0.491 | 69 | 71.6 | 70.4 | 72.8 | |
| CC | 0.002–0.011 | 27 | 70.4 | 68.9 | 72.0 |
| 0.012–0.023 | 30 | 71.3 | 69.8 | 72.8 | |
| 0.024–0.048 | 29 | 71.3 | 69.8 | 72.8 | |
| 0.049–0.491 | 23 | 70.1 | 68.4 | 71.8 | |
aAdjusted by age, BMI, smoking (never, former, current), exercise (none/week, 1–2 times/week, ≥3 times/week), drinking (non-drinker, sometimes, < 150 g/week, ≥150 g/week), and folate intake (quartile). Estimates were calculated with average values of continuous variables
Table 4.
Assessment of interaction between serum CRP concentration and drinking status in global methylation levels in the MTHFR CT/TT group
| 95% confidence interval | 95% confidence interval | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MTHFR (rs1801133) | Drinking status | CRP (mg/dL) | N | Estimated mean methylation (%)a | Lower | Upper | βab | Lower | Upper | P value for trenda |
| CT/TT | Never | 0.002–0.011 | 43 | 69.5 | 67.9 | 71.0 | ||||
| 0.012–0.023 | 33 | 70.5 | 68.9 | 72.2 | ||||||
| 0.024–0.048 | 43 | 71.2 | 69.7 | 72.7 | ||||||
| 0.049–0.491 | 45 | 70.6 | 69.0 | 72.1 | ||||||
| 0.41 | −0.07 | 0.90 | 0.094 | |||||||
| Log-transformed CRP | 0.28 | −0.25 | 0.81 | 0.307 | ||||||
| Former/Current | 0.002–0.011 | 30 | 70.4 | 68.9 | 71.9 | |||||
| 0.012–0.023 | 26 | 70.3 | 68.6 | 72.0 | ||||||
| 0.024–0.048 | 20 | 71.2 | 69.3 | 73.1 | ||||||
| 0.049–0.491 | 24 | 72.8 | 71.1 | 74.6 | ||||||
| 0.83 | 0.23 | 1.44 | 0.007 | |||||||
| Log-transformed CRP | 0.88 | 0.26 | 1.50 | 0.006 | ||||||
aAdjusted by age, BMI, smoking (never, former, current), exercise (none/week, 1–2 times/week, ≥3 times/week), folate intake (quartiles). Estimates were calculated with average values of continuous variables
bAverage relative methylation difference per 1-category increase (included as continuous variable)
Graphical representation of the association of CRP and global methylation is shown in scatter plots (Fig. 1a) of the results in the CT/TT group stratified by BMI, which is potentially the strongest confounder in the data analysis summarized in Table 1 and Additional file 1: Table S1. Removing individual covariates from the model produced the greatest change in the CRP correlation coefficient after removing BMI. As shown in Fig. 1, a weakly positive correlation was observed for the MTHFR CT/TT and BMI < 22.6. The cutoff was the median BMI. Correlation coefficients for CRP were similar those shown above. When the subjects were limited to the former/current drinkers only, the correlation was stronger (Fig. 1b). Additionally, we also measured serum HP antibody and pepsinogen levels, which directly reflect current/former chronic inflammation of the gastric mucosa due to infection, and investigated their association with global methylation levels with and without adjustment by CRP concentration. Results are summarized in Additional file 2: Table S2. Briefly, HP positivity (defined by an antibody level ≥ 10 U/ml) and pepsinogen positivity (defined by a pepsinogen I level < 70 ng/mL and pepsinogen I/II ratios ≤3.0 U/ml) were not significantly associated with global methylation either with or without adjustment. CRP was still associated with global methylation even with these variables (P = 0.009).
Fig. 1.
