Summary
The early detection of relapse following primary surgery for non-small cell lung cancer and the characterization of emerging subclones seeding metastatic sites might offer new therapeutic approaches to limit tumor recurrence. The potential to non-invasively track tumor evolutionary dynamics in ctDNA of early-stage lung cancer is not established. Here we conduct a tumour-specific phylogenetic approach to ctDNA profiling in the first 100 TRACERx (TRAcking non-small cell lung Cancer Evolution through therapy (Rx)) study participants, including one patient co-recruited to the PEACE (Posthumous Evaluation of Advanced Cancer Environment) post-mortem study. We identify independent predictors of ctDNA release and perform tumor volume limit of detection analyses. Through blinded profiling of post-operative plasma, we observe evidence of adjuvant chemotherapy resistance and identify patients destined to experience recurrence of their lung cancer. Finally, we show that phylogenetic ctDNA profiling tracks the subclonal nature of lung cancer relapse and metastases, providing a new approach for ctDNA driven therapeutic studies
Lung cancer is the leading cause of cancer death1–2. Metastatic non-small cell lung cancer (NSCLC) cannot be cured with systemic chemotherapy, yet clinical studies have shown a 5% benefit of post-operative (adjuvant) chemotherapy on overall survival3. This modest survival benefit may reflect a vulnerability of low volume disease within the context of reduced intratumor heterogeneity4. Circulating tumor DNA (ctDNA) detection in plasma following resection of breast5,6 and colorectal7 tumors has been shown to identify patients destined to relapse post-operatively in advance of established clinical parameters. Identifying, monitoring and genomically characterizing residual disease following primary lung cancer surgery may improve outcomes in the adjuvant setting. This would create a therapeutic setting where only patients destined to recur would receive treatment and where intervention could be directed to the evolving tumor subclone that is seeding metastatic recurrence.
Here, we report a bespoke multiplex-PCR NGS approach to ctDNA profiling within the context of the prospective tumor evolutionary NSCLC TRACERx study. We address determinants of ctDNA detection in early-stage NSCLC and investigate the ability of ctDNA to identify and genomically characterize post-operative NSCLC relapse within a tumor phylogenetic framework.
Phylogenetic ctDNA profiling
The TRACERx study monitors the clonal evolution of NSCLC from diagnosis through to death8,9. Using multi-region exome sequencing (M-Seq) derived tumor phylogenetic trees developed through prospective analysis of a 100 patient TRACERx cohort, we conducted a phylogenetic approach to ctDNA profiling in early-stage NSCLC (Fig. 1). Bespoke multiplex-PCR assay-panels were synthesised for each patient, targeting clonal and subclonal single nucleotide variants (SNVs) selected to track phylogenetic tumor branches in plasma (Fig. 1). SNV detection in plasma was established through a calling algorithm employing negative control samples (see Methods). Analytical validation of the multiplex-PCR NGS platform demonstrated a sensitivity of above 99% for the detection of SNVs at frequencies above 0.1% and the specificity of detecting a single SNV was 99.6% (Extended Data Fig. 1a). At least two SNVs were detected in ctDNA from early-stage NSCLCs analyzed in our published discovery cohort data10, demonstrating biological sensitivity of a two SNV threshold for ctDNA detection. Therefore, we prospectively selected a threshold of two detected SNVs for calling a sample ctDNA positive for validation within this study; to minimize type I error when testing up to 30 tumour-specific SNVs per time-point in a single patient (see Extended Data Fig. 1b for justification).
Figure 1. Phylogenetic ctDNA tracking.
Overview of the study methodology. Multi-region sequencing of NSCLC was performed as part of the TRACERx study. PCR assay-panels were designed based on phylogenetic analysis, targeting clonal and subclonal single nucleotide variants to facilitate non-invasive tracking of the patient-specific tumor phylogeny. Assay-panels were combined into multiplex assay-pools containing primers from up to 10 patients. Cell-free DNA was extracted from pre- and post-operative plasma samples and multiplex-PCR performed, followed by sequencing of amplicons. Findings were integrated with M-Seq exome data to track tumor evolution.
Determinants of ctDNA detection in NSCLC
We sought to identify clinicopathological determinants of ctDNA detection in early-stage NSCLC by profiling pre-operative plasma samples in 100 TRACERx patients. Samples from four patients could not be analyzed (see cohort design Extended Data Fig. 2a, patient characteristics Extended Table 1a-c, Supplementary Table 1). Individual assay-panels were designed to target a median of 18 SNVs (range 10 to 22) comprising a median of 11 clonal SNVs (range 2 to 20) and a median of 6 subclonal SNVs (range 0 to 16) per patient (Extended Data Fig. 2b,e).
At least two SNVs were detected in ctDNA pre-operatively in 46 of 96 (48%) early-stage NSCLCs and a single SNV was detected in 12 additional cases (Fig. 2a). Centrally reviewed pathological data revealed that ctDNA detection was associated with histological subtype: 97% (30/31) of lung squamous cell carcinomas (LUSCs) and 71% (5 of 7) of other NSCLC subtypes were ctDNA positive, compared with 19% (11/58) of lung adenocarcinomas (LUADs) (Fig. 2a). 94% (16 of 17) of stage I LUSCs were detected compared with 13% (5 of 39) of stage I LUADs (Extended Data Fig. 3a). Passive release of ctDNA into the circulation may be associated with necrosis11. As expected LUSCs were significantly more necrotic than LUADs12 and ctDNA positive LUADs formed a sub-group of more necrotic tumors compared with ctDNA negative LUADs (Extended Data Fig. 3b). Necrosis, lymph node involvement, lymphovascular invasion, pathological tumor size, Ki67 labelling indices, non-adenocarcinoma histology and total cell-free DNA input predicted ctDNA detection in univariable analyses (Extended Data Fig. 3c). Multivariable analysis revealed non-adenocarcinoma histology, the presence of lympho-vascular invasion and high Ki67 proliferation index as independent predictors of ctDNA detection (Extended Data Fig. 3c). Since FDG-avidity on positron emission tomography (PET) scans correlates with proliferative indices in early-stage NSCLC13,14, we investigated tumor PET FDG-avidity and ctDNA detection. PET FDG-avidity predicted ctDNA detection (area under curve = 0.84, P<0.001, n=92) (Extended Data Fig. 3d). Within LUADs, driver events in KRAS, EGFR or TP53 were not associated with ctDNA detection (Extended Data Fig. 3e).
Figure 2. Clinicopathological predictors of ctDNA detection.
a) Heatmap showing clinicopathological and ctDNA detection data, continuous variables quartiled. Raw data and patient IDs in attached worksheet. b) Detection of clonal and subclonal single nucleotide variants within 46 patients with two or more single nucleotide variants detected in plasma. Histology indicated in panels as LUSC, LUAD and Other.
We analyzed the distribution of clonal and subclonal SNVs in ctDNA positive patients. Clonal SNVs were detected in all 46 ctDNA positive patients and a median of 94% (range 11% to 100%) of clonal SNVs targeted by assay-panels were detected in the ctDNA of these patients. 40 of 46 ctDNA positive patients had subclonal SNVs targeted by assay-panels and subclonal SNVs were detected in 27 (68%) of these patients. A median of 27% (range 0% to 91%) of subclonal SNVs within individual assay-panels were detected in ctDNA positive patients (Fig. 2b). The mean plasma variant allele frequency (VAF) of clonal SNVs was higher than that of subclonal SNVs (Extended Data Fig. 4a, within patient comparison, Wilcoxon signed-rank test, P<0.001, n=27, Supplementary Table 2) supporting the use of clonal alterations as a more sensitive method of ctDNA detection than subclonal alterations10,15.
In ctDNA positive patients, pathologic tumor size correlated with mean clonal plasma VAF (Spearman’s Rho = 0.405, P=0.005, n=46, Extended Data Fig. 4b). CT scan volumetric analyses were evaluated in 37 of 46 ctDNA positive patients (see Extended Data Fig. 4c). Tumor volume correlated with mean clonal plasma VAF (Fig 3a, Spearman’s Rho = 0.63, P<0.001, n=37). A linear relationship between log- transformed volume and log- transformed mean clonal VAF values was observed (Fig. 3a). The line of best fit applied to the data was consistent with the line fitted to NSCLC volumetric data and ctDNA plasma VAFs reported in previously published work16 (Extended Data Fig. 4d). Linear modelling based on the TRACERx data predicted that a primary tumor burden of 10cm3 would result in a mean clonal plasma VAF of 0.1% (95% C.I. 0.06 to 0.18%) (Fig. 3b). Tumor purity was multiplied by tumor volume to control for stromal contamination to determine cancer cell volume corresponding to clonal plasma VAF (Extended Data Fig. 4e). On the assumption that 1cm3 of pure tumor contains 9.4 x 107 cells17, a plasma VAF of 0.1% would correspond to a primary NSCLC malignant burden of 302 million cells (Fig 3b, Extended Data Fig. 4f).
Figure 3. Tumor volume predicts plasma variant allele frequency.
a) Tumor volume (cm3) measured by CT volumetric analysis correlates with mean clonal plasma VAF, n=37, grey vertical lines represent range of clonal VAF, red shading indicates 95% confidence intervals. b) Predicted mean clonal VAF at hypothetical volumes ranging from 1 to 100cm3 based on model in panel a, predicted cancer cell number based on model in extended data 4e. c) Estimated effective subclone size, defined as mean CCF of subclone multiplied by tumor volume and purity, influences subclonal SNV detection. For negative calls, median effective subclone size was 1.70 cm3, range= 0.21-24.11, n=163, for positive calls, median effective subclone size = 4.06 cm3, range = 0.31 – 49.20, n=109. Wilcoxon rank sum test, P<0.001, data from 34 patients (passed volumetric filters with subclonal SNVs represented in assay-panel). d) Estimated effective subclone size correlates with subclonal plasma VAF, n=109 subclonal SNVs, data from 34 patients (passed volumetric filters with detected subclonal SNVs in plasma).
To investigate predictors of subclone detection, detected subclonal SNVs were mapped back to M-seq derived tumor phylogenetic trees. 35 of 57 (61%) shared subclones (identified in more than one tumor region through M-Seq analysis) were identified in ctDNA, compared with 26 of 80 (33%) private subclones (detected in a single tumor region only) (Extended Data Fig 4g). This suggested subclone volume influences subclonal ctDNA detection. Subclone volume was estimated based on mean regional subclone cancer cell fraction and cancer cell volume. Detected subclonal SNVs mapped to subclones with higher estimated volumes than subclones containing undetected SNVs (Fig. 3c) and subclone volume correlated with subclonal SNV plasma VAF (Fig. 3d).
Detecting and characterizing NSCLC relapse
The longitudinal phase of the study aimed to determine if ctDNA profiling with patient-specific assay panels could detect and characterize the branched subclone(s) seeding NSCLC relapse. Pre- and post-surgical plasma ctDNA profiling was performed blinded to relapse status in a sub-group of 24 patients (cohort characteristics, Extended Table 1d-e). This included relapse free patients who had been followed-up for a median of 775 days (range 688 to 945 days, n=10) and confirmed NSCLC relapse cases (n=14) (cohort design, Extended Data Fig. 2c). Additional PCR assays were added to panels in this phase of the study to attempt to improve ctDNA detection in LUADs, a median of 18.5 SNVs (range 12 to 20) were targeted by LUSC assay-panels and a median of 28 SNVs (range 25 to 30) were targeted by LUAD assay-panels (Extended Data Fig. 2d-e).
