Abstract
In clinical trials, information about certain time points may be of interest in making decisions about treatment effectiveness. Rather than comparing entire survival curves, researchers can focus on the comparison at fixed time points that may have a clinical utility for patients. For two independent samples of right-censored data, Klein et al. (2007) compared survival probabilities at a fixed time point by studying a number of tests based on some transformations of the Kaplan-Meier estimators of the survival function. However, to compare the survival probabilities at a fixed time point for paired right-censored data or clustered right-censored data, their approach would need to be modified. In this paper, we extend the statistics to accommodate the possible within-paired correlation and within-clustered correlation, respectively. We use simulation studies to present comparative results. Finally, we illustrate the implementation of these methods using two real data sets.
Keywords: right-censored data, Kaplan-Meier estimator, pseudo-value approach, correlated survival data
1. Introduction
Right-censored survival data often arise in biometrical studies, reliability research, and many other fields. For comparing entire survival curves, many well-established methods have been proposed to test for equality of two survival functions, including weighted logrank tests (Gill, 1980) and weighted Kaplan-Meier tests (Pepe and Fleming 1989). However, rather than comparing entire survival curves, researchers may want to compare survival probabilities at specified time points (or at a single fixed time point). For example, in a chronic disease such as cancer, the 5-year survival rate is often used as an indicator of the severity of the disease and prognosis. Therefore, one may want to compare cancer survival rates in two treatments with the thought that the 5-year survival is sometimes evidence of success against cancer (used to mean a “cure” of the cancer). Patients and physicians often want to know, “Are treatments different at a specified time point?” Therefore, it may be more meaningful to compare the survival probability at a fixed time point.
Moreover, dependent survival times may arise due to paired designs of experiments. In order to reduce the between-subject variability, a number of matched subjects, such as twins, are randomly assigned to different treatments to evaluate effect. Such data are called “paired” right-censored data, where the survival times are independent within groups but dependent between groups (Huster et al., 1989; Dallas and Rao, 2000; Huang and Wolfe 2002).
In some cases, it is difficult or impractical to administer different treatments to the same subject. Therefore, observations are taken from multiple sites or a group of subjects. For example, the failure times of dental implants contributed by each subject tend to correlate, while those from different subjects are independent. The data are considered as “clustered” right-censored data (Jung and Jeong, 2003; Huang and Wolfe, 2002).
To compare the differences between two survival functions for independent right-censored data, one approach is to construct confidence bands for the survival function. Bie et al. (1987), Borgan and Lestøl (1990) and Parzen et al. (1997) showed that the correct coverage probability of confidence bands for the survival function can be improved through the use of suitable transformation of Kaplan-Meier estimators (1958), or equivalently, cumulative hazard functions. An alternative approach to comparing the difference between two groups is to test if two treatment groups have the same survival functions. Pepe and Fleming (1989) used the weighted difference between two Kaplan-Meier estimators and chose a weight function that stabilizes the variance of the test statistic. For comparing two survival probabilities at a fixed point in time, Klein et al. (2007) studied a number of tests based on some transformations of the Kaplan-Meier estimators. They concluded that the tests based on transformed survival functions perform better than the untransformed ones.
For paired right-censored data, Murray (2001) extended the weighted Kaplan-Meier tests to compare the differences between two survival functions. For clustered right-censored data, O'Gorman and Akritas (2004) extended the statistics studied by Gu et al. (1999) to test treatment effect by using Kaplan-Meier estimators. Though Klein et al. (2007) proposed several transformation strategies to compare two survival probabilities at a fixed point in time, the testing methods for comparing paired right-censored data or clustered right-censored data at a fixed time point have yet to be investigated. If we treat them as originating from independent samples, it could lead to misleading results. Therefore, in this paper, the methods described in Klein et al. (2007) are extended to paired right-censored data and clustered right-censored data, respectively. We present systematic studies of the choices of transformations and modify the standard error of their statistics to accommodate possible within-pair correlation and within-cluster correlation.
This paper is organized as follows: Section 2 extends the tests described in Klein et al. (2007) for testing the equality of two survival probabilities at a fixed point in time for paired right-censored data and clustered right-censored data, respectively. Section 3 investigates the accuracy of the asymptotic distributions of the proposed tests and compares the power properties under various alternatives through simulation. In Section 4, we illustrate the implementation of the extended methods through two real data sets. Finally, we offer some concluding remarks.
2. The extended two sample tests
2.1 Paired right-censored data
To compare two survival probabilities for paired right-censored data, let Tik and Cik denote the survival and censoring times of the kth subject of group i for i = 1, 2 and k = 1, 2,…, n where n is the number of pairs. Note Tik is independent of Cik but a dependence is allowed between T1k and T2k and between C1k and C2k. The common marginal survival functions for Tik are denoted by Si (t). When there are some right-censored observations in a data set, one can only observe the random variables Xik = min(Tik, Cik) and δik = I(Tik < Cik), where I(A) is an indicator function of event A, taking value 1 if the event A occurs and value 0 otherwise. Now, let t1 < t2 <… < tD be the distinct ordered failure times based on the pooled data, dij be the number of events at time tj, and yij be the number of subjects at risk in the ith group at time tj. With this notation, the Kaplan-Meier estimators (1958) are given by
If the lifetimes between any two observations are independent, the estimated variances of the Kaplan-Meier estimators are
where
(Greenwood's formula, 1926). In order to test the equality of two survival probabilities, the null hypothesis is specified as
| (1) |
and the alternatives can be specified as
A natural statistic considers the difference between two Kaplan-Meier estimates at time t, that is Ŝ2(t) − Ŝ1(t). Therefore, Klein et al. (2007) studied a number of statistics based on some transformations of the survival function for two independent samples of right-censored data. The test statistic is defined as
where ϕ is a differentiable real-valued function. To accommodate the within-pair dependence, the denominator V(ϕ(Ŝ2(t)) − ϕ(Ŝ1(t))) can be derived as
| (2) |
The first and second terms in (2) correspond to the original variance used in Klein et al. (2007) for independent right-censored data. The third term characterizes the dependence between two Kaplan-Meier estimators. If the two samples are independent, the third term vanishes in this expression. Applying the delta method, the estimated variances can be expressed as
where ϕ′(t) = dϕ(t)/dt. Then, using the results in the Appendix of Murray (2001), an estimate for the covariance part can be represented as
where
where count the number of complete correlated pairs still at risk at times tr and ts in treatment groups 1 and 2, respectively; count the number of individuals from complete pairs who failed at time tr for treatment 1 and failed at time ts for treatment 2; and count the number of complete correlated pairs who failed at time ts for treatment 2 and who are still at risk for failure at time tr for treatment 1.
Since Klein et al. (2007) showed that each transformation of Ŝ2(t)−Ŝ1(t) performs better than the naive test in terms of type I error rates, the same transformations are considered in this study. The first test is based on the naive test, namely ϕ(Ŝi(t)) = Ŝi(t). In addition, testing the null hypothesis (1) is equivalent to the test H0: Λ1(t) = Λ2(t), where Λi is the cumulative hazard function of group i; hence, the second test is based on a logarithmic transformation of the survival function, that is ϕ(Ŝi(t)) = log(Ŝi(t)). The third test is constructed based on ϕ(Ŝi(t)) = log(−log(Ŝi(t))) transformation, since it has been found to be very useful in constructing confidence intervals and confidence bands for survival function (Kalbfeisch and Prentice, 1980). Further, Klein et al. (2007) showed that the test has the best performance in comparing difference for independent right-censored data. The forth test is based on an arcsine-square root transformation, ϕ(Ŝi(t)) = arcsin(Ŝi(t)), which has small sample coverage probabilities for confidence intervals similar to the log-log transformation (Nair, 1984). The final test is the logit transformation, ϕ(Ŝi(t)) = log(Ŝi(t)/(1 − Ŝi (t))) which was considered in Klein et al. (2007).
Under the null hypothesis (1), each of the mentioned tests with different transformations led to an asymptotic standard normal distribution. Therefore, at the α-level, one can conclude that the survival probability is better in the second group, if the test statistic is larger than z1−α, where z1−α is the 100(1 − α) percentile of the standard normal distribution.
