Abstract
Background
Quantifying circulating nucleic-acids is an important new approach to cancer diagnosis/monitoring.
Methods
We compared the suitability of serum versus plasma for measuring microRNAs using RT-qPCR and assessed how pre-analytical variables that can affect circulating-tumor-DNA (ctDNA) quantification in plasma also influence microRNA levels.
Results
Across 62 blood-derived specimens, plasma samples in EDTA, Streck-DNA-plasma and Streck-RNA-plasma tubes showed significantly higher Ct values for multiple housekeeping microRNAs, compared with serum samples. For the EDTA-plasma tubes, this difference was only seen when including the high-speed centrifugation protocol used to optimise ctDNA extraction. In plasma samples derived from blood stored at room temperature for up to 14 days (conditions that typically apply to samples processed for biobanking), levels of endogenous housekeeping microRNAs gradually increased, in parallel with the hemolysis-marker hsa-miR-451a, consistent with release from blood cells/platelets. It was necessary to normalize levels of the housekeeping microRNAs to those of hsa-miR-451a, in order to obtain the stable values needed for referencing test microRNA levels.
Conclusions
Our data indicate that plasma samples prepared for ctDNA extraction are suboptimal for microRNA quantification and require the incorporation of multiple data normalization steps. For prospective studies designed to measure both microRNAs and ctDNA, the most suitable approach would be to obtain both serum (for microRNAs) and plasma (for ctDNA). If only plasma can be collected, we recommend an initial low-speed centrifugation step, followed by aliquoting the supernatant into parallel samples, one for direct microRNA quantification, and the other for a further high-speed centrifugation step to optimize ctDNA retrieval.
Impact
These recommendations will help ‘future-proof’ clinical studies in which quantification of circulating microRNAs is a component.
Keywords: blood, hemolysis, microRNA, plasma, serum
Introduction
New approaches to cancer diagnosis and/or monitoring are based on detecting nucleic acids in blood, including circulating tumor DNA (ctDNA) (1–6) and microRNAs (7,8). Studies using ctDNA have almost exclusively been based on plasma, due to the low yield of ctDNA retrieved from serum (9,10). Stability of ctDNA in plasma is improved using preservatives, for example those in Streck tubes (11–13). However, it is clear that multiple pre-analytical variables must be considered in prospective ctDNA-based clinical trials, including time to plasma isolation and the types of preservative used (14).
Detection of microRNAs is particularly attractive for many non-epithelial and/or pediatric cancers that have a low mutational prevalence (15,16). In testicular malignant germ cell tumors (GCTs), for example, mutations occur in less than half of all cases (17). We previously identified that microRNAs from the miR-371-373 and miR-302/367 clusters were co-ordinately elevated in the serum at the time of malignant GCT diagnosis (18,19) and were also sensitive indicators of disease relapse (20). Subsequent studies, from multiple groups, have all focussed on serum samples and have confirmed the clinical importance of this novel approach to malignant GCT diagnosis (20–30). In consequence, collection of biospecimens including blood for further evaluation of microRNA detection has been embedded in multiple prospective clinical trials across the spectrum of GCTs.
As future studies of circulating nucleic acids are likely to compare or combine microRNAs and ctDNAs for tumor diagnosis/monitoring (31), we sought to address whether plasma would also be a suitable medium for detecting circulating tumor-derived microRNAs. If so, this would allow combinatorial analysis of microRNAs and ctDNAs from the same sample. We also assessed whether pre-analytical factors that can affect ctDNA analysis in plasma samples would also affect microRNA levels in such samples. We compared serum obtained using gel separator tubes with plasma derived from EDTA, Streck DNA and Streck RNA tubes. For the EDTA tubes, we used two-step centrifugation protocols optimized for ctDNA extraction from plasma (32–34). We measured circulating levels of established housekeeping microRNAs and assessed effects on microRNA levels of tube type, time to processing, centrifugation speed and hemolysis. Based on our findings, we recommend protocols for microRNA detection that are optimised for blood samples from biobanks and biological studies linked to clinical trials.
Materials and Methods
Patient samples
In total, we analyzed 62 blood-derived specimens. First, we undertook a comparison study, in which we comprehensively analyzed 27 specimens collected from a cohort of adult male patients (n=7) recruited from a single hospital, via the Cambridge Urology Translational Research and Clinical Trials Biorepository (CUTRACT; cases CUB_001 – CUB_007). This cohort comprised a group with testicular malignant GCTs (n=5) and a control group with non-GCT testicular malignancy (n=2; one relapsed acute myeloid leukemia and one B-cell lymphoma). Second, we performed a time-course study of 26 samples from healthy adult male subjects (n=2), plus nine specimens from a patient with testicular Leydig cell tumor (CUB_008). Details of all specimens, including clinico-pathological information, are provided in Supplementary Table S1. All samples were collected with Local Research Ethics Committee approval (CUB samples reference 03/018; other samples reference 01/128) and informed consent, in accordance with the Declaration of Helsinki.
Sample collection and processing
For the comparison study, blood samples were collected immediately prior to orchiectomy. The tubes used were: 4.9ml Sarstedt S-Monovette Z-Gel (gel separator serum; ‘GSS’), 9ml Sarstedt S-Monovette K3E (EDTA plasma; ‘EP’), 10ml Streck Cell-Free DNA BCT (Streck DNA plasma; ‘SDP’) and 10ml Streck Cell-Free RNA BCT (Streck RNA plasma; ‘SRP’). Six patients provided samples in all four tubes, while one patient (CUB_003) provided samples in three tubes, as an SDP tube was unavailable at the time of patient recruitment. All specimens were centrifuged and processed at room temperature within two hours of venepuncture, according to the recommendations of the manufacturers of each tube. There was no difference in processing times (average or range) for the different tubes in the comparison study. Details of the processing are summarized in Supplementary Table S2. In brief, for GSS tubes, the blood was allowed to clot for 30 minutes (min), then centrifuged at 3,000g for 10min and the separated serum removed from above the gel layer. For EP tubes, the blood was centrifuged at 1,600g for 10min, then the separated plasma was aliquoted and subjected to a secondary spin of 14,400g for 10min, as per standard protocol conditions for preparing plasma for optimal ctDNA analysis (32–34). For SDP and SRP tubes, the blood was initially centrifuged at 300g for 20min and the separated plasma aliquoted into fresh tubes, prior to a secondary spin of 5,000g for 10min. For all samples, the final supernatants were aliquoted into fresh tubes, then stored immediately at -80°C until further analysis.
In the time-course study, we used 2ml SDP tubes, in which levels of RNA recovery and endogenous microRNAs were highly comparable to those in 10ml SDP tubes. Blood samples were collected into 1.2ml EP and 2ml SDP tubes, then stored at room temperature for 0, 2, 4, 7, 10 and 14 days before being processed as above. The samples from one subject (patient CUB_008) were also used to study the effects of centrifugation speed on microRNA recovery. Of two 1.2ml EP tube samples from this patient, one (EP #1) underwent only the first low-speed centrifugation (1,600g) only, whereas the other (EP #2) also underwent the second high-speed centrifugation (14,400g).
