Abstract
The bottom-up self-assembly of protein subunits into supramolecular nanoarchitectures is ubiquitously exploited to recapitulate and expand features of natural proteins to advance nanoscience in medicine. Various chemical and biological re-engineering approaches are available to render given proteins diverse functions. They are, unfortunately, susceptible of compromising protein integrity and stability after extensive modifications. In this study, we introduce a new protein re-engineering method, metal ion assisted interface re-engineering (MAIR), to serve as a robust and universal strategy to extend functions of self-assembly proteins with boosted structural features to advance its diverse biomedical applications. In particular, the MAIR strategy was applied to a widely used natural protein, ferritin, as a model protein to coordinate with copper ions in its mutagenic artificial metal binding domain. Structure directed rational protein mutagenesis was carried out at the C2 interface amino acid residues of the ferritin subunit for metal ion coordination site optimization. Copper binding at the artificial binding pocket was highly specific over the other divalent ions present in physiological fluids, and the structurally embedded copper ion in turn strengthened the overall protein integrity and stability. In the presence of isotopic copper-64, the interface re-engineered ferritin worked as a chelator-free molecular nanoprobe with extraordinarily high specific activity to allow PET imaging of tumors in live animals. We also found that the re-engineered ferritin coordinating with copper ions demonstrates high drug loading capacity of a widely used anti-cancer agent, doxorubicin (DOX), to achieve significant drug retention at the tumor site and enhance tumor regression for improved anti-cancer effects. The MAIR approach, thus, exploited the copper ion to facilitate mutant ferritin derivatives efficient one-step labeling for simultaneous molecular imaging and drug delivery. The reported interface re-engineering strategy provides an unparalleled opportunity to expand protein biofunctions to serve as a new theranostic agent in cancer research.
INTRODUCTION
The self-assembly of polypeptides or protein subunits into specific geometrical architectures has been investigated extensively.1 The profound understanding of protein subunit self-assembly into symmetrical and stable supramolecules has led to the discovery of noncovalent interactions of protein-protein interfaces,2, 3 and provides the basis for protein structure interrogation and manipulation.4,5 Self-assembled proteins with unique properties such as high stability, mono-dispersity, good solubility, and ease of genetic and chemical manipulation, possess high potential in biomedical applications.6–10 Recent attempts of de novo design of protein subunits to self-assemble into nanocages/nanoparticles utilized computer-aided protein programs to precisely predict folding interfaces for designer proteins to advance medicine and materials.6, 11 Nevertheless, such strategies require complicated processes and extensive expertise along with sophisticated computational modeling. Alternatively, re-engineering of available natural or synthetic protein subunits with designated features presents a simpler strategy. In addition, the widely accessible validated protein crystal structure information provides rational guidance on the protein re-engineering processes. This increases the chances of success in protein re-engineering to meet specific purposes, and is more adaptable to a wider research community.
A myriad of re-engineered protein self-assemblies have found various applications. Protein subunits have evolved to self-assemble into nanocages accommodating therapeutic agents, such as oligonucleotides or drugs, for effective drug delivery.12–14 Some protein assemblies support additional protein fusions to assemble into sophisticated architectures. The fusion of up to two copies of green fluorescent proteins to each unit of a rationally designed 60-unit protein icosahedron produced highly bright “standard candles” for fluorescence imaging.4 The fusion of the viral hemagglutinin to ferritin elicited broad immunization responses to H1N1 influenza virus.
Surface modification of assembled proteins also extends the utility of nanostructured proteins. The exterior surface re-engineering of a small heat shock protein nanocage was decorated with a tumor associated macrophage targeting peptide for the detection of atherosclerotic plaque lesions.15 Likewise, a virus capsid with polymer modification on surface modulated the immunogenicity in animals.16 Meanwhile, the re-engineering of interior surfaces of assembling proteins renders them multiple active sites for inorganic nanoparticle synthesis,17 or provides an optimal condition as a nanoreactor for catalytic reactions.18
The cumulative knowledge on protein self-assembly reveals that the interface of assembling subunits has a pivotal role in tuning protein self-assembly through noncovalent bonds and interactions. In expanding the scope of re-engineering for protein assembly, it has been proposed that the interface re-engineering of proteins provide subtle control of the entire protein self-assembly behavior and its functions while maintaining the original protein integrity.19, 20 Several research papers published by Tezcan group reported Zinc ion templated non-self-linking protein forming into dimer or supramolecular assemblies.21, 22 Degrado group in University of Pennsylvania also reported two different protein subunits assembling into heterotetrameric four-helix bundle after making specific amino acid mutations.23 An interface re-engineered protein ensemble could serve as a pH-dependent molecular switch,20 or create a novel protein cube with interface re-engineering of two natural proteins.24 Interface re-engineering of proteins is thus an attractive approach to develop new protein assemblies with novel functions.
In executing interface re-engineering, the interface of subunits usually has restricted space in which only selective small molecules or metal ions can be accommodated without altering protein integrity and overall dimensions. Small molecules usually have limited spatial binding capacities to multiple amino acids, and their interactions with amino acid residues could be attenuated by surrounding interference. In contrast, transition metal ions have high binding affinity to most amino acid side chains. The coordination forces between metal ions and amino acid side chains provide stronger interfacial interactions than hydrogen bonds and van de Waals forces,25 which would constitute the major driving forces in protein self-assembly. Moreover, metal ions compensate the associated entropy expense in the transition of a binary to a ternary system, and impose a geometrical constraint for overall protein assembly. By changing different metal ions with varying affinities to particular amino acids, the interface interactions can be precisely refined upon demand. Meanwhile, some small molecules, such as doxorubicin (DOX), is able to additionally couple with metal ions to form nanocomplexes, such as Cu(II)-DOX,26 Zn(II)-DOX27 and Fe(III)-DOX.28 This unique feature between anti-cancer drug and metal ions expand the scope of MIAR protein re-engineering approaches for simultaneous self-assembly, protein labelling and drug delivery.
Ferritin nanocages have been broadly explored in various fields.9, 13, 16 In biomedicine, they have been used as delivery vehicles for effective anti-cancer drug delivery13, 29 or immunization vaccine against viruses.15 Recent studies have reported the use of ferritins as effective nanoprobes for cancer diagnosis24, 30, immunoassays31, and therapy.26 Most of the applications rely heavily on chemical or biological modification of the original protein structure for desired properties. Such extensive uncontrolled refinement is likely to undermine protein integrity and functions. The substantial surface modification could lead to nanoparticle heterogeneity, structural instability, and batch to batch variation.