As an alternative to the above tests, the pseudo-value approach can be used not only for hypothesis testing but also for obtaining estimates of the model parameters (Andersen et al, 2003; Klein and Andersen, 2005; Klein et al. 2007). Define the pseudo-value by
where (Ŝp(t) is the Kaplan-Meier estimator of the pooled samples and is the Kaplan-Meier estimator of the pooled samples with the kth subject of group i observation removed. When there is no censoring, then θ̂ik is simply the indicator that the kth subject of group i was living at time t. When censoring is present, the pseudo-values are still defined for all individuals and at all times (Perme and Andersen, 2008). Intuitively, the definition of the pseudo-value implies that the pseudo observation can be treated as the individual contribution to the overall Kaplan-Meier estimate. Therefore, as shown in Andersen et al. (2003), these can be used in a generalized linear model to model the effects of covariates on outcome. We consider a generalized linear model with link function g(.) for the pseudo-values, that is
where βT stands for the transpose of β and Zik is a vector of covariates. In our case, Zik is an indicator covariate with value 1 if the patient is in the treatment group and 0 if they are in the control group. Therefore, to model the effects of covariates on outcome, testing the null hypothesis (1) is equivalent to test H0:β = 0 at a fixed time. To estimate regression parameters β, we shall use a generalized estimating equation (GEE) approach (see Liang and Zeger, 1986). Let sik = μ(βTZik) be the inverse function based on g(·) and dμik(β) be the vector of partial derivatives of μik(·) with respect to β. The estimating equation is given by
where Vik(β) is the working covariance matrix. The maximum likelihood estimator of β can be defined as the solution to U(β) =0. Let β̂ be the solution to this equation, using the results from Liang and Zeger (1986), it follows that √n(β̂ − β) is asymptotically normal with mean zero and a covariance that can be estimated consistently by a “sandwich” estimator
where . Therefore, a test based on β̂ and Σ̂ can be used to compare the equality of two survival functions at a fixed time point.
2.2 Clustered right-censored data
For clustered right-censored data, we added a subscript j to indicate the cluster from which each individual was chosen. Let Tijk and Cijk denote the survival and censoring times of the kth subject in cluster j of the ith sample for i = 1, 2, j = 1, 2, ⋯, ni, and k = 1, 2, ⋯, mij. Notice ni is the number of clusters in the ith sample, and mij is the number of subjects in the jth cluster of sample i. Let Tijk and Cijk be independent, and the common marginal survival functions for the subject Tijk in each cluster of the ith group be denoted by Si(t). When there are some right-censored observations, one can only observe the random variables Xijk = min(Tijk, Cijk) and δijk = I(Tijk ≤ Cijk). Now, let t1 < t2 < ⋯ < tD be the distinct, ordered, observed failure times based on the pooled data from the two samples, dijl be the number of failure events of the jth cluster in group i at time tl, and yijl be the number of subjects at risk of the jth cluster in group i at time tl, i = 1,2, j = 1,2, ⋯, ni, l = 1, 2, ⋯, D. The total number of clusters in the two samples is denoted by n.
Using this notation, the Kaplan-Meier estimators (1958) are given by
| (3) |
Although the survival times are correlated in each cluster, Ying and Wei (1994) have shown that the Kaplan-Meier estimator (3) is still consistent and asymptotically normal. For testing the equality of two survival functions, we also considered the test
based on a transformation ϕ of S̃i(t). Since the survival times in the two samples are independent but correlated within each cluster, the variance of ϕ(S̃2(t)) − ϕ(S̃i(t)) is
Therefore, to consider clustered dependence, the martingale technique developed by Ying and Wei (1994) can be applied to derive the variance of S̃i(t). It follows that a valid estimator for the asymptotic variance is
where
, and dNi(u) means the change in the process Ni(u) over a short time interval [t, t + dt) (detailed proof appears in Ying and Wei, 1994). Then, using the delta method, ϕ(S̃i(t)) is asymptotically normal with mean ϕ(Si(t)), and the estimated variance is given by
In clustered right-censored data, we considered the same transformations as described in Section 2.1. Under the null hypothesis (1), each of the above tests has an asymptotic standard normal distribution. Therefore, at the α-level, one can conclude that the survival probability is better in the second group, if the test statistic is larger than z1−α.
Similarly, we discuss another test that is based on a pseudo-value regression technique. Define the j th pseudo-value by
where i = 1, 2, j = 1, 2, ⋯, ni, k = 1, 2, ⋯, mij, and is the total number of observations. Here, S̃p(t) is the Kaplan-Meier estimator of the pooled samples and is the Kaplan-Meier estimator of the pooled samples with the kth subject in cluster j of group i observation removed. The generalized linear model for the pseudo-values is
where Zijk is a vector of covariates. In our case, Zik is an indicator covariate. Therefore, testing the null hypothesis (1) is equivalent to test H0: β = 0 at a fixed time. With clustered observations, the correlation for a given subject must be taken into account. Let sijk = μ(βT Zijk) be the inverse function based on g(·) and dμij(β) = [∂μij1(β)/∂β,…,∂μijmij(β)/∂β]T be the vector of partial derivatives of μij(·) with respect to β. The estimating equation (Liang and Zeger, 1986) is
where Vij(β) is the mij × mij working covariance matrix, θ̃ij = [θ̃ij1, ⋯, θ̃ijmij]T and sij = [sij1, ⋯, sijmij]T. The maximum likelihood estimator of β can be defined as the solution to U(β)=0. Let β̃ be the solution to this equation. Using the results from Liang and Zeger (1986), it follows that √n(β̃ − β) is asymptotic normal with mean zero and a covariance that can be estimated consistently by
where . Therefore, a test based on β̃ and Σ̃ can be used to compare the equality of two survival functions at fixed time point t.
3. Simulation results
To assess the performance of the extended tests, we carried out simulation studies under various scenarios for paired right-censored data and clustered right-censored data, respectively. We denoted NaiP, LogP, LlogP, ArcsP, LogitP as the tests of the naive transformation, the logarithmic transformation, the log-log transformation, the arcsine transformation and the logit transformation of the Kaplan-Meier estimator for paired right-censored data. We denoted NaiC, LogC, LlogC, ArcsC, LogitC as the tests of the above mentioned transformations for clustered right-censored data. Moreover, we also presented the results from the tests proposed by Klein et al. (2007), who assume that data are independent and denoted the related tests as Nai, Log, Llog, Arcs, and Logit.
3.1 Paired right-censored data
To construct the paired right-censored data, we used bivariate exponential distribution to generate survival times through Moran's algorithm (Moran, 1967). Let (V1, V3) and (V2,V4) be mutually independent, but each pair has a bivariate normal distribution with a marginally zero mean, unit variance, and a correlation coefficient √ρ(ρ ≥ 0). We constructed the joint distribution of and . Therefore, T1 and T2 have marginal exponential distributions with failure rates λ1 and λ2, respectively, and the correlation coefficient is ρ. Parameters λ1 were chosen so that in the first group the probability of survival at time point 1 was 0.75. Parameters λ2 were chosen so that λ1 = λ2 for assessing the type I error rates, and the odds ratio of the survival function at time point 1 was 2 or 3 for power comparison. The corresponding correlation between pairs was set as 0.2, 0.5 or 0.7. In addition, the censoring times were also generated from bivariate exponential distribution and the overall censoring fraction in either setup was fixed at 10% and 40%. Total number of pairs was set as 30, 60, or 100. Moreover, for the pseudo-value regression, since an attractive property of the GEE is that the estimator is robust with respect to misspecification of working covariance matrix, we chose an exchangeable working covariance matrix. All tests with nominal level 0.05 were applied to each sample. Empirical rejection probability was obtained based on 2000 simulation runs.
Table 1 shows the empirical type I error rates. All the extended tests preserve reasonable type I error rates as the number of pairs becomes large (>60). As the dependency grows, the performances of the extended tests are much better (close to 0.05) than the tests proposed by Klein et al. (2007). This happens since the extended tests consider the positive within-pair correlation. Moreover, Table 2 shows the power comparison results. As expected, the power of the extended tests increases as the correlation between pairs increase, the odds ratio increases, or the number of pairs increase. By contrast, the power of the tests decreases as the censoring proportion grows.