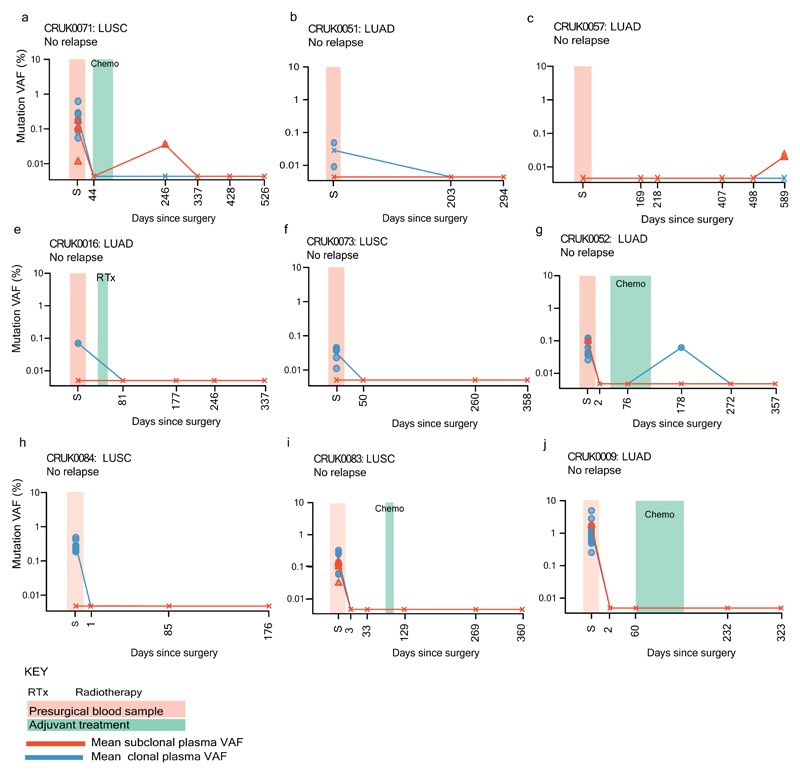
Patients were followed up with three to six monthly clinical assessment and chest radiographs. At least 2 SNVs were detected in 13 of 14 (93%) patients with confirmed NSCLC relapse prior to, or at, clinical relapse (Fig 4a-g, Extended Data Fig. 5). At least two SNVs were detected in 1 of 10 (10%) patients (CRUK0013) with no clinical evidence of NSCLC relapse (Fig. 4h, Extended Data Fig. 6). Excluding a single case where no post-operative plasma was taken prior to clinical relapse (CRUK0041), the median interval between ctDNA detection and NSCLC relapse confirmed on clinically indicated CT imaging (lead-time) was 70 days (range 10 to 346 days). Four of 13 relapse cases exhibited lead-times of more than six months (Fig. 4a-d). In two cases ctDNA detection preceded CT imaging inconclusive for NSCLC relapse by 157 days (CRUK0004, Fig 4b) and 163 days (CRUK0045, Fig 4d). ctDNA profiling reflected adjuvant chemotherapy resistance - CRUK0080, CRUK0004 and CRUK0062 had detectable ctDNA in plasma within 30 days of surgery. The number of detectable SNVs increased in all three cases despite adjuvant chemotherapy, with disease recurring within 1 year of surgery (Fig. 4a-c). In contrast, CRUK0013 had 20 SNVs detectable in ctDNA 72 hours after surgery and 13 SNVs detectable prior to adjuvant chemotherapy; 51 days following completion of adjuvant treatment and at post-operative days 457 and 667 no SNVs were detectable and the patient remains relapse free 688 days post-surgery (Fig. 4h). ctDNA profiling detected intracerebral relapse; CRUK0029 had a PET scan performed 50 days prior to surgery demonstrating normal cerebral appearances. ctDNA remained detectable following surgery, 54 days post-operatively the patient was diagnosed with intracerebral metastasis, no extracranial disease was evident on CT imaging (Fig. 4e).
Figure 4. Post-operative ctDNA detection predicts and characterizes NSCLC relapse.
a-h) Longitudinal cell-free DNA profiling. Circulating tumor DNA (ctDNA) detection in plasma was defined as the detection of two tumor-specific SNVs. Detected clonal (circles, light blue) and subclonal (triangles, colors indicates different subclones) SNVs from each patient-specific assay-panel are plotted on graphs colored by M-Seq derived tumor phylogenetic nodes. Mean clonal (blue) and mean subclonal (red) plasma VAF are indicated on graphs as connected lines. Pre-operative and relapse M-Seq derived phylogenetic trees represented by ctDNA are illustrated above each graph.
We sought to resolve subclonal evolutionary-dynamics associated with NSCLC relapse. Subclonal SNVs displaying plasma VAFs similar to clonal SNVs from clusters confined to a single phylogenetic branch, were detected post-operatively in the ctDNA of four patients who suffered NSCLC relapse (CRUK0004, CRUK0063, CRUK0065 and CRUK0044) (Fig. 4b,f-g, Extended Data Fig 5b). This suggested a relapse process dominated by one subclone represented in our assay-panel. The subclone implicated by ctDNA as driving the relapse in the case of CRUK0004 contained an ERRB2 (HER2) amplification event (>15 copies, triploid background), that may be targetable in NSCLC18 (Fig. 4b). Relapses involving subclones from more than one phylogenetic branch were evident in patients CRUK0080, CRUK0062 (Fig. 4a,c) and CRUK0041 (Extended Data Fig 5c).
Validation of phylogenetic characterization
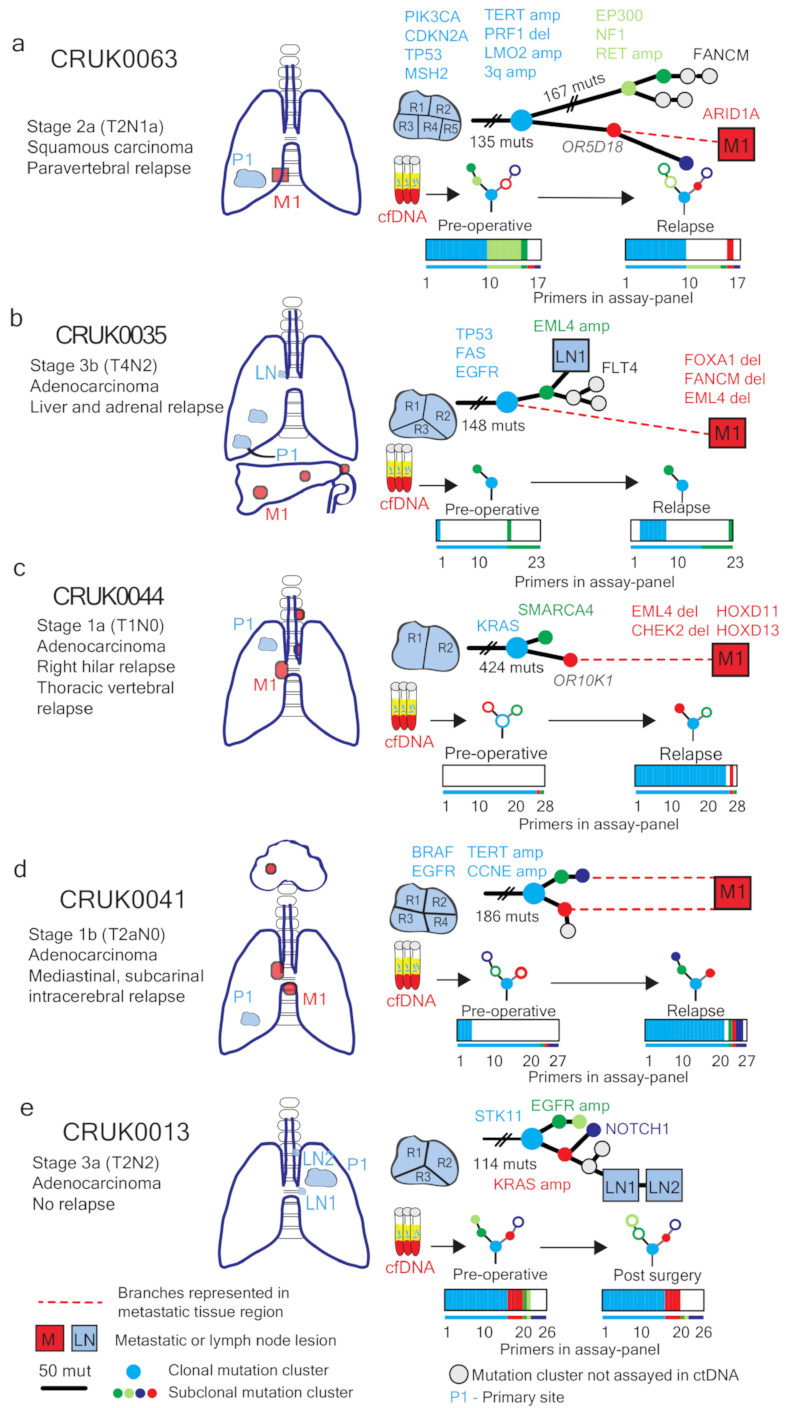
To validate subclonal ctDNA analyses, data acquired from sequencing metastatic tissue at recurrence was integrated with M-seq primary tumor data (for biopsy details, Supplementary Table 3). Patient CRUK0063 suffered para-vertebral relapse of their NSCLC. Post-operative ctDNA analysis revealed the detection of the same subclonal SNV (OR5D18) on four consecutive occasions over a 231-day period (Extended Data Fig. 7a). The OR5D18 SNV traced back to a subclonal cluster private to primary tumor region three (Fig. 5a). CT-guided biopsy tissue was acquired from the para-vertebral metastasis at post-operative day 467. Exome sequencing of relapse tissue revealed the subclonal cluster containing the OR5D18 SNV gave rise to the metastatic subclone (Fig. 5a), this supported ctDNA phylogenetic characterization of relapse. The para-vertebral biopsy contained 88 SNVs not called as present in the primary tumor including an ARID1A stop-gain driver SNV. Re-examination of primary tumor region M-Seq data with a lower SNV calling threshold revealed that 16 of 88 SNVs, including ARID1A, were detectable in primary tumor region three, compared to a maximum of 2 of 88 in other tumor regions (Extended Data Fig. 7b). These data suggest that ctDNA profiling can resolve the primary tumor region from which a low frequency metastatic subclone derives. CRUK0035 developed two liver and one adrenal metastases (Fig. 5b). Sequencing of the metastatic liver deposit revealed that only 109 of 149 SNVs classed as clonal in the primary tumor were detectable in the metastasis. This was suggestive of an ancestral branching event not resolved through primary M-seq analysis (Fig 5b). Post-operative ctDNA profiling identified clonal SNVs present in the liver metastasis biopsy but also revealed SNVs representing a subclone from the primary tumor (Extended Data Fig 7c). This subclone was not present in the metastatic liver deposit (Fig 5b). These data may reflect ctDNA identified from the non-biopsied metastases suggesting multiple metastatic events. CRUK0044 suffered a vertebral and right hilar relapse. Post-operatively the same subclonal SNV (OR10K1), was detected in ctDNA on two occasions 85 days apart (Extended Data Fig. 7d). This SNV was represented in a single subclone detected through sequencing hilar lymph-node metastatic tissue, supporting ctDNA findings (Fig. 5c). CRUK0041 suffered an intracerebral, hilar and subcarinal lymph node relapse. Four subclonal SNVs representing both branches of the tumor phylogenetic tree were detectable in ctDNA at relapse. Consistent with these data, sequencing of subcarinal metastatic tissue revealed the presence of subclonal SNVs mapping to both phylogenetic branches (Fig 5d, Extended Data Fig. 7e). Patient CRUK0013 had detectable ctDNA 3 and 38 days post-operatively. Following adjuvant chemoradiotherapy for lymph-node metastases identified in the pathological specimen, ctDNA levels became undetectable (Fig 4h). Two involved lymph-nodes were sampled for exome analysis together with M-seq of the primary tumor. Four subclonal SNVs detected in ctDNA post-operatively mapped to an ancestral subclone (describing a subclone that existed during the tumor’s evolution) (Extended Data Fig. 7f). This ancestral subclone contained a KRAS amplification (>15 copies, triploid background) and was identified as present in primary tumor and sampled lymph-nodes by M-Seq (Fig. 5e). These data provide phylogenetic characterization of post-operative residual disease that responded to adjuvant chemoradiotherapy (Fig. 4h).
