Abstract
Photolyases (PHRs) and cryptochromes (CRYs) belong to the same family known as blue-light photoreceptors. Although their amino acid sequences and corresponding structures are similar to each other, they exert different functions. PHRs function as an enzyme to repair UV-induced deoxyribonucleic acid (DNA) lesions such as a cyclobutane pyrimidine dimer (CPD) and a (6-4) photoproduct ((6-4)pp), whereas CRYs are a circadian photoreceptor in plants and animals and at the same time they control the photoperiodic induction of flowering in plants. When a new type cryptochrome was identified, it was assumed that another type of CRYs, cryptochrome-DASH (CRY-DASH), which is categorized as a subfamily of photolyase/cryptochrome family, would possess the DNA photolyase activity. However, CRY-DASH had a weak DNA photolyase activity, but the reason for this is still unclear. To clarify the reason, we performed molecular dynamics (MD) simulations for a complex of CPD-PHR or CRY-DASH with damaged double-stranded DNA (dsDNA) and estimated the binding free energy, ΔGbind, between the protein and the damaged dsDNA by using a molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) method. ΔGbind for both proteins were −35 and 57 kcal mol−1, respectively, indicating that the structural stability of CRY-DASH was lower than that of CPD-PHR upon the damaged dsDNA binding. In particular, the number of amino acid residues relevant to the damaged dsDNA binding on the CRY-DASH surface was smaller than that on CPD-PHR. Therefore, the present result suggests that CRY-DASH has a weak DNA photolyase activity because it has a lower binding affinity than CPD-PHR.
Keywords: photolyase/cryptochrome family, duplex DNA binding, molecular mechanics/Poisson-Boltzmann surface area, binding free energy
Significance.
If the UV-induced DNA damages are not repaired, they inflict enormous damage on life. The photolyases have the DNA repair function and the existence of that has been identified in the species from bacteria to plant. The homolog of photolyases, cryptochrome-DASH, has been identified in human. However, cryptochrome-DASH did not indicate the repair quantum yield of equality to photolyases. To clarify that reason, we investigated the duplex DNA binding affinity for photolyases and cryptochrome-DASH by using molecular dynamics simulation and molecular mechanics/Poisson-Boltzmann surface area method. We indicated that the contribution of the protein surface to duplex DNA binding was important.
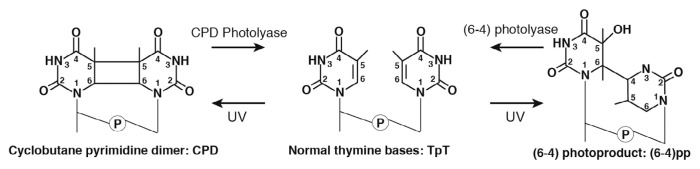
Sunlight is undoubtedly essential for all living organism on Earth, but the UV light in sunlight sometimes exerts adverse effects that cause deoxyribonucleic acid (DNA) damage in genes. Irradiation with a wavelength below 300 nm such as UV-B and UV-C light is extremely harmful to DNA. UV light induces photoexcitation of nucleobases, resulting in covalent bond formations especially between neighboring pyrimidine bases. The covalent bonds in photo-induced DNA lesions are mainly categorized into two types. One is a so-called cyclobutane pyrimidine dimer (CPD) in which two covalent bonds are formed between the pyrimidine rings in the adjacent pyrimidine bases at the 5-position (C5-C5) and at the 6-position (C6-C6) to produce a four membered ring. The other is a so-called (6-4) photoproduct ((6-4)pp), whose covalent bond is formed between the adjacent pyrimidine rings at the 6- and 4-positions (C6-C4) (see Fig. 1) [1–5]. If these photoproducts were to exist in DNA without any repair, they would lead to serious damage to an organism. For example, this damage causes a growth delay in plants, skin cancer in mammals, and so on [1,2–7]. Therefore, to avoid fatal photoinduced damage, all organisms possess diverse DNA repair systems essential for recognizing and removing these photoproducts efficiently.
Figure 1.
Conformational changes from normal thymine bases (TpT) to UV-lesions. (left) cyclobutane pyrimidine dimer (CPD). (right) (6-4) photoproduct. These conformations are induced by UV light absorption, and UV lesions are repaired by photolyases.
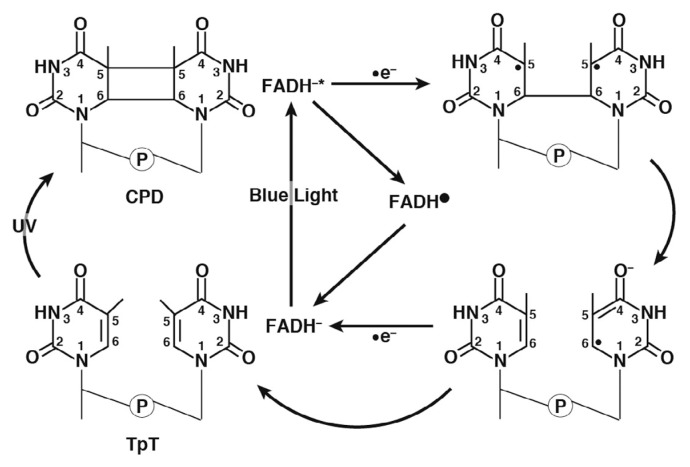
In particular, several plants possess photolyases (PHRs) to repair UV-induced DNA lesions. PHRs involve flavin adenine dinucleotide (FAD) as a cofactor. The oxidation and protonation state of FAD in the ground state is FADH− in PHRs, and DNA repair occurs by a photoinduced electron transfer reaction from FADH− to CPD or (6-4)pp [8–11,12–20]. PHRs are categorized into two types as (a) CPD-PHR and (b) (6-4)pp-PHR, which specifically repair CPD and (6-4)pp, respectively. The repair mechanism of CPD by CPD-PHR illustrated in Figure 2 is briefly explained as follows: (i) PHRs recognize UV-induced DNA lesions and bind to the DNA. (ii) FADH− is excited to FADH−* by blue-light irradiation (wavelengths of 350–450 nm). (iii) An electron transfer from FADH−* to CPD occurs. (iv) The covalent bond (C5-C5) in CPD is first cleaved to provide an electron. (v) Subsequently, another covalent bond (C6-C6) is cleaved. (vi) Finally, the electron returns from a pyrimidine pair to flavin radical (FADH·).
Figure 2.
Repair mechanism of CPD by CPD-PHR.
