Abstract
Background. During the 2014–2015 US influenza season, expanded genetic characterization of circulating influenza A(H3N2) viruses was used to assess the impact of the genetic variability of influenza A(H3N2) viruses on influenza vaccine effectiveness (VE).
Methods. A novel pyrosequencing assay was used to determine genetic group, based on hemagglutinin (HA) gene sequences, of influenza A(H3N2) viruses from patients enrolled at US Influenza Vaccine Effectiveness Network sites. VE was estimated using a test-negative design comparing vaccination among patients infected with influenza A(H3N2) viruses and uninfected patients.
Results. Among 9710 enrollees, 1868 (19%) tested positive for influenza A(H3N2) virus; genetic characterization of 1397 viruses showed that 1134 (81%) belonged to 1 HA genetic group (3C.2a) of antigenically drifted influenza A(H3N2) viruses. Effectiveness of 2014–2015 influenza vaccination varied by influenza A(H3N2) virus genetic group from 1% (95% confidence interval [CI], −14% to 14%) against illness caused by antigenically drifted influenza A(H3N2) virus group 3C.2a viruses versus 44% (95% CI, 16%–63%) against illness caused by vaccine-like influenza A(H3N2) virus group 3C.3b viruses.
Conclusions. Effectiveness of 2014–2015 influenza vaccination varied by genetic group of influenza A(H3N2) virus. Changes in HA genes related to antigenic drift were associated with reduced VE.
Keywords: influenza, genetic characterization, pyrosequencing, influenza vaccine, vaccine effectiveness
(See the editorial commentary by Schotsaert and García-Sastre on page 982.)
The 2014–2015 influenza season was characterized by widespread circulation of influenza A(H3N2) viruses, with high levels of outpatient illness and influenza-associated hospitalization, especially for adults aged ≥65 years [1]. Characterization of influenza A(H3N2) viruses that circulated in the United States indicated that the majority were antigenically different (drifted) from the A(H3N2) vaccine component of 2014–2015 seasonal influenza vaccines for the northern hemisphere [2]. Antigenic relatedness of vaccine strains and circulating viruses is traditionally determined by the hemagglutination inhibition (HI) assay, which measures inhibition of virus-mediated agglutination of red blood cells by reference antisera from ferrets inoculated with vaccine viruses [3]. During the 2014–2015 season, changes in the properties of many influenza A(H3N2) viruses resulted in insufficient hemagglutination activity for characterization by HI [1]. In response, sequencing of hemagglutinin (HA) genes was increasingly used to determine the genetic relatedness of antigenic variants of influenza A(H3N2) viruses [4]. Few studies have evaluated differences in influenza vaccine effectiveness (VE) according to genetic relatedness among influenza viruses [5, 6].
The multisite US Influenza Vaccine Effectiveness Network (Flu VE network) estimates influenza VE each season; interim estimates of VE were 23% (95% confidence interval [CI], 8%–36%) among patients enrolled during 10 November 2014–2 January 2015, when most influenza illnesses were due to influenza A(H3N2) viruses [7]. To improve our ability to characterize the effect of antigenically drifted influenza A(H3N2) viruses on influenza VE, we used a newly developed, high-throughput pyrosequencing assay (unpublished data) to genetically characterize influenza A(H3N2) viruses directly from respiratory specimens collected from patients enrolled at participating sites in the Flu VE network. We report VE for the most common influenza A(H3N2) virus genetic groups circulating in the United States during 2014–2015 and for the subset of influenza A(H3N2) viruses that could not be genetically characterized. In addition, to better understand the genetic variability and geographic and temporal distribution of circulating influenza A(H3N2) viruses in the United States during 2014–2015, we describe the influenza A(H3N2) virus genetic groups among US surveillance viruses during the 2014–2015 influenza season.
MATERIALS AND METHODS
Flu VE Network
Methods of the Flu VE network have been previously described [8–10]. Briefly, patients aged ≥6 months seeking outpatient medical care for an acute respiratory illness within 7 days of illness onset were enrolled at study sites in Michigan, Pennsylvania, Texas, Washington, and Wisconsin during periods of influenza virus circulation at each site. Presence of a high-risk medical condition was defined as documentation of any International Classification of Diseases, Ninth Revision (ICD-9) diagnostic code associated with specified high-risk conditions [11] in the patient's medical record during the 12 months before enrollment. Receipt of 2014–2015 influenza vaccine was confirmed by review of medical records or immunization registries; patients who self-reported vaccination not captured by medical records or registries were assumed to be vaccinated if plausible information on date and location of vaccine receipt was provided. Receipt of prior season (2013–2014) influenza vaccine was obtained from medical records or immunization registries only. We excluded patients vaccinated 0–13 days before illness onset and those aged 6 months–8 years who received only 1 dose of 2014–2015 influenza vaccine, for whom 2 doses were recommended by the Advisory Committee on Immunization Practices [11].
Respiratory specimens (combined nasal and oropharyngeal swab specimens for patients aged ≥2 years; nasal swab specimens only for patients aged 6–23 months) were collected and tested for influenza viruses at Flu VE network laboratories by real-time reverse-transcription polymerase chain reaction (rRT-PCR) [12]. A subset of influenza virus–positive specimens from each network site, including the first 10 positive specimens identified and up to 5 positive specimens identified every 2 weeks during the enrollment period, were sent to National Influenza Surveillance Reference Centers for virus isolation and propagation for antigenic characterization by HI. Given challenges with antigenic characterization of many influenza A(H3N2) viruses, influenza A(H3N2) virus–positive respiratory specimens containing high levels of influenza virus RNA (defined as rRT-PCR cycle threshold values of ≤30) were sent to the Centers for Disease Control and Prevention (CDC; Atlanta, Georgia) for pyrosequencing directly from respiratory specimens. All influenza A(H3N2) virus–positive specimens from the Pennsylvania Flu VE network site were processed by pyrosequencing because, in contrast to other locations, early testing suggested widespread circulation of vaccine-like strains.
Virologic Surveillance
Distribution of influenza A(H3N2) viruses in the United States before and during the 2014–2015 influenza season was examined using virologic surveillance data for influenza A(H3N2) viruses identified by US public health laboratories from 1 March 2014 through 10 April 2015 and submitted to the CDC for virus characterization. The CDC requested that state public health laboratories submit the first 10 influenza virus–positive specimens of any virus type or subtype and up to 5 positive specimens every 2 weeks throughout the season to the CDC. For influenza A(H3N2) viruses that did not achieve sufficient hemagglutination titers for antigenic characterization by HI, genetic characterization was used to determine HA genetic group, and antigenic properties were inferred on the basis of the subset of viruses from the same HA genetic group that could be characterized by HI assays [1].
