Abstract
The mammary gland is composed of a complex cellular hierarchy with unusual postnatal plasticity. The identities of stem/progenitor cell populations, as well as tumour-initiating cells that give rise to breast cancer, are incompletely understood. Here we show that Lgr6 marks rare populations of cells in both basal and luminal mammary gland compartments in mice. Lineage tracing analysis showed that Lgr6+ cells are unipotent progenitors, which expand clonally during puberty but diminish in adulthood. In pregnancy or upon stimulation with ovarian hormones, adult Lgr6+ cells regained proliferative potency and their progeny formed alveoli over repeated pregnancies. Oncogenic mutations in Lgr6+ cells resulted in expansion of luminal cells, culminating in mammary gland tumours. Conversely, depletion of Lgr6+ cells in the MMTV-PyMT model of mammary tumourigenesis significantly impaired tumour growth. Thus, Lgr6 marks mammary gland progenitor cells that can initiate tumours, and cells of luminal breast tumours required for efficient tumour maintenance.
The mammary gland consists of a bi-layered epithelium composed of luminal and basal (myoepithelial) mammary epithelial cells (MECs). Major mammary gland developmental steps occur postnatally, with extensive tissue remodelling during puberty and pregnancy1 governed by stem/progenitor cells2. In vivo lineage tracing studies3–8 have revealed a highly complex mammary epithelial hierarchy. Transformation of different cells within this hierarchy results in phenotypically diverse breast tumours9, although in most cancer models the tumour-initiating cell (TIC) has not yet been identified.
The identity and in vivo properties of adult mammary gland stem cells (MaSCs) are controversial. MaSCs have been described as a basal population able to repopulate the whole mammary gland when transplanted10–13. However, the ability of MaSCs to maintain both basal and luminal compartments postnatally has been disputed3, 6. Unipotent basal and luminal progenitors have been identified6, 11, 13–15, suggesting that postnatal progenitors may be lineage-restricted.
The leucine-rich repeat-containing G-protein coupled receptor 6 (Lgr6) marks stem cells of taste buds16, lung17, and skin18, 19, as well as rare mammary gland cells18. However, whether Lgr6 also marks stem/progenitor cells in the mammary gland was unknown. Here we traced the fate of Lgr6-positive (Lgr6+) cells during postnatal mammary gland formation, homeostasis, pregnancy, and in murine models of basal and luminal breast cancer. We provide evidence that Lgr6+ cells are unipotent progenitors that contribute to alveologenesis and function as TICs for luminal mammary tumours.
Results
Lgr6 marks clonogenic basal and luminal progenitor cells in postnatal mammary gland development
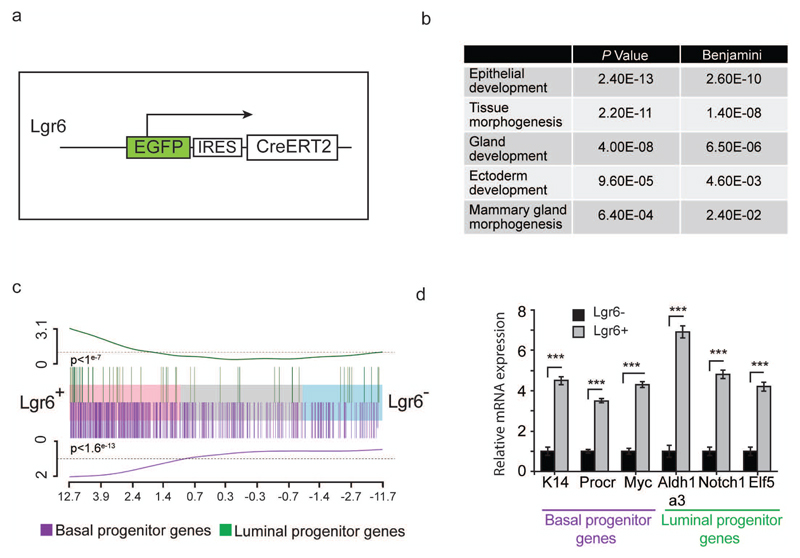
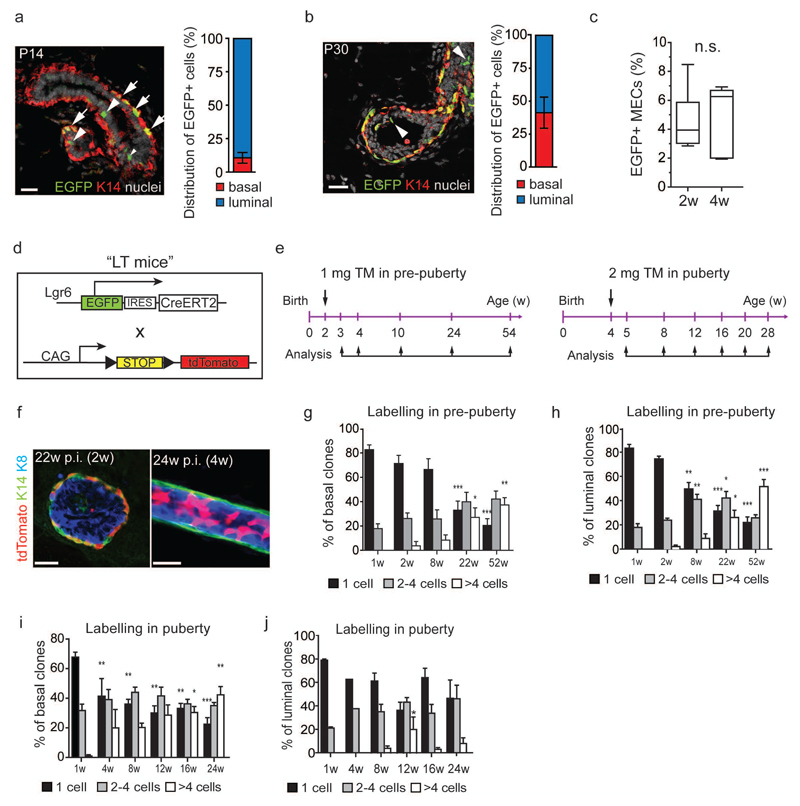
We analysed Lgr6 expression in MECs using Lgr6-EGFP-Ires-CreERT2 reporter mice18 (Lgr6-CreERT2) (Fig. 1a). In 5-week-old virgin Lgr6-CreERT2+/- mice, 2-4% of MECs were EGFP+ (Supplementary Fig. 1a). RNA sequencing of FACS-sorted Lgr6+ and Lgr6- MECs showed differential expression of genes involved in mammary gland development (Fig. 1b). The pool of Lgr6+ cells showed enrichment for previously described gene signatures of both basal and luminal mammary gland progenitor cells (Fig. 1c,d; Supplementary Fig. 1b)4, 20, 21. Consequently, we examined the clonal potential of Lgr6+ cells in pre-pubertal (2-week-old) and pubertal (4-week-old) Lgr6-CreERT2+/- mice. We discovered basal and luminal Lgr6+ cells at both ages (Fig. 2a,b; Supplementary Fig. 1c, 2a). Using FACS, we detected a small fraction (4.5±0.9%) of EGFP+ MECs in pre-puberty (2w, Fig. 2c), containing mostly luminal (89.5±4.0%) and fewer basal epithelial cells (10.5%±4.0%; Fig. 2a). In pubertal mammary glands, we observed more basal (41.2±11.7%) and fewer luminal EGFP+ MECs (58.8±11.7%) compared to pre-pubertal glands (Fig. 2b). However, the overall percentage of EGFP+ cells was not significantly altered (4.6±0.9%, 4w, Fig. 2c).
Figure 1.
Lgr6+ mammary gland cells show gene expression patterns of basal and luminal mammary gland progenitor cells. (a) Description of the Lgr6-CreERT2 allele used for detection and isolation of Lgr6+ mammary gland cells. EGFP: enhanced green fluorescent protein; IRES: internal ribosomal entry site; CreERT2: tamoxifen-inducible Cre recombinase. (b) Table illustrating examples of the gene sets enriched in the Lgr6+ MEC population according to RNA-seq. P values and Benjamini-Hochberg adjusted P-values for difference in enrichment score are shown. (c) Gene enrichment analysis for basal (purple) and luminal (green) progenitor genes in Lgr6+ and Lgr6- cells, showing the fold changes of all genes sequenced in the RNA-Seq experiment running from high on the left (Lgr6+), to low on the right (Lgr6-). The up-regulated basal (purple) or luminal (green) genes from the reported published gene lists are highlighted by the vertical lines. The line at the top and bottom of the barcode shows a cumulative enrichment of each set of genes. (n = 6 mice pooled per replicate). P = 1*10-7 (luminal progenitor), P = 1.58*10-13 (basal progenitor) (by Wilcoxon two-sided gene set test) (d) Q-PCR analysis of mRNA expression between Lgr6+ and Lgr6 -cells (normalized to GAPDH) for genes characteristic of basal and luminal progenitor cells respectively (n = 3 mice). Mean ± s.d., ***P.< 0.001 (t-test). See Supplementary Table 2 for exact p values and source data.
Figure 2.