It is highly desirable to develop a simple one-step strategy to functionalize the ferritin nanocage as a theranostic agent for simultaneous bioimaging and drug delivery. In this study, we report a new protein interface re-engineering approach, MIAR, by using transition metal ions in conjunction with rational protein mutagenesis to refine self-assembly of the ferritin nanocage with enhanced physicochemical properties. The re-engineered nanocage is a new chelator-free molecular imaging nanoprobe with high specific radioactivity for positron emission tomography (PET) imaging of cancer. Meanwhile, such nanocage chelating with Cu(II) improves multiple folds of doxorubicin loading capacity to ultimately broaden the therapeutic index for efficacious anti-cancer therapy. The new interface re-engineered approach revivifies the ferritin nanocage to serve as a quintessential example of a theranostic platform for chelator-free PET imaging and chemotherapy.
RESULTS AND DISCUSSION
Interface re-engineering of ferritin nanocage
Ferritin is a spherical shell protein of about 460 kDa consisting of 24 identical subunits tightly packed in octahedral symmetry with an inner diameter of 8 nm and an outer diameter of 12 nm.19, 32 The key step of interface re-engineering of ferritin is to search for the appropriate positions to install metal ion coordination motifs with low entropy cost in one of the symmetrical C2, C3 and C4 interfaces (Fig. S1). The protein model in Figure S1 shows that the C2 interface has substantially greater interacting surface area than C3 and C4 interfaces, and could be more amenable for structure re-engineering without compromising the protein’s original integrity.19 The amino acids that comprise the individual subunit of ferritin heavy chain fold into three long and one short alpha helix bundles, hence, the mutation sites along the interface of each alpha helix were carefully screened to accommodate Cu2+ ion. It has been reported that the spatial configuration of Cu2+ ion coordination with amino acids at the interface in the square planar format is favorable.33 The introduction of two metal binding histidine residue mutants on each of the two adjacent subunits can meet the Cu2+ coordination requirement to secure the neighboring subunits with high stability. The distance of the two mutation sites in three amino acid intervals in each helix is required to secure Cu2+ coordination at the interface. We used ferritin protein crystal structure (PDB: 1FHA) to identify the entire interface sequences of a single ferritin heavy chain subunit, and rationally mutagenized specific amino acids into histidine residues having high binding affinity to a number of metal ions. We avoided the residues where alternations could potentially devastate the naïve configuration of the C2 interface, which has dominant role upon C3 and C4 interfaces,34 and kept residues at the C2 interface core position unchanged to ensure the correct C2 interface alignment and formation.19
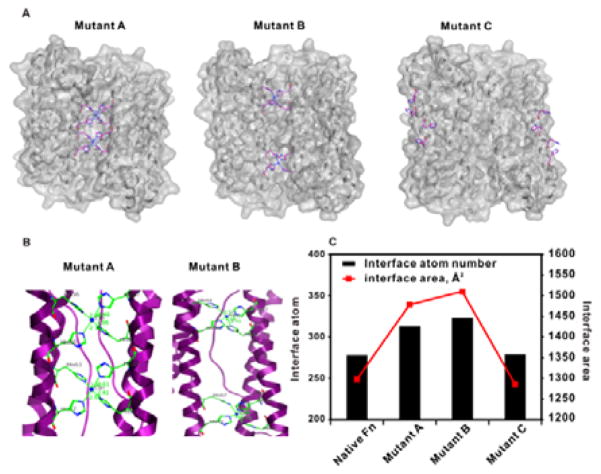
In expediting the mutation process, we employed a programmable stepwise cloning method circumventing the conventional site-directed mutation approach which is intricate and time-consuming. In our method, a total of 5 pairs of 60mer DNA oligos were designed and synthesized, covering the entire span of ferritin subunit sequences with appropriate length of overlapping in the two consecutive sequences (see SI for details). Within five steps of regular PCR reactions, the designed mutation coding DNA sequences were synthesized (Figure S2), following a single step restriction enzyme digestion for complete expression plasmid construction. Table 1 shows the selected two optimal mutants (mutant A and mutant B) amino acid residues, and a control mutant variant (mutant C) on the fourth alpha helix which results in no interaction with the two neighboring subunits. In selecting the appropriate residues as potential mutation sites, we kept the C2 interacting core residues and 2, 3 alpha helix loop residues unchanged, as subtle alternations on those sites could destabilize the subunit assembly and alignment in protein self-assembly. It was recently reported that residue pairs at sites 56 and 60 and sites 63 and 67 facing each other over the interface are appropriate positions to induce mutation for symmetrical ion coordination in their initial screening.19 This is consistent with our results on the high stability of mutant A protein in Cu2+ coordination (Figure 1). We conducted further mutation screening on residues in the same helix, and our results reveal that the residue pairs at sites 52 and 56 and sites 67 and 71 provide an optimal interface for Cu2+ coordination (mutant B) (Figure 1). The mutation pairs on the far end of the fourth helix present almost identical Cu2+ binding affinity as that of native wild-type ferritin (Figure 1C). Computational modeling is an important tool to facilitate prediction of permuting protein structures and ion binding energy calculations. The calculated results (Figure 1C and Table S1) confirm that rational interface re-engineered ferritins (mutant A and B) have much higher Cu2+ binding atoms and areas than those of the native ferritin, and the overall ferritin ensembles (mutant A and B) have enhanced stability with Cu2+ binding at the assembly interface.
Table 1.
Amino acid sequences of native and mutative sites of ferritins
| a.a. position | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Native Fn | N | F | A | K | Y | F | L | H | Q | S | H | E | E | R | E | H | A | E | K | L | M | K | L | Q |
| Mutant A | H | H | H | H | ||||||||||||||||||||
| Mutant B | H | H | H | H |
| a.a. position | 137 | 138 | 139 | 140 | 141 | 142 | 143 | 144 | 145 | 146 | 147 | 148 | 149 | 150 | 151 | 152 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Native Fn | Y | L | N | E | Q | V | K | S | I | K | E | L | G | D | H | V |
| Mutant C | H | H | H | H |
Figure 1.