RNA extraction
RNA was isolated from 200μl of thawed serum or plasma using the miRNeasy serum/plasma kit (Qiagen, Crawley, UK), incorporating MS2 carrier RNA (Roche, Welwyn Garden City, UK) and the exogenous non-human microRNA spike-in, cel-miR-39-3p, as described (20). RNA was eluted in 100μl of nuclease-free water. For the comparison study, 750μl of QIAzol per sample was used. To assess whether the presence of proteins/nucleases within the biospecimens affected RNA retrieval, a subset of samples underwent a further RNA extraction that included a proteinase K digestion step. For this, 15μl proteinase K (minimum concentration >600mAU/ml; Qiagen) was added to 200μl of each sample, then incubated at 60°C for 60 minutes. Subsequently, 750μl of the QIAzol/MS2/cel-miR-39-3p mix was added, following which the protocol proceeded as described (20). The same protocols were used in the time-course study, except that the volume of QIAzol per sample was increased to 1000μl, following an update to the manufacturer’s recommendations.
We also assessed two alternative approaches to RNA extraction from SRP tubes, namely the Plasma/Serum RNA Purification Mini Kit (Norgen BioTek Corp, Thorold, ON, Canada) and the mirVana PARIS isolation kit (Ambion, Warrington, UK). For the Norgen kit, 200μl of circulating sample was used, with the addition of cel-miR-39-3p/MS2 as above, to the lysis buffer A. RNA was eluted using 50μl of nuclease-free water. The standard protocol for the Norgen plasma/serum kit was also adjusted to include a proteinase K digestion step, where 20μl of the proteinase K solution described above was added to lysis buffer A, mixed with the sample and incubated for 60min at 60°C. For the mirVana kit, 400μl of circulating sample was used, with the addition of 5.6 × 108 copies of cel-miR-39-3p spike-in and MS2 carrier RNA to the denaturing solution. In total, 80μl eluate was obtained.
RT-qPCR analysis of microRNA levels
In all experiments, levels of circulating microRNAs were detected using our singleplex RT-qPCR method, as described (20). In short, 5μl of eluted RNA was reverse transcribed using the TaqMan miRNA reverse transcription kit (Life Technologies, Paisley, UK), using the microRNA-specific stem-loop primer from the relevant TaqMan microRNA assay kit (Life Technologies). The final volume of 15μl for each reaction then underwent reverse transcription using a GeneAmp PCR System 9700 (Applied Biosystems, Warrington, UK) at 16°C for 30 min, 42°C for 30 min, followed by a final step of 85°C for 5min. A singleplex final PCR was then performed, as per the manufacturer’s instructions, on a Mastercycler ep_gradient/S realplex (Eppendorf, Stevenage, UK) at 95°C for 10min, followed by 45 cycles of 95°C for 15s and 60°C for one minute, as described (20).
The Ct threshold on the PCR machine was set manually to 2,000 fluorescence units across all PCR plates (20). To exclude non-specific amplification, a no-template control (NTC) was also run for each assay. None of the test samples had expression levels within 2 Ct values of the relevant NTC samples. Samples with Ct values ≥40, or those where the Ct threshold was not reached, were considered non-expressing and arbitrarily assigned a Ct value of 40. The standard deviations (SD) of Ct values for each set of technical triplicate RT-qPCR reactions were calculated. We compared Ct values and SD values between the different sample types. We defined Ct values for sample sets as being suboptimal if mean microRNA expression values were ≥2 Cts greater than the set with the lowest Ct value (i.e. expression levels were ≥4-fold lower). We also defined SD values for sample sets as being suboptimal if they were ≥2. For both Ct and SD assessment, samples that were not suboptimal were deemed to be satisfactory.
We quantified levels of six microRNAs, namely the exogenous spike-in microRNA cel-miR-39-3p (20) (added to each sample immediately prior to RNA extraction); the hemolysis-dependent hsa-miR-451a (35); and the endogenous human reference microRNAs hsa-miR-23a-3p, hsa-miR-30b-5p, hsa-miR-30c-5p and hsa-miR-191-5p, which were previously shown to be readily detectable in both serum and plasma (36,37). In addition, for the time-course study, we quantified levels of three further endogenous microRNAs, again using singleplex RT-qPCR, namely: hsa-miR-130b-3p and hsa-miR-146a-5p, previously shown to be present at stable levels in samples regardless of the degree of hemolysis (38); and hsa-miR-26a-5p, which had also been shown to be stable within the circulation (37).
Hemolysis assessment
In addition to visual inspection of each serum or plasma sample, hemolysis was assessed using two methods (8). First, spectrophotometric analysis of the absorbance of free hemoglobin at 414 nm (A414) (38,39) was performed using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Products, Wilmington, DE, US). Absorbance was tested in triplicate and the mean value recorded. Samples were classified as being hemolyzed if A414 values were >0.2 arbitrary units, as described (38,39). Second, using the RNA extracted from the sample, hsa-miR–451a Ct (35) and delta Ct (hsa-miR–23a–3p minus hsa-miR–451a) hemolysis values were calculated using singleplex RT-qPCR, as described (20). Delta Ct values ≥8 were considered to indicate hemolysis (20).
Statistical analysis
Statistics were performed using GraphPad Prism 6 software (San Diego, California, US). Unless otherwise stated, analyses were performed using either a two-tailed paired t-test (for matched samples) or a two-tailed unpaired t-test with Welch’s correction for unequal variance (for unmatched samples). P-values <0.05 were considered significant.
Results
Comparison study of microRNA levels in serum and plasma samples from different blood collection tubes
In the initial comparison study, we assessed levels of six microRNAs in serum samples prepared in GSS tubes, versus plasma samples prepared in EP, SDP or SRP tubes. Levels of each microRNA were quantified in each sample using technical triplicate RT-qPCR reactions. The SD values for each set of triplicate results were very low for all microRNAs across the GSS, EP and SDP samples. In contrast, the SD values were much higher for the samples in SRP tubes, particularly for hsa-miR-30b-5p and hsa-miR-23a-3p (Supplementary Figure S1).
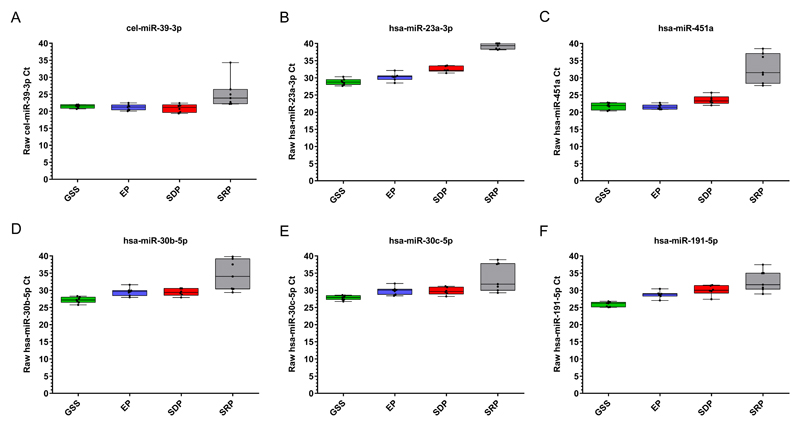
Levels of each individual microRNA in samples from each tube type are shown in Figure 1. In summary:
For the exogenous normalization microRNA cel-miR-39-3p, Ct values and SDs were satisfactory and similar in the GSS, EP and SDP groups (Figure 1, Table 1), indicating acceptable RNA extraction and RT-qPCR efficiency. In contrast, these parameters were suboptimal in the samples in SRP tubes (Table 1), where Ct values were significantly greater than in the EP group (Supplementary Table S3).
