Abstract
Cancer cachexia is associated with muscle weakness and atrophy. We investigated whether 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3), which has previously been shown to increase skeletal myoblast oxygen consumption rate, could reverse the deleterious effects of tumor cell conditioned medium on myoblast function. Conditioned medium from Lewis lung carcinoma (LLC1) cells inhibits oxygen consumption, increases mitochondrial fragmentation, inhibits pyruvate dehydrogenase activity, and enhances proteasomal activity in human skeletal muscle myoblasts. 1α,25(OH)2D3 reverses the tumor cell-mediated changes in mitochondrial oxygen consumption and proteasomal activity, without changing pyruvate dehydrogenase activity. 1α,25(OH)2D3 might be useful in treatment of weakness seen in association with CC.
Keywords: Cancer; Skeletal muscle; Mitochondria; Oxygen consumption; Gene expression; 1α,25-dihydroxyvitamin D3
1. Introduction
Cancer cachexia (CC) is associated with decreased skeletal muscle mass and function, increased morbidity and mortality, and reduced response to chemotherapy and survival following surgery [1,2]. Altered regulation of inflammatory, activin A-myostatin, ubiquitin-proteasome, and autophagy-lysosome pathways is observed in CC in vivo [3–8]. Skeletal muscle mitochondrial volume density, dynamics, and oxidative protein expression are altered in CC models in vivo [9]. Tumor cell-derived mediators such as TNFα, IL-6, TWEAK, myostatin, PTHrp and extra-cellular vesicle HSP have been postulated to play a role in the pathogenesis of CC [3–6,10–15]. Therapy with reagents that block effects of various mediators e.g. anti-IL6 antibody [16], anti-activin/myostatin reagents [10], and anti-TNFα reagents [17,18], however, has not resulted in clinical improvement of CC. Other therapeutic approaches are needed to reverse or mitigate CC. In an earlier report [19], we showed that the active vitamin D metabolite, 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3), increased cellular oxygen consumption rate (OCR), mitochondrial volume and branching in association with changes in expression of mitochondrial pro-fusion and profession proteins and pyruvate dehydrogenase activity in human skeletal muscle myoblasts. The salutary effects of 1α,25(OH)2D3 on skeletal muscle function led us to investigate its ability to block deleterious effects of tumor conditioned medium (CM) on muscle cells. We show that 1α,25(OH)2D3 attenuates the deleterious effects of tumor CM on human skeletal muscle myoblasts.
2. Methods
2.1. Cell culture
Human skeletal muscle cells (Lonza, CC-2561) were grown at 37°C in 5% CO2 in CC-3161 medium [19]. Lewis lung carcinoma-1 cells (LLC1 ATCC® CRL-1642, Manassas, VA 20110 USA) were cultured in Dulbecco's MEM containing 10% FBS. MLE12 cells (ATCC®: CRL-2110) were cultured in HITES medium supplemented with 2% FBS. HEK-293 cells (ATCC®: CRL-1573) were cultured as recommended.
2.2. Measurement of oxygen uptake by cells
An XF24 Extracellular Flux Analyzer (Seahorse Biosciences) was utilized to measure OCR and proton production rate (PPR) as previously described [20]. Cells were seeded on a microplate (10,000 cells/well). At ~80–90% confluence, cells were treated with CM with or without added 1α,25(OH)2D3 (10−8M) or ethanol, which was added 24 h prior to the experiment. LLC1 and MLE12 CM was filtered with 0.45 μM filters prior to use. OCR was measured following sequential addition of oligomycin (0.5 μg/ml), FCCP (1 μM) and rotenone (0.5 μM)/antimycin A (1 μM). Baseline respiration rate, coupled respiration rate, maximal respiration rate and oxidative reserve were calculated [21].
2.3. Assessment of mitochondrial morphology and fragmentation
Myoblasts were plated on 8-well glass-bottom plates (LabTek), treated with LLC1 CM with or without added 1α,25(OH)2D3 (10−8M) or ethanol for 24 h, and incubated with 500 nM Mito-Tracker Green FM (Life Technologies; Ex 490 nm/Em 516 nm) for 5 min at 22 °C. Cells were imaged using a Nikon A1R confocal system [19]. Mitochondrial morphometric analysis was performed as described by Koopman et al. [22,23]. Form factor and aspect ratio were calculated [24]. A decrease in aspect ratio and/or form factor indicates mitochondrial fragmentation [22–24]. For electron microscopy (EM), cells were fixed in Trump's fixative, post-fixed with 1% OsO4, washed in H2O, en-bloc stained with 2% uranyl acetate, dehydrated through an ascending ethanol series, and infiltrated with Embed-812. Sections were stained with lead acetate and observed on a JEOL 1400 electron microscope operated at 80 kV.