Table 1. Empirical type I error rates of tests for paired right-censored data.
| Considering paired correlation | Ignoring paired correlation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ | n | %cens | NaiP | LogP | LlogP | ArcsP | LogitP | Pseudo | Nai | Log | Llog | Arcs | Logit |
|
|
|
||||||||||||
| 0 | 30 | 10 | 0.060 | 0.041 | 0.039 | 0.060 | 0.041 | 0.041 | 0.055 | 0.040 | 0.036 | 0.054 | 0.041 |
| 40 | 0.061 | 0.044 | 0.041 | 0.061 | 0.045 | 0.044 | 0.058 | 0.041 | 0.036 | 0.058 | 0.040 | ||
| 60 | 0.071 | 0.047 | 0.045 | 0.072 | 0.052 | 0.052 | 0.064 | 0.040 | 0.037 | 0.064 | 0.043 | ||
| 60 | 10 | 0.054 | 0.041 | 0.041 | 0.051 | 0.042 | 0.043 | 0.049 | 0.043 | 0.041 | 0.049 | 0.044 | |
| 40 | 0.053 | 0.047 | 0.047 | 0.052 | 0.045 | 0.048 | 0.054 | 0.045 | 0.045 | 0.052 | 0.044 | ||
| 60 | 0.058 | 0.047 | 0.048 | 0.058 | 0.051 | 0.048 | 0.057 | 0.048 | 0.045 | 0.058 | 0.047 | ||
| 100 | 10 | 0.046 | 0.043 | 0.043 | 0.047 | 0.044 | 0.044 | 0.046 | 0.041 | 0.041 | 0.046 | 0.042 | |
| 40 | 0.059 | 0.053 | 0.054 | 0.059 | 0.054 | 0.052 | 0.056 | 0.052 | 0.053 | 0.056 | 0.054 | ||
| 60 | 0.043 | 0.041 | 0.041 | 0.043 | 0.041 | 0.041 | 0.045 | 0.040 | 0.041 | 0.045 | 0.042 | ||
| 0.2 | 30 | 10 | 0.068 | 0.046 | 0.045 | 0.068 | 0.048 | 0.049 | 0.056 | 0.041 | 0.038 | 0.056 | 0.042 |
| 40 | 0.065 | 0.044 | 0.045 | 0.065 | 0.047 | 0.049 | 0.056 | 0.034 | 0.032 | 0.054 | 0.036 | ||
| 60 | 0.064 | 0.043 | 0.043 | 0.068 | 0.047 | 0.041 | 0.058 | 0.036 | 0.031 | 0.054 | 0.034 | ||
| 60 | 10 | 0.058 | 0.050 | 0.051 | 0.058 | 0.052 | 0.054 | 0.049 | 0.040 | 0.038 | 0.048 | 0.051 | |
| 40 | 0.053 | 0.045 | 0.045 | 0.053 | 0.045 | 0.046 | 0.045 | 0.038 | 0.038 | 0.045 | 0.050 | ||
| 60 | 0.051 | 0.043 | 0.044 | 0.051 | 0.045 | 0.045 | 0.045 | 0.038 | 0.037 | 0.045 | 0.041 | ||
| 100 | 10 | 0.054 | 0.052 | 0.052 | 0.053 | 0.052 | 0.052 | 0.048 | 0.041 | 0.040 | 0.047 | 0.051 | |
| 40 | 0.054 | 0.047 | 0.047 | 0.052 | 0.049 | 0.049 | 0.045 | 0.041 | 0.041 | 0.045 | 0.053 | ||
| 60 | 0.052 | 0.045 | 0.047 | 0.052 | 0.048 | 0.047 | 0.047 | 0.043 | 0.044 | 0.047 | 0.044 | ||
| 0.5 | 30 | 10 | 0.069 | 0.045 | 0.045 | 0.069 | 0.049 | 0.053 | 0.040 | 0.025 | 0.022 | 0.039 | 0.024 |
| 40 | 0.061 | 0.040 | 0.040 | 0.061 | 0.046 | 0.047 | 0.037 | 0.022 | 0.019 | 0.038 | 0.023 | ||
| 60 | 0.067 | 0.042 | 0.045 | 0.067 | 0.049 | 0.050 | 0.049 | 0.031 | 0.030 | 0.047 | 0.034 | ||
| 60 | 10 | 0.051 | 0.041 | 0.042 | 0.051 | 0.043 | 0.046 | 0.027 | 0.023 | 0.022 | 0.027 | 0.023 | |
| 40 | 0.057 | 0.048 | 0.049 | 0.058 | 0.051 | 0.052 | 0.032 | 0.027 | 0.027 | 0.032 | 0.027 | ||
| 60 | 0.053 | 0.043 | 0.045 | 0.054 | 0.048 | 0.048 | 0.036 | 0.027 | 0.027 | 0.037 | 0.029 | ||
| 100 | 10 | 0.059 | 0.054 | 0.054 | 0.059 | 0.056 | 0.056 | 0.032 | 0.030 | 0.030 | 0.032 | 0.030 | |
| 40 | 0.054 | 0.047 | 0.047 | 0.054 | 0.049 | 0.049 | 0.034 | 0.030 | 0.030 | 0.034 | 0.032 | ||
| 60 | 0.058 | 0.049 | 0.051 | 0.057 | 0.052 | 0.052 | 0.039 | 0.034 | 0.034 | 0.038 | 0.034 | ||
| 0.7 | 30 | 10 | 0.062 | 0.043 | 0.043 | 0.064 | 0.047 | 0.054 | 0.027 | 0.020 | 0.016 | 0.027 | 0.018 |
| 40 | 0.060 | 0.041 | 0.043 | 0.059 | 0.049 | 0.054 | 0.028 | 0.015 | 0.014 | 0.027 | 0.019 | ||
| 60 | 0.054 | 0.034 | 0.038 | 0.056 | 0.043 | 0.050 | 0.022 | 0.012 | 0.011 | 0.023 | 0.012 | ||
| 60 | 10 | 0.052 | 0.044 | 0.045 | 0.053 | 0.047 | 0.052 | 0.017 | 0.014 | 0.014 | 0.017 | 0.014 | |
| 40 | 0.051 | 0.043 | 0.045 | 0.054 | 0.047 | 0.049 | 0.021 | 0.016 | 0.016 | 0.021 | 0.017 | ||
| 60 | 0.054 | 0.041 | 0.045 | 0.056 | 0.047 | 0.048 | 0.021 | 0.014 | 0.015 | 0.021 | 0.016 | ||
| 100 | 10 | 0.050 | 0.049 | 0.049 | 0.049 | 0.049 | 0.049 | 0.016 | 0.014 | 0.014 | 0.015 | 0.014 | |
| 40 | 0.057 | 0.049 | 0.050 | 0.056 | 0.052 | 0.056 | 0.020 | 0.015 | 0.015 | 0.019 | 0.016 | ||
| 60 | 0.047 | 0.043 | 0.043 | 0.047 | 0.044 | 0.045 | 0.021 | 0.020 | 0.019 | 0.021 | 0.019 | ||
Table 2. Estimated power of several tests for paired right-censored data.
| Paired | Odds ratio=2 | Odds ratio=3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ | n | %cens | NaiP | LogP | LlogP | ArcsP | LogitP | Pseudo | NaiP | LogP | LlogP | ArcsP | LogitP | Pseudo |
|
|
|
|||||||||||||
| 0 | 30 | 10 | 0.273 | 0.223 | 0.226 | 0.267 | 0.238 | 0.235 | 0.551 | 0.499 | 0.515 | 0.547 | 0.525 | 0.521 |
| 40 | 0.230 | 0.182 | 0.200 | 0.228 | 0.205 | 0.192 | 0.484 | 0.401 | 0.448 | 0.473 | 0.455 | 0.432 | ||
| 60 | 0.204 | 0.137 | 0.175 | 0.198 | 0.179 | 0.146 | 0.378 | 0.235 | 0.346 | 0.366 | 0.341 | 0.266 | ||
| 60 | 10 | 0.425 | 0.395 | 0.399 | 0.421 | 0.403 | 0.403 | 0.803 | 0.781 | 0.787 | 0.800 | 0.790 | 0.789 | |
| 40 | 0.405 | 0.366 | 0.382 | 0.401 | 0.383 | 0.377 | 0.743 | 0.705 | 0.731 | 0.740 | 0.731 | 0.720 | ||
| 60 | 0.348 | 0.296 | 0.338 | 0.347 | 0.336 | 0.308 | 0.648 | 0.566 | 0.646 | 0.646 | 0.640 | 0.598 | ||
| 100 | 10 | 0.632 | 0.616 | 0.620 | 0.630 | 0.622 | 0.622 | 0.957 | 0.952 | 0.953 | 0.956 | 0.953 | 0.953 | |
| 40 | 0.559 | 0.537 | 0.547 | 0.557 | 0.549 | 0.542 | 0.923 | 0.914 | 0.920 | 0.922 | 0.920 | 0.915 | ||
| 60 | 0.493 | 0.462 | 0.486 | 0.491 | 0.483 | 0.470 | 0.844 | 0.817 | 0.845 | 0.844 | 0.841 | 0.829 | ||
| 0.2 | 30 | 10 | 0.269 | 0.212 | 0.220 | 0.262 | 0.230 | 0.233 | 0.580 | 0.516 | 0.539 | 0.572 | 0.550 | 0.548 |
| 40 | 0.243 | 0.181 | 0.208 | 0.235 | 0.216 | 0.205 | 0.501 | 0.419 | 0.459 | 0.492 | 0.461 | 0.442 | ||
| 60 | 0.197 | 0.133 | 0.171 | 0.193 | 0.170 | 0.141 | 0.400 | 0.248 | 0.372 | 0.396 | 0.366 | 0.294 | ||
| 60 | 10 | 0.456 | 0.428 | 0.433 | 0.452 | 0.437 | 0.438 | 0.836 | 0.817 | 0.824 | 0.832 | 0.826 | 0.826 | |
| 40 | 0.389 | 0.354 | 0.370 | 0.385 | 0.372 | 0.366 | 0.770 | 0.725 | 0.756 | 0.766 | 0.757 | 0.744 | ||
| 60 | 0.331 | 0.282 | 0.319 | 0.327 | 0.317 | 0.303 | 0.661 | 0.580 | 0.655 | 0.659 | 0.652 | 0.617 | ||
| 100 | 10 | 0.655 | 0.638 | 0.641 | 0.652 | 0.645 | 0.645 | 0.962 | 0.959 | 0.960 | 0.961 | 0.960 | 0.961 | |
