Abstract
High-risk ESBL-producing Enterobacteriaceae (ESBL-E) have been described in wild birds and rodents worldwide. Rats are of special interest not only due to their indicator role for environmental pollution with multi-resistant bacteria but also as possible infection source. Data on the presence of high-risk ESBL-E in urban wildlife from Africa remain scarce, however. Twenty-nine animals from three different rat (Rattus) species were captured in the city of Conakry (Guinea, West Africa) in 2015. Rectal swabs were analyzed for ESBL-E using selective media. Species typing and phenotypic antimicrobial resistance analysis to broad-spectrum beta-lactams and other classes of antimicrobials was performed for Enterobacteriaceae-like isolates using the VITEK®2 system (BioMérieux, Germany). Confirmed ESBL-producing E. coli and K. pneumoniae were whole-genome sequenced and resistance genes, phylogenetic background and genes related to bacterial fitness and virulence were analyzed. In total, six of twenty-nine rats (20%) carried ESBL-E (K. pneumoniae and E. coli). All ESBL-producers were multi-drug resistant with blaCTX−M−15 as the dominating ESBL-type. Interestingly, ESBL-associated clonal lineages E. coli ST38 and K. pneumoniae ST307 were found. The ESBL-plasmid in K. pneumoniae ST307 revealed high sequence similarities to pKPN3-307_TypeC, a >200 kbp IncFII plasmid originating from a human clinical ST307 isolate. This was in contrast to the core genome: the rat isolate was distantly related to the human clinical ST307 isolate (27 SNPs/Mbp). In addition, we identified π-fimbrial, capsule 2, and glycogen synthesis clusters in the rodent ST307 isolate, whose involvement in the adaptation to survival outside the host and in human urinary tracts has been suggested. Our results demonstrate the presence of clinically relevant, ESBL-producing K. pneumoniae ST307 and E. coli ST38 clonal lineages in an urban West African rat population. The human community is likely the initial source of ESBL-E however, rats might function as infection source and transmission hub, accelerated by frequent interactions at a human-wildlife interface.
Keywords: ESBL, rats, clonal spread, MLST, WGS, one health
Introduction
Extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBL-E), especially Klebsiella pneumoniae and Escherichia coli, have been on the rise for years. Diseases these bacteria cause, including diarrhea, septicemia, pneumonia, and urinary tract infections, are not only a cause of child mortality particularly in Africa (Lukac et al., 2015) but are also increasingly difficult to treat due to limitations in antimicrobial therapies (Kanj and Kanafani, 2011).
Besides the threat posed by ESBL-producers, studies report the global spread of K. pneumoniae that produce carbapenemases like KPC, often associated with pandemic high-risk clones predominantly of sequence type (ST) 258 and ST307 (Nordmann et al., 2011; Canton et al., 2012; Campos et al., 2016; Lee et al., 2016; Villa et al., 2017).
While research on ESBL-E has mostly focused on human and veterinary clinical contexts, recent findings have shown their prevalence in extra-clinical settings, such as communities, animals, and the environment suggesting a far broader and constant public health threat (Schaufler et al., 2016). Small mammals and rodents (Bonnedahl et al., 2009; Literak et al., 2010) as well as wild birds have been proposed as indicators for the spread of antimicrobial resistance across different environments (Guenther et al., 2010; Allen et al., 2011). While data on urban wild animals as carriers of ESBL-E in Europe, Asia and North-America exist, the role of urban wildlife in Africa remains rather unclear (Skurnik et al., 2006; Literak et al., 2009). We analyzed urban rats captured in the city of Conakry (Guinea, West Africa) for the occurrence of ESBL-E to detect potential reservoirs outside the clinical setting.
Materials and methods
This study was performed as part of a broader study investigating the role of rodents as potential infection source for various pathogens, in collaboration and under authorization by the Ministry of Livestock and Animal Products (Ministere de l'Elevage et des productions animales) in Conakry, Guinea.
