Abstract
Drosophila is widely used for the dissection of genetic and neuronal mechanisms of behavior. Recently, flies have emerged as a model for investigating the regulation of feeding and sleep. Although typically studied in isolation, increasing evidence points to a fundamental connection between these behaviors. Thus, a system for measuring sleep and feeding simultaneously in a single integrated system is important for interpreting behavioral shifts of either state. Here, we describe the construction and use of the Activity Recording CAFE (ARC), a machine-vision based system for the integrated measurement of sleep and feeding in individual Drosophila. Flies feed on liquid food in a microcapillary and consumption is measured by automated tracking of the liquid meniscus over time. Sleep measurements are obtained from positional tracking of the animals and arousal threshold can be determined by vibrational stimulus response. Using this system, a single computer and experimenter can track diverse behaviors from up to 60 individual flies in a single integrated system. The ARC is efficiently assembled with minimal training and each experiment can be run for up to ~7 days, with a total setup and breakdown time of about 2 hours.
INTRODUCTION
Increasing evidence points to a fundamental relationship between feeding and sleep. In various animal models, restricting food intake drives wakefulness1-3 and manipulating available nutrients modifies sleep on both short and long time scales4-9. Beyond the physical act of feeding, changes in energy storage or metabolism stemming from feeding behavior may also be reflected in sleep. Sleep loss can increase appetite10 and daytime sleepiness has been associated with obesity11.
Drosophila has recently emerged as a model for studying the genetic, environmental, and neuronal basis of sleep—largely due to the ease of high throughput studies at low spatial and monetary cost12-14. Analogous to its mammalian counterpart, fly sleep is characterized by a rise in arousal threshold, a postural shift, homeostatic regulation, and reduced electrical activity in the brain15-17. Fly sleep is also necessary for memory formation and survival18-20. Because of this functional conservation and the powerful genetic tools available for manipulating flies, Drosophila will likely continue to play a large role in dissecting the fundamental relationships between sleep and feeding. However, sleep-feeding studies suffer from a major caveat—the two behaviors are typically not measured together in the same animals. Insights into the direct impact of environmental, genetic, and other manipulations on sleep and feeding—and their interaction—might only be revealed by characterizing both behaviors concurrently in individual animals.
Here, we present a method for the simultaneous measurement of sleep and feeding in adult Drosophila. This method uses high-resolution machine-vision to accurately track the position of individual flies, as well as to follow the meniscus of liquid food-containing capillaries that the animals feed from. Previously, use of the ARC was described for measuring the timing and size of discrete feeding events while simultaneously assessing sleep before and after each event. This allowed us to examine the rapid shifts in sleep which result from a meal4. The primary advantage of the ARC is the coupling of sleep, arousal, and feeding measurement into a single system to reveal direct behavioral correlates and increase experimental throughput. However, individual ARC modules for measuring feeding, sleep, or activity can also be run independently and offer advantages over other independent behavioral measurement systems including ease of chamber customization, increased throughput, and real-time analysis capabilities. Although we previously published an iteration of the ARC that utilized separate cameras and computers for tracking fly movement and food intake4, the upgraded system described here uses a single camera per chamber of 30 flies for real-time measurements of both sleep and feeding. A single computer can typically handle two cameras simultaneously for the study of 60 flies in two chambers.
Comparison with other available methods
The ARC monitors consumption continuously during an experiment by repeatedly sampling liquid food meniscus position rather than making a single measurement at the end21. The system therefore resolves discrete feeding events, facilitating the study of prandial behavior as well as the finer integration of feeding and spatially derived behaviors such as sleep4. In the described setup, the ARC makes measurements from 30 individuals per camera, yet data quality is comparable to that from another recently described capillary-based tool, the Expresso, which is also capable of nanoliter resolution22. Replication of an Expresso experiment shows slightly lower absolute feeding in the ARC, but with similar relative changes in consumption across several diets (Supplementary Fig. 1). The Expresso measures food intake of 5 flies by using LED sensor arrays to measure capillary liquid food levels. Alternative systems for capturing high resolution feeding behavior have focused on quantifying fly-food contact, a correlate of food intake, by using capacitance23,24. Like the Expresso, these systems require independent sensors for each fly, increasing cost per animal. Sensors can also obscure the view of each fly, making it difficult to measure sleep. By assessing the feeding behavior of many flies from a single camera, the ARC substantially reduces cost and increases experimental throughput. It is also notable that the ARC can detect smaller feeding events if the camera is simply moved closer to a subset of capillaries.
The ARC implements object tracking by using JavaGrinders4, an open-source, Java-based framework. Acquisition of frames and image operations are performed with OpenCV25, a real-time computer vision library. While video tracking is notoriously processor intensive—requiring most object trackers to rely on analysis of pre-recorded videos—the ARC alleviates this difficulty by using various computational optimizations that allow for real-time extraction of behavioral properties. Hence, information is saved to a coordinate output file during the experiment, eliminating the need to store large video files and analyze them post-experiment. For most laboratories, this level of data reduction would allow experiments to span days or weeks, rather than restricting studies to briefer periods. The basic ARC does not implement computational methods for classifying specific behaviors such as grooming or wing flicking. However, video (or images) can be optionally stored from the ARC data capture program and analyzed after the fact with machine-learning based analytic systems such as JAABA26. The Java framework also allows for a custom class to override the built-in analysis procedure. This would potentially allow characterization of specific behaviors at run time rather than in post processing with a saved video file.
Although we describe implementation of the ARC for tracking sleep, feeding, and arousal threshold in individual animals, additional JavaGrinders modules are currently available that extend ARC capabilities. The ARC can be configured to track multiple animals in one arena, identify color-labeled objects, combine information from two cameras for 3D spatial localization, and control real-time robotic interfaces in response to animal behavior. One example of these capabilities would be to drive a light stimulus to activate optogenetic tools when flies enter a specific region of an arena, consume food, or perform a certain behavior. With support for input and output to various microcontrollers (e.g., Arduino, Phidget, Raspberry PI GPIO pins, Allen Bradley Programmable Logic Controllers, and Analog Devices SMT), JavaGrinders can communicate with relays, servos, and sensors over a range of communication protocols (e.g., USB, I2C/SPI, and TCP/IP). These features offer a widely adaptable system to explore animal behavior in an adaptive environment not previously accessible in open-source tools. Furthermore, the ARC can be run on single board computers such as the Raspberry Pi 3 Model B or the Hardkernel Odroid. Instructions and fully installed images are available for download on the JavaGrinders ARC blog (http://javagrinders-arc.blogspot.com/2017/01/install-open-source-tools-for-running.html).
Experimental Design
Using the ARC involves 4 major stages: (i) construction and preparation of the ARC chamber and environment, (ii) configuring the computational framework for the data capture software, (iii) running ARC experiments, and (iv) analyzing ARC data.
(i) Construction and preparation of the ARC
While the protocol provided here is a rigid instruction set for building and operating the ARC, the method is highly flexible and can be customized for different uses and conditions. Beginning with its construction, the ARC only requires that animals be individually housed in a chamber which 1) allows full visibility of the animals by a camera and 2) contains an overhead opening in which a capillary containing liquid food can be secured vertically, the bottom of which must be accessible by the animal. The design we provide is an optimization of these steps, meant to maximize throughput and behavioral measurement.