For the endogenous normalization microRNAs hsa-miR-30b-5p, hsa-miR-30c-5p, hsa-miR-191-5p and hsa-miR-23a-3p, Ct values were satisfactory only for serum samples in GSS tubes (Figure 1, Table 1). For the EP and SDP plasma samples, Ct values were suboptimal and significantly higher than in the samples in GSS tubes (Supplementary Table S3). For all four of the normalization microRNAs, the SRP samples showed the highest Ct values, which were significantly greater than in the GSS and EP groups (Supplementary Table S3).
For the hemolysis-associated microRNA hsa-miR-451a, Ct values were satisfactory for the GSS, EP and SDP groups (Figure 1, Table 1), but were again suboptimal in the SRP samples, where they were significantly higher than in each of the other sample groups (Supplementary Table S3).
Figure 1. Levels of individual microRNAs in samples from the four types of blood collection tube in the comparison study.
The plots show Ct values for: A) the exogenous non-human spike-in microRNA cel-miR-39-3p; B)-C) the microRNAs hsa-miR-23a-3p and hsa-miR-451a used for hemolysis assessment; and D)-F) the three established endogenous housekeeping microRNAs hsa-miR-30b-5p, hsa-miR-30c-5p, hsa-miR-191-5p. Key to blood collection tubes: GSS = gel separator serum (green boxes); EP = EDTA plasma (blue); SDP = Streck DNA plasma (red); SRP = Streck RNA plasma (grey). Bar = median, box = interquartile range; whiskers = full range of data.
Table 1. MicroRNA levels in each of the four blood collection tubes from the comparison study.
Comparison of Ct values and SD values between the different sample types. Ct values for sample sets were defined as being suboptimal if mean microRNA expression values were ≥2 Cts greater than the set with the lowest Ct value. SD values for sample sets were defined as suboptimal if they were ≥2. For both Ct and SD assessment, samples that were not suboptimal were deemed to be satisfactory. Key to blood collection tubes: GSS = gel separator serum; EP = EDTA plasma; SDP = Streck DNA plasma; SRP = Streck RNA plasma.
| MicroRNA | Role | GSS | EP | SDP | SRP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean Ct value | SD | Conclusion | Mean Ct value | SD | Conclusion | Mean Ct value | SD | Conclusion | Mean Ct value | SD | Conclusion | ||
| cel-miR-39-3p | Exogenous normalization | 21.43 | 0.49 | Satisfactory | 21.13 | 0.85 | Satisfactory | 20.91 | 1.19 | Satisfactory | 25.23 | 4.29 | Suboptimal |
| hsa-miR-30b-5p | Endogenous normalization | 27.16 | 0.90 | Satisfactory | 29.57 | 1.18 | Suboptimal | 29.44 | 1.04 | Suboptimal | 34.41 | 4.45 | Suboptimal |
| hsa-miR-30c-5p | Endogenous normalization | 27.77 | 0.68 | Satisfactory | 29.93 | 1.16 | Suboptimal | 29.73 | 1.11 | Suboptimal | 33.78 | 4.16 | Suboptimal |
| hsa-miR-191-5p | Endogenous normalization | 25.91 | 0.72 | Satisfactory | 28.76 | 1.00 | Suboptimal | 30.02 | 1.49 | Suboptimal | 32.74 | 3.10 | Suboptimal |
| hsa-miR-23a-3p | Endogenous normalization (hemolysis) | 28.84 | 0.89 | Satisfactory | 30.26 | 1.10 | Satisfactory | 32.47 | 0.86 | Suboptimal | 39.20 | 0.84 | Suboptimal |
| hsa-miR-451a | Hemolysis assessment | 21.73 | 0.96 | Satisfactory | 21.55 | 0.72 | Satisfactory | 23.50 | 1.27 | Satisfactory | 32.90 | 4.30 | Suboptimal |
We next compared mean Ct values for different microRNAs in each of the four tube types (Supplementary Figure S2). Variations in levels of cel-miR-39-3p and those of hsa-miR-30b-5p (Supplementary Figure S2) were lowest for samples in GSS tubes, which showed very tight clustering of Ct values. There was moderate clustering for the EP and SDP samples, but very poor clustering for the SRP samples (Supplementary Figure S2). Identical findings were also seen for hsa-miR-30c-5p, hsa-miR-191-5p and hsa-miR-23a-3p with cel-miR-39-3p. These data indicated that the EP, SDP and SRP tubes resulted in additional technical variation, when compared with GSS tubes.
We attempted to develop a method that would allow satisfactory RNA extraction and RT-qPCR analysis of microRNA levels in plasma from SRP tubes. . Extracting RNA from SRP tubes using the Norgen plasma/serum RNA purification kit reduced variability in levels of all six microRNAs, compared with RNA extracted from SRP tubes using the Qiagen miRNeasy serum/plasma kit (Supplementary Figure S3). However, mean Ct values of all six microRNAs in such SRP/Norgen samples remained ≥2 higher than those obtained with RNA extracted from GSS tubes using the Qiagen miRNeasy kit. Indeed, for the exogenous normalization microRNA cel-miR-39-3p, levels in the SRP/Norgen samples were almost 5 Ct values greater than in the GSS/Qiagen samples. As the eluate from the former was 50μl and the latter 100μl, with 5μl from each being used for PCR, this amounted to ~58-fold less efficient RNA recovery from the original 200μl serum or plasma samples. Such differences in microRNA recovery using the Norgen protocol were not improved by modifications such as the incorporation of a proteinase K digestion step. When RNA was extracted from SRP tubes using the mirVana PARIS kit, expression levels of each of the six microRNAs tested (i.e. those listed in Table 1) were also suboptimal. This large reduction in the efficiency of RNA recovery would be expected to limit the overall sensitivity of a PCR assay for detecting test microRNA biomarkers (particularly those in low abundance), when compared with a serum-based approach. As we were unable to resolve the limitations of SRP tubes for quantifying circulating microRNAs, we did not include them in our subsequent analyses.
Time-course study of the effects of blood storage on microRNA levels in plasma
We studied the effects on microRNA quantification of sample storage in SDP tubes at room temperature for up to 14 days, which is the recommended time limit for sample preservation prior to plasma extraction for ctDNA quantification. We compared our findings with plasma samples stored at room temperature for the same time periods in EP tubes. In total, we analyzed 26 separate samples from two healthy subjects, comparing 14 samples in SDP tubes with 12 in EP tubes (Supplementary Table S1).