Several preceding theoretical studies have mainly investigated the electron transfer (ET) reaction processes for CPD repair [13–15]. These studies proposed the different ET pathways (i.e., a pathway via the adenine moiety of FADH−, a direct pathway from FADH− to CPD, and pathways via the adenine moiety of FADH− and some amino acid residues). The experimental studies also investigated the ET pathways by using femtosecond-resolved transient absorption spectroscopy for the different types of pyrimidine dimers, T< >T, T< >U, U< >T, and U< >U in CPD-PHR. They also proposed an ET pathway via the adenine moiety of FADH− [16,17]. However, the authors stated that the electron passes only through the adenine moiety in FADH−. Although there exists a hot debate on the ET pathways of CPD-PHR, few studies on the binding mechanisms of protein to DNA have been conducted so far.
Besides the studies on PHRs, cryptochromes (CRYs), which belong to the same family as PHRs known as blue-light photoreceptors, were identified in bacteria, algae, plants, fish, and animals as in a diverse group of flavoproteins. CRYs function as UV-A or blue-light receptors and fulfill versatile functions such as flower initiation, circadian clocks, magnetic receptors, and so on [8,19–22]. Although CRYs share homologies in their sequences and have a common FAD cofactor with PHRs, they lack the DNA repair function [23,24]. Cryptochrome-DASH (CRY-DASH), which belongs to one of the subfamilies of the photolyase/cryptochrome family, was identified from Drosophila, Arabidopsis, Synechocystis, and Human. (DASH is named after these four species.) The structure of CRY-DASH resembles those of PHRs [19,25–27]. Five amino acid residues relevant to the active space in E. coli CPD-PHR are Glu274, Trp277, Asn341, Met345, and Trp384, whereas those in Arabidopsis thaliana CRY-DASH are Glu325, Trp328, Asn391, Gln395, and Tyr434, indicating that three of the five amino acid residues commonly exist in both CPD-PHR and CRY-DASH. (Note that the common amino acid residues are denoted with underlines.) On the basis of site directed mutagenesis, it was pointed out that Met345, which is missing in CRY-DASH, might be important for the repair reaction [28]. To prove the assumption, molecular dynamics (MD) simulations were previously performed for the wild-type (WT) of CPD-PHR with double-stranded DNA (dsDNA) and the WT and mutants of CRY-DASH (CRY-DASHQ395A and CRY-DASHQ395M) with single-stranded DNA (ssDNA) [28]. It was found that hydrogen bonds between CPD and Glu were not formed in CRY-DASHWT with ssDNA but were formed in CPD-PHRWT with dsDNA. On the other hand, they were formed in CRY-DASHQ395M, indicating that Met mainly contributes to the formation of these hydrogen bonds. Interestingly, Selby and Sancar have reported that CRY-DASH does not perform the repair function on dsDNA lesions but does perform it on ssDNA lesions in vitro [23]. The latter fact suggests that CRY-DASH itself has the DNA repair ability. Thus, it is highly plausible that a difference between the binding of dsDNA and ssDNA to CRY-DASH plays a key role in the repair function.
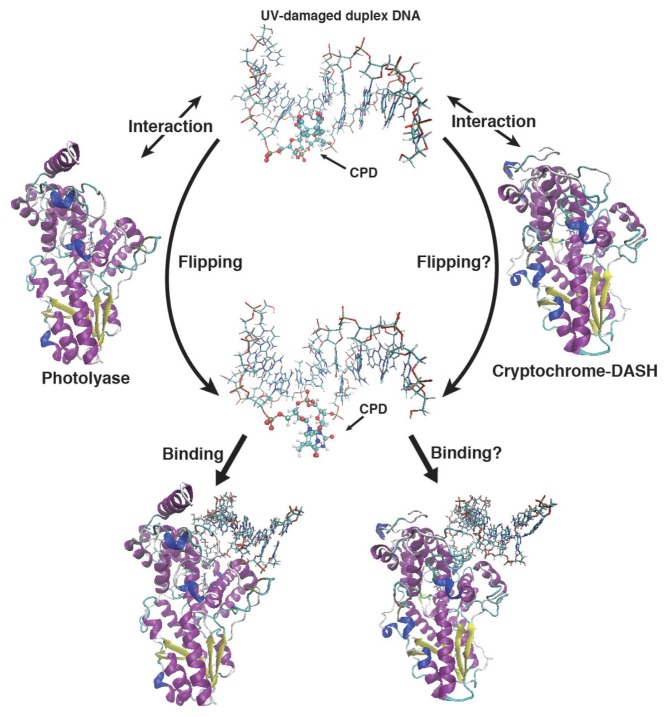
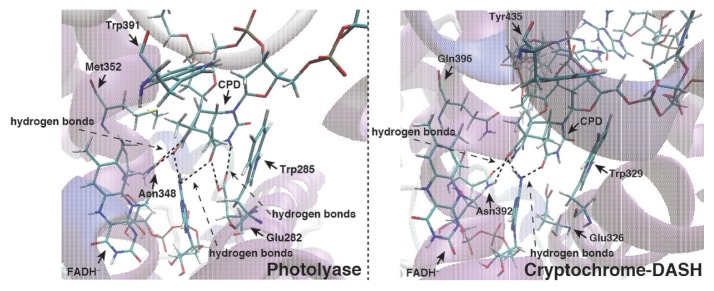
During the past decades, it has not been fully understood why CRY-DASH is unable to express the repair function for UV-induced DNA damage. In previous work, we have investigated the base flipping mechanism of a UV-induced dsDNA lesion using MD simulations with an efficient conformational search algorithm proposed by us [29]. We found that the CPD damaged dsDNA cannot expose the lesion moiety in solution spontaneously. This result indicates that the flipping of the lesion moiety occurs after its binding to CPD-PHR. Judging from the difference between the electrostatic potentials around the DNA binding sites in CRY-DASH and CPD-PHR, CRY-DASH may not induce the flipping of the dsDNA lesion moiety owing to a weaker interaction than with CPD-PHR (see Fig. 3). Because ssDNA lesions are exposed on the outside of the strand without the flipping, it is possible that CRY-DASH may bind to the ssDNA lesions and eventually repair the lesions.
Figure 3.
Duplex DNA binding scheme for CPD-PHR and CRY-DASH. The base flipping of the lesion moiety will occur by the interaction between UV-damaged duplex DNA and protein. But due to the interaction of CRY-DASH is weak, the base flipping might not occur in CRY-DASH. Next, CPD-PHR and CRY-DASH recognize the lesion moiety and bind the duplex DNA. Here the binding affinity of CRY-DASH will be less than that of photolyase.