Two independent Lgr6+ progenitor cell populations contribute to postnatal mammary gland development. (a, b) Confocal images of Lgr6-CreERT2+/- mammary glands at postnatal day 14 (P14) (a) and P30 (b). EGFP+ cells are found in K14+ basal (arrows) and K14- luminal cells (arrowheads). Scale bars: 25 µm. Bar graphs visualise the distribution of EGFP+ cells with respect to mammary epithelial cells (MECs) in (a) 2w-old (n=7 mice pooled from 2 independent experiments) and (b) 4w-old (n=10 mice pooled from 3 independent experiments) Lgr6-CreERT2+/- females as measured by flow cytometry. Mean ± s.e.m. (c) Box-and-whiskers plot showing the frequency of EGFP+ MECs in Lgr6-CreERT2+/- females at 2w and 4w of age (n=7 and n=10 mice pooled from 2 and 3 experiments, respectively). Minima, maxima, median are indicated. n.s., not significant; P = 0.673 (unpaired two-tailed t-test). (d) Lgr6-CreERT2 and Rosa26-tdTomato alleles (“LT mice”) used for lineage tracing. EGFP: enhanced green fluorescent protein; IRES: internal ribosomal entry site; CreERT2: tamoxifen-inducible Cre recombinase; CAG: constitutively active hybrid promoter from cytomegalovirus and chicken ß-actin promoters; STOP: stop cassette flanked by 2 loxP sites (black triangles); tdTomato: tandem dimer tomato red fluorescent protein. (e) Lineage tracing strategy in pre-puberty and puberty. TM, tamoxifen. (f) K14/K8 immunostaining of LT mammary glands 22w post induction (p.i). in pre-puberty (left panel) and 24w p.i. in puberty (right panel). tdTomato+ clones are found in the basal and luminal compartments. Scale bars: 20µm. (g - j) Quantification of tdTomato+ basal (g, i) and luminal (h, j) clone sizes at different time-points post induction. Graphs show the percentage of counted tdTomato+ clones containing 1, 2–4, and >4 cells at each time-point. (Number of analysed clones pooled from n = 3 mice per timepoint: n = 1157, n = 344, n = 542, n = 645, n = 691, respectively, for (g, h); n = 531, n = 1476, n = 1733, n = 163, n = 782, n = 597, respectively, for (i, j). Mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001 (One-way ANOVA). See Supplementary Table 2 for source data for g – j.
To trace the progeny of Lgr6+ cells in vivo, we intercrossed Lgr6-CreERT2 mice with the Rosa26-tdTomato22 reporter strain (“LT mice”, Fig. 2d). We initiated lineage tracing in LT heterozygous females at pre-puberty and puberty (Fig. 2e). 24 hours (h) after tamoxifen administration, we observed rare tdTomato+ MECs (1.7±0.2%, 2w, and 0.9±0.2%, 4w). At both timepoints, the distribution of tdTomato+ MECs corresponded to that of EGFP+ cells (Supplementary Fig. 2b,c) and detection of traced cells was dependent on tamoxifen (Supplementary Fig. 2d,e). Multi-cellular clones of up to 4 cells appeared by 1 week (w) post-induction and larger basal-only, as well as luminal-only, clones appeared thereafter (Fig. 2f; Supplementary Fig. 2f). The proportion of larger multi-cellular clones increased significantly over time (Fig. 2g,h). The proportion of tdTomato+ luminal clones remained greater than that of basal clones (Supplementary Fig. 2g) suggesting that basal and luminal Lgr6+ cells possess similar clonal potential pre-puberty.
Conversely, cells traced from puberty showed a stronger expansion of basal compared with luminal tdTomato+ clones (Fig. 2i,j; Supplementary Fig. 2h). In rare cases, tdTomato+ patches spanned both cell lineages (0.97-1.24%, following labelling in pre-puberty and puberty, respectively) (Supplementary Fig. 2g-i), and some Lgr6+ MECs at puberty co-expressed either Axin2 or Lgr5 mRNA, which may mark bipotential MaSCs. (Supplementary Fig. 2j)3–5. However, apparently mixed clones were observed only after several weeks of tracing (Supplementary Fig 2g,h), suggesting that fusion of adjacent tdTomato+ basal and luminal clones could be responsible for the mixed cell patches. To test whether the Lgr6+ population contains rare, bipotent cells, we transplanted Lgr6+ basal cells into cleared fat pads (Supplementary Fig. 2k). Unlike basal MaSCs, Lgr6+ basal cells were unable to reconstitute the mammary gland, indicating that they cannot adopt a multipotent fate. Our data are therefore consistent with unipotent Lgr6+ basal and luminal progenitors in the early postnatal mammary gland.
Basal and luminal Lgr6+ progenitors contribute to the alveolar network during pregnancy
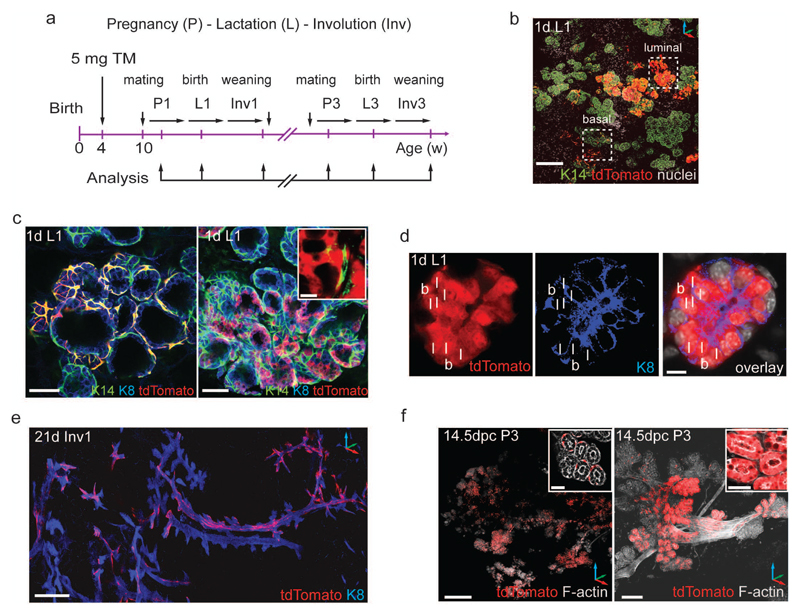
The generation of alveoli during repeated pregnancies requires long-lived, multi-lineage or lineage-restricted progenitor cells3–5, 7, 14, 23. To test the contribution of Lgr6+ progenitors to alveologenesis, we injected 4-week-old LT heterozygous females with tamoxifen and mated them 6w later (Fig. 3a). During pregnancy, tdTomato+ patches were distributed over the whole mammary gland, consisting of basal tdTomato+ cells as well as occasional, multicellular luminal clones in the expanding alveoli (Supplementary Fig. 3a,b). By the start of lactation, vast tdTomato+ alveolar networks had formed (Fig. 3b, Supplementary Fig. 3c,d), mostly composed of labelled basal cells, and less frequently of luminal tdTomato+ alveolar clones not associated with basal tdTomato+ cells (Fig. 3c), suggesting clonal expansion of unipotent basal or luminal progenitors. We rarely detected alveoli incorporating labelled basal and luminal cells (Fig. 3d, Supplementary Fig. 3e).
Figure 3.
Basal and luminal Lgr6+ cells found in puberty are long-lived, lineage-restricted stem cells that contribute clonally to consecutive pregnancies. (a) Scheme illustrating induction of lineage tracing in puberty with analysis in pregnancy (P), lactation (L), and involution (Inv). (b) Confocal z-stack image showing K14 immunostaining of lineage-traced mammary gland on the first day of the first lactation (1d L1). Dashed squares depict basal or luminal tdTomato+ labelled alveolar structures. Scale bar: 100 µm. (c) Alveoli in lactating mammary glands from LT females induced at 4w of age contain multicellular, K14+/tdTomato+ basal clones (left panel) and K8+tdTomato+ luminal clones (right panel). Inset depicts luminal tdTomato+ cells not associated with basal K14+ cells. Scale bars: 50 µm, 10 µm (inset). (d) Confocal image of K8 immunostaining demonstrating alveolus with adjacent basal (b) and luminal (l) tdTomato+ cells. Scale bar: 10 µm. (e) Confocal z-stack image showing K8 immunostaining of lineage traced mammary gland at 21d of involution (21d Inv1). Scale bar: 100 µm. (f) 3D reconstructions of F-actin-stained whole mount mammary glands from Lgr6-CreERT2+/-:Rosa26-tdTomato+/- females at 14.5 dpc of third pregnancy (14.5 dpc P3). Extensive networks of basal (left panel) and luminal (right panel) tdTomato+ clones are sustained over multiple pregnancies. Scale bars: 500 µm (left panel), 50 µm (right panel), 25 µm (insets).
At day (d) 21 of involution, when most of the milk-producing epithelium has regressed, we found tdTomato+ clones lining the mammary ducts and alveolar bud remnants (Fig. 3e, Supplementary Fig. 3f), consisting mostly of basal cells (Supplementary Fig. 3g), and occasionally luminal cells (Supplementary Fig. 3h).
Puberty-labelled LT heterozygous females subjected to multiple pregnancies displayed tdTomato-traced alveolar networks in all mammary glands. We identified large basal or luminal tdTomato+ clones (Fig. 3f), but saw no labelling spanning both compartments.
Thus, we concluded that pubertal Lgr6+ MECs comprise two separate populations of unipotent long-lived and self-renewing mammary gland progenitors clonally contributing to the alveolar network during pregnancy.