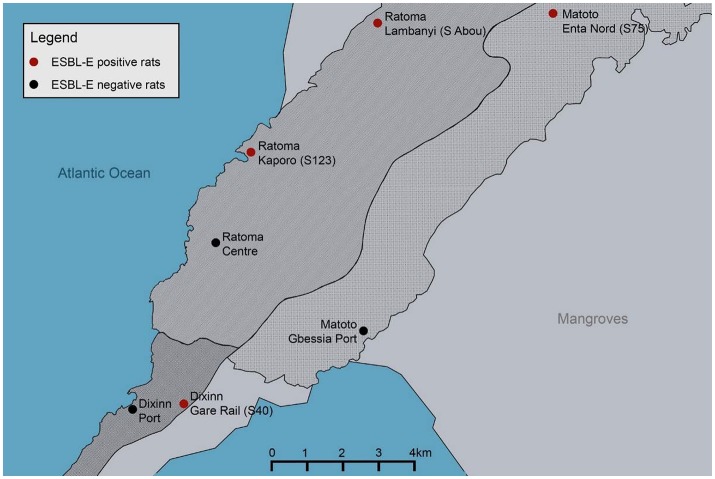
Twenty-nine rats from three different species (Rattus rattus n = 22, Rattus norvegicus n = 6, Cricetomys gambianus n = 1) were randomly captured in Conakry (Guinea, West Africa) between November and December 2015. Animals were trapped with life traps (Sherman LFA live trap; H.B. Sherman Traps, Inc., Tallahassee, FL, USA) inside and outside of households in three different, densely populated districts of Conakry (Dixinn [UTM 28P 645031 1055828], Ratoma [UTM 28P 648851 1063078], Matoto [UTM 28P 657469 1066350]; Figure 1). Trapped rodents were handled using appropriate personal protective equipment and were euthanized with a lethal dose of fluorane according to animal welfare guidelines. Each animal was sampled once and rectal swabs (MASTASWAB, Mast Diagnostics Reinfeld, Germany) were shipped to the Institute of Microbiology and Epizootics, Berlin, Germany.
Figure 1.
Map of Conakry (Guinea, West Africa) marked in red are ESBL-E-positive rat trapping sites, marked in black are ESBL-E-negative rat trapping sites. Red: S40 (Dixinn Gare Rail: 1 positive out of 9 trapped animals [1/9]), S123 (Ratoma Kaporo: 1/10), S75 (Matoto Enta Nord: 2/4), S Abou (Ratoma Lambanyi: 2/2). Black: Dixinn Port (0/2) Matoto Gbessia Port (0/1) Ratoma Centre (0/1).
Selection for cefotaxime-resistant and thus ESBL/AmpC-producing E. coli and K. pneumoniae ssp. pneumoniae was carried out by streaking on CHROMagar™ orientation plates (with and without 4 μg/ml cefotaxime; Mast Diagnostica, Reinfeld, Germany). Enterobacteriaceae-like isolates were species-typed by the automated VITEK®2 system (BioMérieux, Germany), which was also used to determine phenotypic antimicrobial resistance to carbapenems and other classes of antimicrobials (card GN38). In addition, isolates growing on CHROMagarTM that contained cefotaxime were confirmed as ESBL-producers using the phenotypic confirmatory test for ESBL-production (CLSI, 2008). Confirmed E. coli and K. pneumoniae isolates were subjected to whole-genome sequencing (WGS) and plasmid profile analysis as described previously (Schaufler et al., 2016). In brief, WGS was performed using MiSeq Illumina 300 bp paired-end sequencing and a coverage greater than 50 was obtained. After quality control using the NGS tool kit (Patel and Jain, 2012) (70% of bases with a phred score > 20), high quality filtered reads were used for a de novo assembly into contiguous sequences (contigs) using Velvet (Zerbino and Birney, 2008). Assembled draft genomes of the isolates were annotated using RAST (Aziz et al., 2008). WGS data was used for genotypic characterization including determination of the multi-locus sequence type (MLST), resistance genes (ResFinder 2.1), and plasmids (PlasmidFinder1.3); all available on the Center for Genomic Epidemiology website (http://www.genomicepidemiology.org).