Genetic Characterization
Analyses of full-length HA sequences from influenza A(H3N2) viruses collected from January 1 through 1 December 2014 and characterized at the CDC were used to design a high-throughput pyrosequencing assay to screen influenza A(H3N2) viruses for genetic markers associated with 6 major HA genetic groups (3C.2, 3C.2a, 3C.2b, 3C.3, 3C.3a, and 3C.3b). Viral RNA was extracted from clinical specimens in a 96-well plate format, using the MagNA Pure 96 nucleic acid isolation system (Roche Diagnostics, Basel, Switzerland). A fragment of the HA1 coding region, encompassing nucleotides 370–645, was amplified using RT-PCR and subjected to sequencing analysis using a PyroMark Q96ID sequencing platform (Qiagen, Valencia, California), essentially as described previously [13]. Three pyrosequencing primers were used to generate pyrograms for 3 target HA regions encompassing the following nucleotides: (1) 412–431, (2) 456–481, and (3) 559–571. Sequencing results were analyzed using IdentiFire software (Qiagen, Valencia, California) and a purpose-built library containing the signature sequences of target regions. Unique combination of short-signature sequences within each target region allows identification of 3C.2, 3C.2a, 3C.2b, 3C.3, 3C.3a, and 3C.3b clades (protocol for the pyrosequencing assay is available upon request at: fluantiviral@cdc.gov).
In addition, for a subset of viruses, full-length HA sequences were obtained from whole-genome sequencing, performed as previously described [14]. Sequence data were deposited to the Global Initiative on Sharing All Influenza Data. Influenza A(H3N2) viruses were classified into previously described HA genetic groups, based on phylogenetic analysis [15].
2014–2015 Vaccine Components and Genetic Groups
The reference influenza A(H3N2) virus for the 2014–2015 northern hemisphere vaccines was A/Texas/50/2012(H3N2), HA genetic group 3C.1. For antigenic characterization of circulating influenza A(H3N2) viruses, the CDC used a panel of ferret antisera raised against egg- and cell-propagated reference viruses, including A/Texas/50/2012, A/Michigan/15/2014 (group 3C.2a), A/Louisiana/39/2013 (group 3C.2b), A/New York/39/2012 (group 3C.3), A/Pennsylvania/09/2015 (group 3C.3b), and A/Switzerland/9715293/2013 (group 3C.3a), the reference influenza A(H3N2) virus for the 2015–2016 northern hemisphere vaccines.
Determination of VE
Among all patients enrolled in the Flu VE network study, we compared characteristics of patients with genetically characterized influenza A(H3N2) virus infection and all influenza A(H3N2) virus–positive patients to influenza virus–negative patients, excluding patients infected with other influenza viruses. We tested the statistical significance of differences by using the χ2 test for categorical variables and the Wilcoxon Mann–Whitney test for continuous variables. Influenza VE was estimated using a test-negative design [16, 17], which compares the odds of 2014–2015 influenza vaccine receipt among influenza A(H3N2) virus–positive patients (for any influenza A[H3N2] virus and by influenza A[H3N2] virus genetic group) with influenza test-negative patients. VE was estimated as 100% × [1 − adjusted odds ratios (ORs)] from logistic regression models by including network site, patient age, presence of high-risk medical condition, and calendar time as previously described [9]; addition of sex, race/Hispanic ethnicity, and days from illness onset to specimen collection did not change the adjusted OR by ≥5%, the predetermined threshold for inclusion in the model [9]. Logistic regression models for patients aged ≥6 months included patient age (in years, modeled using linear tail-restricted cubic spline functions with 5 knots based on percentiles); models for specific age categories included patient's age in years. We assessed potential effect modification by prior season (2013–2014) vaccination status by including main effects and an interaction term for current and prior season vaccination in logistic regression models [10]. Additional analyses were conducted, with one restricting patients to those enrolled within 4 days of symptom onset and another using an expanded group of patients that included partially vaccinated children. All reported tests were 2-sided, and a P value of <.05 was considered statistically significant; VE was considered statistically significant if 95% CIs excluded 0. Statistical analyses were conducted using SAS for Windows (version 9.3; Cary, North Carolina).
RESULTS
At US Flu VE network sites, 9710 patients aged ≥6 months with acute respiratory illness were enrolled during periods of influenza virus circulation, from 10 November 2014 through 10 April 2015; 1868 (19%) patients tested positive for influenza A(H3N2) virus, while 7390 (75%) patients tested negative for influenza virus. In addition, 4 patients (<1%) were infected with 2009 pandemic influenza A(H1N1) virus, and 399 (4%) were infected with influenza B viruses, including 2 coinfected with influenza A(H3N2) and B viruses. VE against influenza B viruses will be published separately.