The ARC chamber (Fig. 1a) contains half-cylindrical shaped wells to minimize edge dwelling, with a volume substantially larger than the commonly used Drosophila Activity Monitor (DAM) tubes27, allowing the animals more room to explore and exhibit spatial tendencies. The size also allows a camera to track 30 flies, a number comparable to the capacity of a DAM system (32 animals), while still allowing a clear outline of each fly with a FHD camera (1080p resolution). The chambers are vertically oriented to allow visual access to both flies and the food capillaries from a single camera (Fig. 1b). Previously, we found that the vertical orientation does not affect daily sleep patterning of individual flies4. Notably, one ARC controller can operate multiple independent setups; we include a program implementing two tracking cameras (60 flies) called ARCControllerMultiCam.java.
Figure 1. ARC schematic.
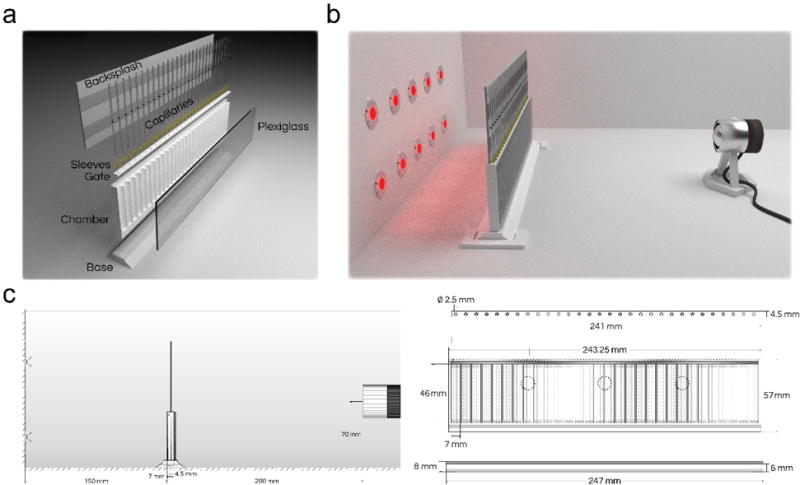
(a) Model of the disassembled ARC components includes the chamber, chamber base, front plexiglass panel, chamber gate, capillary holder sleeves, capillaries, and paper backsplash. (b) Model of the assembled ARC chamber within an incubator with a front facing camera and infrared LED backlighting. (c) Blueprints of the ARC setup showing a side (left) and front (right) profile of the ARC with corresponding measurements. The 3 small circles represent positions for the placement of vibration motors on the back of the chamber.
All parts can be printed in acrylonitrile butadiene styrene (ABS) or polylactic acid (PLA). The chamber material used should be consistent across experiments since there may be differences in fly climbing ability or other behaviors across materials. The ARC chamber, along with the rest of the pieces shown, can be printed using the included .stl files (Supplementary Files). Once printed, the face of the chamber is mounted with a 2-mm thick, clear plexiglass shield to allow visualization of each fly. While plexiglass is sufficiently strong and transparent, other equivalent materials could be used.
At the bottom of each well, agar is dispensed prior to each experiment. This provides flies with ad libitum access to water, eliminating any experimental confounds that could arise from dehydration28. Beneath the chamber, a base plate is used to keep the chamber upright. The base plate should fit snugly to the bottom of the chamber. Above the wells, a gate containing regularly spaced 2.5-mm holes aligned with each well is inserted into the groove above the chamber and pushed into place (Fig. 1c). Each of the holes is fitted with a sleeve to hold 5-μl glass capillaries in place.
A glass capillary containing aqueous food is provided for each fly. To track changes in liquid levels, each capillary also contains a 2-mm oil layer immediately above the aqueous food. This organic phase is made up of dodecane and light mineral oil in a mixture that was determined empirically. Tracking of the oil layer is dependent on an infrared-absorbent, oil-soluble dye that was previously identified by screening several candidates4. Behind the capillaries, a backsplash is attached using a piece of construction paper and double-sided tape (Fig. 1a). This allows the experimenter to keep the capillaries straight and to create a flat contrast between dye bands and the background.
To limit experimental variability, the ARC should be housed in an incubator with controlled light, temperature, and humidity. Drosophila sleep experiments are often conducted at 22-25 °C at a relative humidity of ~60% under a 12-/12-hour light/dark cycle. ARC measurements are dependent only on infrared light and are unaffected by any visible light source used to set the circadian rhythm of the flies. The ARC should be placed on a flat surface between a camera and infrared backlighting. The camera should be facing the wells of the chamber ~200 mm away from the ARC and should have a full view of both the chamber wells and capillaries (Fig. 1b,c). The camera must have its filters replaced with an infrared-pass filter. Alternatively, any USB camera which only senses infrared light can be used.
To detect arousal threshold, an Arduino controller and motor attachment must be set up (Fig. 2a). This system allows the user to deliver an incremental series of vibrational stimuli to test the arousal threshold in addition to the detection of feeding and inactivity-based sleep. Notably, the stimulus delivery from the Arduino is an analog voltage, which can be used to control light, sound, or other stimuli. This allows flexibility in the type of arousal measured for various setups.
Figure 2. Arousal threshold components.
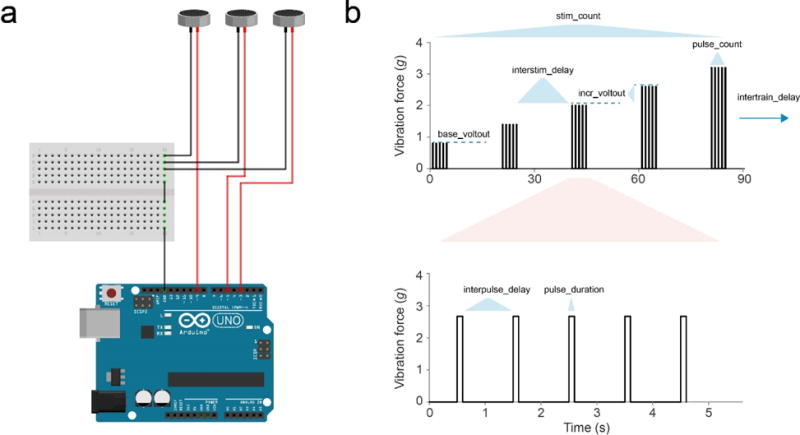
(a) Wiring diagram of the shaft-less vibration motor coupling to the Arduino microcontroller. (b) Plot of stimulus protocol showing vibration force (g) delivered to the back of the ARC chamber over time. Variable names displayed can be altered in ARCController.java document (redrawn from31).
(ii) Assembling the data capture software
The ARC controller can be run by most modern computers; older systems may exhibit lagging or crashing. We recommend a Mac with at least 4+ GB RAM. Use of the two-camera program generally requires 6+ GB RAM. PCs are an acceptable option, but must be running a Linux operating system. Performance, such as the rate of tracking or number of animals, scales with available hardware resources. Thus, increasing the number of cores and/or available RAM will increase experimental capabilities.
Support for the ARC requires installation of several open source libraries as dependencies. The required steps for installing these packages are fairly simple and are listed in detail in the ARC installation blog (http://javagrinders-arc.blogspot.com/2017/01/install-open-source-tools-for-running.html). Specifically, the blog lists installation instructions for Java Standard Edition 8 SDK (http://www.oracle.com/technetwork/java/javase/downloads/jdk8-downloads-2133151.html), Eclipse IDE 3.8+ (http://www.eclipse.org/downloads/packages/eclipse-ide-java-developers/marsr), and MacPorts (https://www.macports.org/install.php) which is used to install OpenCV. The software versions listed have been tested with the current build. Although older versions may not function properly, the blog and provided software will be updated periodically to support future releases of Java and OpenCV.