In the present study, we further investigated the reason why CRY-DASH does not possess the repair function on the dsDNA lesions by using MD simulations for complexes of CPD-PHR or CRY-DASH with the CPD damaged dsDNA, which we hereafter call the CPD-PHR complex or CRY-DASH complex for simplicity. We first modeled a CRY-DASH complex and showed how CRY-DASH binds the damaged dsDNA, because no X-ray structure is available for the CRY-DASH complex. We then compared the obtained structure of the CRY-DASH complex with that of CPD-PHR. Moreover, the binding affinity between them was quantitatively investigated by using a molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) method, and the amino acid residues relevant to the binding were detected using the analyses. In the following section, we explain structure modeling, numerical details of MD simulations, and binding energy analyses. Subsequently, we present the results and discussion. Finally, concluding remarks are described.
Methods
Structure modeling
To model a CRY-DASH complex with damaged dsDNA, we performed the docking simulation using MEGADOCK [30,31]. In Supplementary Text S1, the docking calculation scheme is shown. Here, in our docking simulation using MEGADOCK, we adopted a default parameter set. Among 2000 snapshots, the structure with the best score was chosen for further analyses (S=4152.19). The docking scores are shown in Supplementary Figure S1, and the model structure of the CRY-DASH complex is illustrated in Supplementary Figure S2 (see supporting Supplementary materials for details). Here, for the initial modeling, the X-ray structures of dsDNA with CPD (PDB ID: 1TEZ) [32] and CRY-DASH (PDB ID: 2VTB) [25] were utilized. Here, the –O–CH2–O–part of CPD was replaced with –O–PO2–O–using Winmostar [33].
Molecular dynamics simulations
In the present study, all the MD simulations were performed using the AMBER14 program package [34]. We first performed the geometry optimizations for CPD, FADH−, 7,8-didenethyl-8-hydoroxy-5-deazariboflavin (8-HDF), and methenyltetrahydrofolate (MTHF) with the B3LYP/6-31G(d) level of theory using Gaussian09 [35]. Based on the obtained structures, the force field parameters for the MD simulations were constructed using the antechamber module in AMBER14. In the actual MD simulations, we used these obtained force fields for these unconventional molecules (CPD, FADH−, 8-HDF, and MTHF), the AMBER ff14SB force field [36] for the amino acid residues and nucleic acids, and the TIP3P model for water [37]. Next, we set up the initial state using the tLeap module in AMBER14 for both CPD-PHR and CRY-DASH complexes. Herein, in the case of CPD-PHR, the numbers of atoms for the protein, FAD, 8-HDF, Na+, water, and the damaged dsDNA were 7480, 85, 43, 17, 107052, and 630, respectively, whereas in the case of CRY-DASH, those for the protein, FAD, MTHF, Na+, water, and damaged dsDNA were 8081, 85, 56, 15, 115686, and 630, respectively. Finally, the total numbers of the atoms in each system were 115307 for CPD-PHR and 124553 for CRY-DASH. Subsequently, for each system, we performed energy minimizations under positional constraints with respect to the heavy atoms (the force constant was 10 kcal mol−1 Å−2) for 5000 steps, and performed energy minimizations without the constraints for 10000 steps. Then, the temperature of each system was gradually increased from 0 K to 300 K during 100 ps. In the equilibration processes, MD simulations were carried out with an isobaric isothermal (NPT) ensemble (P=1 atm and T=300 K) during 100 ns. Subsequently, MD simulations were carried out under an NPT ensemble (P=1 atm and T=300 K) during 50 ns in the production runs. The simulation time step was set to 2 fs, and the periodic boundary condition was considered, in which the electrostatic interaction was treated with the particle mesh Ewald (PME) method [38]. Here, the SHAKE algorithm was used for the constraints [39], the Langevin thermostat for the temperature regulation [40], and the Berendsen barostat for the pressure regulation [41]. A snapshot was recorded every 20 ps, and the total snapshots obtained were 25000 after 50 ns for statistical analyses.
Binding free energy calculation
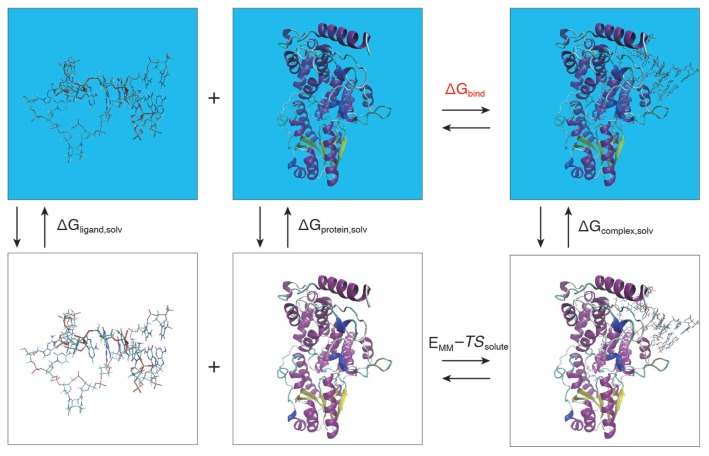
To investigate the bonding affinity, we performed the MM/PBSA method [42,43] for the obtained snapshots. The MM/PBSA method can evaluate the binding free energy, ΔGbind, between a given protein and a target ligand. ΔGbind is estimated by the following equations:
| (1) |
where ΔGcomplex,solv, ΔGprotein,solv, and ΔGligand,solv are the free energy differences for the complex, the protein, and the ligand with or without solvent, respectively. Herein, a subscript “solv” in Eq. (1) represents the aqueous solution. The solvation free energies are calculated as follows;
| (2) |
| (3) |
| (4) |
| (5) |
where X in Eq. (2) represents the complex, the protein, and the ligand. EMM is the molecular mechanics (MM) energy from the force field without the solvent. Eintra consists of three intramolecular contributions, i.e. Ebond, Eangle, and Etorsion. Eelec and EvdW are the intermolecular electrostatic and van der Waals interaction energies, respectively. ΔGsolvation is the solvation free energy, and ΔGsolvation–elec is estimated from the Poisson–Boltzmann method. ΔGnonpolar is estimated from the solvent-accessible surface area (SASA). T and Ssolute are the temperature and the entropy of a solute. We show the relationship for each energy in Figure 4. To evaluate the entropic contribution Ssolute, the normal-mode analysis was performed under the quasi-harmonic approximation [44] implemented in AMBER14. We performed MM/PBSA for 25000 frames from 100 ns to 150 ns in order to estimate averaged value of ΔGbind and its root-mean-square deviation. Then, we obtained partial binding free energy contribution to the amino acid residue Y, , by using Per-residue decomposition protocol in MM/PBSA.py [43]. Per-residue basis decomposition can estimate the contribution of each residue to the total binding free energy [45–49]. To obtain we first divide terms in Eq. (2) into its atomic contribution. The contribution of each atom a to the total electrostatic interaction energy is obtained by
Figure 4.