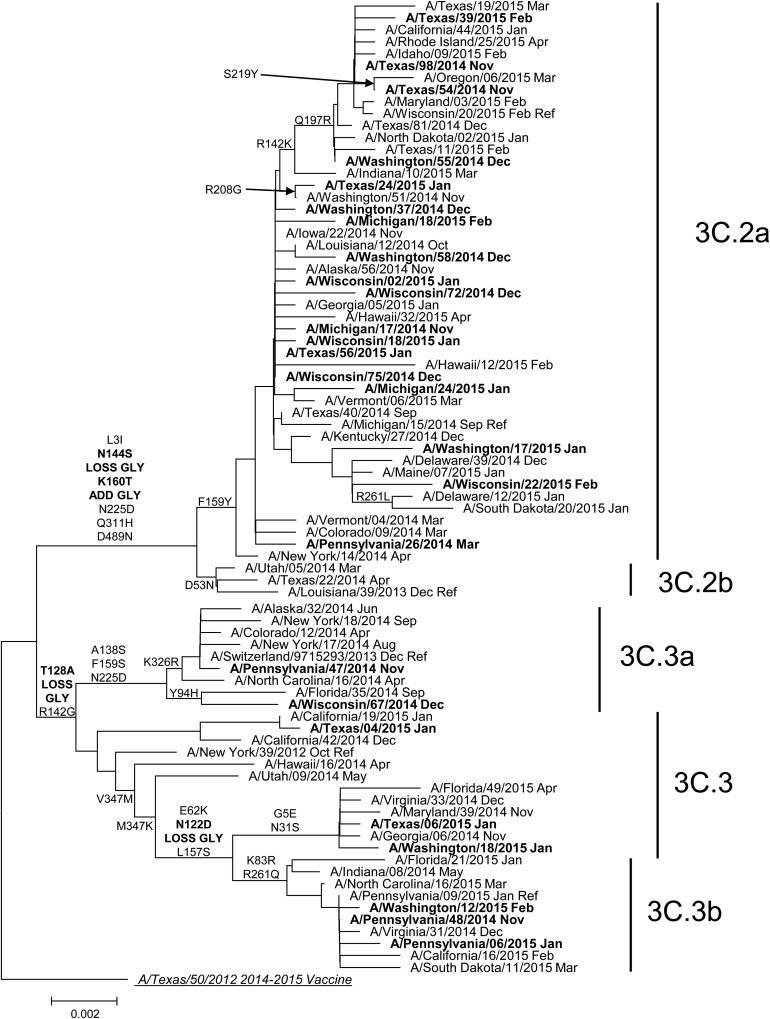
Genetic group was determined by pyrosequencing assay for 1397 of 1464 influenza A(H3N2) virus–positive US Flu VE patient specimens (95%) selected for genetic characterization at the CDC (78% of all influenza A(H3N2) virus–positive cases). Proportions of patients for whom influenza A(H3N2) virus genetic group was determined differed by study site, age, race/Hispanic ethnicity, and self-rated illness severity (P < .05) but were similar by vaccination status (Supplementary Table 1). For 138 specimens characterized by complete HA sequencing and pyrosequencing, results were 100% concordant. Influenza A(H3N2) virus genetic group 3C.2a predominated, accounting for 1134 of 1397 genetically characterized A(H3N2) viruses (81%) from Flu VE network sites. Compared with the 2014–2015 vaccine reference virus A/Texas/50/2012 (group 3C.1; Figure 1), HA sequences from group 3C.2a viruses shared an amino acid change from phenylalanine to tyrosine at position 159 (F159Y), which has been associated with reduced inhibition by ferret antisera raised against the vaccine virus. Of 35 influenza A(H3N2) group 3C.2a viruses from the Flu VE network study characterized by HI, 31 (89%) were poorly inhibited by ferret antisera against the A/Texas/50/2012 vaccine reference virus, in agreement with data from US virologic surveillance (Supplementary Table 2). Among other influenza A(H3N2) viruses from Flu VE network patients, 56 A(H3N2) viruses (4%) belonged to the antigenically drifted genetic group 3C.3a, 48 (3%) to vaccine-like group 3C.3, and 159 (11%) to group 3C.3b. Group 3C.3a viruses, including the 2015–2016 A(H3N2) vaccine reference virus A/Switzerland/9715293/2013 (Figure 1), had a parallel amino acid substitution to group 3C.2a viruses at position 159 (phenylalanine to serine [F159S]) but were considered antigenically similar to group 3C.2a viruses by HI [18]. Group 3C.3b viruses included an amino acid change from lysine to serine at position 157 (L157S) that was not associated with antigenic drift: 10 of 16 A(H3N2) group 3C.3b viruses (63%) from Flu VE network patients were considered antigenically similar to A/Texas/50/2012 by HI, similar to proportions observed among US surveillance isolates (Supplementary Table 2).
Figure 1.
Phylogenetic tree based on hemagglutinin (HA) genes of influenza A(H3N2) viruses collected during March 2014–April 2015 and analyzed at the Centers for Disease Control and Prevention (Atlanta, GA). Phylogenetic tree is based on genetic differences as compared to the A/Texas/50/2012 consensus sequence. Amino acid substitutions delineating major branches are shown. Date of collection follows names of viruses from US public health laboratories participating in US virologic surveillance (normal font) and from patients enrolled at US Influenza Vaccine Effectiveness Network sites (bold font).
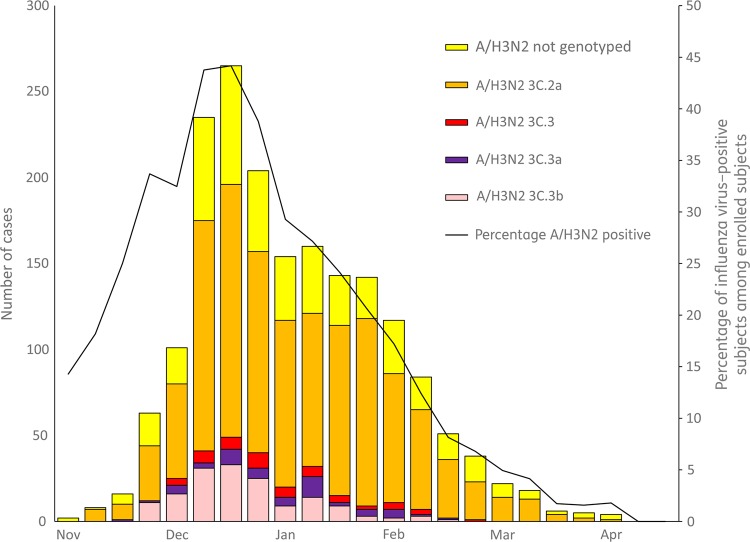
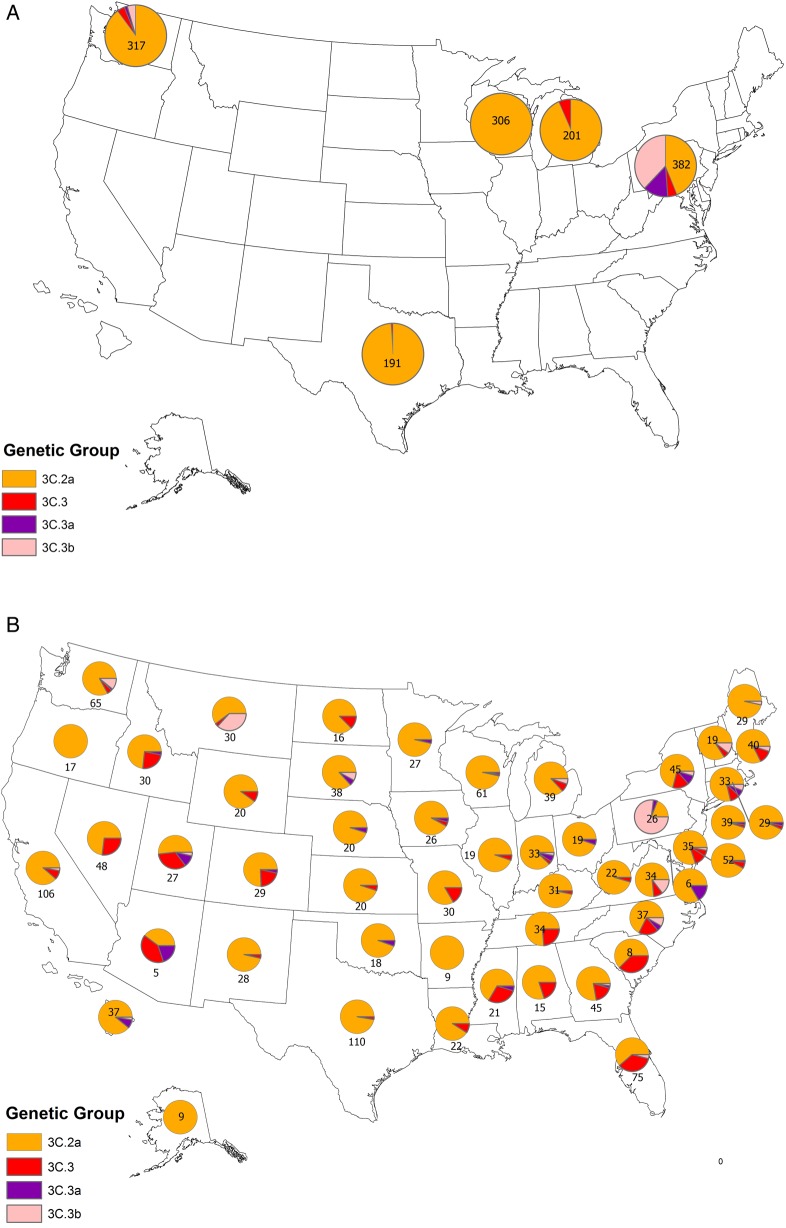