The ARC acquires data from a video feed of the chamber and food capillaries (Fig. 3a). The image areas delineating the entire chamber and set of capillaries are defined by the user. These areas are divided into equally spaced tracks based on the user-defined number of flies or individual enclosures. For animal motion tracking, a reference image of the ARC is taken and used to background subtract subsequent images in real-time, providing an image of pixel differences. This difference should be largest in the image space containing the fly, assuming the animal has moved from its position in the reference image. The tracker then scans each fly track region for the point of greatest difference and, if it is greater than the user-defined threshold, the scanner moves laterally one pixel at a time until a sub-threshold pixel is reached. This threshold scanning continues in a clockwise direction until the outline returns to the starting coordinate (Fig. 3b,c). If the outlined polygon is roughly the size of a fly (also a user-defined parameter), its centroid coordinate is recorded to an output text file. During processing, animal motion is considered as any Euclidean change in the coordinates which exceeds 50% of the animal’s body length coordinate (Fig. 3c).
Figure 3. ARC data capture.
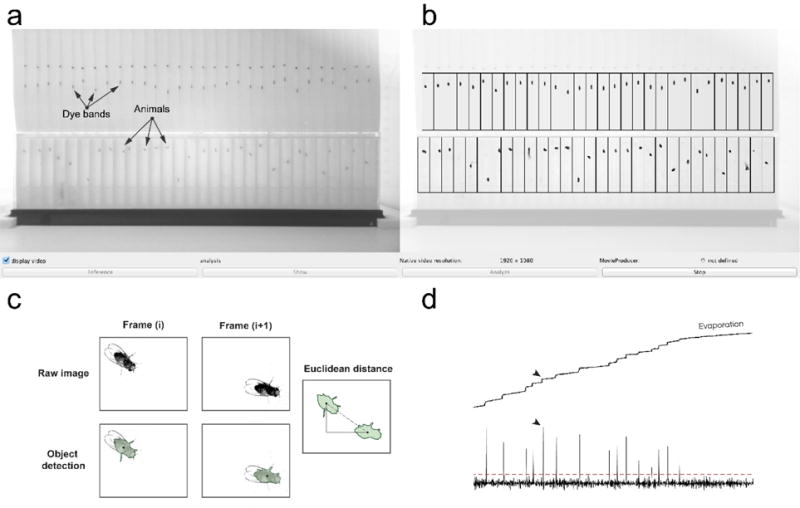
(a) Image of the ARCController.java visual interface. Image is a camera view of dye bands and animals. Black markings above dye bands are calibration marks and should be excluded from tracking region. (b) Visual overlay of user-defined tracking regions over original image. Outlines of detected animals are displayed as black outlines with green fill. (c) Animal motion is calculated using Euclidean distance between coordinates of each frame (panel replicated from Murphy et al., eLife 2016). (d) 24-hour trace of the dye band from one capillary using 1-min averages. Motion events are observed (black arrows) which exceed a statistical cut-off (red dashed line), and are defined as meals. Evaporation is evident between meals as the slight continuous increase in baseline.
A similar algorithm is employed for tracking the vertical position of the liquid food meniscus of the dye band. Since the dye band does not necessarily move from its initial position in the reference image, meniscus tracking is performed using background estimation for each capillary track region. The tracker finds the darkest pixel for each track and then uses auto-detection to outline the object. Any remaining pixels are averaged, with the average value serving as a flat background subtraction value. The meniscus is then tracked using the same method used for animal tracking and is also recorded to the output file. To increase signal-to-noise, average dye-band positions are calculated from 1 minute segments before analysis, although this period length can be changed for higher temporal resolution. Feeding events are statistically identified in post-experiment data analysis (see below) as dye-band object motion events which are at least 4 standard deviations greater than the estimated noise (Fig. 3d).
To track individual dye bands or animals, the parameters establishing the tracking regions within an image and thresholds for determining an object must be set prior to the experiment. The tracking region for the animals is set to encompass the total image space in which any given animal can move. Because animal tracking uses a reference image of the chamber, it is relatively insensitive to dark edges or marks within the image. In contrast, the background estimation used in dye-band tracking requires that the dye is the only dark object in the tracking region. Calibrated 5-μl capillaries are prelabeled with a black mark near the middle of their length. Special attention must be given to loading the dye band below this mark and creating a tracking region which excludes the mark. Alternatively, the marks can be removed by rolling the capillaries in acetone on a hard surface or obtaining unmarked capillaries. Capillaries should be washed following mark removal or before each reuse, as described in the Procedure section.
(iii) Running the ARC
An important part of running a successful ARC experiment is controlling for environmental and developmental effects that might affect the behavior of the animals. General practices to develop a consistent experimental cohort of flies have been described previously29. Briefly, it is important that the animals develop without over-crowding, are housed in similar numbers following eclosion (we generally use 7 to 10 flies per vial), are tested at the same age (we typically assess 5-9 day old adults), and that different test groups are assigned to alternating (or random) compartments in the ARC chamber. This will control for possible effects of microenvironment, including uneven lighting or temperature. Both males and females can be used in the ARC. Egg lay and hatched larvae may be problematic in long-term experiments with mated females, however. Once animals are obtained, the experiments can be carried out by following the steps outlined in the procedure.
Diets can be composed of any water-soluble ingredients. A 2.5% sucrose + 2.5% yeast extract (both w/v) solution is often sufficient for 24 hours of recording without running out of food4,21,30. Other components, including drugs, non-nutritive sweeteners, buffers, preservatives, or other nutritional ingredients can be used in the diet. Low nutrient concentration diets or other manipulations that result in increased food intake may require more frequent capillary changes. ARC runs should be stopped, capillaries replaced, and the ARCController program restarted at each food change. ARCController contains all of the necessary parameters defined by the user for operating the ARC data acquisition, including the frame capture rate and the definition of the tracking regions (Fig. 4).
Figure 4. Flow chart of ARC implementation of the JavaGrinders library.
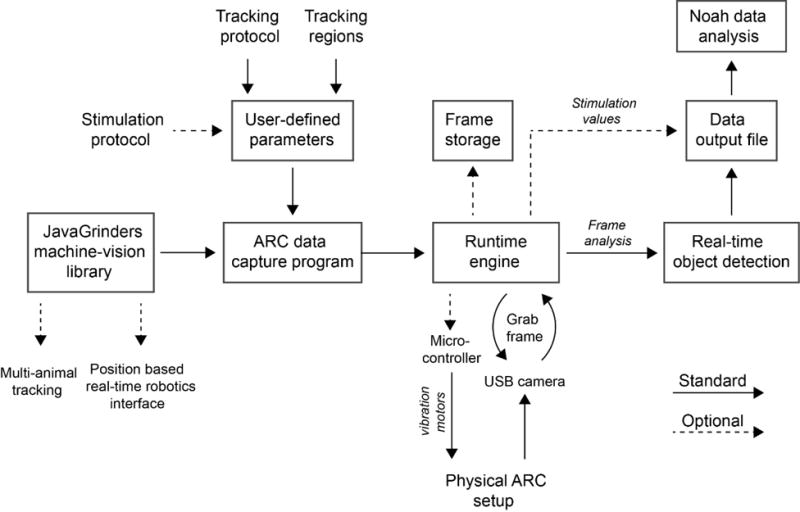
Illustration of instruction and data flow within the ARC. The ARC data capture program employs functions from the JavaGrinders Machine-Vision Library to grab and analyze frames from a USB camera. Pixel coordinate data of animals and food menisci are recorded to a text file and can be analyzed for various behaviors using the Noah program. Vibrational stimuli can optionally be delivered and recorded to the text file.
Prior to recording, flies are typically habituated in the ARC chamber for 20-24 hours. The capillaries do not need to be loaded with the dye-labeled oil during habituation since measurements are not typically made during this period. Habituation time can vary or be ignored to fit experimental needs. Depending on the incubator humidity, the agar at the bottom of each well dries out within 5-7 days, providing a practical time limit to long-term studies. If necessary, flies could be extracted from individual wells by mouth pipette and ARC measurements continued in another chamber with fresh agar.