Figure 5. Phylogenetic trees incorporating relapse tissue sequencing data.
Phylogenetic trees based on mutations found in primary and metastatic tissue (a-d), or primary tumor and lymph node biopsies (e). Colored nodes in phylogenetic trees indicate cancer clones harboring mutations assayed for in ctDNA, grey indicates a clone not assayed. Branch length is proportional to number of mutations unless crossed. Dashed red lines show branches leading to metastatic relapse. Colored bars below show the number of assays per sample detected preoperatively and at relapse (a-d) or in the absence of relapse, post-surgery (e). Thin colored bar shows number of assays in total. Colors match clones on the phylogenetic trees.
ctDNA profiling in the metastatic setting
Patient CRUK0063 underwent examination through the PEACE post-mortem study 24 hours following death. M-Seq data from the six post-mortem tumor regions (para-aortic, para-vertebral and lung metastases, day 857), the para-vertebral relapse biopsy (day 467) and five primary tumor regions (day 0) were combined to infer the phylogenetic structure of this patient’s NSCLC (Fig. 6a). All seven metastatic tumor regions arose from a single ancestral subclone represented by phylogenetic cluster 8. Six metastatic regions shared a later phylogenetic origin, cluster 12 (Fig. 6b). The single tumor region not containing phylogenetic cluster 12 was sampled from the para-aortic metastasis at autopsy and contained a private subclone represented by phylogenetic cluster 9 (Fig. 6b).
Figure 6. ctDNA tracking of lethal cancer subclones in CRUK0063.
Phylogenetic analysis of one relapse biopsy (day 467) and five metastatic biopsies (post mortem) a) To-scale phylogenetic tree of CRUK0063 including M-seq based on metastatic and primary tumor regions. Branch length is proportional to number of mutations in each subclone. b) Phylogenetic trees matching metastatic lesions, colored nodes represent mutation clusters found at each site and assayed for in ctDNA. Open circles represent mutation clusters not detected in ctDNA. c) Tracking plot showing mean VAF of identified mutation clusters in ctDNA. Size of dots indicates number of assays detected. Colors correspond to mutation clusters and match panels a) and b).
We designed a bespoke ctDNA assay-panel to retrospectively track metastatic subclonal burden. 20 clonal SNVs and a median of 8 subclonal SNVs (range 4 to 15) in each of 9 metastatic subclonal clusters were targeted by the assay-panel (Extended Data Fig. 8). Since 103 variants per time-point were profiled, SNV detection thresholds were increased to maintain platform specificity (see Methods). This resulted in a predicted false positive rate (FPR) of 0.0011 translating to a 10.7% risk of a single false-positive SNV at each time point and a 0.5% risk of 2 false-positive SNVs at each time point when testing 103 SNVs.
Two clonal SNVs were detected by the 103 SNV assay-panel at day 151 post-surgery (Fig 6c, Extended Data Fig. 8), 189 days prior to the time point ctDNA was detected using the 19 SNV assay-panel in Figure 4f. At day 242 a single subclonal SNV was detected from phylogenetic cluster 8 (Fig 6c, Extended Data Fig. 8), within the context of a 10.7% false-positive risk a single SNV call could represent type I error. At day 466, following clinical-relapse at the thoracic para-vertebral site, 18 of 20 SNVs mapping to phylogenetic clusters (8,11 and 12) were detected in ctDNA, these subclonal clusters were shared between six of seven metastatic sites (Fig 6b-c, Extended Data Fig. 8). Single SNVs from two private subclones (phylogenetic cluster 5 and 9) were also detectable in ctDNA at day 466 (Fig 6c, Extended Data Fig. 8). These subclones were not identified in the CT guided para-vertebral biopsy taken at day 467 (Fig. 6b). The mean plasma VAF of the SNVs detected in phylogenetic clusters 11, 8, 12, 9 and 5 mirrored their proximity to the clonal cluster (light blue) in the M-Seq derived phylogenetic tree (Fig. 6a,c). This suggested a tiered burden of subclonal disease concordant with M-seq phylogenetic inferences. Mean clonal VAF fell in response to palliative radiotherapy and chemotherapy, but at day 767 increased (Fig. 6c). Single SNVs mapping to phylogenetic clusters 5 and 9 and two SNVs mapping to phylogenetic cluster 2 were now detectable in ctDNA 90 days before death (Fig. 6a-c, Extended Data Fig 8). These three phylogenetic clusters represented subclones private to the para-aortic metastases (Fig 6. a-b). Consistent with these data significant para-aortic progression was observed at post-mortem compared with most recent CT imaging performed 112 days before death - which showed no evidence of para-aortic disease.
Discussion
In summary, we find predictors of ctDNA detection in early-stage NSCLC characterized by non-adenocarcinoma histology, necrosis, increased proliferative indices and lymphovascular invasion. Triple negative breast cancers display necrosis19, high proliferative indices20,21 and are associated with increased ctDNA levels compared with other breast cancer subtypes6 suggesting extension of these observations beyond NSCLC.
Tumour volume correlated with the mean plasma VAF of clonal SNVs in ctDNA-positive NSCLCs (Fig 3a.). A primary NSCLC tumour volume of 10 cm3 predicted a ctDNA plasma VAF of 0.1%. The sensitivity of the multiplex-PCR NGS platform was in excess of 99% at VAFs of 0.1% and above, suggesting optimum platform sensitivity with tumour burdens in excess of 10 cm3. Low-dose CT lung screening can identify lung nodules with diameters from 4mm22. Assuming a spherical nodule this would translate to a tumor volume of 0.034cm3. Based on the relationship between tumor volume and ctDNA plasma VAF observed in this study a tumor volume of 0.034cm3 would equate to a plasma VAF of 1.8 x 10-4 % (95% CI, 9.8 x 10-6 to 0.0033%), at the extreme of detection limits of current ctDNA platforms23. Sensitivity of clonal SNV ctDNA directed early NSCLC screening may therefore be constrained by tumor size using current technologies.
A limitation to targeted ctDNA profiling is cost, estimated at $1750 per patient for sequencing a single tumor region, synthesis of a patient-specific assay-panel and profiling of five plasma samples. Adjuvant platinum-based chemotherapy in NSCLC improves cure rates following surgery in only 5% of patients and 20% patients receiving chemotherapy experience acute toxicities3. There is a need to increase adjuvant therapy efficacy and better target its use. Bespoke ctDNA profiling can characterize the subclonal dynamics of relapsing NSCLC and identify adjuvant chemotherapy resistance. These findings indicate that drug development guided by ctDNA platforms to identify residual disease, define adjuvant treatment response and target emerging subclones prior to clinical recurrence in NSCLC, with appropriate CLIA validation, are now feasible.
Methods
Patients and samples
The cohort of 100 patients evaluated within this study comprises the first 100 patients prospectively analyzed by the lung TRACERx study (Clinicaltrials.gov no: NCT01888601, approved by an independent Research Ethics Committee, 13/LO/1546) and mirrors the prospective 100 patient cohort by Jamal-Hanjani M et al9. All surgically resected primary tumor samples were macroscopically reviewed by a pathologist. Spatially separated tumor regions, documented by photography, were collected and snap frozen in liquid nitrogen for subsequent DNA extraction. Relapse tissue samples, excess to diagnostic requirements, were acquired via clinical procedures including CT guided biopsy and endoscopic bronchial ultrasound guided biopsy. Fresh tissue for research was snap frozen immediately following acquisition for subsequent DNA extraction. Post mortem examination was performed through the PEACE study within 24 hours of death (Clinicaltrials.gov no: NCT03004755, approved by an independent Research Ethics Committee, 13/LO/0972). Details on relapse tissue sampling available in Supplementary Table 3. Informed consent was obtained from all subjects in this study.
Tissue microarray creation and Ki67 immunohistochemistry
Tissue microarrays (TMAs) were created of 100 NSCLC cases for Ki67+ immunohistochemistry. Representative tumor areas were defined by examination of H&E stained sections from all 100 tissues blocks. From each NSCLC case two 2mm cores were selected from different regions within each specimen and re-embedded in recipient blocks, resulting in a TMA of 200 cores with four normal lung cores as negative control. 2-5μm sections from tissue-microarrays containing tumor were cut. Immunohistochemistry with anti-Ki67 monoclonal antibody (Dilution 1:100; clone MIB-1; DAKO Agilent Technologies LDA, UK Limited, Stockport, Cheshire SK8 3GR, UK) was performed using BenchMark Ultra (Ventana/Roche). The percentage of Ki67 positive cells were averaged across two tumor sections for each case. Detection was performed using the peroxidase-based detection reagent conjugate (OptiView DAB IHC Detection Kit; Ventana Medical Systems, Inc).
Central histopathological review
Digital images of tumor sections from all cases were reviewed in detail centrally by at least one pathologist, and in cases of uncertainty, by two. Percentage of necrosis and the presence of lymphovascular invasion were all evaluated on digital images from scanned diagnostic slides blinded to the ctDNA detection status of the patient in question.
