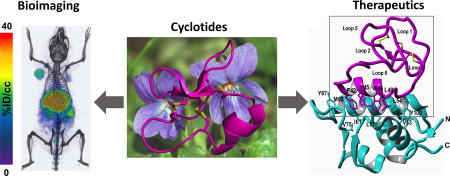
Abstract
Cyclotides are fascinating microproteins (≈30–40 residues long) with a unique head-to-tail cyclized backbone, stabilized by three disulfide bonds forming a cystine knot. This unique topology makes them exceptionally stable to chemical, thermal and biological degradation compared to other peptides of similar size. Cyclotides have been also found to be highly tolerant to sequence variability, aside from the conserved residues forming the cystine knot, able to cross cellular membranes and modulate intracellular protein-protein interactions both in vitro and in vivo. These properties make them ideal scaffolds for many biotechnological applications. This article provides and overview of the properties of cyclotides and their applications as molecular imaging agents and peptide-based therapeutics.
Keywords: cyclotide, cystine knot, circular protein, drug design, bioimaging agents, MCoTI-I, kalata B1
TOC
The success of protein-based therapeutics is revolutionizing drug development. Peptide and protein-based therapeutics can target with high selectivity and specificity defective protein-protein interactions involved in human disease. Despite their success, however, there are still numerous stability and delivery issues associated with their use as therapeutic agents. For example, monoclonal antibodies -one of the most successful protein-based therapeutics with several blockbuster drugs on the market and many more in clinical development- can only target extracellular molecular targets due to their inability to cross biological membranes. They are also extremely expensive to produce and are not bioavailable due to their susceptibility to proteolytic degradation. These issues have led to the exploration of alternative protein scaffolds as a source for novel types of protein-based therapeutics.1,2
Special attention has been recently given to the use of highly constrained poly-peptides as extremely stable and versatile scaffolds for production of high affinity ligands for specific protein capture and/or development of therapeutics.3,4 Cyclotides represent a new emerging family of large plant-derived backbone-cyclized polypeptides (≈30–40 amino acids long) that share a 3 disulfide-stabilized core characterized by an unusual knotted structure (Fig. 1).5 Cyclotides have several characteristics that make them ideal as drug development and bioimaging tools (see recent reviews6,7). For example, they are remarkably stable to chemical, thermal, and proteolytic degradation due to the circular topology and cystine knot.8 Their relative small size makes them also readily accessible by chemical synthesis and can also be encoded within standard cloning vectors, and expressed in bacteria or animal cells (see recent reviews on the production of cyclotides9). Cyclotides from the trypsin inhibitor subfamily, can cross the cellular membranes of mammalian cells through different endocytic pathways10,11 being able to target intracellular protein interactions both in vitro and in vivo.12 The molecular/structural determinants of membrane crossing for the MCoTI-based cyclotides has not been completely elucidated yet, although it is likely related to their high positive charge at physiological pH. Finally, cyclotides have been shown to be orally active.13 For example, the first cyclotide to be discovered, kalata B1, is an orally effective uterotonic,8 and several other kalata B1-based cyclotides have also shown to be orally active.13,14 Altogether, these characteristics make the cyclotide scaffold an ideal substrate for molecular evolution strategies to enable generation and selection of novel peptide-based diagnostics, therapeutics and research tools. This article provides a brief overview of their properties and their use as molecular frameworks for the design of peptide-based diagnostic and therapeutic tools.
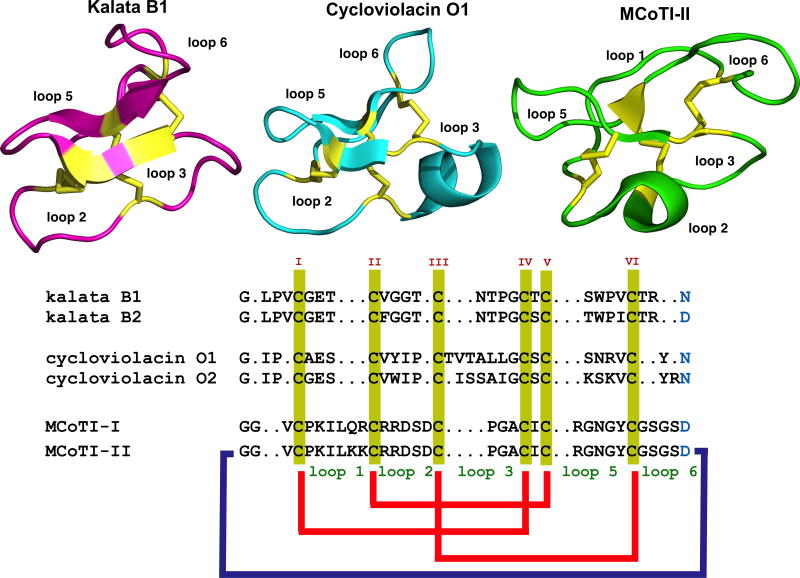
Figure 1.
Sequence alignment and structures of different cyclotides belonging to the Möbius (kalata B1, pdb: 1NB1), bracelet (cycloviolacin O1, pdb: 1NBJ) and trypsin inhibitor (MCoTI-II, pdb: 1IB9) subfamilies. Conserved Cys and Asp/Asn (required for cyclization) residues are marked in yellow and light blue, respectively. Disulfide connectivities and backbone-cyclization are shown in red and a dark blue line, respectively. The six Cys residues are labeled with roman numerals whereas loops connecting the different Cys residues are designated with arabic numerals. Molecular graphics were created using PyMol.
Discovery and distribution
The first cyclotide cyclotide was discovered in the late 1960s by Gran when studying an indigenous medicine in central Africa that was used to facilitate childbirth.15 This traditional remedy was based on a tea obtained from the plant Oldelandia affinis from the Rubiaceae family.16 Analysis of the tea revealed that the main component with uterotonic activity was a peptide around 30 residues long that was named kalata B1. Due to the limitations of the protein chemistry techniques available in the early 1970s, it was not until 1995 when the Cys-knot backbone-cyclized nature of kalata B1 was first elucidated (Fig. 1).8 Around the same time, other macrocyclic peptides of similar size, sequence and structure that kalata B1, were also isolated from plants of the Rubiaceae and Violaceae families (Fig. 1).17–19 These findings lead to the definition of the cyclotide family of microproteins in 1999 based on the sequence and structure homology of these new discovered type of backbone-cyclized Cys-knotted polypeptides.20 Cyclotides have been also found in plants from the Cucurbitaceae, Fabaceae, Solanaceae and Apocynaceae families in addition to Rubiaceae and Violaceae, with the latter two families providing the majority of the cyclotides known thus far (Fig. 2).21
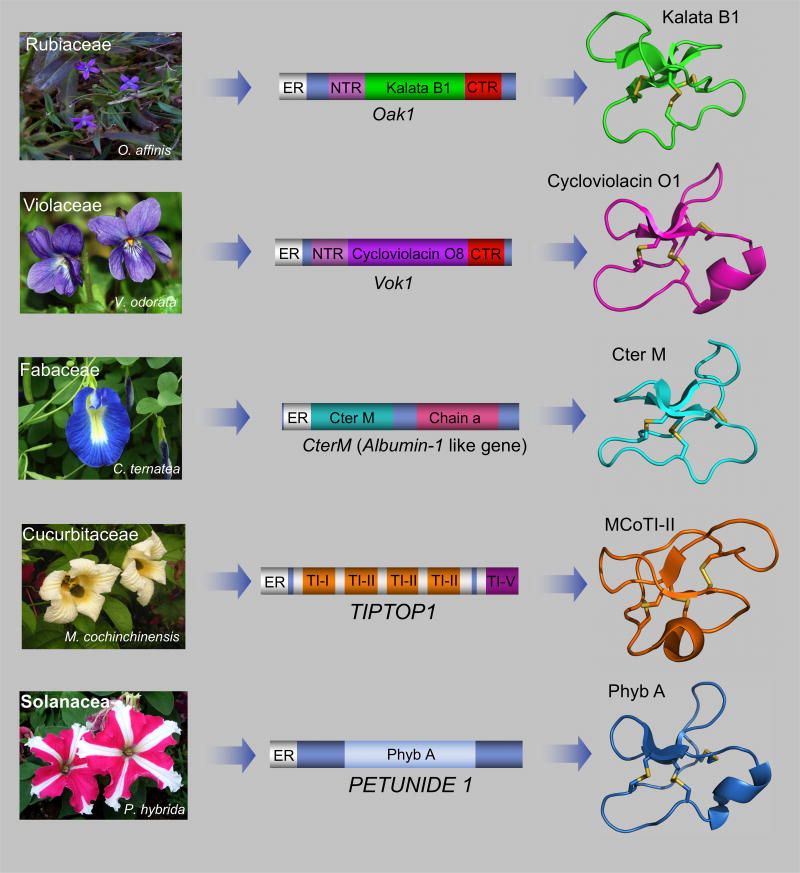
Figure 2.
Genetic origin of cyclotides in plants. Some plants from the Rubiaceae and Violaceae families have dedicated genes to the production of cyclotides. These genes encode protein precursors containing an ER signal peptide, an N-terminal pro-region, the N-terminal repeat (NTR), the mature cyclotide domain and a C-terminal flanking region (CTR).61 Cyclotides recently isolated from C. ternatea (Fabaceae family), are produced from precursor proteins containing an ER signal peptide immediately followed by the cyclotide domain, which is flanked at the C-terminus by a peptide linker and the albumin a-chain. The cyclotide domain, in this case, replaces the albumin-1 b-chain.5 The trypsin inhibitor subfamily of cyclotides are produced from TIPTOP proteins, which contain a tandem series of cyclic trypsin inhibitors terminating with an acyclic trypsin inhibitor.38 The protein precursors for cyclotides from the Solanaceae family are encoded in genes similar to those found in the Rubiaceae and Violaceae plants with dedicated precursor proteins that have an ER signal, a pro-region, the linear peptide precursor, and end with a hydrophobic tail.63 Molecular graphics were created using PyMol.