(iv) Analyzing ARC data
Once the ARC is finished running, there will be a text file created by ARCController.java containing coordinates of each tracked dye band and fly. The first column of the dataset is a timestamp, followed by a y-value for each dye band, and then finally an x-y pair for each animal track (Supplementary Fig. 2). If the startArduinoStimulator(); is active, the output file will also contain a stimulus record as an additional column appended to the end of the file. This file can be directly loaded and processed by the Noah analysis program. After loading the file, many forms of analysis can be performed on the data set using intuitive user prompts and labeled output files. This includes binning behaviors by a specified time, spatial mapping, statistical behavioral compilation, and arousal threshold analysis. Alternatively, the regular structure of this data output is highly suitable for the use of custom analysis software or porting into existing analysis software. A sample data set of 30 Canton-S males recorded over 2 days is included (Supplementary Files).
Level of expertise
Construction of the ARC is simple enough for most graduate or capable undergraduate students. Installation of the data capture and analysis software requires some familiarity with computers, particularly software installation, using the terminal, and folder structures; the installation is straightforward when using the provided instructions. Executing an ARC experiment can be managed by an undergraduate student with minimal training.
MATERIALS
REAGENTS
Flies (control strain or genotype of interest). We have used Canton-S for the data shown here.
Dodecane (CH3(CH2)10CH3; Sigma-Aldrich, cat. no. 297879) ! CAUTION Dodecane can cause respiratory sensitization if inhaled.
Light mineral oil (Sigma-Aldrich, cat. no. 330779)
Copper (II) 5,9,14,18,23,27,32,36-octabutoxy-2,3-naphthalocyanine (Sigma-Aldrich, cat. no. 415286)
Chloroform (Sigma-Aldrich, cat. no. C2432) ! CAUTION Chloroform readily evaporates and can affect the central nervous system through inhalation, ingestion, or absorption through the skin. Use gloves when handling this substance. Work with this substance in a well-ventilated area or under fume hood.
Sucrose (Sigma-Aldrich, cat. no. 57-50-1)
Bacto Yeast Extract (BD, cat. no. 212750)
Agar (BD, cat. no. 214030)
SOFTWARE
Python 3.4+
Noah.py (included in Supplementary Files)
Java Standard Edition 8 Development Kit
Eclipse IDE for Java
COMPUTER
Any modern Mac or PC (PC must be running Linux OS) with 4+ GB RAM (6+ for 2 cameras) for data capture
PC for data analysis
Microsoft LifeCam 1080p (IR filter must be replaced with IR pass filter) or any infrared-sensitive USB camera
3D PRINT DESIGN FILES
(all .stl files are included in Supplementary Files)
ARC_chamber.stl
ARC_base.stl
ARC_gate.stl
ARC_cameramount.stl
ARC_cornerbracket.stl
ARC_chamberpiece1.stl and ARC_chamberpiece2.stl (if chamber is printed as halves)
EQUIPMENT
5-μl borosilicate glass capillaries (VWR, cat. no. 53432-706)
Parafilm (Sigma-Aldrich, cat. no. P7793-1EA)
Kimwipes (Fisher Scientific, cat. no. 06-666)
Double-sided tape (Staples, cat. no. 504829)
White construction paper (Staples, cat. no. 826292)
3D printer and filament (3D prints can alternatively be ordered online through various agents)
Infrared 850 nm LED reel (Environmental Lights, Product No. irrf850-390-reel)
Infrared Polyester Pass Filter (B&H Photo, # LE87C44)
200-μl pipette tips
0.2-μm cellulose acetate syringe filter (VWR, cat. no. 28145-477)
OPTIONAL EQUIPMENT FOR AROUSAL THRESHOLD ANALYSIS
Arduino Uno microcontroller
Shaft-less vibrating motors (Precision Microdrives, model 312-110)
USB A/B type cable
REAGENT SETUP
MENISCUS LABELING DYE
To make the meniscus marking dye, prepare 4 mL of 3:1 light mineral oil: dodecane solution. Add 10 mg of the copper dye reagent and vortex thoroughly. Centrifuge at 1,000 rpm for 5 seconds and collect supernatant for use. Store at room temperature away from light and heat for up to 2 years.
LIQUID DIET PREPARATION
For a standard liquid diet, dissolve 2.5 g sucrose and 2.5 g Bacto yeast extract in distilled water to a final volume of 100 mL. A hotplate with magnetic stirrer be used. When cool, sterile filter using a 0.2-μm cellulose acetate syringe filter. Aliquots can be stored at −20 °C for at least 1 year. Preparation may vary depending on diet.
AGAR PREPARATION
Combine 1 g agar with 100 mL distilled water. Dissolve agar completely by boiling on a hotplate with magnetic stirrer. After removing from the hotplate, wait until agar is warm to the touch (~50 °C) before adding to ARC chamber, as described below. Use freshly prepared agar for each experiment to avoid microbial contamination.
EQUIPMENT SETUP
PRINTING CHAMBER
Parts can either be printed using acrylonitrile butadiene styrene (ABS) or polylactic acid (PLA) filament. Δ CRITICAL Different materials may affect fly climbing or other behaviors. If flies have trouble climbing PLA, print chambers in ABS. If printing of the chamber consistently leads to edge warping, print the 2 pieces separately (ARC_chamberpiece1.stl and ARC_chamberpiece2.stl in Supplementary Files) and fuse post-print using chloroform.
CONSTRUCTION OF THE CHAMBER
Cut a piece of 2-mm thick acrylic plexiglass to 50 × 244 mm, or large enough to cover the well and gate region of the chamber while leaving the solid bottom of the chamber exposed for insertion into the base. Clamp the plexiglass to the chamber and apply a small amount of chloroform to all edges where the two materials meet. Be careful not to use too much chloroform. A proper amount will draw in between surfaces, while an excess will spill onto glass and create a cloudy view into the chamber. Allow the materials to fuse for at least 1 hour in a well-ventilated area or fume hood. The gate should be sanded gently until it can be moved in and out of the chamber sliding port without excessive force. To make sleeves which hold the capillaries in the gate, cut the narrow end of a 200-μl pipette tip with a razor blade until a capillary can fit through it using a small amount of force. Then cut the wide end so the section of the pipette tip is approximately 5 mm in length. Capillaries should not be able to slide freely through the sleeve.
PREPARATION OF THE RECORDING ENVIRONMENT
If the camera used is not already infrared sensitive, it must be converted by opening the housing and replacing the infrared filter with an infrared pass filter. The infrared filter, which can be removed with a razor blade, is the small glass lens with a shiny green tint found close to the actual sensor. For the pass filter, a lens-shaped piece of polyester infrared pass sheet is cut and fit over the exterior glass lens of the camera housing.
The infrared light source should be placed ~150 mm behind the chamber (Fig. 1c). Ideally, the lighting should be placed on the metal surface of the incubator wall or some other material that can serve as a heat sink. Alternatively, a fan can be placed perpendicular to the chamber to prevent local differences in temperature. While the number and distribution of infrared sources may vary depending on the brand of LEDs and of the camera used, infrared backlighting should be evenly distributed across the chamber and sufficiently intense to easily distinguish flies and dye bands from their respective backgrounds. We suggest several parallel strips of 850 nm infrared lighting reel (Environmental Lights, see Materials) of a width equal to the chamber. The camera should be mounted in the printed camera mount and secured to the incubator approximately 200 mm from the camera mount. If the lighting and filtering have been set up correctly, the camera should show a dark image under normal (visible) light when infrared lights are off.