Central radiology review & volume estimation
92 of 96 anonymized diagnostic PET-CT were retrospectively reviewed by a Nuclear Medicine Physician, blinded to the initial PET-CT reports. Scan images were not available in three cases (CRUK0025, CRUK0039 and CRUK0023) and in one case a pre-operative PET-CT was not performed (CRUK0082). CT and PET images were matched and fused into transaxial, coronal and sagittal images and reviewed on a dedicated PET/CT software visualizer (AW 4.1/4.2 GE medical systems). The semi-quantitative parameter Standardized Uptake Value (SUV) max for the primary tumor mass was calculated and recorded along with SUVmax of mediastinal background uptake. Tumor-to-background ratio (TBR) was calculated based on SUVmax of the tumor divided by mediastinal background uptake24,25. Tumor volume was determined based on tumor CT scans. CT slices of the primary tumor were measured with 3D Slicer applying the “lung algorithm window” settings, tumor contours were segmented on each axial CT slice. These steps were performed by an experienced resident (W.L.B.), and all contours were confirmed and edited where necessary, by a radiologist with 14 years of experience in cancer imaging (F.M.F.). Effective tumor volume was defined as tumor volume multiplied with the mean purity of the tumor based on M-seq, purity estimates derived from ASCAT analysis as described9. Effective subclone size was defined as mean cancer cell fraction (CCF) across the regions of the mutation cluster multiplied by tumor volume and mean tumor purity.
DNA extraction & quantification
For cell-free DNA (cfDNA) extraction, blood samples were collected in K2 EDTA tubes. Samples were processed within 2 hours of collection by double spinning of blood first at 10 minutes at 1000g then plasma 10 minutes at 2000g. Plasma was stored in 1ml aliquots at – 80°C. Up to 5ml of plasma per case was available for this study (range 1-5ml, median 5 ml). The entire volume of plasma was used for cfDNA extraction. cfDNA was extracted using the QIAamp Circulating Nucleic Acid kit (Qiagen) and eluted into 50 µl DNA Suspension Buffer (Sigma). The purified cfDNA was stored at -20°C until use. Genomic DNA was extracted from each tumor region as described9. Every cfDNA sample was QCed and quantified on the Bioanalyzer High Sensitivity (Agilent) using a standard curve generated from pre-quantified mono-nucleosomal DNA samples. Plasma cfDNA consists of a main mono-nucleosomal peak (~160 bp); for some samples, di-nucleosomal and tri-nucleosomal peaks are visible (at ~320 bp and ~500 bp, respectively). The library prep method used selectively amplifies the mono-nucleosomal fraction of cfDNA.
Exome sequencing and processing
Whole exome sequencing was performed on DNA purified from tumor tissue and normal blood as described9, with the exception of CRUK0063_BR_T1-R1. This capture was performed according to the manufacturer’s 200 ng DNA protocol (Agilent). Annotated SNV calls are available in Supplementary Table 3 in Jamal-Hanjani et al. 20179. For this study, one relapse sample was acquired through metastatic tissue biopsies from each of four patients (CRUK0035, CRUK0041, CRUK0044, CRUK0063). Additionally, six metastatic samples were acquired at post mortem examination of CRUK0063. Genomic DNA was purified from all tissue samples, and processed through the TRACERx bioinformatics pipeline as described9. Annotated SNV calls are available in Supplementary Table 4.
SNV assay design
The Natera assay design pipeline was used to generate forward and reverse PCR primers for all somatic SNVs detected in tumor samples. The assays were combined into pools such that any primer pair in a pool is not predicted to form primer dimers. In this way 10 balanced pools were created, each containing the assays for 10 patients’ SNVs. For each patient, assays were prioritized such that, 1) assays covering driver SNVs had highest priority, and 2) there was uniform sampling of phylogenetic tree. For the longitudinal cohort, up to 10 extra assays were generated for adenocarcinoma samples. SNV assays were ordered from IDT (Coralville, IA) as individual oligos in 96-well plates, desalted and normalized to 100 µM each. The oligos were pooled according to the pooling strategy previously described10 and each pool was QC-ed by running the multiplex PCR and sequencing protocol using one plasma cfDNA library from a healthy subject. For each pool, the sequencing data was analyzed to determine the amount of primer-dimer reads and to identify drop-out assays. Primers contributing to dimer formation were removed from each final pool.
Analytical validation
Synthetic spikes representing twenty SNVs randomly selected from Pool 1 were designed and synthesized (IDT, Coralville, IA) as 160 bp oligos with the respective SNV placed in the middle (position 80). These synthetic spikes were mixed at equimolar ratios and used to prepare a library. This library was titrated into a library prepared from mono-nucleosomal DNA (10,000 copies) from a normal cell line (AG16778 from Coriell, Camden, NJ). The library of 20 synthetic spikes was titrated into the mononucleosomal DNA library at 2.5%, 0.5%, 0.25%, 0.1%, 0.05% and 0% (each in triplicate), and 0.01%, 0.005% and 0.001% (each in quadruplicate. Because preparing spiked samples at such low levels is either subject to sampling noise (0.01% spikes into 10,000 genomic copies background is equivalent to one mutant copy), or is not possible (at levels less than 0.01%), samples were mixed as libraries. Following library mixing and sequencing, data was analyzed to detect all the targets in Pool 1 using the same parameters as used for the patient samples. Targets that had a depth of read less than the threshold were not analyzed. The measured VAF of each spike for the samples with 2.5% nominal input was used to calculate an input correction factor (measured VAF/2.5%); that was applied to the other inputs of the corresponding spike titration series. The measured VAF differed from the nominal input most likely because the mononucleosomal fragmentation pattern is not entirely random. Because of this, the actual input levels differ from the nominal input levels, and the sensitivity is measured based on corrected input intervals (chosen such that there are a meaningful number of samples in each interval). Sensitivity of >99% at SNV input frequencies down to 0.1% was achieved (199 SNVs detected out of 201 eligible positive positions), with a specificity of 99.6% for all negative SNV positions (19 false positive SNVs called out of 5099 eligible positions).
Plasma SNV mPCR-NGS workflow
Forty microliters of the extracted cfDNA from each case was used as input into library preparation using the Natera Library Prep Kit. All purified libraries were QC-ed on the LabChip GX 5k DNA chip. Successful libraries had a single peak at ~250 bp. The amplified libraries were then analyzed by mPCR-NGS. Optimal mPCR conditions were as described. Each PCR assay pool was used to amplify the SNV targets from the 10 corresponding samples and 20 negative control samples (plasma libraries prepared from healthy subjects). Each amplicon pool was sequenced on one Illumina HiSeq 2500 Rapid Run with 50 cycles paired-end reads using the Illumina Paired End v1 kit with an average target DOR of ~40,000 per assay.
Plasma SNV calling algorithm
The set of SNVs covered by the assays in a pool were considered as target SNVs for the corresponding sequencing run. Target assays with <1000 reads in the plasma samples were considered failed and were not analyzed further. At each SNV position, an error model was built using all of the 20 negative control samples plus the cancer samples that were not expected to contain that particular SNV (based on tumor-tissue sequencing). Samples with high plasma VAF (>20%) among the putative negatives were considered to have possible germline mutation and were excluded from the error model. A confidence score was calculated for each target SNV based on the error model. A positive plasma SNV call was made if the confidence score for that mutation in the corresponding plasma sample passed our confidence threshold. The SNVs called positive in plasma samples that were not expected to contain the given SNVs were considered ‘false positive’, the false positive rate under these conditions was 0.24%. Notably, there was no difference in depth of read between called and not called SNVs (Extended Data Fig 1c). New assays were designed for CRUK0063 based on M-seq of metastatic biopsy retrieved at day 467 and of metastatic lesions harvested post mortem. A total of 103 assays were designed compared to 19 based on the primary tumor alone. An updated error threshold was designed to control for false positives by using the original threshold to make SNV calls on the negative samples in the run; the rate of calls were measured and defined as false-positives. This false positive rate was then applied to the number of eligible positions in the positive samples. This was repeated with more stringent thresholds until the expected number of false positives in the eligible positions becomes ~1. All multiplex PCR-NGS ctDNA SNV assays are available in Supplementary Table 5 (Baseline, pre-operative cohort assays), Supplementary Table 6 (Longitudinal Assays), and Supplementary Table 7 (Extended Longitudinal Assays for CRUK0063).
Cross-platform validation using generic PCR-NGS panel section
Cross-platform validation was performed in 28 patients with M-Seq confirmed SNV(s) within one or more hotspots targeted by a generic multiplex PCR-NGS panel (Extended Table 2a-b, Supplementary Table 8). 20ng of isolated cfDNA was used for library preparation using the Oncomine™ Lung cfDNA Assay (ThermoFisher Scientific), according to the manufacturer’s instructions. Automated template preparation and chip loading was conducted on the Ion Chef™ instrument using the Ion 520™ & Ion 530™ Kit-Chef (ThermoFisher Scientific). Ultimately, samples were sequenced on Ion 530™ chips using the Ion S5™ System (ThermoFisher Scientific). Sequencing data was accessed on the Torrent suite v5.2.2. Reads were aligned against the human genome (hg19) using Alignment v4.0-r77189, and variants were called using the coverage Analysis v4.0-r77897 plugin. All 18 bespoke-panel ctDNA negative patients had no tumor SNVs detectable in plasma pre-operatively by the generic panel supporting biological specificity of the bespoke targeted approach, 7 of 10 bespoke-panel ctDNA positive patients had tumor SNVs detected in plasma by the generic panel (Extended Table 2a-b). SNVs detected by hotspot panel not identified by M-Seq are displayed in Extended Table 2c.
Processing and phylogenetic analysis of relapse and primary tumor multiregion whole exome data
Biopsies from multiple regions from the primary tumor (n=327), metastatic biopsies (n=4) and matching blood germline samples (n=100) were subjected to multi-region whole exome sequencing and analysis including estimation of copy number, purity and ploidy, and phylogenetic tree construction as described9. Briefly, phylogenetic analysis was performed based on CCF determined for SNVs and clustered across tumor regions using a modified version of Pyclone9 into clusters with similar CCF values, filtered and processed as described9. Mutation clusters are assumed to represent tumor subclones, either current or ancestral, and are used as input for construction of phylogenetic relationship. Phylogenetic trees were primarily constructed using the published tool CITUP (0.1.0)26. However, in a small number of cases, including all relapse/autopsy cases, manual tree construction was required and performed as described9. Complete detail of primary tumor tree construction can be found in Jamal-Hanjani et al. 20179. Relapse tree construction was performed as follow: CRUK0063: clustering was performed twice, once across 5 primary tumor regions and once across 5 primary, 1 relapse, and 6 autopsy regions. To ensure consistency, when deriving a phylogenetic tree based on all tumor regions, CCF clusters based on clustering only the primary tumor regions were maintained for mutations not involved in metastatic relapse. A phylogenetic tree was constructed based on 17 mutation clusters. CRUK0035: Clustering primary tumor regions with the relapse region revealed one cluster private to the relapse, and one cluster shared with the relapse and all other regions. CRUK0044: Clustering primary tumor regions with the relapse region revealed a cluster private to the relapse, descended from a cluster private to region 1 in the primary tumor. CRUK0041: Clustering primary tumor regions with the relapse region revealed cluster 4 as private to the relapse. This cluster must have evolved from cluster 3 only found in the relapse and in region 4. A private cluster 6 in region 4 must have evolved from cluster 4. However, this conflicts with clusters 2 and 5, found in the relapse and regions 1-3, but not region 4. This can be reconciled by assuming a polyclonal relapse, seeded primarily from regions 1-3, but with some contribution from cluster 3, private to region 4. Cluster data is available in Supplementary Table 4 under “PyClonePhyloCluster”.
