Abstract
The transcription factor E4bp4/Nfil3 has been shown to have a critical role in the development of all innate lymphoid cell types including Natural Killer (NK) cells. Here, we show that post-translational modifications (PTMs) of E4bp4 by either SUMOylation or phosphorylation have profound effects on both E4bp4 function and NK cell development. We examined the activity of E4bp4 mutants lacking PTMs and found that Notch1 was a novel E4bp4 target gene. We observed that abrogation of Notch signalling impeded NK cell production and the total lack of NK cell development from E4bp4-/- progenitors was completely rescued by short exposure to Notch peptide ligands. This work reveals both novel mechanisms in NK cell development by a transcriptional network including E4bp4 with Notch and that E4bp4 is a central hub to process extrinsic stimuli.
Introduction
Natural Killer (NK) cells are innate lymphoid cells (ILCs) that act at the interface between innate and adaptive immunity (1). They play a central role in the immune system via the production of cytokines and their ability to kill both virus-infected cells and cancer cells (2). Haematopoietic stem cells (HSCs) in the bone marrow (BM) give rise to NK cells through a number of distinct development stages including common lymphoid progenitors (CLPs), NK cell progenitors (NKPs), immature NK cells (iNK) and mature NK cells (mNK). NK cells require extrinsic factors such as IL-15, and stromal cells for their development and maturation but also require the expression of a small number of critical transcription factors. Of those transcription factors identified to date, the only one known to be essential for commitment to the NK cell lineage is the basic leucine zipper protein E4bp4 (also known as Nfil3). In the E4bp4-/- mouse the production of NKPs, iNK and mNK cells in the BM is completely compromised (3–5). E4bp4 is expressed in CLPs and it can regulate the expression of other genes important for NK cell development (5–7). In particular, E4bp4 binds regulatory regions of the Id2 and Eomes genes and induces their expression (5, 7). E4bp4’s critical function in NK cells is specific to the early stages of the developmental pathway as ablation of E4bp4 in peripheral mNK cells does not affect NK cell numbers or their response to cytomegalovirus infection (8).
As well as NK cells, E4bp4 is essential for the development of all other ILC subsets and is required for the correct formation of Peyer’s patches (6, 9–11). In the absence of E4bp4, the number of ILCs is greatly reduced in the small intestine (all subsets), colon (all subsets), lung (ILC1 and ILC2) and fat tissue (ILC2) (6, 9, 10). Additionally, E4bp4 was found to be required for the development of the earliest ILC progenitor, confirming its central role in the commitment to all innate lymphocyte lineages (6, 7, 11). Studies of E4bp4-/- mice have also revealed its indispensable role in multiple immunological processes including the production of cytokines by CD4+ T helper cells (12, 13) and the differentiation of TH17 cells (14).
Very little is known about how either extrinsic or intrinsic stimuli influence E4bp4. Cytokines have been shown to regulate E4bp4 expression principally, IL-7 via STAT5 signalling in ILC progenitors (7) and IL-15 in NK cells (5). The IL-15 signalling pathway in NK cells has been shown to regulate E4bp4 expression via the PDK1 and mTOR kinases, which are both essential for NK cell proliferation and development (15, 16). Extrinsic stimuli can modulate the expression of E4bp4, but to date, there is no evidence suggesting that the activity of the E4bp4 protein itself can be regulated.
In this study, we demonstrate that two post-translational modifications (PTMs), SUMOylation and phosphorylation, regulate E4bp4 activity. Both PTMs modify E4bp4 at multiple residues and regulate its function with a similar outcome. Mutant versions of E4bp4 were created lacking SUMOylation or phosphorylation sites and when expressed in haematopoietic progenitor cells (HPCs) led to the production of much greater numbers of NK cells compared to the wild-type (WT) form of E4bp4. The transcriptional activity of E4bp4 was also significantly increased in the absence of either SUMOylation or phosphorylation sites. In particular, E4bp4 PTM mutants promoted the expression of Notch1 to a greater extent than the WT-form of E4bp4, revealing a potential role for Notch signalling in E4bp4-directed NK cell development. We show that Notch1 is a transcriptional target of E4bp4 and that abrogation of Notch signalling can impede NK cell production. Remarkably, brief exposure to Notch ligand can completely rescue NK cell development in E4bp4-/- HPCs. Our results indicate that the action of E4bp4 in NK cells is highly regulated and can potentially be strongly influenced by extrinsic stimuli which has important implications for the expansion of NK cells e.g. for use in immunotherapy.
Materials and Methods
Mice
E4bp4-/- mice were generated as previously described (3), all those used were on a C57BL/6 background, between six and twelve weeks old and matched for age and gender. Rbpjflox/flox mice (17) were on a FVB background. All animal husbandry and experimental procedures were carried out according to UK Home Office regulations and local guidelines.
Analysis of SUMOylation in vivo using 6His-SUMO HeLa cells
The protocol used was adapted from Tatham et al, 2009 (26). The HeLa cell lines 6His-SUMO-1, 6His-SUMO-2, 6His-SUMO-3 and parental HeLa cells were transfected with pCMV-E4BP4 or pCMV (empty vector). After an input sample was removed, the remaining cells were lysed in 6 M Guanidinium-HCl, before Ni2+ affinity purification. Ni2+ NTA agarose beads (Qiagen) were incubated with cell lysates overnight (O/N) at 4°C. Samples were washed with 8 M Urea and His-tagged proteins were eluted with 200 mM imidazole.
Immunoprecipitation of FLAG-E4bp4
E4bp4 cDNA was cloned into the pCMV-script vector (Promega) using primers to incorporate a 5’ FLAG tag after the start codon. HeLa cell lines 6His-SUMO1, 6His-SUMO2 and 6His-SUMO3 were transfected with pCMV-FLAG-E4bp4 or pCMV (empty vector). Cells were lysed using a two-step lysis protocol (18) and lysates were incubated with anti-FLAG M2 Affinity Gel (Sigma Aldrich) O/N at 4°C. Samples were centrifuged and the supernatant was removed. Each sample was washed with TBS (50 mM Tris-HCl, 50 mM NaCl, pH 7.4) before elution of purified material by Laemmli sample buffer.
Western blotting
Cell lysates and protein samples were mixed 1:1 with Laemmli sample buffer and reduced by boiling in 5% β-mercaptoethanol. Samples were separated on 8% polyacrylamide gels, transferred to PVDF membranes and membranes were probed with primary antibodies against: E4bp4 (C18; Santa Cruz Biotech), SUMO2/3 (AbCam), FLAG (M2; Sigma Aldrich), 6X-His (4D11; AbCam), α-Tubulin (DM1A; eBioscience), RanGAP1 (AbCam), Histone H3 (AbCam). Appropriate HRP-conjugated secondary antibodies (AbCam) were used with Western Lightning® Plus-ECL detection reagents (Perkin Elmer) to determine chemiluminescence. Images of exposed blots were digitally acquired using the ChemiDoc™ XRS+ system (Bio-Rad).
Site directed mutagenesis
Single base pair mutations were made in the E4bp4 cDNA (K10R, K116R, K219R, K337R, K394R, S286A, S301A, and S353A), in the pCMV-script expression vector, using the QuikChange® XL site-directed mutagenesis kit (Agilent) and appropriately designed primers. Each mutant was also cloned into the pMSCV-IRES-hCD2 retroviral expression vector (5). The 5X-SUMO and S286-353A mutants were also cloned into the lentiviral expression vector pCSGW.
Mass spectrometry
293T cells were transfected with pCMV-FLAG-E4bp4 and pCDNA3-VP35 (19) using Lipofectamine 2000 (Life Technologies). The presence of the Ebola virus VP35 protein, helped to enhance recombinant protein expression (19). E4bp4 was immunoprecipitated from whole cell lysates using anti-FLAG M2 Affinity Gel (Sigma Aldrich). Bound material was eluted using 150 ng/µl FLAG peptide (Sigma Aldrich). Purified E4bp4 was concentrated using vacuum centrifugation and resuspended in 100 mM ammonium bicarbonate pH 8. Samples were reduced with 5 mM dithiothreitoland 14 mM iodoacetamide was used to label reduced cysteines. Proteomics-grade trypsin (Promega) was used to digest the E4bp4 protein for 6 h at 37 °C. For SUMOylated peptide analysis samples were sequentially digested with GluC (Roche) for 6 h at RT.