Lgr6+ progenitors contribute minimally to adult mammary gland homeostasis and are continuously lost over time
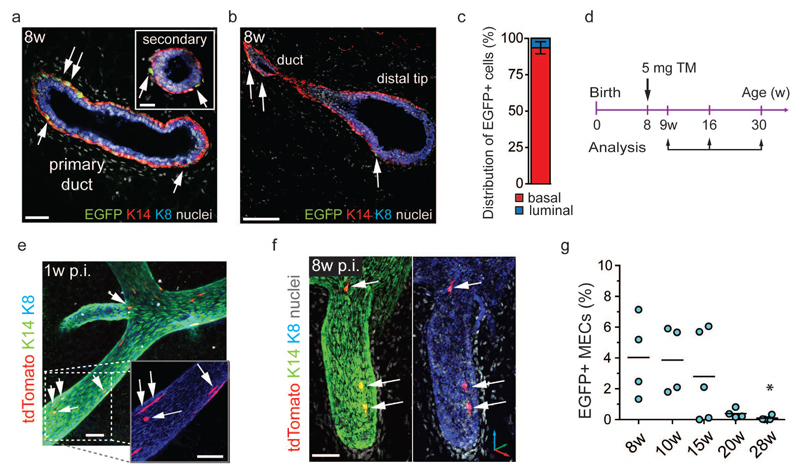
Consistent with the increased presence of basal clones at puberty, in 8-week-old virgin Lgr6-CreERT2+/- females, EGFP expression was almost exclusively detected in basal MECs (93.5±4.2%, Fig. 4a-c). EGFP+ cells were found in primary and secondary ducts, including the distal tips of the ducts (Fig. 4a, b) with a frequency (4.7±2.9%) comparable to that of younger mice (Fig. 2c). To test the clonal capacity of these cells in the virgin adult, we lineage-traced 8-week-old LT heterozygous females, labelling approximately 1.1% of MECs (Fig. 4d, Supplementary Fig. 4b, c). 1w post-tamoxifen, tdTomato labelled mostly single cells with occasional multicellular clones (Fig. 4e). 8w post-tamoxifen, we detected very few basal tdTomato+ clones (Fig. 4f) and after 22w, multi-cellular tdTomato+ clones had disappeared. The frequencies of tdTomato+ MECs (0.28–0.60%) and EGFP+ MECs (0.15%) also decreased (Fig. 4g, Supplementary Fig. 4d, e). Thus, Lgr6+ cells do not play an obvious role in mammary gland homeostasis in adult virgin mice and their numbers decrease with age.
Figure 4.
Lgr6+ progenitors contribute minimally to adult mammary gland homeostasis and are continuously lost over time. (a, b) Confocal image of a mammary gland thick section from 8w old LT virgin stained for K14 and K8. K14+/EGFP+ basal cells (arrows) are found in primary and secondary ducts (a) and in the distal tips of the mammary ducts (b). Scale bars: (a) 50 µm, 20 µm (inset), (b) 50µm. (c) Bar graph showing the distribution of EGFP+ cells over the total MEC population (n = 6 mice pooled from 2 independent experiments). Mean ± s.e.m. (d) Time-points of tamoxifen administration and analysis to trace Lgr6+ cells during tissue homeostasis. (e, f) Confocal reconstructions of K8/K14 immunostained mammary gland ducts from LT female 1w (e) and 8w (f) post-induction. Arrows indicate K14+/tdTomato+ single cells and bi-cellular clones. Scale bars: 50 µm (e), 50 µm (f). (g) Percentage of EGFP+ MECs in Lgr6-CreERT2+/- females from 8w to 28w of age (n = 4, n = 4, n = 5, n = 4, and n = 4 mice, pooled from 2 independent experiments for 8w, 10w, and 1 experiment for 15w, 20w, 28w). Lines indicate mean. *P = 0.0106 (One-way ANOVA). See Supplementary Table 2 for source data.
Mostly quiescent Lgr6-expressing cells can be reactivated by pregnancy or hormone treatment
To test the capacity of Lgr6+ cells to expand in the adult, we analysed pregnant females lineage-traced from 8w (Fig. 5a). Surprisingly, tdTomato+ clones formed vast alveolar networks over the whole mammary gland at mid-pregnancy, with tdTomato+ myoepithelial cells in ducts and alveoli (Fig. 5b). Furthermore, we observed luminal-only labelled alveoli (Fig. 5c), possibly derived from the small fraction of luminal EGFP+ cells found in nulliparous adult mice (Fig. 4c). Thus, pregnancy seems to re-awaken the clonogenic potential of both adult Lgr6+ MEC populations.
Figure 5.
Mostly quiescent Lgr6-expressing cells can be reactivated by pregnancy or hormone treatment. (a) Scheme illustrating induction of lineage tracing in 8w-old LT females and analysis over pregnancy. Pregnancy (P); lactation (L); involution (Inv). (b, c) 3D reconstructions of K14/K8 immunostained mammary gland from LT female at 14.5 dpc P1. Scale bars: 50 µm (b), 40 µm (c). (d) Quantification of alveoli containing basal and luminal tdTomato+ cells when traced into the first lactation (lact.) after tamoxifen administration to LT females at 8w of age. (1316 counted alveoli pooled from 3 mice:). (e) 3D reconstruction of mammary duct from LT female 3w p.i. and 2w post-implantation of 17 β -estradiol and progesterone (E/P) pellets. Expansion of luminal tdTomato+ clones (arrows). Scale bar: 50 µm. (f) Distribution of tdTomato+ cells over MECs 2w post-treatment with 17 β-estradiol (E, n = 3 mice) and 17β-estradiol/progesterone (E/P, n = 4 mice), compared to tamoxifen-treated control (C) mice (n = 3 mice). Mean ± s.e.m. *P < 0.038 (One-way ANOVA). (g) Lgr6+ luminal cells in mammary duct of P56 Lgr6-CreERT2+/- female staining positive for oestrogen receptor-α (ERα) (arrows). Scale bar: 25 µm. See Supplementary Table 2 for source data for f.
As with lineage tracing from puberty, we observed sparse mixed-lineage patches of tdTomato+ cells in alveoli (Supplementary Fig. 4f) and in the main mammary ducts (Supplementary Fig. 4g) during the first pregnancy. However, over 97% of labelled alveoli were basal or luminal only tdTomato+ (Fig. 5d) and subsequent pregnancies revealed no alveoli labelled in both epithelial compartments.
To determine whether hormone stimulation activates Lgr6+ MECs in the adult, we treated females, labelled at 10w, with subcutaneous 17β-estradiol (E) and progesterone (P) pellets for 14 days. Although the frequencies of basal or luminal Lgr6+ MECs had not increased, we observed an expansion of luminal tdTomato+ clones either with E/P or with E alone (Fig. 5e, f). In line with this, we discovered luminal Lgr6+ cells co-expressing oestrogen receptor alpha (ERα) (Fig. 5g). Thus, pharmacologic stimulation with ovarian hormones induces the preferential expansion of luminal Lgr6+ progenitors.
Lgr6 is expressed in human breast cancers and its up-regulation correlates with lower disease-free survival
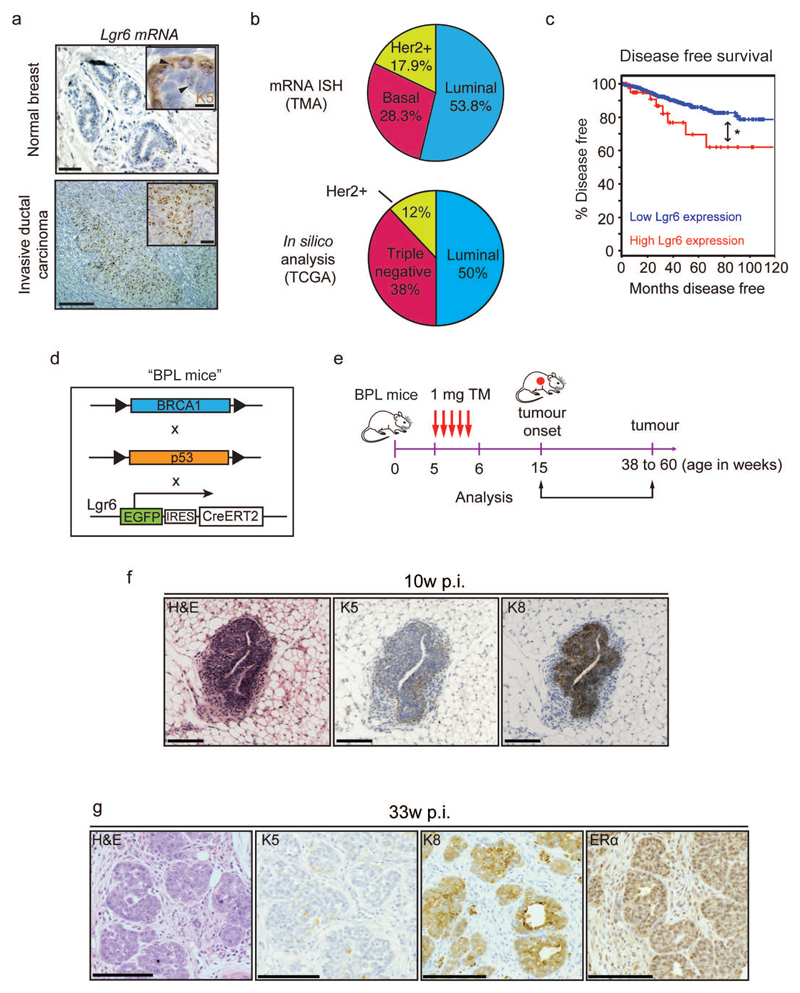
Tumour histological classification is based on expression of distinct hormonal receptors: ERα and/or progesterone receptor 1 (luminal), human epidermal growth factor receptor 2 (HER2-positive) or the absence of these markers (basal-like/triple-negative)24–26. In situ hybridization on over 500 human breast cancers detected Lgr6 expression in 49% of tumours (Fig. 6a). Of these, around half were luminal, in agreement with publically available data for breast cancers with Lgr6 up-regulation27, 28 (Fig. 6b). Disease-free survival in patients with high Lgr6 expression was significantly decreased over all tumour types compared to patients with low Lgr6 expression (Fig. 6c), suggesting that Lgr6 overexpression correlates with tumour aggressiveness.
Figure 6.