Even though most of the cyclotides have been isolated from the plants of the coffee and violet families, the distribution of cyclotides within these two families is quite different. For example, only around 5% of plants from the coffee family that have been analyzed so far have shown to contain cyclotides; in contrast all of the plants studied from the violet family have been found to contain cyclotides.21 Cyclotides are usually distributed across all tissues, including flowers, leaves, stems, roots, and in some cases even seeds, with a single plant typically producing multiple cyclotides (≈10–160).22–24
Initially, the discovery of novel cyclotides was almost exclusively based on their isolation from the plant followed by chemical characterization of the corresponding peptides.25 The use of more modern chemical approaches has reduced the amount of plant material required for a full characterization of the cyclotides contained in the sample when compared to the early methods. For example, with the use of MALDI-TOF/TOF and LC-MS/MS techniques is not uncommon to discover novel cyclotides from plant samples as small as 1 cm2 of leaf tissue.26–28 Since naturally-occurring cyclotides are ribosomally produced from genetically encoded protein precursors the use of in silico screening approaches can be used for the identification of cyclotides in plants.29In a recent study, the use of in silico transcriptomic and proteomic screening identified 164 cyclotides in Viola tricolor, which led to the authors of this study to estimate the total number of cyclotides in the Violaceae family alone to be around 150,000.30 The CyBase database was recently created to allow easy access to the large number of cyclotide sequences that have been found thus far.31 This database can be accessed through a publicly available website (http://CyBase.org.au) and contains above 300 known cyclotides as well as useful proteomic tools for their analysis.
Structure
All naturally-occurring cyclotides are 27–37 amino acid long, contain six Cys residues and are backbone-cyclized (Fig. 1). The Cys residues are forming three disulfide bonds that adopt Cys-knot topology, where the disulfides CysI-CysIV and CysII-CysV form a ladder arrangement with the disulfide CysIII-CysVI running through them (Fig. 3A). The highly interlocked structure of this cyclic cysteine knot (CCK) motif makes the cyclotide backbone very compact and rigid.32 Accordingly, cyclotides are extremely stable compounds that are highly resistant to thermal and chemical denaturation as well as biological degradation.33,34 The fact that the first cyclotide, kalata B1, which was able to remain structurally intact and biologically active after being extracted by boiling water to make a medicinal tea, is proof of the extraordinary stability of these type of cyclic polypeptides.
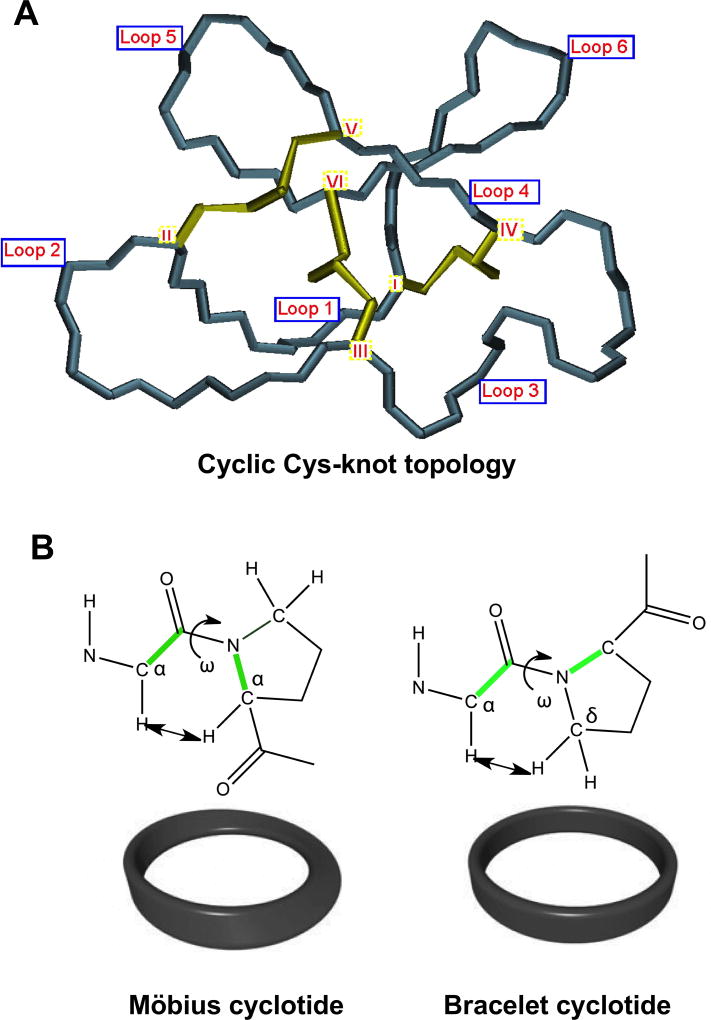
Figure 3.
Structural characteristics of the cyclic cysteine know (CCK) topology found in all cyclotides. A. Detailed three-dimensional structure of the cyclic cystine knot (CCK) topology and the connecting loops found in cyclotides. The six Cys residues are labeled with roman numerals whereas loops connecting the different Cys residues are designated with arabic numerals. B. Möbius cyclotides contain a cis-Pro residue in loop 5 that induces a local 180° backbone twist, whereas bracelet cyclotides do not possess this structural feature.
Cyclotides have been classified into three subfamilies known as the Möbius, bracelet, and trypsin inhibitor cyclotide subfamilies.35 Although all the subfamilies share the same cyclic Cys-knot topology, the composition of the loops is different in each subfamily. For example, Möbius cyclotides, such as kalata B1, contain a cis-Pro bond at loop 5 resulting in a slight twist of the backbone, while bracelet cyclotides do not have it (Fig. 3B). Bracelet cyclotides are by far the more abundant making up approximately two-thirds of the sequenced cyclotides known thus far.36 These cyclotides are slightly larger and more structurally diverse than Möbius cyclotides. Despite its abundance in nature, bracelet cyclotides are more difficult to fold in vitro than either Möbius or trypsin inhibitor cyclotides, which makes them more challenging to obtain by synthetic methods using standard peptide synthesis protocols. Accordingly, bracelet cyclotides are less used in the development of the biotechnological applications that will be described later in this review.
The trypsin inhibitor subfamily of cyclotides consists of only a small number cyclotides isolated from the seeds of the plant Momocordica cochinchinesis (Cucurbitaceae family).37,38 They are potent trypsin inhibitors (Ki ≈ 20 – 30 pM) and do not share significant sequence homology with the other cyclotides beyond the presence of the three-cystine bridges that adopt a similar backbone-cyclic cystine-knot topology (Fig. 1). These cyclotides are more sequence-related to linear cystine-knot squash trypsin inhibitors and for this reason are sometimes referred as cyclic knottins.39
More recently, a novel type of cyclotides possessing novel sequence features, including a lysine-rich nature, have been isolated from two species of Australasian plants from the Violaceae family.40 However, there is still limited information on how easy these novel cyclotides can be prepared by chemical means, which should dictate its potential to be used as useful molecular scaffolds for biotechnological applications.
Biological activities of naturally-occurring cyclotides
The biological function of the naturally-occurring cyclotides of the Möbius and bracelet sub-families in plants seems to be primarily as host-defense agents as deduced from their activity against insects.5,41–43 For example there are numerous reports in the literature showing that cyclotides can efficiently inhibit the growth and development of nematodes and trematodes,44–46 and of mollusks.47
The biological activity of these type of cyclotides involves the interaction with cellular membranes disrupting their normal function. For example, ingestion of cyclotides by the larvae of Lepidopteran species disrupts their midgut membranes.48 The mechanism of how these type of cyclotides work has been widely studied and for example, for kalata B1, is well established that the first step on the interaction with the membranes involves specific binding of the cyclotide to the phosphatidylethanolamine phospholipids present in the cellular membrane. This step compromises the integrity of the membrane causing the formation of pores and/or leakage of cell contents (see a recent review in this topic49).
In addition to their insecticidal and nematocidal activities, cyclotides have been also shown antimicrobial and anti-tumor activities. The cyclotides from the Möbius and bracelet sub-families have shown to possess amphipathic properties due to the presence hydrophobic and hydrophilic patches located in different regions of their surface, resembling to some extent the nature of classical antimicrobial peptides. The cyclotide balata B1 shows antimicrobial activity against Gram-negative and Gram-positive bacteria.50 Similar antimicrobial activities have been also found in cyclotide from Hedyota biflora (Rubiaceae family)51,52 and Clitoria ternatea (Fabaceae family).53 The most active cyclotide tested so far is the bracelet cyclotide cycloviolacin O2, which was shown to have antimicrobial activity against Staphylococcus aureus in a mouse infection model.54 Despite these encouraging results, it should be noted that the antimicrobial activity of most cyclotides when tested in vitro seems to depend on the buffer composition occurring only under non-physiological conditions when low ionic strength buffers are used. This seriously limits its potential on the design of effective antimicrobial therapeutics. Several cyclotides have been described to possess selective cytotoxicity against several cancer cell lines, including primary cancer cell lines, when compared to normal cells.55–57 For example, a recent publication described the cytotoxic activity of cyclotide vingo 5 from Viola ignobilis in HeLa cells.58 In addition, three new cyclotides isolated from Hedyotis diffusa, a Chinese medicinal plant from the Rubiaceae family, have been shown to induce apoptosis, and inhibit proliferation and migration of several prostate cancer cell lines.59 The most active cyclotide of the three, cyclotide DC3, inhibited tumor growth in a mouse xenograft model.