Phosphorylated peptides were enriched using TiO2 (GL sciences) as previously described (20) and phosphopeptides were eluted with 150 mM ammonium hydroxide and 50% acetonitrile (v/v) (Millipore). Peptides were chromatographically resolved on an Ultimate 3000 RS-LC-nano System (Dionex), with an Acclaim PepMap100, C18 stationary phase (Thermo Fisher). Real-time tandem mass spectra were acquired on an LTQ Velos Pro linear ion trap (Thermo Scientific). Initial phosphopeptide identification from the LC–MS/MS data was performed using a Sequest search in Proteome discoverer 1.3 (Thermo Fisher) against the Mouse Uniprot database (accessed: 19/08/14) including dynamic side-chain modifications including phosphorylation (+79.966) on serine, threonine, and tyrosine residues. Putative phosphopeptides were then validated using an algorithm for phosphorylation site identification (21) and manually assessed for accuracy.
The SUMOylated peptides were analysed using a mixture of data-dependent acquisitions and targeted MS/MS scans to search for peptides containing putative sites of SUMOylation based on a SUMO tag of GGTQQFV. Specifically, targeted MS/MS scans were always collected for the following m/z values, which correspond to E4bp4 peptides predicted to have SUMOylated lysines: K10 m/z 2+ = 531.99; K116 m/z 2+ = 796.49; K219 m/z 2+ = 853.54; K337 m/z 2+ = 825.52).
In vitro development of NK cells from transduced lineage negative bone marrow cells
Lin- BM cells were isolated from mouse leg bones and cultured in DMEM supplemented with 10% FCS (Stemcell Technologies), 50 μM β-mercaptoethanol (Gibco), 10 ng/ml Flt3L (PeproTech), 10 ng/ml IL-7 (PeproTech), and 100 ng/ml SCF (PeproTech). After 48 h cells were transduced by spinfection at 700 g and 20°C for 45 min with 8 μg/ml Polybrene. Cells were transduced with pMSCV-IRES-hCD2, containing either WT or mutant forms of E4bp4. Transduced cells were cultured for 72 h before being resuspended in α-MEM supplemented with 20% FCS, β-mercaptoethanol, and 30 ng/ml IL-15 (PeproTech) and re-plated onto OP9 stromal cells for a further 7 days of culture.
For experiments involving Notch1 signalling, Lin- BM cells were cultured on OP9, OP9-DL1 or plates pre-coated with rDLL1 (R&D Systems) or rDLL4 (R&D Systems). Plates were pre-coated with 10 µg/ml rDLL1/rDLL4 for 3 h at RT. Cells were incubated in α-MEM supplemented with 10% FCS, β-mercaptoethanol, 1 mM Sodium Pyrvuate, 25 mM HEPES and for the first 7 days with Flt3L, IL-7, and SCF. Cells were incubated for another 7 days on either on OP9 or OP9-DL1 in the presence of IL-15.
Rbpjflox/flox Lin- BM cells were lentivirally transduced on the day of isolation with pCSGW-Cre (or empty vector) that co-expresses hCD2. Cells were transduced by spinfection and were cultured for two days in the presence of Flt3L, IL-7, and SCF. Cells were either transferred onto rDLL4-coated plates for three more days of culture or transferred directly on OP9. Cells were cultured on OP9 for 7 days with IL-15. For flow cytometry analysis, all cells were gated for hCD2 expression to identify the population transduced with lentivirus.
Flow cytometry and cell sorting
Cells were stained with the following antibodies, all of which were anti–mouse and from eBioscience: CD2 (RM2-5), CD3 (17A2), CD11b (M1/70), CD19 (1D3), CD127 (A7R34), B220 (RA3-6B2), Gr1 (RB6-8C5), NK1.1 (PK136), Ter119 (TER119), NKp46 (29A1.4) and anti–human CD2 (RPA-2.10). The lineage cocktail contained B220, CD2, CD11b, Gr1, NK1.1, and Ter119. Cells were analysed on a Fortessa system (Becton Dickinson), sorted using FACSAria (Becton Dickinson) and data analysis was performed using FlowJo.
Cycloheximide time course
3T3 cells were stably transduced with pMSCV-E4bp4-hCD2 (or E4bp4 mutant versions) and after 48 h cells were bulked sorted for high hCD2 expression. Transduced cell lines were incubated with 50 µg/ml cycloheximide (Sigma-Aldrich) for 0-16 h. Nuclear extracts were prepared for all samples using NE-PER extraction regents (Pierce).
Luciferase assay
E4bp4 transcriptional activity was analysed using the pGL3-E4bp4-CS vector (22), containing the Firefly luciferase reporter gene and the Dual-Luciferase® reporter assay system (Promega). 3T3 cells were transfected with pMSCV-E4bp4-hCD2, pGL3-E4bp4-CS (or empty vector) and pRL-CMV (normalisation control). After 48 h, cells were lysed and the Dual-Luciferase® reporter assay system was used to determine Luciferase activities.
Quantitative PCR
MNK-1 cells (23) were transduced with pCSGW-E4bp4 (or E4bp4 5X SUMO or E4bp4 S286-353A mutants) as previously described (5). RNA was isolated using the RNeasy mini kit (Qiagen) and cDNA was synthesised using 1st Strand cDNA synthesis kit (Roche). QPCR was performed using TaqMan (Life Technologies) assays for Hprt1 (Mm00446968_m1), E4bp4 (Nfil3; Mm00600292_s1), Eomes (Mm01351985_m1), Gata3 (Mm00484683_m1), Notch1 (Mm00435249_m1) and T-bet (Tbx21; Mm00450960_m1). Samples were analysed using an Applied Biosystems 7500 Fast Real-Time PCR system. Ct values from samples were compared with a standard curve made from a known concentration of plasmid DNA (Eomes, T-bet, Gata3) or cDNA from a known number of murine splenocytes (Notch, Hprt1). The expression of all genes was normalised to Hprt1.
Chromatin immunoprecipitation
Regulatory regions of Notch1 were searched for putative E4bp4-binding sites (T(T/G)A(T/C)GTAA) using MatInspector (Genomatix). MNK-1 cells were transduced with a lentivirus expressing FLAG-E4bp4 and ChIP experiments were performed as previously described (5). Briefly, protein-DNA complexes were immunoprecipitated with IgG (EMD Millipore), M2 antibody to FLAG (Sigma-Aldrich), or polyclonal E16 antibody to E4bp4 (Santa Cruz Biotechnology, Inc.). Purified DNA was amplified using SYBR Select master mix (Life Technologies) and primers designed to recognise putative E4bp4-binding regions. The primers used were Notch1A forward primer (5’–3’) ctatatttttgccttgacagctaaagg & reverse primer (5’–3’) gaagtacgaagcatgcttgc producing an amplicon of 168bp, Notch1B cacatctgtgagctatttttgg & gactgactaaactaacattcccac 170bp, Notch1C ctcagaaactggcctcaagc & cacttgcagtcaggcgttc 144bp, Notch1D cacgccatcttaaagagctc & gtaaccaactgcactcttctcc 135bp, Notch1E caccaagaattcccaggag & gagtgcagtcacgtgctgac 144bp and Notch1 F ctcagactctctcggtaagtgtc & cgtgtggagctactctggc 160bp.
Results
The E4bp4 transcription factor is SUMOylated
To investigate how E4bp4 protein function might be regulated, we performed a yeast-two-hybrid screen to try to identify binding partners for the E4bp4 protein. Eleven proteins received multiple hits in the screen, but the protein with the highest number of positive identifications was PIAS1 (Supplemental Table 1). PIAS1 is a small ubiquitin-like modifier (SUMO) E3 ligase required for the addition of post-translational SUMO modifications (24), suggesting that E4bp4 may be post-translationally SUMOylated.