Lgr6+ cells are present in human breast carcinoma and are able to initiate tumours in mouse models of breast cancer. (a) In situ hybridisation (ISH) analysis of Lgr6 mRNA expression on human breast carcinoma tissue microarrays (TMA) compared to normal healthy breast tissue. Keratin 5 (K5) co-staining demonstrates the presence of basal and luminal Lgr6+ mammary gland cells (inset, upper panel). Scale bars: 100µm, 20µm (insets) (b) Prevalence of Lgr6 expression or up-regulation within the three categories of human breast tumours as determined by ISH on tissue microarrays (n = 550 tumours; upper pie chart) and in silico analysis of breast cancer gene expression data available from The Cancer Genome Atlas (TCGA; lower pie chart) (cBioportal, RNASeq, and ref59. (c) Disease free survival in n = 1093 patients with Lgr6 up-regulation compared to control patients (cBioportal, RNASeq, and ref 59). *P = 0.042 (log-rank test). (d) Description of the BPL mouse strain, derived from breeding of BRCA1f/f, p53f/f, and Lgr6-CreERT2 mice. (e) Treatment schedule of 5w-old BPL mice with tamoxifen and subsequent tumour analysis. (f) Immunohistochemistry of tumours from BPL mice 10 weeks post-injection with tamoxifen, stained with haematoxylin-eosin (HE) and probed for markers against basal (K5+) and luminal (K8+) cells. Scale bars: 100µm. (g) Immunohistochemistry of tumours from BPL mice 33 weeks post-injection stained with HE and probed for K5, K8 and ERα. Scale bars: 100 µm.
Lgr6+ progenitors are potent TICs
To test whether Lgr6+ cells can function as breast cancer TICs, we inactivated Trp53 and Brca1, two of the most commonly mutated breast tumour suppressors29, in Lgr6+ cells by generating Brca1loxP/loxP:p53loxP/loxP:Lgr6-EGFP-CreERT2 (BPL) mice (Fig. 6d). Gene inactivation was triggered in 5-week-old mice by tamoxifen injection (Fig. 6e). Initial K8-positive tumour lesions were detected 10w post-injection (Fig. 6f). Subsequently, BPL animals developed mammary gland tumours expressing the luminal markers K8 and ERα (Fig. 6g), demonstrating that oncogenic mutations in Lgr6+ cells can result in luminal cancer.
We assessed the tumour-initiating potential of Lgr6+ cells in a second breast cancer model by deleting the F-box and WD repeat domain containing 7 (Fbxw7) tumour suppressor (commonly down-regulated in luminal breast cancer30, 31) in combination with activation of oncogenic K-RasG12D in Lgr6+ cells (FRL mice) (Supplementary Fig. 5a,b). Oncogenic activation of Ras signalling occurs in over 20% of human breast carcinomas32, 33. FRL animals showed rapid tumour onset, with luminal lesions appearing 3w post-tamoxifen (Supplementary Fig. 5c). 8w post-tamoxifen, mammary gland carcinomas expressing K8 and ERα were present (Supplementary Fig. 5d). In both FRL and BPL breast cancer models, 100% of analysed tumours arising from Lgr6+ cells were positive for K8, and despite the relatively low frequency of Lgr6+ MECs at 5 weeks of age, the tumour penetrance was high (BPL 60%, FRL 100%) (Supplementary Fig. 5e). Lgr6+ cells are therefore potent TICs for luminal mammary tumours.
Rare Lgr6+ cells contribute clonally to luminal, but not basaloid, mammary tumours
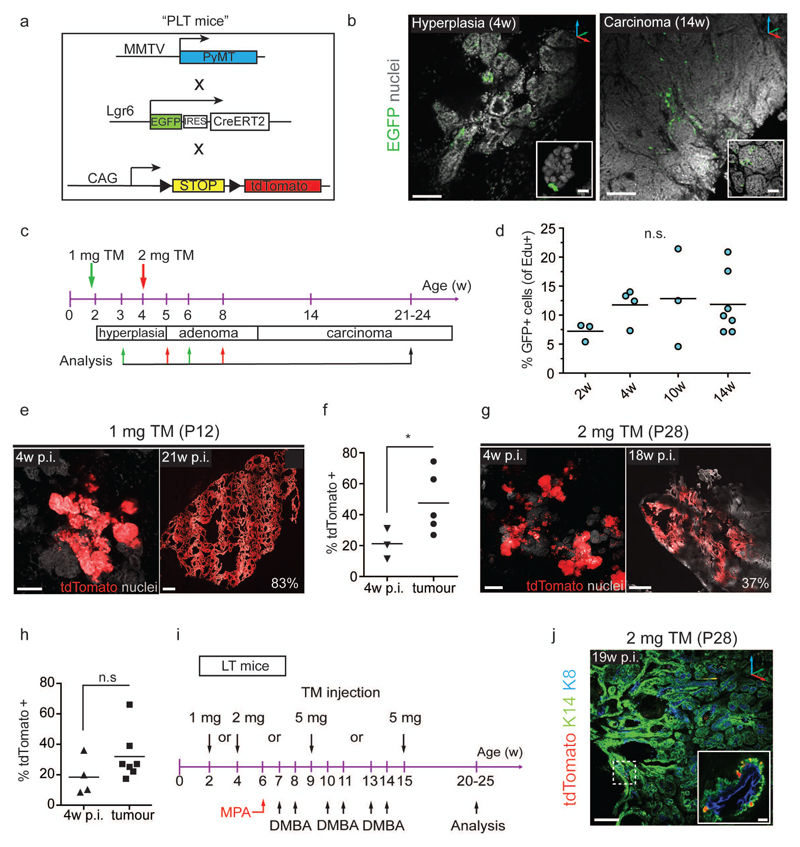
Mouse mammary tumour virus promoter-driven Polyoma virus middle T antigen (MMTV-PyMT) is a well-established model of luminal breast cancer34, resulting in adenocarcinomas whose gene expression profile resembles that of luminal progenitor cells20. To investigate whether these tumours contain Lgr6+ cells, we crossed the MMTV-PyMT strain with Lgr6 lineage-tracer mice to create the PLT model (Fig. 7a). We identified rare Lgr6+ cells (2.3%±1.0% to 4.7%±1.0%) in normal and hyperplastic glands, and in tumours (Fig. 7b). We traced Lgr6+ cell progeny in developing MMTV-PyMT tumours after labelling PLT mice during pre-puberty and puberty (Fig. 7c). At 12d of age, small, isolated lesions were visible in PyMT+/- mammary glands (Supplementary Fig. 6a,b). Therefore, we administered tamoxifen between 9 and 12d of age to label Lgr6+ cells before detectable tumour formation, resulting in a recombination efficiency similar to that seen in LT animals (1.5% of MECs). Hyperplastic regions at 2w expressed PyMT protein independently of Lgr6 expression (Supplementary Fig. 6c,d).
Figure 7.
Lgr6+ cells contribute to luminal, but not basal, mammary tumours (a) Description of MMTV-PyMT, Lgr6-CreERT2, and Rosa26-tdTomato alleles (“PLT mice”) for detection and lineage tracing of Lgr6+ cells and induction of mammary tumours. MMTV: murine mammary tumour virus promoter; PyMT: polyoma virus middle T oncogene. (b) Representative confocal z-stack images showing rare EGFP+ mammary gland cells in PL females. Left: Hyperplastic mammary gland of 4w-old female. Scale bars: 100 µm; 25 µm (inset). Right: Mammary carcinoma of 14w-old female. Scale bars: 100 µm; 50 µm (inset). (c) Scheme illustrating tamoxifen administration in pre-puberty (green arrows) and puberty (red arrows) and analysis time-points to assess the contribution of Lgr6+ cells to MMTV-PyMT-induced mammary tumours. (d) Frequency of EdU+ cells with respect to EGFP+ cells in mammary glands of PL females of different ages. Lines indicate means. (n = 4 mammary glands pooled from n = 3 mice/time-point). Mean ± s.e.m.; n.s., not significant; P = 0.542 (One-way ANOVA). (e) Confocal z-stack images demonstrating expansion of tdTomato+ clones over time after induction at P12. Scale bars: 100 µm (left), 500 µm (right). Percentage indicates extent of tdTomato-labelled tumour area. (f) Quantification of tdTomato+ mammary gland area or tumour area 4w p.i. at P12 (n = 3 mice) and at carcinoma stage (21w – 24w, n = 5 mice). Lines indicate means. *P < 0.048. (unpaired two-tailed t-test) (g) Confocal z-stack images demonstrating expansion of tdTomato+ clones over time after induction at P28. Scale bars: 100 µm (left), 500 µm (right). (h) Quantification of tdTomato+ mammary gland area or tumour area 4w p.i. at P28 (n = 4 mice) and at carcinoma stage (21w – 24w, n = 7 mice). Lines indicate mean. n.s., not significant; p = 0.166 (unpaired two-tailed t-test). (i) Scheme summarizing the protocol used to study the contribution of Lgr6+ cells to MPA/DMBA induced carcinogenesis. MPA: Medroxyprogesterone acetate; DMBA: 7,12-Dimethylbenz[a]anthracene. (j) 3D reconstruction of K14/K8 immunostaining of MPA/DMBA mammary tumour from LT female induced at 4w. Only sporadic K14+/tdTomato+ cells are observed (dashed box and inset). Scale bars: 200 µm, 10 µm (inset). See Supplementary Table 2 for source data for d, f, h.
48h after tamoxifen injection at 4w, we observed tdTomato+ cells adjacent to EGFP+/tdTomato+ cells (Supplementary Fig. 6e), indicating quickly dividing Lgr6+ cells. By administering 5-ethynyl-2′-deoxyuridine (EdU) to Lgr6-CreERT2+/-:PyMT+/- mice of different ages, we confirmed a minor, but stable, population (between 7.2±0.9% and 12.8±4.9%) of cycling EGFP+ cells in tumours (Fig. 7d, Supplementary Fig. 6f).
Multicellular tdTomato-expressing clones expanded over time when labelled at tumour onset (P12) and could occupy large areas of the tumour in fully-grown carcinomas (Fig. 7e,f). When labelled after tumour initiation (P28), tdTomato+ clones continued to expand (Fig. 7g,h). Very few tdTomato+ cells retained expression of Lgr6, or expressed the gene de novo (Supplementary Fig. 6g) and no tdTomato+ cells were found in vehicle-treated control animals (Supplementary Fig. 6h), indicating that rare Lgr6+ cells can expand from the earliest stages of tumour development and contribute to large portions of tumour mass.