| 40 | 0.594 | 0.567 | 0.581 | 0.592 | 0.584 | 0.576 | 0.942 | 0.731 | 0.939 | 0.941 | 0.939 | 0.936 | ||
| 60 | 0.497 | 0.463 | 0.492 | 0.498 | 0.493 | 0.495 | 0.865 | 0.840 | 0.867 | 0.865 | 0.850 | 0.850 | ||
| 0.5 | 30 | 10 | 0.324 | 0.265 | 0.276 | 0.319 | 0.292 | 0.300 | 0.646 | 0.586 | 0.605 | 0.635 | 0.615 | 0.626 |
| 40 | 0.254 | 0.198 | 0.226 | 0.250 | 0.231 | 0.228 | 0.575 | 0.478 | 0.540 | 0.566 | 0.547 | 0.536 | ||
| 60 | 0.206 | 0.119 | 0.176 | 0.201 | 0.182 | 0.155 | 0.422 | 0.266 | 0.400 | 0.418 | 0.393 | 0.325 | ||
| 60 | 10 | 0.503 | 0.473 | 0.481 | 0.499 | 0.488 | 0.491 | 0.908 | 0.895 | 0.900 | 0.905 | 0.902 | 0.902 | |
| 40 | 0.442 | 0.410 | 0.426 | 0.440 | 0.430 | 0.426 | 0.844 | 0.809 | 0.835 | 0.844 | 0.838 | 0.830 | ||
| 60 | 0.359 | 0.319 | 0.355 | 0.364 | 0.356 | 0.346 | 0.719 | 0.645 | 0.721 | 0.719 | 0.715 | 0.691 | ||
| 100 | 10 | 0.721 | 0.706 | 0.714 | 0.720 | 0.715 | 0.717 | 0.990 | 0.988 | 0.989 | 0.988 | 0.988 | 0.988 | |
| 40 | 0.655 | 0.637 | 0.646 | 0.651 | 0.647 | 0.646 | 0.966 | 0.960 | 0.965 | 0.966 | 0.965 | 0.964 | ||
| 60 | 0.547 | 0.515 | 0.544 | 0.547 | 0.544 | 0.530 | 0.905 | 0.878 | 0.906 | 0.905 | 0.904 | 0.895 | ||
| 0.7 | 30 | 10 | 0.357 | 0.287 | 0.306 | 0.350 | 0.322 | 0.345 | 0.723 | 0.659 | 0.687 | 0.715 | 0.695 | 0.715 |
| 40 | 0.288 | 0.220 | 0.253 | 0.287 | 0.264 | 0.266 | 0.624 | 0.534 | 0.600 | 0.622 | 0.608 | 0.601 | ||
| 60 | 0.243 | 0.170 | 0.219 | 0.243 | 0.225 | 0.215 | 0.456 | 0.296 | 0.440 | 0.457 | 0.434 | 0.386 | ||
| 60 | 10 | 0.621 | 0.591 | 0.604 | 0.619 | 0.609 | 0.620 | 0.954 | 0.947 | 0.951 | 0.953 | 0.952 | 0.953 | |
| 40 | 0.534 | 0.493 | 0.517 | 0.535 | 0.521 | 0.522 | 0.881 | 0.858 | 0.878 | 0.881 | 0.877 | 0.872 | ||
| 60 | 0.426 | 0.361 | 0.419 | 0.429 | 0.419 | 0.407 | 0.784 | 0.709 | 0.786 | 0.786 | 0.784 | 0.763 | ||
| 100 | 10 | 0.809 | 0.802 | 0.803 | 0.809 | 0.807 | 0.807 | 0.998 | 0.988 | 0.998 | 0.998 | 0.998 | 0.998 | |
| 40 | 0.746 | 0.728 | 0.743 | 0.746 | 0.744 | 0.744 | 0.987 | 0.986 | 0.987 | 0.988 | 0.987 | 0.987 | ||
| 60 | 0.623 | 0.588 | 0.624 | 0.627 | 0.623 | 0.612 | 0.936 | 0.915 | 0.941 | 0.938 | 0.938 | 0.932 | ||
To summarize all the simulation results from Table 1 and Table 2, we followed Klein et al. (2007) and applied analysis of variance (ANOVA) techniques concerned with both type I and type II error rates. For the type I error rate, outcome variable, Y, was defined as the percent rejection rate minus the nominal level of 5. Therefore, good performance of the test is implied by numerically small estimates for the expectation E(Y). In addition, we considered four different factors, namely TEST, CORR, NUM, and CEN, which represent the test methods, the correlation, the number of pairs, and the censoring proportion, respectively. In such a setting, TEST has 6 levels, CORR has 4 levels, NUM has 3 levels, and CEN has 2 levels. We considered fitting models without an intercept as:
| (4) |
| (5) |
| (6) |
| (7) |
respectively. Table 3 shows the average deviations from the nominal 5 percent level of six tests. That is, we calculated the average estimated type I error rate and then subtracted 0.05. Table 4 shows the average rejection rates for six tests using ANOVA by using model (4)-(7).
Table 3.
Average deviations from nominal 5 percent level of six tests using ANOVA (for paired right-censored data).
| TEST | NaiP | LogP | LlogP | ArcsP | LogitP | Pseudo | |
|---|---|---|---|---|---|---|---|
| NUM | 30 | 1.33 | -0.70 | -0.74 | 1.34 | -0.35 | -0.11 |
| 60 | 0.36 | -0.51 | -0.44 | 0.38 | -0.35 | -0.13 | |
| 100 | 0.41 | -0.08 | -0.05 | 0.36 | 0.06 | 0.09 | |
| CORR | 0.0 | 0.55 | -0.52 | -0.58 | 0.50 | -0.48 | -0.47 |
| 0.2 | 0.87 | -0.27 | -0.25 | 0.82 | -0.12 | -0.02 | |
| 0.5 | 0.85 | -0.42 | -0.38 | 0.87 | -0.10 | 0.05 | |
| 0.7 | 0.53 | -0.52 | -0.42 | 0.58 | -0.15 | 0.23 | |
| CEN | 10% | 0.69 | -0.42 | -0.43 | -0.68 | -0.25 | -0.06 |
| 40% | 0.71 | -0.44 | -0.39 | 0.70 | -0.18 | -0.04 | |
| 60% | 0.60 | -0.68 | -0.54 | 0.67 | -0.27 | -0.27 | |
| 0.70 | -0.43 | -0.41 | 0.69 | -0.21 | -0.05 |
Table 4.
Average rejection rates for six tests using ANOVA for different NUM, CORR and CENS (for paired right-censored data).
| TEST | NaiP | LogP | LlogP | ArcsP | LogitP | Pseudo | |
|---|---|---|---|---|---|---|---|
| NUM | 30 | 38.26 | 31.62 | 34.43 | 37.63 | 35.34 | 35.16 |
| 60 | 60.71 | 57.79 | 59.21 | 60.46 | 59.48 | 59.24 | |
| 100 | 76.85 | 74.43 | 76.28 | 76.73 | 76.39 | 76.25 | |
| CORR | 0.0 | 53.21 | 49.76 | 51.07 | 52.85 | 51.45 | 50.84 |
| 0.2 | 54.98 | 49.56 | 52.75 | 54.52 | 53.14 | 52.67 | |
| 0.5 | 60.23 | 56.71 | 58.36 | 59.86 | 58.82 | 58.78 | |
| 0.7 | 66.02 | 62.44 | 64.39 | 65.86 | 64.87 | 65.25 | |
| CEN | 10% | 61.47 | 58.43 | 59.30 | 61.09 | 59.84 | 60.15 |
| 40% | 55.75 | 50.80 | 53.99 | 55.45 | 54.30 | 53.62 | |
| 60% | 47.05 | 40.16 | 46.01 | 46.93 | 45.77 | 43.14 | |
| 58.61 | 54.62 | 56.64 | 58.27 | 57.67 | 56.88 |
From Table 3, we see that all the transformed tests seem better than the untransformed (naive) test based on our simulation. Moreover, the last row of Table 3 shows the marginal effects of TEST from model (7). It is evident that the test based on the arcsine-square root transformation (ArcsP) tends to have slightly elevated type I error rates while the other tests are slightly conservative. The pseudo-value regression performs best since the average deviations from the nominal 5 percent level are close to 0. In addition, from Table 4, we see that the power increases with the sample size and paired correlation. Conversely, the power decreases with the censoring proportion. In the last row of Table 4, we show the marginal effects of TEST from model (7). Although the naive test (NaiP) and the arcsine transformation (ArcsP) have higher power, they are anti-conservative. Consequently, in summary, pseudo-value regression is a satisfactory method for comparing paired right-censored data at a fixed point based on the simulation.