Although, these results are promising, the therapeutic index (i.e. the ratio between the dose required for therapeutic effects versus toxic effects on normal cells) of cytotoxic cyclotides is not optimal yet indicating that will require optimization before they can be developed into effective anti-cancer agents.
It is worth noting, that recent studies on identification of the molecular targets of labor-accelerating kalata B7 and analogs were shown to be the G protein-coupled oxytocin and vasopressin V1a receptors.60
Biosynthesis of cyclotides
Naturally-occurring cyclotides are enzymatically processed from ribosomally produced precursor proteins. In many cases, cyclotides possess dedicated genes that encode multiple copies of the same cyclotide, and in others, mixtures of different cyclotide sequences.61
The first genes encoding cyclotide precursor proteins were found in the plant O. affinis (Rubiaceae family) for the kalata cyclotides (Fig. 2).41 The gene encodes a protein containing an endoplasmic reticulum (ER)-targeting sequence, a pro-region, a highly conserved N-terminal repeat (NTR) region, a mature cyclotide domain, and a hydrophobic C-terminal tail (Fig. 4).62Similar genes have also discovered in other plants from the Violacea and Rubiaceae,64,6 and more recently also in plants from the Solanaceae, Fabaceae and Cucurbitaceae families (Fig. 2).5,38,53,63 These new genes provide novel protein precursor architectures, indicating a high diversity in the way cyclotides are produced in nature.
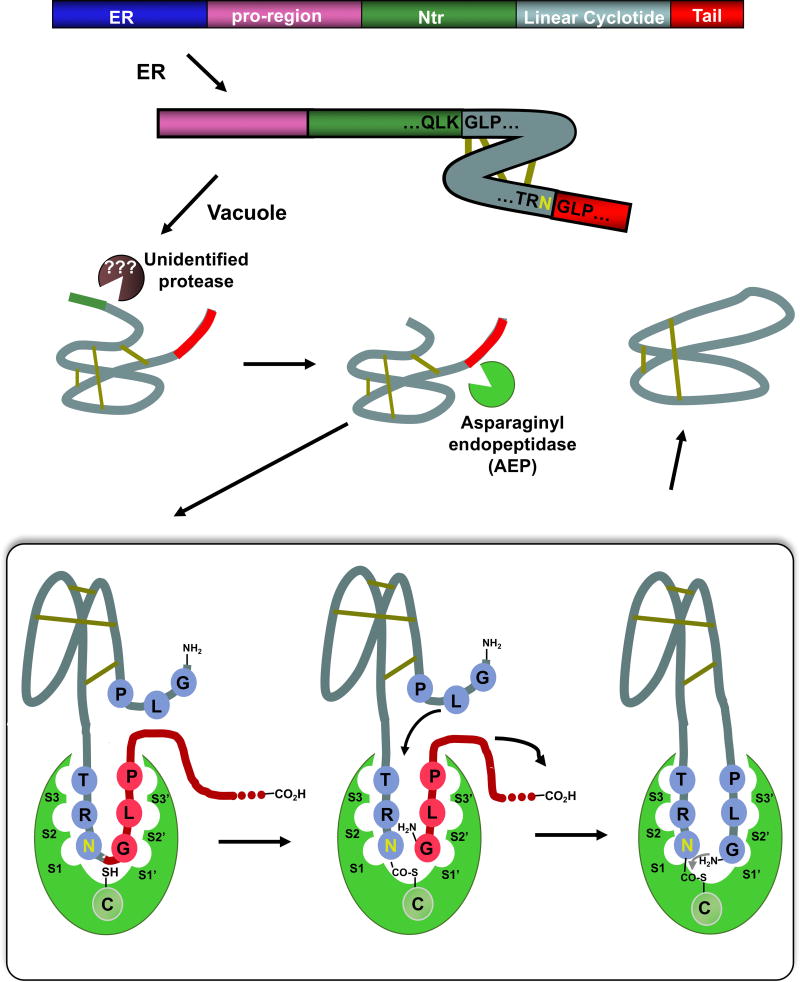
Figure 4.
Scheme summarizing the proposed mechanism for the biosynthesis of cyclotide kalata B1. It is now well accepted that the cyclization step is mediated by an asparaginyl endopeptidase (AEP), a common Cys protease found in plants. The cyclization takes place at the same time as the cleavage of the C-terminal pro-peptide from the cyclotide precursor protein through a transpeptidation reaction. The transpeptidation reaction involves an acyl-transfer step from the acyl-AEP intermediate to the N-terminal residue of the cyclotide domain.61 The kalata B1 protein precursor contains an ER signal peptide, an N-terminal pro-region, the N-terminal repeat (NTR), the mature cyclotide domain and a C-terminal flanking region (tail, also known as CTR).
The post-translational modifications involved in the biosynthesis of cyclotides is not fully understood yet (Fig. 4). Recent studies, however, have indicated that asparaginyl endopeptidase (AEP)-like ligases are a key element in the C-terminal cleavage and cyclization of cyclotides. The transpeptidation reaction involves an acyl-transfer step from the acyl-AEP intermediate to the N-terminal residue of the cyclotide domain.64,65 AEPs are Cys proteases that are very abundant in plants, where they specifically cleave the peptide bond at the C-terminus of Asn and, less efficiently, Asp residues. All the cyclotide precursor proteins identified so far contain a well-conserved Asn/Asp residue at the C-terminus of the cyclotide domain in loop 6. Two AEP-like ligases have been identified so far, rOaAEP1b and butelase-1, which were cloned from the cyclotide-producing plants O. affinis and C. ternatea, respectively.66,67 These ligases have shown efficient capabilities in cyclizing various peptides, including linear cyclotide precursors containing the C-terminal recognitions sequence and even D-peptides.66,68–71 Despite the tremendous progress done on understanding at the mechanistic level of how cyclotides are produced in plants, there is still is not too much known about the N-terminal cleavage process and the protease involved at that step.
Chemical synthesis of cyclotides
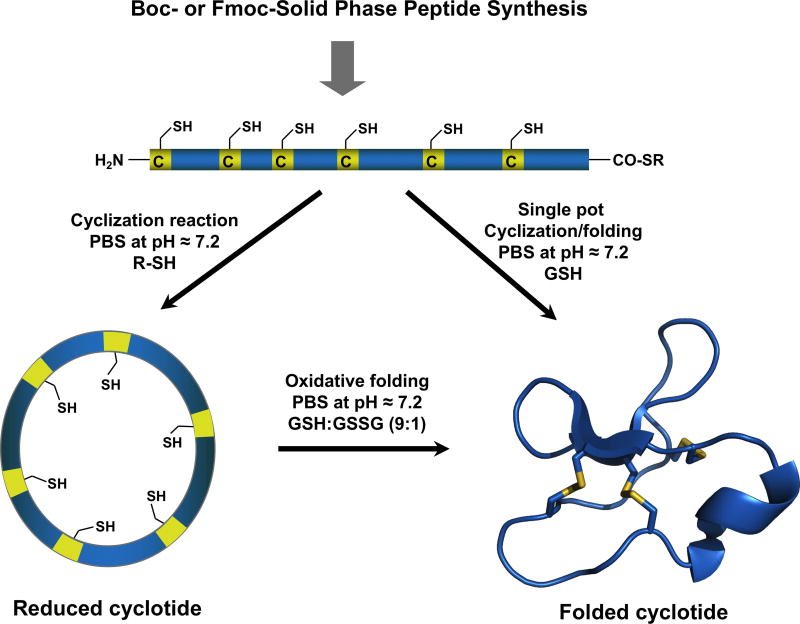
Due to the relatively small size of cyclotides (≈30–40 amino acids long), the corresponding linear precursors can be readily synthesized by chemical methods using solid-phase peptide synthesis (SPPS) (for a recent review in this topic see9). The backbone cyclization step of the linear precursor can be easily accomplished in aqueous buffers under physiological conditions by using an intramolecular version of native chemical ligation (NCL).72 The only requirement is for the linear precursor to contain an N-terminal cysteine and an α-thioester group at the C-terminus (Fig. 5).9 Peptide α-thioesters can be easily generated using standard solid-phase peptide synthesis approaches using either Boc- or Fmoc-based chemistry.9 The linear precursor once cleaved from the resin can be cyclized and folded sequentially. The cyclization and folding steps can be also carried out in a ‘single pot’ reaction by using glutathione (GSH) as a thiol additive.73 This approach has successfully been used to chemically generate many native and engineered cyclotides.6
Figure 5.
Chemical synthesis of cyclotides by making use of an intramolecular native chemical ligation (NCL). This method requires the chemical synthesis of a linear precursor bearing an N-terminal Cys residue and an α-thioester moiety at the C-terminus. The linear precursor can be cyclized first under reductive conditions and then folded using a redox buffer containing reduced and oxidized glutathione (GSH).9 Alternatively, the cyclization and folding can be efficiently accomplished also in a ‘single pot’ reaction when the cyclization is carried out in the presence of reduced GSH as the thiol cofactor.9
Chemoenzymatic cyclization of the corresponding synthetic linear precursors using AEP-like ligases has been also described for the synthesis of cyclotides.66,67 Intriguingly, these enzymes do not require the cyclotide linear precursor to be natively folded for the cyclization to proceed efficiently.66
The serine protease trypsin has also been used to produce several cyclotides based on the naturally occurring trypsin inhibitor cyclotide MCoTI-II.74 In this case, folded linear cyclotide precursors bearing the P1 and P1′ residues at the C- and N-termini respectively were used as a viable substrate for trypsin-mediated cyclization. The use of trypsin-mediated cyclization provides a very efficient route for obtaining cyclotides with trypsin inhibitory properties with yields close to 92% for cyclotide MCoTI-II.74 It should be noted, however, that the introduction of mutations that affect the binding to the proteolytic enzyme may affect the cyclization yield.9 The transpeptidase like sortase A (SrtA) enzyme has been also used for the production of cyclotides from synthetic linear precursors.75 Due to the SrtA sequence requirements to work efficiently, the heptapeptide motif (LPVTGGG) is left at the ligation site, which should be taken into consideration when producing biologically active cyclotides.