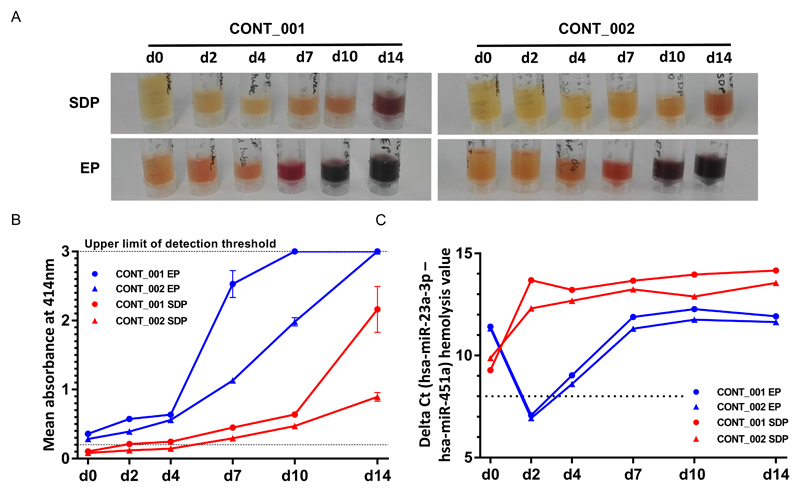
Levels of hemolysis were lower in the SDP samples compared with the EP samples at matched time-points, as indicated by sample inspection (Figure 2A) and by spectrophotometric measurement of free hemoglobin absorption at 414nm (A414) (Figure 2B). Using the standard A414 threshold of >0.2 arbitrary units (38,39), the EP samples were hemolyzed within 1 hour of collection (i.e. the d0 samples). They showed an abrupt increase in hemolysis from d4, consistently reaching the upper limit of detection threshold by d14. In contrast, hemolysis was not consistently present in the SDP samples until d7. Assessment of hemolysis using microRNA delta Ct values (hsa-miR-23a-3p – hsa-miR-451a) indicated hemolysis at almost all time-points in both sample types, albeit with occasional variation in the values observed (Figure 2C).
Figure 2. Levels of hemolysis in plasma samples from the time-course study.
A) Appearance of the plasma samples derived from the SDP and EP tubes taken from two healthy control subjects (CONT_001 and CONT_002) at the six time-points [day (d) 0, d2, d4, d7, d10 and d14]. The darker the sample color, the greater the degree of hemolysis. B) Spectrophotometric analysis of the plasma samples in A), showing mean absorbance of free hemoglobin at 414nm (A414). The lower dotted line indicates the threshold (0.2) above which samples were classified as hemolyzed, while the upper dotted line (3.0) indicates the upper limit of detection of the spectrophotometer. C) Hemolysis assessment by delta Ct (hsa-miR-23a-3p – hsa-miR-451a) values. The dotted line indicates the Ct value threshold (8) above which samples were classified as hemolyzed. Key: EP = EDTA plasma (blue lines); SDP = Streck DNA plasma (red lines). Error bars = SD.
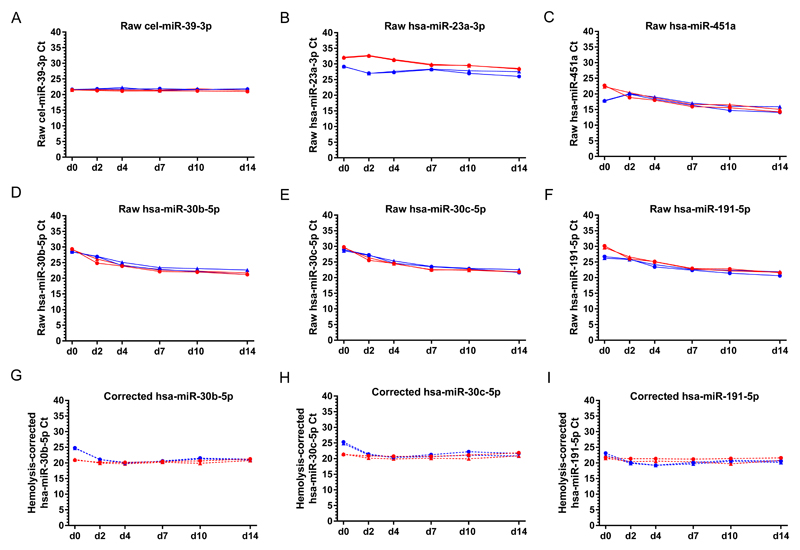
Next, we investigated changes in levels of individual microRNAs in plasma obtained from the SDP versus the EP tubes over the 14 day time-course (20). As cel-miR-39-3p was spiked-in immediately prior to RNA extraction for each time-point, it was not surprising that we saw no difference in levels of this microRNA (Figure 3A). In contrast, levels of all six endogenous housekeeping microRNAs tested [hsa-miR-30b-5p, hsa-miR-30c-5p, hsa-miR-191-5p (Figure 3); hsa-miR-26a-5p, hsa-miR-130b-3p, hsa-miR-146a-5p (Supplementary Figure S4)] all showed similar gradual reductions in Ct values over the time-course, indicating gradual increases in expression levels in the blood from which the plasma samples were derived. Similar changes were seen in the red blood cell derived microRNA hsa-miR-451a, suggesting that the elevations in endogenous housekeeping microRNAs were related to hemolysis. Indeed, normalization of levels of the six endogenous housekeeping microRNAs to those of hsa-miR-451a resulted in stabilization of microRNA measurements over the time-course (Figure 3 and Supplementary Figure S4). These findings were confirmed in a further set of SDP samples obtained from a patient with testicular Leydig cell tumor (CUB_008; Supplementary Table S1, Supplementary Figure S5).
Figure 3. MicroRNA levels in plasma samples from the time-course study.
Graphs show Ct values for the plasma samples derived from the SDP and EP tubes taken from two healthy control subjects (CONT_001 and CONT_002) at the six time-points [day (d) 0, d2, d4, d7, d10 and d14]. Panels A)-F) show raw Ct values (solid lines) for: A) the exogenous non-human spike-in microRNA cel-miR-39-3p; B)-C) the microRNAs hsa-miR-23a-3p and hsa-miR-451a used for hemolysis assessment; and D-F) the three established endogenous housekeeping microRNAs hsa-miR-30b-5p, hsa-miR-30c-5p and hsa-miR-191-5p. Panels G)-I) show the hemolysis-corrected Ct values (dotted lines) for the three endogenous housekeeping microRNAs shown in D)-F). Key: EP = EDTA plasma (blue lines); SDP = Streck DNA plasma (red lines). Note that error bars are not visible as they fall within each respective data-point.