2.4. Assessment of mitochondrial protein expression
We assessed changes in mitochondrial protein expression using Western blotting with antibodies (from Abcam, unless otherwise noted) directed against the following mitochondrial proteins or complexes: VDAC1/Porin (ab15895), PDH antibody cocktail (ab110416), PDH E1-α-subunit, phospho-293 (ab177461), PDP2 (ab133982), PDK4 (ab71240), total OXPHOS (ab110411), Mfn-1 (ab57602), Mfn-2 (ab56889), OPA1 (ab42364), Drp-1 (ab56788), and Fis1 (sc-98900, Santa Cruz Biotechnology) as described [19].
2.5. Assessment of mitochondrial and nuclear DNA
Total DNA was prepared from treated myoblasts was used to measure mitochondrial genes ND1 and ND6 and nuclear genes BECN1 and NEB using a NovaQUANT™ human mitochondrial to nuclear DNA ratio kit (EMD Millipore).
2.6. Measurement of pyruvate dehydrogenase in cell homogenates
Pyruvate dehydrogenase (PDH) was measured in 96-well plates with a colorimetric assay (BioVision, Milpitas, CA).
2.7. Measurement of proteasomal activity
Chymotrypsin-like proteasomal activity was measured with the Proteasome-Glo kit from Promega (G8660). Myoblasts were scraped from a T175 fiask, re-suspended in growth medium, and added to a 96-well white-walled plate at a seeding density of 10,000 cells/ well. At 80–90% confluence, cells were treated with LLC1 CM or non-CM with or without added 1α,25(OH)2D3 (10−8M) or ethanol for 24 h prior to the experiment. 100 μL of Proteasome-Glo cell-based reagent was added to 100uL of sample and incubated for 10 min. Luminescence was measured on a SpectroMax M2e.
2.8. Preparation of libraries
mRNA-seq libraries were prepared using 200 ng of total RNA with a TruSeq RNA Sample Prep Kit v2 (Illumina) as described previously [19]. Libraries were sequenced at approximately 75 million reads/sample following Illumina's protocol using the Illumina cBot and HiSeq 3000/4000 PE cluster kit. The flow cells were sequenced as 100 X 2 paired end reads on an Illumina HiSeq 4000 using Hiseq 3000/4000 sequencing kits and HCS v3.3.20 data collection software. Base-calling was performed using Illumina's RTA version 2.5.2.
2.9. mRNA-seq data analysis
Processing of the mRNA-seq data was performed using MAP-RSeq workflow (v1.2.0.0) [25] and RSeQC software (v2.3.2) [26]. Paired-end reads were aligned by TopHat (v2.0.12) against the hg19 and mm10 genome build for human and mouse samples [27]. Gene counts were generated using FeatureCount software (v1.4.4); the gene annotation files were obtained from Illumina. Differential expression analysis was performed with edgeR v2.6.2 to identify genes with altered expression between treatment groups [28]. A cutoff for false discovery rate–adjusted p-value was set at 0.01. MetaSecKB (http://bioinformatics.ysu.edu/secretomes/animal/index.php) was used to determine the secretome from differentially expressed genes.
2.10. Pathway analysis
Pathway enrichment analysis was performed with Ingenuity Pathway Analysis program (IPA, Ingenuity Systems; cut-off P value = .05).
2.11. Analysis of genes that encode mitochondrial proteins
Mitochondrial proteins were identified based on a compendium from MitoCarta [29].
2.12. Statistical methods
Statistical differences between samples were analyzed using Student's two-tailed t-test, assuming equal variance. A P value of <0.05 was regarded as statistically significant.
2.13. Data sharing
All sequencing data in this report have been deposited in Gene Expression Omnibus database, accession number GEO103550.