Computational schemes of the binding free energies based on MM/PBSA. The free energies colored in black are directly calculated, while the free energy of interest colored in red is indirectly did using the thermodynamic cycle of other free energies.
| (6) |
where qa and qb are atomic partial charge of the atom a and b, rab is the distance between them. Similarly, one half of the pairwise energy for van der Waals interaction energy between protein and ligand, , to avoid double counting. Using the SASA of each atom a, a non-polar part of solvent effects on binding free energy is represented as
| (7) |
where SASAa,protein and SASAa,liand is equal to zero depending on which component the atom belong to. γ is set to 0.0072 kcal mol−1 Å−2 in AMBER 14. To calculate the contribution of atom a to the electrostatic part of solvent effects, the generalized Born (GB) approach is used. The contribution of atom a is given by;
| (8) |
| (9) |
where κ is the Debye-Hückel screening parameter. ɛω is a dielectric constant for the solvent (80). αa and αb are the effective Born radii of atoms a and b, respectively. Using these contributions to each atom, the partial binding free energy contribution to the amino acid residue Y is evaluated as
| (10) |
Note here that the entropic and intra-molecular contributions appearing in Eps. (2) and (3) are neglected in this analysis.
Results and Discussion
Comparison of the active site of CPD-PHR and CRY-DASH
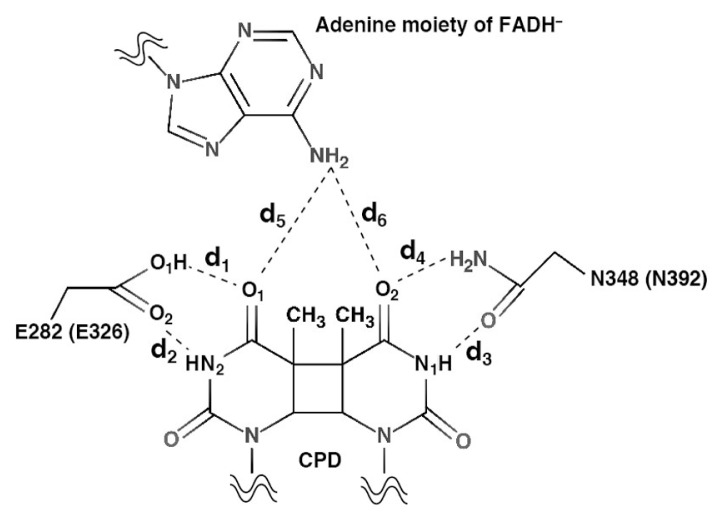
We compared the energy minimized structures starting from an X-ray structure of the CPD-PHR complex and from a model CRY-DASH complex (see Supplementary Fig. S3). We investigated the atomic distances between CPD and Glu282 (Glu326), Asn348 (Asn392), and FADH− for CPD-PHR (CRY-DASH), where the definition of them is given in Figure 5. These distances for CPD-PHR, d1, d2, d3, d4, d5, and d6 were 3.37, 4.98, 2.96, 2.76, 3.02, and 3.53 Å, while those for CRY-DASH were 2.80, 5.24, 2.93, 2.85, 2.86, and 3.91 Å. Therefore, it is confirmed that the model structure of CRY-DASH at the active site resembles that of CPD-PHR. Furthermore, total root mean square deviations (RMSDs) measured from initial structures for CPD-PHR and CRY-DASH were 1.98±0.09 and 3.15±0.08 Å, respectively (see Supplementary Fig. S4). The partial RMSDs of the protein and DNA for CPD-PHR were 1.75±0.07 and 3.25±0.32 Å, while those for CRY-DASH were 2.63±0.05 Å and 5.12±0.24 Å. Although RMSD of CRY-DASH was large compared to that of CPD-PHR, these were considered to be the equilibrium states because of maximum fluctuation being within 5%. On the other hand, RMSDs measured from averaged structures of CPD-PHR and CRY-DASH obtained by MD simulations were 1.32±0.01 and 1.37±0.08 Å, respectively (see Supplementary Fig. S5). Judging from these RMSD values with smaller fluctuations, obtained trajectories are sufficiently relaxed from the initial models. In the structures with the lowest RMSD value with respect to the averaged structure, the distances of d5 and d6 were 3.67 and 3.17 Å for CPD-PHR (RMSD=1.08 Å) and 3.27 and 3.19 Å for CRY-DASH (RMSD=1.13 Å), respectively. In addition to the hydrogen bonds between Asn348 and CPD, other hydrogen bonds were formed between Glu282 and CPD in CPD-PHR complex, while they were not found in the CRY-DASH complex (see Fig. 6). These average distances for CPD-PHR (CRY-DASH), d1, d2, d3, d4, d5, and d6 were 2.85±0.12 Å (4.84±0.86 Å), 3.59±0.61 Å (3.53±0.50 Å), 3.27±0.28 Å (3.10±0.20 Å), 2.88±0.12 Å (2.90±0.13 Å), 3.50±0.34 Å (3.40±0.36 Å), and 3.10±0.24 Å (3.20±0.28 Å), respectively. These results indicate that the hydrogen bonds between the 3′ side of CPD and Asn are stable for each system, whereas the bond between the 5′side of CPD and Glu is unstable in CRY-DASH complex. Therefore, the active site of CRY-DASH is less stable than that of CPD-PHR, and the electron transfer reaction is less likely to occur on CRY-DASH.
Figure 5.
Definition of bond distance among amino acid residue, FADH−, and CPD. E282 (E326) and N348 (N392) are the amino acid residue in CPD-PHR (CRY-DASH). d1, d2, d3, d4, d5, and d6 are the distances between O1 of CPD and O1 of E282 (E326), between N2 of CPD and O2 of E282 (E326), between N1 of CPD and O of N348 (N392), between O2 of CPD and N of N348 (N392), between O1 of CPD and N of FADH−, and between O2 of CPD and N of FADH−, respectively
Figure 6.