Comparison of methods of hemolysis assessment in plasma samples
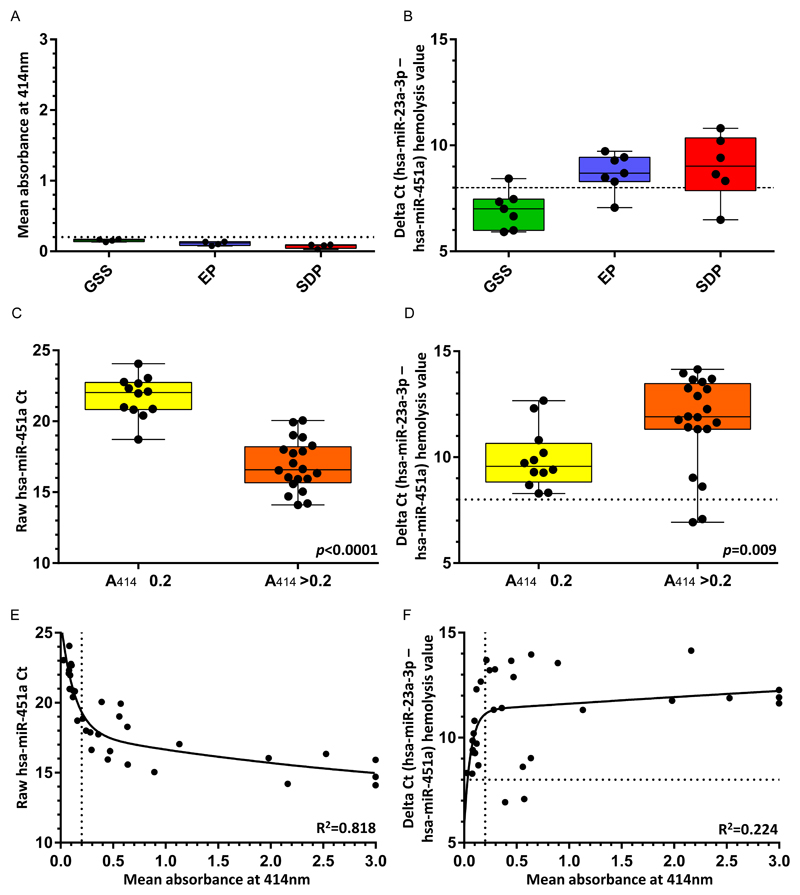
When quantifying test microRNAs, it is important also to measure hemolysis, in order to exclude any confounding effects of microRNAs released from blood cells (40). We investigated whether previously established parameters for measuring hemolysis in serum using microRNA quantification (8,20,41) could also be applied to the plasma samples in our study. The established delta Ct (hsa-miR-23a-3p - hsa-miR-451a) hemolysis method was applied to the samples from the comparison study in which there was no evidence of hemolysis by spectrophotometry (n=20) (Figure 4A). A delta Ct value indicating hemolysis (delta Ct >8) was seen in only one of seven GSS serum samples but in six of seven EP and five of six SDP plasma samples (Figure 4B). The false positive results in the plasma samples were attributable to significantly lower detection levels (higher Ct values) of the endogenous housekeeping gene hsa-miR-23a-3p in the EP and SDP samples, compared with the GSS samples (p=0.018 and p=0.0002, respectively) (Figure 1E, Supplementary Table S3).
Figure 4. Comparison of methods of hemolysis quantification.
A)-B) Measurement of hemolysis in samples from the comparison study using: mean absorbance of free hemoglobin at 414nm (A414), as assessed by spectrophotometry (A); and delta Ct (hsa-miR-23a-3p - hsa-miR-451a) values (B). C)-D) Raw Ct hsa-miR-451a values (C) and delta Ct (hsa-miR-23a-3p - hsa-miR-451a) values (D) in 32 EP and SDP samples from both the comparison and time-course studies, identified by spectrophotometry as non-hemolyzed (yellow box; A414 values ≤0.2) or hemolyzed (orange box; A414 values >0.2). E)-F) Associations of raw Ct hsa-miR-451a values (E) and delta Ct (hsa-miR-23a-3p - hsa-miR-451a) values (F) with A414 absorbance for the 32 plasma samples analyzed in C) and D), including the lines of best fit. Key: GSS = gel separator serum (green box); EP = EDTA plasma (blue); SDP = Streck DNA plasma (red). Bar = median, box = interquartile range; whiskers = full range of data. Hemolysis thresholds are indicated by the dotted lines, as defined in the legend to Figure 2.
When analysing 32 available EP and SDP samples from across the comparison and time-course studies, only 20 (62%) were classified as hemolyzed by spectrophotometry (Figures 4C/D), whereas 30 (94%) were classified as hemolyzed by using the delta Ct value (Figure 4D). The samples that were classified as hemolyzed by spectrophotometry showed higher delta Ct values (p=0.009, unpaired t-test; Figure 4D) and lower raw Ct values for hsa-miR-451a (p<0.0001, unpaired t-test; Figure 4C), compared with the samples that were classified as non-hemolyzed by spectrophotometry. The A414 absorbance values strongly fitted a non-linear association with the raw hsa-miR-451a Ct values (R2=0.818; Figure 4E). However, they only weakly fitted a non-linear association with the delta Ct (hsa-miR-23a-3p – hsa-miR-451a) values (R2=0.224; Figure 4F), in keeping with the limited accuracy of hsa-miR-23a-3p quantification in the plasma samples. The dynamic range of the A414 absorbance values was greater than those of the raw hsa-miR-451a values (Figure 4E) and delta Ct (hsa-miR-23a-3p – hsa-miR-451a) values (Figure 4F), further indicating the limited utility of microRNA quantification for assessing hemolysis in the plasma samples.