3. Results
3.1. The inhibitory effects of LLC1 CM on mitochondrial oxygen uptake in human skeletal muscle myoblasts are mitigated by the addition of 1α,25(OH)2D3
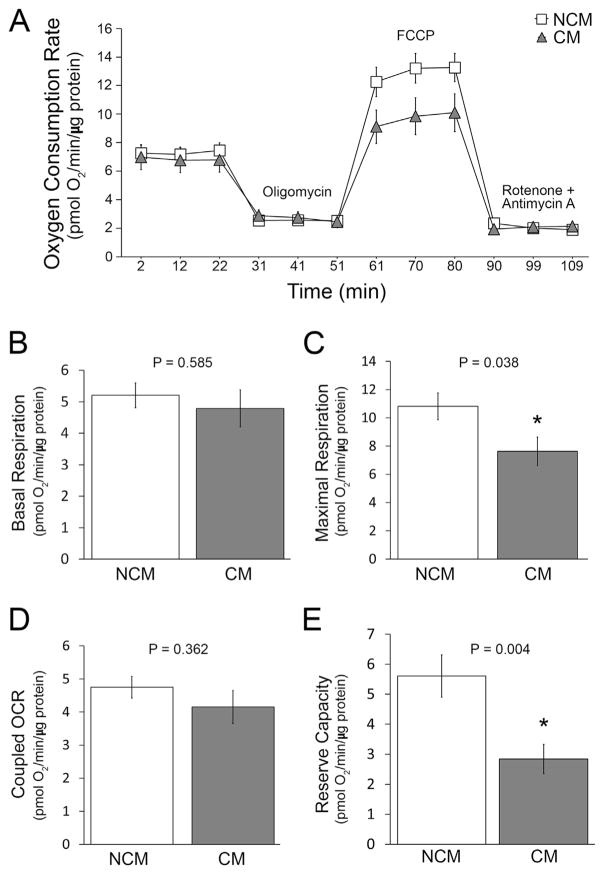
Mitochondrial OCR in human myoblasts following the addition of LLC1 CM (25% (v/v) for a period of 24 h) was compared to OCR in cells treated with LLC1 culture medium alone (non-CM) (Fig. 1, panels A–E) or to the OCR in cells treated with CM from MLE12 cells. Maximal respiration and reserve capacity were inhibited by the addition of LLC1 CM compared to non-CM (P =.038 and .004, respectively). Coupled OCR tended to decrease (Fig. 1D). Basal respiration was unchanged (P =.585). Similar data were obtained when the effects of CM from highly tumorigenc LLC1 cells and less invasive MLE12 CM were compared. Basal respiration, maximal respiration, coupled OCR, and reserve capacity were all reduced (P =.012, .013, 0.003, and 0.025, respectively) by LLC1 CM when compared to MLE12 CM. Interestingly, the effects of LLC1 CM were not observed in human embryonic kidney cells (HEK293). In association with the decrease in OCR, the proton production rate (PPR) in human myoblasts increased following treatment of myoblasts with LLC1 CM compared to LLC1 non-CM (13.194 ± 1.06 pmole and 8.171 ± 0.783 H+/min/μg protein following oligomycin treatment of cells (P =.003), and 8.674 ± 0.560 and 4.541 ± 0.703 pmol H+/min/μg protein (P < .001).
Fig. 1.
Oxygen consumption rate (OCR) was measured in human myoblasts following the addition of LLC1 non-conditioned medium (NCM, □) or LLC1 conditioned medium (CM, ▲) for 24 h. A. OCR in myoblasts in the presence or absence of indicated reagents. B: Basal respiration. C: maximal respiration. D: coupled OCR. E: reserve capacity. * = statistically significant change.
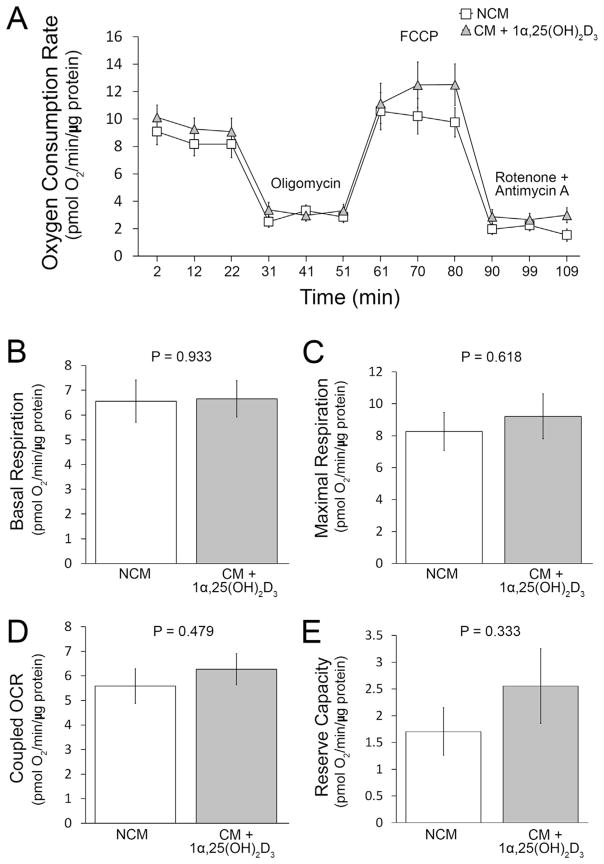
To assess whether the addition of 1α,25(OH)2D3 to LLC1 CM mitigates the inhibition of mitochondrial OCR, we added 10−8M 1α,25(OH)2D3 to LLC1 CM prior to the addition of CM to myoblast cultures. There are no significant differences between basal, maximal, coupled OCR or reserve capacity between LLC1 non-CM or LLC1 CM + 1α,25(OH)2D3 treated human skeletal muscle myoblasts (Fig. 2).
Fig. 2.