(A) Computational simulation of three interface re-engineered ferritin variants with copper chelation. (B) Magnified protein ribbon structures of two potent permutated ferritin variants coordinated with copper. The amino acid residues associated with copper coordination are labelled. (C) Computational calculation of interface atom number and interface area of different ferritin variants.
Characterization of Cu2+ coordination with interface re-engineered ferritin nanocages
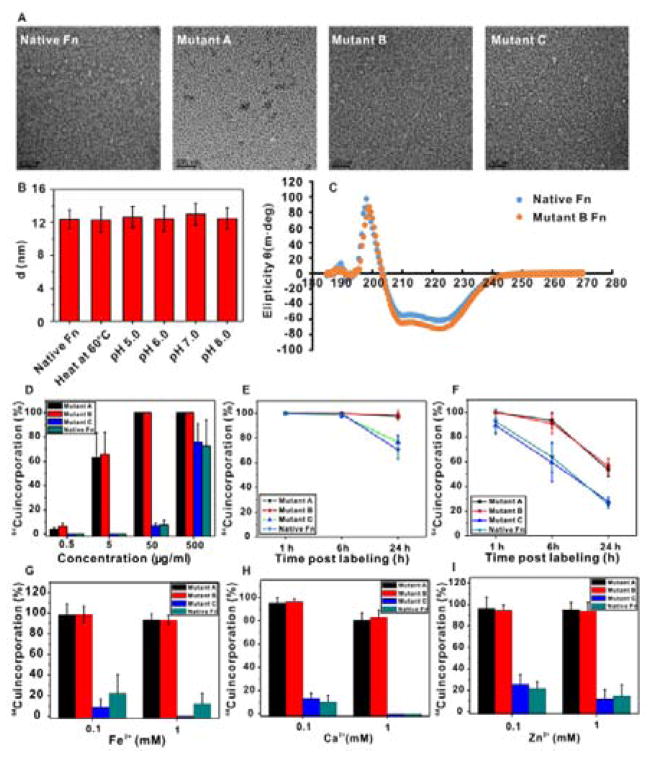
Denaturing SDS-PAGE gel electrophoresis results showed that both the E. Coli expressed and size-exclusion column purified ferritin subunit analogues presented identical molecular weight at 21 kDa with high purity (Figure S3). All re-engineered ferritin subunits can self-assemble into homogeneous spherical hollow nanocages as shown by TEM imaging (Figure 2A), and both mutants A and B showed high stability in buffers of various pH values in the absence of Cu2+ ion (Figure 2B). We further used circular dichroism (CD) spectroscopy to study the secondary structure of our targeted mutant B in PBS without Cu2+ ion (Figure 2C). It was shown that mutant B had identical secondary structure as native ferritin at physiological pH (pH = 7.2). Ferritin in general is a family of proteins that can tolerate a wide range of fusion peptide/protein modifications,24, 35, 36 whereas the stability of the modified proteins is usually in question. Our results indicate that the interface re-engineered subunits retain the self-assembly feature for nanocage ensembles without compromising secondary structure integrity.
Figure 2.
(A) TEM images of ferritin variants with uranyl acetate negative staining. Scale bar=100 nm. (B) Dynamic light scattering study of the nanocage size of mutant B ferritin under heating (60 °C) and different pH values in buffers. (C) Circular dichroism spectra of native ferritin and mutant B ferritin in PBS (0.5 mg/mL). (D) Radioactive 64Cu incorporation ratio of ferritin variants under different concentrations. (E) 64Cu incorporation stability study of ferritin variants in mouse serum over 24 h. (F) 64Cu incorporation stability study of ferritin variants in the presence of strong metal chelator, EDTA, in mouse serum over 24 h. (G–I) Divalent metal ion competition with 64Cu in ferritin variants.
We then investigated the coordination of Cu2+ with self-assembling protein analogues. The radioisotopic 64Cu2+ was used in our study to facilitate sensitive and quantitative readout over the subtle amino acid residue variations in protein analogues. The chelator, 2,2′-bipyridine, was firstly used to prepare metal ion free ferritin nanocages before adding 64Cu2+ into protein solutions. The incorporation of 64Cu2+ was over 65% at 5 μg/mL of mutant A and B proteins, whereas the native ferritin had marginal 64Cu2+binding capacity at the same protein concentration (Figure 2D). Higher protein concentrations (50 μg/mL) led to over 95% 64Cu2+ incorporation for mutants A and B, while native ferritin showed less than 10% 64Cu2+ binding, presumably through the non-specific binding of 64Cu2+ with amino acid residues, such as histidine, and to the hydrophobic domains within the ferritin structure.37 The stability test of 64Cu2+ coordinated ferritin derivatives in mouse serum demonstrated the high affinity of 64Cu2+ for mutants A and B over 24 h, whereas native ferritin and mutant control showed dramatic 64Cu2+ loss within the same time frame (Figure 2E). Ethylenediaminetetraacetic acid (EDTA) has fairly high stability constant to a number of divalent transition metal ions,38 including Cu2+. In our study, we discovered that the high affinity of 64Cu2+ for mutants A and B is able to resist competitive binding from EDTA for at least 6 h, and over 50% 64Cu2+ remained complexed after 24 h of co-incubation. In contrast, mutant C and native ferritin had dramatic loss of 64Cu2+ over the incubation period (Figure 2E).
Next, we calculated the number of copper binding pockets of the ferritin derivatives. After incubation with 64Cu2+, various concentrations of cold Cu2+ were added to the ferritin-64Cu2+ solution. The copper binding pocket numbers were calculated by the formula: (64Cu2+) × Yield/Moles (Ferritin). Table S2 summarizes all experimental parameters and the calculated results. The mutant A has a maximum of 18 accessible Cu2+ binding pockets in the assembled protein nanocage, and mutant B has 20 copper binding pockets. The experimental results are highly consistent with the computational modeling calculation which predicts that mutant B possesses slightly larger interfacial areas and lower binding entropy in the presence of Cu2+ at the designed interface coordination domain (Figure 1). In contrast, mutant C and native ferritin have limited Cu2+ binding capacities. To further confirm the highly specific binding of Cu2+ to the re-engineered mutants A and B over other divalent transition metals, a competitive binding assay was conducted. In the presence of 1 mM of cold divalent metal ions in solution, the 64Cu2+ incorporation ratio remained unchanged in mutants A and B, whereas mutant C and native ferritin had significant 64Cu2+ replacement by other divalent metals at 0.1 mM concentration.