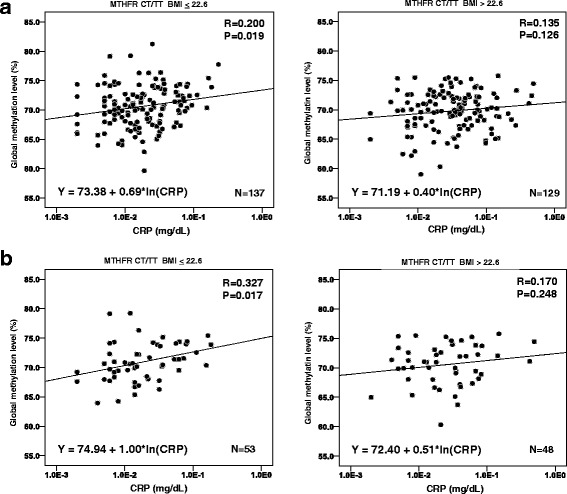
a Scatter plots for the correlation between CRP and global methylation in the CT/TT group stratified by the median of BMI. The scale of CRP was log-transformed. Formulas are calculated by single regression with natural logarithm of CRP and intercept. b Scatter plots for the correlation between CRP and global methylation in the CT/TT and former/current drinker group stratified by the median of BMI
Discussion
The study results show that serum CRP concentration was positively correlated with the global DNA methylation level of peripheral blood leukocytes. As mentioned in the introduction, it is widely known that chronic inflammation caused by infection or other factors is associated with aberrant DNA methylation, followed by cancer development [8, 13]. With respect to gastrointestinal inflammation, a potential association reported is that of the activation of reactive oxygen/nitrogen species with an increase in the expression of DNA methyltransferase (DNMT) [29, 30]. However, as DNA methylation increased by only 0.43% per quartile category for CRP in this study, the association should be considered weak even though it was significant. Few studies have assessed the association of methylation of peripheral leukocytes and CRP, but they reported no association between global LINE-1 methylation level and CRP [31, 32]. Thus, our finding needs to be interpreted with caution, especially because they can be explained by differences in white blood cell population [33]. Nevertheless, the SNP-and alcohol-associated interactions shown in Tables 3 and 4 indicate that the association we observed cannot be explained by these differences only.
Interestingly, we found that MTHFR genotype appears to modify the statistical association of CRP with global DNA methylation. The missense rs1801133 polymorphism (p.Ala222Val) in the MTHFR gene is known to affect enzyme activity, and its heterozygote CT and homozygotes TT have only around 65% and 30% of normal activity, respectively [34]. The loss of activity of MTHFR can lead to lower DNA methylation level through a dysfunction of conversion of 5,10-methylenetetrahydrofolate (5,10-MTHF) to 5-methyltetorahydrofolate (5-MTHF), which is a methyl donor for DNA methylation [35]. In a previous study, the MTHFR TT homozygote showed a lower methylation level when plasma folate level was low, whereas a significant positive correlation was not observed in the wild-type group (CC homozygote) [19]. That report also showed that MTHFR wild-type individuals maintain high methylation levels no matter how low plasma folate levels are. These findings suggested that variation in MTHFR activities can augment the variation of DNA methylation induced by other DNA methylation determinants (“epimutagens”). In our previous study, alcohol intake modified the association between folate intake level and global methylation level [15]. Alcohol consumption interferes with folate metabolism, which in turn disturbs methyl DNA synthesis. This suggests that modification of folate metabolism varies the regulation of DNA methylation by other epigenetically effective factors. With respect to the influence of alcohol, although there are some reports stating that excessive intake of alcohol is associated with peripheral blood global hypermethylation [36, 37], we did not observe a clear association between these aspects in our studies (15 and this study).
Regarding an association with diseases, a decrease in MTHFR activity does not necessarily facilitate cancer development. Zhou et al. reported that the homozygote of the minor allele of the rs1801133 polymorphism of the MTHFR (TT) gene was associated with reduced colorectal cancer risk [38], and low folate and high alcohol intake was more harmful for colorectal cancer incidence in the MTHFR wild type (CC) group than in TT group [39–44]. On the contrary, TT mutation was reported to be a risk factor for breast cancer incidence, and the association was strengthened by low folate intake or high alcohol intake [45–50]. In our present study, the MTHFR CT/TT group showed a stronger association between DNA methylation and CRP level than the other groups, particularly with alcohol consumption (Tables 2, 3 and 4). This suggests that different regulation patterns of DNA methylation (determined by MTHFR mutation, inflammation, alcohol consumption, folate intake etc.) are associated with different risks of disease. Although regulation of DNA methylation and its consequences are not monotonous, these past and our present findings suggest that the effects of potential epimutagens differ among individuals depending on genetic and other environmental status.
In addition, BMI had a strong influence as a confounding factor in this study. The background pertaining to confounding by BMI is considered as follows: there is an intermediate correlation between BMI and CRP, with the coefficient R being 0.265 (P < 0.001), and BMI is negatively correlated with global methylation, as shown in Additional file 1: Table S1. There are some reports stating that global DNA methylation is negatively correlated with numerical indicators of obesity, including BMI; this is consistent with our findings [51, 52].