OCR was measured in myoblasts following the addition of LLC1 non-conditioned medium (NCM, □) or LLC1 conditioned medium to which was added 1α,25(OH)2D3 (CM + 1α,25(OH)2D3, ▲) for period of 24 h. A. OCR was measured in human skeletal muscle myoblasts in the presence or absence of re-agents as indicated. B: Basal respiration. C: maximal respiration. D: coupled OCR. E: reserve capacity.
3.2. The effects of LLC1 CM on mitochondrial morphology in cultured human skeletal muscle myoblasts are mitigated by the addition of 1α,25(OH)2D3 to LLC1 CM
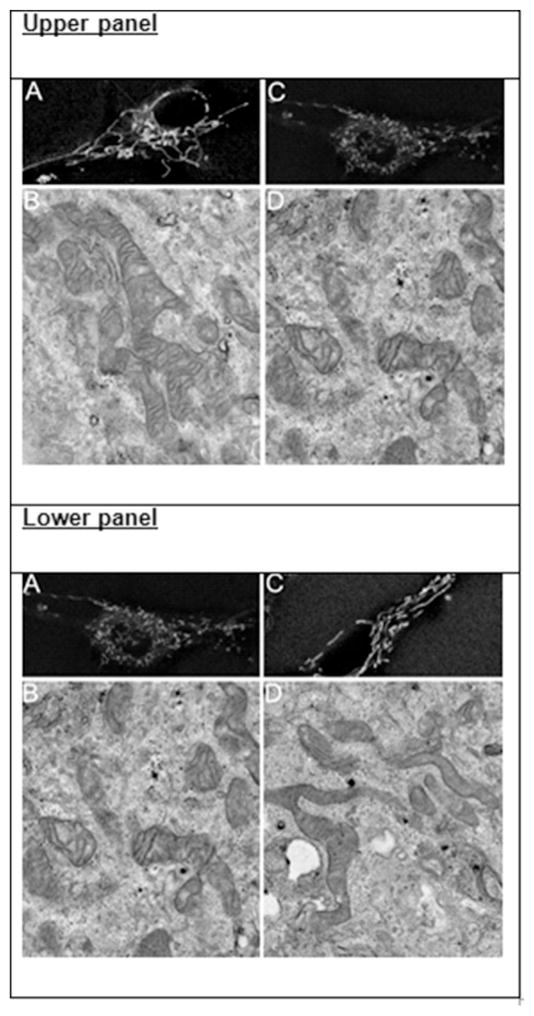
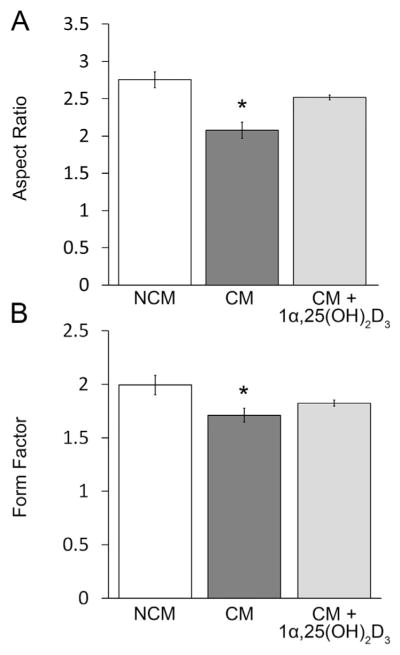
We imaged labeled cells using confocal microscopy following the addition of LLC1 CM or LLC1 non-CM. MitoTracker Green labeled myoblast mitochondria normally appear elongated and filamentous following the addition of LLC1 non-CM to human skeletal muscle myoblasts (Fig. 3A, upper panel), a finding confirmed with EM (Fig. 3B, upper panel). In contrast, following the addition of LLC1 CM to myoblasts, mitochondria appeared rounded and fragmented, a finding also confirmed by EM (Fig. 3C and D, upper panel). Aspect ratio and form factor analysis of mitochondria in myoblasts treated with LLC1 non-CM or LLC1 CM demonstrated a decrease in the aspect ratio and form factor in myoblasts treated with LLC1 CM (P < .001, Fig. 4A and B).
Fig. 3.
Upper panel: A Morphology of labeled myoblast mitochondria following addition of LLC1 non-CM to cells. B: Morphology of mitochondria in myoblasts treated with LLC1 non-CM using EM. C: Morphology of labeled myoblast mitochondria following addition of LLC1 CM to cells. D: Morphology of mitochondria in myoblasts treated with LLC1 CM using EM. Note presence of filamentous mitochondria in myo-blasts treated with LLC1 non-CM, panels A and B. Following treatment of myoblasts with LLC1 CM, panels C and D, appear fragmented.