Cellular interaction of the interface re-engineered ferritin nanocage and cytotoxicity of DOX@mutant B
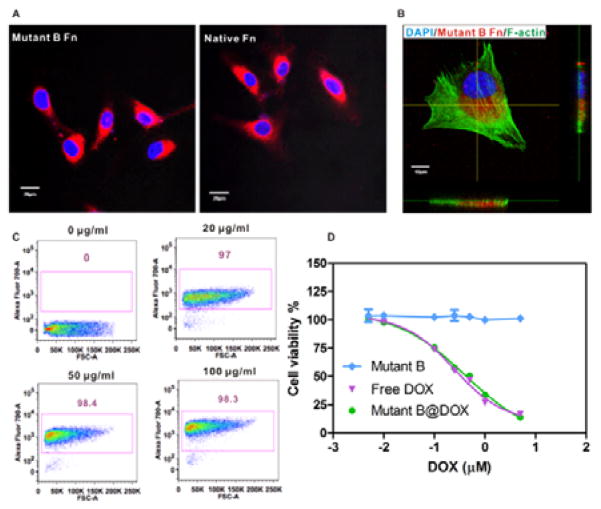
The improved physicochemical features of mutant B over the native ferritin and mutant A in terms of high structural stability and Cu2+ binding affinity make it an optimal nanoprobe for highly sensitive PET imaging. We thus used mutant B as a quintessential example to investigate its interaction with cancer cells. It was reported that ferritins are highly favorable to interact with various cancer cells and enter into the cytosol through transferrin receptor mediated binding and internalization.14, 29 Confocal laser scanning microscopy (CLSM) imaging results showed that various cancer cells readily take up mutant B (labeled with Cy5.5 on protein surface) into the cytoplasm after 4 h incubation (Figure S6). In our subsequent studies, we selected a malignant glioblastoma cell line, U87MG, as the cancer cell model for in vitro and in vivo studies. The CLSM imaging results indicated that mutant B and native ferritin had similar cellular uptake behavior (Figure 3A). This is a critical indication that the interface re-engineering of subunits does not markedly alter surface properties of ferritin nanocages in their interaction with cancer cell surface receptors. The 3D CLSM imaging analysis showed that the mutant B was only distributed in the cytoplasm (Figure 3B) with extremely high uptake rate (>97%), independent of protein concentrations (Figure 3C). The cytotoxicity of mutant B@DOX was further investigated via MTT assay. The mutant B@DOX was incubated with U87MG cells for 48 h, and free DOX was used as a control. As given in Figure 3D, the half maximal inhibitory concentration (IC50) of mutant B@DOX (0.44 μM) is comparable to that of free DOX (0.37 μM). Importantly, the mutant B nanocage alone shows no disturbance on cancer cell viability (Figure 3D and S7). These data verified that the mutant B@DOX can be used as a promising candidate for cancer therapy.
Figure 3.
(A) Cancer cell (U87MG) uptake of ferritin variants labeled with Cy5.5 fluorophore. Scale bar = 20 μm (B) Magnified confocal laser scanning imaging (CLSM) (Z-section) of intracellular uptake of mutant B ferritin after 2 h incubation. Scale bar = 10 μm. Blue: nucleus counterstained with DAPI; Red: Cy5.5 labeled mutant B ferritin; Green: F-actin stained with Alexa488 conjugated phalloidin. (C) Flow cytometry analysis of cellular uptake of mutant B ferritin at different concentrations. (D) MTT assay of mutant B ferritin with cancer cells (n = 6).
In vivo distribution of the interface re-engineered ferritin nanocage
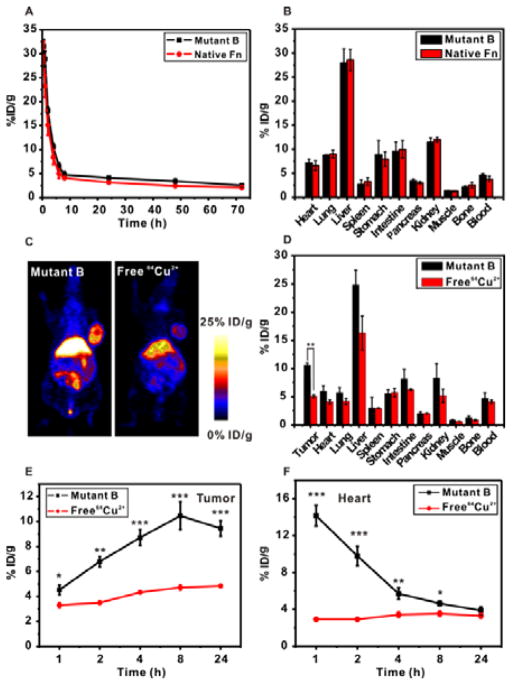
The pharmacokinetics (PK) and distribution of nanomaterials in living animals are two key factors determining their suitability as probes for molecular imaging or as delivery vehicles for effective drug delivery.39 We investigated the biodistribution and blood circulation of the re-engineered ferritin nanocage (mutant B) in comparison with the native ferritin in healthy mice. Our results showed that mutant B had very similar PK and distribution behaviors as those of the native ferritin (Figure 4A and B and Table 2), suggesting that the interface re-engineering strategy on ferritin subunit retained the original surface properties of the self-assembled protein in the blood. Most nanomaterials rely heavily on surface modification with poly(ethylene glycol) (PEG) to prolong circulation and minimize macrophage uptake in vivo. For example, bacteriophage MS2 capsids underwent extensive external PEGylation to obtain long blood circulation time along with chemical modification of the capsid interior with chelators to allow for PET imaging.40 The modification of protein cage interior and exterior surfaces fundamentally changes the surface property of the original protein, and has the risk of compromised protein assembly integrity. On top of that, the intensive chemical modification usually requires additional purification steps, which seriously hampers the protein yield and quality. In sharp contrast, our re-engineering method avoids surface PEGylation, while retains the nanocage for comparable blood circulation time (t1/2 = 72 h for mutant B) as that of the original ferritin. Meanwhile, the re-engineered protein nanocage (mutant B) stably sequesters 64Cu2+ within the structure for PET imaging.
Figure 4.