Limitations of this study are as follows. First, since the study was conducted under a cross-sectional design, we cannot elucidate the causality of the observed associations. Second, although serum CRP concentration is an established predictive factor for many kind of diseases, including cancer and cardiovascular diseases [21–24, 53], particularly when assayed by the high-sensitivity method (see Materials and Methods), it is easily changed by acute infection or other cause of inflammation. To avoid misclassification and weakened associations, serum CRP concentration should ideally be measured in the steady state. Third, repeated comparisons for multiple subgroups were performed, which might have led to over-fitted and false-positive results by chance. Several associations we observed can be considered weak in spite of their statistical significance. As we did not investigate white blood cell population, change with systemic inflammation cannot be excluded. Finally, sample size was limited, and the study lacks sufficient statistical power to detect small effects and statistical interaction.
Conclusion
Overall, our study suggests that systemic inflammation suggested by higher serum CRP is weakly associated with the global DNA hypermethylation of peripheral blood leukocytes. However, this association was more clearly seen among individuals carrying the minor allele of the MTHFR rs1801133 missense SNP (known as C677T), which has been shown to have induce reduced enzymatic activity and to cause global hypomethylation when folate level is low. Our results and previous findings also suggest that global DNA methylation is determined by many factors, including folate level, degree of systemic inflammation, metabolic status, and food and beverages intake, and that these items interact in a complex way. A comprehensive understanding of these interactions between acquired and genetic factors is critical to the realization of “tailor-made cancer prevention”.
Additional files
Table S1. Details of the models for the assessment of CRP and global methylation in all subjects. (DOCX 19 kb)
Table S2. Assessment of association between HP infection and global methylation levels with/without adjustment with life-style factors, folate intake and serum CRP concentration. (DOCX 20 kb)
Acknowledgments
We are deeply grateful to Dr. Hiroe Ono for conducting the DNA methylation assay, and Dr. Hiromi Sakamoto for her helpful advice on this assay. We also thank Ms. Yoko Odaka and Misuzu Okuyama for technical assistance.
Funding
This study was supported by Grants-in-Aid for the Third Term Comprehensive Ten-Year Strategy for Cancer Control (H18-3jigan-ippan-001 and H21-3jigan-ippan-002, S Tsugane) and for the Research on Applying Health Technology from the Ministry of Health, Labour and Welfare of Japan (H23-jitsuyoka(gan)-ippan-002, T Yoshida and M Iwasaki); by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO) (10–41, T Yoshida); and by a Health and Labour Sciences Research Grant for Research on the Risk of Chemical Substances from the Ministry of Health, Labour and Welfare of Japan (H14-shokuhin kagaku-015 and H17-kagaku-014, S Tsugane and M Iwasaki).
Availability of data and materials
The data that support the findings of this study are available on request from the corresponding author.
Abbreviations
- BMI
Body mass index
- CRP
C-reactive-protein
- HP
H. pylori
- LUMA
Luminometric Methylation Assay
Authors’ contributions
All authors were involved with the study concept and design. MI, YK, SY, HO, HN, RK, and ST participated in the acquisition of data. TY measured global DNA methylation level and genotyping. MN, MI, TY, and ST contributed to the analysis and interpretation of data. MN and MI conducted the statistical analyses and MN wrote the manuscript. All authors participated in the interpretation of results and critical revision of the manuscript for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from 405 matched pairs. The study protocol was approved by the institutional review boards of the National Cancer Center (2010–188) and the Institute of Medical Science, The University of Tokyo (26-23), Tokyo, Japan.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4089-z) contains supplementary material, which is available to authorized users.
Contributor Information
Masanori Nojima, Email: nojima@ims.u-tokyo.ac.jp.
Motoki Iwasaki, Phone: +81-3-3542-2511, Email: moiwasak@ncc.go.jp.
Yoshio Kasuga, Email: ykasuga@hosp.nagano-matsushiro.or.jp.
Shiro Yokoyama, yokoyama@med.email.ne.jp.
Hiroshi Onuma, Email: inadaclinic@krd.biglobe.ne.jp.
Hideki Nishimura, Email: nisimura@hospital.nagano.nagano.jp.
Ritsu Kusama, Email: kusamadayo@ai.wakwak.com.
Teruhiko Yoshida, Email: tyoshida@ncc.go.jp.
Shoichiro Tsugane, Email: stsugane@ncc.go.jp.