Once the position of the camera and LED lighting are set, the chamber position must be determined. To check this, view a live image on the computer screen by using native webcam software or the ARCController software (described below). While viewing the live image, place the chamber between the camera and infrared lighting such that 1) the entire chamber and at least the liquid-filled section of the capillaries just fit in the camera view and 2) the edges of the chamber are square in the image. Once these conditions are met, mark the edges of the chamber base. Remove the chamber and, in place of the markings, secure two corner brackets to ensure consistent chamber placement (Fig. 1b).
CONSTRUCTION OF THE STIMULUS DELIVERY SYSTEM
To use the ARC for stimulus response measurements, a microcontroller setup needs to be included. Notably, this setup is optional, and is not required for normal motion-based sleep or feeding measurements. For the default stimulus response setup, attach an Arduino Uno to the data capture computer using a USB A/B type cable. On the Arduino, load the jArduino firmware (included with the Supplementary Files or on GitHub, https://github.com/SINTEF-9012/JArduino; instructions are found in the readme.txt file). Notably, the GitHub source contains a bug that may prevent loading to the board. The bug is fixed in the version provided in the Supplementary Files, which is recommended. Attach one wire of a Precision Microdrives shaft-less vibration motor to a PWM pin on the board. These will be marked by a ~ symbol and are usually pins 3, 5, 6, 9, 10, and 11, but may vary on different board layouts. Finally, connect the vibrator’s ground wires to the Arduino’s ground (GND) pin (Fig. 2a). The parameters passed to the Arduino for delivering stimuli (Fig. 2b) are listed in the ARC runtime program and are based on prior work31.
VIBRATION MOTOR ATTACHMENT
While there are many ways in which the vibration motors can be temporarily attached to the chambers, we found that a small dot of hot glue applied by a glue gun worked well and was easily removed before washing the chambers. An example setup using three motors secured to the chamber is shown (Fig. 1c).
SOFTWARE INSTALLATION
To install the necessary open source libraries as dependencies, follow the instructions on the ARC blog (http://javagrinders-arc.blogspot.com/2017/01/install-open-source-tools-for-running.html). Once the necessary components are installed, the ARC data capture program, ARCController.java, can be run within an integrated developer environment (IDE). The Eclipse IDE allows users to access and change the ARCController settings and optimize tracking conditions. To set up the IDE, open the Eclipse application and, when prompted, choose a default workspace (typically in the Documents folder). Next, copy the JavaGrinders_ARC folder (from the Supplementary Files or download and unzip from the ARC installation blog) into the Eclipse workspace folder. In Eclipse, open the package explorer by selecting menu item Window → Show View → Package Explorer. From the File menu, select New → Java Project. In the Java Project Wizard, enter Project name “JavaGrinders_ARC” and click Finish. If the ARC project is successfully imported into the package explorer, navigate to JavaGrinders_ARC → src → _ARC and select “ARCController.java”. Select menu item Navigate → Open to access all of the necessary parameters for operating the ARC data acquisition program, including the frame capture rate, definitions for the tracking regions, and the stimulus protocol. Default parameters are provided and their optimized settings are described below. To test the camera, select menu item Run → Run. If the program finds a video camera, it will start the tracker and open the main window.
SETTING AND OPTIMIZING TRACKING PARAMETERS
To define tracking regions for the animals and the capillaries, coordinates need to be determined. The program will take a given user-defined rectangle and sub-divide it into an equal number of tracks, defined by the value of kNAnimals (30 for the standard ARC chamber). To obtain coordinates for each rectangle, first place an ARC chamber in the incubator within the mounting brackets. Next, run ARCController.java. After defining the output file name and location, a pane with two windows will appear. The left is the live image from the camera and the right is an analysis window, showing object tracking data overlaid on the image. Click “Reference” to capture a reference image of your chamber. The reference button should only be used to find appropriate parameters and not during a run, as it can modify the adaptive background determination performed for the food tracking. At the top of the Tracker window, select Objects → Screen info → Rectangle. A single rectangle can now be drawn by clicking and dragging on the live image. Once you have drawn the rectangle, which should define the outer edges of the entire area containing the fly enclosures, the following coordinates will be displayed in the Eclipse console window as: new Rectangle (ARCAnimalLeft, ARCAnimalTop, ARCAnimalWidth, ARCAnimalHeight). These 4 parameters should be entered and saved into the ARCController.java program. Once these parameters are set, close the current tracker window and re-run ARCController.java to check that your defined tracking window aligns well with the chamber.
For tracking of the capillary dye bands, use the same method for defining the fly enclosures to define the capillary tracking region. Importantly, this region should include as much of the food-containing segment of the capillaries as possible while excluding any existing markings on the capillaries and parts of the chamber that are darker than the dye band. An example of how the chamber and dye tracking regions should look is shown (Fig. 3a,b). The parameters derived from the rectangle drawn around the capillaries will be displayed in the Eclipse console window as: new Rectangle (ARCFoodLeft, ARCFoodTop, ARCFoodWidth, ARCFoodHeight). As for the fly tracking, these 4 parameters should be entered and saved into ARCController.java. Once these parameters are set, again close and re-run ARCController.java to check that the animal and capillary tracking windows align well with the chamber.
The variables used by the program to identify flies are: kAnimalPxThresh, which defines the threshold pixel difference for outlining objects; kAnimObjSize, which creates a range of acceptable object size; and kAnimIsDkObj, which tells the program whether the fly is a dark object on a light background (true) or vice versa (false). Lowering kAnimalPxThresh will track objects that have little contrast from their background, but may also increase false positive tracks. If an animal is not within the acceptable kAnimObjSize, the tracker records a missed read. To optimize kAnimalPxThresh, place an ARC loaded with animals into the brackets and run ARCController.java. Click Objects → Get Pixel Value and click on the darkest region of the background within your animal track region. Record the pixel value written to the console. Next, find a stationary animal and record the pixel value of its center. Subtracting the animal pixel value from the background pixel value will give you an approximation of kAnimalPxThresh. Change the value and restart ARCController.java. Click Reference and then Show. If the red representations of the animals in the analysis window are smaller than the animals, decrease kAnimalPxThresh, and vice versa. Repeat until the analysis window shows the correct size for the animals. To determine an appropriate animal size (kAnimObjSize), click Analysis → Get Coordinates for → Rectangle, and draw a rectangle around a fly and use the area output written to the console.
Once all parameters are set and saved, re-run ARCController.java and only click Show. If the flies and dye bands are faithfully represented in red color on the right panel of the tracking window, click Analyze. Data are now being saved to the output file in real time. The animals and dye bands should then be identified as green polygons in the analysis window (Fig. 3b). Notably, JavaGrinders uses background subtraction, so if a fly has not moved since its position during the reference image, then it will not show up immediately in the right panel. This is not an issue, since the Noah analysis software will backfill initial empty coordinates with the first identified position. The capillaries use adaptive thresholding and should all show up immediately. The program will then collect data until Stop is clicked or the user-defined recording duration is reached.
To measure arousal threshold, the stimulus protocol needs to be turned on. In ARCController.java find the line containing the phrase “//startArduinoStimulator();”. Delete the “//” at the beginning of the line to activate the stimulus protocol. ARCController contains default stimulus protocol values which were successfully employed previously4. However, the specific stimuli delivery can be tailored to individual experimental setups. The default parameters deliver a stimulus protocol once every hour (Fig. 2b). To modify this protocol, edit the values in ARCController.java found immediately below the line which reads “public static void runArduinoStimulator() {”.