Active sites strucutres of CPD-PHR and CRY-DASH with the lowest RMSD values. Dotted lines correspond to the bond among amino acid residures, FADH−, and CPD. And dotted arrows represent the position the hydogen bond is formed.
The preceding studies by Essen and Klar proposed that the proton transfer would occur from Glu283 to CPD during the repair reaction [4]. Later, Liu and co-workers used CPD-PHR mutant changed from Glu to Ala and observed the repair efficiency change upon the mutation. According to their result, the repair quantum yields dramatically decreased from the WT (0.82) to the mutant (0.40) [17]. Previously, Sato and co-workers theoretically indicated that these hydrogen bonds were not formed in the CRY-DASH and ssDNA with CPD [28]. Therefore, these hydrogen bonds play an important role in the UV lesion repair reaction deduced from the averaged structural analyses performed here.
Estimated binding free energy and partial contributions of amino acid residues on the active site
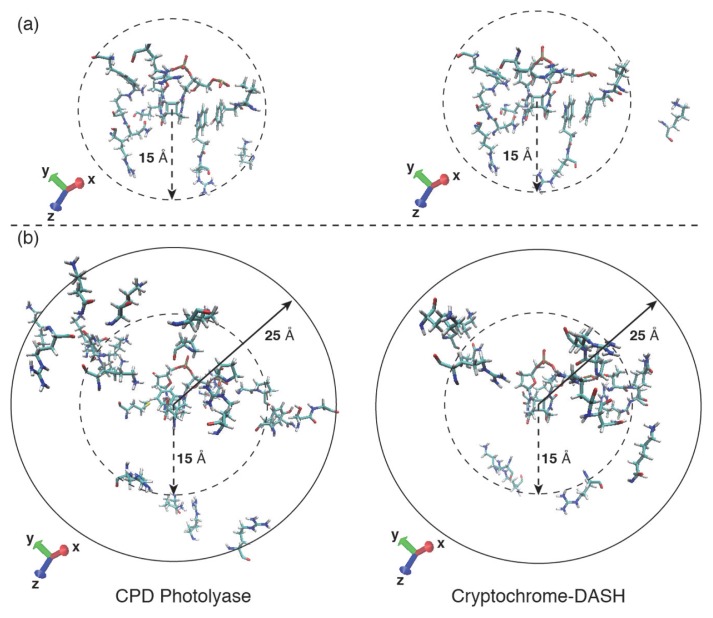
We elucidated the important amino acid residues for DNA binding using MM/PBSA. ΔGbind for the CPD-PHR and CRY-DASH complex were −35±14 and 57±19 kcal mol−1, respectively. Note here that the previously study investigated the ΔGbind for CPD-PHR, and that value was −32 kcal mol−1 [50], which is in reasonably good accordance with the former value. The binding affinity of CRY-DASH to the damaged dsDNA was evidently lower than that of CPD-PHR, indicating that the CRY-DASH complex seemed to be unstable. In Table 1, we listed the common amino acid residues in each protein with large contributions to the DNA binding. Especially, the common amino acid residues nearby the active site were as follows: Val147 (194), Tyr148 (195), Lys160 (205), Arg231 (277), Trp285 (329), Arg286 (330), Asn348 (392), Arg349 (393), Arg351 (395), Arg403 (447), and Phe405 (449), where the numbers in parenthesis are the corresponding amino acid residue numbers in CRY-DASH. As shown in Figure 7(a), these common amino acid residues were often located around the active site (≤15 Å from CPD). On the other hand, the non-common amino acid residues with large contribution to the DNA binding were listed in Table 2. As shown in Figure 7(b), some non-common amino acid residues were located on the protein surface (>15 Å from CPD). Moreover, the number of the amino acid residues that distribute to protein surface in CPD-PHR was larger than that in CRY-DASH, indicating binding ability to the DNA depends strongly on the surface amino acid residues. As described in the Introduction, the Met352 of CPD-PHR contributed favorably to dsDNA binding (−1.9 kcal mol−1), whereas the Gln396 at the same position in CRY-DASH did not (1.5 kcal mol−1). Judging again from the results, a mutation from Met to Gln may result in a reduction of photolyase activity on the dsDNA lesions.
Table 1.
Decomposed binding free energy on a per-residue basis of the common amino acid residues largely contributing to the DNA binding,
| CPD-PHR | CRY-DASH | ||
|---|---|---|---|
| Val147 | −3.3±0.8 | Val194 | −3.2±0.6 |
| Tyr148 | −3.6±0.9 | Tyr195 | −4.2±0.6 |
| Lys160 | −1.4±0.3 | Lys205 | −1.6±0.3 |
| Arg231 | −3.4±0.9 | Arg277 | −3.1±1.4 |
| Trp285 | −2.3±0.6 | Trp329 | −2.5±0.6 |
| Arg286 | −1.7±0.3 | Arg330 | −3.2±0.5 |
| Asn348 | −1.9±0.7 | Asn392 | −3.5±0.7 |
| Arg349 | −8.4±2.1 | Arg393 | −8.8±1.9 |
| Arg351 | −2.4±0.5 | Arg395 | −3.5±0.5 |
| Arg403 | −3.1±2.5 | Arg447 | −9.6±1.5 |
| Phe405 | −3.8±1.5 | Phe449 | −1.3±0.4 |
Amino acid residue/Per-residue free energy decomposition [kcal/mol]
Figure 7.
Amino acid residues which have the partial binding free energy contribution lower than −1.0 kcal mol−1.
(a) The common amino acid residues in both CPD-PHR and CRY-DASH are Val147 (194), Tyr148 (195), Lys160 (205), Arg231 (277), Trp285 (329), Arg286 (330), Asn348 (392), Arg349 (393), Arg351 (395), Arg403 (447), Phe405 (449). Here, the numbers in parenthesis are the residue numbers in CRY-DASH. (b) The non-common amino acid residues in CPD-PHR are Arg10, Arg106, Ser139, Gly140, Tyr145, Gly149, Pro150, Asn154, Lys247, Arg273, Arg277, Arg305, Met352, Lys360, Arg367, Trp391, Pro399, Pro401, Asn406, Ser409, Gln410, Lys412, Lys413, Lys434, Arg459, Lys461, Lys464, and Lys471, and those in CRY-DASH are Thr196, Gln197, Arg199, Lys200, Arg259, Kys274, Lys288, Arg334, Lys339, Arg351, Arg404, Asn441, Pro443, Arg444, Lys453, Asn457, and Lys496.