3.2 Clustered right-censored data
To assess the performance of the extended tests for clustered right-censored data, we conducted simulation studies under various scenarios. In each cluster, the Clayton-Oakes model (Clayton and Cuzick, 1985; Oakes, 1989),
was used to generate correlated survival times for the jth cluster of the ith sample, where Sijk(t) was the kth marginal survival function of the jth cluster in the ith group and parameter ζ was used to control the strength of the correlation among subjects in each cluster (details appear in Cai and Shen, 2000). Large ζ induces smaller intracluster correlation and ζ = 2.0, 0.5, and 0.214 were employed corresponding to the Kendall tau (τ) of 0.2, 0.5 and 0.7, respectively. The total number of clusters of the two samples was set to be 30, 60, and 100. Cluster size mij was set to be 2.
Table 5 displays the empirical type I error rates. It is apparent that all the extended tests maintain reasonable type I error rates as sample size increases. However, the type I error rates of the tests developed in Klein et al. (2007) are anti-conservative when correlation exists. This happens because ignoring positive cluster correlation results in an underestimation of the true variance. Table 6 shows the power comparison results. The power of all of the tests considered here decrease as the correlation among subjects in each cluster increases. That's because the denominator of the test statistics considers the positive clustered correlation and has higher variance covariance estimate. As expected, the power of the extended tests increases as the sample size increases and odds ratio grows; and the power of extended tests decreases as the censoring proportion increases.
Table 5. Empirical type I error rates of tests for clustered right-censored data.
| Considering clustered correlation | Ignoring clustered correlation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| τ | n | %cens | NaiC | LogC | LlogC | ArcsC | LogitC | Pseudo | Nai | Log | Llog | Arcs | Logit |
|
|
|
||||||||||||
| 0 | 30 | 10 | 0.082 | 0.069 | 0.058 | 0.078 | 0.063 | 0.063 | 0.056 | 0.043 | 0.038 | 0.054 | 0.040 |
| 40 | 0.081 | 0.069 | 0.065 | 0.081 | 0.069 | 0.067 | 0.063 | 0.046 | 0.042 | 0.062 | 0.047 | ||
| 60 | 0.119 | 0.083 | 0.081 | 0.112 | 0.085 | 0.074 | 0.077 | 0.037 | 0.050 | 0.075 | 0.055 | ||
| 60 | 10 | 0.067 | 0.058 | 0.056 | 0.060 | 0.059 | 0.061 | 0.055 | 0.047 | 0.047 | 0.051 | 0.048 | |
| 40 | 0.067 | 0.060 | 0.062 | 0.059 | 0.063 | 0.060 | 0.057 | 0.049 | 0.048 | 0.055 | 0.050 | ||
| 60 | 0.070 | 0.056 | 0.056 | 0.071 | 0.059 | 0.056 | 0.053 | 0.036 | 0.046 | 0.055 | 0.045 | ||
| 100 | 10 | 0.057 | 0.053 | 0.055 | 0.053 | 0.055 | 0.054 | 0.052 | 0.047 | 0.047 | 0.050 | 0.047 | |
| 40 | 0.059 | 0.057 | 0.056 | 0.061 | 0.058 | 0.053 | 0.057 | 0.051 | 0.051 | 0.056 | 0.052 | ||
| 60 | 0.069 | 0.060 | 0.064 | 0.071 | 0.065 | 0.048 | 0.059 | 0.049 | 0.055 | 0.060 | 0.056 | ||
| 0.2 | 30 | 10 | 0.069 | 0.055 | 0.052 | 0.064 | 0.054 | 0.059 | 0.064 | 0.043 | 0.039 | 0.061 | 0.043 |
| 40 | 0.078 | 0.059 | 0.055 | 0.075 | 0.058 | 0.067 | 0.066 | 0.046 | 0.041 | 0.064 | 0.046 | ||
| 60 | 0.098 | 0.066 | 0.062 | 0.091 | 0.077 | 0.075 | 0.067 | 0.041 | 0.041 | 0.063 | 0.044 | ||
| 60 | 10 | 0.061 | 0.051 | 0.055 | 0.056 | 0.056 | 0.057 | 0.070 | 0.056 | 0.056 | 0.068 | 0.057 | |
| 40 | 0.067 | 0.058 | 0.058 | 0.063 | 0.059 | 0.064 | 0.073 | 0.058 | 0.058 | 0.070 | 0.062 | ||
| 60 | 0.070 | 0.056 | 0.061 | 0.070 | 0.061 | 0.066 | 0.067 | 0.048 | 0.053 | 0.065 | 0.054 | ||
| 100 | 10 | 0.060 | 0.055 | 0.054 | 0.054 | 0.054 | 0.046 | 0.064 | 0.060 | 0.060 | 0.064 | 0.062 | |
| 40 | 0.058 | 0.053 | 0.052 | 0.048 | 0.053 | 0.044 | 0.062 | 0.057 | 0.057 | 0.061 | 0.058 | ||
| 60 | 0.059 | 0.050 | 0.055 | 0.061 | 0.057 | 0.048 | 0.060 | 0.049 | 0.052 | 0.059 | 0.055 | ||
| 0.5 | 30 | 10 | 0.074 | 0.048 | 0.056 | 0.065 | 0.060 | 0.071 | 0.097 | 0.076 | 0.074 | 0.095 | 0.077 |
| 40 | 0.095 | 0.063 | 0.067 | 0.091 | 0.073 | 0.083 | 0.108 | 0.079 | 0.070 | 0.103 | 0.074 | ||
| 60 | 0.115 | 0.067 | 0.066 | 0.097 | 0.072 | 0.082 | 0.101 | 0.058 | 0.056 | 0.084 | 0.061 | ||
| 60 | 10 | 0.058 | 0.048 | 0.051 | 0.056 | 0.053 | 0.054 | 0.089 | 0.078 | 0.077 | 0.086 | 0.078 | |
| 40 | 0.064 | 0.049 | 0.052 | 0.053 | 0.055 | 0.053 | 0.095 | 0.084 | 0.084 | 0.095 | 0.085 | ||
| 60 | 0.073 | 0.056 | 0.067 | 0.077 | 0.067 | 0.059 | 0.084 | 0.065 | 0.073 | 0.086 | 0.075 | ||
| 100 | 10 | 0.052 | 0.045 | 0.050 | 0.051 | 0.051 | 0.050 | 0.082 | 0.078 | 0.076 | 0.081 | 0.079 | |
| 40 | 0.062 | 0.052 | 0.054 | 0.047 | 0.055 | 0.051 | 0.092 | 0.087 | 0.086 | 0.091 | 0.088 | ||
| 60 | 0.062 | 0.047 | 0.056 | 0.062 | 0.057 | 0.052 | 0.077 | 0.065 | 0.067 | 0.077 | 0.069 | ||
| 0.7 | 30 | 10 | 0.085 | 0.056 | 0.057 | 0.080 | 0.063 | 0.086 | 0.131 | 0.109 | 0.098 | 0.125 | 0.100 |
| 40 | 0.073 | 0.045 | 0.048 | 0.071 | 0.054 | 0.077 | 0.121 | 0.090 | 0.076 | 0.114 | 0.084 | ||
| 60 | 0.101 | 0.066 | 0.067 | 0.092 | 0.077 | 0.082 | 0.106 | 0.066 | 0.059 | 0.098 | 0.077 | ||
| 60 | 10 | 0.063 | 0.049 | 0.052 | 0.052 | 0.053 | 0.060 | 0.112 | 0.101 | 0.101 | 0.109 | 0.102 | |
| 40 | 0.054 | 0.043 | 0.049 | 0.053 | 0.050 | 0.052 | 0.112 | 0.095 | 0.095 | 0.111 | 0.099 | ||
| 60 | 0.072 | 0.053 | 0.062 | 0.074 | 0.065 | 0.075 | 0.108 | 0.077 | 0.085 | 0.108 | 0.090 | ||
| 100 | 10 | 0.062 | 0.051 | 0.054 | 0.058 | 0.058 | 0.049 | 0.125 | 0.118 | 0.117 | 0.124 | 0.119 | |
| 40 | 0.056 | 0.049 | 0.048 | 0.049 | 0.051 | 0.056 | 0.108 | 0.100 | 0.100 | 0.107 | 0.101 | ||
| 60 | 0.057 | 0.048 | 0.053 | 0.059 | 0.054 | 0.060 | 0.092 | 0.078 | 0.081 | 0.091 | 0.082 | ||
Table 6. Estimated power of several tests for clustered right-censored data.