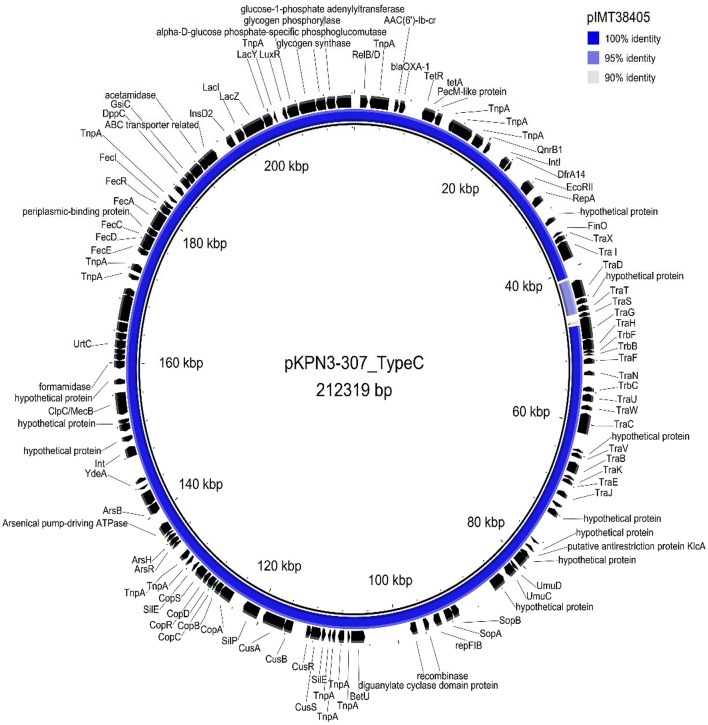
The plasmid sequence (KY271406) of pKPN3-307_TypeC (Villa et al., 2017) originating from a clinical ST307 isolate from Italy was mapped to the whole-genome of the ST307 isolate IMT38405 using Geneious 6.6. Plasmid contents of both genomes were then compared using Blast Ring Image Generator (BRIG) (Alikhan et al., 2011) (Figure 2).
Figure 2.
Circular visualization of plasmid pIMT38405 of K. pneumoniae ST307 resulting from a mapping of Illumina reads against plasmid pKPN3-307_TypeC (Villa et al., 2017) as a reference sequence using Blast Ring Image Generator (BRIG) (Alikhan et al., 2011).
Genes related to bacterial fitness (π-fimbrial cluster, capsule 2 Enterobacter, glycogen synthesis cluster; (Villa et al., 2017)) were analyzed accordingly. The number of SNPs in the core genome between isolates of the same ST was calculated using Harvest Suite 1.0 (Parsnp) and MEGA 6.0 (http://www.megasoftware.net/).
Accession numbers
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the Bioproject number PRJNA421654 (PJMH00000000 [IMT38402], PJMI00000000 [IMT38403], PJMJ00000000 [IMT38404], PJMK00000000 [IMT38405], PJML00000000 [IMT38406], PJMM00000000 [IMT38444]).
Results and discussion
We isolated coliform-appearing bacteria from 93% of all rats (27/29). Twenty percent (6/29) carried ESBL-E (three K. pneumoniae [IMT38444, IMT38403, IMT38405] and three E. coli [IMT38402, IMT38404, IMT38406]).