Lower panel: A: Morphology of labeled myoblast mitochondria following addition of LLC1 CM to cells. B: Morphology of mitochondria in myoblasts treated with LLC1 CM using EM. C: Morphology of labeled myoblast mitochondria following addition of LLC1 CM + 1α,25(OH)2D3, 10−8M to cells. D: Morphology of mitochondria in myoblasts treated with LLC1 CM + 1α,25(OH)2D3, 10−8M using EM. Note presence of fragmented mitochondria in myoblasts treated with LLC1 CM, panels A and B. Following treatment of myoblasts with LLC1 CM + 1α,25(OH)2D3, 10−8M, panels C and D, mitochondria , appear filamentous.
Fig. 4.
A. Aspect ratio of labeled myoblast mitochondria B. Form factor of labeled myoblast mitochondria following addition of LLC1 non-conditioned medium (NCM), LLC1 conditioned medium (CM) or LLC1 conditioned medium (CM) + 1α,25(OH)2D3,10−8M.
To examine whether the addition of 1α,25(OH)2D3 prevents the fragmentation of mitochondria seen by the addition of LLC1 CM, we added 10−8M 1α,25(OH)2D3 to CM prior to the addition of LLC1 CM to myoblasts. Following the addition of 1α,25(OH)2D3 to LLC1 CM, mitochondrial morphology appeared more normal with less mitochondrial fragmentation (Fig. 3A–D, lower panel). These data were confirmed by quantitative assessment of mitochondrial morphology. No differences in aspect ratio and form factor were quantitatively apparent (Fig. 4A, B). These results suggest that 1α,25(OH)2D3 restores mitochondrial morphology in human skel etal muscle myoblasts when added to LLC1 CM.
Concomitant with changes in mitochondrial morphology, the expression of the fission mediator, FIS1, assessed during whole transcriptome sequencing (GEO103550) was increased (FDR = 0.044) following treatment of myoblasts with LLC1 CM, whereas, expression of the fusion mediators DRP-1, OPA1, MFN 1 and MFN 2 expression decreased (FDR = 0.0003, FDR = 0.0147, FDR = 0.06, and FDR = 0.0005, respectively). The mRNA expression data for FIS1 were confirmed by Western blot analysis using a FIS1 antibody. Addition of 1α,25(OH)2D3 to LLC1 CM did not change expression levels of FIS1, DRP2, OPA1 or MFN1 and MFN2 in myo-blasts compared to LLLC1 CM alone, suggesting the normalization of mitochondrial morphology by 1α,25(OH)2D3 is not associated with changes in known mediators of mitochondrial fission and fusion.
There were no changes in the amount of mitochondrial DNA relative to the amount of cellular DNA (ND1+ND6/BECN1+NEFD) in cells treated with LLC1 CM, LLC1 CM along with 1α,25(OH)2D3 or LLC1 non CM (P values: LLC1 CM vs. LLC1 CM + 1α,25(OH)2D3,10−8M = 0.46, LLC1 non-CM vs. LLC1 CM = 0.34, LLC1 non-CM vs. LLC1 CM + 1α,25(OH)2D3, 10−8M = 0.50).
3.3. 1α,25(OH)2D3 partially reverses the inhibitory effect of LLC1 CM on pyruvate dehydrogenase (PDH)
LLC1 CM inhibits PDH activity (LLC1 CM = 21.63 ± 0.32 nmoles of NADH/min/milligram protein vs. non-CM = 25.52 ± 0.74 nmoles of NADH/min/milligram protein (N = 5), P < .0014) (Fig. 1A, supplementary data). The decrease in PDH activity is associated with an increase in the amount of inactive S293 phospho-PDH in myoblasts assessed by Western blotting (P =.05, Fig. 1B, supplementary data). The decrease in PDH activity is associated with a decrease in PDHA1 mRNA expression (FDR = 0.043, Fig. 1C, supplementary data). There is a 40-fold increase in the expression of PDK4 mRNA (FDR = 0.001, Fig. 1D, supplementary data) and a decrease in the expression of PDP2 mRNA (FDR = 0.039, Fig. 1E, supplementary data). SIRT4 mRNA expression is increased (FDR = 0.003, Fig. 1F, supplementary data). The addition of 1α,25(OH)2D3 to LLC1 CM suppresses PDK4 expression and tends to increase the expression of PDP2 and phospho-PDH. These changes are not, however, associated with a statistically significant increase in PDH activity (LLC1 CM = 21.63 ± 0.32 nmole NADH/min/mg protein vs LLC1 CM + 1α,25(OH)2D3 = 21.46 ± 0.19 n mole NADH/minutes/mg protein, P =.66).
3.4. The inhibitory effect of LLC1 CM on proteasomal activity in myoblasts is mitigated by the addition of 1α,25(OH)2D3 to LLC1 CM
Proteasome activity in human skeletal muscle myoblasts is enhanced by incubation of cells with LLC1 CM. These effects are corrected by the addition of 1α,25(OH)2D3 to LLC1 CM (Supplemental Fig. 2).