Table 2.
Decomposed binding free energy on a per-residue basis of the non-common amino acid residues largely contributing to the DNA binding,
| CPD-PHR | CRY-DASH | ||||||
|---|---|---|---|---|---|---|---|
| Arg10 | −1.2±0.2 | Arg367 | −1.1±0.2 | Thr196 | −3.4±0.8 | Lys453 | −7.2±2.8 |
| Arg106 | −1.0±0.2 | Trp391 | −2.0±0.7 | Gln197 | −1.8±1.0 | Asn457 | −2.2±1.2 |
| Ser139 | −1.1±1.2 | Pro399 | −2.9±0.5 | Arg199 | −3.4±0.4 | Lys496 | −1.5±0.2 |
| Gly140 | −1.6±0.8 | Pro401 | −4.2±0.8 | Lys200 | −1.7±1.8 | ||
| Tyr145 | −1.7±0.9 | Asn406 | −2.0±1.3 | Arg259 | −1.2±0.3 | ||
| Gly149 | −2.1±0.5 | Ser409 | −2.9±2.1 | Lys274 | −2.4±0.2 | ||
| Pro150 | −2.5±0.6 | Gln410 | −4.8±2.7 | Lys288 | −1.8±0.4 | ||
| Asn154 | −1.3±0.5 | Lys412 | −1.8±0.4 | Arg334 | −1.8±0.4 | ||
| Lys247 | −1.2±0.2 | Lys413 | −6.2±2.2 | Lys339 | −2.6±0.5 | ||
| Arg273 | −1.2±0.2 | Lys434 | −1.0±0.1 | Arg351 | −3.6±0.4 | ||
| Arg277 | −2.0±0.3 | Arg459 | −1.2±0.2 | Arg404 | −4.0±0.4 | ||
| Arg305 | −2.6±0.4 | Lys461 | −1.6±0.3 | Asn441 | −1.2±0.3 | ||
| Met352 | −1.9±0.5 | Lys464 | −2.3±1.2 | Pro443 | −3.1±0.6 | ||
| Lys360 | −2.1±0.7 | Lys471 | −4.8±2.2 | Arg444 | −10.3±1.2 | ||
Amino acid residues/Per-residue free energy decomposition [kcal/mol]
Contribution of amino acid residues to duplex DNA binding on the protein surface
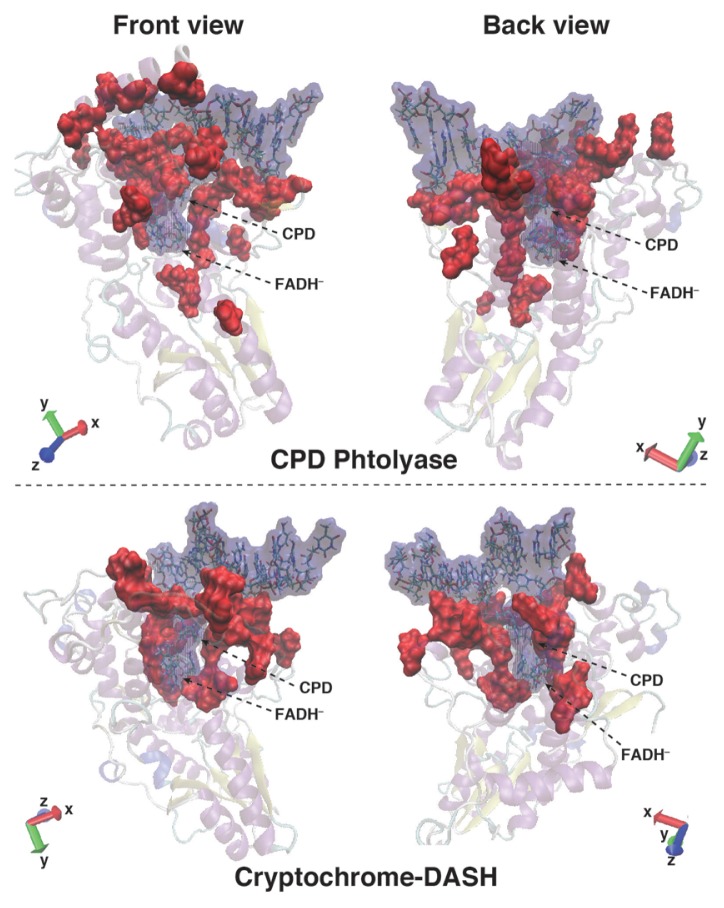
Large differences between CPD-PHR and CRY-DASH were observed for the amino acid residues on the protein surface attributed to the attractive interaction with dsDNA. We showed the contact surface between dsDNA and protein in Figure 8. The contact area between CRY-DASH and dsDNA is less than that of CPD-PHR. As shown in the left upper panel of Figure 8, some amino acid residues enfold dsDNA in CPD-PHR to tightly bind to the dsDNA. However, in CRY-DASH, the numbers of the amino acid residues that bound to dsDNA is less than that in CPD-PHR. Therefore, the contact area between CRY-DASH and dsDNA is narrower than that of CPD-PHR.
Figure 8.
Contact surface between protein and DNA. Blue and red color surfaces are duplex DNA with FADH−, and the amino acid residues that largely contribute to the DNA binding, respectively.
As shown in Supplementary Figure S6, an α helix was formed both in CPD-PHR and CRY-DASH, whereas the loop structure found in CPD-PHR, which tightly binds to dsDNA, was missing in CRY-DASH. The amino acid residues that comprise the α helix in CPD-PHR were Phe405, Asn406, Pro407, Ala408, Ser409, Gln410, Ala411, Lys412, and Lys413, whereas those in CRY-DASH were Phe449, Ser450, Ile451, Pro452, Lys453, Gln454, Ala455, Gln456, and Asn457. As shown in Supplementary Figure S7 (see the licorices colored with red), some of the amino acid residues in the α helix significantly contributed to the dsDNA legion binding. For example, the binding free energies with DNA were −3.8 (Phe405), −2.0 (Asn406), −3.0 (Ser409), −4.8 (Gln410), −1.8 (Lys412), and −6.2 (Lys413) kcal mol−1 for CPD-PHR and −1.3 (Phe449), −7.2 (Lys453), and −2.2 (Asn457) for CRY-DASH, respectively. The binding affinity of CRY-DASH to the dsDNA was obviously less than that of CPD-PHR in total. Therefore, the reason why the damaged dsDNA cannot bind to CRY-DASH is attributed to the insufficient number of amino acid residues mainly in the α helix that contribute to the stability of damaged dsDNA binding.