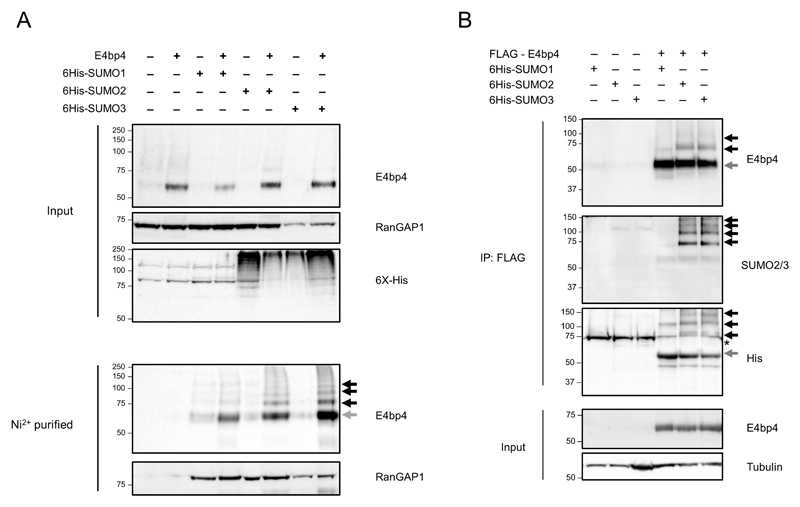
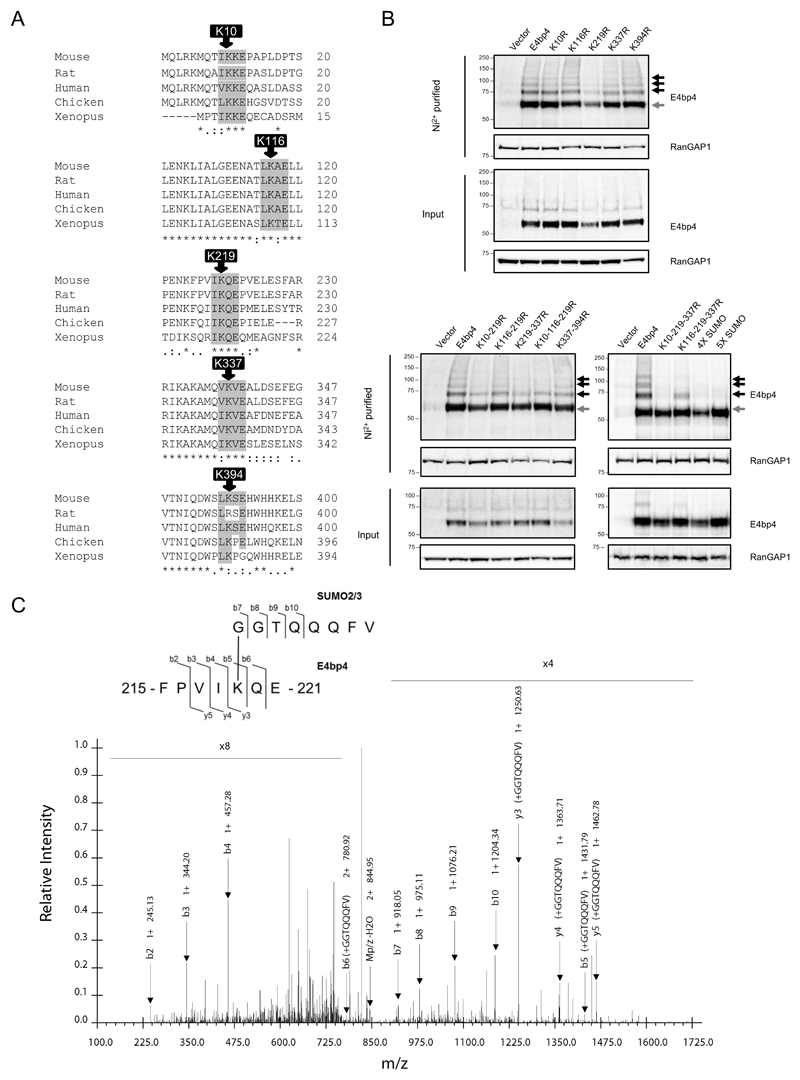
SUMO proteins are reversible post-translational protein modifiers and mammals express four SUMO isoforms, designated SUMO1 to SUMO4 (25). Mature SUMO2 and SUMO3 proteins differ by only three amino acids and are functionally homologous, whilst SUMO4 cannot be efficiently processed in mammalian cells and is not thought to be functional (25). E4bp4 was expressed in HeLa cells stably expressing 6His-SUMO1, 6His-SUMO2 and 6His-SUMO3 (26) and, following enrichment of all SUMOylated proteins by Ni2+ affinity chromatography, higher molecular weight forms of E4bp4 were observed (Fig. 1A). Each SUMO conjugate adds 10-15 kDa onto the apparent molecular weight of a protein, so the higher molecular weight forms of E4bp4 correspond to multiply SUMOylated versions of the protein. FLAG tagged E4bp4 was also expressed in each 6His-SUMO HeLa cell line and purified using anti-FLAG affinity resin and the same multiple higher molecular weight forms of E4bp4 were observed in the presence of 6His-SUMO2 and 6His-SUMO3 (Fig. 1B). E4bp4 was examined for the presence of the SUMOylation consensus motif ψ-K-x-E in its primary amino acid sequence (Fig. 2A). Five potential modification sites were discovered that were all highly conserved across a range of species (Fig. 2A). To establish if any of these sites were SUMOylated, the central lysine residue at each site was individually mutated to an arginine in the E4bp4 cDNA by site-directed mutagenesis. This mutation abolishes any SUMO modification at that site while maintaining the structural integrity of the protein (26). To assess the effect of the mutations, each SUMO mutant was expressed in the 6His-SUMO2 HeLa cells (Fig. 2B). E4bp4-K219R was the only individual mutant to affect the SUMOylation of E4bp4, but it did not fully remove all E4bp4 SUMOylation (Fig. 2B). All multi-site mutants affected SUMOylation of E4bp4, in particular, the 5X-SUMO mutant lacking all 5 putative SUMOylation sites, had no higher molecular weight forms of E4bp4 (Fig. 2B).
Figure 1.
E4bp4 is SUMOylated. (A) E4bp4 was expressed in HeLa cells stably expressing 6His-SUMO1, 6His-SUMO2 or 6His-SUMO3. Protein extracts were purified by Ni2+ affinity chromatography under denaturing conditions and analysed by Western blot. Input samples were lysed in Laemlli sample buffer and directly compared. (B) FLAG-E4bp4 was expressed in 6His-SUMO1, 6His-SUMO2 and 6His-SUMO3 HeLa cells and anti-FLAG antibody was used to immunoprecipitate E4bp4 before Western blot analysis. Grey arrows indicate unmodified E4bp4 and black arrows indicate SUMO modified forms of E4bp4 with higher molecular weights. Both are representative of three independent experiments.
Figure 2.
Mapping E4bp4 SUMOylation sites. (A) Sites of potential E4bp4 SUMO modification based on the presence of the Ψ-K-x-E motif. Asterisks highlight perfectly conserved residues and dots highlight partially conserved residues. Position of last amino acid in sequence indicated. (B) Mutant versions of E4bp4 lacking SUMOylation sites (lysine to arginine point mutations) were expressed in 6His-SUMO2 HeLa cells and protein extracts were purified by Ni2+ affinity chromatography under denaturing conditions and analysed by Western blots. Grey arrows highlight unmodified E4bp4 and black arrows highlight SUMO modified forms of E4bp4 with higher molecular weights, representative of three independent experiments. (C) Mass spectrometry identification of E4bp4 peptide SUMO2/3 modified at K219. FLAG-E4bp4 was expressed in 293T cells, purified from whole cell lysate using anti-FLAG immunoprecipitation and subjected to sequential digest by trypsin and Glu-C. E4bp4 peptides were purified and analysed by LC-MS/MS and SUMOylated peptides were identified by the presence of a GGTQQQFV modification on a lysine side chain. Annotated CID tandem mass spectra of +2 ion at m/z 853.95, with schematic representation of the identified peptide shown with detected b and y ions labelled from the fragmentation of E4bp4 peptide and SUMO tag.
To confirm the presence of SUMO modifications, purified E4bp4 protein was analysed by mass spectrometry (MS). Studying SUMO modifications by MS is challenging as SUMOylated forms of a protein are generally low in abundance and standard trypsin cleavage results in long SUMO peptide ‘tails’ remaining conjugated to target peptides, making them difficult to detect in standard MS (27). We established a system where FLAG epitope-tagged E4bp4 protein was expressed in 293T cells, purified by immunoprecipitation and sequentially digested with both trypsin and Glu-C. This novel double digest strategy aimed to produce short E4bp4 peptides with reduced SUMO isopeptide side chains on modified peptides. Using this approach a SUMO modified peptide was predicted to have a –GGTQQQFV side chain attached to a modified lysine. MS/MS analysis readily identified an E4bp4 peptide with a SUMO modification at K219, further confirming the presence of this PTM (Fig. 2C). These data demonstrated that the E4bp4 protein has SUMO modifications, leading to the question: do these modifications have an effect on E4bp4’s function as a transcription factor in NK cell development?