Statistical data analysis
Analysis was performed in the R statistical environment version 3.2.3 and SPSS version 24. All statistical tests were two-sided unless expressly stated. Multivariate logistic regression used detection of ctDNA (the dependent variable) classified as detection of 2 or more patient-specific variants in ctDNA and the covariates listed in Supplementary Table 1. All predictors were entered simultaneously into the regression. All continuous independent variables were found to be linearly related to the logit of the dependent variable (assessed via the Box-Tidwell procedure). The logistic regression model was statistically significant, X2(10) = 81.35, P<0.001 and the Hosmer-Lemeshow P value was 0.9858 indicating that the model was not a poor fit. To determine the ability of PET TBR to predict whether or not tumor ctDNA was identified in plasma, PET TBR estimates were analyzed by ROC curve analysis against binary detection of ctDNA in plasma at baseline based on at least two variants detected, significance test based on Wilcoxon rank sum test. For analysis involving longitudinally detected variants (Figure 4, Extended Figure 5), only subclonal variants from pyclone clusters present in phylogenetic trees were displayed, this did not affect ctDNA detection status of any time-points. In non-relapse cases presented in Extended Data Fig 6 all detected subclonal SNVs were plotted. To determine the relationship between tumor volume and ctDNA VAF, ctDNA assays against clonal SNVs were selected. For each patient, the mean ctDNA VAF of the clonal SNVs was determined as baseline for 38/46 patients with at least 2 SNVs detected in plasma. As detailed in Extended Fig. 4c, 9/46 patients were not included in the analysis: CRUK0036 had no pre-op CT scan available, CRUK0087 and CRUK0096 had a large cavity inside the primary cancer, CRUK0099 had a collapsed lung making volume assessment inaccurate, CRUK0100, CRUK0077, CRUK0052 had a CT slice spacing of > 5 mm, and finally CRUK0088 and CRUK0091 had a total tumor volume < 3.5 cm3. Linear regression was performed on log-transformed mean VAF and tumor volume. The log transformation was justified as it symmetrized the residuals in the model. An independent analysis was performed where tumor volume was multiplied with tumor purity to estimate the cancer cell volume. The same log transformation and analysis was applied to data acquired from Newman et al.16, where ctDNA VAF was determined based on CAPP-seq analysis with matched tumor volume data available. To analyze clone size versus ctDNA VAF for subclonal SNVs, the mean CCF of the mutations within a subclonal mutation cluster was multiplied with tumor volume, and as a second independent analysis, with tumor purity.
Extended Data
Extended Data Figure 1. Multiplex-PCR next-generation sequencing platform analytical validation.
a) Analytical validation of the multiplex-PCR NGS platform was performed by spiking synthetic single nucleotide variants into control cell-free DNA. Sensitivity and specificity of the platform at different spike concentrations was ascertained, 95% binomial confidence interval displayed as error bars. b) Specificity of ctDNA detection based on a 1 SNV and 2 SNV call threshold taking into account parallel testing of multiple SNVs. c) The median depth of read across a position did not vary depending on whether an SNV position was called or not called using the platform error-model. Wilcoxon Test, P=0.786, median depth of read at uncalled positions = 45,777 (n=3,745), range: 0 to 146774, median depth of read at called positions = 45,478, range= 1,354 to 152,974 (n=1,124). Whiskers represent 1.5 times the interquartile range, 2-sided test.
Extended Data Figure 2. Study construction and assay-panel design.
a) The pre-operative study phase cohort consisted of 100 TRACERx patients present in the first 100 patient TRACERx cohort in April 2016. Pre-operative plasma samples were profiled in 96 patients for reasons listed. bi and ii) Contents of patient-specific assay-panels designed in the pre-operative study phase. c) The longitudinal study phase cohort consisted of patients with confirmed NSCLC relapse and patients without relapse. d) Contents of patient-specific assay-panels designed in the longitudinal phases of this study. e) Single nucleotide variant type targeted.
Extended Data Figure 3. Clinicopathological predictors of ctDNA detection.
a) 96 patients in pre-operative cohort stratified by pathological TNM stage. b) LUSCs and ctDNA positive LUADs are significantly more necrotic that ctDNA negative LUADs. Significant differences in necrosis between groups: LUSCs (median necrosis 40%) (n=31), ctDNA positive LUADs (median necrosis 15%) (n=11) and ctDNA negative LUADs (median necrosis 2%) (n=47), Kruskal-Wallis test, P<0.001, 2-sided pairwise comparisons were performed using Dunn’s procedure with Bonferroni correction. c) Univariate (left) and multivariate analyses (right) were performed, by logistic regression to determine significant predictors of ctDNA detection in early-stage NSCLC. ctDNA detection was defined as detection of two or more SNVs in pre-operative plasma samples. Details regarding multivariable analysis methodology are in methods. d) Receiver operating characteristic curve (ROC) analysis of pre-operative PET scan FDG-avidity (normalized as tumor background ratio (TBR), see methods), as a predictor of ctDNA detection (92/96 PET scans were available for central review). Median PET TBR of detected tumors = 9.01, n=45. Median PET TBR of undetected tumors= 3.64, n=47. P-value based on Wilcoxon Rank Sum Test. e) LUAD subtype analyses based on ctDNA detection and the presence of an EGFR, KRAS or TP53 driver mutation.
Extended Data Figure 4. Predictors of plasma variant allele frequency.
a) Plasma variant allele frequencies of SNVs detected in plasma in 46 patients who were ctDNA positive (two or more SNVs detected). Clonal (blue) and subclonal (red) variant allele frequencies indicated, mean shown as horizontal line. Driver variants shown as triangles. b) Mean clonal VAF correlated with maximum tumor size measured in post-surgical specimen (pathological size, n=46) grey vertical bars represent range of clonal variant allele frequency. Shaded red background indicates 95% confidence interval. c) Filtering steps taken to define a group of ctDNA positive patients with volumetric data considered adequate to model tumor volume and plasma variant allele frequency. d) Scatter plot showing mean clonal VAF relative to tumor volume for TRACERx (blue dots and fitted blue line, n=37) and VAF relative to volume for previously published data based on CAPP-seq analysis of ctDNA (orange dots and orange fitted line, n=9). Orange shaded background indicates 95% confidence interval based on CAPP-seq data. e) Mean clonal VAF correlated with tumor volume × tumor purity (cancer cell volume), n=37. Shaded red background indicates 95% confidence interval. f) Association between number of cancer cells and VAF of clonal SNVs in plasma based on linear modelling of Extended Data Fig 4f. g) Detected subclonal SNVs were mapped back to M-Seq derived tumor phylogenetic trees (process illustrated in graphic). Detected private subclones (subclones identified within only a single tumor region) are coloured red. Shared subclones (subclones detected in more than one tumor regions) are light blue. Subclonal nodes were sized based on the maximum recorded cancer cell fraction (CCF). The top row of phylogenetic trees represent subclonal nodes targeted by primers within that patient’s assay panel, the bottom row represent subclonal nodes detected in ctDNA, within this row grey subclonal nodes represent subclones not detected in ctDNA.
Extended Data Figure 5. Longitudinal ctDNA profiling, remaining relapse cases.
a) Kaplan-Meier curve demonstrate relapse free survival for patients in whom ctDNA was detected versus patients in whom ctDNA was not detected. b-h) Longitudinal cell-free DNA profiling. Circulating tumor DNA (ctDNA) detection in plasma was defined as the detection of two tumor-specific SNVs. Relapse was based on imaging-confirmed NSCLC relapse, imaging performed as clinically indicated. Detected clonal (circles, light blue) and subclonal (triangles, colors indicates different subclones) SNVs from each patient-specific assay-panel are plotted on graphs colored by M-Seq derived tumor phylogenetic nodes. Mean clonal (blue) and mean subclonal (red) VAF are indicated on graphs. Pre-operative and relapse M-Seq derived phylogenetic trees represented by ctDNA are illustrated above each graph in cases where subclonal SNVs were detected.
Extended Data Figure 6. Longitudinal ctDNA profiling, non-relapse cases.
a-j) Detected clonal (circles, light blue) and subclonal (red triangles) SNVs from each patient-specific assay-panel are plotted on graphs. Mean clonal (blue) and mean subclonal (red) VAF are indicated on graphs.
Extended Data Figure 7. Heatmaps illustrating detection of SNVs in bespoke panel at each sampled time point.
a, c-f) Bespoke assay panels for CRUK0063, CRUK0035, CRUK0044, CRUK0041 and CRUK0013. Colors indicate originating subclonal cluster based on the phylogenetic trees above the heatmap. Light blue indicates clonal mutation cluster. Full panel with cluster color shown below each heatmap. Filled squares indicates detection of a given variant in plasma ctDNA. Y-axis shows day of sampling, y-axis labels appended with [R] indicates day of clinical relapse. b) Re-examination of primary tumor regions from CRUK0063 with lowered threshold to potentially identify SNVs private to the sequenced relapse biopsy. 16/88 variants were found at very low VAF in region 3, indicating this region from the primary likely gave rise to the metastasis.
Extended Data Figure 8. Heatmap illustrating detection of SNVs in bespoke panel based on M-seq of metastatic tumor regions for patient CRUK0063 for all sampled time points.
Colors indicate originating subclonal cluster based on the phylogenetic trees above the heatmap. Light blue indicates clonal mutation cluster. Full panel with cluster color shown below each heatmap. Filled squares indicates detection of a given variant in plasma ctDNA. Y-axis shows day of sampling.
Extended Table 1a. Patient characteristics.
Clinical characteristics 96 patient pre-operative cohort
table of clinical characteristics describing the 96 patient pre-operative cohort
| Characteristic | Total | |
|---|---|---|
| Age | <60 | 19 |
| ≥60 | 77 | |
| Sex | Male | 60 |
| Female | 36 | |
| ECOG PS | 0 | 49 |
| 1 | 47 | |
| Histology | Adenocarcinoma | 58 |
| Squamous cell carcinoma | 31 | |
| Carcinosarcoma | 2 | |
| Large cell carcinoma | 1 | |
| Adenosquamous carcinoma | 3 | |
| Large cell neuroendocrine carcinoma | 1 | |
| TNM stage | Ia | 24 |
| Ib | 35 | |
| IIa | 12 | |
| IIb | 11 | |
| IIIa | 13 | |
| IIIb | 1 | |
| Lymph node metastasis | Yes | 24 |
| No | 72 | |
| Pleural involvement | Yes | 27 |
| No | 69 | |
| Vascular invasion | Yes | 41 |
| No | 55 | |
| Resection margin | R0 | 91 |
| R1 | 5 | |
| Smoking status | Never smoked | 11 |
| Recent ex-smoker | 30 | |
| Ex-smoker | 48 | |
| Current smoker | 7 | |
| Ethnicity | White British | 85 |
| White-other | 4 | |
| White-Irish | 4 | |
| Caribbean | 3 | |
Extended Table 1b.
demonstrating distribution of stage and whether the patient received adjuvant chemotherapy
| No adjuvant therapy | Adjuvant therapy | ||
|---|---|---|---|
| TNM Stage | Ia | 24 | 0 |
| Ib | 31 | 4 | |
| IIa | 3 | 9 | |
| IIb | 4 | 7 | |
| IIIa | 6 | 7 | |
| IIIb | 0 | 1 | |
Extended Table 1c.