(A) Pharmacokinetics of 64Cu coordinated mutant B ferritin and native ferritin in healthy balb/c mice over 72 h (n = 3/group). (B) Biodistribution of 64Cu coordinated mutant B ferritin and native ferritin in different major organs in healthy balb/c mice at 24 h postinjection (n = 3/group). (C) PET images of 64Cu coordinated mutant B ferritin (left panel) and free 64Cu2+ (right panel) in U87MG tumor mice. (D) Biodistribution of 64Cu coordinated mutant B ferritin and free 64Cu2+ in U87MG tumor mice at 24 h postinjection (n = 3/group). **P<0.01. (E) Quantitative analysis of tumor uptake of 64Cu coordinated mutant B ferritin and free 64Cu2+ at different time points (n = 3/group). *P<0.05; **P<0.01; ***P<0.001. (F) Quantitative analysis of signals from the heart of 64Cu coordinated mutant B ferritin and free 64Cu2+ in U87MG tumor mice at different time points (n = 3/group). *P<0.05; **P<0.01; ***P<0.001.
Table 2.
Pharmacokinetics of 64Cu coordinated mutant B and native ferritin nanoprobes in healthy balb/c mice (n=3/group)
| Group | AUC Infusion (%ID/mL·hr) | AUC 0.72 (%ID/mL·hr) | CL (mL/hr) | T1/2 (hr) | V_SS (mL) |
|---|---|---|---|---|---|
| Mutant | 648.21328 | 367.30096 | 0.154315197 | 72.2 | 13.97749 |
| WT | 556.08956 | 292.65964 | 0.179831144 | 81.91 | 18.47092 |
It was found that free 64Cu2+ ions are likely to accumulate in the liver and bladder.40 In our research, we found that free 64Cu2+ also has affinity to tumor associated tissues (Figure 4C).41 Nevertheless, 64Cu2+ labeled re-engineered ferritin nanocage (mutant B) showed strikingly 2.5-fold higher tumor retention than the free 64Cu2+ after 8 h, and remained 2-fold higher after 24 h (Figure 4C–E). In addition, the free 64Cu2+ remained at a constant level in the circulation, while the mutant B coordinated 64Cu2+ presented a typical fast elimination in the first 4 h, followed by a gradual excretion in the next 20 h (Figure 4F). The mutant B coordinated 64Cu2+ and free 64Cu2+ demonstrate drastically different PK and distribution behavior. We conclude that the PET results from 64Cu2+ represent the nanoprobe behavior with minimal, if any, free 64Cu2+ interference. We have confirmed that the coordination of 64Cu2+ with mutant B is in a chemically stable status, 64Cu2+ labeled re-engineered ferritin nanocage is thus a robust nanoprobe for PET imaging.
PET imaging of tumor using interface re-engineered ferritin nanoprobe
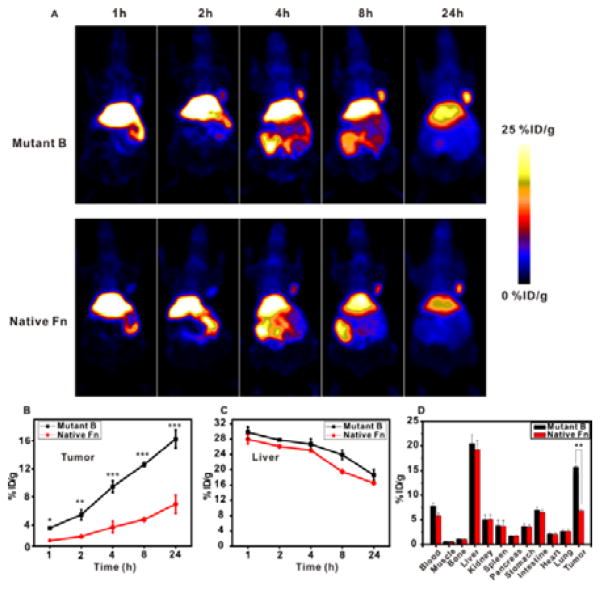
We used the human glioblastoma cell line, U87MG, as a tumor model to evaluate the new re-engineered ferritin nanoprobe. To inject an equivalent dose of radioactivity (150 μCi), 20 times lower amount of mutant B ferritin nanoprobe was needed relative to the native ferritin. The lower protein usage would minimize the immunogenicity and avoid non-specific distribution in animals. Interestingly, 64Cu2+ labeled mutant B resulted in consistently higher tumor uptake than that of native ferritin over 24 h (Figure 5A). Quantitative analysis indicated that the interface re-engineered mutant B nanoprobe had 16 %ID/g at the tumor site after 24 h, while the native ferritin had only 5 %ID/g in the same time frame (Figure 5B). It is also of note that the observed 5 %ID/g tumor site radioactivity could be attributed collectively to the released free 64Cu2+ from the native ferritin nanocage and the loosely incorporated 64Cu2+ within the native ferritin. This possible heterogeneity incorporating free 64Cu2+ and coordinated 64Cu2+ makes it difficult for data interpretation. Meanwhile, the liver uptakes of 64Cu2+ are almost identical in permuted and native ferritins (Figure 5C). Therefore, the highly specific tumor accumulation in PET imaging was obtained by the re-engineered mutant B nanoprobe (Figure 5).
Figure 5.
(A) PET imaging of 64Cu coordinated mutant B ferritin (upper panel) and native ferritin (lower panel) of U87MG tumor mice at different time points. An equivalent of 150 μCi radioactivity was injected into each mouse. (B) Quantitative analysis of tumor accumulation of 64Cu coordinated mutant B and native ferritin nanoprobes at different time points (n = 3/group). (C) Quantitative analysis of liver retention of 64Cu coordinated mutant B and native ferritin nanoprobes at different time points (n = 3/group). (D) Biodistribution of 64Cu coordinated mutant B and native ferritin nanoprobes at 24 h postinjection (n = 3/group). **P<0.05.