Recombinant expression of cyclotides
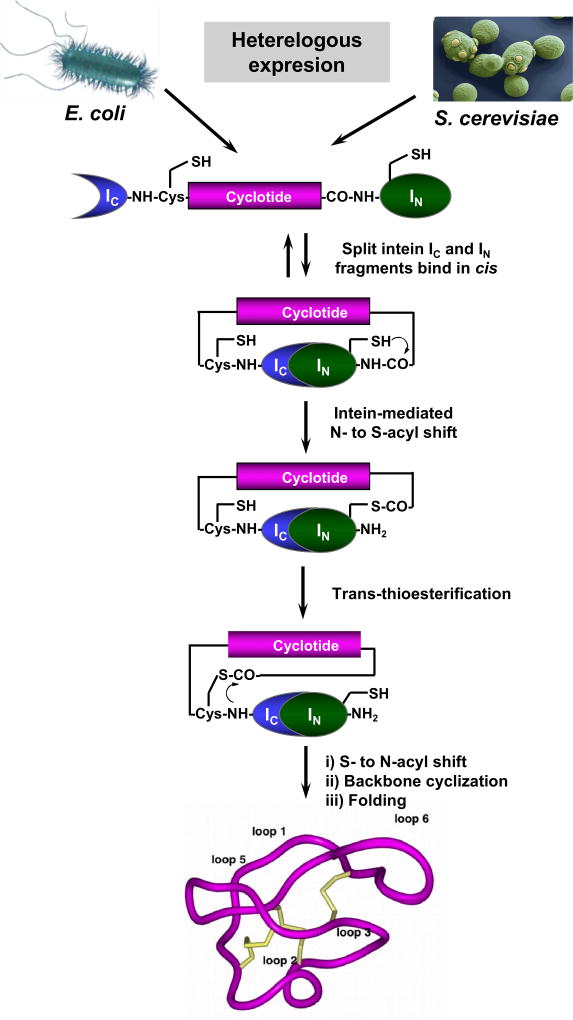
The discovery of intein-mediated protein splicing both in cis and trans has made possible the generation of backbone-cyclized polypeptides using standard expression systems (for a recent review in this topic see9) (Fig. 6). Our group pioneered the use of intein-mediated backbone cyclization for the biosynthesis of fully folded cyclotides inside bacterial cells by making use of an intramolecular version of intein-mediated ligation (also called Expressed Protein Ligation (EPL)).9
Figure 6.
Heterologous expression of cyclotides by protein trans-splicing (PTS).76–78 To facilitate the backbone cyclization the linear cyclotide precursor is fused in-frame at the C- and N-termini directly to the IN and IC polypeptides of the Npu DnaE split intein. This approach has been used for the generation of MCoTI-cyclotides, where the native Cys residue located at the beginning of loop 6 was used to facilitate backbone cyclization. During the intramolecular processing of this construct the N- and C-termini of the linear cyclotide precursor are linked together through a native peptide bond through a transpeptidation reaction mediated by the self-processing domains of the split intein. This approach has been successfully used to produce bioactive cyclotides in eukaryotic and prokaryotic expression systems.76–78
More recently, our group made use of intein-mediated protein trans-splicing (PTS) for the efficient production of naturally-occurring and engineered cyclotides in prokaryotic and eukaryotic expression systems (Fig. 6).76,77 This approach is quite efficient, providing in-cell production of folded cyclotides that can reach intracellular concentrations in the range of 20–40 μM. In E. coli, this level of expression equals to ≈10 mg of folded cyclotide per 100 g of wet cells for cyclotide MCoTI-I.78
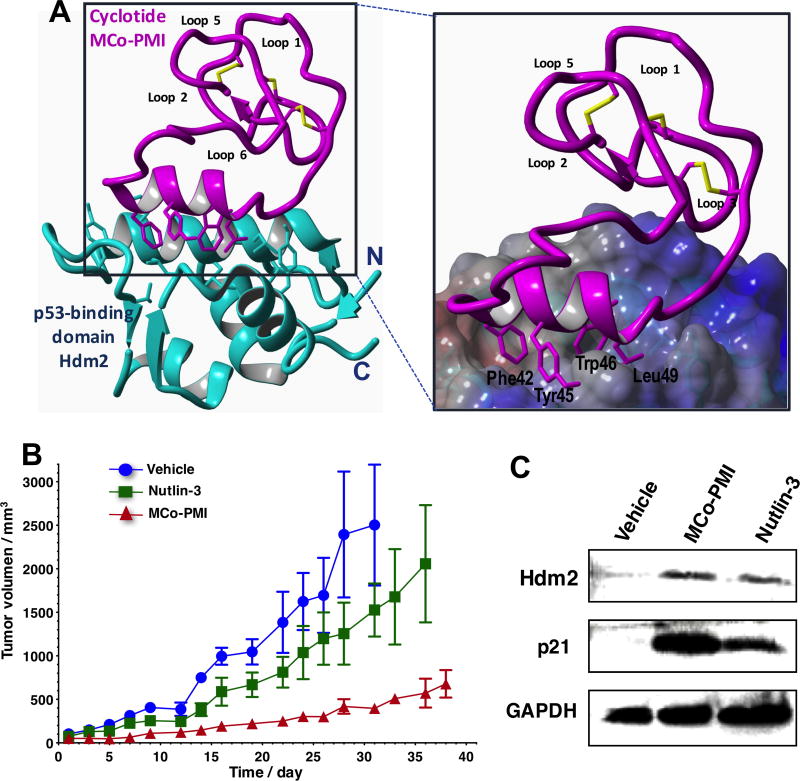
Efficient in-cell production of cyclotide allows the generation of large genetically-encoded libraries of cyclotides inside live cells that can be rapidly screened for the selection of novel sequences able to modulate or inhibit the biological activities of selected biomolecular targets. In addition, it also facilitates the production of cyclotides labeled with NMR active isotopes such as 15N and/or 13C in a very inexpensive fashion. The production of 15N and/or 13C-labeled cyclotides makes possible the use of heteronuclear NMR spectroscopy to study structure-activity relationships of any biologically active cyclotides and their molecular targets. For example, this was recently demonstrated in the structural studies carried out on a cyclotide engineered to bind the p53 binding domain of the E3-ligases Hdm2 and HdmX (Fig. 7A).12
Figure 7.
Structure and in vivo activity of the first cyclotide designed to antagonize an intracellular protein-protein interaction in vivo. A. Solution structure of the engineered cyclotide MCo-PMI (magenta) and its intracellular molecular target, the p53 binding domain of oncogene Hdm2 (blue). The cyclotide binds with low nM affinity to both the p53-binding domains of Hdm2 and HdmX. B. Cyclotide MCo-PMI activates the p53 tumor suppressor pathway and blocks tumor growth in a human colorectal carcinoma xenograft mouse model. HCT116 p53+/+ xenografts mice were treated with vehicle (5% dextrose in water), nutlin 3 (10 mg/kg) or cyclotide (40 mg/kg, 7.6 mmol/kg) by intravenous injection daily for up to 38 days. Tumor volume was monitored by caliper measurement. C. Tumors samples were also subjected to SDS-PAGE and analyzed by western blotting for p53, Hdm2 and p21, indicating activation of p53 on tumor tissue.
Engineered cyclotides with novel biological activities
The unique properties associated with the cyclotide scaffold make this molecular framework extremely valuable in the development of novel peptide-based therapeutics (see Table 1).34,79 As mentioned earlier, the CCK topology found in cyclotides provides them with a highly rigid and compact structure that is extremely resistant to physical, chemical and biological degradation. In addition, the cyclotide scaffold is highly tolerant to mutations making it an ideal molecular framework for molecular evolution and grafting for production of novel cyclotides with new biological activities. Cyclotides from the trypsin inhibitor subfamily are also not toxic to mammalian cells in vitro up to concentrations of 100 µM12 or in vivo with daily doses of cyclotides of up to 40 mg/kg,12 and have been shown to be able to cross cellular membranes10,11 for targeting intracellular cytosolic protein-protein interactions.12 The trypsin inhibitory properties of these cyclotides can be easily removed by mutating the residue Lys4 located in the loop 1 to a non-positively charged residue. The pharmacologic potential of engineered cyclotides grafted with biological active peptide sequences was first demonstrated in two early studies aimed to develop novel anti-cancer80,81 and anti-viral peptide-based therapeutics.74 Molecules with anti-angiogenic activity have potential applications in cancer treatment due to the fact the tumor growth is usually associated with unregulated angiogenesis. In this early case, an Arg-rich peptide antagonist for the interaction of vascular endothelial growth factor A (VEGF-A) and its receptor was grafted into several loops of cyclotide kalata B1 producing cyclotides with anti-VEGF activity.80 The most active cyclotide was able to block VEGF-A receptor binding with an IC50 of ≈ 12 μM, which although is the first example of a successful functional redesign of a cyclotide, its biological activity would still need to be improved by several orders of magnitude for a potential pharmacological application in vivo. A similar approach was also used recently to target the bradykinin and melanocortin 4 receptors for pain and obesity management, respectively.13,82 The kalata B1-based bradykinin antagonist was shown to be orally active13 highlighting the potential of the cyclotide scaffold for the development of orally-bioavailable peptide-based therapeutics. The cyclotide MCoTI-I from the trypsin inhibitor subfamily has been also employed for the design of a potent CXCR4 antagonist.83 This cytokine receptor is associated with multiple types of cancers where its overexpression/activation promotes metastasis, angiogenesis, and tumor growth and/or survival.84
Table 1.