While the ARC is designed to eliminate the need for video/image storage, an option for saving frames at a given rate is included. To use this, remove the “//” at the beginning of the following lines in ARCController.java.
//ImgProducer theProducer = new ImgProducer(null,theARCTracker.nativeWidth,theARCTracker.nativeHeight,60000);
//theProducer.getItsRenderer().setPlotTargetStyle(FrameRenderer.TargetStyle_RAWIMAGE);
//theProducer.setSubRect(kFoodArea);
//theARCTracker.setVideoProducer(theProducer);
The value of 60000 (highlighted in gray) denotes the time in milliseconds between the storage of images. Notably, storing these images is processor intensive and can increase the risk of older machines crashing.
INSTALLING DATA ANALYSIS SOFTWARE
Download python 3.x on a PC (currently Noah.py only runs on the PC version of python) in order to run Noah.py. When running Noah for data analysis, it will prompt for the entry of several values regarding the tracking conditions. First, it will request the number of pixels per cm in the tracking setup. To obtain this value, measure the width of the chamber in cm (using a ruler) and in pixels by using the Rectangle tool in the ARCController.java program, as described above (the third parameter provides the user-drawn rectangle width in pixels). Use these numbers to derive pixels per cm. The length of a fly is assumed to be ~0.3 cm for filtering motion events less than half a body length, which has been found to be appropriate for video-based fly motion tracking32. A calibration preset in Noah determines the volume of food consumed by using a cm-to-μl conversion based on the recommended 5-μl capillaries. Both the fly length and conversion factor can be changed in the constants section of Noah.py, which is found in the first 60 lines of the script. Python scripts can be edited by right-clicking the program icon and selecting Edit with IDLE. The conversion factor for capillaries can be determined by loading several capillaries with 5 μl of water, measuring the liquid height, and calculating the average cm/μl.
PROCEDURE
(see Supplementary Video 1 for an overview of the procedure)
Chamber setup ● TIMING ~20 min
-
1|
Boil agar (see Reagent Setup) and allow to cool until flask is warm to the touch.
-
2|
Using a 1000-μl pipettor, pipette 300 μl of 1% (w/v) agar into each fly compartment. Angle the pipettor against the back wall (not the transparent front cover) during pipetting. Allow 2+ hours to cool and solidify. Just prior to the experiment, slide a chamber gate into place, using a small dot of hot glue to secure if the gate feels loose.
Loading animals for habituation ● TIMING ~40 min
-
3|
Thaw a liquid diet and fill appropriate number of capillaries (30 per ARC chamber) with habituation diet. Wipe away excess fluid on the exterior of each capillary. Capillaries can be filled and maintained on a “capillary holding box” which is made by wrapping a small plastic container, such as a 1000-μl pipette tip container, with a strip of double-sided tape. Capillaries are filled approximately halfway to two-thirds full.
-
4|
Using a mouth aspirator, gently aspirate a fly through the gate hole for each compartment. Quickly place the gate sleeve into the hole followed by a habituation capillary to block the fly from exiting. Repeat until all wells are loaded.
-
5|
Attach capillary backsplash to the capillaries. If experiment requires arousal threshold measurement, attach shaft-less vibration motors by applying a small dot of hot glue to the back of the motors and then pressing to the chamber (these can be easily removed with a flat metal object following the experiment).
-
6|
Place chamber in the recording incubator inside brackets and allow flies to habituate before preparing dye-labeled capillaries. Flies are typically habituated under these conditions for 20-24 hours, but this can be changed according to the experiment.
Preparing/changing dye-labeled capillaries ● TIMING ~45 min
-
7|
Place ~40 μl of the marking dye on a small piece of parafilm. Quickly touch the tip of a capillary to the dye solution, allowing a small amount to load. Gently blot the capillary tip on a kimwipe until the dye band is ~2 mm in height. Quickly place the same capillary end into liquid diet and allow dye band to rise until it is about halfway to two-thirds up the capillary or ~5 mm below any external marks that might obscure the meniscus. Δ CRITICAL If the dye band is visually split (e.g., due to an air bubble), discard capillary. This split can cause major tracking issues. Maintain capillaries on a holding box (see step 3) and repeat capillary filling until the needed number of capillaries are made. ? TROUBLESHOOTING
-
8|
Take the chamber out of the recording incubator and swap out all habituation capillaries with dye band-labeled capillaries. The backsplash should be kept on during this step. Δ CRITICAL Do not remove more than 3 capillaries at any time. This will ensure the backsplash stays in place and that animals do not escape through the sleeves. Make sure capillaries are relatively straight and at the same height. Return the chamber to the recording incubator.
-
9|
Open ARCController.java in the Eclipse compiler. If you are not running an arousal threshold experiment, comment the line containing “StartArduinoStimulator();“ by placing “//” at the start of the line. Run ARCController.java by pressing the green play button at the top of the eclipse window. Name your output file. Δ CRITICAL If you name an output file with a name already used in the environment, the file will be overwritten. If you have not already set the tracking regions and optimized the tracking parameters, refer to the SETTING AND OPTIMIZING TRACKING PARAMETERS section under EQUIPMENT SETUP. If you have, proceed to the next step. ? TROUBLESHOOTING
-
10|
Click show. If the tracking analysis window outlines the animals and dye bands well, click “Analyze”. If they do not, continue to optimize the tracking parameters until tracking is accurate. Stop software when experiment is over, or stop and start again for capillary changes (typically once per day). Use a different output file name for each continuous period of data collection. ? TROUBLESHOOTING
-
11|
When experiment is done, disassemble chamber. Remove sleeves and soak in 70% ethanol for at least 1 hour before rinsing with distilled water and drying.
Cleaning chamber and capillaries ● TIMING ~10 min
-
12|
Use water pick to remove agar from wells. Scrub each well with a pipe cleaner using unscented soapy water, followed by a thorough rinse. Allow chambers to dry and store until next experiment. If they are going to be reused, capillaries can be washed by flushing them under running distilled water to clear remaining liquid, boiling in distilled water for 30 minutes, and then using a vacuum system to suck the capillaries dry. However, this may lead to dye streaking if done improperly.
-
13|
Rinse sleeves with distilled water and allow to dry. Store in a clean container at room temperature until next experiment.
Processing ARC file ● TIMING ~2 min
-
14|
Move the file generated by the ARC to a PC running python. Run python and select option 1 by typing 1 and pressing enter. Click and drag the text file onto the Noah.py window. The address of the file will then appear. If it does not, type the address of the file manually (e.g. C:\Users\UserName\Desktop\outputfile.txt) and press enter.
-
15|
Feeding, tracking, and stimulus delivery data are now ready to be used by Noah for various analyses. The software is prompt based, in which the program indicates the desired input at each stage. To use any analysis feature, type the number given in the menu for the corresponding analysis feature. For instance, to obtain feeding data bins, select option 5. Input the desired bin size in following to the prompt “Enter Bin Size(mins):”. You can then add bins for meal frequency, meal size, and satiety by typing “y” in response to each request (e.g. “Add bins for frequency (y/n): ”). The file will then automatically be written to the environment in which the program is in as a comma separated values sheet. The output file name is automatically tied to the original file name (e.g., “outputfile.txt” yields feedBins_binSize_outputfile.csv). All other analyses work in a similar fashion.
● TIMING
Steps 1-2, Preparing the chamber, 20 min
Steps 3-6, Loading animals for habituation, 40 min
Steps 7-11, Preparing/changing dye-labeled capillaries, 45 min
Steps 12-13, Cleaning chamber, 10 min
Steps 14-15, Processing ARC file, 2 min
TROUBLESHOOTING
See Table 1 for troubleshooting guidance.
Table 1.