Prior to the enrollment period at Flu VE network sites, the proportion of influenza A(H3N2) viruses belonging to HA group 3C.2a that were submitted to the CDC from US surveillance laboratories increased from 38% (of 92 viruses characterized) in March 2014 to 73% (of 327) in November 2014. At Flu VE network sites, the proportion of group 3C.2a viruses increased during the 2014–2015 season; peak numbers of group 3C.2a–positive patients were enrolled in December, corresponding to peak case enrollment (Figure 2). While group 3C.2a viruses predominated, distribution of genetic group varied by Flu VE network site (Figure 3A), with greater circulation of vaccine-like strains at the Pennsylvania site. Similar variability in the geographic distribution of HA genetic group was observed among influenza A(H3N2) viruses collected by US public health laboratories during November 2014—April 2015 (Figure 3B).
Figure 2.
Numbers of polymerase chain reaction (PCR)–confirmed infections with influenza A(H3N2) viruses by genetic group and percentage influenza virus positivity among patients with medically-attended acute respiratory illness, by week of symptom onset, US Influenza Vaccine Effectiveness Network, 10 November 2014–10 April 2015.
Figure 3.
Geographic distribution of genetic groups of influenza A(H3N2) viruses in the United States and from patients enrolled in the US Influenza Vaccine Effectiveness Network study. A, Viruses from influenza A(H3N2) virus–positive patients enrolled from 10 November 2014 through 10 April 2015 (n = 1397). Pie charts present the proportional distribution of hemagglutinin (HA) genetic group, based on the number of genetically characterized influenza A(H3N2) viruses from patients enrolled at each study site. B, Viruses identified by US public health laboratories from 10 November 2014 through 10 April 2015 and submitted to the Centers for Disease Control and Prevention for genetic characterization (n = 1633). Pie charts present the proportional distribution of HA genetic group, based on the number of genetically characterized influenza A(H3N2) viruses from each state or territory.
Estimates of VE
A total of 1817 influenza A(H3N2) virus–positive patients and 7079 influenza virus test–negative patients from the Flu VE network study contributed to VE estimates (Table 1), after excluding 399 influenza B virus–positive patients, 175 incompletely vaccinated patients aged 6 months to 8 years, 149 patients vaccinated 0–13 days before illness onset, and 40 influenza virus test–negative patients with illness onset outside the period when influenza cases were identified at each study site. Compared with influenza virus test–negative patients, higher percentages of influenza A(H3N2) virus–positive patients were aged ≥65 years, white non-Hispanic, reported excellent or very good general health, enrolled within 2 days of illness onset, and reported feverishness during current illness. Influenza virus test–negative patients were more likely to report cigarette smoking or household exposure to smoke and to live in households with ≥1 child aged <12 years.
Table 1.