All ESBL-producers were multi-drug resistant (Table 1). BlaCTX−M−15 genes occurred in all strains with the exception of one blaCTX−M−14- (IMT38444) and one blaCTX−M−9-carrying strain (K. pneumoniae IMT38403). Since blaCTX−M−15 is the most prevalent ESBL-gene in human samples worldwide (Ewers et al., 2012) it seems likely that humans may act as a reservoir for antimicrobial resistance genes, which can be transmitted to rats and other animals (Gakuya et al., 2001). MLST identified three different STs for K. pneumoniae (ST307, ST502, and ST896) and two for E. coli (ST38 and ST4684) (Table 1).
Table 1.
Main characteristics of rat ESBL-producing K. pneumoniae and E. coli.
| Sample number | Bacterial species | Sampling point (UTM, city district) | Rat species | ST | Phenotypic resistance | Beta-lactam resistance genes | |
|---|---|---|---|---|---|---|---|
| MAIN CHARACTERISTICS | |||||||
| IMT38444 | K. pneumoniae | S 123 (28P 648851 1063078, Ratoma Kaporo) | R. rattus | ST-896 | CEF, TET, GEN, SXT, TOB | blaCTX−M−14, blaSHV−11−like, blaTEM−1B | |
| IMT38405 | K. pneumoniae | S 40 (28P 646660 1055837, Dixinn Gare Rail) | R. rattus | ST-307 | CEF, TET, ENR, GEN, TOB, SXT | blaCTX−M−15, blaOXA−1, blaSHV−28, blaTEM−1B | |
| IMT38403 | K. pneumoniae | S 75 (28P 657469 1066350, Matoto Enta Nord) | R. rattus | ST-502 | CEF, CAP, TET, SXT | blaCTX−M−9, blaDHA−1, blaSHV−62 | |
| IMT38402 | E. coli | S 75 (28P 657469 1066350, Matoto Enta Nord) | R. norvegicus | ST-38 | CEF, TET, SXT | blaCTX−M−15, blaTEM−1B | |
| IMT38406 | E. coli | S Abou (n.a., Ratoma Lambanyi) | R. rattus | ST-4684 | CEF, TET, SXT | blaCTX−M−15 | |
| IMT38404 | E. coli | S Abou (n.a., Ratoma Lambanyi) | R. rattus | ST-4684 | CEF, TET, SXT | blaCTX−M−15 | |
| Sample number | Aminoglycosides | Fluoroquinolones | Tetracyclines | Chloramphenicol | Sulfametoxazole/ Trimethoprim | Plasmid content | pMLST summary |
| ADDITIONAL GENOTYPIC RESISTANCE AND PLASMIDS | |||||||
| IMT38444 | aac(3)-IId-like, strA, strB | oqxA-like, oqxB-like, QnrS1 | tet(A), tet(D) | sul1, sul2, dfrA1 | IncFII, IncFIB(K), ColRNAI | IncF[K9:A-:B-] | |
| IMT38405 | aac(3)-IIa-like, strA-like, strB, aac(6')Ib-cr | oqxA-like, oqxB-like, QnrB66-like | tet(A) | catB3-like | sul2, dfrA14-like | IncFIB(K), IncFII(K) | IncF[K7:A-:B-] |
| IMT38403 | aadA5, strA-like | QnrB4 | tet(B)-like | catA1-like | sul1, dfrA1, dfrA15 | IncFIA(HI1), IncFIB(K), IncHI1B, IncR, ColRNAI | IncF[F-:A13*:B-] |
| IMT38402 | strA, strB | QnrS1 | tet(A) | catB1-like | sul2, dfrA14-like | IncFIB(K), IncFIB(AP001918), ColRNAI | IncF[F-:A-:B53] |
| IMT38406 | strA-like, strB | QnrS1 | tet(A)-like | sul2, dfrA14-like | ColRNAI | ||
| IMT38404 | strA-like, strB-like | QnrS1 | tet(A)-like | sul2, dfrA14-like | ColRNAI | ||
IMT, Institut für Mikrobiologie und Tierseuchen; ST, sequence type; CEF, cefotaxime; CHL, chloramphenicol; ENR, enrofloxacin; GEN, gentamicin; SXT, sulfamethoxazole; TET, tetracycline; TOB, tobramycin. ColRNAI is a small colicin plasmid, n.a. non available.