Details regarding timing of pre-operative blood sample
Demonstrating the time-points at which pre-operative plasma was acquired for patients within the cohort
| Days pre-surgery | Number | Details |
|---|---|---|
| Within 24 hours | 91 | |
| 24-72 hours | 2 | CRUK0051, 0003 |
| 8 days | 2 | CRUK0073, 0096 |
| 31 days | 1 | CRUK0089 |
Extended Table 1d.
Clinical characteristics 24 patient longitudinal sub-cohort
table of clinical characteristics describing 24 patient longitudinal cohort
| Characteristic | Total | |
|---|---|---|
| Age | <60 | 5 |
| ≥60 | 19 | |
| Sex | Male | 16 |
| Female | 8 | |
| ECOG PS | 0 | 12 |
| 1 | 12 | |
| Histology | Adenocarcinoma | 16 |
| Squamous cell carcinoma | 8 | |
| TNM stage | Ia | 3 |
| Ib | 7 | |
| IIa | 3 | |
| IIb | 7 | |
| IIIa | 3 | |
| IIIb | 1 | |
| Lymph node metastasis | Yes | 9 |
| No | 15 | |
| Pleural involvement | Yes | 7 |
| No | 17 | |
| Vascular invasion | Yes | 12 |
| No | 12 | |
| Resection margin | R0 | 23 |
| R1 | 1 | |
| Smoking status | Never smoked | 1 |
| Recent ex-smoker | 5 | |
| Ex-smoker | 16 | |
| Current smoker | 2 | |
| Ethnicity | White British | 21 |
| White-other | 2 | |
| Caribbean | 1 | |
Extended Table 1e.
demonstrating distribution of stage in the longitudinal cohort and whether the patient received adjuvant chemotherapy.
| No adjuvant therapy | Adjuvant therapy | ||
|---|---|---|---|
| TNM Stage | Ia | 3 | 0 |
| Ib | 6 | 1 | |
| IIa | 0 | 3 | |
| IIb | 2 | 5 | |
| IIIa | 1 | 2 | |
| IIIb | 0 | 1 | |
Table 2a. Cross platform validation using a generic approach to ctDNA profiling.
Bespoke panel detected NSCLCs - cross platform validation
a) 7/10 (70%) of bespoke-panel ctDNA positive patients had tumor SNVs detectable in plasma preoperatively by a generic hotspot PCR-NGS lung panel (Lung Oncomine, Thermofisher). The three bespoke-panel ctDNA positive patients undetected by the generic panel had mean clonal plasma variant allele frequencies lower than the 0.1% plasma variant allele frequency (VAF) limit of detection reported for the generic panel (shaded yellow). b) Based on CT volumetric assessment of each patient’s primary tumor we predicted plasma VAF corresponding to a tumor of that size (see Figure 3 and Methods for details of approach). This allowed us to infer platform sensitivities for each patient within the bespoke-panel non-detected cohort. Six LUADs (shaded green; CRUK0037, CRUK0051, CRUK0004, CRUK0039, CRUK0025 and CRUK0048) had tumor volumes approximating to a plasma VAF of more than 0.1%. This suggested that these tumors resided within the top platform sensitivity bracket of both the generic and bespoke-panel ctDNA platforms. No ctDNA was detected by either platform in these cases, suggesting biological specificity of the bespoke-panel.
| Bespoke-panel | Generic-panel | |||||
|---|---|---|---|---|---|---|
| Case | Volume cm3 | Plasma VAF (mean clonal) | ctDNA positive | Histology | Hotspot SNVs tumor | Hotspot SNVs detected |
| CRUK0029 | 38.51 | 2.10 | Yes | LUAD | 1 | 1 |
| CRUK0009 | 69.01 | 1.71 | Yes | LUAD | 1 | 1 |
| CRUK0062 | 58.48 | 1.41 | Yes | LUSC | 1 | 1 |
| CRUK0081 | 16.41 | 0.21 | Yes | LUSC | 1 | 1 |
| CRUK0089 | 17.39 | 0.16 | Yes | LUSC | 1 | 1 |
| CRUK0022 | 17.20 | 0.08 | Yes | LUAD | 1 | 0 |
| CRUK0067 | 6.64 | 0.07 | Yes | LUSC | 1 | 0 |
| CRUK0052 | 43.69 | 0.06 | Yes | LUAD | 2 | 1 |
| CRUK0064 | 9.24 | 0.05 | Yes | LUSC | 1 | 0 |
| CRUK0034 | 10.59 | 0.01 | Yes | LUAD | 1 | 1 |
| 2 b - Bespoke panel non-detected NSCLCs - cross platform validation | ||||||
| Bespoke-panel | Generic-panel | |||||
| Case | Volume cm3 | Predicted plasma VAF | ctDNA positive | Histology | Hotspot SNVs tumor | Hotspot SNVs detected |
| CRUK0037 | 197.42 | 2.96 (1.01 to 8.67) | No | LUAD | 1 | 0 |
| CRUK0051 | 27.28 | 0.32 (0.21 to 0.49) | No | LUAD | 1 | 0 |
| CRUK0004 | 23.30 | 0.27 (0.18 to 0.41) | No | LUAD | 1 | 0 |
| CRUK0039 | 21.68 | 0.25 (0.16 to 0.38) | No | LUAD | 2 | 0 |
| CRUK0025 | 19.06 | 0.22 (0.14 to 0.33) | No | LUAD | 2 | 0 |
| CRUK0048 | 17.00 | 0.19 (0.12 to 0.29) | No | LUAD | 2 | 0 |
| CRUK0026 | 7.45 | 0.08 (0.04 to 0.14) | No | LUAD | 1 | 0 |
| CRUK0030 | 7.28 | 0.07 (0.04 to 0.14) | No | LUAD | 2 | 0 |
| CRUK0057 | 5.95 | 0.06 (0.03 to 0.12) | No | LUAD | 1 | 0 |
| CRUK0018 | 4.65 | 0.04 (0.02 to 0.10) | No | LUAD | 1 | 0 |
| CRUK0027 | 4.61 | 0.04 (0.02 to 0.10) | No | LUAD | 1 | 0 |
| CRUK0007 | 4.18 | 0.04 (0.02 to 0.09) | No | LUAD | 1 | 0 |
| CRUK0049 | 3.61 | 0.03 (0.01 to 0.08) | No | LUAD | 1 | 0 |
| CRUK0035 | 3.31 | 0.03 (0.01 to 0.08) | No | LUAD | 1 | 0 |
| CRUK0058 | 2.76 | 0.03 (0.01 to 0.07) | No | LUAD | 1 | 0 |
| CRUK0021 | 2.70 | 0.02 (0.01 to 0.07) | No | LUAD | 2 | 0 |
| CRUK0093 | 0.73 | 0.01 (0.001 to 0.03) | No | LUSC | 2 | 0 |
| CRUK0014 | 0.90 | 0.01 (0.002 to 0.03) | No | LUAD | 1 | 0 |
| Multiplex-PCR NGS | ||
| Targeted panel | >99% sensitivity at 0.1% VAF and above | Platform sensitivities predicted based on tumor volume and analytical validation data in Extended Data 1 |
| 84% sensitivity at 0.05% to 0.1% VAF | ||
| 46 % sensitivity 0.01% to 0.05% VAF | ||
| 4.2% sensitivity <0.01% | ||
| Generic panel | 90% sensitivity at 0.1% VAF and above | Oncomine lung panel sensitivity data reported at https://www.thermofisher.com/order/catalog/product/A31149 |
Table 2 c.
c) Hotspot SNVs not identified in tumor tissue through exome sequencing were identified in plasma of 9 of 28 patients by the generic panel. This suggested non-tumor origin of cell-free DNA, platform non-specificity or an evolving minor subclone or second primary.
Variants detected by generic PCR-NGS hotspot panel not detected in M-Seq analysis of tumor
| (unfiltered) | (unfiltered) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Gene | Location | Position | Ref | Variant | AA change | Plasma VAF | DOR | ctDNA positive | Combined exome VAF | Germline VAF |
| CRUK0052 | PIK3CA | chr3 | 178936091 | G | A | p.E545K | 0.81 | 60360 | Yes | ND | ND |
| CRUK0052 | PIK3CA | chr3 | 178952085 | A | G | p.H1047R | 0.12 | 52325 | Yes | 0.075 | ND |
| CRUK0062 | PIK3CA | chr3 | 178936091 | G | A | p.E545K | 0.97 | 89616 | Yes | 0.016 | ND |
| CRUK0062 | PIK3CA | chr3 | 178952085 | A | G | p.H1047R | 0.05 | 79205 | Yes | 0.005 | ND |
| CRUK0062 | TP53 | chr17 | 7577556 | C | A | p.C242F | 0.05 | 93383 | Yes | ND | ND |
| CRUK0089 | TP53 | chr17 | 7577121 | G | A | p.R273C | 0.06 | 59849 | Yes | 0.168 | ND |
| CRUK0004 | PIK3CA | chr3 | 178936091 | G | A | p.E545K | 0.59 | 73941 | No | 0.081 | ND |
| CRUK0018 | PIK3CA | chr3 | 178936091 | G | A | p.E545K | 4.44 | 99159 | No | ND | ND |
| CRUK0018 | PIK3CA | chr3 | 178952085 | A | G | p.H1047R | 0.81 | 77806 | No | 0.044 | ND |
| CRUK0021 | PIK3CA | chr3 | 178952085 | A | G | p.H1047R | 0.11 | 50107 | No | ND | ND |
| CRUK0027 | PIK3CA | chr3 | 178952085 | A | G | p.H1047R | 0.11 | 65449 | No | ND | ND |
| CRUK0037 | PIK3CA | chr3 | 178952085 | A | G | p.H1047R | 0.09 | 51071 | No | ND | ND |
| CRUK0058 | KRAS | chr12 | 25398284 | C | A | p.G12V | 3.44 | 63090 | No | 0.124 | ND |
ND - non detected
DOR - depth of read
Combined exome VAF (unfiltered) - Variant allele frequency across all tumor regions analysed (without call filters).
Supplementary Material
Supplementary information is available in the online version of the paper
Acknowledgements
We dedicate this manuscript to the memory of Roberto Macina. We thank Samantha Navarro and Antony Tin for facilitating the PEACE ctDNA analysis. We thank the members of the TRACERx and PEACE consortia for participating in this study. C.S. is Royal Society Napier Research Professor. This work is supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001169,FC001202), the UK Medical Research Council (FC001169,FC001202), and the Wellcome Trust (FC001169,FC001202). CS is funded by Cancer Research UK (TRACERx and CRUK Cancer Immunotherapy Catalyst Network), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rosetrees Trust, NovoNordisk Foundation (ID 16584), the Prostate Cancer Foundation, the Breast Cancer Research Foundation, the European Research Council (THESEUS) and Support was provided to CS by the National Institute for Health Research, the University College London Hospitals Biomedical Research Centre and the Cancer Research UK University College London Experimental Cancer Medicine Centre. P.V.L. is a Winton Group Leader in recognition of the Winton Charitable Foundation’s support towards the establishment of the Francis Crick Institute.