Characteristics of Patients Enrolled in the US Flu Vaccine Effectiveness Network Study, by Influenza Virus–Specific Real-Time Reverse-Transcription Polymerase Chain Reaction (rRT-PCR) Resulta
| Variable | Influenza A(H3N2) Virus–Positive Cases |
Influenza Virus–Negative Controls (n = 7078) | P Valueb | |
|---|---|---|---|---|
| Genetically Characterized (n = 1397) | All (n = 1817) | |||
| Site | <.001 | |||
| Michigan | 201 (14.4) | 307 (16.9) | 1152 (16.3) | |
| Pennsylvania | 382 (27.3) | 424 (23.3) | 1024 (14.5) | |
| Texas | 191 (13.7) | 280 (15.4) | 1318 (18.6) | |
| Washington | 317 (22.7) | 416 (22.9) | 2272 (32.1) | |
| Wisconsin | 306 (21.9) | 390 (21.5) | 1312 (18.5) | |
| Age | <.001 | |||
| 6 mo–8 y | 327 (23.4) | 396 (21.8) | 1946 (27.5) | |
| 9–17 y | 229 (16.4) | 306 (16.8) | 950 (13.4) | |
| 18–49 y | 389 (27.9) | 531 (29.2) | 2206 (31.2) | |
| 50–64 y | 205 (14.7) | 281 (15.5) | 1118 (15.8) | |
| ≥65 y | 247 (17.7) | 303 (16.7) | 858 (12.1) | |
| Male sex | 601 (43.0) | 767 (42.2) | 2969 (42.0) | .84 |
| Race/ethnicity | <.001 | |||
| White, non-Hispanic | 1081 (77.7) | 1397 (77.1) | 5182 (73.4) | |
| Black, non-Hispanic | 108 (7.8) | 133 (7.3) | 552 (7.4) | |
| Hispanic | 82 (5.9) | 119 (6.6) | 685 (9.7) | |
| Other, non-Hispanic | 120 (8.6) | 162 (9.0) | 673 (9.5) | |
| Any high-risk condition | 521 (37.3) | 671 (36.9) | 2636 (37.2) | .81 |
| Interval from onset to specimen collection | <.001 | |||
| 0–2 d | 698 (50.0) | 839 (46.2) | 1904 (26.9) | |
| 3–4 d | 488 (35.0) | 638 (35.1) | 2771 (39.1) | |
| 5–7 d | 211 (15.1) | 340 (18.7) | 2403 (34.0) | |
| Reported general health statusc | .01 | |||
| Excellent/very good | 1026 (76.1) | 1342 (74.2) | 4991 (70.6) | |
| Good | 282 (20.3) | 368 (20.3) | 1639 (23.2) | |
| Fair/poor | 83 (6.0) | 99 (5.5) | 442 (6.3) | |
| Self/household exposure to smokec | 176 (12.6) | 229 (12.6) | 1248 (17.7) | <.001 |
| ≥1 child <12 y of age in householdc | 522(37.6) | 708 (39.0) | 2828 (40.0) | .30 |
| Reported fever | 1132 (81.2) | 1459 (80.4) | 3876 (54.9) | <.001 |
| Reported current health assessment score, median (IQR)d | 50 (40–70) | 50 (40–70) | 60 (45–75) | <.001 |
| Vaccination status 2014–2015 | .03 | |||
| ≥1 doses | 749 (53.6) | 939 (51.7) | 3866 (54.6) | |
| 0 doses | 648 (46.4) | 878 (48.3) | 3212 (45.4) | |
Data are no. (%) of subjects, unless otherwise indicated.
Abbreviation: IQR, interquartile range.
a Exclusions from total enrollment: 2009 pandemic influenza A(H1N1) virus positive (n = 4), influenza B virus positive (n = 397), coinfections with influenza A(H3N2) and B viruses (n = 2), influenza A virus positive of undetermined subtype (n = 19), indeterminate rRT-PCR result (n = 7).
b The χ2 statistic was used to assess differences between the numbers of persons with influenza A(H3N2) virus–positive and influenza virus–negative test results with respect to the distributions of site, age group, sex, race/ethnicity, presence of any high risk condition, interval from illness onset to specimen collection, general health status, smoke exposure in self/household, presence of children ages <12 years in the household, and vaccination status. The Wilcoxon Mann–Whitney test was used to assess differences with respect to the distribution of the current health assessment. A P value of <.05 is statistically significant.
c Data were missing for 14 enrollees.
d Possible values range from 1 (the worst) to 100 (the best). Data were missing for 16 enrollees.
In all, 4811 patients (54%) included in the analysis had received 2014–2015 influenza vaccine, including 939 of 1817 influenza A(H3N2) virus–positive patients (52%) and 3866 of 7078 influenza virus–negative patients (55%); VE against influenza A(H3N2) virus–associated illness was 7% (95% CI, −5% to 17%; Table 2). Estimated VE against influenza A(H3N2) virus–associated illness was 14% (95% CI, −3% to 28%) among patients enrolled prior to 2 January 2015 and −2% (95% CI, −20% to 14%) among those enrolled after 2 January 2015.
Table 2.
Adjusted Vaccine Effectiveness (VE) Against Influenza A(H3N2) Virus–Associated Illness, Overall and by Genetic Group, Among Patients Aged ≥6 Months Enrolled at US Influenza Vaccine Effectiveness Network Sites, 2014–2015
| Genetic Group, Age | Influenza A(H3N2)–Positive Cases, Proportion (%) Vaccinated | Influenza Virus–Negative Controls, Proportion (%) Vaccinated | VE, % (95% CI)a |
|---|---|---|---|
| Overallb | |||
| All ages | 939/1817 (51.7) | 3866/7078 (54.6) | 7 (−5 to 17) |
| 6 mo–8 y | 160/396 (40.4) | 1013/1946 (52.1) | 20 (−3 to 37) |
| 9–49 y | 355/837 (42.4) | 1387/3156 (44.0) | −5 (−24 to 12) |
| ≥50 y | 424/584 (72.6) | 1466/1976 (74.2) | 9 (−14 to 28) |
| Genetic group 3C.2a | |||
| All ages | 597/1101 (54.2) | 3866/7078 (54.6) | 1 (−14 to 14) |
| 6 mo–8 years | 103/243 (42.4) | 1013/1946 (52.1) | 16 (−13 to 37) |
| 9–49 y | 217/486 (44.7) | 1387/3156 (44.0) | −15 (−41 to 7) |
| ≥50 y | 277/372 (74.5) | 1466/1976 (74.2) | 8 (−21 to 30) |
| Genetic group 3C.3b | |||
| All ages | 56/156 (35.9) | 3866/7078 (54.6) | 44 (16 to 63) |
| 6 mo–8 y | 4/36 (11.1) | 1013/1946 (52.1) | NR |
| 9–49 y | 25/78 (32.1) | 1387/3156 (44.0) | 35 (−13 to 63) |
| ≥50 y | 27/42 (64.3) | 1466/1976 (74.2) | NR |
| Genetic group 3C.3a | |||
| All ages | 31/55 (56.4) | 3866/7078 (54.6) | −48 (−169 to 19) |
| Genetic group 3C.3 | |||
| All ages | 27/47 (57.5) | 3866/7078 (54.6) | 1 (−87 to 48) |
Abbreviations: CI, confidence interval; NR, not reported.
a VE was estimated as 100% × [1 − odds ratio], where the odds ratio is calculated as the ratio of the odds being vaccinated among subjects with influenza A(H3N2) virus–associated illness to the odds of being vaccinated among influenza virus–negative subjects. Odds ratios were estimated using logistic regression. If the 95% CI excludes 0, the results are considered statistically significant. Models were adjusted for site, age, any high-risk condition, and calendar time (biweekly intervals).
b Includes influenza A(H3N2) viruses for which the genetic group was not determined.