Importantly, Lgr6+ cells contributed to the maintenance of fully developed MMTV-PyMT tumours, as demonstrated by labelling at 14w (Supplementary Fig. 6i). Similar to lineage tracing at earlier stages, we were able to detect multicellular tdTomato+ clones as early as 1w post-induction (Supplementary Fig. 6j), which expanded until the mice were sacrificed 7-10 weeks later (Supplementary Fig. 6k). Lgr6+ MECs therefore contribute to increase the cell mass of MMTV-PyMT-driven tumours throughout neoplastic development.
Since Lgr6+ cells are present in both MEC compartments, we asked whether they are also involved in basal mammary tumour formation. Transformation of basal cells by medroxy-progesterone acetate (MPA) and di-methylbenz(a)anthracene (DMBA) treatment results in basaloid and mixed lineage tumours35,36. We induced lineage tracing in Lgr6-CreERT2+/-:Rosa26-tdTomato+/- females before, during, and after MPA/DMBA treatment (Fig. 7i). Although basal and luminal Lgr6+ cells were clearly detectable in MPA/DMBA-treated mammary glands (Supplementary Fig. 6l), we did not discover expanding tdTomato+ clones in the resulting tumours, but only some K14+/tdTomato+ cells (Fig. 7j). This result suggests that neither basal nor luminal Lgr6+ MECs are involved in MPA/DMBA-induced basal tumourigenesis.
Lgr6+ cells sustain tumour growth and display characteristics of TICs
The tumour-forming capacity of Lgr6+ cells, together with their contribution to tumour expansion, suggests that they have TIC properties. To assess whether Lgr6+ cells are resistant to chemotherapy, another TIC characteristic, we treated labelled tumour cells from PLT mice in organoid culture with doxorubicin (Supplementary Fig. 7a). The proportion of tdTomato+ cells within labelled organoids increased after doxorubicin treatment when compared to untreated cells (Supplementary Fig. 7b,c), suggesting that Lgr6+ progeny are more resistant to chemotherapy. To directly test the requirement for Lgr6+ cells in MMTV-PyMT tumours, we assessed the consequences of Lgr6+ cell depletion, using MMTV-PyMT:Lgr6-EGFP-CreERT2 (PL) mice crossed to a Rosa26-LSL-diptheria toxin receptor (DTR) strain to create the PDL model (Fig. 8a). We treated PDL animals every other week between two and ten weeks of age with alternate injections of tamoxifen and DT (Fig. 8b). Depletion of Lgr6+ cells by diphtheria toxin (DT) injection was efficient, indicated by a reduction in EGFP+ cells and reduced tumour cell proliferation in PDL animals compared with controls (Fig. 8c,d).
Figure 8.
Lgr6+ cells contribute to tumour maintenance and show enhanced chemotherapy resistance in the MMTV-PyMT model of breast carcinoma. (a) Description of PDL mice derived from crossing of the MMTV-PyMT, Rosa26-LSL-DTR, and Lgr6-CreERT2 strains. (b) Scheme of PDL mouse treatment with tamoxifen and DT. (c) Immunohistochemistry of mammary tumours from PD and PDL mice treated with tamoxifen and diphtheria toxin (DT), and stained with haematoxylin-eosin (HE) and probed for EGFP and the proliferation marker ki67. Scale bars: 100µm. (d) Quantification of EGFP+ and ki67+ cells from PL and PDL mammary tumours. (n = 300 cells pooled from 10 different fields in 5 biological replicates). Mean ± s.d. *P < 0.05; **P < 0.01; ***P < 0.001 (t-test). (e) Analysis of tumour onset in PDL and PL mice after tamoxifen and DT treatment (n = 6 mice per group). The line indicates the mean. *P = 0.048 (f) HE staining on tumours from PDL and PL mice after tamoxifen and DT treatment. Scale bars: 500µm (1x), 50µm (40x). (g) Scheme of treatment to assess the role of Lgr6+ cells in subcutaneous tumour xenografts. (h) Analysis of tumour size 5 weeks post-transplantation (n = 5 mice per group). The line indicates the mean. *P = 0.0401. See Supplementary Table 2 for source data for e, h.
Tumour onset was delayed in PDL mice (Fig. 8e), confirming that Lgr6+ cells contribute to tumour outgrowth. Importantly, depletion of Lgr6+ cells resulted in a dramatic change in tumour histology. Control PL tumours showed a solid architecture, morphologically undifferentiated tumour cells, and a high mitotic index, as reported for the MMTV-PyMT model. In contrast, PDL tumours had a dominant dilated ductal and acinar phenotype with well-differentiated elements, fewer undifferentiated cells, and less mitotic activity than controls (Fig. 8f).
To further explore the capacity of Lgr6+ cells to maintain tumour growth, we isolated tumour cells from untreated PDL animals. Application of DT to cultured primary tumour cells resulted in decreased Lgr6 mRNA levels, consistent with substantial depletion of Lgr6+ cells (Supplementary Fig. 7d,e). DT-treated cells had a reduced ability to form spheres compared with untreated cells (Supplementary Fig. 7f). To test the requirement for Lgr6+ cells in vivo, we injected Nu/Nu mice with equal numbers of DT-treated and untreated cells (Fig. 8g). Tumours derived from DT-treated cells were significantly smaller (Fig. 8h) and showed a differentiated tumour morphology similar to that observed in DT-treated PDL animals (Supplementary Fig. 7g). These observations show that Lgr6+ cells promote sphere-forming capability, are resistant to chemotherapy, sustain tumour growth and are necessary to recreate the normal tumour histology, all characteristics of TICs.
Discussion
Our study describes multiple functions of Lgr6+ cells in the mammary gland during postnatal development, pregnancy, and cancer (Supplementary Fig. 8).
We identified two epithelial mammary gland progenitor populations marked by Lgr6 that contribute to organ development, in the basal and luminal compartments, respectively. The proportion of Lgr6+ cells remains stable during postnatal development, but declines in adulthood, correlating with reduced clonal expansion in virgin mice. Pre-puberty, most Lgr6+ cells are luminal, but the basal Lgr6+ population expands more during puberty and by adulthood, the majority of Lgr6+ cells are basal. Lgr6+ cells contribute to ductal morphogenesis during postnatal development, and although they don’t play an obvious role in tissue homeostasis, adult Lgr6+ cells can ‘reactivate’ and sustain alveologenesis throughout multiple pregnancies.
Our clonal analysis showed that Lgr6+ cell progeny are almost exclusively restricted to a single compartment. Adjacent basal and luminal lineage-traced cells were undetectable in the few first weeks post-labelling in pre-puberty and puberty. Rare mixed-lineage tdTomato+ patches were observed only when traced into adulthood or alveologenesis in pregnancy, periods of massive epithelial expansion37, 38. Thus, they could represent fusion of expanding basal and luminal clones. Although use of a single-colour reporter limited our ability to resolve individual, neighboring basal and luminal Lgr6+ clones, the multicolour reporter R26R-Confetti39 did not recombine with Lgr6-EGFP-IRES-CreERT2 in our hands. However, the late appearance of mixed-lineage tdTomato+ patches and their absence in consecutive pregnancies makes it unlikely that they are derived from bipotential progenitor cells.3–5.
If rare Lgr6+ cells possessed bipotential properties, these might overlap with Axin2+ cells, for which such a lineage switch was shown2, 5. Although a few Lgr6+ cells stained positive for Axin2 mRNA, Lgr6+ cells, unlike Axin2+ basal cells, failed to reconstitute mammary glands when transplanted into emptied fat pads (Supplementary Fig. 2k). Thus, while our observations do not exclude the existence of bipotential MaSCs5, 40, our data support a unipotent progenitor model for Lgr6+ MECs, which is further substantiated by recent data41.
Luminal Lgr6+ cells contribute to alveologenesis independently of basal SCs, and are reminiscent of hormone receptor-negative, parity-identified MECs (PI-MECs)14, 15. On the other hand, their high clonogenic capabilities at peak oestrogen levels (in puberty and pregnancy) resemble the typical proliferative pattern of hormone receptor expressing cells42, 43. The existence of ERα-positive luminal progenitor cells is disputed44, but has been described based on label-retaining experiments23, 45, 46 and lineage tracing of Notch3-expressing MECs8. Like luminal Lgr6+ cells, Notch3+ progenitors are heterogeneous for ERα expression, undergo a temporal state of quiescence, and contribute to alveologenesis8. Hence, Lgr6 expression marks luminal progenitor cells with differing ER signaling abilities, which contribute to duct formation and alveologenesis.
Lgr6+ progenitors can function as cells of origin of mammary tumours, but interestingly, two different oncogenic combinations targeted to Lgr6+ cells, p53/Brca1 loss and K-Ras activation/Fbxw7 loss, only gave rise to luminal tumours. The combination of p53 and Brca1 loss typically results in tumours with a basal phenotype, with only a minor proportion of luminal tumours47–49, but only K8-positive lesions were observed in our model. Brca1/p53 tumours originate from the luminal compartment and then switch to a basal phenotype50. It is therefore possible that the Lgr6+ population is incapable of such a switch and, when mutated for Brca1 and p53, gives rise to luminal tumours. This interpretation raises the possibility that the cell of origin, but not the combination of oncogenes, is the primary determinant of tumour type in these models.
Consistent with Lgr6+ progenitor cells and bipotential MaSCs being separate populations, Lgr6+ cell progeny did not contribute to basaloid DMBA/MPA-induced mammary tumours35, in which basal MaSCs are the presumed targets of chemical transformation36, 51, 52. In contrast, fate mapping showed that luminal Lgr6+ progenitor cells contribute to early tumour outgrowth in PyMT-induced luminal tumours. The MMTV promoter is predominantly active in the luminal compartment53, 54 and most pre-pubertal Lgr6+ MECs are of a luminal nature. These observations are supported by recent studies 55–57 showing that lineage-restricted luminal stem cells can initiate mammary tumours.