The TRACERx consortium members and affiliations
The TRACERx (Tracking Non-small Cell Lung Cancer Evolution) study (Clinicaltrials.gov no: NCT01888601) is sponsored by University College London (UCL/12/0279) and has been approved by an independent Research Ethics Committee (13/LO/1546). TRACER is funded by Cancer Research UK (grant number C11496/A17786) and coordinated through the Cancer Research UK & UCL Cancer Trials Centre.
Charles Swanton1,2,5, Mariam Jamal-Hanjani1, Christopher Abbosh1, Selvaraju Veeriah1, Seema Shafi1, Justyna Czyzewska-Khan1, Diana Johnson1, Joanne Laycock1, Leticia Bosshard-Carter1, Gerald Goh1, Rachel Rosenthal1, Pat Gorman1, Nirupa Murugaesu1, Robert E Hynds1,3, Gareth Wilson1,2, Nicolai J Birkbak1,2, Thomas B K Watkins2, Nicholas McGranahan1,2, Stuart Horswell2, Maise Al Bakir2, Eva Grönroos2, Richard Mitter2, Mickael Escudero2, Aengus Stewart2, Peter Van Loo2, Andrew Rowan2, Hang Xu2, Samra Turajlic2,4, Crispin Hiley2, Jacki Goldman2, Richard Kevin Stone2, Tamara Denner2, Nik Matthews2, Greg Elgar2, Sophia Ward2, Jennifer Biggs2, Marta Costa2, Sharmin Begum2, Ben Phillimore2, Tim Chambers2, Emma Nye2, Sofia Graca2, Maise Al Bakir2, Kroopa Joshi1, Andrew Furness1, Assma Ben Aissa1, Yien Ning Sophia Wong1, Andy Georgiou1, Sergio Quezada1, John A Hartley1, Helen L Lowe1, Javier Herrero1, David Lawrence5, Martin Hayward5, Nikolaos Panagiotopoulos5, Shyam Kolvekar5, Mary Falzon5, Elaine Borg5, Teresa Marafioti5, Celia Simeon5, Gemma Hector5, Amy Smith5, Marie Aranda5, Marco Novelli5, Dahmane Oukrif1, Ayse U Akarca1, Sam M Janes5, Ricky Thakrar5, Martin Forster5, Tanya Ahmad5, Siow Ming Lee5, Dionysis Papadatos-Pastos5, Dawn Carnell5, Ruheena Mendes5, Jeremy George5, Neal Navani5, Asia Ahmed5, Magali Taylor5, Junaid Choudhary5, Yvonne Summers6, Raffaele Califano6, Paul Taylor6, Rajesh Shah6, Piotr Krysiak6, Kendadai Rammohan6, Eustace Fontaine6, Richard Booton6, Matthew Evison6, Phil Crosbie6, Stuart Moss6, Faiza Idries6, Leena Joseph6, Paul Bishop6, Anshuman Chaturved6, Anne Marie Quinn6, Helen Doran6, Angela leek7, Phil Harrison7, Katrina Moore7, Rachael Waddington7, Juliette Novasio7, Fiona Blackhall8, Jane Rogan7, Elaine Smith6, Caroline Dive9, Jonathan Tugwood9, Ged Brady9, Dominic G Rothwell9, Francesca Chemi9, Jackie Pierce9, Sakshi Gulati9, Babu Naidu10, Gerald Langman10, Simon Trotter10, Mary Bellamy10, Hollie Bancroft10, Amy Kerr10, Salma Kadiri,10 Joanne Webb10, Gary Middleton10, Madava Djearaman10, Dean Fennell11, Jacqui A Shaw11, John Le Quesne11, David Moore11, Anne Thomas11, Harriet Walter11, Joan Riley11, Luke Martinson11, Apostolos Nakas12, Sridhar Rathinam12, William Monteiro13, Hilary Marshall13, Louise Nelson12, Jonathan Bennett12, Joan Riley12, Lindsay Primrose12, Luke Martinson12, Girija Anand14, Sajid Khan15, Anita Amadi16, Marianne Nicolson17, Keith Kerr17, Shirley Palmer17, Hardy Remmen17, Joy Miller17, Keith Buchan17, Mahendran Chetty17, Lesley Gomersall17, Jason Lester18, Alison Edwards18, Fiona Morgan19, Haydn Adams19, Helen Davies19, Malgorzata Kornaszewska20, Richard Attanoos21, Sara Lock22, Azmina Verjee22, Mairead MacKenzie23, Maggie Wilcox23, Harriet Bell24, Natasha Iles24, Allan Hackshaw24, Yenting Ngai24, Sean Smith24, Nicole Gower24, Christian Ottensmeier25, Serena Chee25, Benjamin Johnson25, Aiman Alzetani25, Emily Shaw25, Eric Lim26, Paulo De Sousa26, Monica Tavares Barbosa26, Alex Bowman26, Simon Jordan26, Alexandra Rice26, Hilgardt Raubenheimer26, Chiara Proli26, Maria Elena Cufari26, John Carlo Ronquillo26, Angela Kwayie26, Harshil Bhayani26, Morag Hamilton26, Yusura Bakar26, Natalie Mensah26, Lyn Ambrose26, Anand Devaraj26, Silviu Buderi26, Jonathan Finch26, Leire Azcarate26, Hema Chavan26, Sophie Green26, Hillaria Mashinga26, Andrew G Nicholson26, 27, Kelvin Lau28, Michael Sheaff28, Peter Schmid28, John Conibear28, Veni Ezhil29, Babikir Ismail29, Melanie Irvin-sellers29, Vineet Prakash29, Peter Russell30, Teresa Light30, Tracey Horey30, Sarah Danson31, Jonathan Bury31, John Edwards31, Jennifer Hill31, Sue Matthews31, Yota Kitsanta31, Kim Suvarna31, Patricia Fisher31, Allah Dino Keerio31, Michael Shackcloth32, John Gosney32, Pieter Postmus32, Sarah Feeney32, Julius Asante-Siaw32, Tudor Constatin33, Raheleh Salari33, Nicole Sponer33, Ashwini Naik33, Bernhard Zimmermann33, Matthew Rabinowitz33, Hugo J.W.L. Aerts34, Stefan Dentro35, Christophe Dessimoz36,37,38.
Affiliations
1. Cancer Research UK Lung Cancer Centre of Excellence, University College London Cancer Institute, United Kingdom
2. The Francis Crick Institute, United Kingdom
3. Lungs for Living, UCL Respiratory, University College London, United Kingdom
4. The Royal Marsden Hospital, United Kingdom
5. University College London Hospitals NHS Foundation Trust, United Kingdom
6. University Hospital of South Manchester, United Kingdom
7. Manchester Cancer Research Centre Biobank, United Kingdom
8. Christie NHS Foundation Trust, Manchester, United Kingdom
9. Cancer Research UK Manchester Institute, United Kingdom
10. Heart of England NHS Foundation Trust, Birmingham, United Kingdom
11. Cancer Studies and Molecular Medicine, University of Leicester, United kingdom
12. Leicester University Hospitals, United Kingdom
13. National Institute for Health Research Leicester Respiratory Biomedical, Research Unit, United Kingdom
14. North Middlesex Hospital, United Kingdom
15. Royal Free Hospital, United Kingdom
16. Barnet Hospital, United Kingdom
17. Aberdeen Royal Infirmary, United Kingdom
18. Velindre Cancer Centre, Cardiff, Wales, United Kingdom
19. Cardiff & Vale University Health Board, Cardiff, Wales, United Kingdom
20. University Hospital Of Wales Heath Park, Cardiff, Wales, United Kingdom
21. Department of Pathology, University Hospital of Wales and Cardiff University, Heath Park, Cardiff, Wales, United Kingdom
22. The Whittington Hospital NHS Trust, United Kingdom
23. Independent Cancer Patients Voice, United Kingdom
24. Cancer Research UK & UCL Cancer Trials Centre, United Kingdom
25. University Hospital Southampton NHS Foundation Trust, United Kingdom
26. Royal Brompton and Harefield NHS Foundation Trust, United Kingdom
27. National Heart and Lung Institute, Imperial College, United Kingdom
28. Barts Health NHS Trust, United Kingdom
29. Ashford and St. Peter's Hospitals NHS Foundation Trust, United Kingdom
30. The Princess Alexandra Hospital NHS Trust, United Kingdom
31. Sheffield Teaching Hospitals NHS Foundation Trust, United Kingdom
32. Liverpool Heart and Chest Hospital NHS Foundation Trust, United Kingdom
33. Natera Inc., 201 Industrial Road, Suite 410, San Carlos, CA 94070
34. Dana-Farber Cancer Institute, Brigham & Women’s Hospital, Harvard Medical School, 450 Brookline Ave, JF518, Boston, MA 02115-5450, USA
35. Wellcome Trust Sanger Institute, Hinxton, CB10 1SA, United Kingdom
36. Bioinformatics Group, Department of Computer Science, University College London
37. University of Lausanne
38. Swiss Institute of Bioinformatics
The PEACE consortium members and affiliations
The PEACE (Posthumous Evaluation of Advanced Cancer Environment) Study (Clinicaltrials.gov no: NCT03004755), is sponsored by University College London (UCL/13/0165) has been approved by an independent Research Ethics Committee (13/LO/0972). PEACE is funded by Cancer Research UK (C416/A21999) and coordinated through the Cancer Research UK & UCL Cancer Trials Centre.