3.5. Identification of potential mediators of changes in myoblast OCR secreted by LLC1 cells
We analyzed the expression of mRNAs for secreted proteins from LLC1 and MLE12 cell lines. A total of 609 mRNAs were differentially regulated between LLC1 and MLE12 cells. Supplementary Table 1 shows results of up-regulated or down-regulated mRNAs encoding secreted proteins of significance. Up-regulated mRNAs encoding proteins that could potentially alter muscle metabolism include those for Gdf15, Il11; Il27, Il34, Pthlh; Tgfβi, TgfBr3, CCl2, CCl28, CCl7, and Tnfrsf18.
4. Discussion
Cancer cachexia is associated with muscle weakness and increased morbidity, mortality and reduced response to chemotherapy and surgical interventions [1,2,30,31]. In the current report we demonstrate that lung cancer cell CM from the highly tumorigenic cell line Lewis lung carcinoma 1 cell line [32], directly reduces cellular OCR in human skeletal muscle myoblasts when compared to non-CM and CM from a distal respiratory epithelial cell line (MLE12) with low metastatic potential and a low propensity to cause cachexia [33]. We observed that mitochondria appeared more fragmented when treated with LLC1 CM. Mitochondrial fragmentation is associated with a reduction in mitochondrial O2 consumption [34–38] and it is plausible that the observed reduction in OCR following treatment of skeletal muscle cells with LLC1 CM is at least partly due to an increase in mitochondrial fragmentation. The increased expression of mRNA and protein for FIS1, a mediator of mitochondrial fission [34,39,40] and the reduced expression of mRNAs for the mitochondrial fusion mediators, OPA1 and MFN2 [35,41,42] might account for the increase in mitochondrial fragmentation.
Earlier, we demonstrated increases in cellular OCR in human skeletal muscle myoblasts following treatment with the active metabolite of vitamin D, 1α,25(OH)2D3 [19]. We hypothesized that 1α,25(OH)2D3 might ameliorate the deleterious effects of LLC1 tumor medium on mitochondrial performance. We observed an increase in cellular OCR when 1α,25(OH)2D3 was added to LLC1 CM prior to its being used to treat human skeletal muscle myoblasts. Additionally, we observed that mitochondrial fragmentation induced by LLC1 CM in human skeletal muscle myoblasts was attenuated by the addition of 1α,25(OH)2D3 to LLC1 CM. The less fragmented appearance of myoblast mitochondrial morphology induced by 1α,25(OH)2D3 upon addition to LLC1 CM is likely to be associated with increased mitochondrial OCR. The change in fragmentation cannot be accounted for by changes in the expression of known fission or fusion mediators as concentrations of these mediators were minimally changed by the addition of 1α,25(OH)2D3 addition to LLC1 CM.
The activity of mitochondrial enzymes was altered by LLC1 CM and partially reversed by 1α,25(OH)2D3. Pyruvate dehydrogenase which catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA and links the glycolytic pathway to Krebs's cycle was inhibited by LLC1 CM [43,44]. The decrease in activity induced by LLC1 cell medium is associated with a decrease in the expression of PDHA1 mRNA and an increase in inactive phospho-PDH concentrations. Elevated phospho-PDH concentrations were associated with an increase in the expression of PDK4, and a decrease in the expression of PDP2 mRNA. SIRT4 mRNA expression is also increased. SIRT4 enzymatically hydrolyzes lipoamide cofactors from the E2 component (dihydrolipoyllysine acetyltransferase (DLAT)) of the PDH complex, diminishing PDH activity [45]. The addition of 1α,25(OH)2D3 to LLC1 CM partially reversed the deleterious effect of LLC1 CM on PDH in human skeletal muscle myoblasts. 1α,25(OH)2D3 suppressed PDK4 expression and reduced the amount of inactive phospho-PDH.
To identify mediators of altered mitochondrial function and morphology secreted by tumor cells, we carried out WTSS of highly tumorigenic LLC1 cells and minimally invasive MLE12 cells. Messenger RNAs encoding secreted proteins were analyzed to identify substances that were secreted only by invasive LLC 1 cells. mRNAs encoding a large number of secreted proteins were expressed in much higher amount in LLC1 cells. The protein products encoded by such mRNAs included proteins that could potentially mediate cancer cell cachectic effects and proteins associated with increased metastatic potential. We identified several potential mediators of cachexia including Gdf15, a secreted ligand of the TGFβ superfamily, members of the interleukin family of proteins (IL11, Il27, Il33, and Il34) and parathyroid hormone-related protein (PTHrP) based on the expression of larger amounts mRNA in LLC1 cells vs. MLE12 cells. Interleukin 6 has been implicated as a mediator of cachexia in a variety of clinical situations. Parathyroid hormone related protein which activates the PTH receptor has been implicated in recent studies as a mediator of CC [15]. Several of the mRNAs that are increased in LLC1 cells, encode secreted proteins that may be relevant to the metastasis of tumors (for example MMP mRNAs). Isolation of the relevant proteins with testing of their biological properties in model systems will be helpful in identifying the main mediators of CC.