Lgr6+ cells also play an important role in conferring tumour aggressiveness in the MMTV-PyMT model, since Lgr6+ cell-depleted tumours were more differentiated and less proliferative. Lgr6+ cells are self-sustaining, fuel tumour growth, and strongly affect the composition of the tumour, characteristics of TICs. As current efforts to target TICs often rely on very broad and non-specific markers58, characterisation of this rare Lgr6+ cell pool will benefit research into TIC-targeting therapies.
Methods
Mouse Lines
Lgr6-EGFP-IRES-CreERT2 18 mice were a kind gift from Hans Clevers (Hubrecht Institute, The Netherlands); Brca1LoxP/LoxP; p53LoxP/LoxP 60; Fbxw7LoxP/LoxP 61; KrasLSL-G12D 62; MMTV-PyMT 34; Rosa26-LSL-tdTomato 22; Rosa26-iDTR 63 mouse lines have been previously described. These lines were crossed to obtain the different genotypes. Control mice used for the Lgr6-positive cell depletion experiments shown in Figure 8 were mice lacking either the Cre recombinase or the Diphtheria toxin receptor allele. For this study, we used only female mice, which were between 12 days and 9 weeks of age at the start of experiments.
For tracing experiments, a single dose of 1 mg tamoxifen (Sigma) in 50 μl sunflower oil (Sigma) was injected intraperitoneally in 2 week-old mice. Four week-old females received 1.5 mg or 2 mg tamoxifen in 75 μl or 100 μl volume, respectively. These doses were reported not to interfere with normal mammary epithelial cell proliferation 3.
For Cre induction at later time-points, we injected 5 mg tamoxifen in 250 μl oil. For depletion experiments, mice were injected with 100 μg g-1 body weight tamoxifen once daily for 3 consecutive days once every two weeks between the ages of 2 and 10 weeks. Tamoxifen (Sigma-Aldrich) was dissolved in peanut oil (Sigma-Aldrich).
We controlled for potential tamoxifen effects on mammary gland development and as shown by others3, 56, 64, tamoxifen administration transiently delayed the outgrowth of the epithelium during puberty. However, this delay in outgrowth was reversed by the time the mice reached adulthood.
Mice for depletion experiments were injected intraperitoneally with 12ng g-1 body weight Diphtheria toxin (DT) (Sigma-Aldrich) once daily for 3 consecutive days once every two weeks between the ages of 2 and 10 weeks.
Study Approval
Mouse experiments were carried out with the approval of the London Research Institute’s Ethical Review Committee according to the UK Animals (Scientific Procedures) Act 1986, or in accordance to the ethical guidelines and after approval from the ethics committee of the “The Board of Agriculture, Experimental Animal Authority, Stockholm, Sweden”.
EdU labelling
EdU (Invitrogen) was injected intraperitoneally at 0.2 mg 10 g-1 body weight and mammary glands were harvested 3h after injection. EdU incorporation was visualised using a Click-iT Imaging kit (Invitrogen).
MPA/DMBA treatment
Mice were anaesthetised using ketamine-xylazine, and slow-release (90 days) MPA pellets (50 mg; Innovative Research of America) were subcutaneously implanted on the right flank. DMBA (100 μl, 10 mg ml-1 in sunflower oil; Sigma) was administered six times by oral gavage over the next 8 weeks as described in Fig. 7i.
Pharmacological stimulation with ovarian hormones
Mice were anaesthetised with isoflurane (Baxter), and slow-release (90 days) MPA and/or 17β-estradiol pellets (50 mg and 0,72 mg, respectively; Innovative Research of America) were subcutaneously implanted in the right flank.
Immunofluorescence and immunohistochemistry staining
Mice were euthanized by cervical dislocation and the mammary glands were dissected out. Dissected mammary glands were pre-fixed in 4% paraformaldehyde for 30–60 min. at room temperature to preserve native fluorescence. Tissues were washed with phosphate-buffered saline (PBS), embedded in optimal cutting temperature compound (OCT; Tissue-Tek) and stored at -80°C.
For whole mount immunostaining, mammary glands were thawed and washed twice with PBS for 1h. The tissue was permeabilized with 2 washes of PBS0.5T (PBS + 0,5% v/v Triton X-100) for 1h. For immunostaining, the samples were incubated overnight at 4°C with primary antibodies diluted in PBD0.2T (PBS + 1%BSA + 1% v/v DMSO + 0.2% v/v Triton X-100). The next day, they were washed 4 times with PBD0.2T for 1h. Tissues were incubated with secondary antibodies plus DAPI (4',6-diamidino-2-phenylindole; Roche) or TO-PRO-3 (Life Technologies) at 4°C overnight. After 4 washes with PBD0.2T for 1h, tissues were optically cleared in 80% glycerol in PBS at room temperature. The stained mammary glands were mounted between 2 cover slips and sealed with silicone. Mammary glands from pregnant mice were cut into smaller pieces with scalpels prior to mounting.
For whole mount staining of the F-actin cytoskeleton, mammary glands were permeabilized with PBS0.5T for 2h and incubated overnight at 4°C with Alexa 647-linked phalloidin (1:100; Invitrogen) and DAPI diluted in PBS0.5T, followed by optical clearing with 80% glycerol.
For confocal analysis of thick tissue sections, mammary gland sections of 100–150 µm were cut using a cryostat (Leica Biosystems) and OCT was washed off with warm (37°C) PBS. The sections were permeabilized for 1h with PBS0.5T and incubated overnight at 4°C with primary antibodies diluted in PBD0.2T. The next day, they were washed 4 times with PBD0.2T for 15 min. Tissues were incubated with secondary antibodies at 4°C overnight and washed 4 times with PBD0.2T for 15 min. DAPI or TO-PRO-3 were added to the last washing step. The stained mammary gland sections were mounted in ProLong Gold (Life Technologies) between 2 cover slips.
For immunohistochemistry, mammary glands were fixed overnight in 10 % neutral buffered formalin, then washed with 70 % ethanol, processed and embedded into paraffin. Sections were cut 4 µm thick and deparaffinized in Histo-Clear (National Diagnostics), rinsed in ethanol and washed in water and PBS. Sections were stained with H&E or with antibodies for immunohistochemistry. Endogenous peroxidase was blocked with 3% hydrogen peroxide for 5 min followed by washes with PBS. Epitope blocking was performed for 1 h at room temperature (RT) with blocking solution: 1 % bovine serum albumin (Sigma-Aldrich), 5 % normal donkey serum (EMD-Millipore) and 0.4 % Triton X-100 (Sigma-Aldrich) diluted in PBS. Sections were then incubated overnight at 4ºC with primary antibodies diluted in blocking solution. Next day, sections were washed with PBS and incubated for 2 h at RT with secondary antibodies (Life Technologies) diluted in blocking solution, then washed again and staining was visualized with 3.3’-diaminobenzidine (DAB) followed by counterstaining with Mayer’s haematoxylin. The sections were then dehydrated and mounted.
Antibodies used were against Keratin 8 (DSHB; TROMA-I, 1:500), Keratin 14 (Abcam; Ab9220, 1:500), GFP (Abcam; Ab13970, 1:250), Keratin 5 (Abcam; Ab52635, 1:500), Estrogen receptor alpha (Santa Cruz; sc-542, 1:200), Ki67 (Abcam; Ab16667, 1:200), CD49f (BD Pharmingen, 555734, 1:100), anti-Middle T oncogene (Santa Cruz; sc-53481, 1:100). All secondary antibodies were biotinylated, horse radish peroxidase (HRP)-conjugated, or Alexa-conjugated antibodies (Molecular Probes, 1:330): goat anti-rat A405, goat anti-rabbit A488, goat anti-chicken A488, donkey anti-rabbit A488, goat anti-rat A546, goat anti-rabbit A647, goat anti-rat A647.
Microscopic imaging
Confocal images were acquired using Zeiss LSM710, LSM780 or LSM880 confocal microscopes (Carl Zeiss Microscopy). Water immersion lenses (20x, W Plan-Apochromat, 1.0 numerical aperture; and 40x, C-Apochromat, 1.2 numerical aperture) were used to record optical sections at 1,024 x 1,024 or 2048 x 2048 pixels and data were processed using Zeiss AxioVision or ZEN software. Images were converted to RGB images with ImageJ software.
Quantification immunohistochemistry
For Ki67 quantification, 10 regions were randomly chosen per slide at 40x magnification and from at least 3 different animals. The number of Ki67-positive cells was counted against the total number of cells in each region.
Tissue preparation for histological analysis and in situ hybridization (ISH)
Tissues were fixed overnight in 4% paraformaldehyde and paraffin-embedded according to standard techniques.
For haematoxylin and eosin staining as well as for ISH, paraffin-embedded mammary glands were cut into 4 µm sections using a microtome (Leica Biosystems). Haematoxylin and eosin staining was performed using a standard protocol.
In situ hybridization
In situ hybridization for Lgr6 RNA transcripts was performed using the RNAscope 2.0 High Definition assay and for double ISH against Lgr6 and Axin2 RNA or Lgr6 and Lgr5 RNA, we used the RNAscope 2-Plex Chromogenic kit (Advanced Cell Diagnostics, Inc., Hayward, CA, USA). All assays were performed according to the manufacturer’s instructions using specific probe sets against murine Lgr6 (Probe: Mm-Lgr6), murine Lgr5 (Probe: Mm-Lgr5-C2), and murine Axin2 (Probe: Mm-Axin2-C2). As positive and negative controls, we used the RNAscope Mm-Polr2a (312471) and dapB (310043) control probe sets.
We performed RNA in situ hybridization for Lgr6 mRNA expression in mammary glands from untreated and tamoxifen-treated mice. According to our experiments, tamoxifen administration does not obviously affect the expression pattern of Lgr6 mRNA.