Plasmid profiling and bioinformatics analysis revealed the presence of large plasmids (>100 kbp) and incompatibility (Inc) types (e.g., IncFII, IncFIA, IncFIB) often associated with ESBL-plasmids in the K. pneumoniae strains (Lee et al., 2016). One E. coli strain harbored an IncFIB(K) and IncFIB(AP001918)-type plasmid (ST38, IMT38402, Table 1). Large plasmids, however, were absent in both ST4684 E. coli strains (IMT38404 and IMT38406) suggesting a chromosomal ESBL-gene location. This was supported by WGS results of both E. coli strains as (i) the blaCTX−M genes were detected on very large contigs (>250 kbp), (ii) BLAST analysis revealed mostly chromosomal hits for the CTX-M-carrying contigs, and (iii) no ESBL-associated Inc-types were identified. The mobile element Tn3 was found 300 bp downstream of CTX-M-15 and ISEc9 (ISEcp1-like element) directly upstream in an otherwise chromosomal background, which reinforced our hypothesis.
The chromosomal integration of ESBL-genes in E. coli from wild birds has previously been reported by Guenther et al. (2017). However, in this previous work, ST38 carried chromosomally-encoded ESBLs, contrarily to our current findings. The comparison of these wild bird ST38 to our rat ST38 isolates revealed a high number of SNPs (>1800 SNPs/Mbp) demonstrating their phylogenetically distinct character. E. coli ST38 is a prevalent human clinical pathogen but has also been reported on samples from wildlife as mentioned above (Turton et al., 2016).
Interestingly, K. pneumoniae ST307 has emerged worldwide in different locations and has been described as a putative high-risk clone associated with the production of carbapenemase KPC-2 and/or CTX-M-15 (Zhang et al., 2016; Kim et al., 2017). It has been frequently reported in human clinical samples from Italy, Korea, Pakistan, and Morocco and in pets from Japan. Although in small numbers detected, some of the rat strains resemble common pathogenic STs suggesting spill-over of clinical strains into an urban rat population in West Africa or vice versa.
Interestingly, comparison of the K. pneumoniae ST307 (IMT38405) ESBL-plasmid (~210 kbp) with a recently published K. pneumoniae WGS of an isolate with the same ST (Kim et al., 2017; Villa et al., 2017) isolated from a human clinical sample demonstrated high plasmid similarities (pKPN3-307_TypeC, KY271406)(Figure 2). Approximately 180 kbp of the rat isolate's plasmid sequence was 100% identical to the 212 kbp reference plasmid KPN3-307_TypeC. The remaining 30 kbp showed a similarity of 95%. Differences were only observed for one IS6 insertion family element (IS6100) upstream of the tra region. SNPs were found in finO and several genes from the tra operon.
When comparing the whole-genome sequence of IMT38405 to three recently published K. pneumoniae ST307 (Civ4, KH43, KL49; Villa et al., 2017), we detected between 155 and 432 SNPs in total (27–78 SNP/Mbp), excluding a recent clonal spread. Considering the SNPs Mbp−1 ratio of clonal EHEC strains during the outbreak (1.8 SNPs/Mbp) in Germany in 2011, our SNPs Mbp−1 ratio is tenfold higher, which indicates several years of separate evolution (Grad et al., 2012; de Been et al., 2014).
However, the occurrence of almost identical plasmids underlines the importance of plasmid-driven spread and transmission of antimicrobial resistances independent of the host's core genome.