SUMOylation of the E4bp4 transcription factor influences NK cell development
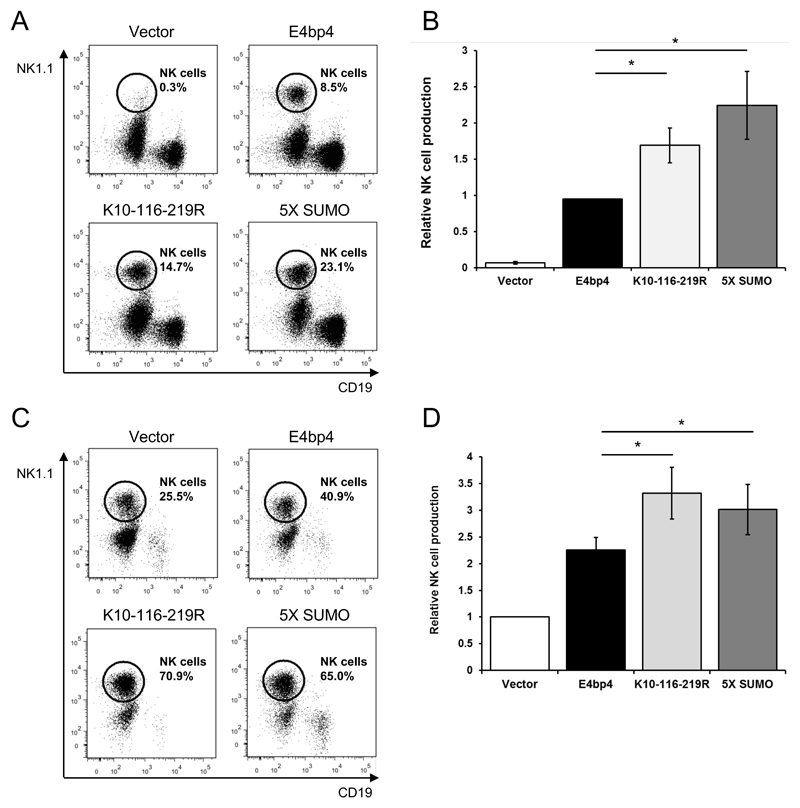
SUMOylation can affect the function of a transcription factor in different ways: cellular localisation; interactions with other proteins or; ability to regulate the expression of target genes (24). As E4bp4 is critical for the development of NK cells, we postulated that SUMOylation may regulate this function. Therefore, we compared the ability of E4bp4 to promote NK cell development with and without SUMO modifications. Lineage negative (Lin-) BM cells were isolated from E4bp4-/- mice and transduced with a retrovirus expressing either the WT-form of E4bp4 or one of the E4bp4 mutants lacking SUMO modification sites. WT-form E4bp4 rescued NK cell development from E4bp4-/- Lin- BM cells as previously reported (3, 5), however, the number of NK cells produced was significantly higher when the cells expressed E4bp4 SUMOylation mutants (Fig. 3A-3C). As the number of NK cells produced can vary between assays, the percentage of NK cells produced by each mutant was normalised to the positive control condition (i.e. WT-form E4bp4) (Fig. 3B). The same assay was also performed using E4bp4+/+ Lin- BM cells, as previous studies had shown that E4bp4 is a limiting factor for NK cell development in WT HPCs (3, 5). In the E4bp4+/+ Lin- BM cells, expression of WT-form E4bp4 increased the level of NK cell production compared to the empty vector, but again the expression of E4bp4 SUMO mutants led to significantly greater levels of NK cell production (Fig. 3C, 3D). These findings showed that SUMOylation can influence the function of E4bp4 and when SUMOylation is removed the activity of E4bp4, as measured by NK cell output, is substantially increased.
Figure 3.
The removal of SUMOylation sites of E4bp4 promotes NK cell production. Lin- BM cells were isolated from E4bp4-/- (A) or E4bp4+/+ (C) mouse BM and cultured for two days in the presence of IL-7, Flt3-L and SCF before retroviral transduction with a MSCV-IRES-hCD2 construct. Transduced cells were cultured for three days, before being transferred onto OP9 stromal cells and cultured in the presence of IL-15 for 7 days. (A, C) Flow cytometry analysis identified hCD2+ (transduced) cells and the presence of NK1.1+ CD19- NK cells. (B, D) Relative levels of NK cell production between E4bp4 mutants normalised to WT-form E4bp4 (B) or empty vector (D). Data are representative of four independent experiments for each mutant. Error bars show SEM. *, P < 0.05.
E4bp4 is multiply phosphorylated and these modifications influence NK cell development
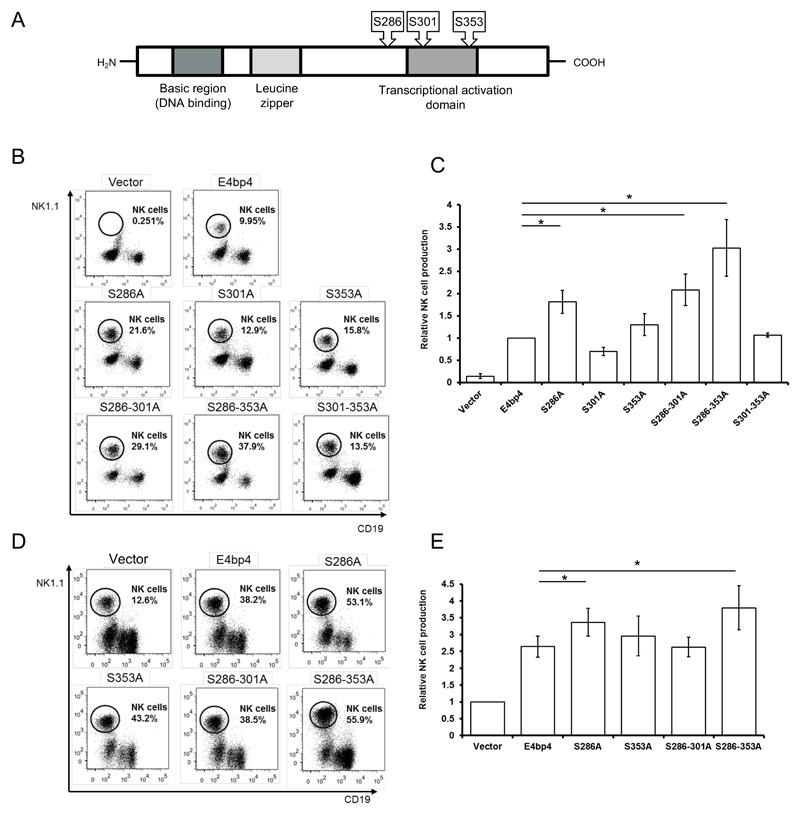
Previously, E4bp4 was reported to be phosphorylated when expressed in chick pineal gland and it was proposed that this PTM might regulate transcriptional activity (28). However, this study did not determine which residues of E4bp4 are phosphorylated. To address this, we purified FLAG-E4bp4 expressed in 293T cells and digested the protein using trypsin for LC-MS/MS analysis. The tandem MS conclusively revealed that E4bp4 has three phosphorylation sites at serines 286, 301 and 353 (Fig. 4A) (Supplemental Fig. 1-3). Each of these serine residues were mutated to alanine to abolish any phosphorylation but maintain protein conformation (29). As described for SUMOylation, we sought to determine if phosphorylation regulates the function of E4bp4. Each phosphorylation mutant was expressed by retroviral expression vector in E4bp4-/- Lin- BM cells and these were cultured in conditions promoting NK cell development. The ability of the phosphorylation mutants to rescue NK cell development was compared to the WT-form of E4bp4 and several of the mutants were found to promote significantly higher levels of NK cell production (Fig. 4B, 4C). In particular, the S286A mutant produced double the number of NK cells as WT-form E4bp4 (Fig. 4B). The phosphorylation mutants were also transduced into E4bp4+/+ Lin- BM cells and NK cell production was greatly enhanced in the cells transduced with the phosphorylation mutants compared to those transduced with the WT-form of E4bp4, particularly with the S286A and S286-353A mutants (Fig. 4D, 4E). The phenotype observed when phosphorylation sites were mutated replicated that described above for SUMOylation. Therefore, both PTMs negatively regulate the function of E4bp4 in NK cells and manipulating the PTMs of E4bp4 provides a potentially simple mechanism to control NK cell production.
Figure 4.
The removal of phosphorylation sites of E4bp4 promotes NK cell production. (A) Schematic representation of the E4bp4 protein showing the positions of phosphorylation sites identified by LC-MS/MS. Conserved domain structure of E4bp4 shown (not to scale), which has been identified through sequence homology and mutational studies. (B, D) Lin- BM cells were isolated from E4bp4-/- (B) or E4bp4+/+ (D) mouse BM and cultured for two days in the presence of IL-7, Flt3-L and SCF before retroviral transduction with a MSCV-IRES-hCD2 construct. Transduced cells were cultured for three days, before being transferred onto OP9 stromal cells and cultured in the presence of IL-15 for 7 days. Flow cytometry analysis identified hCD2+ (transduced) cells and the presence of NK1.1+ CD19- NK cells. (C, E) Relative levels of NK cell production between E4bp4 mutants normalised to WT-form E4bp4 (C) or empty vector (E). Data are representative of three independent experiments for each mutant. Error bars show SEM. *, P < 0.05.