Breast cancer tissue microarrays (TMAs)
The following commercially available TMAs were used for analysis: BR1503b, BR1505b, BR1921a, BRC961, BR8017, BR1006, BR728 (all US Biomax, Inc.). TMAs were hybridized with an Lgr6 probe using RNAscope. Mm-Polr2a (312471) and Hs-UBC-Human (310041) were used as positive controls and dapB (310043) was used as negative control. Tumour cores showing staining for the UBC and Polr2a, but not dapB, were evaluated for Lgr6 expression.
Disease free survival analysis
The RNAseq data of 1100 invasive breast carcinoma patients was obtained from the cBio Cancer Genomics Portal (http://cbioportal.org), and ref. 59 http://dx.doi.org/10.1016/j.cell.2015.09.033. Disease-free survival analysis was then performed using the cBioPortal web tool set with a z-score threshold of ± 3.
RNA sequencing (RNA-Seq)
Lgr6-positive and negative cells from two different replicates, each composed of 6 animals, were FACS-sorted and RNA recovered by MagMAX Total RNA isolation kit (Life Technologies). RNA concentrations were determined with a Qubit fluorometer (ThermoFisher) as 7-25 pg/ul before analysis of RNA quality on a Bioanalyzer (Agilent) (RIN 5-8). cDNA (50-75ng/μl) was generated from 5μl RNA (NuGEN/Next-Gen), sheared (Covaris S-series), and cDNA fragments were cleaned up (Zymo Research) before 10ng were used to generate an amplified cDNA library (NuGEN Ovation) over 18 PCR cycles. cDNA was sequenced on a HiSeq 2500 system (Illumina) in paired-end 50 mode.
RNA-Seq analysis
Gene level abundance estimates were generated using RSEM (v1.2.22)65 by mapping 50bp paired-end reads against the Mus musculus mm10 Refseq transcriptome and reference genome with the STAR (v2.4.2a)66 aligner. Normalisation factors were calculated using the MedianNorm function from the Bioconductor package EBSeq (v1.8.0). The posterior probability of differential gene expression was assessed using the EBTest (maxround=5) and GetDEResults (FDR=0.01) functions also from EBSeq.
Expression profiles of normalised abundance estimates were presented as heatmaps. A count of 1 was added to all normalised read counts prior to converting to a log base 2 scale. Genes were clustered using a euclidean distance metric and complete agglomeration method. Genewise counts were converted to z-scores by subtracting the mean and dividing by the standard deviation to aid visualisation.
A Wilcoxon two-sided gene set test was used to look for enrichment of known luminal (n=55) and basal (n=469) progenitor genes within a rank ordered list of 15902 gene posterior fold changes 4, 20, 21.
Tumour sphere cultivation and treatment
Mammary glands from PDL mice were dissected, mashed gently with a MACS dissociator (Miltenyi Biotec) and incubated in HBSS with 300U ml-1 collagenase (Sigma) plus 300 μg hyaluronidase (300U ml-1 activity) (Sigma) and digested for 30m at 37°C. Red blood cells were lysed and samples digested with Trypsin for 3 min at 37°C before being filtered with a 70 by 40 μm-mesh. Cells (9 to 15*106) were then plated onto collagen (Sigma-Aldrich) coated 150mm dishes and incubated overnight at 37°C in DMEM F12 (Gibco) plus L-glut, 2% FBS, 1% penicillin/streptomycin, 10 μg/mL insulin, 20 μg/mL EGF (Peprotech). Media was replaced 24h later with DMEM F12 (50 mL), 1mL B27 (STEMCELL) (50x), 20 ng/mL EGF and FGF (Peprotech), 4 μg/mL heparin, 1% penicillin/streptomycin. Cells were then treated with 4-hydroxytamoxifen (Sigma-Aldrich) at a final concentration of 0.25 μM for 3 days. After washing away the 4-hydroxytamoxifen, cells were treated with or without diphtheria toxin (DT) (25 ng/mL) for 3 days. After treatment, cells were harvested and equal numbers of treated and non-treated cells were re-plated by serial dilution onto ultra-low attachment plates. Colony formation was assessed 8 to 10 days after plating.
Tumour organoid cultivation and treatment
Briefly, mammary gland tissue was isolated and collagenase digested as described for the isolation of tumour spheres. The digestion mixture was washed 3 times with PBS and organoids were separated from single cells by 4 differential centrifugation steps67. Organoids were plated in growth factor reduced matrigel (BD Biosciences) on ultra-low attachment cell culture dishes (Corning) and cultivated in murine organoid media (MEBM, Lonza) containing B27 growth supplement (STEMCELL), 20 ng ml-1 rhEGF and rhbFGF, 4µg ml-1 heparin (Santa Cruz), and 1% penicillin/streptomycin.
Organoids were treated for 48 hrs with 2 µM doxorubicin (Sigma). After treatment, the organoids were washed 3 times with PBS and allowed to recover for 12 days.
FACS
Cells for FACS were isolated from mammary glands as described above for the tumour sphere isolation protocol. Three to eight million cells were incubated in 250 μl 2% FBS/PBS with primary antibodies for 30min on ice, with gentle agitation by flicking every 5 min. Primary antibodies were washed with 2% FBS/PBS, and cells incubated for 30 min with secondary antibodies on ice in the dark. Primary antibodies employed were: anti-CD24-PeCy7 (BD Biosciences #101822, 1:50), anti-CD29-Alexa-647 (BD Biosciences #102214, 1:100), anti-CD45-Pacific blue (BD Biosciences #103126, 1:100), anti-CD31-Pacific blue (BD Biosciences #102422, 1:100), anti-Ter119-Brilliant violet (BD Biosciences #562774, 1:100), biotinylated anti-CD45, anti-CD31, anti-Ter119 (clones 30F-11, 390, TER-119; eBioscience: catalogue #13-0451, #13-0311, #13-5921, all 1:100), anti-CD24-PerCP-Cy5.5 (clone M1/69; BD Pharmingen: catalogue #562360, 1:50), anti-CD29-Pe-Cy7 (clone HMb1-1; eBioscience, catalogue: #25-0291-82, 1:100), and Streptavidin-V450 (BD Biosciences, catalogue #560797, 1:100). DAPI or SytoxRed were added to exclude dead cells. Data analysis and cell sorting was performed on a FACSAria sorter and LSRFORTESSA II flow cytometer using the FACS DiVa software (BD Biosciences). LIN-population was isolated by exclusion of CD451/CD311/CD140α-positive cells. Cell populations were quantified as an average (+/- s.d.) of at least 3 experiments (employing at least 3 animals per group per experiment). Data were analysed using FlowJo software (FLOWJO, LLC).
Cleared fat pad transplantation
EGFP+ or EGFP- lin- CD29hiCD24+ cells were sorted into HBSS/2%FBS, spun down and resuspended in a mixture of HBSS/2%FBS and matrigel (1:1). The cells were injected into the cleared #4 fat pads of 21d-old NOD/SCID females.
Quantitative RT-PCR
RNA was isolated from dissected mammary gland cells or FACS-sorted cells using Qiagen Micro-RNA kit (Qiagen) or the MagMAX Total RNA isolation kit (Life Technologies). The cDNA was generated using the Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics) and was used for qPCR SYBR-green (Life Technologies) detection using the primers listed in Supplemental Table 1.
Quantification of the number and size of clones traced in the mammary gland
Three independent mice were analysed for each time point. For each traced mammary duct area, the size and keratin expression (K14+ or K8+) of tdTomato+ clones was recorded. The clones were grouped according to size (1 cell, 2–4 cells, >4 cells) and their frequency was calculated versus the total number of traced clones.
We defined clones as single tdTomato+ cells or clusters of one or more cells that contacted each other 3. We excluded extensively traced areas, in which the discrimination between basal and luminal cells was impeded.
Quantification of tdTomato+ tumour area
For each analysed mammary gland or tumour, 3–4 100 µm thick longitudinal sections were stained with TO-PRO3 and whole tissue sections were recorded by confocal microscopy. ImageJ software was used to outline and measure the total tissue area and the tdTomato+ tissue area and the percentage of the tdTomato+ area was calculated.
Statistics and Reproducibility
Data were analysed using GraphPad Prism software (GraphPad Software, Inc.) and are presented as the mean with s.e.m or s.d. (as indicated in figure legends). For box-and-whisker plots, data are presented as the median with highest and lowest values. Multiple groups were tested using analysis of variance (ANOVA) and comparisons between 2 groups were performed using t-tests. P < 0.05 was considered statistically significant. Asterisks indicate levels of significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001)
EdU detection was repeated at least three times per labelling timepoint. Immunohistological and immunefluorescence stainings were repeated at least three times. In situ hybridization was repeated at least three times for each probe. FACS analysis and tumour sphere experiments were performed at least three times.
Data availability
RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE75648. Previously published RNAseq data of 1100 invasive breast carcinoma patients were obtained from the cBio Cancer Genomics Portal (http://cbioportal.org) and ref. 59.
Source data for Fig. 1d, Fig. 2g – j, Fig. 4g, Fig. 5f, Fig. 7d, f, h, Fig. 8e, h, and Supplementary Fig. 7c, e, f have been provided as Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgements
We thank Åsa Bergström, Maryam Saghafian, and Raoul Kuiper for technical help and support, and Elin Tüksammel for mouse husbandry, genotyping, and surgery. We are grateful to Hans Clevers, Hubrecht Institute, The Netherlands, for providing the Lgr6-EGFP-Ires-CreERT2 mouse line. We also thank Pia-Maria Blaas, and Emilio Casanova for critical reading of the manuscript. Finally, we thank Catherine Cremona for her terrific assistance in writing the manuscript.