Troubleshooting
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
|
Setup |
jArduino firmware will not load to board with the following error message: error: no matching function for call to ‘JArduino::init_JArduino()’ |
Serial connection is not synchronized between computer and Arduino. | Go into the JArduinoFirmware.ino file and replace the following line: void setup() { //initialize the JArduino protocol_JArduino.init_JArduino(); } with void setup(){ //initialize the JArduino protocol uint16_t baud = 9600;_JArduino.init_JArduino(baud); } |
| 7 | Meniscus-labeling dye bands are frequently splitting when loading capillaries. | Excess time between loading dye band and liquid food allows a bubble to form at base of capillary. | Load capillary faster or increase the % of dodecane in the dye solution to prevent dye from climbing walls of capillaries. |
| 9 | ARCTracker failed to connect to USB camera at startup with error message: java.lang.Exception: FrameCapture Exception in OpenCVRecordProc: no valid frame during initialization |
USB camera is being used by another program from OpenCV. | Unplug the USB camera, reboot the computer, and plug the USB camera back in. |
| 10 | Meniscus labeling dye bands are streaking (or disappearing) during experiment. | There is not enough surfactant to keep dye band from sticking to the wall of the capillary. | Increase the % of dodecane in the dye solution and make sure the dye is centrifuged thoroughly to remove non-dissolved particulates. |
| 10 | The liquid food is evaporating too quickly. | There is low humidity in the chamber or the gate does not seal the chamber well. | Either create a tighter fit between the gate and chamber by using a new gate which is slightly larger, increase the humidity in the incubator housing the ARC, or lower the incubator temperature within an acceptable experimental range. |
| 10 | Liquid diet is totally consumed before 24 hours. | Some diets or manipulations will cause this to happen because the fly is too hungry or because evaporation is too high. | Stop ARCTracker and change capillaries mid-day, then restart ARCTracker. It may be best to do this switch at Zeitgeber time = 12 hours, when animals are already quite active, to limit disruption of any natural behavior. |
| 10 | Animal tracking has high error rate | This could result from several optical traits within the ARC system, but is most likely caused by poor contrast between the chamber and flies. | Increase backlighting by using more infrared LEDs. If this is not an option, the tracking parameters can be altered to limit the number of false tracks obtained. To do this, run the ARCTracker program with incremental increases or decreases in kAnimPxThresh. Using the “Show” button can verify the accuracy of your tracking parameters. |
| 10 | Dye-band tracking has high error rate. | Backlighting is insufficient or there is another object in the dye-band tracking space. | Add lighting or change USB camera settings until lighting provides strong contrast between the dye bands and background. Search the image carefully for any specks or discolorations on the capillary backsplash paper. Make sure the capillary markings are outside of the defined track areas. |
| 10 | Selecting “Show” outlines objects clearly in analysis window, but objects are not being tracked after starting analysis. | This is likely because objects are being identified, but are not close enough to the specified size of the object. | Check the approximate size in pixels of the dye bands and animals. Change the kAnimObjSize or kFoodObjSize to reflect the size of the animals or dye bands. |
ANTICIPATED RESULTS
Data obtained from a standard ARC experiment can be compared to the sample data set that used 2.5% sucrose + 2.5% yeast extract food (Fig. 5). Circadian profiles should show heightened activity and reduced sleep near Zeitgeber time 0 and 12 hours (when the visible light source turns on and off, respectively). Rhythmicity should be evident in individuals and in averaged cohort data (Fig. 5a,b). Feeding should also show rhythmicity33, however the peak timing and intensity can vary substantially depending on the diet, genotype, or the amount of time the animal has been habituated in the chamber. Integration of sleep and feeding should show an increase in sleep following meals (Fig. 5c), however, the intensity of postprandial sleep is also dependent on the diet used4. Analysis of postprandial sleep is studied using individual meals rather than animals to capture the variability in meal size and nutrient consumption within individuals4. However, gross trends are nearly indistinguishable when using individual meals or animals. Spatial analysis reveals that flies spend more time near the food capillary at the top of the chamber than the agar at the bottom (Fig. 5d). Unhealthy flies or animals under other manipulations may show deviations from this pattern. If the experiment contains arousal threshold measurements, these values should share a similar profile to that of sleep (Fig. 5e). Furthermore, arousal threshold surrounding meals should show that consumption temporarily raises arousal threshold (Fig. 5f). Poor data quality or unhealthy animals may result in less obvious trends in circadian behavior, spatial tendency, or postprandial sleep; although there is no consensus on how health affects these behaviors.
Figure 5. Expected data from the ARC.
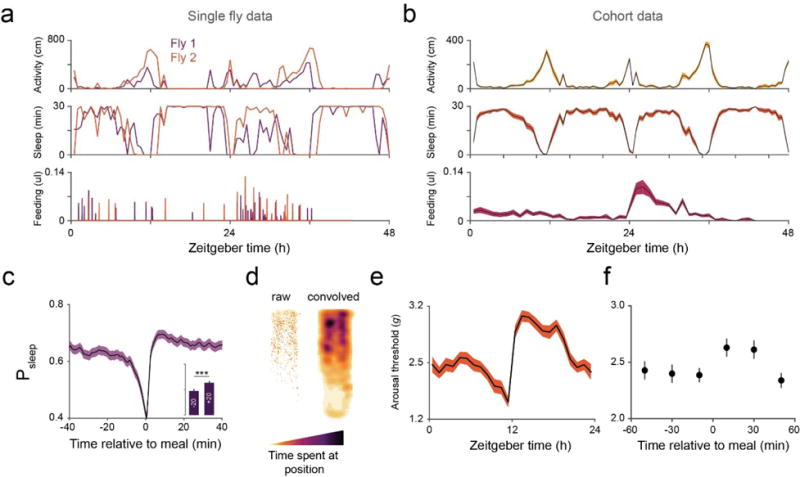
(a) Individual activity, sleep, and feeding profiles for 2 individuals over a 48-hour period. (b) Activity, sleep and feeding averaged from a cohort (n = 30 flies, shaded line represents mean ± s.e.m.). (c) Postprandial sleep plot of individual meals from a cohort. Inset graph shows average Psleep (± s.e.m.) in the 20 min before (−20) and after (+20) meals (axis scaled to the parent graph). n = 486 events from 30 flies; ***, p < 0.001, Wilcoxon matched-pairs sign rank test. (d) Heat map of fly location over the 48-hour period. n = 30 flies. (e) Circadian rhythm of arousal threshold has a similar profile to sleep. (f) Animal stimulus response at 0–20, 20–40, and 40–60 min before and after meals. Arousal events are filtered for prior time inactive to control for sleep depth (0–5 mins shown, minimum 1 sec inactivity). Circles represent mean ± s.e.m.; n = 316 meals from 60 flies. All flies were Canton-S males.
Supplementary Material
Supplementary Figure 1: Comparison of data between Expresso and ARC. (a) Total ingestion volume and calorie intake for 33 minutes on the indicated diet following 24 hours of starvation, measured in the Expresso. n = 20 male flies. Expresso data were extracted from22. (b) Replication of Expresso experiment using the ARC. n = 15 Canton-S males. Box and whiskers represent mean with 95% confidence intervals; blue circles represent mean ± s.e.m.
Supplementary Figure 2: ARC data output structure. Mock data portraying 7 reads from the ARC. Each row represents a single read and each column is organized by the repeating structure shown. Labels (red) do not appear in the output file, whereas data (black) will appear in a tab-delimited format. The final column (Stimulus Delivered) will only appear in the data file if the optional arousal threshold module is run.