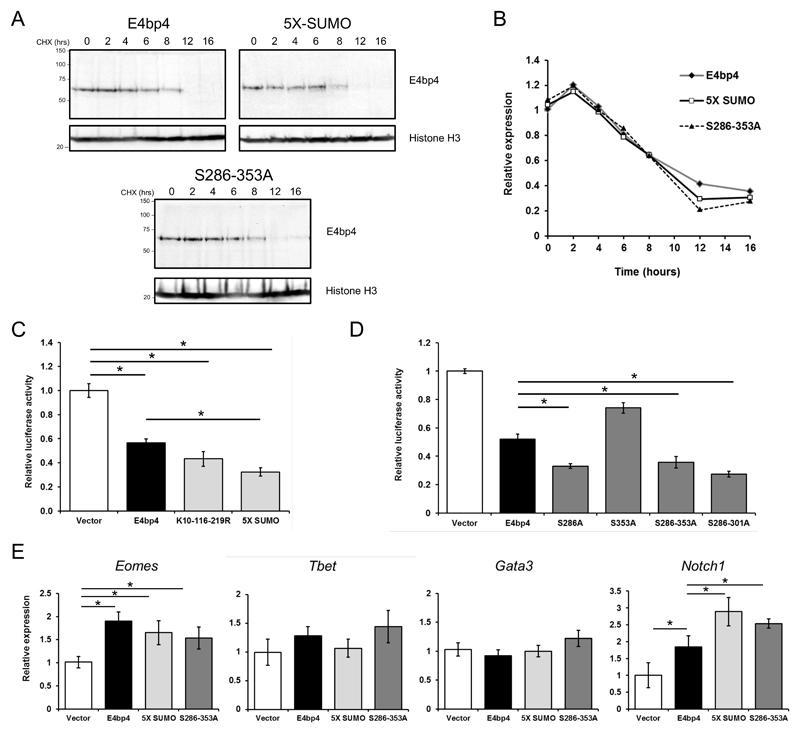
SUMOylation and phosphorylation do not affect the stability of E4bp4
SUMOylation and phosphorylation can both affect the stability of a transcription factor and influence its proteasomal degradation (24, 29). As the absence of both PTMs enhanced the function of E4bp4 during NK cell development, we tested if the mutant versions of E4bp4, lacking PTM sites, had altered stability. Using a cycloheximide time course assay on cell lines stably expressing the 5X-SUMO, S286-353A mutants or the WT-form of E4bp4, the stability of each form of E4bp4 was compared. Both mutants had very similar half-lives to WT E4bp4 with protein levels reduced by almost half after 8 hours of cycloheximide treatment (Fig. 5A, 5B).
Figure 5.
Post-translational modifications regulate E4bp4’s transcriptional activity, but not its stability. (A) 3T3 cells stably expressing E4bp4, 5X-SUMO and S286-353A were treated with cycloheximide (CHX, 50 µg ml-1) for the times indicated. Western blotting was used to compare nuclear extracts. (B) Quantification of relative E4bp4 expression from Western blot signals using densitometry. Level of E4bp4 is represented in arbitrary units after signals were normalised relative to those of Histone H3. (C, D) Transcriptional reporter assay using a Firefly Luciferase gene downstream of E4bp4 consensus binding sites. Relative luciferase activity was measured after 48 h from 3T3 cells transfected with MSCV-E4bp4-IRES-hCD2 and Firefly luciferase construct. Data are representative of ten independent experiments for each mutant. (E) MNK-1 cells were transduced with either vector control, E4bp4, 5X-SUMO or S286-353A. Expression of Eomes, Gata3, Tbet and Notch1 were determined by QPCR. Data are representative of three independent experiments for each mutant. Error bars show SEM. *, P < 0.05.
The transcriptional activity of E4bp4 can be regulated by SUMOylation and phosphorylation
E4bp4 was first identified as a transcriptional repressor and has been shown to repress the expression of numerous target genes in vivo e.g. Il-13 in TH2 cells (12). However, E4bp4 has likewise been found to transactivate the expression of various target genes, including Id2 and Eomes in NK cells (5). We postulated that SUMOylation and phosphorylation might influence the ability of E4bp4 to control target gene expression. To analyse the effect on gene transcription, we used a luciferase reporter gene assay as readout. We co-transfected cells with an E4bp4 expression vector and a plasmid with three E4bp4 DNA binding sequences upstream of the pGL3 promoter luciferase reporter. E4bp4 was found to act as a transcriptional repressor in this context (Fig. 5C). The WT-form of E4bp4 consistently led to a 50% decrease in luciferase expression, as previously reported (22), but the E4bp4 5X-SUMO mutant promoted even greater transcriptional repression than the WT-form of E4bp4 (Fig. 5C). A similar observation was observed for the E4bp4 phosphorylation mutants, particularly those containing a S286A mutant residue (Fig. 5D). These data indicate the the E4bp4 PTM mutants influence transcription via E4bp4-binding consensus sequences in an episomal context. Based on these findings, we decided to test, in a NK cell context, if the E4bp4 PTM mutants had any effect on the expression of endogenous genes known to strongly influence lymphoid cell development.
We transduced the mouse NK cell line, MNK-1 (7, 23) with the WT-form of E4bp4, the 5X-SUMO and S286-353A mutants. As seen previously, E4bp4 promoted Eomes expression in these cells (5) and a similar level of increase in expression was seen with both E4bp4 mutants (Fig. 5E). The presence of E4bp4 did not affect the transcription of Gata3 or Tbet (Fig. 5E). However, E4bp4 was found to promote the expression of Notch1 and, strikingly, an even greater increase in expression was seen with the 5X-SUMO and S286-353A mutants (Fig. 5E). This provided further evidence that both SUMOylation and phosphorylation negatively regulate the transcriptional activation effect of E4bp4. Our data show that removing these PTMs of the E4bp4 proteins makes it either a more potent transcriptional activator or more potent transcriptional repressor depending on context. This observation provides an insight into the mechanism by which the mutant forms of E4bp4 enhance NK cell production to a greater extent than WT-form E4bp4 and suggests a previously unknown mechanism of NK cell development, namely, that E4bp4 might act on the Notch signalling pathway.
E4bp4 can act through Notch to promote NK cell development
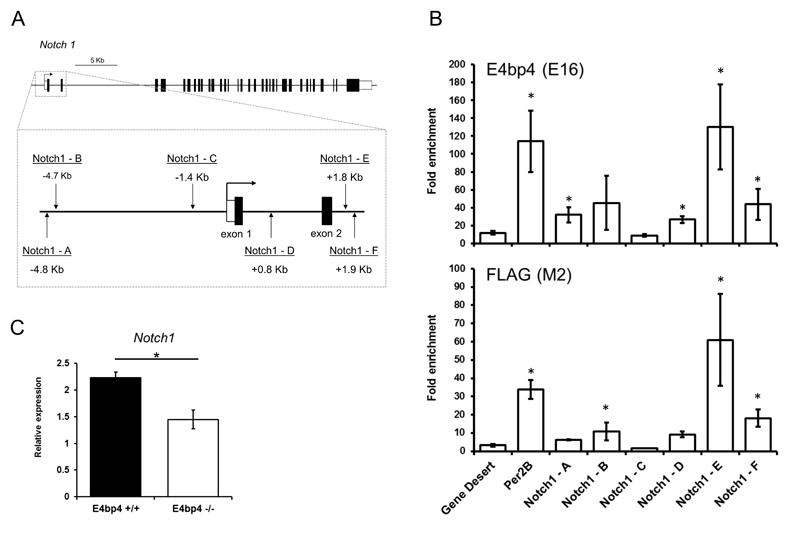
As E4bp4 seemed to transactivate the expression of Notch1, we looked to see if this was a direct effect and hence might potentially influence NK cell production. It has previously been suggested that Notch1 could have a role in the early stages of murine NK cell development as transient Notch signalling was shown to induce the development of NK cells from Pax5-/- pro-B cells and murine HSCs (30, 31). We first wanted to establish if Notch1 is a direct transcriptional target of E4bp4. Using chromatin immunoprecipitation (ChIP) we analysed whether E4bp4 could directly bind to the regulatory region of the Notch1 gene in vivo. We transduced MNK-1 cells with FLAG-tagged E4bp4 and protein-chromatin complexes were precipitated by either IgG, anti-FLAG or anti-E4bp4 antibodies. We searched the regulatory regions around the transcriptional start site (TSS) of Notch1 and identified six putative E4bp4 binding sites (Fig. 6A). E4bp4 binding was found to be massively enriched at predicted site E, to an even greater degree than the previously best characterised E4bp4 binding regulatory region found in the Per2B gene (32). In addition, sites A and B upstream of the TSS and sites D and F downstream of the TSS were also enriched in the E4bp4 immunoprecipitated samples (Fig. 6B). This indicated that E4bp4 binds to regulatory regions of Notch1 that could enhance its transcriptional activation. To further test the hypothesis that E4bp4 can regulate the expression of Notch1, we postulated that a loss of E4bp4 should influence Notch1 expression in HPCs in vivo. We compared the expression of Notch1 in E4bp4+/+ and E4bp4-/- Lin- BM cells and found that, in the absence of E4bp4, Notch1 expression was indeed significantly reduced as we have shown previously for another E4bp4 target gene, Id2 (3) (Fig. 6C).