This work was supported by grants from Marie Skłodowska-Curie actions (Project # 297639) and the Wenner Gren Foundation (to L.B.), the Swedish Cancer Society (to M.G., R.T.), the German Research Foundation DFG (to M.G., DFG Ge 2386/1-1), the German Academic Exchange Service DAAD (to D.B.), the Knut and Alice Wallenbergs Foundation (to I.D.), the Breast Cancer Theme group at the Karolinska Institutet (to A.A., R.T.), the Swedish Research Council (to R.T.), the Center for Innovative Medicine in the Department of Biosciences and Nutrition, Karolinska Institutet (to R.T.), and the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001039), the UK Medical Research Council (FC001039), and the Wellcome Trust (FC001039)
This study was performed partly at the Francis Crick Institute in London, partly at the Wallenberg Institute for Regenerative Medicine Flow Cytometry Facility, and Regenerative Medicine, Karolinska Institutet, Huddinge, Sweden, and partly at the Live Cell Imaging Unit/Nikon Center of Excellence in the Department of Biosciences and Nutrition, which receives funding from the Knut and Alice Wallenberg Foundation, the Swedish Research Council, the Center for Innovative Medicine and the Jonasson donation to the School of Technology and Health, Kungliga Tekniska Högskolan, Huddinge, Sweden.
Footnotes
Author contributions
L.Blaas. and F.P. designed and performed experiments, analysed data and wrote the manuscript. H. A. M. performed immunostaining. A. B. A. performed mouse experiments, in situ hybridization and immunostaining. E.J.R. generated the FRL mice. M.G., A.M. performed experiments and analysed data. D.B. conducted confocal imaging and analysed data. I.D. planned and set up flow cytometry experiments. B.SD. and R.S. performed immunohistochemistry and in situ hybridization. R.M. performed RNAseq analysis. L.Bhaw and A. K. S. performed RNA sequencing. J.J. provided analytical tools. G.S. performed immunopathology evaluations of mammary gland tumours. I.M. assisted with MMTV-PyMT tumour cell isolation and culture, and data interpretation. R.T. and A.B. supervised the project and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Macias H, Hinck L. Mammary gland development. Wiley Interdiscip Rev Dev Biol. 2012;1:533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inman JL, Robertson C, Mott JD, Bissell MJ. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142:1028–1042. doi: 10.1242/dev.087643. [DOI] [PubMed] [Google Scholar]
- 3.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- 5.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 6.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 7.dos Santos CO, et al. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7123–7130. doi: 10.1073/pnas.1303919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafkas D, et al. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. The Journal of cell biology. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skibinski A, Kuperwasser C. The origin of breast tumor heterogeneity. Oncogene. 2015;34:5309–5316. doi: 10.1038/onc.2014.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 11.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 13.Prater MD, et al. Mammary stem cells have myoepithelial cell properties. Nature cell biology. 2014;16:942–950. doi: 10.1038/ncb3025. 941-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang TH, et al. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast cancer research : BCR. 2014;16:R1. doi: 10.1186/bcr3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner KU, et al. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 16.Ren W, et al. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:16401–16406. doi: 10.1073/pnas.1409064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oeztuerk-Winder F, Guinot A, Ochalek A, Ventura JJ. Regulation of human lung alveolar multipotent cells by a novel p38alpha MAPK/miR-17-92 axis. Embo J. 2012;31:3431–3441. doi: 10.1038/emboj.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 19.Fullgrabe A, et al. Dynamics of Lgr6(+) Progenitor Cells in the Hair Follicle, Sebaceous Gland, and Interfollicular Epidermis. Stem Cell Reports. 2015;5:843–855. doi: 10.1016/j.stemcr.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim E, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast cancer research : BCR. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodilla V, et al. Luminal progenitors restrict their lineage potential during mammary gland development. PLoS biology. 2015;13:e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaanta AS, Virtanen C, Selfors LM, Brugge JS, Neel BG. Evidence for a multipotent mammary progenitor with pregnancy-specific activity. Breast cancer research : BCR. 2013;15:R65. doi: 10.1186/bcr3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prat A, Ellis MJ, Perou CM. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol. 2012;9:48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldhirsch A, et al. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 27.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scata KA, El-Deiry WS. p53, BRCA1 and breast Cancer chemoresistance. Adv Exp Med Biol. 2007;608:70–86. doi: 10.1007/978-0-387-74039-3_5. [DOI] [PubMed] [Google Scholar]
- 30.Wei G, Wang Y, Zhang P, Lu J, Mao JH. Evaluating the prognostic significance of FBXW7 expression level in human breast cancer by a meta-analysis of transcriptional profiles. J Cancer Sci Ther. 2012;4:299–305. doi: 10.4172/1948-5956.1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akhoondi S, et al. Inactivation of FBXW7/hCDC4-beta expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast cancer research : BCR. 2010;12:R105. doi: 10.1186/bcr2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright KL, et al. Ras Signaling Is a Key Determinant for Metastatic Dissemination and Poor Survival of Luminal Breast Cancer Patients. Cancer Res. 2015;75:4960–4972. doi: 10.1158/0008-5472.CAN-14-2992. [DOI] [PubMed] [Google Scholar]
- 33.Wallace MD, et al. Comparative oncogenomics implicates the neurofibromin 1 gene (NF1) as a breast cancer driver. Genetics. 2012;192:385–396. doi: 10.1534/genetics.112.142802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin Y, et al. Characterization of medroxyprogesterone and DMBA-induced multilineage mammary tumors by gene expression profiling. Molecular carcinogenesis. 2005;44:42–50. doi: 10.1002/mc.20119. [DOI] [PubMed] [Google Scholar]
- 36.Oshima R, Kim S, Roopra A, Alexander CM. A Phenotypic Mouse Model of Basaloid Breast Tumors. PloS one. 2012;7:e30979. doi: 10.1371/journal.pone.0030979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giraddi RR, et al. Stem and progenitor cell division kinetics during postnatal mouse mammary gland development. Nat Commun. 2015;6:8487. doi: 10.1038/ncomms9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richert MM, Schwertfeger KL, Ryder JW, Anderson SM. An atlas of mouse mammary gland development. J Mammary Gland Biol Neoplasia. 2000;5:227–241. doi: 10.1023/a:1026499523505. [DOI] [PubMed] [Google Scholar]
- 39.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Visvader JE, Clevers H. Tissue-specific designs of stem cell hierarchies. Nature cell biology. 2016;18:349–355. doi: 10.1038/ncb3332. [DOI] [PubMed] [Google Scholar]
- 41.Wuidart A, et al. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016;30:1261–1277. doi: 10.1101/gad.280057.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyamala G, Chou YC, Cardiff RD, Vargis E. Effect of c-neu/ErbB2 expression levels on estrogen receptor alpha-dependent proliferation in mammary epithelial cells: implications for breast cancer biology. Cancer Res. 2006;66:10391–10398. doi: 10.1158/0008-5472.CAN-06-0321. [DOI] [PubMed] [Google Scholar]
- 43.Mastroianni M, et al. Wnt signaling can substitute for estrogen to induce division of ERalpha-positive cells in a mouse mammary tumor model. Cancer letters. 2010;289:23–31. doi: 10.1016/j.canlet.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arendt LM, Kuperwasser C. Form and Function: how Estrogen and Progesterone Regulate the Mammary Epithelial Hierarchy. J Mammary Gland Biol Neoplasia. 2015;20:9–25. doi: 10.1007/s10911-015-9337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boras-Granic K, Dann P, Wysolmerski JJ. Embryonic cells contribute directly to the quiescent stem cell population in the adult mouse mammary gland. Breast cancer research : BCR. 2014;16:487. doi: 10.1186/s13058-014-0487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Booth BW, Smith GH. Estrogen receptor-alpha and progesterone receptor are expressed in label-retaining mammary epithelial cells that divide asymmetrically and retain their template DNA strands. Breast cancer research : BCR. 2006;8:R49. doi: 10.1186/bcr1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burga LN, et al. Loss of BRCA1 leads to an increase in epidermal growth factor receptor expression in mammary epithelial cells, and epidermal growth factor receptor inhibition prevents estrogen receptor-negative cancers in BRCA1-mutant mice. Breast cancer research : BCR. 2011;13:R30. doi: 10.1186/bcr2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tung N, et al. Prevalence and predictors of loss of wild type BRCA1 in estrogen receptor positive and negative BRCA1-associated breast cancers. Breast cancer research : BCR. 2010;12:R95. doi: 10.1186/bcr2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tung N, et al. Estrogen receptor positive breast cancers in BRCA1 mutation carriers: clinical risk factors and pathologic features. Breast cancer research : BCR. 2010;12:R12. doi: 10.1186/bcr2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 51.Joshi PA, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 52.Schramek D, et al. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature. 2010;468:98–102. doi: 10.1038/nature09387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner KU, et al. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic research. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 54.Malhotra GK, et al. The role of Sox9 in mouse mammary gland development and maintenance of mammary stem and luminal progenitor cells. BMC developmental biology. 2014;14 doi: 10.1186/s12861-014-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koren S, et al. PIK3CA induces multipotency and multi-lineage mammary tumours. Nature. 2015;525:114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- 56.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 57.Bao L, Cardiff RD, Steinbach P, Messer KS, Ellies LG. Multipotent luminal mammary cancer stem cells model tumor heterogeneity. Breast cancer research : BCR. 2015;17:137. doi: 10.1186/s13058-015-0615-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen K, Huang YH, Chen JL. Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta pharmacologica Sinica. 2013;34:732–740. doi: 10.1038/aps.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ciriello G, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jandke A, et al. The F-box protein Fbw7 is required for cerebellar development. Dev Biol. 2011;358:201–212. doi: 10.1016/j.ydbio.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 62.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 63.Buch T, et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 64.Shehata M, van Amerongen R, Zeeman AL, Giraddi RR, Stingl J. The influence of tamoxifen on normal mouse mammary gland homeostasis. Breast Cancer Res. 2014;16:411. doi: 10.1186/s13058-014-0411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Developmental cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE75648. Previously published RNAseq data of 1100 invasive breast carcinoma patients were obtained from the cBio Cancer Genomics Portal (http://cbioportal.org) and ref. 59.
Source data for Fig. 1d, Fig. 2g – j, Fig. 4g, Fig. 5f, Fig. 7d, f, h, Fig. 8e, h, and Supplementary Fig. 7c, e, f have been provided as Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.