Summary of work published in engineered cyclotides with novel biological activities leading to therapeutic and bioimaging applications.
| Cyclotide | Biological activity | Loop Modified |
Application | Ref. |
|---|---|---|---|---|
|
Möbius subfamily
| ||||
| Kalata B1 | VEGF-A antagonist | 2, 3, 5 & 6 | Anti-angiogenic, potential anti-cancer activity | 80 |
| Kalata B1 | Dengue NS2B-NS3 Protease inhibitor | 2 & 5 | Anti-viral for Dengue virus infections | 94 |
| Kalata B1 | Bradikynin B1 receptor antagonist | 6 | Chronic and inflammatory pain | 13 |
| Kalata B1 | Melanocortin 4 receptor Agonist | 6 | Obesity | 82 |
| Kalata B1 | Neuropilin-1/2 antagonist | 5 & 6 | Inhibition of endothelial cell migration and angiogenesis | 95 |
| Kalata B1 | Immunomodulator | 5 & 6 | Protecting against multiple sclerosis | 89 |
| Kalata B1 | Immunomodulator | 4 | Protecting against multiple Sclerosis | 14 |
|
| ||||
| Trypsin inhibitor subfamily | ||||
|
| ||||
| MCoTI-I | CXCR4 antagonist | 6 | Anti-metastatic and anti-HIV PET-CT imaging | 73,92,96 |
| MCoTI-I | p53-Hdm2/HdmX Antagonist | 6 | Anti-tumor by activation of p53 pathway | 12 |
| MCoTI-II | FMDV 3C protease Inhibitor | 1 | Antiviral for foot-and-mouth disease | 74 |
| MCoTI-II | β-Tryptase inhibitor | 3, 5 & 6 | Inflammation disorders | 87 |
| MCoTI-II | β-Tryptase inhibitor Human elastase inhibitor | 1 | Inflammation disorders | 81 |
| MCoTI-II | CTLA-4 antagonist | 1,3 & 6 | Immunotherapy for cancer | 97 |
| MCoTI-II | Tryptase inhibitor | 1 | Anticancer | 98 |
| MCoTI-II | VEGF receptor agonist | 6 | Wound healing and cardiovascular damage | 99 |
| MCoTI-I | α-Synuclein-induced cytotoxicity inhibitor | 6 | Parkinson’s disease Validate phenotypic screening of genetically-encoded cyclotide libraries | 77 |
| MCoTI-II | BCR-Abl kinase Inhibitor | 1 & 6 | Chronic myeloid leukemia Attempt to graft both a cell penetrating peptide and kinase inhibitor | 100 |
| MCoTI-I | MAS1 receptor agonist | 6 | Lung cancer and myocardial infarction | 101 |
| MCoTI-II | SET antagonist | 6 | Potential anticancer | 90 |
| MCoTI-II | FXIIa and FXa inhibitors | 1 & 6 | Antithrombotic and cardiovascular disease | 102 |
| MCoTI-II | Thrombospondin-1 (TSP-1) agonist | 6 | Microvascular endothelial cell migration inhibition Anti-angiogenesis | 103 |
| MCoTI-II | Antiangiogenic | 5 & 6 | Anti-cancer | 104 |
The trypsin inhibitor subfamily of cyclotides has been also used for the design for protease inhibitors with potential pharmacological relevance. Proteases are well-recognized drug targets as they are involved many human diseases including inflammatory diseases, cancer, cardiovascular and neurodegenerative conditions.85,86 For example, mutated versions of the cyclotide MCoTI-II were transformed into potent and selective foot-and-mouth-disease (FMDV) 3C protease, β-tryptase and human leukocyte elastase inhibitors.74,81,87 β-Tryptase and human leukocyte elastase are validated targets for inflammatory disorders.
In a recent work, a point mutated version of cyclotide kalata B1 was reported to have oral activity in a mouse model of multiple sclerosis.88 The pharmacological potential of grafted cyclotides in the context of multiple sclerosis has been also explored by grafting peptide sequences from the MOG35-55 epitope onto the cyclotide kalata B1.89
Without any question, one of the most exciting features of the cyclotide scaffold is that some cyclotides, those from the trypsin inhibitor subfamily, can penetrate cells and access the cytosolic cellular fraction. This exceptional property found in some cyclotides makes possible the cellular delivery of biologically active cyclotides to target intracellular protein-protein interactions. In a recent report, the cyclotide MCoTI-I was engineered to produce a potent inhibitor for the interaction between p53 and the proteins Hdm2/HdmX (Fig. 7). 12 The engineered cyclotide, MCo-PMI, was able to bind with low nanomolar affinity to both Hdm2 and HdmX, showed high stability in human serum, and was cytotoxic to wild-type p53 cancer cell lines by activating the p53 tumor suppressor pathway both in vitro and in vivo (Fig. 7).12 This report constitutes the first example of an engineered cyclotide being able to target an intracellular protein-protein interaction in an animal model of human colon carcinoma therefore highlighting the therapeutic potential of MCoTI-cyclotides for targeting intracellular protein-protein interactions. An identical approach but using cyclotide MCoTI-II instead was also reported to antagonize in vitro the SET protein, which is overexpressed in some human cancers.90
MCoTI-grafted cyclotides have been also used to inhibit α-synuclein-induced cytotoxicity when expressed in yeast S. cerevisiae.77 α-Synuclein is a small lipid-binding protein that is prone to misfolding and aggregation, that has been linked to Parkinson’s disease by genetic evidence and its abundance in the Parkinson’s disease-associated intracellular aggregates known as Lewy bodies, and therefore it is a validated therapeutic target for Parkinson’s disease.
Given the good in vivo biological activity of some MCoTI-cyclotides, the biodistribution and potential to cross the blood brain barrier of cyclotides MCoTI-I/II have been recently studied.91,92 These two reports indicated that MCoTI-cyclotides are distributed predominantly to the serum and kidneys, confirming that they are stable in serum and suggesting that they are eliminated from the blood through renal clearance.91,92 In addition, cyclotide MCoTI-II showed no significant uptake into the brain.91
Screening of cyclotide-based libraries
As discussed earlier, in-cell production of natively folded cyclotides makes possible the production of large libraries of genetically-encoded cyclotides, potentially containing billions of members. The generation of this tremendous molecular diversity should allow the selection of strategies that mimic the evolutionary processes found in nature to select novel cyclotide sequences able to target specific molecular targets. As a proof of principle, a genetically-encoded library based on cyclotide MCoTI-I was generated to mutate every single amino acid in loops 1, 2, 3, 4 and 5 in order to explore the effects on folding and trypsin binding activity of the resulting mutants.93 The results obtained in this early work showed that most of the mutants were able to fold efficiently therefore emphasizing the high plasticity and sequence tolerance of MCoTI-based cyclotides.93
Cyclotide-based libraries have been also used for phenotypic screening in eukaryotic cells.77 In this work, an engineered cyclotide (MCoCP4) that was designed to reduce toxicity of human α-synuclein in live yeast cells was selected by phenotypic screening from cells transformed with a mixture of plasmids encoding MCoCP4 and inactive cyclotide MCoTI-I in a ratio of 1 to 50,000. These results show the potential for performing phenotypic screening using genetically encoded cyclotide-based libraries in eukaryotic cells for the rapid selection of novel bioactive cyclotides. Moreover, heterologous production of cyclotides in eukaryotic expression systems should also allow the introduction different post-translational modifications not available in bacterial expression systems, like phosphorylation and/or glycosylation.
In addition to the use of biological expression systems for screening purposes, the development of efficient methods for the chemical synthesis, cyclization and folding of cyclotide-based libraries also allows to perform high throughput screening on chemically-generated libraries of cyclotides.73 A recent report showed that bioactive folded MCoTI-based cyclotides can be efficiently produced in parallel using a ‘tea-bag’ approach in combination with high efficient cyclization-folding protocols.73 The approach described in this work also included an efficient purification procedure to rapidly remove non-folded or partially folded cyclotides from the cyclization-folding crude. This method can be used for the purification of cyclotide mixtures, therefore making it compatible with the synthesis of amino acid and positional scanning libraries to perform efficient screening of large chemical-generated libraries.
Cyclotides as molecular imaging probes
Developing suitable diagnostic molecular tools is key for early detection and monitoring in the successful treatment of many diseases, including cancer.1,6 The technological advances in imaging instrumentation during the past two decades has dramatically increased our bioimaging capabilities and has also increased the need for improved molecular imaging agents. Optimal imaging agents should provide high affinity to provide high selectivity over healthy tissue, rapid clearance from healthy tissue to reduce background signal, and high chemical and biological stability.