Figure 6.
Notch1 is a transcriptional target of E4bp4 in NK cells. (A) Notch1 locus showing location of predicted E4bp4 binding sites identified through the presence of the E4bp4 minimal consensus binding sequence TTA(T/C)(G/A)TAA(C/T) (Cowell, 2002). Filled boxes = exons; clear boxes = UTR regions. (B) ChIP analysis of E4bp4 binding at the Notch1 loci in chromatin from MNK-1 cells stably transduced with FLAG-E4bp4. E16 is a polyclonal antibody to E4bp4 and M2 is a monoclonal antibody to FLAG. Per2B was used as a positive control51 and gene desert as a negative control. Data is representative of three biological replicates. (C) QPCR analysis of Notch1 expression in Lin- BM cells. Data is representative of six biological replicates. Error bars show SEM. *, P < 0.05.
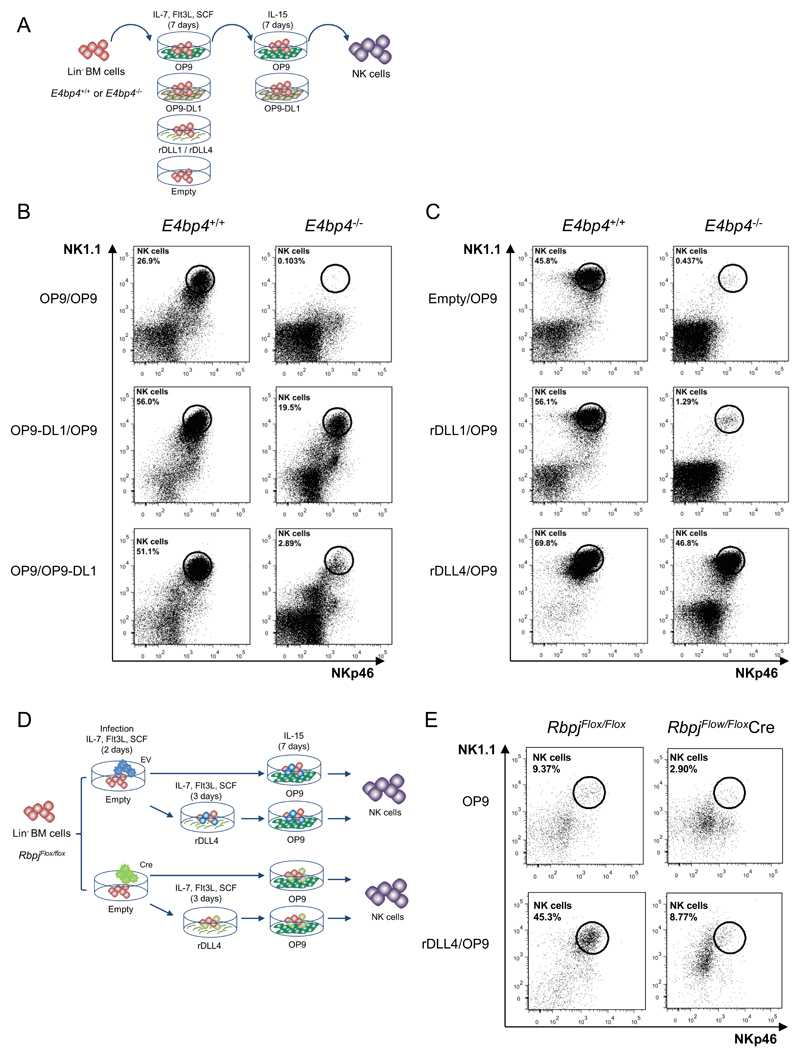
As E4bp4 appeared to regulate Notch1 expression, we speculated that enhanced Notch signalling, at an early stage, could potentially affect the development of NK cells from E4bp4-/- HPCs. To study NK cell development from HPCs in the presence or absence of Notch signalling, we cultured Lin- BM cells on either OP9 stromal cells or OP9-DL1 cells (that express the Notch ligand delta-like 1) (Fig. 7A). Subsequently, cells were transferred onto fresh OP9 and cultured in the presence of IL-15 (Fig. 7A). When E4bp4-/- cells were grown on control OP9, no NK cell development was observed, but when the E4bp4-/- cells were grown on OP9-DL1 for the first part of the culture, remarkably, NK cell development was rescued (Fig. 7B). The same result was not observed when the cells were grown on OP9-DL1 for the second part of the culture (Fig. 7B). To eliminate the influence of stromal cells, tissue culture plates were coated with either recombinant delta-like ligand 1 (rDLL1) or rDLL4 protein. These plates were used for the first 7-day period of the culture (Fig. 7B). Delta-like ligands have previously been shown to efficiently induce Notch signalling when immobilised onto plastic surfaces (33). When cultured on empty plates, the E4bp4-/- Lin- BM cells did not develop into NK cells, but when grown on rDLL4-coated plates the production of NK cells could be dramatically rescued even in the absence of the critical transcription factor for NK cell development (Fig. 7C). Only a very partial rescue was observed when the cells were cultured on rDLL1-coated plates (Fig. 7C). Additionally, when E4bp4+/+ HPCs were cultured on rDLL4-coated plates, the level of NK cell production was increased compared to cells grown on uncoated plates (Fig. 7C).
Figure 7.
Notch signalling at an early stage can induce NK cell development in the absence of E4bp4. (A) Schematic representation of culture system used to induce Notch signalling and promote NK cell development from Lin- BM cells. (B) Flow cytometry analysis of NK cell production following cultivation on indicated combination of OP9 or OP9-DL1 stromal cells or (C) on empty plates or plates coated with recombinant rDLL1/rDLL4. Lin- BM cells were first cultured for 7 days in IL-7, Flt3L, SCF before transferring to new plates for culture in IL-15. Data is representative of four (B) or two (C) biological replicates. (D) Schematic representation of two-stage culture of NK cell development from Lin- BM cells following from Cre-mediated deletion of Rbpj gene following lentiviral transduction. (E) Flow cytometry analysis of NK cell production derived from Lin- BM isolated from Rbpj floxed mice and transduced with either empty or Cre-expressing lentiviral vector. Cells were cultivated either on OP9 stromal cells alone or on rDLL4-coated plates before transfer on to OP9 as indicated. Data shown is for cells gated on hCD2 expression.
Notch signalling has a pivotal role in T cell lineage commitment and recent evidence has identified a crucial role for Notch signalling in ILC development (34–38). We tested if abrogation of Notch signalling would have any direct influence on NK cell development by using a method engendering Cre-mediated deletion of the Rbpj gene (39). Recombination signal-binding protein JK (RBP-J) is a transcriptional cofactor critical for the expression of target genes activated by the Notch signalling pathway (17). HPCs isolated from Rbpjflox/flox mice were transduced by a lentivirus co-expressing Cre recombinase and truncated human CD2. The human CD2 expression served to mark all transduced cells. Following transduction, HPCs were cultured on OP9 cells with IL-15 or first cultured on rDLL4-coated plates for 3 days before transfer to OP9 plus IL-15 (Fig. 7D). Much reduced numbers of mature NK cells developed from those HPCs subject to Cre-deletion (Fig. 7E). This differential effect was greatly accentuated by pre-incubation with rDLL4 that selectively enhances NK cell development from HPCs (Fig. 7E). These data suggest that Notch signalling can play a role in the early stages of NK cell development and this action is as an integral part of the E4bp4-mediated transcriptional network that controls NK cell production.