Effect of centrifugation speed on microRNA levels in plasma
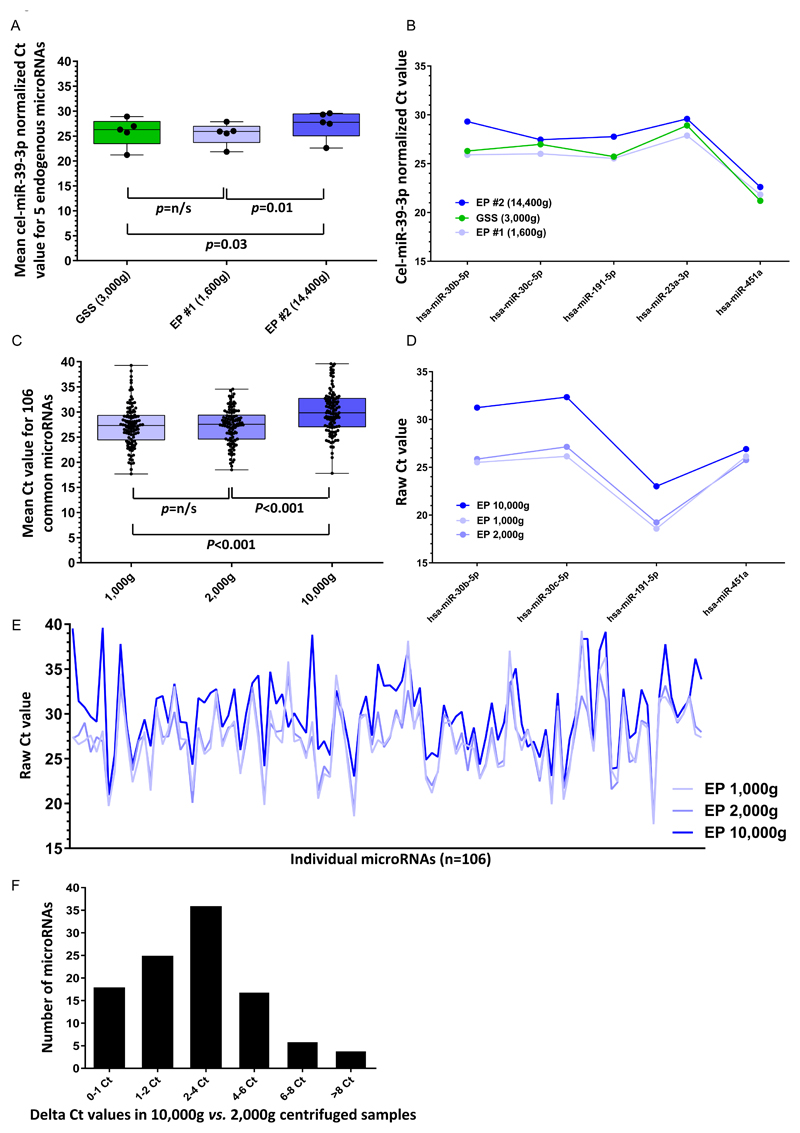
We sought to determine whether the differences observed between the serum and plasma samples were due to inherent features of the sample types or to the different processing protocols used. We examined the effect of centrifugation speed on microRNA recovery from plasma, using samples extracted from EP tubes, in comparison with serum samples prepared from GSS tubes. We first used samples from patient CUB_008, to compare the effect of a single low-speed centrifugation step (1,600g; EP #1) with the standard dual centrifugation that is typically used when preparing plasma for ctDNA analysis (1,600g then 14,400g; EP #2). The mean expression level of five microRNAs (namely hsa-miR-30b-5p, -30c-5p, -191-5p, -23a-3p and -451a) was significantly lower (higher Ct values) in the EP #2 sample, compared with the EP #1 (p=0.03) and the GSS sample (p=0.01) (Figure 5A). When analyzing each microRNA individually, levels were also lower in the EP #2 sample compared with the EP #1 and GSS samples (Figure 5B). There were no differences between the EP #1 and GSS samples in the mean expression levels (Figure 5A), nor in the levels of each microRNA individually (Figure 5B).
Figure 5. Effect of centrifugation speed on microRNA levels in plasma samples.
A) Ct values (cel-miR-39-3p-normalized) for five endogenous microRNAs (hsa-miR-30b-5p, hsa-miR-30c-5p, hsa-miR-191-5p, hsa-miR-23a-3p and hsa-miR-451a) in the GSS, EP #1 and EP #2 samples from patient CUB_008; B) Individual Ct values (cel-miR-39-3p-normalized) for the five endogenous microRNAs assessed in A). Key for A)-B): GSS = gel separator serum (green box); EP #1 = EDTA plasma (low-speed centrifugation only, light blue); EP #2 = EDTA plasma (including high-speed centrifugation, blue). C) Mean Ct values for 106 endogenous microRNAs in EP samples processed using different centrifugation speeds (42). D)-E) Individual raw Ct values for four of the five endogenous microRNAs from panels A) and B) (D) and all 106 microRNAs (E) in EP samples processed using different centrifugation speeds. F) Differences in Ct values for all 106 microRNAs in EP samples centrifuged at 10,000g vs. 2,000g. Key for C)-F): EP 1,000g = EDTA plasma samples with low-speed centrifugation at 1,000 g (light-blue); EP 2,000g = EDTA plasma samples with low-speed centrifugation at 2,000 g (mid-blue); EP 10,000g = EDTA plasma samples with high-speed centrifugation at 10,000 g (dark-blue). In panels A) and C), bar = median, box= interquartile range; whiskers = full range of data.
We extended these findings by interrogating data from a published global microRNA profiling study of EP samples (from breast cancer patients) that had been centrifuged at various speeds (42). For the 106 microRNAs with Ct values <35 [listed in (42)], mean levels were significantly lower in EP samples that underwent high-speed centrifugation (10,000g), compared with EP samples that underwent low-speed centrifugations (either 2,000g or 1,000g; p<0.001 for both comparisons) (Figure 5C). There was no significant difference in mean microRNA expression levels between the 1,000g and 2,000g centrifugation samples (Figure 5C). Levels of individual microRNAs were again lower (higher Ct values) in the samples that had received high-speed centrifugation, compared with the low-speed samples (Figure 5D). The differences in expression levels were particularly large for the three endogenous housekeeping genes hsa-miR-30b-5p, hsa-miR-30c-5p, hsa-miR-191-5p, but were also seen for hsa-miR-451a (data on hsa-miR-23a-3p levels were not available in the published dataset). Across the 106 microRNAs, high-speed centrifugation had a highly unpredictable effect on microRNA levels compared with low-speed centrifugation (Figure 5E). Ct values were almost always higher in the high-speed samples, albeit with rare exceptions (Figure 5E). The difference in expression levels ranged from zero to >8 Ct values, with a modal difference of 2-4 Ct values (Figure 5F).
Discussion
Biospecimens collected by tissue banks increasingly include blood-derived samples for quantification of circulating nucleic acids. Most effort to date has focussed on studying pre-analytical variables that affect ctDNA quantification (11–14). Here, we sought to establish how such variables affect microRNA levels and to generate a recommended protocol for collecting and processing blood-derived biospecimens, in order to maximise the yield of recovered microRNAs. Our comparison study demonstrated that serum samples were better suited for microRNA studies than the plasma samples tested, as the latter showed greater technical variation, with consistently lower levels of endogenous housekeeping microRNAs. The differences we observed were not attributable to the method used for microRNA extraction, as the Qiagen miRNeasy serum/plasma kit was independently demonstrated to provide the highest yield of microRNAs from plasma samples (42). The difference between serum and plasma from EP tubes (Figure 1) is likely to relate to the two-spin processing used for preparing the latter, a method that is typically adopted to optimize ctDNA extraction. Samples processed from EP tubes with a low-speed centrifugation step only did not show significant differences in microRNA levels compared with serum samples (Figure 5A/B), indicating that plasma is not necessarily an inferior substrate for microRNA extraction, if appropriate protocols are adopted. We found Streck RNA (SRP) tubes to be unsuitable for microRNA studies, despite using various RNA extraction methods and protocol modifications, including the incorporation of a proteinase K digestion step.
The uniform clotting process inherent in preparing serum removes a large proportion of protein from a whole blood sample. One practical benefit of this is that the layer of protein obtained when extracting RNA from serum is relatively small. While endogenous microRNAs are released into the serum during the clotting process, for example from platelets, the overall pipeline involved in serum processing is straightforward, and most importantly, provides very similar levels of endogenous housekeeping microRNAs across samples. As plasma lacks the clotting step, the protein layer obtained during RNA extraction is large, leading to smaller yields of supernatant. This may necessitate repeated sample processing in order to obtain adequate volumes of eluate for microRNA quantification, potentially exacerbating the technical variations in measuring microRNA levels in plasma.