VE estimates were similar when restricted to 1359 infections with genetically characterized influenza A(H3N2) viruses (6% [95% CI, −7% to 18%]) or 458 infections with uncharacterized influenza A(H3N2) viruses (11% [95% CI, −10% to 27%]), but they differed by HA genetic group. Among all patients aged ≥6 months, estimated VE against illness due to infection with HA genetic group 3C.2a viruses was 1% (95% CI, −14% to 14%]) versus 44% (95% CI, 16%–63%) against illness due to group 3C.3b viruses (Table 2). Among patients infected with viruses from HA genetic groups 3C.3 or 3C.3a, VE estimates were not statistically significant. No statistically significant interaction was observed between current (2014–2015) and prior (2013–14) season vaccination status, for any influenza A(H3N2) virus–related illness, or for genetic groups of influenza A(H3N2) viruses. Inclusion of partially vaccinated children and restriction of analysis to patients enrolled within 4 days of symptom onset resulted in similar VE estimates (data not shown).
DISCUSSION
The 2014–2015 influenza season in the United States was notable for the early and widespread circulation of influenza A(H3N2) viruses, the majority of which belonged to an emergent HA genetic group 3C.2a and showed substantial antigenic drift from the A(H3N2) component of 2014–2015 northern hemisphere vaccines [1]. Expanded genetic characterization of influenza A(H3N2) viruses from surveillance sites in the United States identified variability in geographic and temporal distribution of genetic groups of influenza A(H3N2) viruses. Development of a pyrosequencing assay, with high throughput, provided genetic group information on a large number of influenza A(H3N2) viruses from Flu VE network sites, permitting strain-specific estimates that revealed differences in VE caused by specific HA genetic groups of influenza A(H3N2) viruses. Low VE against genetic group 3C.2a viruses was consistent with antigenic drift. Low or null effectiveness for 2014–2015 influenza vaccines against influenza A(H3N2) virus–associated illness was also reported in estimates from other surveillance networks in which influenza A(H3N2) group 3C.2a viruses predominated, including networks in Canada [6, 19] and the United Kingdom [20]. In contrast, we found evidence of modest (44%) VE against influenza A(H3N2) viruses in group 3C.3b, which were characterized as antigenically similar to the 2014–2015 vaccine reference virus A/Texas/50/2012. Viruses from the 3C.3b genetic group appeared to be more common in some regions of the country, including the Pennsylvania Flu VE network site. Although vaccination provided some protection against influenza A(H3N2) viruses belonging to group 3C.3b, the predominance of the drifted 3C.2a viruses in the United States resulted in low overall VE against medically attended influenza-associated illness during the 2014–2015 season [21].
Differences in VE have been observed during seasons with mixed circulation of viruses antigenically matched and mismatched to vaccine components [22, 23], with the 2014–2015 season having a very high proportion of circulating influenza A(H3N2) viruses that were antigenically mismatched to the vaccine. During seasons in which antigenically drifted viruses circulate, VE has not consistently been associated with the degree of antigenic similarity as measured by the HI assay [23]. The addition of genetic characterization of a large number of influenza A(H3N2) viruses from patients enrolled in the Flu VE network study during the 2014–2015 season provided the opportunity to examine differences in VE by genetic group. This analysis suggests that protection against influenza A(H3N2) viruses may be related to vaccine protection against specific virus genetic groups and the distribution of those viruses in the community. In addition, genetic characterization of influenza A(H3N2) viruses from US laboratories provided insight into the rapid spread of antigenically mismatched viruses from a single genetic group, as well as local variability in the distribution of HA genetic groups antigenically matched to vaccine. Genetic data also provided better representation of antigenic mismatch between vaccine and circulating influenza A(H3N2) viruses, many of which could not be characterized using the HI assay. In the future, studies to assess VE against distinct groups of influenza viruses may augment data from traditional methods to identify potential vaccine candidates and ultimately lead to improved vaccines [5].
The 2014–2015 influenza season highlighted challenges with formulation of influenza vaccines that require frequent updates because of rapid changes in circulating viruses, as well as limitations inherent in the current methods to select vaccine virus candidates. In February 2014, at the time of virus strain selection for 2014–2015 northern hemisphere vaccines, >90% of characterized influenza A(H3N2) viruses were found to be antigenically similar to the 2013–2014 vaccine reference virus, A/Texas/50/2012 [15]. The WHO recommended no change in the influenza A(H3N2) vaccine strain for the 2014–2015 season [15]. In March 2014, after the 2014–15 vaccine strain had been selected, antigenic drift variants belonging to HA genetic groups 3C.3a and 3C.2a were detected. During May–September, 2014, the proportion of these drift variants increased rapidly, with a predominance of HA genetic group 3C.2a viruses in the United States, and <50% of influenza A(H3N2) viruses characterized by HI were inhibited by ferret reference antisera against A/Texas/50/2012 [24]. These virologic findings, together with epidemiologic and human serologic data, resulted in a change in the recommended A(H3N2) vaccine component to an A/Switzerland/9715293/2013-like virus from HA group 3C.3a for the 2015 southern hemisphere season [18].
Results from the Flu VE network indicated that differences in VE may have resulted from a small degree of genetic change associated with substantial antigenic differences within related HA genetic groups of influenza A(H3N2) viruses. Amino acid substitutions at specific HA antigenic sites have been associated with major antigenic changes (referred to as cluster transitions) in influenza A(H3N2) viruses since their emergence in humans in 1968 [25]. Influenza A(H3N2) genetic group 3C.2a viruses possessed multiple amino acids changes in HA relative to the A/Texas/50/2012 reference vaccine virus, including the F159Y change near the receptor-binding site [26]. In addition, the majority of viruses in HA genetic group 3C.2a contain an amino acid substitution at residue 160 that adds a glycosylation site at residues 158–160, which may shield antigenic sites from neutralizing antibodies [27]. Interestingly, influenza A(H3N2) genetic group 3C.3a viruses, including the 2015–2016 northern hemisphere vaccine reference virus A/Switzerland/9715293/2013, contain a different amino acid at residue 159 that nonetheless may have resulted in antigenic properties similar to those of 3C.2a viruses [18]. In contrast, HA genetic group 3C.3b viruses, which do not have antigenically important amino acid substitutions like that at residue 159, were characterized as antigenically similar to A/Texas/50/2012, consistent with the observation of modest VE. Small numbers of infections with HA group 3C.3 and 3C.3a viruses limited our ability to estimate VE against illness due to these genetic groups. During October 4—28 November 2015, the majority of 3C.2a viruses characterized were well inhibited by ferret reference antisera raised against A/Switzerland/9715293/2013, indicating better antigenic match between 2015–2016 vaccine and circulating influenza A(H3N2) viruses [28]. However, the effectiveness of influenza vaccines against circulating influenza A(H3N2) viruses will need to be monitored closely because of the likelihood of continuing drift among circulating strains.