Discussion
E4bp4 is a critical regulator of NK cell and ILC development, as well as being pivotal to a variety of other immunological processes. E4bp4 has a profound effect on NK cell production despite there being only a relatively small increase in E4bp4 mRNA levels during NK cell development (5). Little is known about any mechanisms that exist to control the activity of E4bp4 protein. Here, we demonstrate that the E4bp4 protein has multiple SUMO modifications and is predominantly modified by the SUMO2/3 isoform. Several large scale proteomic screens have been performed looking to identify cellular SUMOylation targets and three of these studies identified the same modified E4bp4 SUMO modified peptide as identified here, further confirming K219 as a site an endogenous site of SUMO modification (40). Our MS analysis of the E4bp4 protein also revealed it is phosphorylated at three sites; S286, S301 and S353. These findings confirm the results of previous large-scale phosphoproteomic screens that had identified those sites as potential E4bp4 phosphorylation sites (41–43).
E4bp4 plays a central role in NK cell development where its expression in CLPs is required to commit developing cells to the NK lineage (5). We have shown that the SUMOylation and phosphorylation sites of E4bp4 have a dramatic influence both on the activity of E4bp4 and on NK cell development. When comparing the activity of WT-form E4bp4 to mutant forms that lack SUMOylation or phosphorylation sites, the mutant forms were found to consistently promote greater levels of NK cell production. The two types of PTM identified here, act in a similar manner to negatively regulate the activity of E4bp4 during NK cell production. E4bp4 was previously demonstrated to be a limiting factor for NK cell development, as transduction of E4bp4+/+ Lin- BM cells with E4bp4 caused increased levels of NK cell production (3, 5). Transducing E4bp4+/+ cells with mutant forms of E4bp4 also increased NK cell production but to a greater extent than the WT-form E4bp4. This demonstrated that SUMOylation and phosphorylation both negatively regulate the activity of E4bp4, as removing the sites of these modifications increases E4bp4 activity and ultimately NK cell production. As E4bp4 is critical for NK cell development, it is highly likely that its activity is tightly controlled by multiple mechanisms as aberrant activity could lead to defective haematopoiesis. There are no other well characterized examples where the PTM of a single transcription factor can have such a dramatic effect on a complex biological process such as lineage development. It would be extremely useful to be able to detect in vivo SUMOylation and, for that matter, phosphorylation of the E4bp4 protein within individual cell nuclei. However, this is not yet technically feasible in the absence of high affinity antibodies that recognise SUMOylated or phosphorylated residues in a context specific to E4bp4. An additional problem is that endogenous protein levels of E4bp4 are very low and all post-translational modifications are dynamic, therefore, the modified forms of E4bp4 protein at any one time in cell nuclei are only a tiny fraction of the total E4bp4 protein present.
SUMOylation and phosphorylation were both found to suppress the transcriptional activity of E4bp4 regardless of whether it was activating or repressing transcription. We compared the effect of WT-form E4bp4 and the PTM mutant forms on the expression of transcription factors known to regulate lymphoid commitment. The remarkable outcome of this comparison was that Notch1 expression was upregulated in the presence of WT-form E4bp4 but that expression was significantly further enhanced in the presence of both the E4bp4 5X-SUMO mutant and the S286-353A phosphorylation mutant. This allowed us to propose a previously unknown mechanism namely that E4bp4 may promote NK cell development via the Notch signalling pathway.
Notch signalling activated by extrinsic ligands was already suggested to have a role in the development of both murine and human NK cells (30, 31, 44–46). It most likely to be required only transiently during the early phase of NK cell development, as prolonged signalling induces T cell development (47). We show that Cre-mediated deletion of Rbpj in HPCs results in impaired NK cell development, which is the first report of an intrinsic role for Notch in NK cell development. However, it is important to highlight that both from our data and from previous data that NK cell development is not critically dependent on Notch signalling, but that its activation significantly enhances NK cell development. In particular, mice with conditional deletion of Notch1 in their haematopoietic cells do not have significantly reduced numbers of NK cells (48). This could be because the E4bp4-mediated transcriptional network in the conditional knockout cells remains intact and can compensate for the lack of Notch1. Additionally, although early stages of NK cell development may be somewhat impaired with Notch1 deleted, normal homeostatic processes could lead to the accumulation of normal numbers of peripheral NK cells in steady state conditions.
Like Notch, E4bp4 is required during early lymphocyte development and must be expressed in CLPs for them to commit to the NK lineage (5), however, it is dispensable for the maturation and functionality of mature NK cells (8). E4bp4 and Notch1 are also important for the development of other innate lymphoid cell types, for example, lymphoid tissue inducer (LTi) cells, where Notch signalling is required to engage the LTi developmental pathway but also needs to be turned off later to avoid diversion to T cell fate (36). We found that E4bp4 transcriptionally regulates Notch1 as it binds directly to regulatory regions of the Notch1 gene and in the absence of E4bp4, Notch1 expression was reduced in HPCs. E4bp4 most prominently bound to a region 1.8 Kb upstream of the TSS of Notch1 in a similar manner to two other transcription factors, DLX5 and ERβ, that are known to regulate Notch1 expression (49). Strikingly, increased Notch signalling during the early part of NK cell development alone was sufficient to completely rescue the development of NK cells from E4bp4-/- progenitor cells. This strongly suggests that Notch1 acts downstream of E4bp4 during NK cell development. This rescue was only achieved when Notch ligands were present during the early stage of NK cell development. The rescue from E4bp4-/- progenitors was achieved using both DLL1 expressing OP9 stromal cells and rDLL1 and rDLL4 proteins immobilised on plastic plates. The rDLL4 had a much greater effect than the rDLL1 on NK cell development as DLL4 binds Notch1 with much higher affinity than DLL1 (50).
As Notch1 ligands are expressed in the bone marrow microenvironment (51) it appears that the availability of Notch signalling at the appropriate time could drive NK cell development. Similarly to E4bp4, Notch1 has also been found to regulate the expression of Eomes (52), which could be a further means by which Notch1 enhances NK cell development via the E4bp4-regulated pathway. We suggest that both the critical role established for E4bp4 in ILC development and the data shown here for NK cells is indicative of a central mechanism linking extrinsic signals via E4bp4 to direct transcriptional control of all ILC production.
We have previously shown that thymic NK cells can develop in the thymus independently of the presence of E4bp4 (53). However, the starting cells in those experiments were DN1 thymocytes so they would have already received substantial Notch signalling in the thymus. The results presented here potentially explain why the thymic NK cells do not require E4bp4 to develop. It could well be that the thymic environment provides an “early” Notch rescue analogous to that shown in Figure 7B that abrogates the requirement for E4bp4 expression.
Control of E4bp4 function by PTMs and extrinsic stimuli has significant implications for the production of human NK cells for use in immunotherapy. The production of NK cells from various sources (e.g. induced-pluripotent stem cells and umbilical cord blood stem cells) involves the use of cytokines and stromal cells to commit the cells to the NK lineage, but influencing E4bp4 activity through its PTMs in these cells could provide a simple strategy to enhance the process. It has recently been reported that NK cells develop from adipose tissue-derived stem cells following transduction with E4bp4 (54). The manipulation of E4bp4 activity is likely to have a very important role in producing future NK cell immunotherapeutic products. This approach, in time, may even allow direct mobilisation of NK cell production in vivo as immunotherapy.
Supplementary Material
Acknowledgements
We are very grateful to Professor Ron Hay, University of Dundee for the kind gift of the 6His-SUMO HeLa cells lines. We are also very grateful to Professor Kristjan Jessen, University College London for the gift of Rbpj floxed mice. We thank R. Subramanian for animal husbandry and Dr Steve Ley, Francis Crick Institute, Mill Hill Laboratory, London for the kind gift of VP35 protein expression construct. We also thank our colleague Dr. Ilaria Nisoli for kind help during some experimental procedures.
This work was supported by a BBSRC DTP studentship (TK) and a Medical Research Council Grant (G0901737) (HJMB).