| Clustered | Odds ratio=2 | Odds ratio=3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| τ | n | %cens | NaiC | LogC | LlogC | ArcsC | LogitC | Pseudo | NaiC | LogC | LlogC | ArcsC | LogitC | Pseudo |
|
|
|
|||||||||||||
| 0 | 30 | 10 | 0.286 | 0.249 | 0.249 | 0.277 | 0.256 | 0.342 | 0.599 | 0.549 | 0.562 | 0.588 | 0.567 | 0.621 |
| 40 | 0.273 | 0.227 | 0.236 | 0.260 | 0.243 | 0.306 | 0.522 | 0.459 | 0.482 | 0.509 | 0.485 | 0.548 | ||
| 60 | 0.243 | 0.184 | 0.190 | 0.227 | 0.203 | 0.252 | 0.443 | 0.370 | 0.384 | 0.424 | 0.392 | 0.415 | ||
| 60 | 10 | 0.467 | 0.437 | 0.439 | 0.459 | 0.444 | 0.519 | 0.831 | 0.811 | 0.817 | 0.826 | 0.819 | 0.848 | |
| 40 | 0.412 | 0.384 | 0.394 | 0.410 | 0.398 | 0.470 | 0.758 | 0.727 | 0.743 | 0.752 | 0.741 | 0.787 | ||
| 60 | 0.314 | 0.282 | 0.285 | 0.307 | 0.290 | 0.355 | 0.637 | 0.595 | 0.602 | 0.627 | 0.609 | 0.652 | ||
| 100 | 10 | 0.635 | 0.618 | 0.622 | 0.633 | 0.624 | 0.683 | 0.959 | 0.955 | 0.957 | 0.958 | 0.957 | 0.969 | |
| 40 | 0.585 | 0.563 | 0.574 | 0.582 | 0.575 | 0.631 | 0.933 | 0.926 | 0.930 | 0.932 | 0.928 | 0.944 | ||
| 60 | 0.442 | 0.417 | 0.423 | 0.437 | 0.426 | 0.495 | 0.802 | 0.779 | 0.784 | 0.798 | 0.787 | 0.817 | ||
| 0.2 | 30 | 10 | 0.270 | 0.224 | 0.235 | 0.260 | 0.238 | 0.325 | 0.545 | 0.477 | 0.508 | 0.535 | 0.514 | 0.576 |
| 40 | 0.254 | 0.205 | 0.222 | 0.250 | 0.225 | 0.309 | 0.522 | 0.459 | 0.482 | 0.509 | 0.485 | 0.548 | ||
| 60 | 0.233 | 0.173 | 0.187 | 0.222 | 0.192 | 0.247 | 0.436 | 0.360 | 0.383 | 0.417 | 0.389 | 0.405 | ||
| 60 | 10 | 0.397 | 0.367 | 0.376 | 0.391 | 0.377 | 0.460 | 0.790 | 0.763 | 0.775 | 0.785 | 0.778 | 0.819 | |
| 40 | 0.386 | 0.356 | 0.366 | 0.382 | 0.370 | 0.444 | 0.705 | 0.667 | 0.692 | 0.702 | 0.693 | 0.743 | ||
| 60 | 0.286 | 0.247 | 0.264 | 0.281 | 0.266 | 0.325 | 0.595 | 0.540 | 0.563 | 0.586 | 0.566 | 0.614 | ||
| 100 | 10 | 0.574 | 0.552 | 0.559 | 0.570 | 0.563 | 0.634 | 0.930 | 0.922 | 0.927 | 0.929 | 0.927 | 0.940 | |
| 40 | 0.549 | 0.521 | 0.537 | 0.547 | 0.540 | 0.600 | 0.889 | 0.875 | 0.885 | 0.889 | 0.886 | 0.913 | ||
| 60 | 0.431 | 0.396 | 0.405 | 0.426 | 0.408 | 0.478 | 0.789 | 0.765 | 0.774 | 0.784 | 0.777 | 0.800 | ||
| 0.5 | 30 | 10 | 0.226 | 0.177 | 0.191 | 0.218 | 0.200 | 0.286 | 0.462 | 0.379 | 0.413 | 0.446 | 0.423 | 0.492 |
| 40 | 0.234 | 0.169 | 0.197 | 0.226 | 0.202 | 0.280 | 0.434 | 0.349 | 0.396 | 0.421 | 0.399 | 0.461 | ||
| 60 | 0.206 | 0.140 | 0.158 | 0.194 | 0.165 | 0.225 | 0.379 | 0.278 | 0.312 | 0.356 | 0.322 | 0.345 | ||
| 60 | 10 | 0.321 | 0.291 | 0.299 | 0.315 | 0.304 | 0.399 | 0.705 | 0.660 | 0.689 | 0.700 | 0.690 | 0.738 | |
| 40 | 0.333 | 0.291 | 0.308 | 0.327 | 0.311 | 0.393 | 0.651 | 0.593 | 0.634 | 0.645 | 0.634 | 0.691 | ||
| 60 | 0.280 | 0.229 | 0.253 | 0.273 | 0.254 | 0.321 | 0.533 | 0.479 | 0.502 | 0.524 | 0.508 | 0.549 | ||
| 100 | 10 | 0.502 | 0.477 | 0.491 | 0.501 | 0.494 | 0.562 | 0.893 | 0.873 | 0.885 | 0.891 | 0.886 | 0.914 | |
| 40 | 0.469 | 0.438 | 0.462 | 0.468 | 0.462 | 0.536 | 0.838 | 0.811 | 0.834 | 0.836 | 0.833 | 0.866 | ||
| 60 | 0.368 | 0.329 | 0.350 | 0.365 | 0.352 | 0.404 | 0.728 | 0.694 | 0.713 | 0.724 | 0.713 | 0.753 | ||
| 0.7 | 30 | 10 | 0.218 | 0.156 | 0.171 | 0.209 | 0.180 | 0.263 | 0.422 | 0.327 | 0.364 | 0.402 | 0.375 | 0.451 |
| 40 | 0.217 | 0.148 | 0.174 | 0.209 | 0.181 | 0.273 | 0.404 | 0.292 | 0.344 | 0.388 | 0.353 | 0.426 | ||
| 60 | 0.197 | 0.127 | 0.141 | 0.177 | 0.147 | 0.202 | 0.378 | 0.273 | 0.296 | 0.349 | 0.310 | 0.342 | ||
| 60 | 10 | 0.308 | 0.271 | 0.284 | 0.302 | 0.289 | 0.374 | 0.650 | 0.594 | 0.621 | 0.640 | 0.626 | 0.683 | |
| 40 | 0.307 | 0.253 | 0.276 | 0.298 | 0.283 | 0.373 | 0.597 | 0.531 | 0.574 | 0.589 | 0.576 | 0.638 | ||
| 60 | 0.241 | 0.193 | 0.216 | 0.235 | 0.218 | 0.283 | 0.529 | 0.461 | 0.488 | 0.517 | 0.496 | 0.545 | ||
| 100 | 10 | 0.438 | 0.415 | 0.423 | 0.436 | 0.426 | 0.501 | 0.827 | 0.808 | 0.815 | 0.824 | 0.818 | 0.856 | |
| 40 | 0.429 | 0.392 | 0.412 | 0.423 | 0.415 | 0.504 | 0.788 | 0.756 | 0.781 | 0.816 | 0.781 | 0.829 | ||
| 60 | 0.369 | 0.322 | 0.339 | 0.361 | 0.345 | 0.429 | 0.705 | 0.667 | 0.684 | 0.700 | 0.688 | 0.730 | ||
As follows, we also apply ANOVA techniques to summarize all the simulation results in Table 5 and Table 6. Table 7 shows the average deviations from the nominal 5 percent level of six tests and Table 8 shows the average rejection rates for the six tests using ANOVA by using model (4)-(7) for clustered right-censored data.
Table 7.