There is need for additional therapeutic approaches for CC because of a lack of effective therapeutic modalities for its treatment [10,16–18]. We found that 1α,25(OH)2D3 inhibited the deleterious effects of tumor cell CM on skeletal muscle cells by reversing altered mitochondrial function, morphology and enzyme activity. Our data suggest that 1α,25(OH)2D3 may be useful in the treatment of weakness seen in tumor cachexia.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants 1R01DK107870-01A1, DK066013 (to R. K.), HL126451 (to G. C. S.), the Fred and A Katherine B. Andersen Foundation.
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrc.2018.01.092.
Footnotes
Conflict of interest statement
None of the authors has any conflicts of interest.
References
- 1.Fearon K, Strasser F, Anker SD, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 2011;12:489–495. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- 2.van Vugt JL, Levolger S, Coelen RJ, et al. The impact of sarcopenia on survival and complications in surgical oncology: a review of the current literature. J Surg Oncol. 2015;112:681–682. doi: 10.1002/jso.24064. [DOI] [PubMed] [Google Scholar]
- 3.Catalano MG, Fortunati N, Arena K, et al. Selective up-regulation of tumor necrosis factor receptor I in tumor-bearing rats with cancer-related cachexia. Int J Oncol. 2003;23:429–436. [PubMed] [Google Scholar]
- 4.Ebrahimi B, Tucker SL, Li D, Abbruzzese JL, et al. Cytokines in pancreatic carcinoma: correlation with phenotypic characteristics and prognosis. Cancer. 2004;101:2727–2736. doi: 10.1002/cncr.20672. [DOI] [PubMed] [Google Scholar]
- 5.Costelli P, Muscaritoli M, Bonetto A, et al. Muscle myostatin signalling is enhanced in experimental cancer cachexia. Eur J Clin Invest. 2008;38:531–538. doi: 10.1111/j.1365-2362.2008.01970.x. [DOI] [PubMed] [Google Scholar]
- 6.Strassmann G, Fong M, Kenney JS, et al. Evidence for the involvement of interleukin 6 in experimental cancer cachexia. J Clin Invest. 1992;89:1681–1684. doi: 10.1172/JCI115767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schersten T, Lundholm K. Lysosomal enzyme activity in muscle tissue from patients with malignant tumor. Cancer. 1972;30:1246–1251. doi: 10.1002/1097-0142(197211)30:5<1246::aid-cncr2820300516>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 8.McClung JM, Judge AR, Powers SK, et al. p38 MAPK links oxidative stress to autophagy-related gene expression in cachectic muscle wasting. Am J Physiol Cell Physiol. 2010;298:C542–C549. doi: 10.1152/ajpcell.00192.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carson JA, Hardee JP, VanderVeen BN. The emerging role of skeletal muscle oxidative metabolism as a biological target and cellular regulator of cancer-induced muscle wasting. Semin Cell Dev Biol. 2016;54:53–67. doi: 10.1016/j.semcdb.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov. 2015;14:58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 11.Oka M, Yamamoto K, Takahashi M, et al. Relationship between serum levels of interleukin 6, various disease parameters and malnutrition in patients with esophageal squamous cell carcinoma. Cancer Res. 1996;56:2776–2780. [PubMed] [Google Scholar]
- 12.Benny Klimek ME, Aydogdu T, Link MJ, et al. Acute inhibition of myostatin-family proteins preserves skeletal muscle in mouse models of cancer cachexia. Biochem Biophys Res Commun. 2010;391:1548–1554. doi: 10.1016/j.bbrc.2009.12.123. [DOI] [PubMed] [Google Scholar]
- 13.Mittal A, Bhatnagar S, Kumar A, et al. The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J Cell Biol. 2010;188:833–849. doi: 10.1083/jcb.200909117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, Liu Z, Ding H, et al. Tumor induces muscle wasting in mice through releasing extracellular Hsp70 and Hsp90. Nat Commun. 2017;8:589. doi: 10.1038/s41467-017-00726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kir S, White JP, Kleiner S, et al. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature. 2014;513:100–104. doi: 10.1038/nature13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayliss TJ, Smith JT, Schuster M, et al. A humanized anti-IL-6 antibody (ALD518) in non-small cell lung cancer. Expet Opin Biol Ther. 2011;11:1663–1668. doi: 10.1517/14712598.2011.627850. [DOI] [PubMed] [Google Scholar]
- 17.Jatoi A, Dakhil SR, Nguyen PL, et al. A placebo-controlled double blind trial of etanercept for the cancer anorexia/weight loss syndrome: results from N00C1 from the North Central Cancer Treatment Group. Cancer. 2007;110:1396–1403. doi: 10.1002/cncr.22944. [DOI] [PubMed] [Google Scholar]