On the other hand, we found some amino acid residues with a highly repulsive contribution in the CRY-DASH complex. In particular, the binding energies of Asp442, Glu445, and Asp446 with DNA were 4.7, 20.9, and 19.3 kcal mol−1, respectively. In CPD-PHR, the amino acid residues at the same position are replaced by Asp398, Pro401, and Leu402, the binding energies with DNA were 2.3, −4.3, and −0.7 kcal mol−1, respectively. Here the amino acid residues (Asp and Glu) with a positive value indicate a negative charge, and the DNA is also a negative charge. Therefore, one of the reasons why the CRY-DASH complex is unstable might be because of this strong repulsive interaction between the negatively charged amino acid residues with the backbone of dsDNA.
Judging from the above analyses, if the relevant amino acid residues in CRY-DASH were mutated to adequate amino acid residues that largely contribute to DNA binding in CPD-PHR, it is expected that the binding affinity of dsDNA with CRY-DASH might increase and result in sufficient DNA repair ability. To prove the hypothesis, we need to perform the same analyses presented here for mutants with amino acid residues that significantly contribute to the binding of dsDNA. In future works, the analyses of CRY-DASH mutants will be carried out.
Conclusion
To elucidate the reason why CRY-DASH cannot repair dsDNA in vivo, we calculated the binding free energies of CRY-DASH and CPD-PHR with dsDNA, ΔGbind, by using the MM/PBSA method. The total ΔGbind of CRY-DASH was positive, whereas that of CPD-PHR was negative, indicating that the complex of CRY-DASH with the damaged dsDNA is unstable. Furthermore, we decomposed ΔGbind into the contributions from individual amino acid residues. According to the analyses, it was found that the amino acid residues around the active site had a large contribution to ΔGbind in both CPD-PHR and CRY-DASH. However, the Met352 and Trp391 of CPD-PHR contributed favorably to the DNA binding, whereas the Gln395 and Tyr435 of CRY-DASH did not. It was also found that the differences in individual binding energies between CPD-PHR and CRY-DASH originated mainly from the amino acid residues on the protein surface. In particular, the number of amino acid residues of the α helix in CRY-DASH that contributed to the DNA binding was less than that of CPD-PHR, and some amino acid residues in the CRY-DASH complex contributed to a highly repulsive, negative charge for the damaged dsDNA binding. These facts might be some of the major reasons why CRY-DASH cannot repair the UV-induced DNA damage of dsDNA.
Supplementary Information
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research of the Innovative Areas “Photosynergetics” (No. JP26107004), the Research Fellowship for Young Scientists (No. JP15J03797), and the Grant-in-Aid for Young Scientists (A) (No. JP16H06164) from the Japan Society for the Promotion of Science (JSPS). The computations were performed at the Research Center for Computational Science (RCCS), Institute of Molecular Science (IMS); the Advanced Center for Computing and Communication (ACCC), RIKEN; the Institute of Solid State Physics (ISSP), University of Tokyo; and the Center for Computational Sciences (CCS), University of Tsukuba. This research was partially supported by the Initiative on Promotion of Supercomputing for Young or Women Researchers, Information Technology Center, the University of Tokyo. Computational resources were provided under the Collaborative Research Program for Young/Women Scientists by the Academic Center for Computing and Media Studies, Kyoto University and the “TSUBAME Encouragement Program for Young/Female Users” of the Global Scientific Information and Computing Center, Tokyo Institute of Technology.
Footnotes
Conflicts of Interest
All the authors declare that they have no conflict of interest.
Author Contributions
RS, RH, and YS directed this project. RS, RH, and YS wrote the manuscript. RS performed the theoretical calculations and data analysis. YS and RH assisted in the calculations.
References
- 1.Taylor J-S. DNA, sunlight, and skin cancer. J Chem Educ. 1990;67:835–841. [Google Scholar]
- 2.Teramura AH. Effects of ultraviolet-B radiation on the growth and yield of crop plants. Physiol Plant. 1983;58:415–427. [Google Scholar]
- 3.de Gruijl FR, Roza L. Photoreactivation in humans. J Photochem Photobiol B, Biol. 1991;10:367–371. doi: 10.1016/1011-1344(91)80022-a. [DOI] [PubMed] [Google Scholar]
- 4.Li YF, Kim S-T, Sancar A. Evidence for lack of DNA photoreactivating enzyme in humans. Proc Natl Acad Sci USA. 1993;90:4389–4393. doi: 10.1073/pnas.90.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornman JF, Teramura AH. Effects of ultraviolet-B radiation on terrestrial plants. In: Young AR, Björn LO, Moan J, Nultsch W, editors. Environmental UV-Photobiology. Springer; Boston, MA: 1993. pp. 427–471. [Google Scholar]
- 6.Vink AA, Roza L. Biological consequences of cyclobutane pyrimidine dimers. J Photochem Photobiol B. 2001;65:101–104. doi: 10.1016/s1011-1344(01)00245-7. [DOI] [PubMed] [Google Scholar]
- 7.Hitomi K, Arvai AS, Yamamoto J, Hiomi C, Teranishi M, Hirouchi T, et al. Eukaryotic class II cyclobutane pyrimidine dimer photolyase structure reveals basis for improved ultraviolet tolerance in plants. J Biol Chem. 2012;287:12060–12069. doi: 10.1074/jbc.M111.244020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 9.Essen L-O. Photolyases and cryptochromes: common mechanisms of DNA repair and light-driven signaling? Curr Opin Struct Biol. 2006;16:51–59. doi: 10.1016/j.sbi.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Essen L-O, Klar T. Light-driven DNA repair by photolyases. Cell Mol Life Sci. 2006;63:1266–1277. doi: 10.1007/s00018-005-5447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sancar A. Structure and function of photolyase and in vivo enzymology: 50th anniversary. J Biol Chem. 2008;283:32153–32157. doi: 10.1074/jbc.R800052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antony J, Medvedev DM, Stuchebrukhov AA. Theoretical study of electron transfer between the photolyase catalytic cofactor FADH− and DNA thymine Dimer. J Am Chem Soc. 2000;122:1057–1065. [Google Scholar]
- 13.Medvedev D, Stuchebrukhov AA. DNA repair mechanism by photolyase: electron transfer path from the photolyase catalytic cofactor FADH− to DNA thymine dimer. J Theor Biol. 2001;210:237–248. doi: 10.1006/jtbi.2001.2291. [DOI] [PubMed] [Google Scholar]