A further consideration when evaluating blood-derived biospecimens is the stability of microRNAs during prolonged storage of unprocessed blood. Such conditions typically apply to the blood specimens from which plasma samples are obtained for biobanking, for example those from a multi-center clinical trial. In plasma samples derived from blood stored over a period of up to 14 days at room temperature, we observed increases in levels of the endogenous housekeeping microRNAs hsa-miR-30b-5p, hsa-miR-30c-5p and hsa-miR-191-5p. These were associated with parallel changes in the hemolysis marker hsa-miR-451a, suggesting that the microRNAs were likely to have been released into the plasma from red blood cells and/or platelets, either through active shedding or passively as a result of hemolysis. As a result, stabilization of the levels of housekeeping microRNAs in plasma samples over the 14 day time-course required correction for levels of hsa-miR-451a.
An additional consequence of the variations in levels of housekeeping microRNAs in the plasma samples studied is that hemolysis could not reliably be indicated by the delta Ct (hsa-miR-23a-3p - hsa-miR-451a) microRNA quantification method. This is an important contrast with serum samples, where hemolysis can be quantified accurately using this microRNA method (8,20). In our comparison study, the majority of the plasma samples were falsely classified as showing hemolysis by the delta Ct method. This was predominantly due to reduced levels of the housekeeping microRNA hsa-miR-23a-3p, rather than alterations in the levels of the direct hemolysis marker hsa-miR-451a. In our time-course study, delta Ct hemolysis levels in the plasma samples increased then plateaued during a period of ongoing hemolysis (which was indicated by sample inspection and spectrophotometric measurement of hemoglobin absorbance). Normalization of hsa-miR-23a-3p levels to those of hsa-miR-451a, as required for other housekeeping microRNAs in stored plasma samples, would not be appropriate when using the delta Ct (hsa-miR-23a-3p - hsa-miR-451a) method for hemolysis quantification.
Processing of plasma samples for ctDNA quantification typically involves a double centrifugation process, which includes a high-speed (≥10,000g) second spin, designed to remove background genomic DNA contamination (32–34). Our experimental findings and re-analysis of published data (42) show that such a two-step process significantly alters the profile of circulating microRNAs detected in plasma samples, compared with plasma samples processed by low-speed centrifugation only and also with serum samples. These findings are consistent with a previous global microRNA profiling study showing that the two-step method, including high-speed centrifugation, resulted in lower microRNA levels in plasma when compared with serum (43). Our analyses indicate that the alterations in plasma microRNA levels produced by high-speed centrifugation are highly variable and unpredictable. The microRNAs affected most may predominantly be present in relatively large structures in the plasma, such as platelets or large extracellular vesicles, that are removed by the high-speed centrifugation step (42).
Taken together, our data allow us to make recommendations for the design of biological studies linked to clinical trials, and/or retrospective studies of samples in biorepositories, where quantification of circulating microRNAs is a requirement. Our findings indicate that serum is the optimum biospecimen for quantifying circulating microRNAs, as it is subject to the least pre-analytical technical variation. The optimal approach for studies measuring both circulating microRNAs and ctDNA would therefore be to collect both serum (for microRNAs) and plasma (for ctDNA). If only plasma can be collected, then immediate processing is recommended, in order to avoid the technical variations that affect microRNA levels after storage and/or transport of plasma at room temperature. We further recommend that such plasma processing should involve a low-speed centrifugation step only, followed by aliquoting the supernatant into parallel samples prior to storage at -80°C. Such aliquots could then be used directly for microRNA quantification, or subjected to a further high-speed (≥10,000g) centrifugation step to optimise ctDNA retrieval (42).
For retrospective analysis of microRNA levels in plasma samples from biorepositories, particularly those prepared for ctDNA extraction, we recommend normalization of housekeeping microRNA levels using cel-miR-39-3p (correcting for technical differences in RNA recovery) and hsa-miR-451a (correcting for the effects of hemolysis), and also the use of multiple housekeeping microRNAs for referencing the levels of test microRNAs. These steps will improve the likelihood of avoiding biologically inconsistent or even contradictory results, as has recently been highlighted for various urological tumors (7). Despite these steps, our data indicate that the results of retrospective microRNA quantification work in such plasma samples should be interpreted with caution.
In our opinion, these technical considerations are of vital importance to the design of future clinical trials and/or retrospective studies of biobank samples. Our recommendations are practicable and scalable. They will help future-proof clinical studies in which quantification of circulating microRNAs is a component.
Supplementary Material
Acknowledgements
We thank the CUTRACT team for their assistance with this project and specifically Sara Stearn and Anne George. The urology bio-repository is funded by the CRUK Cambridge Centre urology malignancy programme. We are grateful to Laura Esposito, Emma Bell and Charlie Massie for technical advice.
Financial support: We acknowledge grant funding from The St.Baldrick’s Foundation (M.J. Murray/N. Coleman), The Isaac Newton Trust (M.J. Murray) and Great Ormond Street Hospital Children’s Charity/Children with Cancer UK (N. Coleman/M.J. Murray). We also thank the Max Williamson Fund (M.J. Murray/N. Coleman) for financial support.