We identified K. pneumoniae ST307 chromosomally-encoded genes, whose involvement in the adaptation to survival outside the host and in the human urinary tract has been previously suggested: the π-fimbrial cluster enabling bacterial mobility, and a serum resistance-conferring capsule 2 (Villa et al., 2017). A plasmid-encoded glycogen synthesis cluster (Villa et al., 2017), which may contribute to environmental survival through energy advantages and thus possibly to the bacterial spread between different hosts and settings, was also present. Carriage of these genes might be involved in the success of KPC-2 producing K. pneumoniae ST307 but probably predates the uptake of carbapenemase-plasmids as we found the same genes in our CTX-M-15-producing isolates.
In summary, our results demonstrate the presence of clinically relevant, ESBL-producing K. pneumoniae and E. coli clones in an urban West African rat population. Given the sanitary conditions in this region, rats could act either as indicators or environmental reservoirs of ESBL-E with a risk of transmission to humans with a putative public health impact. Clearly, more in-depth data are needed to define the risk of ESBL-E transmission by rats.
Author contributions
SG, KS, FL, and KN conceived and designed the experiments; KN, FL, AD, LK, LV, and KB collected the data and samples; KS performed laboratory analysis; KS, SG, LW, and TS analyzed the data; KS and SG wrote the article. All authors have read and approved the final draft of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Ministry of Livestock and Animal Products for their assistance. We would like to thank Silvia Rath and the Laboratoire Guineo-Allemande for their excellent logistic support. Our gratitude goes to the local veterinary authorities for their valuable help during field work.
Footnotes
Funding. Field work was funded by the Konsortium des Deutschen Zentrum für Infektionsforschung (DZIF, FKZ 01KA1403).
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S. E., Boerlin P., Janecko N., Lumsden J. S., Barker I. K., Pearl D. L., et al. (2011). Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in southern Ontario, Canada. Appl. Environ. Microbiol. 77, 882–888. 10.1128/AEM.01111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnedahl J., Drobni M., Gauthier-Clerc M., Hernandez J., Granholm S., Kayser Y., et al. (2009). Dissemination of Escherichia coli with CTX-M Type ESBL between humans and yellow-legged gulls in the South of France. PLoS ONE 4:e5958. 10.1371/journal.pone.0005958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos A. C., Albiero J., Ecker A. B., Kuroda C. M., Meirelles L. E. F., Polato A., et al. (2016). Outbreak of Klebsiella pneumoniae carbapenemase-producing K pneumoniae:a systematic review. Am. J. Infect. Control 44, 1374–1380. 10.1016/j.ajic.2016.03.022 [DOI] [PubMed] [Google Scholar]
- Canton R., Akova M., Carmeli Y., Giske C. G., Glupczynski Y., Gniadkowski M., et al. (2012). Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clinic. Microbiol. Infect. 18, 413–431. 10.1111/j.1469-0691.2012.03821.x [DOI] [PubMed] [Google Scholar]
- CLSI (2008). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard - 3rd Edn. Wayne, IL: CLSI. [Google Scholar]
- de Been M., Lanza V. F., de Toro M., Scharringa J., Dohmen W., Du Y., et al. (2014). Dissemination of Cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 10:e1004776. 10.1371/journal.pgen.1004776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C., Bethe A., Semmler T., Guenther S., Wieler L. H. (2012). Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18, 646–655. 10.1111/j.1469-0691.2012.03850.x [DOI] [PubMed] [Google Scholar]
- Gakuya F. M., Kyule M. N., Gathura P. B., Kariuki S. (2001). Antimicrobial susceptibility and plasmids from Escherichia coli isolated from rats. East Afr. Med. J. 78, 518–522. 10.4314/eamj.v78i10.8960 [DOI] [PubMed] [Google Scholar]