Investigation of tumor treatment by mutant B@DOX in vivo
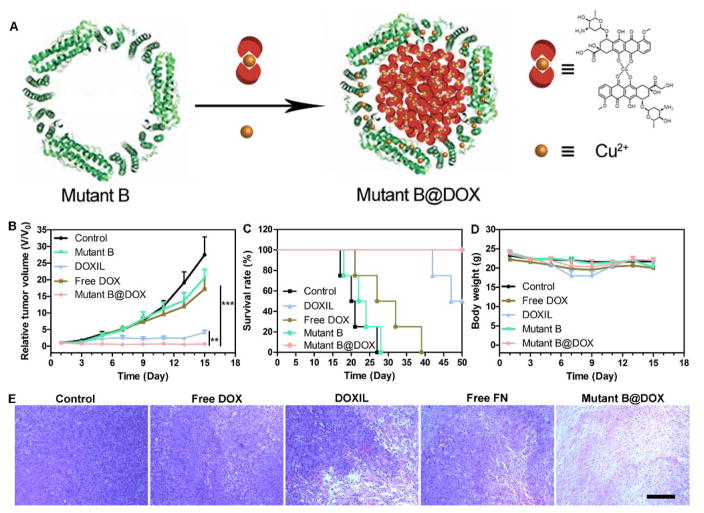
Encouraged by the promising MTT result in vitro, the antitumor ability of mutant B@DOX was further investigated in a U87MG xenograft tumor model in vivo. When the volume of the tumor reached 80 mm3, the tumor-bearing nude mice were divided into five groups randomly with the following treatment: mutant B@DOX (12.5 mg/kg of mutant B and 2 mg/kg of DOX), free DOX (2 mg/kg), DOXIL® (2 mg/kg), mutant B (12.5 mg/kg), and saline every other day for three times. As given in Figure 6A, both mutant B@DOX and DOXIL® groups were more efficacious than all the other groups. Interestingly, the mutant B@DOX group exhibited the best tumor inhibition, which may be due to the prolonged circulation time of mutant B in the blood (Figure 5). Moreover, the mutant B@DOX can promote the tumor mice survival rate (Figure 6B), and no mice died after 50 days. Conversely, all mice died within 28 days in the control group. The body weight was also measured from various groups. There was no obvious body weight loss during the whole period of experiments in the mutant B@DOX group (Figure 6C). The body weight of animals treated by DOXIL® and free DOX decreased slightly. In addition, hematoxylin-eosin (H&E) stained images of major organs were given in Figure S9. Compared with the control group, all the treatment groups showed no obvious systemic toxicity. The H&E stained images of tumor sections from different groups were shown in Figure 6D. Compared with all the other groups, greater tumor cell apoptosis and necrosis can be detected after the administration of mutant B@DOX. The above results demonstrated that the mutant B@DOX can suppress tumor growth effectively and prolong survival time of tumor bearing mice.
Figure 6.
(A) Schematic illustration of Mutant B@DOX preparation. Cu2+ was coordinated with DOX first, then the DOX-Cu2+ complexes were encapsulated into the Mutant B ferritin. (B) Tumor growth curves under various treatments. (C) Survival rates of tumor xenograft mice from different groups. (D) The body weight of mice from different groups. (E) H&E stained tumor histological sections from different groups on the 7th day. Scale bar = 200 μm. **P<0.05, ***P<0.01.
CONCLUSION
There have been prodigious efforts to control the self-assembly behaviors of proteins to design advanced protein architectures, or incorporate novel functions for various applications. The combination of divalent transition metal coordination with interface re-engineered mutant protein assembling subunits provides a simple and robust strategy to diversify the original protein structures. Importantly, the interface re-engineering approach simultaneously strengthens the protein ensemble’s structural stability, lowers the interfacial entropy, and expands its function. The widely used self-assembled ferritin nanocage coordinated with divalent transition metal ion, copper, was used as a prototypical example to demonstrate the advantages of the interface re-engineering strategy to enhance self-assembly protein stability and biofunctionality. Moreover, the copper ion can promote the DOX loading efficiency in the mutant ferritin. The as-prepared theranostic nanoplatform enhanced specific PET imaging of tumor and inhibited the tumor growth efficiently. We envision that this one-step labeling and DOX encapsulated re-engineering ferritin will provide a new approach to engineer existing proteins for nanomedicine applications.
MATERIALS AND METHODS
Reagents
All chemical reagents were purchased from Sigma-Aldrich. Primers were purchased from Integrated DNA Technologies. Deionized (DI) water with resistivity of 18.0 MΩ was obtained from a Millipore water purification system.
Cell lines
HT29, SKOV3, U87MG, A549, PC3, and MDA-MB-231 cell lines were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured in different media with 10% FBS (HT29 and SKOV3 cells were cultured in McCoy’s 5A media, U87MG and A549 cells were cultured in MEM media, PC3 cells were cultured in F-12 media, and MDA-MB-231 cells were cultured in L-15 media. BL21 (DE3) competent cells were bought from Thermo Fisher Scientific.
Experimental procedures
Preparation of ferritin and ferritin mutants
Specific amino acid mutation sites were introduced in a ferritin coding sequence. A total of three ferritin mutants were prepared. All the ferritin and ferritin mutants were expressed in a vector with a backbone of pRSFDuet-1. Construction of ferritin mutants was performed according to a step by step polymerase chain reaction (PCR) synthetic method. A total of 10 primers were used for mutant preparation (see the supporting information for primer details).
After five rounds of PCR, ferritin coding sequences were inserted into the vectors by NcoI and XhoI double digestion. After confirmation by sequencing, ferritin mutant plasmids were transformed into BL21 (DE3) cells. For ferritin expression, 1 L LB media containing ferritin E. coli was grown at 37 ºC until OD600 0.6 was attained, followed by induction with 1 mM IPTG at 37 ºC for 4 h. Bacteria were collected by centrifugation at 7000 × g for 10 min, followed by sonication in PBS. The supernatant was collected after centrifugation at 13000 × g for 10 min, and then incubated at 60 ºC with a water bath for 10 min. After centrifugation at 13000 × g for 10 min, the supernatant was purified with a HiLoad Superdex 200 PG (GE) column. The eluted proteins were used without further purification.
Characterization of ferritin and ferritin mutants
Transmission electron microscopy (TEM) images were obtained on a FEI Tecnai 12 (120 kV) transmission electron microscope. For TEM analysis, samples were prepared by coating 20 μL solution onto carbon coated copper grids, followed by 20 μL uranyl acetate (1%) negative staining.
Preparation of ferritin and ferritin mutants for metal ion binding
Ferritins used for metal ion binding were dialyzed into sodium acetate (50 mM pH 5.0) first. Then, 5 mM sodium dithionite and 1 mM 2, 2′-bipyridine were added to the ferritin solution to facilitate iron removal. After final dialysis into sodium acetate (50 mM pH 5.0), the proteins were ready for use.