- 18.Jatoi A, Ritter HL, Dueck A, et al. A placebo-controlled, double-blind trial of infliximab for cancer-associated weight loss in elderly and/or poor performance non-small cell lung cancer patients (N01C9) Lung Canc. 2010;68:234–239. doi: 10.1016/j.lungcan.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan ZC, Craig TA, Folmes CD, et al. 1alpha,25-Dihydroxyvitamin D3 regulates mitochondrial oxygen consumption and dynamics in human skeletal muscle cells. J Biol Chem. 2016;291:1514–1528. doi: 10.1074/jbc.M115.684399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folmes CD, Arrell DK, Zlatkovic-Lindor J, et al. Metabolome and metaboproteome remodeling in nuclear reprogramming. Cell Cycle. 2013;12:2355–2365. doi: 10.4161/cc.25509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folmes CD, Martinez-Fernandez A, Perales-Clemente E, et al. Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cell. 2013;31:1298–1308. doi: 10.1002/stem.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koopman WJ, Distelmaier F, Esseling JJ, et al. Computer-assisted live cell analysis of mitochondrial membrane potential, morphology and calcium handling. Methods. 2008;46:304–311. doi: 10.1016/j.ymeth.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Koopman WJ, Visch HJ, Smeitink JA, et al. Simultaneous quantitative measurement and automated analysis of mitochondrial morphology, mass, potential, and motility in living human skin fibroblasts. Cytometry Part A: J Int Soc Anal Cytol. 2006;69:1–12. doi: 10.1002/cyto.a.20198. [DOI] [PubMed] [Google Scholar]
- 24.Aravamudan B, Kiel A, Freeman M, et al. Cigarette smoke-induced mitochondrial fragmentation and dysfunction in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2014;306:L840–L854. doi: 10.1152/ajplung.00155.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalari KR, Nair AA, Bhavsar JD, et al. MAP-RSeq: mayo analysis pipeline for RNA sequencing. BMC Bioinf. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 27.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan BH, Birdsell LA, Martin L, et al. Sarcopenia in an overweight or obese patient is an adverse prognostic factor in pancreatic cancer. Clin Canc Res. 2009;15:6973–6979. doi: 10.1158/1078-0432.CCR-09-1525. [DOI] [PubMed] [Google Scholar]
- 31.Lieffers JR, Bathe OF, Fassbender K, et al. Sarcopenia is associated with postoperative infection and delayed recovery from colorectal cancer resection surgery. Br J Canc. 2012;107:931–936. doi: 10.1038/bjc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertram JS, Janik P. Establishment of a cloned line of Lewis Lung Carcinoma cells adapted to cell culture. Canc Lett. 1980;11:63–73. doi: 10.1016/0304-3835(80)90130-5. [DOI] [PubMed] [Google Scholar]
- 33.Wikenheiser KA, Vorbroker DK, Rice WR, et al. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci U S A. 1993;90:11029–11033. doi: 10.1073/pnas.90.23.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 36.Kao SH, Yen MY, Wang AG, et al. Changes in mitochondrial morphology and bioenergetics in human lymphoblastoid cells with four novel OPA1 mutations. Invest Ophthalmol Vis Sci. 2015;56:2269–2278. doi: 10.1167/iovs.14-16288. [DOI] [PubMed] [Google Scholar]
- 37.Powers SK, Wiggs MP, Duarte JA, et al. Mitochondrial signaling contributes to disuse muscle atrophy. Am J Physiol Endocrinol Metab. 2012;303:E31–E39. doi: 10.1152/ajpendo.00609.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romanello V, Sandri M. Mitochondrial biogenesis and fragmentation as regulators of protein degradation in striated muscles. J Mol Cell Cardiol. 2013;55:64–72. doi: 10.1016/j.yjmcc.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 39.James DI, Parone PA, Mattenberger Y, et al. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 40.Lee YJ, Jeong SY, Karbowski M, et al. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Detmer SA, Ewald AJ, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 43.Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem Soc Trans. 2003;31:1143–1151. doi: 10.1042/bst0311143. [DOI] [PubMed] [Google Scholar]
- 44.Patel MS, Korotchkina LG. Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans. 2006;34:217–222. doi: 10.1042/BST20060217. [DOI] [PubMed] [Google Scholar]
- 45.Mathias RA, Greco TM, Oberstein A, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.