Footnotes
Conflict of interest statement: the authors have nothing to disclose
References
- 1.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 2.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 3.Gray ES, Rizos H, Reid AL, Boyd SC, Pereira MR, Lo J, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6(39):42008–18. doi: 10.18632/oncotarget.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nature communications. 2015;6:8760. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 6.Schreuer M, Meersseman G, Van Den Herrewegen S, Jansen Y, Chevolet I, Bott A, et al. Quantitative assessment of BRAF V600 mutant circulating cell-free tumor DNA as a tool for therapeutic monitoring in metastatic melanoma patients treated with BRAF/MEK inhibitors. J Transl Med. 2016;14:95. doi: 10.1186/s12967-016-0852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fendler A, Stephan C, Yousef GM, Kristiansen G, Jung K. The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol. 2016;13(12):734–52. doi: 10.1038/nrurol.2016.193. [DOI] [PubMed] [Google Scholar]
- 8.Murray MJ, Huddart RA, Coleman N. The present and future of serum diagnostic tests for testicular germ cell tumours. Nat Rev Urol. 2016;13(12):715–25. doi: 10.1038/nrurol.2016.170. [DOI] [PubMed] [Google Scholar]
- 9.Anker P, Lyautey J, Lederrey C, Stroun M. Circulating nucleic acids in plasma or serum. Clin Chim Acta. 2001;313(1–2):143–6. doi: 10.1016/s0009-8981(01)00666-0. [DOI] [PubMed] [Google Scholar]
- 10.Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18(1):65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- 11.Kang Q, Henry NL, Paoletti C, Jiang H, Vats P, Chinnaiyan AM, et al. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem. 2016;49(18):1354–60. doi: 10.1016/j.clinbiochem.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Parpart-Li S, Bartlett B, Popoli M, Adleff V, Tucker L, Steinberg R, et al. The Effect of Preservative and Temperature on the Analysis of Circulating Tumor DNA. Clin Cancer Res. 2017;23(10):2471–77. doi: 10.1158/1078-0432.CCR-16-1691. [DOI] [PubMed] [Google Scholar]
- 13.Toro PV, Erlanger B, Beaver JA, Cochran RL, VanDenBerg DA, Yakim E, et al. Comparison of cell stabilizing blood collection tubes for circulating plasma tumor DNA. Clin Biochem. 2015;48(15):993–8. doi: 10.1016/j.clinbiochem.2015.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Dessel LF, Beije N, Helmijr JC, Vitale SR, Kraan J, Look MP, et al. Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions. Mol Oncol. 2017;11(3):295–304. doi: 10.1002/1878-0261.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litchfield K, Summersgill B, Yost S, Sultana R, Labreche K, Dudakia D, et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nature communications. 2015;6:5973. doi: 10.1038/ncomms6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray MJ, Halsall DJ, Hook CE, Williams DM, Nicholson JC, Coleman N. Identification of MicroRNAs From the miR-371~373 and miR-302 Clusters as Potential Serum Biomarkers of Malignant Germ Cell Tumors. Am J Clin Pathol. 2011;135(1):119–25. doi: 10.1309/AJCPOE11KEYZCJHT. [DOI] [PubMed] [Google Scholar]
- 19.Palmer RD, Murray MJ, Saini HK, van Dongen S, Abreu-Goodger C, Muralidhar B, et al. Malignant germ cell tumors display common microRNA profiles resulting in global changes in expression of messenger RNA targets. Cancer Res. 2010;70(7):2911–23. doi: 10.1158/0008-5472.CAN-09-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray MJ, Bell E, Raby KL, Rijlaarsdam MA, Gillis AJ, Looijenga LH, et al. A pipeline to quantify serum and cerebrospinal fluid microRNAs for diagnosis and detection of relapse in paediatric malignant germ-cell tumours. Br J Cancer. 2016;114(2):151–62. doi: 10.1038/bjc.2015.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belge G, Dieckmann KP, Spiekermann M, Balks T, Bullerdiek J. Serum levels of microRNAs miR-371-3: a novel class of serum biomarkers for testicular germ cell tumors? Eur Urol. 2012;61(5):1068–9. doi: 10.1016/j.eururo.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 22.Dieckmann KP, Radtke A, Spiekermann M, Balks T, Matthies C, Becker P, et al. Serum Levels of MicroRNA miR-371a-3p: A Sensitive and Specific New Biomarker for Germ Cell Tumours. Eur Urol. 2017;71(2):213–20. doi: 10.1016/j.eururo.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 23.Dieckmann KP, Spiekermann M, Balks T, Flor I, Loning T, Bullerdiek J, et al. MicroRNAs miR-371-3 in serum as diagnostic tools in the management of testicular germ cell tumours. Br J Cancer. 2012;107(10):1754–60. doi: 10.1038/bjc.2012.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillis AJ, Rijlaarsdam MA, Eini R, Dorssers LC, Biermann K, Murray MJ, et al. Targeted serum miRNA (TSmiR) test for diagnosis and follow-up of (testicular) germ cell cancer patients: A proof of principle. Mol Oncol. 2013;7:1083–92. doi: 10.1016/j.molonc.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray MJ, Coleman N. Testicular cancer: a new generation of biomarkers for malignant germ cell tumours. Nat Rev Urol. 2012;9(6):298–300. doi: 10.1038/nrurol.2012.86. [DOI] [PubMed] [Google Scholar]
- 26.Rijlaarsdam MA, van Agthoven T, Gillis AJ, Patel S, Hayashibara K, Lee KY, et al. Identification of known and novel germ cell cancer-specific (embryonic) miRs in serum by high-throughput profiling. Andrology. 2015;3(1):85–91. doi: 10.1111/andr.298. [DOI] [PubMed] [Google Scholar]
- 27.Spiekermann M, Belge G, Winter N, Ikogho R, Balks T, Bullerdiek J, et al. MicroRNA miR-371a-3p in serum of patients with germ cell tumours: evaluations for establishing a serum biomarker. Andrology. 2015;3(1):78–84. doi: 10.1111/j.2047-2927.2014.00269.x. [DOI] [PubMed] [Google Scholar]
- 28.Spiekermann M, Dieckmann KP, Balks T, Bullerdiek J, Belge G. Is Relative Quantification Dispensable for the Measurement of MicroRNAs as Serum Biomarkers in Germ Cell Tumors? Anticancer Res. 2015;35(1):117–21. [PubMed] [Google Scholar]
- 29.Syring I, Bartels J, Holdenrieder S, Kristiansen G, Muller SC, Ellinger J. Circulating Serum miRNA (miR-367-3p, miR-371a-3p, miR-372-3p and miR-373-3p) as Biomarkers in Patients with Testicular Germ Cell Cancer. J Urol. 2015;193(1):331–7. doi: 10.1016/j.juro.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 30.van Agthoven T, Looijenga LH. Accurate primary germ cell cancer diagnosis using serum based microRNA detection (ampTSmiR test) Oncotarget. 2016 doi: 10.18632/oncotarget.10867. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Y, Xu P, Mi Y, Wang W, Pan X, Wu X, et al. Plasma MiRNA alterations between NSCLC patients harboring Del19 and L858R EGFR mutations. Oncotarget. 2016;7(34):54965–72. doi: 10.18632/oncotarget.10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiu RW, Poon LL, Lau TK, Leung TN, Wong EM, Lo YM. Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma. Clin Chem. 2001;47(9):1607–13. [PubMed] [Google Scholar]
- 33.El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta. 2013;424:222–30. doi: 10.1016/j.cca.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Swinkels DW, Wiegerinck E, Steegers EA, de Kok JB. Effects of blood-processing protocols on cell-free DNA quantification in plasma. Clin Chem. 2003;49(3):525–6. doi: 10.1373/49.3.525. [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207(7):1351–8. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59(1):S1–6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 37.Murray MJ, Raby KL, Saini HK, Bailey S, Wool SV, Tunnacliffe JM, et al. Solid Tumors of Childhood Display Specific Serum microRNA Profiles. Cancer Epidemiol Biomarkers Prev. 2015;24(2):350–60. doi: 10.1158/1055-9965.EPI-14-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirschner MB, Edelman JJ, Kao SC, Vallely MP, van Zandwijk N, Reid G. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front Genet. 2013;4:94. doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6(9):e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5(3):492–7. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell E, Watson HL, Bailey S, Murray MJ, Coleman N. A Robust Protocol to Quantify Circulating Cancer Biomarker MicroRNAs. Methods Mol Biol. 2017;1580:265–79. doi: 10.1007/978-1-4939-6866-4_18. [DOI] [PubMed] [Google Scholar]
- 42.Page K, Guttery DS, Zahra N, Primrose L, Elshaw SR, Pringle JH, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One. 2013;8(10):e77963. doi: 10.1371/journal.pone.0077963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7(7):e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.