Cytotoxicity Assay
Different cells lines (HT29, U87MG, NIH3T3, PC3 and HEK293) were used in the cytotoxicity assay. Cells were seeded into 96 well plates at density of 1×104 cells per well and cultured in their appropriate media. 24 h later, Different concentrations of ferritin (1, 10, 50, 80 and 100 μg/mL) were added into each well and incubated at 37 ºC for 24 h. After washing with PBS, 5 μ MTT (10 μg/mL) solution was added into each well and incubated for another 3 h. 100 μL DMSO was added into each well to dissolve the intracellular formazan crystals. A plate reader was used to measure the absorbance at 570 nm.
DOX loading procedure in mutant B
DOX (400 μL, 1 mg/mL) was incubated with CuCl2 (10 mg/mL) for 2 h, then mutant B (1 mL, 2 mg/mL) was added into the above solution for another 6 h. Next, the mutant B@DOX was purified by an Amersham Biosciences PD-10 desalting column to remove the free DOX and Cu2+. The content of DOX was quantified by HPLC at a wavelength of 480 nm. The DOX loading content was calculated by the formula: loading content (%) = DOX/(DOX+mutant B) × 100. The DOX encapsulation efficiency is 6.9 %, and the DOX loading content was 13.8 %.
Animal Models
All animal studies involved in this project were conducted according to a protocol approved by the National Institutes of Health Animal Care and Use Committee (NIH-ACUC). 6–7-week-old athymic nude mice purchased from Harlan were subcutaneously injected with 2×106 human glioblastoma U87MG cells on the right flank. The animals were ready for PET imaging studies and tumor therapy experiments when tumor size reached about 200 and 80 mm3, respectively.
64Cu2+ binding study
64Cu2+ (500 μCi) was added into different concentrations of ferritins (0.5, 5, 50 and 500 μg/mL) to evaluate the binding ability of ferritin and ferritin mutants. After 1 h incubation, instant thin layer chromatography (iTLC) was used to evaluate the labeling efficiency of different ferritins.
Metal ion competition study
After 1 h incubation of 64Cu2+ (500 μCi) and ferritins, different metals (Ca2+, Fe2+, Zn2+) of different concentrations (0.1 and 1 mM) were added into ferritin solutions (1 mg/mL). iTLC was used to test the binding between 64Cu2+ and ferritins.
MicroPET Imaging
U87MG tumor bearing mice were injected intravenously with ferritins (10 μg for native ferritin and 0.5 μg for mutant B) 150 μCi 64Cu2+. PET scanning and imaging analysis were carried out on a micro PET scanner (Siemens Inveon) at 1, 2, 4, 8, and 24 h post injection. 3-dimensional regions of interest (ROIs) were drawn on the organs and tumors with decay-corrected whole body coronal images. The percentage injected dose per gram (%ID/g) was then calculated according to the ROI readings.
Biodistribution
Mice were sacrificed after the 24 h PET scanning. Organs of interest and tumors were collected and weighed. The radioactivity was measured with a gamma counter (Beckman 8000). The uptake of 64Cu was calculated in %ID/g according to the prepared standards.
Pharmacokinetics study
300 μCi 64Cu2+ -ferritin complex was injected into mice through the tail vein. Mice blood samples were collected at different time points (0.5, 1, 2, 4, 6, 8, 24, 48 and 72 h) after injection. Blood samples were weighed and radioactivity was measured with a gamma counter. The %ID/g was also calculated for comparison.
Tumor therapy study
The U87MG tumor-bearing mice were divided into five groups. The in vivo treatment studies were performed by tail vein injection with PBS, DOX (2 mg/kg), Doxil® (2 mg/kg), mutant B (12.5 mg/kg), and mutant B@DOX (12.5 mg/kg of mutant B and 2 mg/kg of DOX) every other day for three times. The body weight and tumor size were monitored every other day. The tumor volumes were calculated by the formula: V=length (mm) × width (mm)2/2.
Supplementary Material
Acknowledgments
This work was supported by the Major State Basic Research Development Program of China (2017YFA0205201, 2014CB744503, and 2013CB733802), the National Natural Science Foundation of China (NSFC) (81422023, 51273165, 81371596, and U1505221), and the intramural research program (IRP), National Institutes of Health (NIH).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Conflict of Interest
The authors declare no competing financial interest.
References
- 1.Fegan A, White B, Carlson JC, Wagner CR. Chem Rev. 2010;110:3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, Thornton JM. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Zhang F, Wang Z, Liu Y, Fu X, Jin A, Yung BC, Chen W, Fan J, Yang X, Niu G, Chen X. J Am Chem Soc. 2016;138:15027–15034. doi: 10.1021/jacs.6b09014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsia Y, Bale JB, Gonen S, Shi D, Sheffler W, Fong KK, Nattermann U, Xu C, Huang PS, Ravichandran R, Yi S, Davis TN, Gonen T, King NP, Baker D. Nature. 2016;535:136–139. doi: 10.1038/nature18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zandi R, Reguera D, Bruinsma RF, Gelbart WM, Rudnick J. Proc Natl Acad Sci U S A. 2004;101:15556–15560. doi: 10.1073/pnas.0405844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe S, Hirata K, Ueno T, Morino K, Shimizu N, Yamamoto M, Takata M, Yashima E, Watanabe Y. J Am Chem Soc. 2009;131:6958–6960. doi: 10.1021/ja901234j. [DOI] [PubMed] [Google Scholar]
- 7.Uchida M, Flenniken ML, Allen M, Willits DA, Crowley BE, Brumfield S, Willis AF, Jackiw L, Jutila M, Young MJ, Douglas T. J Am Chem Soc. 2006;128:16626–16633. doi: 10.1021/ja0655690. [DOI] [PubMed] [Google Scholar]