Supplementary Files: Compressed archive of ARC software. Archive contains 7 .stl files for 3-D printing of ARC components, jArduino firmware, JavaGrinders framework and ARC modules (ARCController and ARCControllerMultiCam), Noah analysis program (python-based), and a sample ARC data set of 30 Canton-S males over 2 days.
Supplementary Video 1: Video guide to using the ARC.
EDITORIAL SUMMARY.
The construction and use of the Activity Recording CAFE (ARC), a machine-vision based system for the integrated measurement of sleep and feeding in individual Drosophila, is described.
Acknowledgments
We thank Jack Jacobs and Amita Sehgal for comments on the manuscript. This work was funded by the National Institutes of Health (R21DK092735 to W.W.J.).
Footnotes
TWEET #neuroscience #drosophila #machinevision #behavior
AUTHOR CONTRIBUTIONS
All authors contributed to the conception, development, and testing of the ARC; K.R.M., R.H., and W.W.J. wrote the manuscript; all of the authors reviewed and revised the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.MacFadyen UM, Oswald I, Lewis SA. Starvation and human slow-wave sleep. J Appl Physiol. 1973;35:391–394. doi: 10.1152/jappl.1973.35.3.391. [DOI] [PubMed] [Google Scholar]
- 2.Dario AJ, Lopes PR, Freitas CG, Paschoalini MA, Marino-Neto J. Electrographic patterns of postprandial sleep after food deprivation or intraventricular adrenaline injections in pigeons. Brain Res Bull. 1996;39:249–254. doi: 10.1016/0361-9230(95)02115-9. [DOI] [PubMed] [Google Scholar]
- 3.Keene AC, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010;20:1209–1215. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy KR, et al. Postprandial sleep mechanics in Drosophila. eLife. 2016;5 doi: 10.7554/eLife.19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linford NJ, Chan TP, Pletcher SD. Re-patterning sleep architecture in Drosophila through gustatory perception and nutritional quality. PLoS Genet. 2012;8:e1002668. doi: 10.1371/journal.pgen.1002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catterson JH, et al. Dietary modulation of Drosophila sleep-wake behaviour. PLoS ONE. 2010;5:e12062. doi: 10.1371/journal.pone.0012062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells AS, Read NW, Uvnas-Moberg K, Alster P. Influences of fat and carbohydrate on postprandial sleepiness, mood, and hormones. Physiol Behav. 1997;61:679–686. doi: 10.1016/s0031-9384(96)00519-7. [DOI] [PubMed] [Google Scholar]
- 8.Landstrom U, Knutsson A, Lennernas M, Soderberg L. Laboratory studies of the effects of carbohydrate consumption on wakefulness. Nutr Health. 2000;13:213–225. [PubMed] [Google Scholar]
- 9.Grandner MA, Kripke DF, Naidoo N, Langer RD. Relationships among dietary nutrients and subjective sleep, objective sleep, and napping in women. Sleep Med. 2010;11:180–184. doi: 10.1016/j.sleep.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koban M, Sita LV, Le WW, Hoffman GE. Sleep deprivation of rats: the hyperphagic response is real. Sleep. 2008;31:927–933. [PMC free article] [PubMed] [Google Scholar]
- 11.Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: Sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann Intern Med. 2004;141:846–850. doi: 10.7326/0003-4819-141-11-200412070-00008. [DOI] [PubMed] [Google Scholar]
- 12.Cirelli C, Bushey D. Sleep and wakefulness in Drosophila melanogaster. Ann N Y Acad Sci. 2008;1129:323–329. doi: 10.1196/annals.1417.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Artiushin G, Sehgal A. The Drosophila circuitry of sleep-wake regulation. Curr Opin Neurobiol. 2017 doi: 10.1016/j.conb.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenspan RJ, Tononi G, Cirelli C, Shaw PJ. Sleep and the fruit fly. Trends Neurosci. 2001;24:142–145. doi: 10.1016/s0166-2236(00)01719-7. [DOI] [PubMed] [Google Scholar]
- 15.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 16.Hendricks JC, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/S0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 17.van Alphen B, Yap MH, Kirszenblat L, Kottler B, van Swinderen B. A dynamic deep sleep stage in Drosophila. J Neurosci. 2013;33:6917–6927. doi: 10.1523/JNEUROSCI.0061-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–1576. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 20.Berry JA, Cervantes-Sandoval I, Chakraborty M, Davis RL. Sleep facilitates memory by blocking dopamine neuron-mediated forgetting. Cell. 2015;161:1656–1667. doi: 10.1016/j.cell.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deshpande SA, et al. Quantifying Drosophila food intake: comparative analysis of current methodology. Nat Methods. 2014;11:535–540. doi: 10.1038/nmeth.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yapici N, Cohn R, Schusterreiter C, Ruta V, Vosshall LB. A taste circuit that regulates ingestion by integrating food and hunger signals. Cell. 2016;165:715–729. doi: 10.1016/j.cell.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ro J, Harvanek ZM, Pletcher SD. FLIC: high-throughput, continuous analysis of feeding behaviors in Drosophila. PLoS ONE. 2014;9:e101107. doi: 10.1371/journal.pone.0101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itskov PM, et al. Automated monitoring and quantitative analysis of feeding behaviour in Drosophila. Nat Commun. 2014;5:4560. doi: 10.1038/ncomms5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradski G. The OpenCV library. Doctor Dobbs J. 2000;25:120–126. [Google Scholar]
- 26.Kabra M, Robie AA, Rivera-Alba M, Branson S, Branson K. JAABA: interactive machine learning for automatic annotation of animal behavior. Nat Methods. 2013;10:64–67. doi: 10.1038/nmeth.2281. [DOI] [PubMed] [Google Scholar]
- 27.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Locomotor activity level monitoring using the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.prot5518. [DOI] [PubMed] [Google Scholar]
- 28.Ja WW, et al. Water- and nutrient-dependent effects of dietary restriction on Drosophila lifespan. Proc Natl Acad Sci. 2009;106:18633–18637. doi: 10.1073/pnas.0908016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilestro GF. Video tracking and analysis of sleep in Drosophila melanogaster. Nat Protoc. 2012;7:995–1007. doi: 10.1038/nprot.2012.041. [DOI] [PubMed] [Google Scholar]
- 30.Ja WW, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faville R, Kottler B, Goodhill GJ, Shaw PJ, van Swinderen B. How deeply does your mutant sleep? Probing arousal to better understand sleep defects in Drosophila. Sci Rep. 2015;5:8454. doi: 10.1038/srep08454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donelson NC, et al. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS ONE. 2012;7:e37250. doi: 10.1371/journal.pone.0037250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Comparison of data between Expresso and ARC. (a) Total ingestion volume and calorie intake for 33 minutes on the indicated diet following 24 hours of starvation, measured in the Expresso. n = 20 male flies. Expresso data were extracted from22. (b) Replication of Expresso experiment using the ARC. n = 15 Canton-S males. Box and whiskers represent mean with 95% confidence intervals; blue circles represent mean ± s.e.m.
Supplementary Figure 2: ARC data output structure. Mock data portraying 7 reads from the ARC. Each row represents a single read and each column is organized by the repeating structure shown. Labels (red) do not appear in the output file, whereas data (black) will appear in a tab-delimited format. The final column (Stimulus Delivered) will only appear in the data file if the optional arousal threshold module is run.
Supplementary Files: Compressed archive of ARC software. Archive contains 7 .stl files for 3-D printing of ARC components, jArduino firmware, JavaGrinders framework and ARC modules (ARCController and ARCControllerMultiCam), Noah analysis program (python-based), and a sample ARC data set of 30 Canton-S males over 2 days.
Supplementary Video 1: Video guide to using the ARC.