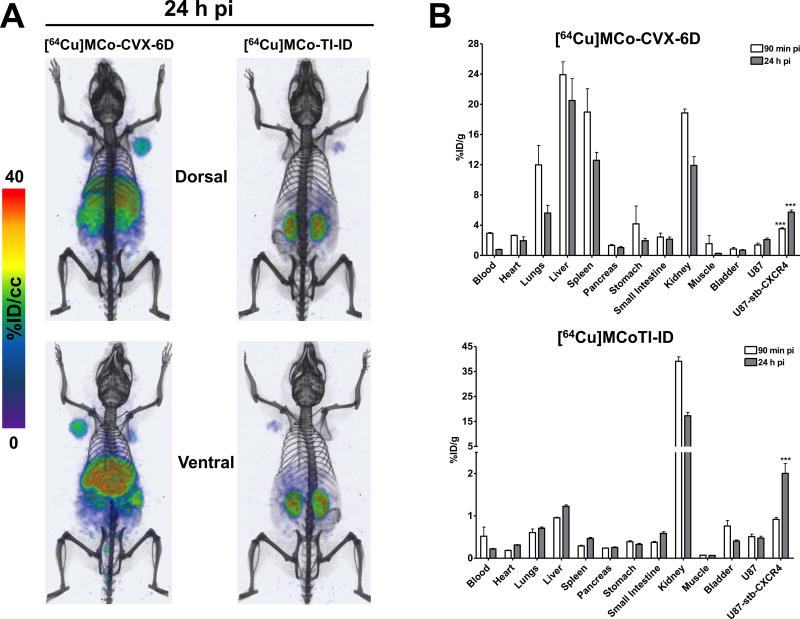
Properly functionalized engineered linear squash trypsin inhibitors, which share sequence homology and structure with the trypsin inhibitor subfamily of cyclotides but are not backbone-cyclized, have been shown to be excellent bioimaging and detection tools in cancer (recently reviewed in 1). Integrin-binding variants based on the Ecballium elaterium trypsin inhibitor II (EETI) have been extensively used as bio-imaging agents. They have been shown to provide significant tumor accumulation and low imaging signals in kidney, liver, and other organs. More recently, the cyclotide MCoTI-I from the trypsin inhibitor subfamily has been also shown to be an excellent bioimaging tool to visualize CXCR4-overexpressing cancer cells in a mouse model (Table 1 and Fig. 8).92 In this work, a [64Cu]-DOTA-labeled version of cyclotide MCo-CVX-6D, which is a potent CXCR4 antagonist with a remarkable in vivo resistance to biological degradation in serum, was used for the efficient detection of tumors containing CXCR4-expressing cells in mice using positron emission tomography–computed tomography (PET-CT) (Fig. 8A).92. The biodistribution of [64Cu]MCo-CVX-6D confirmed the detection of tumors with high CXCR4 expression and demonstrated favorable kinetics by PET imaging (Fig. 8B). In contrast, the [64Cu]-DOTA-labeled version of cyclotide MCoTI-I showed major accumulation in the kidneys which is consistent with renal clearance of this cyclotide (Fig. 8B). Rapid and non-invasive detection of CXCR4 overexpression in all malignant lesions in entirety should provide unprecedented opportunities to stratify patients for CXCR4-based therapies as well as testing efficacy of the corresponding treatment.
Figure 8.
Use of a CXCR4-targeting cyclotide as a bioimaging tool for detecting CXCR4-overexpressing tumor cells in animal models.92 A. Distribution of [64Cu]MCo-CVX-6D (CXCR4-targetign cyclotide) and [64Cu]MCoTI-ID (a DOTA-labeled variant of native trypsin inhibitor cyclotide MCoTI-I) in NOD/SCID mice bearing U87 and U87-stb-CXCR4 tumors with PET-CT. Representative PET-CT volume rendered images of mice (n=3) injected with 250 µCi of [64Cu]MCo-CVX-6D or [64Cu]MCoTI-ID recorded 24 h post tracers injection demonstrating high accumulation of [64Cu]MCo-CVX-6D in the U87-stb-CXCR4 tumor and [64Cu]MCoTI-ID showed mostly kidney uptake, suggesting renal clearance for the non-CXCR4-targetign cyclotide. B. Ex vivo evaluation of [64Cu]MCo-CVX-6D and [64Cu]MCoTI-ID distribution and specificity in NOD/SCID mice bearing U87 and U87-stb-CXCR4 tumors. Ex vivo biodistribution analysis was performed at 90 min and 24 h after post tracers injection.
Concluding remarks
We can say with confidence that cyclotides are now becoming to be a well-studied family of micro-proteins that given their unique properties are also starting to gain acceptance as molecular scaffolds for the potential design of novel peptide-based therapeutics and diagnostic tools. Their unique knotted arrangement of three disulfide bonds and circular backbone topology provides a unique molecular framework that confers exceptional resistance to thermal/chemical denaturation, and enzymatic degradation. Cyclotides can also cross mammalian cellular membranes to target protein-protein interactions in vitro but also and more importantly in animal models. The fact that cyclotides can target extracellular as well as intracellular targets highlights the high stability of the Cys-knot to be degraded/oxidized under complex biological conditions. The relative small size of cyclotides facilitates their chemical synthesis allowing the introduction of chemical modifications such as non-natural amino acids and PEGylation to improve their pharmacological properties. Moreover, cyclotides are amenable to substantial sequence variation and can be produced in several heterologous expression systems making them ideal substrates for molecular evolution strategies to enable generation and selection of compounds with optimal binding and inhibitory characteristics. All these unique properties make them promising leads or frameworks for the design of novel peptide-based therapeutics as well as bioimaging reagents.
No cyclotides have reached human clinical trials yet, however, the results obtained with several bioactive cyclotides in animal models may hint that this could occur in a not too distant future. One the main challenges that affect cyclotides, if they want to compete with small-molecule therapeutics, is their oral bioavailability. Although some cyclotides have proven to be orally active, there is still little information about their oral bioavailability. It is anticipated, however, that more studies on the biopharmaceutical properties of these exciting new micro-proteins may be available very soon.
Acknowledgments
This work was supported by National Institutes of Health Research Grant R01-GM113363, Department of Defense Congressionally Directed Medical Research Programs in Lung Cancer Grant LC150051, BROAD Medical Research Program-Crohn's & Colitis Foundation of America Grant #483566, Lupus Research Institute and Whittier Foundation.
References
- 1.Kintzing JR, Cochran JR. Engineered knottin peptides as diagnostics, therapeutics, and drug delivery vehicles. Curr Opin Chem Biol. 2016;34:143–150. doi: 10.1016/j.cbpa.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Kintzing JR, Filsinger Interrante MV, Cochran JR. Emerging Strategies for Developing Next-Generation Protein Therapeutics for Cancer Treatment. Trends Pharmacol Sci. 2016;37(12):993–1008. doi: 10.1016/j.tips.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang CC, Chen JJ, Yang PC. Multifunctional transcription factor YY1: a therapeutic target in human cancer? Expert Opin Ther Targets. 2006;10(2):253–266. doi: 10.1517/14728222.10.2.253. [DOI] [PubMed] [Google Scholar]
- 4.Camarero JA. Legume cyclotides shed light on the genetic origin of knotted circular proteins. Proc Natl Acad Sci U S A. 2011;108(25):10025–10026. doi: 10.1073/pnas.1107849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poth AG, Colgrave ML, Lyons RE, Daly NL, Craik DJ. Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc Natl Acad Sci U S A. 2011;108(25):1027–1032. doi: 10.1073/pnas.1103660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould A, Camarero JA. Cyclotides: Overview and biotechnological applications. Chembiochem. 2017 doi: 10.1002/cbic.201700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craik DJ, Du J. Cyclotides as drug design scaffolds. Curr Opin Chem Biol. 2017;38:8–16. doi: 10.1016/j.cbpa.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 8.Saether O, Craik DJ, Campbell ID, Sletten K, Juul J, Norman DG. Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry. 1995;34(13):4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Bi T, Camarero JA. Chemical and biological production of cyclotides. Adv Bot Res. 2015;76:271–303. doi: 10.1016/bs.abr.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras J, Elnagar AY, Hamm-Alvarez SF, Camarero JA. Cellular uptake of cyclotide MCoTI-I follows multiple endocytic pathways. J Control Release. 2011;155(2):134–143. doi: 10.1016/j.jconrel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales L, Henriques ST, Kerr MC, et al. Identification and characterization of a new family of cell-penetrating peptides: cyclic cell-penetrating peptides. J Biol Chem. 2011;286(42):36932–36943. doi: 10.1074/jbc.M111.264424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Y, Majumder S, Millard M, et al. In Vivo Activation of the p53 Tumor Suppressor Pathway by an Engineered Cyclotide. J Am Chem Soc. 2013;135(31):11623–11633. doi: 10.1021/ja405108p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong CT, Rowlands DK, Wong CH, et al. Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew Chem Int Ed Engl. 2012;51(23):5620–5624. doi: 10.1002/anie.201200984. [DOI] [PubMed] [Google Scholar]
- 14.Thell K, Hellinger R, Sahin E, et al. Oral activity of a nature-derived cyclic peptide for the treatment of multiple sclerosis. Proc Natl Acad Sci U S A. 2016;113(15):3960–3965. doi: 10.1073/pnas.1519960113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gran L. Oxytocic principles of Oldenlandia affinis. Lloydia. 1973;36(2):174–178. [PubMed] [Google Scholar]
- 16.Gran L. On the effect of a polypeptide isolated from "Kalata-Kalata" (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol Toxicol (Copenh) 1973;33(5):400–408. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 17.Gustafson KR, Sowder RC, Louis E, Henderson LE, et al. Circulins A and B. Novel human immunodeficiency virus (HIV)-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J Am Chem Soc. 1994;116:9337–9338. [Google Scholar]
- 18.Witherup KM, Bogusky MJ, Anderson PS, et al. Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J Nat Prod. 1994;57(12):1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 19.Schöpke T, Hasan Agha MI, Kraft R, Otto A, Hiller K. Hämolytisch aktive komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci Pharm. 1993;61:145–153. [Google Scholar]