- 8.Comellas-Aragones M, Engelkamp H, Claessen VI, Sommerdijk NA, Rowan AE, Christianen PC, Maan JC, Verduin BJ, Cornelissen JJ, Nolte RJ. Nat Nanotechnol. 2007;2:635–639. doi: 10.1038/nnano.2007.299. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Huang P, Jacobson O, Wang Z, Liu Y, Lin L, Lin J, Lu N, Zhang H, Tian R, Niu G, Liu G, Chen X. ACS Nano. 2016;10:3453–3460. doi: 10.1021/acsnano.5b07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin X, Xie J, Zhu L, Lee S, Niu G, Ma Y, Kim K, Chen X. Angew Chem Int Ed. 2011;50:1569–1572. doi: 10.1002/anie.201006757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher JM, Harniman RL, Barnes FR, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, Miles MJ, Sessions RB, Verkade P, Woolfson DN. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Worsdorfer B, Woycechowsky KJ, Hilvert D. Science. 2011;331:589–592. doi: 10.1126/science.1199081. [DOI] [PubMed] [Google Scholar]
- 13.Liang M, Fan K, Zhou M, Duan D, Zheng J, Yang D, Feng J, Yan X. Proc Natl Acad Sci U S A. 2014;111:14900–14905. doi: 10.1073/pnas.1407808111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan K, Cao C, Pan Y, Lu D, Yang D, Feng J, Song L, Liang M, Yan X. Nat Nanotechnol. 2012;7:459–464. doi: 10.1038/nnano.2012.90. [DOI] [PubMed] [Google Scholar]
- 15.Uchida M, Kosuge H, Terashima M, Willits DA, Liepold LO, Young MJ, McConnell MV, Douglas T. ACS Nano. 2011;5:2493–2502. doi: 10.1021/nn102863y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raja KS, Wang Q, Gonzalez MJ, Manchester M, Johnson JE, Finn MG. Biomacromolecules. 2003;4:472–476. doi: 10.1021/bm025740+. [DOI] [PubMed] [Google Scholar]
- 17.Nam YS, Park H, Magyar AP, Yun DS, Pollom TS, Jr, Belcher AM. Nanoscale. 2012;4:3405–3409. doi: 10.1039/c2nr30115f. [DOI] [PubMed] [Google Scholar]
- 18.Maity B, Fujita K, Ueno T. Curr Opin Chem Biol. 2015;25:88–97. doi: 10.1016/j.cbpa.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 19.Huard DJ, Kane KM, Tezcan FA. Nat Chem Biol. 2013;9:169–176. doi: 10.1038/nchembio.1163. [DOI] [PubMed] [Google Scholar]
- 20.Dalmau M, Lim S, Wang SW. Nano Lett. 2009;9:160–166. doi: 10.1021/nl8027069. [DOI] [PubMed] [Google Scholar]
- 21.Salgado EN, Brodin JD, To MM, Tezcan FA. Inorg Chem. 2011;50:6323–6329. doi: 10.1021/ic200746m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salgado EN, Lewis RA, Faraone-Mennella J, Tezcan FA. J Am Chem Soc. 2008;130:6082–6084. doi: 10.1021/ja8012177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. J Mol Biol. 2002;321:923–938. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 24.Lai YT, Reading E, Hura GL, Tsai KL, Laganowsky A, Asturias FJ, Tainer JA, Robinson CV, Yeates TO. Nat Chem. 2014;6:1065–1071. doi: 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jockusch RA, Lemoff AS, Williams ER. J Am Chem Soc. 2001;123:12255–12265. doi: 10.1021/ja0106873. [DOI] [PubMed] [Google Scholar]
- 26.Zhen Z, Tang W, Chen H, Lin X, Todd T, Wang G, Cowger T, Chen X, Xie J. ACS Nano. 2013;7:4830–4837. doi: 10.1021/nn305791q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai X, Luo Y, Zhang W, Du D, Lin Y. ACS Appl Mater Interfaces. 2016;8:22442–22450. doi: 10.1021/acsami.6b04933. [DOI] [PubMed] [Google Scholar]
- 28.Jabłońska-Trypuć A, Świderski G, Krętowski R, Lewandowski W. Molecules. 2017;22:1106. doi: 10.3390/molecules22071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Fang CJ, Ryan JC, Niemi EC, Lebron JA, Bjorkman PJ, Arase H, Torti FM, Torti SV, Nakamura MC, Seaman WE. Proc Natl Acad Sci U S A. 2010;107:3505–3510. doi: 10.1073/pnas.0913192107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Liang M, Li X, Fan K, Xiao J, Li Y, Shi H, Wang F, Choi HS, Cheng D, Yan X. ACS Nano. 2016;10:4184–4191. doi: 10.1021/acsnano.5b07408. [DOI] [PubMed] [Google Scholar]
- 31.Lee EJ, Ahn KY, Lee JH, Park JS, Song JA, Sim SJ, Lee EB, Cha YJ, Lee J. Adv Mater. 2012;24:4739–4744. 4730. doi: 10.1002/adma.201200728. [DOI] [PubMed] [Google Scholar]
- 32.Lawson DM, Artymiuk PJ, Yewdall SJ, Smith JM, Livingstone JC, Treffry A, Luzzago A, Levi S, Arosio P, Cesareni G, et al. Nature. 1991;349:541–544. doi: 10.1038/349541a0. [DOI] [PubMed] [Google Scholar]
- 33.Salgado EN, Lewis RA, Mossin S, Rheingold AL, Tezcan FA. Inorg Chem. 2009;48:2726–2728. doi: 10.1021/ic9001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Orner BP. Int J Mol Sci. 2011;12:5406–5421. doi: 10.3390/ijms12085406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanekiyo M, Wei CJ, Yassine HM, McTamney PM, Boyington JC, Whittle JR, Rao SS, Kong WP, Wang L, Nabel GJ. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ke Z, Abe S, Ueno T, Morokuma K. J Am Chem Soc. 2012;134:15418–15429. doi: 10.1021/ja305453w. [DOI] [PubMed] [Google Scholar]
- 37.Boyd D, Vecoli C, Belcher DM, Jain SK, Drysdale JW. J Biol Chem. 1985;260:11755–11761. [PubMed] [Google Scholar]
- 38.Boija S, Almesaker A, Hedenstrom E, Bylund D, Edlund H, Norgren M. J Mass Spectrom. 2014;49:550–556. doi: 10.1002/jms.3372. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Niu G, Chen X. Pharm Res. 2014;31:1358–1376. doi: 10.1007/s11095-013-1103-7. [DOI] [PubMed] [Google Scholar]
- 40.Farkas ME, Aanei IL, Behrens CR, Tong GJ, Murphy ST, O’Neil JP, Francis MB. Mol Pharm. 2013;10:69–76. doi: 10.1021/mp3003754. [DOI] [PubMed] [Google Scholar]
- 41.Guo W, Sun X, Jacobson O, Yan X, Min K, Srivatsan A, Niu G, Kiesewetter DO, Chang J, Chen X. ACS Nano. 2015;9:488–495. doi: 10.1021/nn505660r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.