References
- 1.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 2.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki Y, Nojima M, Suzuki H, Yasui H, Maruyama R, Yamamoto E, et al. Genomic vulnerability to LINE-1 hypomethylation is a potential determinant of the clinicogenetic features of multiple myeloma. Genome Med. 2012;4:101. doi: 10.1186/gm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu X, Gammon MD, Hernandez-Vargas H, Herceg Z, Wetmur JG, Teitelbaum SL, et al. DNA methylation in peripheral blood measured by LUMA is associated with breast cancer in a population-based study. FASEB J. 2012;26:2657–2666. doi: 10.1096/fj.11-197251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friso S, Udali S, Guarini P, Pellegrini C, Pattini P, Moruzzi S, et al. Global DNA hypomethylation in peripheral blood mononuclear cells as a biomarker of cancer risk. Cancer Epidemiol Biomark Prev. 2013;22:348–355. doi: 10.1158/1055-9965.EPI-12-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS One. 2012;7:e34615. doi: 10.1371/journal.pone.0034615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuchiba A, Iwasaki M, Ono H, Kasuga Y, Yokoyama S, Onuma H, et al. Global methylation levels in peripheral blood leukocyte DNA by LUMA and breast cancer: a case-control study in Japanese women. Br J Cancer. 2014;110:2765–2771. doi: 10.1038/bjc.2014.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- 9.Liu Y, Ding J, Reynolds LM, Lohman K, Register TC, De La Fuente A, et al. Methylomics of gene expression in human monocytes. Hum Mol Genet. 2013;22:5065–5074. doi: 10.1093/hmg/ddt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds LM, Taylor JR, Ding J, Lohman K, Johnson C, Siscovick D, et al. Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nat Commun. 2014;5:5366. doi: 10.1038/ncomms6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maegawa S, Gough SM, Watanabe-Okochi N, Lu Y, Zhang N, Castoro RJ, et al. Age-related epigenetic drift in the pathogenesis of MDS and AML. Genome Res. 2014;24:580–591. doi: 10.1101/gr.157529.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet. 2014;10:e1004458. doi: 10.1371/journal.pgen.1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto E, Toyota M, Suzuki H, Kondo Y, Sanomura T, Murayama Y, et al. LINE-1 hypomethylation is associated with increased CpG island methylation in helicobacter pylori-related enlarged-fold gastritis. Cancer Epidemiol Biomark Prev. 2008;17:2555–2564. doi: 10.1158/1055-9965.EPI-08-0112. [DOI] [PubMed] [Google Scholar]
- 14.Abu-Remaileh M, Bender S, Raddatz G, Ansari I, Cohen D, Gutekunst J, et al. Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res. 2015;75:2120–2130. doi: 10.1158/0008-5472.CAN-14-3295. [DOI] [PubMed] [Google Scholar]
- 15.Ono H, Iwasaki M, Kuchiba A, Kasuga Y, Yokoyama S, Onuma H, et al. Association of dietary and genetic factors related to one-carbon metabolism with global methylation level of leukocyte DNA. Cancer Sci. 2012;103:2159–2164. doi: 10.1111/cas.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–48. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- 17.Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, et al. LUMA (LUminometric methylation assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–1995. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki M, Ono H, Kuchiba A, Kasuga Y, Yokoyama S, Onuma H, et al. Association of postmenopausal endogenous sex hormones with global methylation level of leukocyte DNA among Japanese women. BMC Cancer. 2012;12:323. doi: 10.1186/1471-2407-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iso H, Noda H, Ikeda A, Yamagishi K, Inoue M, Iwasaki M, et al. The impact of C-reactive protein on risk of stroke, stroke subtypes, and ischemic heart disease in middle-aged Japanese: the Japan public health center-based study. J Atheroscler Thromb. 2012;19:756–766. [PubMed] [Google Scholar]
- 21.Toriola AT, Grankvist K, Agborsangaya CB, Lukanova A, Lehtinen M, Surcel HM. Changes in pre-diagnostic serum C-reactive protein concentrations and ovarian cancer risk: a longitudinal study. Ann Oncol. 2011;22:1916–1921. doi: 10.1093/annonc/mdq694. [DOI] [PubMed] [Google Scholar]