- Grad Y. H., Lipsitch M., Feldgarden M., Arachchi H. M., Cerqueira G. C., FitzGerald M., et al. (2012). Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011 (vol 109, pg 3065, 2012). Proc. Natl. Acad. Sci. U.S.A. 109, 5547–5547. 10.1073/pnas.1121491109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther S., Grobbel M., Heidemanns K., Schlegel M., Ulrich R. G., Ewers C., et al. (2010). First insights into antimicrobial resistance among faecal Escherichia coli isolates from small wild mammals in rural areas. Sci. Tot. Environ. 408, 3519–3522. 10.1016/j.scitotenv.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Guenther S., Semmler T., Stubbe A., Stubbe M., Wieler L. H., Schaufler K. (2017). Chromosomally encoded ESBL genes in Escherichia coli of ST38 from Mongolian wild birds. J. Antimicrob. Chemother. 72, 1310–1313. 10.1093/jac/dkx006 [DOI] [PubMed] [Google Scholar]
- Kanj S. S., Kanafani Z. A. (2011). Current concepts in antimicrobial therapy against resistant gram-negative organisms: extended-spectrum beta-lactamase producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae, and multidrug-resistant Pseudomonas aeruginosa. Mayo Clin. Proc. 86, 250–259. 10.4065/mcp.2010.0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. O., Song S. A., Yoon E. J., Shin J. H., Lee H., Jeong S. H., et al. (2017). Outbreak of KPC-2-producing Enterobacteriaceae caused by clonal dissemination of Klebsiella pneumoniae ST307 carrying an IncX3-type plasmid harboring a truncated Tn4401a. Diagn. Microbiol. Infect. Dis. 87, 343–348. 10.1016/j.diagmicrobio.2016.12.012 [DOI] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park K. S., Kim Y. B., Jeong B. C., Lee S. H. (2016). Global dissemination of Carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Literak I., Dolejska M., Cizek A., Djigo C. A. T., Konecny A., Koubek P. (2009). Reservoirs of antibiotic-resistant Enterobacteriaceae among animals sympatric to humans in senegal: extended-spectrum beta-lactamases in bacteria in a black rat (Rattus rattus). Afr. J. Microbiol. Res. 3, 751–754. [Google Scholar]
- Literak I., Dolejska M., Janoszowska D., Hrusakova J., Meissner W., Rzyska H., et al. (2010). Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 76, 8126–8134. 10.1128/AEM.01446-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac P. J., Bonomo R. A., Logan L. K. (2015). Extended-spectrum beta-lactamase-producing Enterobacteriaceae in children: old foe, emerging threat. Clin. Infect. Dis. 60, 1389–1397. 10.1093/cid/civ020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Naas T., Poirel L. (2011). Global Spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. K., Jain M. (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619. 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufler K., Semmler T., Wieler L. H., Wohrmann M., Baddam R., Ahmed N., et al. (2016). Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410–another successful pandemic clone? FEMS Microbiol Ecol 92:fiv155. 10.1093/femsec/fiv155 [DOI] [PubMed] [Google Scholar]
- Skurnik D., Ruimy R., Andremont A., Amorin C., Rouquet P., Picard B., et al. (2006). Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57, 1215–1219. 10.1093/jac/dkl122 [DOI] [PubMed] [Google Scholar]
- Turton J. F., Doumith M., Hopkins K. L., Perry C., Meunier D., Woodford N. (2016). Clonal expansion of Escherichia coli ST38 carrying chromosomally-integrated OXA-48 Carbapenemase gene. J. Med. Microbiol. 65, 538–546. 10.1099/jmm.0.000248 [DOI] [PubMed] [Google Scholar]
- Villa L., Feudi C., Fortini D., Brisse S., Passet V., Bonura C., et al. (2017). Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genomics 3:e000110. 10.1099/mgen.0.000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E. (2008). Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhou K., Zheng B., Zhao L., Shen P., Ji J., et al. (2016). High prevalence of ESBL-producing Klebsiella pneumoniae causing community-onset infections in China. Front. Microbiol. 7:1830. 10.3389/fmicb.2016.01830 [DOI] [PMC free article] [PubMed] [Google Scholar]