- 20.Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294(5):1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 21.Gruber CW, Elliott AG, Ireland DC, et al. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20(9):2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez JF, Gagnon J, Chiche L, et al. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39(19):5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 23.Trabi M, Craik DJ. Tissue-specific expression of head-to-tail cyclized miniproteins in Violaceae and structure determination of the root cyclotide Viola hederacea root cyclotide1. Plant Cell. 2004;16(8):2204–2216. doi: 10.1105/tpc.104.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trabi M, Svangard E, Herrmann A, et al. Variations in cyclotide expression in viola species. J Nat Prod. 2004;67(5):806–810. doi: 10.1021/np034068e. [DOI] [PubMed] [Google Scholar]
- 25.Gran L. Isolation of oxytocic peptides from Oldenlandia affinis by solvent extraction of tetraphenylborate complexes and chromatography on Sephadex LH-20. Lloydia. 1973;36(2):207–208. [PubMed] [Google Scholar]
- 26.Hashempour H, Koehbach J, Daly NL, Ghassempour A, Gruber CW. Characterizing circular peptides in mixtures: sequence fragment assembly of cyclotides from a violet plant by MALDI-TOF/TOF mass spectrometry. Amino Acids. 2013;44(2):581–595. doi: 10.1007/s00726-012-1376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koehbach J, Attah AF, Berger A, et al. Cyclotide discovery in Gentianales revisited--identification and characterization of cyclic cystine-knot peptides and their phylogenetic distribution in Rubiaceae plants. Biopolymers. 2013;100(5):438–452. doi: 10.1002/bip.22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farhadpour M, Hashempour H, Talebpour Z, et al. Microwave-assisted extraction of cyclotides from Viola ignobilis. Anal Biochem. 2016;497:83–89. doi: 10.1016/j.ab.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Velasquez JE, van der Donk WA. Genome mining for ribosomally synthesized natural products. Curr Opin Chem Biol. 2011;15(1):11–21. doi: 10.1016/j.cbpa.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellinger R, Koehbach J, Soltis DE, Carpenter EJ, Wong GK, Gruber CW. Peptidomics of Circular Cysteine-Rich Plant Peptides: Analysis of the Diversity of Cyclotides from Viola tricolor by Transcriptome and Proteome Mining. J Proteome Res. 2015;14(11):4851–4862. doi: 10.1021/acs.jproteome.5b00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulvenna JP, Wang C, Craik DJ. CyBase: a database of cyclic protein sequence and structure. Nucleic Acids Res. 2006;34(Database issue):D192–194. doi: 10.1093/nar/gkj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Backbone Dynamics of Cyclotide MCoTI-I Free and Complexed with Trypsin. Angew Chem Int Ed Engl. 2010;49(39):7030–7034. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43(20):5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 34.Garcia AE, Camarero JA. Biological activities of natural and engineered cyclotides, a novel molecular scaffold for peptide-based therapeutics. Curr Mol Pharmacol. 2010;3(3):153–163. doi: 10.2174/1874467211003030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weidmann J, Craik DJ. Discovery, structure, function, and applications of cyclotides: circular proteins from plants. J Exp Bot. 2016;67(16):4801–4812. doi: 10.1093/jxb/erw210. [DOI] [PubMed] [Google Scholar]
- 36.Wang CK, Kaas Q, Chiche L, Craik DJ. CyBase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008;36(Database issue):D206–210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heitz A, Hernandez JF, Gagnon J, et al. Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry. 2001;40(27):7973–7983. doi: 10.1021/bi0106639. [DOI] [PubMed] [Google Scholar]
- 38.Mylne JS, Chan LY, Chanson AH, et al. Cyclic peptides arising by evolutionary parallelism via asparaginyl-endopeptidase-mediated biosynthesis. Plant Cell. 2012;24(7):2765–2778. doi: 10.1105/tpc.112.099085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiche L, Heitz A, Gelly JC, et al. Squash inhibitors: from structural motifs to macrocyclic knottins. Curr Protein Pept Sci. 2004;5(5):341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- 40.Ravipati AS, Henriques ST, Poth AG, et al. Lysine-rich Cyclotides: A New Subclass of Circular Knotted Proteins from Violaceae. ACS Chem Biol. 2015;10(11):2491–2500. doi: 10.1021/acschembio.5b00454. [DOI] [PubMed] [Google Scholar]
- 41.Jennings C, West J, Waine C, Craik D, Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc Natl Acad Sci U S A. 2001;98(19):10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jennings CV, Rosengren KJ, Daly NL, et al. Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: do Mobius strips exist in nature? Biochemistry. 2005;44(3):851–860. doi: 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- 43.Pinto MF, Fensterseifer IC, Migliolo L, et al. Identification and structural characterization of novel cyclotide with activity against an insect pest of sugar cane. J Biol Chem. 2012;287(1):134–147. doi: 10.1074/jbc.M111.294009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colgrave ML, Kotze AC, Huang YH, O'Grady J, Simonsen SM, Craik DJ. Cyclotides: natural, circular plant peptides that possess significant activity against gastrointestinal nematode parasites of sheep. Biochemistry. 2008;47(20):5581–5589. doi: 10.1021/bi800223y. [DOI] [PubMed] [Google Scholar]
- 45.Colgrave ML, Kotze AC, Ireland DC, Wang CK, Craik DJ. The anthelmintic activity of the cyclotides: natural variants with enhanced activity. Chembiochem. 2008;9(12):1939–1945. doi: 10.1002/cbic.200800174. [DOI] [PubMed] [Google Scholar]
- 46.Malagon D, Botterill B, Gray DJ, et al. Anthelminthic activity of the cyclotides (kalata B1 and B2) against schistosome parasites. Biopolymers. 2013;100(5):461–470. doi: 10.1002/bip.22229. [DOI] [PubMed] [Google Scholar]
- 47.Plan MR, Saska I, Cagauan AG, Craik DJ. Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail) J Agric Food Chem. 2008;56(13):5237–5241. doi: 10.1021/jf800302f. [DOI] [PubMed] [Google Scholar]
- 48.Barbeta BL, Marshall AT, Gillon AD, Craik DJ, Anderson MA. Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc Natl Acad Sci U S A. 2008;105(4):1221–1225. doi: 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Troeira Henriques S, Craik DJ. Cyclotide Structure and Function: The Role of Membrane Binding and Permeation. Biochemistry. 2017;56(5):669–682. doi: 10.1021/acs.biochem.6b01212. [DOI] [PubMed] [Google Scholar]
- 50.Tam JP, Lu YA, Yang JL, Chiu KW. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci U S A. 1999;96(16):8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen GK, Zhang S, Wang W, Wong CT, Nguyen NT, Tam JP. Discovery of a linear cyclotide from the bracelet subfamily and its disulfide mapping by top-down mass spectrometry. J Biol Chem. 2011;286(52):44833–44844. doi: 10.1074/jbc.M111.290296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong CT, Taichi M, Nishio H, Nishiuchi Y, Tam JP. Optimal oxidative folding of the novel antimicrobial cyclotide from Hedyotis biflora requires high alcohol concentrations. Biochemistry. 2011;50(33):7275–7283. doi: 10.1021/bi2007004. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen GK, Zhang S, Nguyen NT, et al. Discovery and characterization of novel cyclotides originated from chimeric precursors consisting of albumin-1 chain a and cyclotide domains in the fabaceae family. J Biol Chem. 2011 doi: 10.1074/jbc.M111.229922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fensterseifer IC, Silva ON, Malik U, et al. Effects of cyclotides against cutaneous infections caused by Staphylococcus aureus. Peptides. 2015;63:38–42. doi: 10.1016/j.peptides.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 55.Lindholm P, Göransson U, Johansson S, et al. Cyclotides: A novel type of cytotoxic agents. Mol Cancer Ther. 2002;1:365–369. [PubMed] [Google Scholar]
- 56.Svangard E, Goransson U, Hocaoglu Z, et al. Cytotoxic cyclotides from Viola tricolor. J Nat Prod. 2004;67(2):144–147. doi: 10.1021/np030101l. [DOI] [PubMed] [Google Scholar]
- 57.Herrmann A, Burman R, Mylne JS, et al. The alpine violet, Viola biflora, is a rich source of cyclotides with potent cytotoxicity. Phytochemistry. 2008;69(4):939–952. doi: 10.1016/j.phytochem.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 58.Esmaeili MA, Abagheri-Mahabadi N, Hashempour H, Farhadpour M, Gruber CW, Ghassempour A. Viola plant cyclotide vigno 5 induces mitochondria-mediated apoptosis via cytochrome C release and caspases activation in cervical cancer cells. Fitoterapia. 2016;109:162–168. doi: 10.1016/j.fitote.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 59.Hu E, Wang D, Chen J, Tao X. Novel cyclotides from Hedyotis diffusa induce apoptosis and inhibit proliferation and migration of prostate cancer cells. Int J Clin Exp Med. 2015;8(3):4059–4065. [PMC free article] [PubMed] [Google Scholar]
- 60.Koehbach J, O'Brien M, Muttenthaler M, et al. Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design. Proc Natl Acad Sci U S A. 2013;110(52):21183–21188. doi: 10.1073/pnas.1311183110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Craik DJ, Malik U. Cyclotide biosynthesis. Curr Opin Chem Biol. 2013;17(4):546–554. doi: 10.1016/j.cbpa.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 62.Arnison PG, Bibb MJ, Bierbaum G, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poth AG, Mylne JS, Grassl J, et al. Cyclotides associate with leaf vasculature and are the products of a novel precursor in petunia (Solanaceae) J Biol Chem. 2012;287(32):27033–27046. doi: 10.1074/jbc.M112.370841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saska I, Gillon AD, Hatsugai N, et al. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282(40):29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 65.Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA. Biosynthesis of circular proteins in plants. Plant J. 2008;53(3):505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- 66.Nguyen GK, Wang S, Qiu Y, Hemu X, Lian Y, Tam JP. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol. 2014;10(9):732–738. doi: 10.1038/nchembio.1586. [DOI] [PubMed] [Google Scholar]