- 22.Dossus L, Jimenez-Corona A, Romieu I, Boutron-Ruault MC, Boutten A, Dupre T, et al. C-reactive protein and postmenopausal breast cancer risk: results from the E3N cohort study. Cancer Causes Control. 2014;25:533–539. doi: 10.1007/s10552-014-0355-9. [DOI] [PubMed] [Google Scholar]
- 23.Ko YJ, Kwon YM, Kim KH, Choi HC, Chun SH, Yoon HJ, et al. High-sensitivity C-reactive protein levels and cancer mortality. Cancer Epidemiol Biomark Prev. 2012;21:2076–2086. doi: 10.1158/1055-9965.EPI-12-0611. [DOI] [PubMed] [Google Scholar]
- 24.Shiels MS, Pfeiffer RM, Hildesheim A, Engels EA, Kemp TJ, Park JH, et al. Circulating inflammation markers and prospective risk for lung cancer. J Natl Cancer Inst. 2013;105:1871–1880. doi: 10.1093/jnci/djt309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh H, Iwasaki M, Hanaoka T, Kasuga Y, Yokoyama S, Onuma H, et al. Serum organochlorines and breast cancer risk in Japanese women: a case-control study. Cancer Causes Control. 2009;20:567–580. doi: 10.1007/s10552-008-9265-z. [DOI] [PubMed] [Google Scholar]
- 26.Ma E, Iwasaki M, Kobayashi M, Kasuga Y, Yokoyama S, Onuma H, et al. Dietary intake of folate, vitamin B2, vitamin B6, vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer: a case-control study in Japan. Nutr Cancer. 2009;61:447–456. doi: 10.1080/01635580802610123. [DOI] [PubMed] [Google Scholar]
- 27.Ishihara J, Inoue M, Kobayashi M, Tanaka S, Yamamoto S, Iso H, et al. Impact of the revision of a nutrient database on the validity of a self-administered food frequency questionnaire (FFQ) J Epidemiol. 2006;16:107–116. doi: 10.2188/jea.16.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsubono Y, Takamori S, Kobayashi M, Takahashi T, Iwase Y, Iitoi Y, et al. A data-based approach for designing a semiquantitative food frequency questionnaire for a population-based prospective study in Japan. J Epidemiol. 1996;6:45–53. doi: 10.2188/jea.6.45. [DOI] [PubMed] [Google Scholar]
- 29.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 30.Na HK, Woo JH. Helicobacter pylori induces Hypermethylation of CpG Islands through upregulation of DNA methyltransferase: possible involvement of reactive oxygen/nitrogen species. J Cancer Prev. 2014;19:259–264. doi: 10.15430/JCP.2014.19.4.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, et al. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics. 2010;5:222–228. doi: 10.4161/epi.5.3.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang FF, Santella RM, Wolff M, Kappil MA, Markowitz SB, Morabia A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics. 2012;7:606–614. doi: 10.4161/epi.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardenas A, Allard C, Doyon M, Houseman EA, Bakulski KM, Perron P, et al. Validation of a DNA methylation reference panel for the estimation of nucleated cells types in cord blood. Epigenetics. 2016; in press [DOI] [PMC free article] [PubMed]
- 34.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 35.Stern LL, Mason JB, Selhub J, Choi SW, Genomic DNA. Hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomark Prev. 2000;9:849–853. [PubMed] [Google Scholar]
- 36.Semmler A, Heese P, Stoffel-Wagner B, Muschler M, Heberlein A, Bigler L, Prost JC, Frieling H, Kornhuber J, Banger M, Bleich S, Hillemacher T, Linnebank M. Alcohol abuse and cigarette smoking are associated with global DNA hypermethylation: results from the German investigation on neurobiology in alcoholism (GINA) Alcohol. 2015;49:97–101. doi: 10.1016/j.alcohol.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Kim DS, Kim YH, Lee WK, Na YK, Hong HS. Effect of alcohol consumption on peripheral blood Alu methylation in Korean men. Biomarkers. 2016;21:243–248. doi: 10.3109/1354750X.2015.1134661. [DOI] [PubMed] [Google Scholar]
- 38.Zhou D, Mei Q, Luo H, Tang B, Yu P. The polymorphisms in methylenetetrahydrofolate reductase, methionine synthase, methionine synthase reductase, and the risk of colorectal cancer. Int J Biol Sci. 2012;8:819–830. doi: 10.7150/ijbs.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, Stampfer MJ, Giovannucci E, Artigas C, Hunter DJ, Fuchs C, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098–1102. [PubMed] [Google Scholar]