Average deviations from nominal 5 percent level of six tests using ANOVA (for clustered right-censored data).
| TEST | NaiC | LogC | LlogC | ArcsC | LogitC | Pseudo | |
|---|---|---|---|---|---|---|---|
| NUM | 30 | 2.96 | 0.80 | 0.73 | 0.56 | 1.17 | 2.16 |
| 60 | 1.26 | 0.20 | 0.44 | 0.65 | 0.60 | 0.76 | |
| 100 | 1.83 | 0.19 | 0.29 | 0.26 | 0.44 | 0.04 | |
| CORR | 0.0 | 1.88 | 1.10 | 0.87 | 1.53 | 1.12 | 0.97 |
| 0.2 | 1.55 | 0.52 | 0.43 | 1.00 | 0.57 | 0.62 | |
| 0.5 | 1.75 | 0.08 | 0.50 | 1.05 | 0.78 | 1.03 | |
| 0.7 | 1.55 | -0.12 | 0.13 | 1.05 | 0.48 | 1.33 | |
| CEN | 10% | 1.58 | 0.32 | 0.42 | 1.06 | 0.66 | 0.92 |
| 40% | 1.78 | 0.48 | 0.55 | 1.25 | 0.82 | 1.06 | |
| 60% | 3.04 | 0.90 | 1.25 | 2.80 | 1.63 | 1.47 | |
| 1.68 | 0.40 | 0.48 | 1.16 | 0.74 | 0.98 |
Table 8.
Average rejection rates for six tests using ANOVA for different NUM, CORR and CENS (for clustered right-censored data).
| TEST | NaiC | LogC | LlogC | ArcsC | LogitC | Pseudo | |
|---|---|---|---|---|---|---|---|
| NUM | 30 | 31.80 | 25.29 | 27.66 | 30.67 | 28.29 | 35.67 |
| 60 | 48.86 | 44.98 | 46.79 | 48.27 | 47.08 | 53.62 | |
| 100 | 65.24 | 63.14 | 63.34 | 62.22 | 64.47 | 69.26 | |
| CORR | 0.0 | 55.50 | 52.54 | 53.38 | 54.88 | 53.64 | 58.90 |
| 0.2 | 51.76 | 48.23 | 49.70 | 51.24 | 49.97 | 55.93 | |
| 0.5 | 45.57 | 40.90 | 43.33 | 44.95 | 43.65 | 50.15 | |
| 0.7 | 47.71 | 36.19 | 38.66 | 51.13 | 39.19 | 46.42 | |
| CEN | 10% | 50.23 | 46.47 | 47.80 | 49.56 | 48.23 | 54.40 |
| 40% | 47.04 | 42.47 | 44.73 | 46.54 | 45.00 | 51.30 | |
| 60% | 39.13 | 33.75 | 35.40 | 37.96 | 35.93 | 40.75 | |
| 48.63 | 44.46 | 46.27 | 48.05 | 46.61 | 52.85 |
As shown in Table 7, comparison of the tests shows that all transformed tests seem better than the naive test based on our simulation. All the tests tend to be slightly anti-conservative. The tests based on the log and loglog transformation perform better in comparison. In addition, from Table 8, we see that the power increases with sample size but decreases with the correlation. In summary, considering both type I and type II error rates and given that the test based on the loglog transformation had a higher average rejection rate than log transformation. We suggest that using loglog transformation is a satisfactory method for clustered right-censored data.
4. Examples
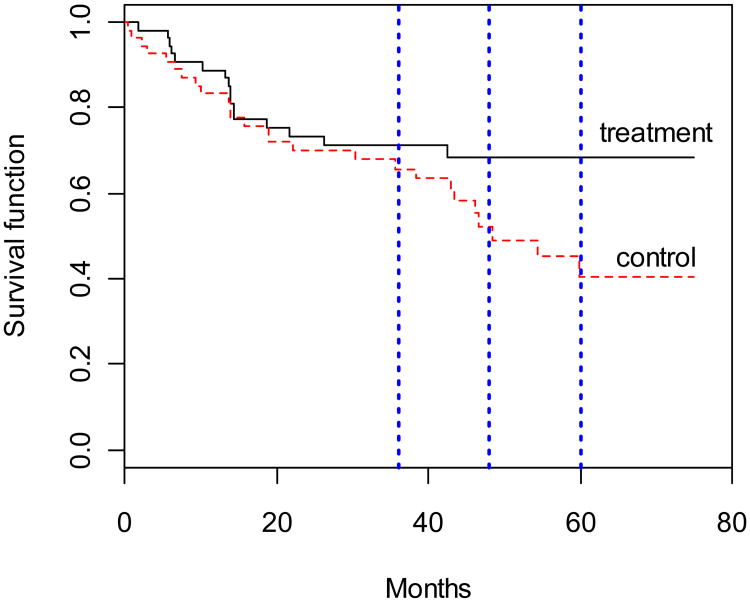
For paired right-censored data, the extended tests are applied to the Diabetic Retinopathy Study (DRS) analyzed by Huster et al. (1989). Diabetic retinopathy is a complication associated with diabetes mellitus consisting of abnormalities in the microvasculature within the retina of the eye. There are 1742 patients in this study. Huster et al. (1989) used a subset, 50% sample of the high-risk patients as defined by DRS criteria, to demonstrate their proposed method. A total of 54 patients with juvenile diabetes are investigated in this study for purpose of illustration. Each patient had one eye randomized to argon laser treatment and the other eye received a xenon arc photocoagulator. Patients were followed in order to detect vision loss, with survival times defined as the initiation of treatment to blindness (i.e. visual acuity below 5/200 for two visits in a row). Since time to vision loss is positively correlated within individuals, paired right-censored data arise. In such a case, the primary goal is to understand the effectiveness of laser photocoagulation in delaying the onset of blindness in patients with diabetic retinopathy. We compare the survival probability at 36, 48, and 60 months, respectively.
The sample censoring rate is 61%. The Kaplan-Meier estimates of the marginal survival functions for the treatment group and control group are displayed in Figure 1 and the testing results are listed in Table 9. The vertical dash lines mark the three time points, respectively. The plot shows a significant visual difference at 60 months. The p-values of the tests compared in Table 9 are all less than the pre-specified significance level of 0.05. This implies that for diabetic patients the laser treatment produced a higher 5-year (60-months) survival rate. In addition, the plot shows a less significant visual difference at 48 months. The extended methods consider within-pair correlation yield p-values that are less than 0.05. This implies that ignoring dependence deflates the significance level because of within-pair correlation.
Fig. 1.
Estimated survival curves for the Diabetic Retinopathy Study.
Table 9.
One-sided p-values of several tests for two studies.
| Diabetic retinopathy study (paired right-censored data) | |||||||
|---|---|---|---|---|---|---|---|
| months | method | naive | log | loglog | arcsine | logit | pseudo |
| 36 | Klein | 0.275 | 0.276 | 0.276 | 0.275 | 0.275 | |
| Extended | 0.229 | 0.230 | 0.230 | 0.229 | 0.229 | 0.227 | |
| 48 | Klein | 0.053 | 0.060 | 0.056 | 0.055 | 0.056 | |
| Extended | 0.027* | 0.033* | 0.028* | 0.027* | 0.028* | 0.030* | |
| 60 | Klein | 0.005* | 0.012* | 0.006* | 0.006* | 0.007* | |
| Extended | 0.002* | 0.007* | 0.002* | 0.002* | 0.003* | 0.007* | |
|
| |||||||
| Otology study (clustered right-censored data) | |||||||
| months | method | naive | log | loglog | arcsine | logit | pseudo |
|
| |||||||
| 12 | Klein | 0.014* | 0.020* | 0.016* | 0.016* | 0.018* | |
| Extended | 0.020* | 0.029* | 0.022* | 0.022* | 0.025* | 0.038* | |
p-value is less than 0.05.
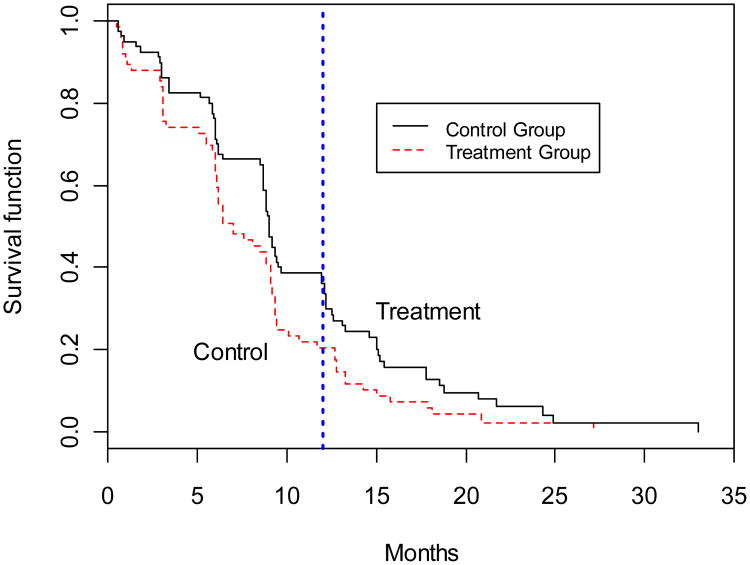
Secondly, we applied the extended tests for clustered right-censored data to the otology study conducted by Le and Lindgren (1996). This study enrolled 78 children, aged 6 months to 8 years who had been diagnosed as having otitis media in both ears and who had received ventilating tubes as a surgical intervention. These children were randomized to either a no-treatment group (n1 = 38) or a post-surgery treatment group (n2 = 40). They received regular follow-up care to determine if their tubes were functioning. The aim of the study was to determine whether the tube life in the post-surgery treatment group was longer than that in the control group. We compared the survival probability at 12 months, using the lifetimes of tubes noted in the original study to estimate the survival probability. The Kaplan-Meier estimates of survival functions for the two groups are displayed in Figure 2, and the testing results are listed in Table 9.