Charles Swanton1,2,3, Mariam Jamal-Hanjani1,2,3, Christopher Abbosh1,2,3, Kai-Keen Shiu3, John Bridgewater3, Daniel Hochauser1,3, Peter Van Loo2, Sergio Quezada1, Stephan Beck1, Peter Parker2, Henning Walczak1, Tariq Enver1, Mary Falzon3, Ian Proctor3, Ron Sinclair3, Chi-wah Lok3, Marco Novelli3, Teresa Marafioti3, Elaine Borg3, Miriam Mitchison3, Giorgia Trevisan3, Mark Lynch3, Sebastian Brandner3, Faye Gishen23, Adrian Tookman23,24, Paddy Stone3, Caroline Sterling3, James Larkin4,5, Samra Turajlic2,4,5, Gert Attard4,5, Ros Eeles4,5, Chris Foster4,5, Steve Bova25, Andrea Sottoriva4,5, Simon Chowdhury6, Chandra Ashish6, James Spicer6, Mark Stares1,2,3, Joanna Lynch4,6, Carlos Caldas18,19, James Brenton18, Rebecca Fitzgerald20, Merche Jimenez-Linan7, Elena Provenzano7, Alison Cluroe7, Grant Stewart26, Colin Watts7, Richard Gilbertson19, Ultan McDermott7,8, Simon Tavare27, Tim Maughan28, Ian Tomlinson21, Peter Campbell8, Iain McNeish17, Andrew Biankin17, Antony Chambers17, Sioban Fraser17, Karin Oien17,Matt Krebs9, Fiona Blackhall10, Yvonne Summers10, Caroline Dive9, Richard Marais9, Louise Carter10, Daisuke Nonaka10, Anne Marie Quinn10, Nathalie Dhomen9, Dean Fennell11, John Le Quesne11, David Moore11, Jacqui Shaw11, Babu Naidu12, Shobhit Baijal12, Bruce Tanchel12, Gerald Langman12, Martin Collard12, Peter Cockcroft12, Joanne Taylor12, Hollie Bancroft12, Amy Kerr12, Gary Middleton12, Joanne Webb12, Salma Kadiri12, Dr Peter Colloby12, Bernard Olisemeke12, Rodelaine Wilson12, Christian Ottensmeier22, David Harrison29, Massimo Loda13, Adreinne Flanagan14, Maggie Wilcox15, Mairead McKenzie15, Allan Hackshaw16, Jonathan Lederman16, Abby Sharp16, Laura Farrelly16
Affiliations
1. Cancer Research UK Lung Cancer Centre of Excellence, University College London Cancer Institute, United Kingdom
2. The Francis Crick Institute, United Kingdom
3. University College London Hospitals NHS Foundation Trust, United Kingdom
4. The Royal Marsden Hospital, United Kingdom
5. The Institute of Cancer Research, London, United Kingdom
6. Guy’s and St Thomas’, NHS Foundation Trust, London, United Kingdom
7. Addenbrooke’s Hospital, NHS Foundation Trust, Cambridge, United Kingdom
8. Wellcome Trust Sanger Institute, Hinxton, United Kingdom
9. Cancer Research UK Manchester Institute, United Kingdom
10. Christie NHS Foundation Trust, Manchester, United Kingdom
11. Leicester University Hospitals, United Kingdom
12. Heart of England NHS Foundation Trust, Birmingham, United Kingdom
13. Division of Cancer Studies, King’s College London, United Kingdom
14. Royal National Orthopaedic Hospital, NHS Foundation Trust, London, United Kingdom
15. Independent Cancer Patients’ Voice, London, United Kingdom
16. Cancer Research UK & UCL Cancer Trials Centre, United Kingdom
17. Wolfson Wohl Cancer Research Centre, University of Glasgow
18. Cancer Research UK Cambridge Centre, Cambridge, United Kingdom
19. Department of Oncology, University of Cambridge, Cambridge, United Kingdom
20. Medical Research Council (MRC) Cancer Unit, Hutchison-MRC Research Centre and University of Cambridge, Cambridge, United Kingdom
21. Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, United Kingdom
22. University of Southampton and Southampton University Hospitals, Southampton
23. Royal Free Hospital, London, United Kingdom
24. Marie Curie Hospice, Hampstead, United Kingdom
25. Prostate Cancer Research Centre, University of Tampere, Finland
26. Department of Surgery, University of Cambridge, Cambridge, United Kingdom
27. Cancer Research UK Cambridge Institute, University of Cambridge, Cambridge, United Kingdom
28. Cancer Research UK/ Medical Research Council Oxford Institute for Radiation Oncology, Department of Oncology, University of Oxford, Oxford, United Kingdom
29. University of St Andrews, Fife, United Kingdom
Footnotes
Authorship contribution statement
C.A., N.J.B., G.A.W., M.J-H., T.C., R.S., and J.L-Q. contributed equally to this work. C.A. and C.S. co-wrote the manuscript. C.A., M.J.H., and C.S. conceived study design. C.A., N.J.B., G.A.W. and R.R. integrated clinicopathological data, exome data and ctDNA data. M.R. B.G.Z, J.L., T.C., R.S., E.K., N.S., D.H., A.N. and A.P., conducted and analysed multiplex-PCR NGS experimental work. N.J.B, G.A.W, T.B.K.W, M.A.B, R.R., and N.M. conducted M-Seq analyses of exome data. J.L-Q, T.M. and D.A.M. conducted pathological review. F.F., R.E. and F.Z. conducted radiological review of PET scans. H.J.W.L.A., W.L.B., F.M.F. and N.J.B. conducted radiomic analyses. S.V., D.J., J.L., S.S., J.C-K., A.R., T.C., D.O. and A.U.A. conducted TRACERx sample processing. G.E., S.W., N.M. and G.A.W. conducted exome sequencing. L.M., J.R. and J.S. conducted ctDNA cross-platform validation. M.J.H., C.D., J.S. and C.S. designed study protocols. C.H., S.L.M., M.D.F., T.A., M.Fa., E.B., D.L., M.H., S.K., N.P., S.M.J., R.T., A.A., F.B., Y.S., R.S., L.J., A.M.Q, P.C., B.N., G.M., G.L., S.T., M.N., H.R., K.K., M.C., L.G., D.F., A.N., S.R., G.A., S.K., P.R., V.E., B.I., M.I-S., V.P., J.L., M.K., R.A., H.A., H.D., S.L. are clinical members of TRACERx study sites. J.H. and H.L. run the UCL GCLP facility. A.H., H.B., N.I. and Y.N. were involved in study oversight. J.A.S., J.L-Q., Z.S., E.G., S.K., S.T., M.A.B, R.F.S., J.H., A.S., S.Q., P.V.L., C.D. and J.L. gave advice and reviewed the manuscript. A.H. gave statistical advice. C.S. provided overall study oversight.
Author information
The authors declare competing financial interests: details are available in the online version of the paper.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability Statement
Sequence data has been deposited at the European Genome-Phenoma Archive (EGA), which is hosted by the The European Bioinformatics Institute (EBI) and the Centre for Genomic Regulation (CRG), under accession numbers EGAS00001002247 (primary tumor data) and EGAS00001002415 (metastatic tumor data). Further information about EGA can be found on https://ega-archive.org, “The European Genome-phenome Archive of human data consented for biomedical research”27.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Pignon J-P, Tribodet H, Scagliotti GV, Douillard J-Y, Shepherd FA, Stephens RJ, et al. Lung Adjuvant Cisplatin Evaluation: A Pooled Analysis by the LACE Collaborative Group. Journal of Clinical Oncology. 2008;26(21):3552–9. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 4.Landau Dan A, Carter Scott L, Stojanov P, McKenna A, Stevenson K, Lawrence Michael S, et al. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell. 152(4):714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaver JA, Jelovac D, Balukrishna S, Cochran RL, Croessmann S, Zabransky DJ, et al. Detection of Cancer DNA in Plasma of Patients with Early-Stage Breast Cancer. Clinical Cancer Research. 2014;20(10):2643–50. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Science Translational Medicine. 2015;7(302):302ra133–302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 7.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Science Translational Medicine. 2016;8(346):346ra92–ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamal-Hanjani M, Hackshaw A, Ngai Y, Shaw J, Dive C, Quezada S, et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol. 2014;12(7):e1001906. doi: 10.1371/journal.pbio.1001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamal-Hanjani M. TRACERx – Tracking Non-Small Cell Lung Cancer Evolution. New England Journal of Medicine. 2017 (accepted, in press) [Google Scholar]
- 10.Jamal-Hanjani M, Wilson GA, Horswell S, Mitter R, Sakarya O, Constantin T, et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Annals of Oncology. 2016;27(5):862–7. doi: 10.1093/annonc/mdw037. [DOI] [PubMed] [Google Scholar]
- 11.D LA, Jr, Bardelli A. Liquid Biopsies: Genotyping Circulating Tumor DNA. Journal of Clinical Oncology. 2014;32(6):579–86. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caruso R, Parisi A, Bonanno A, Paparo D, Quattrocchi E, Branca G, et al. Histologic coagulative tumour necrosis as a prognostic indicator of aggressiveness in renal, lung, thyroid and colorectal carcinomas: A brief review. Oncology Letters. 2012;3(1):16–8. doi: 10.3892/ol.2011.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vesselle H, Schmidt RA, Pugsley JM, Li M, Kohlmyer SG, Vallières E, et al. Lung Cancer Proliferation Correlates with [F-18]Fluorodeoxyglucose Uptake by Positron Emission Tomography. Clinical Cancer Research. 2000;6(10):3837–44. [PubMed] [Google Scholar]
- 14.Higashi K, Ueda Y, Yagishita M, Arisaka Y. FDG PET measurement of the proliferative potential of non-small cell lung cancer. The Journal of Nuclear Medicine. 2000;41(1):85. [PubMed] [Google Scholar]
- 15.Murtaza M, Dawson S-J, Pogrebniak K, Rueda OM, Provenzano E, Grant J, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nature Communications. 2015;6:8760. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–54. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Monte U. Does the cell number 109 still really fit one gram of tumor tissue? Cell Cycle. 2009;8(3):505–6. doi: 10.4161/cc.8.3.7608. [DOI] [PubMed] [Google Scholar]
- 18.Peters S, Zimmermann S. Targeted therapy in NSCLC driven by HER2 insertions. Translational Lung Cancer Research. 2014;3(2):84–8. doi: 10.3978/j.issn.2218-6751.2014.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2005;19(2):264–71. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 20.Keam B, Im S-A, Kim H-J, Oh D-Y, Kim JH, Lee S-H, et al. Prognostic impact of clinicopathologic parameters in stage II/III breast cancer treated with neoadjuvant docetaxel and doxorubicin chemotherapy: paradoxical features of the triple negative breast cancer. BMC Cancer. 2007;7:203. doi: 10.1186/1471-2407-7-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee J, Han SW, Oh DY, Kim JH, Im SA, Han W, et al. The clinicopathologic characteristics and prognostic significance of triple-negativity in node-negative breast cancer. BMC Cancer. 2008;8:307. doi: 10.1186/1471-2407-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Team TNLSTR. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. New England Journal of Medicine. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotech. 2016;34(5):547–55. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofheinz F, Butof R, Apostolova I, Zophel K, Steffen IG, Amthauer H, et al. An investigation of the relation between tumor-to-liver ratio (TLR) and tumor-to-blood standard uptake ratio (SUR) in oncological FDG PET. EJNMMI Res. 2016;6(1):19. doi: 10.1186/s13550-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butof R, Hofheinz F, Zophel K, Stadelmann T, Schmollack J, Jentsch C, et al. Prognostic Value of Pretherapeutic Tumor-to-Blood Standardized Uptake Ratio in Patients with Esophageal Carcinoma. J Nucl Med. 2015;56(8):1150–6. doi: 10.2967/jnumed.115.155309. [DOI] [PubMed] [Google Scholar]
- 26.Malikic S, McPherson AW, Donmez N, Sahinalp CS. Clonality inference in multiple tumor samples using phylogeny. Bioinformatics. 2015;31(9):1349–56. doi: 10.1093/bioinformatics/btv003. [DOI] [PubMed] [Google Scholar]
- 27.Lappalainen I, Almeida-King J, Kumanduri V, Senf A, Spalding JD, Ur-Rehman S, et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat Genet. 2015;47(7):692–5. doi: 10.1038/ng.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.