- 40.Chen J, Giovannucci E, Kelsey K, Rimm EB, Stampfer MJ, Colditz GA, et al. A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res. 1996;56:4862–4864. [PubMed] [Google Scholar]
- 41.Ulrich CM, Kampman E, Bigler J, Schwartz SM, Chen C, Bostick R, et al. Colorectal adenomas and the C677T MTHFR polymorphism: evidence for gene-environment interaction? Cancer Epidemiol Biomark Prev. 1999;8:659–668. [PubMed] [Google Scholar]
- 42.Ma J, Stampfer MJ, Christensen B, Giovannucci E, Hunter DJ, Chen J, et al. A polymorphism of the methionine synthase gene: association with plasma folate, vitamin B12, homocyst(e)ine, and colorectal cancer risk. Cancer Epidemiol Biomark Prev. 1999;8:825–829. [PubMed] [Google Scholar]
- 43.Lightfoot TJ, Barrett JH, Bishop T, Northwood EL, Smith G, Wilkie MJ, et al. Methylene tetrahydrofolate reductase genotype modifies the chemopreventive effect of folate in colorectal adenoma, but not colorectal cancer. Cancer Epidemiol Biomark Prev. 2008;17:2421–2430. doi: 10.1158/1055-9965.EPI-08-0058. [DOI] [PubMed] [Google Scholar]
- 44.Matsuo K, Ito H, Wakai K, Hirose K, Saito T, Suzuki T, et al. One-carbon metabolism related gene polymorphisms interact with alcohol drinking to influence the risk of colorectal cancer in Japan. Carcinogenesis. 2005;26:2164–2171. doi: 10.1093/carcin/bgi196. [DOI] [PubMed] [Google Scholar]
- 45.Lin WY, Chou YC. Wu MH, Huang HB, Jeng YL, Wu CC et al: the MTHFR C677T polymorphism, estrogen exposure and breast cancer risk: a nested case-control study in Taiwan. Anticancer Res. 2004;24:3863–3868. [PubMed] [Google Scholar]
- 46.Shrubsole MJ, Gao YT, Cai Q, Shu XO, Dai Q, Hebert JR, et al. MTHFR polymorphisms, dietary folate intake, and breast cancer risk: results from the shanghai breast cancer study. Cancer Epidemiol Biomark Prev. 2004;13:190–196. doi: 10.1158/1055-9965.EPI-03-0273. [DOI] [PubMed] [Google Scholar]
- 47.Ericson U, Sonestedt E, Ivarsson MI, Gullberg B, Carlson J, Olsson H, et al. Folate intake, methylenetetrahydrofolate reductase polymorphisms, and breast cancer risk in women from the Malmo diet and cancer cohort. Cancer Epidemiol Biomark Prev. 2009;18:1101–1110. doi: 10.1158/1055-9965.EPI-08-0401. [DOI] [PubMed] [Google Scholar]
- 48.He JM, Pu YD, Wu YJ, Qin R, Zhang QJ, Sun YS, et al. Association between dietary intake of folate and MTHFR and MTR genotype with risk of breast cancer. Genet Mol Res. 2014;13:8925–8931. doi: 10.4238/2014.October.31.7. [DOI] [PubMed] [Google Scholar]
- 49.Platek ME, Shields PG, Marian C, McCann SE, Bonner MR, Nie J, et al. Alcohol consumption and genetic variation in methylenetetrahydrofolate reductase and 5-methyltetrahydrofolate-homocysteine methyltransferase in relation to breast cancer risk. Cancer Epidemiol Biomark Prev. 2009;18(9):2453. doi: 10.1158/1055-9965.EPI-09-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maruti SS, Ulrich CM, Jupe ER, White E. MTHFR C677T and postmenopausal breast cancer risk by intakes of one-carbon metabolism nutrients: a nested case-control study. Breast Cancer Res. 2009;11:R91. doi: 10.1186/bcr2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perng W, Mora-Plazas M, Marín C, Rozek LS, Baylin A, Villamor E. A prospective study of LINE-1 DNA methylation and development of adiposity in school-age children. PLoS One. 2013;8:e62587. doi: 10.1371/journal.pone.0062587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de la Rocha C, Pérez-Mojica JE, León SZ, Cervantes-Paz B, Tristán-Flores FE, Rodríguez-Ríos D, et al. Associations between whole peripheral blood fatty acids and DNA methylation in humans. Sci Rep. 2016;6:25867. doi: 10.1038/srep25867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaptoge S, Di Angelantonio E, Pennells L, Wood AM, White IR, Gao P, et al. C-reactive protein, fibrinogen, and cardiovascular disease prediction. N Engl J Med. 2012;367:1310–1320. doi: 10.1056/NEJMoa1107477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Details of the models for the assessment of CRP and global methylation in all subjects. (DOCX 19 kb)
Table S2. Assessment of association between HP infection and global methylation levels with/without adjustment with life-style factors, folate intake and serum CRP concentration. (DOCX 20 kb)
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