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 2.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 4.Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, Look AT, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Male V, Nisoli I, Kostrzewski T, Allan DS, Carlyle JR, Lord GM, Wack A, Brady HJ. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med. 2014;211:635–642. doi: 10.1084/jem.20132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geiger TL, Abt MC, Gasteiger G, Firth MA, O'Connor MH, Geary CD, O'Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, Sun JC. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu W, Domingues RG, Fonseca-Pereira D, Ferreira M, Ribeiro H, Lopez-Lastra S, Motomura Y, Moreira-Santos L, Bihl F, Braud V, Kee B, et al. NFIL3 Orchestrates the Emergence of Common Helper Innate Lymphoid Cell Precursors. Cell Rep. 2015;10:2043–2054. doi: 10.1016/j.celrep.2015.02.057. [DOI] [PubMed] [Google Scholar]
- 8.Firth MA, Madera S, Beaulieu AM, Gasteiger G, Castillo EF, Schluns KS, Kubo M, Rothman PB, Vivier E, Sun JC. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J Exp Med. 2013;210:2981–2990. doi: 10.1084/jem.20130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz GT, Carotta S. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, Hooper LV. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife. 2014;3:e04406. doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashiwada M, Cassel SL, Colgan JD, Rothman PB. NFIL3/E4BP4 controls type 2 T helper cell cytokine expression. EMBO J. 2011;30:2071–2082. doi: 10.1038/emboj.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motomura Y, Kitamura H, Hijikata A, Matsunaga Y, Matsumoto K, Inoue H, Atarashi K, Hori S, Watarai H, Zhu J, Taniguchi M, et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat Immunol. 2011;12:450–459. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, Rothman PB, Takahashi JS, Hooper LV. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727–730. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcais A, Cherfils-Vicini J, Viant C, Degouve S, Viel S, Fenis A, Rabilloud J, Mayol K, Tavares A, Bienvenu J, Gangloff YG, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat Immunol. 2014;15:749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang M, Li D, Chang Z, Yang Z, Tian Z, Dong Z. PDK1 orchestrates early NK cell development through induction of E4BP4 expression and maintenance of IL-15 responsiveness. J Exp Med. 2015;212:253–265. doi: 10.1084/jem.20141703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 18.Klenova E, Chernukhin I, Inoue T, Shamsuddin S, Norton J. Immunoprecipitation techniques for the analysis of transcription factor complexes. Methods. 2002;26:254–259. doi: 10.1016/S1046-2023(02)00029-4. [DOI] [PubMed] [Google Scholar]
- 19.Gantke T, Boussouf S, Janzen J, Morrice NA, Howell S, Mühlberger E, Ley SC. Ebola virus VP35 induces high-level production of recombinant TPL-2-ABIN-2-NF-kappaB1 p105 complex in co-transfected HEK-293 cells. Biochem J. 2013;452:359–365. doi: 10.1042/BJ20121873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li QR, Ning ZB, Tang JS, Nie S, Zeng R. Effect of peptide-to-TiO2 beads ratio on phosphopeptide enrichment selectivity. J Proteome Res. 2009;8:5375–5381. doi: 10.1021/pr900659n. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, DiMaggio PA, Jr, Perlman DH, Zakian VA, Garcia BA. Novel phosphorylation sites in the S. cerevisiae Cdc13 protein reveal new targets for telomere length regulation. J Proteome Res. 2013;12:316–327. doi: 10.1021/pr300408v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeung J, O'Sullivan E, Hubank M, Brady HJ. E4BP4 expression is regulated by the t(17;19)-associated oncoprotein E2A-HLF in pro-B cells. Br J Haematol. 2004;125:560–567. doi: 10.1111/j.1365-2141.2004.04953.x. [DOI] [PubMed] [Google Scholar]
- 23.Allan DS, Kirkham CL, Aguilar OA, Qu LC, Chen P, Fine JH, Serra P, Awong G, Gommerman JL, Zúñiga-Pflücker JC, Carlyle JR. An in vitro model of innate lymphoid cell function and differentiation. Mucosal Immunol. 2014;8:340–351. doi: 10.1038/mi.2014.71. [DOI] [PubMed] [Google Scholar]
- 24.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 26.Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- 27.Da Silva-Ferrada E, Lopitz-Otsoa F, Lang V, Rodriguez MS, Matthiesen R. Strategies to Identify Recognition Signals and Targets of SUMOylation. Biochem Res Int. 2012;2012:875148. doi: 10.1155/2012/875148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doi M, Okano T, Yujnovsky I, Sassone-Corsi P, Fukada Y. Negative control of circadian clock regulator E4BP4 by casein kinase Iepsilon-mediated phosphorylation. Curr Biol. 2004;14:975–980. doi: 10.1016/j.cub.2004.05.043. [DOI] [PubMed] [Google Scholar]
- 29.Holmberg CI, Tran SE, Eriksson JE, Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci. 2002;27:619–627. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]
- 30.Carotta S, Brady J, Wu L, Nutt SL. Transient Notch signaling induces NK cell potential in Pax5-deficient pro-B cells. Eur J Immunol. 2006;36:3294–3304. doi: 10.1002/eji.200636325. [DOI] [PubMed] [Google Scholar]
- 31.Rolink AG, Balciunaite G, Demoliere C, Ceredig R. The potential involvement of Notch signaling in NK cell development. Immunol Lett. 2006;107:50–57. doi: 10.1016/j.imlet.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Ohno T, Onishi Y, Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucl Acids Res. 2007;35:648–655. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. Immobilization of Notch ligand, Delta-1, is required for induction of Notch signaling. J Cell Sci. 2000;113:4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 34.Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Possot C, Schmutz S, Chea S, Boucontet L, Louise A, Cumano A, Golub R. Notch signaling is necessary for adult, but not fetal, development of RORgt(+) innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- 36.Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORgt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, Radtke F. Transcription factor RORa is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, McKenzie AN, Carotta S, Nutt SL, Belz GT. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. Notch-RBPJ signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 40.Hendricks IA, D'Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;10:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 43.Weintz G, Olsen JV, Frühauf K, Niedzielska M, Amit I, Jantsch J, Mages J, Frech C, Dölken L, Mann M, Lang R. The phosphoproteome of toll-like receptor-activated macrophages. Mol Syst Biol. 2010;6:371. doi: 10.1038/msb.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeHart SL, Heikens MJ, Tsai S. Jagged2 promotes the development of natural killer cells and the establishment of functional natural killer cell lines. Blood. 2005;105:3521–3527. doi: 10.1182/blood-2004-11-4237. [DOI] [PubMed] [Google Scholar]
- 45.Bachanova V, McCullar V, Lenvik T, Wangen R, Peterson KA, Ankarlo DE, Panoskaltsis-Mortari A, Wagner JE, Miller JS. Activated notch supports development of cytokine producing NK cells which are hyporesponsive and fail to acquire NK cell effector functions. Biol Blood Marrow Transplant. 2009;15:183–194. doi: 10.1016/j.bbmt.2008.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haraguchi K, Suzuki T, Koyama N, Kumano K, Nakahara F, Matsumoto A, Yokoyama Y, Sakata-Yanagimoto M, Masuda S, Takahashi T, Kamijo A, et al. Notch activation induces the generation of functional NK cells from human cord blood CD34-positive cells devoid of IL-15. J Immunol. 2009;182:6168–6178. doi: 10.4049/jimmunol.0803036. [DOI] [PubMed] [Google Scholar]
- 47.Rothenberg EV. Transcriptional control of early T and B cell developmental choices. Annu Rev Immunol. 2014;32:283–321. doi: 10.1146/annurev-immunol-032712-100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radtke F, Ferrero I, Wilson A, Lees R, Aguet M, MacDonald HR. Notch1 deficiency dissociates the intrathymic development of dendritic cells and T cells. J Exp Med. 2000;191:1085–1094. doi: 10.1084/jem.191.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brooks YS, Ostano P, Jo SH, Dai J, Getsios S, Dziunycz P, Hofbauer GF, Cerveny K, Chiorino G, Lefort K, Dotto GP. Multifactorial ERbeta and NOTCH1 control of squamous differentiation and cancer. J Clin Invest. 2014;124:2260–2276. doi: 10.1172/JCI72718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrawes MB, Xu X, Liu H, Ficarro SB, Marto JA, Aster JC, Blacklow SC. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J Biol Chem. 2013;288:25477–25489. doi: 10.1074/jbc.M113.454850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber JM, Calvi LM. Notch signaling and the bone marrow hematopoietic stem cell niche. Bone. 2010;46:281–285. doi: 10.1016/j.bone.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho OH, Shin HM, Miele L, Golde TE, Fauq A, Minter LM, Osborne BA. Notch regulates cytolytic effector function in CD8+ T cells. J Immunol. 2009;182:3380–3389. doi: 10.4049/jimmunol.0802598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crotta S, Gkioka A, Male V, Duarte JH, Davidson S, Nisoli I, Brady HJ, Wack A. The transcription factor E4BP4 is not required for extramedullary pathways of NK cell development. J Immunol. 2014;192:2677–2688. doi: 10.4049/jimmunol.1302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ning H, Lei HE, Xu YD, Guan RL, Venstrom JM, Lin G, Lue TF, Xin Z, Lin CS. Conversion of adipose-derived stem cells into natural killer-like cells with anti-tumor activities in nude mice. PLoS ONE. 2014;9:e106246. doi: 10.1371/journal.pone.0106246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.