Fig. 2.
Estimated survival curves for the otology study.
In this data set, the sample censoring rate is 7.7%. At the 0.05 significance level, the p-values of these tests show that the lifetime of the post-surgery treatment is longer than the lifetime of the control group at 12 months. Although the test results are identical to the methods proposed by Klein et al. (2007), the p-values of each extended test is larger than the test proposed by Klein et al. (2007), since our tests considering the clustering effect.
5. Concluding remarks
In this paper, we extended the methods proposed by Klein et al. (2007) to compare the differences between two survival functions at a fixed time point for paired right-censored data and clustered right-censored data, respectively. We derived the variance of the statistics to accommodate the possible within-pair correlation and within-cluster correlation. Moreover, the pseudo-value regression approach is also used to model the effects of covariates on the outcome for hypothesis testing. Note that for clustered right-censored data, the tests are valid under unequal intracluster correlation coefficients or an unequal number of units in each cluster. Overall speaking, for paired right-censored data, if there is a positive correlation within each pair, ignoring positive dependence leads to more conservative tests, while for clustered data, ignoring (the positive) dependence leads to more liberal tests. Other studies support this conclusion. For example, Wang and Fygenson (2009) demonstrate the importance of separately accounting for intra-subject factor effect and the between-subject factor effect for the framework of a semi-parametric quantile regression model. In addition, this paper also presents two examples of applications on a diabetic retinopathy study for paired right-censored data and an otology study for clustered right-censored data. Although there is no significant difference between the tests based on different transformations, we showed that the proposed test is applicable for testing the survival probabilities at a fixed time point.
Faced with the problem of including some other covariates to compare the survival curves at a fixed time, one approach is to base the test on a stratified version of one of the tests discussed in Section 2. Suppose the population can be divided into g strata. For paired right-censored data, let Ŝig(t) be the Kaplan-Meier estimators in the g stratum for sample i, where g = 1,2,…, m, i = 1,2.; in this case, the stratified test can be defined as
| (8) |
Similarly, for clustered right-censored data, simply change Ŝ1g(t) to S̃ig(t) in (8), and the variance part can be computed as describe in Section 2.
Computer codes written in FORTRAN for this type of analyses are available from the first author upon request. To compare the equality of two independent samples of survival functions, Logan et al. (2008) formulated the problem as testing for differences in survival curves after a prespecified time point when researchers anticipate that the survival curves appear to cross at some time point. In our future work, we will investigate tests based on different transformations for testing this hypothesis H0: S1(t) = S0(t), for t ≥ t0 where t0 can be prespecified. In addition, in order to derive a sample size formula, we will consider a specific alternative hypothesis and derive the theoretical property of the test statistic.
Acknowledgments
The authors thank the Editor and the reviewers for their helpful comments. These led to considerable improvements in this paper. In addition, we wish to thank editorial assistants, Yvonne Poindexter and Lynne Berry, for their editorial work on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen PK, Klein JP, Rosthøj S. Generalized linear models for correlated pseudo-observations with applications to multi-state models. Biometrika. 2003;90:15–27. [Google Scholar]
- Bie O, Borgan Ø, Liestøl K. Confidence intervals and confidence bands for the cumulative hazard rate function and their small sample properties. Scandinavian Journal of Statistics. 1987;14:221–233. [Google Scholar]
- Borgan Ø, Liestøl K. A note on confidence intervals and bands for the survival curve based on transformations. Scandinavian Journal of Statistics. 1990;17:35–41. [Google Scholar]
- Cai J, Shen Y. Permutation tests for comparing marginal survival functions with clustered failure time data. Statistics in Medicine. 2000;19:2963–2973. doi: 10.1002/1097-0258(20001115)19:21<2963::aid-sim593>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Clayton D, Cuzick J. Multivariate generalizations of the proportional hazards model (with discussion) Journal of the Royal Statistical Society, Series A. 1985;148:82–117. [Google Scholar]
- Dallas MJ, Rao PV. Testing equality of survival functions based on both paired and unpaired censored data. Biometrics. 2000;56:154–159. doi: 10.1111/j.0006-341x.2000.00154.x. [DOI] [PubMed] [Google Scholar]
- Gill RD. Censoring and stochastic integrals, Tract 124. The Mathematical Centre; Amsterdam: 1980. [Google Scholar]
- Greenwood M. The natural duration of cancer, reports on public health and medical subjects. Vol. 33. London: H.M. Stationery Office; 1926. [Google Scholar]
- Gu M, Follmann D, Geller NL. Monitoring a general class of two-sample survival statistics with applications. Biometrika. 1999;86:45–57. [Google Scholar]
- Huang X, Wolfe RA. A frailty model for informative censoring. Biometrics. 2002;58:510–520. doi: 10.1111/j.0006-341x.2002.00510.x. [DOI] [PubMed] [Google Scholar]
- Huster WS, Brookmeyer R, Self SG. Modeling paired survival data with covariates. Biometrics. 1989;45:145–156. [PubMed] [Google Scholar]
- Jung SH, Jeong JH. Rank tests for clustered survival data. Lifetime Data Analysis. 2003;9:21–33. doi: 10.1023/a:1021869803601. [DOI] [PubMed] [Google Scholar]
- Kalbfeisch JD, Prentice RL. The statistical analysis of failure time data. New York: Wiley; 1980. [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation form incomplete observations. Journal of the American Statistical Association. 1958;53:457–481. [Google Scholar]
- Klein JP, Andersen PK. Regression modeling of competing risks data based on pseudo-values of the cumulative incidence function. Biometrics. 2005;61:223–229. doi: 10.1111/j.0006-341X.2005.031209.x. [DOI] [PubMed] [Google Scholar]
- Klein JP, Logan B, Harhoff M, Anderson PK. Analyzing survival curves at a fixed point in time. Statistics in Medicine. 2007;26:4505–4519. doi: 10.1002/sim.2864. [DOI] [PubMed] [Google Scholar]
- Le CT, Lindgren BR. Duration of ventilating tubes: A test of comparing two clustered samples of censored data. Biometrics. 1996;52:328–334. [PubMed] [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Logan BR, Klein JP, Zhang MJ. Comparing treatments in the presence of crossing survival curves. An application to bone marrow transplantation. Biometrics. 2008;64:733–740. doi: 10.1111/j.1541-0420.2007.00975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran PAP. Testing for correlation between non-negative variates. Biometrika. 1967;54:385–394. [PubMed] [Google Scholar]
- Murray S. Using weighted Kaplan-Meier statistics in nonparametric comparisons of paired censored survival outcomes. Biometrics. 2001;57:361–368. doi: 10.1111/j.0006-341x.2001.00361.x. [DOI] [PubMed] [Google Scholar]
- Nair VN. Confidence bands for survival functions with censored data: a comparative study. Technometrics. 1984;26:265–275. [Google Scholar]
- Oakes D. Bivariate survival models induced by frailties. Journal of the American Statistical Association. 1989;84:487–493. [Google Scholar]
- O'Gorman JT, Akritas MG. Nonparametric models and methods for designs with dependent censored data: Part II. Journal of Nonparametric Statistics. 2004;16:613–622. doi: 10.1111/j.0006-341x.2001.00088.x. [DOI] [PubMed] [Google Scholar]
- Parzen MI, Wei LJ, Ying Z. Simultaneous confidence intervals for the difference of two survival functions. Scandinavian Journal of Statistics. 1997;24:309–314. [Google Scholar]
- Pepe MS, Fleming TR. Weighted Kaplan-Meier statistics: a class of distance tests for censored survival data. Biometrics. 1989;45:497–507. [PubMed] [Google Scholar]
- Perme MP, Andersen PK. Checking hazard regression models using pseudo-observations. Statistics in Medicine. 2008;27:5309–5328. doi: 10.1002/sim.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Fygenson M. Inference for censored quantile regression models in longitudinal studies. Annals of Statistics. 2009;37:756–781. [Google Scholar]
- Ying Z, Wei LJ. The Kaplan-Meier estimate for dependent failure time observations. Journal of Multivariate Analysis. 1994;50:17–29. [Google Scholar]