- 14.Prytkova TR, Beratan DN, Skourtis SS. Photo-selected electron transfer pathways in DNA photolyase. Proc Natl Acad Sci USA. 2007;104:802–807. doi: 10.1073/pnas.0605319104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazawa Y, Nishioka H, Yura K, Yamato T. Discrimination of class I cyclobutane pyrimidine dimer photolyase from blue light photoreceptors by single methionine residue. Biophys J. 2008;94:2194–2203. doi: 10.1529/biophysj.107.119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao Y-T, Saxena C, Wang L, Sancar A, Zhong D. Direct observation of thymine dimer repair in DNA by photolyase. Proc Natl Acad Sci USA. 2005;102:16128–16132. doi: 10.1073/pnas.0506586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Tan C, Guo X, Kao Y-T, Li J, Wang L, et al. Dynamics and mechanism of cyclobutanine pyrimidine dimer repair by DNA photolyase. Proc Natl Acad Sci USA. 2011;108:14831–14836. doi: 10.1073/pnas.1110927108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Guo X, Tan C, Li J, Kao Y-T, Wang L, et al. Electron tunneling pathways and role of adenine in repair of cyclobutane pyrimidine dimer by DNA photolyase. J Am Chem Soc. 2012;134:8104–8114. doi: 10.1021/ja2105009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, et al. Identification of a new cryptochrome class: structure, function, and evolution. Mol Cell. 2003;11:59–67. doi: 10.1016/s1097-2765(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 20.Partch CL, Sancar A. Photochemistry and photobiology of cryptochrome blue-light photopigments: the search for a photocycle. Photochem Photobiol. 2005;81:1291–1304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- 21.Lin C, Todo T. The cryptochromes. Genome Biol. 2005;6:220–229. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 23.Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci USA. 2006;103:17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller M, Carell T. Structural biology of DNA photolyases and cryptochromes. Curr Opin Struct Biol. 2009;19:277–285. doi: 10.1016/j.sbi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Haung Y, Baxter R, Smith BS, Partch CL, Colbert CL, et al. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for Photolyase activity. Proc Natl Acad Sci USA. 2006;103:17701–17706. doi: 10.1073/pnas.0608554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klar T, Pokorny R, Moldt J, Batschauer A, Essen L-O. Cryptochrome 3 from arabidopsis thiliana: structural and functional analysis of its complex with a folate light antenna. J Mol Biol. 2007;366:954–964. doi: 10.1016/j.jmb.2006.11.066. [DOI] [PubMed] [Google Scholar]
- 27.Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen L-O. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA. 2008;105:21023–21027. doi: 10.1073/pnas.0805830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato R, Kitoh-Nishioka H, Ando K, Yamato T. Computational study on the roles of amino acid residues in the active site formation mechanism of blue-light photoreceptors. Chem Phys Lett. 2015;633:247–251. [Google Scholar]
- 29.Sato R, Harada R, Shigeta Y. Theoretical analyses on a flipping mechanism of UV-induced DNA damage. Biophys Physiobiol. 2016;13:311–319. doi: 10.2142/biophysico.13.0_311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohue M, Matsuzaki Y, Uchikoga N, Ishida T, Akiyama Y. MEGADOCK: an all-to-all protein-protein interaction prediction system using tertiary structure data. Protein Pept Lett. 2014;21:766–778. doi: 10.2174/09298665113209990050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohue M, Shimoda T, Suzuki S, Matsuzaki Y, Ishida T, Akiyama Y. MEGADOCK 4.0: an ultra-high-performance protein-protein docking software for heterogeneous supercomputers. Bioinformatics. 2014;30:3281–3283. doi: 10.1093/bioinformatics/btu532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mees A, Klar T, Gnau P, Hennecke U, Eker APM, Carell T, et al. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 33.Senda N. Winmostar v. 6.016. Delphi; [Google Scholar]
- 34.Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, et al. AMBER 14. University of California; San Francisco: 2014. [Google Scholar]
- 35.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 09, Revision D.01. Gaussian Inc; Wallingford, CT: 2009. [Google Scholar]
- 36.Malter JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorgensen WL, Chandrasekhar J, Nadura JD. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 38.Darden T, York D, Pedersen L. Particle mesh ewald: An N-log(N) method for ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 39.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 40.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford University; New York: 1987. [Google Scholar]
- 41.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 42.Massova I, Kollman PA. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect Drug Discover Des. 2000;18:113–135. [Google Scholar]
- 43.Miller BRIII, McGee TDJr, Swails JM, Homeyer N, Gohlke H, Roitberg AE. MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 44.Brooks BR, Janežič D, Karplus M. Harmonic analysis of large systems. I. Methodology. J Comput Chem. 1995;16:1522–1542. [Google Scholar]
- 45.Gohlke H, Kiel C, Case DA. Insights into protein-protein binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J Mol Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad R, Brandsdalm BO, Michaud-Soret I, Willassen NP. Ferric uptake regulator protein: binding free energy calculations and per-residue free energy decomposition. Proteins. 2008;75:373–386. doi: 10.1002/prot.22247. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Zhang S, Liu X, Zhang Q. Insights into drug resistance of mutations D30N and I50V to HIV-1 protease inhibitor TMC-114: free energy calculation and molecular dynamic simulation. J Mol Model. 2010;16:459–468. doi: 10.1007/s00894-009-0553-7. [DOI] [PubMed] [Google Scholar]
- 48.Khosa S, Frieg B, Mulnaes D, Kleinschrodt AH, Gohlke H, Smits HJ. Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Sci Rep. 2016;6:18679–18692. doi: 10.1038/srep18679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cele FN, Ramesh M, Soliman ME. Per-residue energy decomposition pharmacophore model to enhance virtual screening in drug discovery: a study for identification of reverse transcriptase inhibitors as potential anti-HIV agents. Drug Des Devel Ther. 2016;10:1365–1377. doi: 10.2147/DDDT.S95533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada D, Dokainish HM, Iwata T, Yamamoto J, Ishikawa T, Todo T, et al. Functional conversion of CPD and (6-4) photolyases by mutation. Biochemistry. 2016;55:4173–4183. doi: 10.1021/acs.biochem.6b00361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.