Our results are subject to several limitations. Influenza A(H3N2) virus–positive specimens from study participants submitted for characterization had lower rRT-PCR cycle threshold values, possibly corresponding with higher viral loads [29]. Antigenic properties were inferred from a subset of viruses characterized by HI. However, antigenic characterization of viruses from the Flu VE network agreed with virologic surveillance data. Second, the validity of the test-negative design depends upon accurate classification of influenza status [30]. In addition to the use of the highly specific rRT-PCR assay, patients in the Flu VE network study were enrolled within 7 days of illness onset, when viral shedding was highest, decreasing the likelihood of false-negative results. Last, patients at higher risk of influenza-related illness have higher rates of influenza vaccination; while estimates adjust for potential confounding variables, residual confounding may bias estimates. However, the test-negative design, by testing patients presenting for care, reduces potential selection bias related to care-seeking behavior [16, 17].
During 2014–2015, several genetically related influenza A(H3N2) viruses circulated in the United States, but one group predominated over time. We found that influenza vaccination in 2014–2015 did not reduce the likelihood of influenza illness caused by the predominant genetic group of influenza A(H3N2) viruses but prevented some disease due to influenza A(H3N2) viruses in less prevalent genetic groups [21]. Overall low effectiveness of influenza vaccines may have contributed to high rates of influenza-associated hospitalizations observed during the season [1], despite vaccination of approximately two thirds of the US population aged ≥65 years [31]. Expanded availability of high-throughput platforms for nucleotide sequencing and genetic characterization of larger numbers of influenza viruses may provide more complete and timely information on emergent influenza viruses to inform vaccine strain selection and help evaluate protection provided by vaccination.
Supplementary Material
Notes
Acknowledgments. We thank the study participants at each of the 5 Centers for Disease Control and Prevention (CDC) US Flu VE Network sites; Joyce Benoit, Erika Kiniry, Lawrence Madziwa, Matt Nguyen, and C. Hallie Phillips, Group Health Research Institute; G.K. Balasubramani, Arlene C. Bullotta, Rina Chabra, Heather Eng, Samantha Ford, Edmund Garofolo, Jennifer Gray, Robert Hickey, Philip Iozzi, Barbara Kevish, Donald B. Middleton, Krissy K. Moehling, Christopher Olbrich, Jonathan M. Raviotta, Evelyn C. Reis, Charles R. Rinaldo, Edmund M. Ricci, Sandra Sauereisen, Sean Saul, Terri Sax, Michael Susick, Leonard Urbanski, Stephen Wisniewski, University of Pittsburgh; Joshua Petrie, Caroline Cheng, Casey Martens, EJ McSpadden, Anne Kaniclides, Ryan Malosh, Emily Martin, Samantha Harrison, Kajal Magal, Brian Nixon, Jessica Obidike, Mallory Theisen, Emily Valice, Kevin Zhang, University of Michigan School of Public Health; Lois Lamerato and Heather Lipkovich, Henry Ford Health System; Donald Wesson, Michael Reis, Madhava Beeram, Jessica Pruszynski, Lydia Clipper, Archana Nangrani, Kempapura Murthy, Anne Robertson, Patricia Sleeth, Virginia Gandy, Teresa Ponder, Mary Kylberg, Hope Gonzales, Martha Zayed, Deborah Furze, Baylor Scott and White Health; Vasanthi Avadhanula, Alan Jewell, Kirtida Patel and Sneha Thaker, Baylor College of Medicine; and Jennifer Meece, Jennifer King, Deanna Cole, Sandy Strey, Donna David, Jackie Salzwedel, Carla Rottscheit, Sarah Koptizke, Laurel Verhagen, Gregg Greenwald, Phillip Bertz, Lynn Ivacic, Hannatu Amaza, Braiden Andersen, Elizabeth Armagost, Yvonne Cerne, Kathleen Cushman, Shelia Drowatzky, Keith Gilge, William Gillaspie, Krista Herkert, Deborah Hilgemann, Tara Johnson, Bryan Joosse, Alex Krenzke, Kelly Mathews, Madalyn Minervini, Vicki Moon, Suellyn Murray, Rebecca Pilsner, DeeAnn Polacek, Zoe Retzlaff, Kirsten Schultz, Teresa Schultz, Adam Smith, Kristja Vittallo, Jane Wesely, Kelly Wirkus, and Bobbi Bradley, Marshfield Clinic Research Foundation.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Financial support. This work was supported by the CDC (cooperative agreements U01IP000466, U01IP000467, U01IP000471, U01IP000473, and U01IP000474).