- 67.Harris KS, Durek T, Kaas Q, et al. Efficient backbone cyclization of linear peptides by a recombinant asparaginyl endopeptidase. Nat Commun. 2015;6:10199. doi: 10.1038/ncomms10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernath-Levin K, Nelson C, Elliott AG, et al. Peptide macrocyclization by a bifunctional endoprotease. Chem Biol. 2015;22(5):571–582. doi: 10.1016/j.chembiol.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 69.Hemu X, Qiu Y, Nguyen GK, Tam JP. Total Synthesis of Circular Bacteriocins by Butelase 1. J Am Chem Soc. 2016;138(22):6968–6971. doi: 10.1021/jacs.6b04310. [DOI] [PubMed] [Google Scholar]
- 70.Nguyen GK, Hemu X, Quek JP, Tam JP. Butelase-Mediated Macrocyclization of d-Amino-Acid-Containing Peptides. Angew Chem Int Ed Engl. 2016;55(41):12802–12806. doi: 10.1002/anie.201607188. [DOI] [PubMed] [Google Scholar]
- 71.Nguyen GK, Qiu Y, Cao Y, Hemu X, Liu CF, Tam JP. Butelase-mediated cyclization and ligation of peptides and proteins. Nat Protoc. 2016;11(10):1977–1988. doi: 10.1038/nprot.2016.118. [DOI] [PubMed] [Google Scholar]
- 72.Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of Proteins by Native Chemical Ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 73.Aboye T, Kuang Y, Neamati N, Camarero JA. Rapid parallel synthesis of bioactive folded cyclotides by using a tea-bag approach. Chembiochem. 2015;16(5):827–833. doi: 10.1002/cbic.201402691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, Tate EW. Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org Biomol Chem. 2008;6(8):1462–1470. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- 75.Jia X, Kwon S, Wang CI, et al. Semienzymatic cyclization of disulfide-rich peptides using Sortase A. J Biol Chem. 2014;289(10):6627–6638. doi: 10.1074/jbc.M113.539262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jagadish K, Borra R, Lacey V, et al. Expression of fluorescent cyclotides using protein trans-splicing for easy monitoring of cyclotide-protein interactions. Angew Chem Int Ed Engl. 2013;52(11):3126–3131. doi: 10.1002/anie.201209219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jagadish K, Gould A, Borra R, et al. Recombinant Expression and Phenotypic Screening of a Bioactive Cyclotide Against alpha-Synuclein-Induced Cytotoxicity in Baker's Yeast. Angew Chem Int Ed Engl. 2015;54(29):8390–8394. doi: 10.1002/anie.201501186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jagadish K, Camarero JA. Recombinant Expression of Cyclotides Using Split Inteins. Methods Mol Biol. 2017;1495:41–55. doi: 10.1007/978-1-4939-6451-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Henriques ST, Craik DJ. Cyclotides as templates in drug design. Drug Discov Today. 2010;15(1–2):57–64. doi: 10.1016/j.drudis.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 80.Gunasekera S, Foley FM, Clark RJ, et al. Engineering stabilized vascular endothelial growth factor-A antagonists: synthesis, structural characterization, and bioactivity of grafted analogues of cyclotides. J Med Chem. 2008;51(24):7697–7704. doi: 10.1021/jm800704e. [DOI] [PubMed] [Google Scholar]
- 81.Thongyoo P, Bonomelli C, Leatherbarrow RJ, Tate EW. Potent inhibitors of beta-tryptase and human leukocyte elastase based on the MCoTI-II scaffold. J Med Chem. 2009;52(20):6197–6200. doi: 10.1021/jm901233u. [DOI] [PubMed] [Google Scholar]
- 82.Eliasen R, Daly NL, Wulff BS, Andresen TL, Conde-Frieboes KW, Craik DJ. Design, synthesis, structural and functional characterization of novel melanocortin agonists based on the cyclotide kalata B1. J Biol Chem. 2012;287(48):40493–40501. doi: 10.1074/jbc.M112.395442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aboye TL, Ha H, Majumder S, et al. Design of a novel cyclotide-based CXCR4 antagonist with anti-human immunodeficiency virus (HIV)-1 activity. Journal of medicinal chemistry. 2012;55(23):10729–10734. doi: 10.1021/jm301468k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol. 2004;14(3):171–179. doi: 10.1016/j.semcancer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 85.Culp E, Wright GD. Bacterial proteases, untapped antimicrobial drug targets. J Antibiot (Tokyo) 2017;70(4):366–377. doi: 10.1038/ja.2016.138. [DOI] [PubMed] [Google Scholar]
- 86.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278(1):16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 87.Sommerhoff CP, Avrutina O, Schmoldt HU, Gabrijelcic-Geiger D, Diederichsen U, Kolmar H. Engineered cystine knot miniproteins as potent inhibitors of human mast cell tryptase beta. J Mol Biol. 2010;395(1):167–175. doi: 10.1016/j.jmb.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 88.Thell K, Hellinger R, Schabbauer G, Gruber CW. Immunosuppressive peptides and their therapeutic applications. Drug Discov Today. 2014;19(5):645–653. doi: 10.1016/j.drudis.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang CK, Gruber CW, Cemazar M, et al. Molecular grafting onto a stable framework yields novel cyclic peptides for the treatment of multiple sclerosis. ACS Chem Biol. 2014;9(1):156–163. doi: 10.1021/cb400548s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.D'Souza C, Henriques ST, Wang CK, et al. Using the MCoTI-II Cyclotide Scaffold To Design a Stable Cyclic Peptide Antagonist of SET, a Protein Overexpressed in Human Cancer. Biochemistry. 2016;55(2):396–405. doi: 10.1021/acs.biochem.5b00529. [DOI] [PubMed] [Google Scholar]
- 91.Wang CK, Stalmans S, De Spiegeleer B, Craik DJ. Biodistribution of the cyclotide MCoTI-II, a cyclic disulfide-rich peptide drug scaffold. J Pept Sci. 2016;22(5):305–310. doi: 10.1002/psc.2862. [DOI] [PubMed] [Google Scholar]
- 92.Lesniak WG, Aboye T, Chatterjee S, Camarero JA, Nimmagadda S. In vivo Evaluation of an Engineered Cyclotide as Specific CXCR4 Imaging Reagent. Chemistry. 2017 doi: 10.1002/chem.201702540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Austin J, Wang W, Puttamadappa S, Shekhtman A, Camarero JA. Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I. Chembiochem. 2009;10(16):2663–2670. doi: 10.1002/cbic.200900534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao Y, Cui T, Lam Y. Synthesis and disulfide bond connectivity-activity studies of a kalata B1-inspired cyclopeptide against dengue NS2B-NS3 protease. Bioorg Med Chem. 2010;18(3):1331–1336. doi: 10.1016/j.bmc.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 95.Getz JA, Cheneval O, Craik DJ, Daugherty PS. Design of a cyclotide antagonist of neuropilin-1 and -2 that potently inhibits endothelial cell migration. ACS Chem Biol. 2013;8(6):1147–1154. doi: 10.1021/cb4000585. [DOI] [PubMed] [Google Scholar]
- 96.Aboye TL, Ha H, Majumder S, et al. Design of a novel cyclotide-based CXCR4 antagonist with anti-human immunodeficiency virus (HIV)-1 activity. J Med Chem. 2012;55(23):10729–10734. doi: 10.1021/jm301468k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maass F, Wustehube-Lausch J, Dickgiesser S, et al. Cystine-knot peptides targeting cancer-relevant human cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) J Pept Sci. 2015;21(8):651–660. doi: 10.1002/psc.2782. [DOI] [PubMed] [Google Scholar]
- 98.Quimbar P, Malik U, Sommerhoff CP, et al. High-affinity cyclic peptide matriptase inhibitors. J Biol Chem. 2013;288(19):13885–13896. doi: 10.1074/jbc.M113.460030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chan LY, Gunasekera S, Henriques ST, et al. Engineering pro-angiogenic peptides using stable, disulfide-rich cyclic scaffolds. Blood. 2011;118(25):6709–6717. doi: 10.1182/blood-2011-06-359141. [DOI] [PubMed] [Google Scholar]
- 100.Huang YH, Henriques ST, Wang CK, et al. Design of substrate-based BCR-ABL kinase inhibitors using the cyclotide scaffold. Sci Rep. 2015;5:12974. doi: 10.1038/srep12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aboye T, Meeks CJ, Majumder S, Shekhtman A, Rodgers K, Camarero JA. Design of a MCoTI-Based Cyclotide with Angiotensin (1-7)-Like Activity. Molecules. 2016;21(2) doi: 10.3390/molecules21020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Swedberg JE, Mahatmanto T, Abdul Ghani H, et al. Substrate-Guided Design of Selective FXIIa Inhibitors Based on the Plant-Derived Momordica cochinchinensis Trypsin Inhibitor-II (MCoTI-II) Scaffold. J Med Chem. 2016;59(15):7287–7292. doi: 10.1021/acs.jmedchem.6b00557. [DOI] [PubMed] [Google Scholar]
- 103.Chan LY, Craik DJ, Daly NL. Cyclic thrombospondin-1 mimetics: grafting of a thrombospondin sequence into circular disulfide-rich frameworks to inhibit endothelial cell migration. Biosci Rep. 2015;35(6) doi: 10.1042/BSR20150210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chan LY, Craik DJ, Daly NL. Dual-targeting anti-angiogenic cyclic peptides as potential drug leads for cancer therapy. Sci Rep. 2016;6:35347. doi: 10.1038/srep35347. [DOI] [PMC free article] [PubMed] [Google Scholar]