Potential conflicts of interest. R. K. Z. has received grants from Sanofi Pasteur, Pfizer, and Merck. M. P. N. received grants from Pfizer and Merck. A. S. M. has received grants from the CDC and Sanofi Pasteur and has served as a consultant for GSK, Sanofi Pasteur, Novavax, Novartis, and Protein Sciences. S. E. O. has received grants from the CDC. E. A. B. and H. Q. M. have received grants from the CDC and MedImmune. M. G. received grants from the CDC during the conduct of the study and grants from MedImmune/AstraZeneca outside the submitted work. P. A. P. served on the speaker bureau at Medimmune, and within the past year he served as a scientific advisor on influenza for AstraZeneca. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Appiah GD, Blanton L, D'Mello T et al. . Influenza activity - United States, 2014–2015 season and composition of the 2015–16 influenza vaccine. MMWR Morb Mort Wkly Rep 2015; 64:583–90. [PMC free article] [PubMed] [Google Scholar]
- 2. D'Mello T, Brammer L, Blanton L et al. . Update: influenza activity--United States, 28 September 2014–21 February 2015. MMWR Morb Mort Wkly Rep 2015; 64:206–12. [PMC free article] [PubMed] [Google Scholar]
- 3. Hensley SE. Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr Opin Virol 2014; 8:85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klimov AI, Garten R, Russell C et al. . WHO recommendations for the viruses to be used in the 2012 Southern Hemisphere Influenza Vaccine: epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from February to September 2011. Vaccine 2012; 30:6461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Skowronski DM, Chambers C, Sabaiduc S et al. . Integrated sentinel surveillance linking genetic, antigenic, and epidemiologic monitoring of influenza vaccine-virus relatedness and effectiveness during the 2013–2014 influenza season. J Infect Dis 2015; 212:726–39. [DOI] [PubMed] [Google Scholar]
- 6. Skowronski DM, Chambers C, Sabaiduc S et al. . A perfect storm: impact of genomic variation and serial vaccination on low influenza vaccine effectiveness during the 2014-2015 season. Clin Infect Dis 2016; 63:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flannery B, Thaker SN, Clippard J et al. . Interim estimates of 2013–2014 seasonal influenza vaccine effectiveness - United States, February 2014. MMWR Morb Mort Wkly Rep 2014; 63:137–42. [PMC free article] [PubMed] [Google Scholar]
- 8. Gaglani M, Pruszynski J, Murthy K et al. . Influenza vaccine effectiveness against 2009 pandemic influenza A(H1N1) virus differed by vaccine type during 2013–2014 in the United States. J Infect Dis 2016; 213:1546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McLean HQ, Thompson MG, Sundaram ME et al. . Influenza vaccine effectiveness in the United States during 2012–2013: variable protection by age and virus type. J Infect Dis 2015; 211:1529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ohmit SE, Thompson MG, Petrie JG et al. . Influenza vaccine effectiveness in the 2011–2012 season: protection against each circulating virus and the effect of prior vaccination on estimates. Clin Infect Dis 2014; 58:319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grohskopf LA, Olsen SJ, Sokolow LZ et al. . Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP) -- United States, 2014–2015 influenza season. MMWR Morb Mort Wkly Rep 2014; 63:691–7. [PMC free article] [PubMed] [Google Scholar]
- 12. Belongia EA, Kieke BA, Donahue JG et al. . Effectiveness of inactivated influenza vaccines varied substantially with antigenic match from the 2004–2005 season to the 2006–2007 season. J Infect Dis 2009; 199:159–67. [DOI] [PubMed] [Google Scholar]
- 13. Levine M, Sheu TG, Gubareva LV, Mishin VP. Detection of hemagglutinin variants of the pandemic influenza A (H1N1) 2009 virus by pyrosequencing. J Clin Microbiol 2011; 49:1307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou B, Donnelly ME, Scholes DT et al. . Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza A viruses. J Virol 2009; 83:10309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Recommended composition of influenza virus vaccines for use in the 2014–2015 northern hemisphere influenza season. Wkly Epidemiol Rec 2014; 89:93–104. [PubMed] [Google Scholar]
- 16. Foppa IM, Haber M, Ferdinands JM, Shay DK. The case test-negative design for studies of the effectiveness of influenza vaccine. Vaccine 2013; 31:3104–9. [DOI] [PubMed] [Google Scholar]
- 17. Jackson ML, Nelson JC. The test-negative design for estimating influenza vaccine effectiveness. Vaccine 2013; 31:2165–8. [DOI] [PubMed] [Google Scholar]
- 18. Recommended composition of influenza virus vaccines for use in the 2015 southern hemisphere influenza season. Wkly Epidemiol Rec 2014; 89:441–52. [PubMed] [Google Scholar]
- 19. Skowronski DM, Chambers C, Sabaiduc S et al. . Interim estimates of 2014/15 vaccine effectiveness against influenza A(H3N2) from Canada's Sentinel Physician Surveillance Network, January 2015. Euro Surveill 2015; 20:pii:21022. [DOI] [PubMed] [Google Scholar]
- 20. Pebody RG, Warburton F, Ellis J et al. . Low effectiveness of seasonal influenza vaccine in preventing laboratory-confirmed influenza in primary care in the United Kingdom: 2014/15 mid-season results. Euro Surveill 2015; 20:21025. [PubMed] [Google Scholar]
- 21. Flannery B, Clippard J, for the U.S. Flu VE Network. End-of-season influenza vaccine effectiveness estimates for the 2014–15 season: US Influenza Vaccine Effectiveness (Flu VE) Network. http://www.cdc.gov/vaccines/acip/meetings/meetings-info.html. Accessed 26 May 2016.
- 22. Darvishian M, Bijlsma MJ, Hak E, van den Heuvel ER. Effectiveness of seasonal influenza vaccine in community-dwelling elderly people: a meta-analysis of test-negative design case-control studies. Lancet Infect Dis 2014; 14:1228–39. [DOI] [PubMed] [Google Scholar]
- 23. Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis 2012; 12:36–44. [DOI] [PubMed] [Google Scholar]
- 24. Blanton L, Brammer L, Smith S et al. . Update: influenza activity -- United States and worldwide, 18 May–20 September 2014. MMWR Morb Mort Wkly Rep 2014; 63:861–4. [PMC free article] [PubMed] [Google Scholar]
- 25. Koel BF, Burke DF, Bestebroer TM et al. . Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013; 342:976–9. [DOI] [PubMed] [Google Scholar]
- 26. Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep 2015; 12:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 2014; 6:1294–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith S, Blanton L, Kniss K et al. . Update: influenza activity - United States. MMWR Morb Mort Wkly Rep 2015; 64:1342–8. [DOI] [PubMed] [Google Scholar]
- 29. Spencer S, Chung J, Thompson M et al. . Factors associated with real-time RT-PCR cycle threshold values among medically attended influenza episodes. J Med Virol 2015; 88:719–23. [DOI] [PubMed] [Google Scholar]
- 30. Jackson ML, Rothman KJ. Effects of imperfect test sensitivity and specificity on observational studies of influenza vaccine effectiveness. Vaccine 2015; 33:1313–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. CDC. Flu vaccination coverage, United States, 2014–15 influenza season. http://www.cdc.gov/flu/fluvaxview/coverage-1415estimates.htm. Accessed 26 May 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.